Abstract
TFIID is a cornerstone of eukaryotic gene regulation. Distinct TFIID complexes with unique subunit composition exist and several TFIID subunits are shared with other complexes, conveying intricate cellular decision making to control subunit allocation and functional assembly of this essential transcription factor. However, the underlying molecular mechanisms remain poorly understood. Here, we used quantitative proteomics to examine TFIID submodules and assembly mechanisms in human cells. Structural and mutational analysis of the cytoplasmic TAF5-TAF6-TAF9 submodule identified novel interactions crucial for TFIID integrity, and for allocating TAF9 to TFIID or the SAGA co-activator complex. We discover a key checkpoint function for the chaperonin CCT, which specifically associates with nascent TAF5 for subsequent handover to TAF6-TAF9 and ultimate holo-TFIID formation. Our findings illustrate at the molecular level how multisubunit complexes are crafted in the cell, involving checkpoint decisions facilitated by a chaperone machine.
Introduction
Eukaryotic gene transcription by RNA polymerase II (pol II) is controlled by a plethora of proteins, preassembled into complexes including basal transcription factors1,2. TFIID is first to bind the core promoter, nucleating the pre-initiation complex (PIC). In humans, TFIID comprises the TATA-binding protein (TBP) and 13 TBP-associated factors (TAFs). Cryo-electron microscopy (cryo-EM) provided important insights into TFIID architecture and promoter binding3,4. Distinct TFIID complexes were identified, containing paralogues of TAFs and TBP, with key roles in cellular differentiation1,2,5. Human TFIID subunits TAF9, TAF10 and TAF12 are also present in SAGA (Spt-Ada-Gcn5-acetyltransferase)6, a co-activator complex which, like TFIID, is globally required for efficient pol II transcription7,8. In yeast TAF5 and TAF6 are also shared with SAGA, while human SAGA contains the paralogues TAF5L and TAF6L9. Sharing of subunits is not unique to TFIID and SAGA but occurs frequently in complexes controlling transcription and chromatin1,2,6–11. Faithful subunit allocation and regulated complex assembly thus must be essential for proper cell development and function, however, the underlying molecular mechanisms and factors involved remain elusive. Chaperones are known to facilitate the folding of key proteins in essential processes, such as kinase-mediated cellular signalling, proteasome assembly or nucleosome turnover11–15. The CCT (chaperonin containing TCP1) complex was shown to mediate ATP-dependent folding of many substrates including actin, tubulin and recently the CRLCSA DNA repair factor16–18. CCT adopts a barrel-like shape, with two hetero-octameric rings stacked on top of each other creating two folding chambers18.
Here, we utilized targeted quantitative mass spectrometry (qMS)19,20 to systematically probe TAF interactions in the nucleus and cytoplasm of human cells, to discover molecular mechanisms of TFIID formation. We identified a novel cytoplasmic TFIID submodule comprising TAF5, TAF6 and TAF9. The crystal structure of this complex revealed intricate TAF-TAF interactions, setting the stage for mutational studies. Intriguingly, our analyses reveal a key checkpoint function of CCT, providing unprecedented insight into early steps of holo-TFIID assembly regulated by this chaperonin.
Results
TFIID and SAGA submodules in the cytoplasm
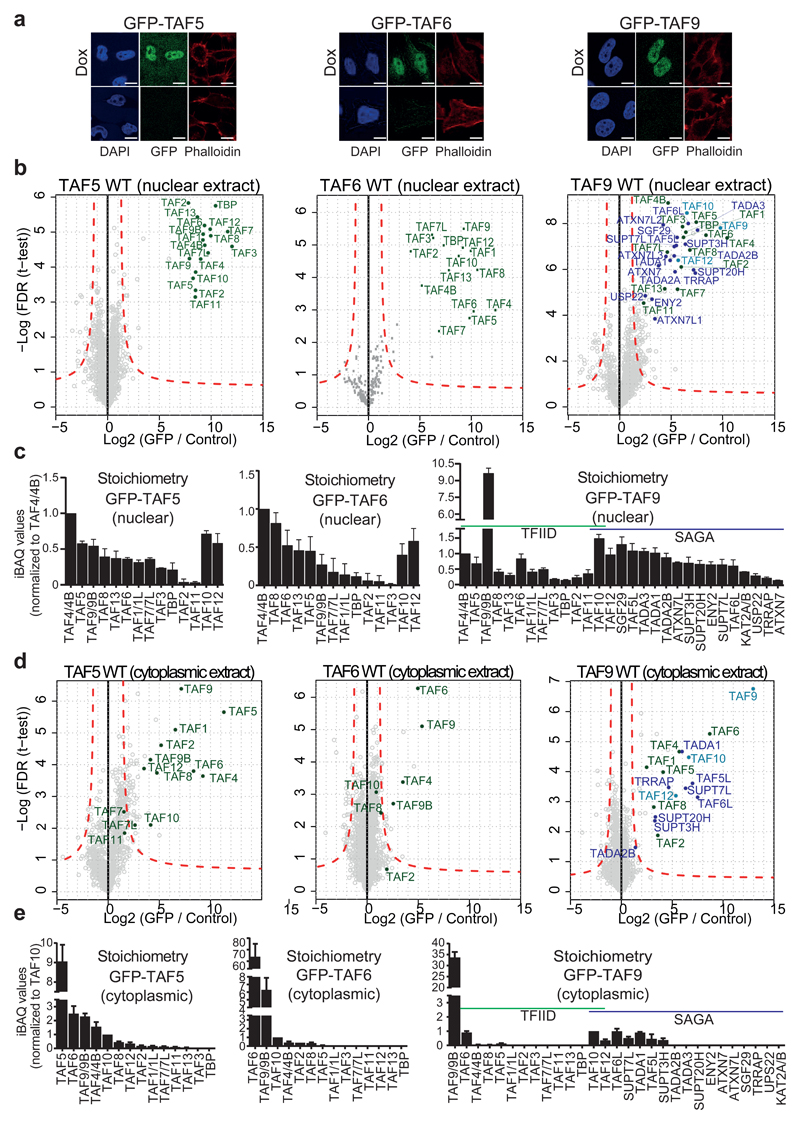
The presence of stable, partial TFIID assemblies in cells suggests formation of the holo-complex from discrete, functional submodules3,21–23. TAF5 is the presumed central scaffold within TFIID and was shown to interact with TAF6 and TAF93,24. TAF6 and TAF9 form a heterodimer stabilized by pairing of their histone-fold domains (HFDs)25. We created stable doxycycline (Dox) inducible HeLa-derived cell lines26 expressing TAF5, TAF6 or TAF9 as N-terminal green fluorescent protein (GFP) fusions. As expected, all GFP-TAF proteins predominantly localized to the nucleus (Fig. 1a). GFP-TAF5 expression levels were close to endogenous, while GFP-TAF6 was elevated. α-GFP immunoblotting and quantitative RT-PCR showed that GFP-TAF9 levels were in between GFP-TAF5 and GFP-TAF6 (Supplementary Fig. 1). We examined integration of GFP-TAF proteins in TFIID by GFP co-immunoprecipitation (co-IP) from nuclear extracts of Dox-treated cells followed by intensity-based absolute quantification (iBAQ) quantitative mass spectrometry (qMS)27. All GFP-TAF proteins enriched the complete holo-TFIID complex (Fig. 1b,c). TAF9 is shared between TFIID and SAGA6, and all SAGA subunits were likewise enriched in nuclear GFP-TAF9 co-IPs. We determined the relative abundance of TFIID subunits, evidencing similar stoichiometries in all experiments. Moreover, comparable levels of SAGA and TFIID were present in the GFP-TAF9 co-IPs (Fig. 1c). Our results demonstrate that GFP-tagged TAF5 and TAF6 are efficiently integrated into TFIID, while, as expected, GFP-TAF9 integrates in both TFIID and SAGA.
Figure 1. TFIID and SAGA submodules in the cytoplasm.
a, Cellular localization of GFP-TAF5, GFP-TAF9 and GFP-TAF6 by confocal microscopy; scale bar = 10 μm. b, GFP-TAF5 and GFP-TAF6 enrich all TFIID subunits in co-IPs from nuclear extracts; GFP-TAF9 enriches TFIID and SAGA subunits. Subunits unique to TFIID are colored in green, subunits unique to SAGA in purple, shared subunits in blue. Dashed red lines denote threshold between background and significant enrichment (two-tailed t-test; FDR = 1%; S0 =1). Each data point is plotted as average of technical triplicates. c, Relative abundance of TFIID and SAGA subunits from nuclear extracts are shown, normalized to TAF4/4B. Each bar represents an average of technical triplicates. Error bars = s.d. of mean. d, GFP-TAF5 and GFP-TAF6 enrich a subset of TFIID subunits in co-IPs from the cytoplasm; GFP-TAF9 enriches both TFIID and SAGA subunits. Each data point is plotted as average of technical triplicates. e, Relative abundance of TFIID and SAGA subunits in the cytoplasmic co-IPs. Each bar represents an average of technical triplicates. Error bars = s.d. of mean. Source data for c and e are available online.
Next, we analysed GFP-TAFs purified from the corresponding cytoplasmic extracts (Fig. 1d,e). We found that TAF6 and TAF9 co-purified efficiently with GFP-TAF5 revealing the presence of a stable heterotrimeric complex in the cytoplasm. We also found significant levels of TAF4, TAF8, TAF10 and TAF12, reminiscent of stable TFIID subassemblies in recombinant reconstitution experiments3. Cytoplasmic GFP-TAF6 predominantly co-purified TAF9. Cytoplasmic GFP-TAF9 co-IPs showed enrichment of TAF6, and the SAGA subunits TAF5L, TAF6L, TADA1, SUPT3H and SUPT7L. TAF10 and TAF12, like TAF9, are shared between TFIID and SAGA, and were also present. In SAGA, TAF10 and TAF12 are known to form specific heterodimers with SUPT7L and TADA1, respectively. Thus, the SAGA module we observed may represent a putative core-SAGA complex mirroring a core-TFIID complex originally identified in D. melanogaster3,21. Taken together, our data provide evidence for the presence of discrete TFIID and SAGA submodules in the cytoplasm, which likely represent building blocks of the respective holo-complexes.
Crystal structure of TAF5-TAF6-TAF9
Our qMS experiments revealed TAF5-TAF6-TAF9 as a prevalent TFIID submodule in the cytoplasm. Limited proteolysis of full-length TAF5-TAF6-TAF9 identified a sample suitable for crystallization (Supplementary Figs. 2, 3 and Supplementary Table 1). Crystals comprising a TAF5 construct spanning the N-terminal domain (NTD) and WD40 repeat domain (TAF5194-800), bound to an extended TAF6-TAF9 heterodimer (TAF61-92; TAF91-120), diffracted to high resolution (Supplementary Fig. 4). Phases were obtained with tantalum bromide clusters and the structure refined to 2.5 Å (Table 1) resulting in excellent electron density (Supplementary Fig. 4e-h). The final model includes residues 207-800 of TAF5 (residues 194-206, 381-416 and 748-753 were not modeled due to poor definition), residues 6-92 of TAF6 and residues 5-120 of TAF9.
Table 1. Data collection, phasing and refinement statistics.
| Native (PDB 6F3T)a |
Ta6Br12a | |
|---|---|---|
| Data collection | ||
| Space group | I23 | I23 |
| Cell dimensions | ||
| a, b, c (Å) | 339.05, 339.05, 339.05 | 338.16, 338.16, 338.16 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Peak | ||
| Wavelength | 1.0044 | 1.25439 |
| Resolution (Å)b | 79.92-2.50 (2.60-2.50)b | 48.81-3.80 (3.90-3.80)b |
| Rmerge | 0.1358 (3.048) | 0.173 (1.17) |
| I/σ(I) | 8.67 (0.64) | 9.48 (2.08) |
|
CC1/2 Completeness (%) |
0.995 (0.188) 99.03 (91.70) |
0.996 (0.676) 100.00 (100.00) |
| Redundancy | 4.69 (4.15) | 5.78 (5.70) |
| Refinement | ||
| Resolution (Å) | 79.92-2.50 | |
| No. reflections | 219090 | |
| Rwork / Rfree | 19.73 / 22.00 | |
| No. atoms | ||
| Protein | 24768 | |
| Ion (Cl-) | 7 | |
| Water | 391 | |
| B factors | ||
| Protein | 88.6 | |
| Ligand/ion | 79.6 | |
| Water | 68.8 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.004 | |
| Bond angles (°) | 1.06 |
A single crystal was used to collect the native and the Ta6Br12 dataset.
Values in parentheses are for highest-resolution shell.
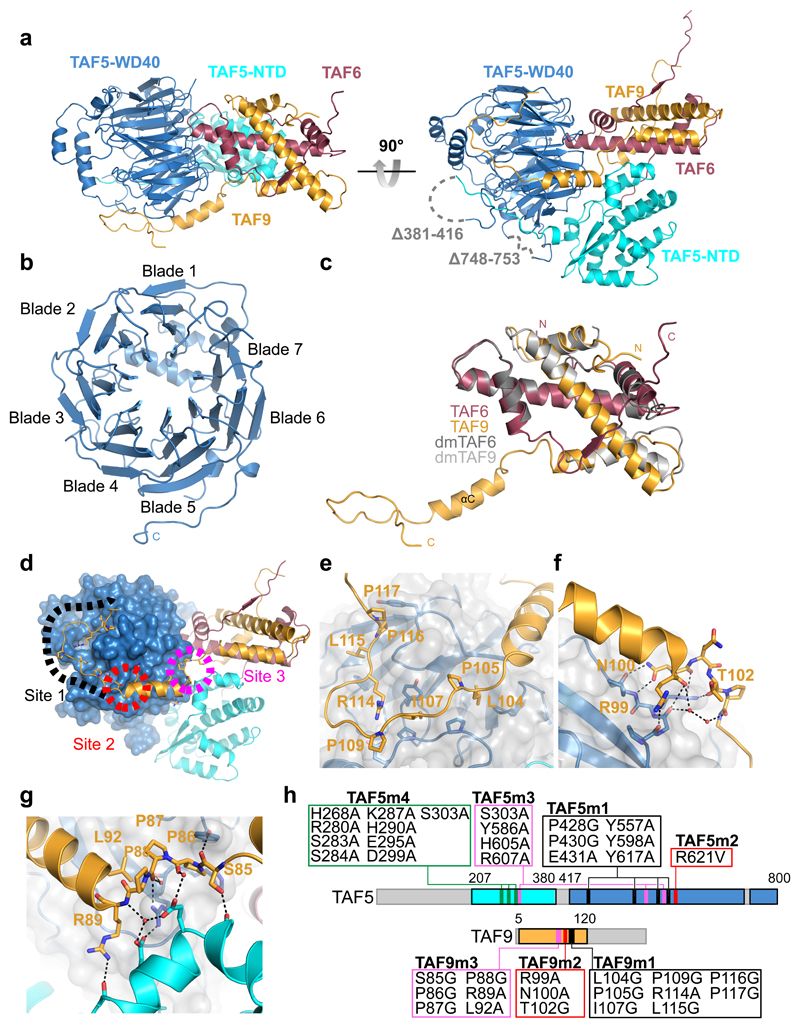
The crystal structure displays a compact, triangular shape (Fig. 2a), with the TAF6-TAF9 HFD heterodimer sandwiched between the NTD and WD40 repeat domain of TAF5, giving rise to an intertwined architecture based on TAF5 as the central scaffold. The TAF5 NTD was crystallized previously in isolation28. Significant differences are apparent in the TAF5-TAF6-TAF9 complex, notably, the N-terminal α-helix 7 of the NTD is rearranged owing to intimate interactions with the TAF5 WD40 repeat domain (Supplementary Fig. 5). The WD40 repeat adopts a seven-bladed β-propeller with pronounced α-helical insertions in blades 1 and 7. TAF5 engages extensively with the central α-helix of the TAF6 HFD via its NTD and the bottom cavity of the WD40 domain (Fig. 2b). The HFDs of TAF6 and TAF9 adopt a similar structure as in the D. melanogaster TAF6-TAF9 HFD pair29.
Figure 2. Crystal structure of TAF5-TAF6-TAF9 complex.
a, The TAF5-TAF6-TAF9 complex is shown in a cartoon representation in two views. The TAF6-TAF9 HFD heterodimer is intimately wedged in between the TAF5 NTD and WD40 repeat domain. TAF5 NTD and WD40 repeat domain are colored in cyan and blue, respectively. TAF6 is colored in red, TAF9 is colored in orange. Disordered loops are drawn as grey dashed lines. b, The TAF5 WD40 repeat domain is viewed from its bottom cavity, adopting a 7-bladed β-propeller. c, Human TAF6-TAF9 superimposed on the TAF6-TAF9 HFD dimer from D. melanogaster (PDBID 1TAF 29). D. melanogaster proteins are colored in grey, human TAF in red and human TAF9 in orange. Note the extended C-terminal domain in human TAF9. d, Overview of the structure with the three major interactions anchoring TAF9 to TAF5.e-g, Zoom-in showing TAF9 C-terminal loop (e), hydrogen-bond network at C-terminus of helix αC (f), and triple proline-turn (g). h, Mutations to probe TAF5-TAF9 interfaces. Colors as in (d); green indicates mutated region in TAF5 NTD targeting TAF5-TAF6-TAF9 interface.
Our human TAF9 construct comprises an extended C-terminal region, which is completely resolved in the electron density (Fig. 2c). This TAF9 C-terminal domain adopts a novel, distinct conformation crucial for TAF5-TAF6-TAF9 complex integrity, wrapping around the TAF5 WD40 repeat domain like a clamp with three major anchor points (Fig. 2d). The TAF9 C-terminal loop region attaches tightly to the surface of the TAF5 WD40 domain involving residues L104, I107 and L115 (Fig. 2e). The second anchor is formed by the TAF9 αC helix packing laterally against the TAF5-WD40 repeat domain, engaging in multiple hydrogen bonds mediated by R99, N100 and T102 (Fig. 2f). Moreover, a triple proline turn formed by TAF9 residues P86-P88, interacts with both the TAF5 NTD and WD40 repeat domain (Fig. 2g). This remarkable multitude of protein-protein interactions combine to a total buried surface area of 2713 Å2 in the TAF5-TAF6-TAF9 complex.
TAF5-TAF9 interactions dictate TFIID assembly
Our crystal structure revealed unexpected, intricate interactions by TAF9, which we next probed in cells by qMS, using a series of mutants (Fig. 2h). Four TAF9 mutants (TAF9m1-4) were designed with the aim to disrupt the TAF5-TAF9 interface, while maintaining proper folding of the proteins. TAF9m1 targeted hydrophobic side chains of the α-helix inserted in hydrophobic pockets on the TAF5 WD40 surface as well as prominent salt bridges in between the TAFs. In TAF9m2, we abolished the intricate network of hydrogen bonds between TAF5 and TAF9. TAF9m3 was designed to destabilize the triple proline-turn in the C-terminal extension and abolish the TAF5S303-TAF9R89 interaction. Finally, TAF9m4 combined all of these mutations.
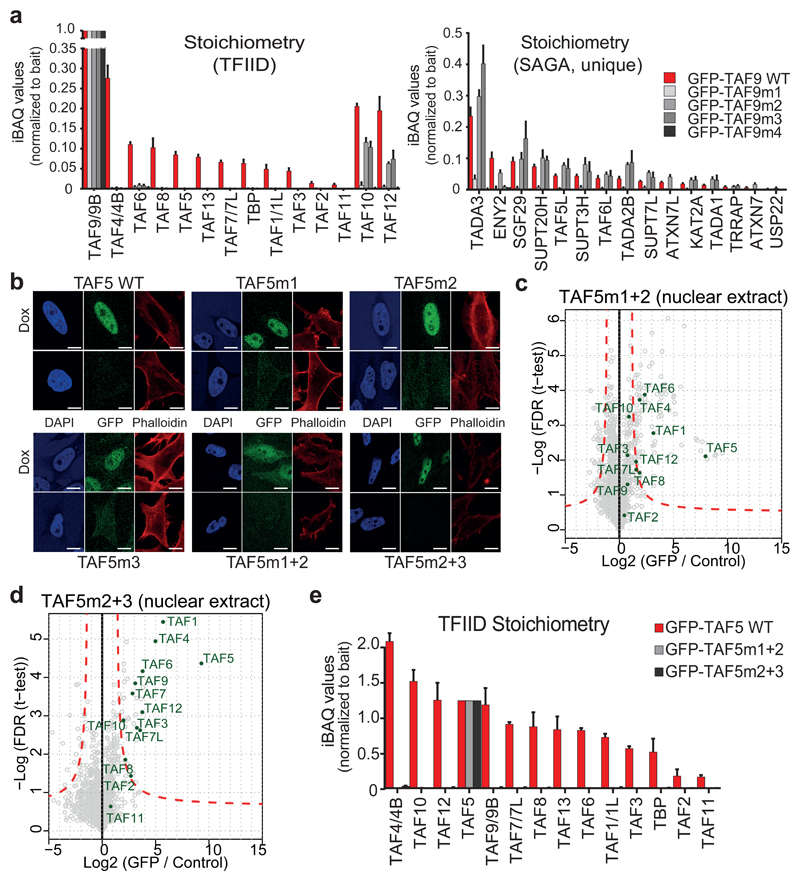
GFP-TAF9 mutants were expressed and co-IPs of nuclear extracts subjected to qMS (Fig. 3a, Supplementary Fig. 6a-d). TAF9m1 did not incorporate into TFIID, while retaining a low, but significant interaction with TAF6. While strongly reduced as compared to wild-type, TAF9m1 incorporation into a (partial) SAGA complex was indicated by the significantly enriched SAGA subunits TADA3, TADA2B, SUPT7L, SUPT20H, KAT2A and TRRAP (Supplementary Fig. 6a). TAF9m2 disrupted TFIID formation to a similar extent as TAF9m1, while maintaining pairing with TAF6. This TAF9 mutant had no noticeable effect on SAGA. Likewise, TAF9m3 compromised TFIID assembly. Interestingly, TAF9m3 efficiently enriched most subunits of SAGA, with the exception of the subunits forming the deubiquitination (DUB) module9, suggesting a role for the region mutated in TAF9m3 in mediating DUB integration. Finally, TAF9m4 targeting all TAF9-TAF5 interfaces abolished both TFIID and SAGA formation completely and only associated with TAF6. In human SAGA, TAF9 pairs with the TAF6 paralogue TAF6L. Of note, TAF6L was not observed in TAF9m1 (and TAF9m4) co-IPs. Our results demonstrate that mutation of TAF9 residues critical for TAF5-TAF9 interaction completely disrupt TFIID formation. In contrast, the TAF9 mutations affect SAGA formation to a much lower extent, implying that the conformation adopted by TAF9 and its interactions may be significantly different in TFIID as compared to SAGA.
Figure 3. TAF5-TAF9 interfaces probed by mutagenesis and quantitative proteomics.
a, Stoichiometry plots of TFIID and SAGA subunits from GFP-TAF9 co-IPs are shown, normalized to bait protein. Each bar represents an average of technical triplicates. Error bars = s.d. of mean. b, Confocal fluorescence microscopy reveals nuclear localization of all GFP-TAF5 proteins studies except for GFP-TAF5m1+2 which is also present in the cytoplasm; scale bar = 10 μm. c-e, Nuclear Co-IPs of GFP-TAF5, GFP-TAF5m1+2 and GFP-TAF5m2+3 evidence compromised TFIID assembly by the mutants. Each data point in volcano plots is plotted as average of technical triplicates. Dashed red lines denote threshold between background and significant enrichment (two-tailed t-test; FDR = 1%; S0 =1). Each bar in stoichiometry plot represents an average of technical triplicates. Error bars = s.d. of mean. n = 2 independent experimental replicates for the representative GFP-TAF5m1+2 sample. Source data for a and e are available online.
We introduced mutations in TAF5 reciprocal to those in the TAF9 mutants for validation (Fig 2h, Fig. 3b-e, Supplementary Fig. 6e-g, Supplementary Fig. 7). First, we included mutations in TAF5, targeting each of the three distinct interface regions individually. Co-IPs and qMS of GFP-tagged versions showed that the three individual TAF5 mutants did not wholly disrupt TFIID but that the relative abundance of TFIID subunits was reduced as compared to wild-type, indicating less efficient TFIID formation (Supplementary Fig. 6f). This is not due to reduced protein levels, since all TAF5 mutants are expressed at levels higher than wild-type (Supplementary Fig. 6g). We next prepared TAF5 mutants, each combining two sets of mutations. Both TAF5m1+2 (P428G, P430G, Q431A, Y557A, Y598A, Y617A, R621V) and TAF5m2+3 (S303A, Y586A, H605A, R607A, R621V), were completely defective in TFIID formation (Fig. 3c-e).
Our mutational analyses compellingly validate the atomic interactions observed in our crystal structure, confirming their central importance for TFIID assembly and integrity in vivo. Moreover, our results with mutants of TAF9 argue for distinct interactions of this shared subunit within TFIID and SAGA, implicating TAF9 structural dynamics in the formation pathways of the holo-complexes.
Chaperonin CCT interacts with the WD40 domain of TAF5
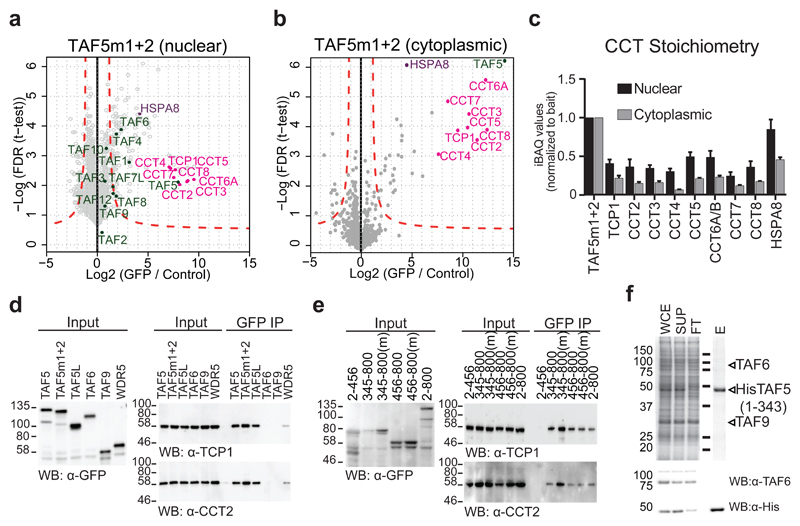
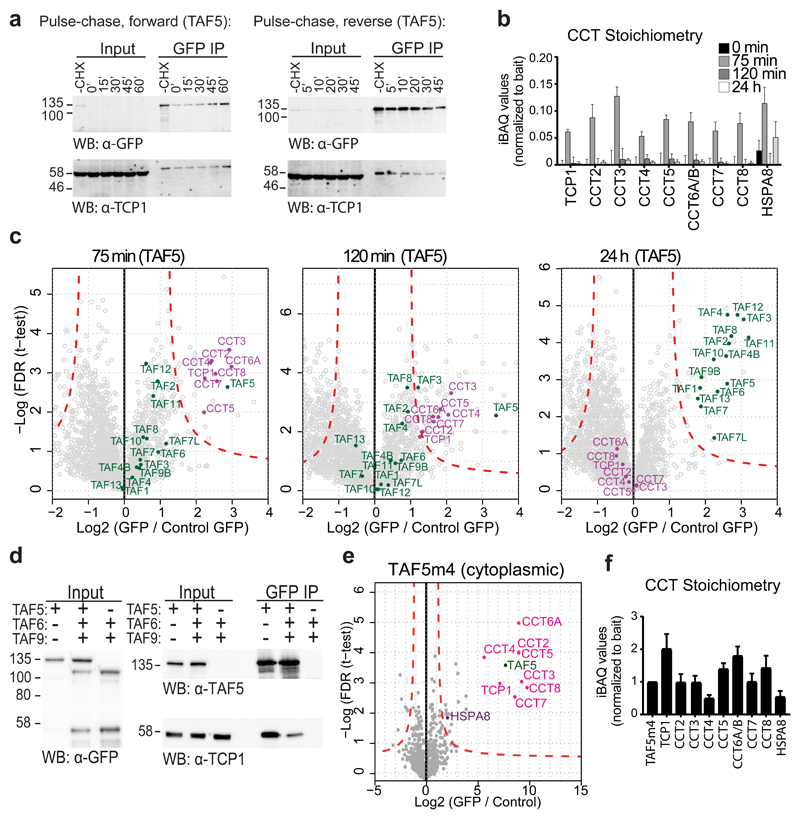
Cells expressing GFP-TAF5m1+2 exhibit marked GFP signal in the cytoplasm, in contrast to predominantly nuclear fluorescence of the other mutant and wild-type GFP-TAF5 cell lines (Fig. 3b). Intriguingly, TAF5m1+2 co-IPs revealed eight highly enriched proteins in close to equimolar amounts, which we identified as the subunits of the CCT complex (Fig. 4a,b). CCT assists folding of about 10% of the proteome, including many WD40 repeat proteins16, 17, 30. In addition, strong enrichment of the HSPA8 protein was observed (Fig. 4a,b), suggesting that HSPA8 is involved in TAF5 delivery to the CCT chaperonin31. We reasoned that CCT may facilitate folding of the TAF5 C-terminal WD40 repeat domain to allow handover to the TAF6-TAF9 heterodimer, as an essential step in holo-TFIID formation. Our qMS experiments show that TAF5m1+2 is deficient in accreting TFIID subunits including TAF6-TAF9 (Fig. 4a-c). We speculated that the TAF5m1+2 protein may be retained in the CCT folding chamber, possibly due to impaired interactions with TAF6-TAF9. We asked whether our observation only related to this particular mutant. Careful inspection confirmed the presence of CCT, albeit in lower amounts, in all GFP-TAF5 mutant co-IPs (Supplementary Fig. 8a,b). Intriguingly, CCT was likewise detected in co-IPs of wild-type GFP-TAF5 from the cytoplasm, but it was absent in the nuclear extract, suggesting transient CCT interactions with newly-synthesized TAF5 (Supplementary Fig. 8b). Similar co-IPs of GFP-tagged TBP, TAF1, TAF6, TAF7 and TAF9 did not contain any CCT subunit (data not shown). We conclude that the CCT interaction is highly specific for the TAF5 subunit of TFIID.
Figure 4. Chaperonin CCT engages TAF5.
a-c GFP-TAF5m1+2 enriches all subunits of CCT in virtually stoichiometric ratios. Chaperonin subunits are colored in magenta, TFIID subunits in green, HSPA8 in purple. Relative stoichiometry is normalized to bait. Each data point in volcano plots is plotted as average of technical triplicates. Dashed red lines denote threshold between background and significant enrichment (two-tailed t-test; FDR = 1%; S0 =1). Each bar in stoichiometry plot represents an average of technical triplicates. Error bars = s.d. of mean. n = 2 independent experimental replicates for the representative GFP-TAF5m1+2 nuclear sample. d, GFP-TAF interactions with CCT in whole cell extract from transient transfection experiments by immunoblot. Input represents 1% of the protein sample used in the co-IPs. Hallmark CCT subunits TCP1 and CCT2 were probed. Technical replicates n = 3. e, Transient transfection experiments with whole cell extract GFP-fusions of WT or m1+2 (m) TAF52-456 (ΔWD40), TAF5345-800 (ΔNTD) and TAF5456-800 (WD40) probed by co-IPs and immunoblot. Technical replicates n = 2. f, Talon pull-down of co-expressed TAF6, TAF9 and truncated TAF5 analyzed by SDS-PAGE (left) and immunoblot (right). WCE stands for whole cell extract, SUP for cleared lysate, FT for flowthrough, E for eluted fraction His for oligohistidine tag. Source data for c are available online. Uncropped blot/gel images are shown in Supplementary Data Set 1.
We used transiently transfected HEK293T cells to substantiate our findings. Overexpressing GFP-TAF5, GFP-TAF5m1+2 or SAGA-specific GFP-TAF5L, strongly enriched CCT in co-IPs, unlike GFP-tagged TAF6 and TAF9 or WDR5, a transcriptional regulator with a comparable WD40 repeat domain (Fig. 4d), confirming that TAF5 and its close paralogue TAF5L are bona fide CCT substrates. Note that WDR5 can be produced in E. coli implying that WDR5 folding does not depend on a eukaryotic chaperone32. Transient overexpression of TAF5 fragments confirmed that the WD40 domain is mediating the CCT interaction and that the NTD did not interact with CCT subunits (Fig. 4e). Recombinant reconstitution also confirmed that the TAF5-TAF6-TAF9 complex is dependent on the TAF5 WD40 domain (Fig. 4f).
CCT hands over nascent TAF5 to TAF6-TAF9 for holo-TFIID assembly
CCT-TAF5 complex formation would presumably occur following translation in the cytoplasm. We tested this in pulse-chase experiments in our Dox-inducible cell lines (Fig. 5a-c, Supplementary Fig. 9a,b). Prior to adding Dox, cycloheximide (CHX) was applied to block translation, resulting in mRNA accumulation. In the chase phase, CHX block and Dox were removed and nascent GFP-TAF5 sampled over time, revealing progressive CCT binding to newly-synthesized GFP-TAF5 in co-IP experiments (Fig. 5a). GFP-TAF5m1+2 yielded comparable results; GFP-TAF7 was included as a negative control (Supplementary Fig. 9b). In reverse pulse-chase experiments, prolonged Dox-induction enabled GFP-TAF5 protein accumulation, followed by CHX addition to inhibit new protein synthesis. We observed time-dependent disassociation of GFP-TAF5 from CCT. In contrast, TAF5m1+2 remained bound to the chaperonin. Mass spectrometry analysis of GFP-TAF5 induced cells showed that newly-synthesized TAF5 enriched all CCT subunits (Fig. 5b), with CCT binding peaking at 75 min before declining, corroborating the transient nature of the interaction. Notably, co-IPs at later time points progressively enriched TAFs and TBP, while CCT disappeared into background, consistent with TAF5 release from CCT and incorporation into holo-TFIID complex (Fig. 5c).
Figure 5. CCT is required for holo-TFIID assembly.
a, CCT interaction with newly-synthesized GFP-TAF5 was probed by pulse-chase experiments. Input represents 5% of the protein sample used in co-IPs. b, c, Pulse-chase experiment sampled at different time-points. CCT binding to TAF5 peaks at 75 min after inducing GFP-TAF5 expression. Relative abundance was normalized to bait. Baseline was taken at t=0. CCT subunits are colored in magenta and TFIID subunits in green. Each data point in volcano plots is plotted as average of technical triplicates. Dashed red lines denote threshold between background and significant enrichment (two-tailed t-test; FDR = 1%; S0 =1). Each bar in stoichiometry plot represents an average of technical triplicates. Error bars = s.d. of mean. d, Transient co-transfection experiments with GFP-TAF5, GFP-TAF6 and GFP-TAF9 probed by co-IPs and immunoblot. TAF6-TAF9 is required for releasing TAF5 from CCT. Technical replicates n = 2. Uncropped blot/gel images are shown in Supplementary Data Set 1. e, f, qMS analysis of GFP-TAF5m4 co-IPs from cytoplasmic fraction. Stoichiometry plot is normalized to bait. Each data point in volcano plots is plotted as average of technical triplicates. Each bar in stoichiometry plot represents an average of technical triplicates. Error bars = s.d. of mean. Source data for b and f are available online.
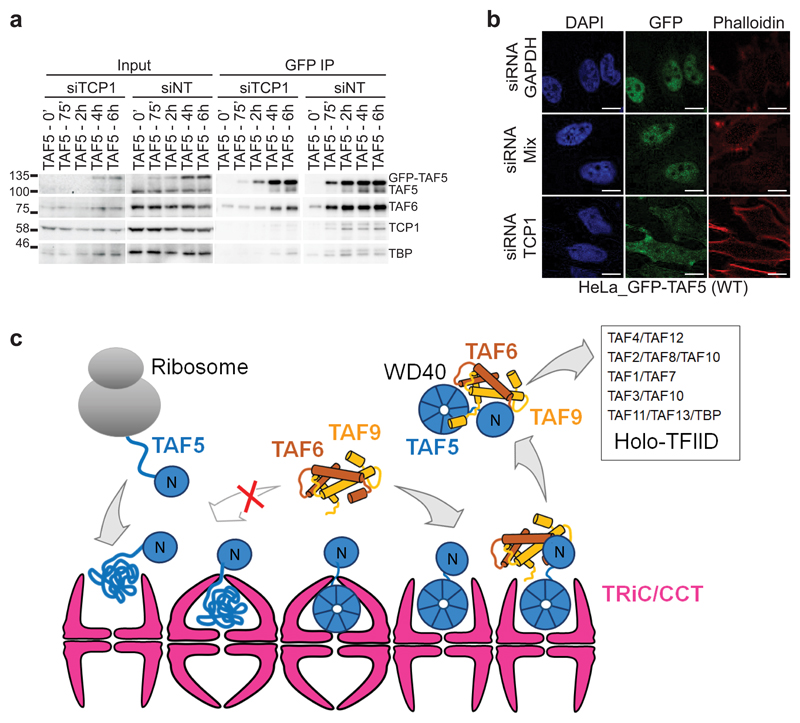
Our model implies that handover of TAF5 from CCT to TAF6-TAF9 would be controlled by the levels of TAF6-TAF9 heterodimer and be dependent on the initial encounter of TAF6-TAF9 with the NTD of TAF5. Indeed, transient co-expression experiments of GFP-tagged TAF5, TAF6 and TAF9 in combinations underscored that TAF6-TAF9 is required for TAF5 release from CCT (Fig. 5d). We designed a TAF5 mutant in the NTD (TAF5m4), predicted to disrupt the interaction with TAF6-TAF9 (Fig. 2h and Supplementary Fig. 7). Co-IP of this mutant followed by qMS showed that TAF5m4 does not form TFIID submodules and that it is retained in the CCT complex (Fig. 5e,f). These data indicate that the CCT complex acts as an essential checkpoint in TFIID assembly. Next, we investigated the effect of siRNA mediated knockdown of TCP1, which affects CCT function16. TCP1 knockdown in GFP-TAF5 induced cells markedly reduced co-purification of TAF6 as well as TBP, indicating that CCT action is a prerequisite to prime TAF5 for TAF6-TAF9 binding and incorporation into TFIID (Fig. 6a). Consistent with this, TCP1 knockdown impacted cellular localisation, with a significant portion of GFP-TAF5 now retained in the cytoplasm (Fig. 6b).
Figure 6. CCT chaperonin-assisted early events in TFIID assembly.
a, siRNA-mediated knockdown of CCT subunit TCP1 impedes GFP-TAF5 integration into TFIID. Control cells were treated with a non-targeting siRNA (siNT). Input represent 2% of the protein sample used in each co-IP. Technical replicates n = 2. Uncropped blot/gel images are shown in Supplementary Data Set 1. b, TCP1 knockdown affects nuclear localization of GFP-TAF5; scale bar = 10 μm. Technical replicates n = 2. c, Newly- synthesized TAF5 is captured by the chaperonin (left). The TAF5 N-terminal domain folds autonomously, Folding of the WD40 repeat domain depends on CCT. Release of readily folded TAF5 from the chaperonin requires binding of TAF6-TAF9, resulting in a discrete TAF5-TAF6-TAF9 complex in the cytoplasm. Binding of further TAFs and TBP completes functional holo-TFIID assembly.
Discussion
Taken together, we provide here unique, molecular level insight into early events in regulated assembly of a basal transcription factor, TFIID. Our results provide compelling evidence that the chaperonin CCT associates with newly-synthesized TAF5 in the cytoplasm of cells, acting as an essential checkpoint for functional TFIID holo-complex formation (Fig. 6c, Supplementary Video). Our data demonstrate that CCT intervention is a prerequisite for forming a TFIID submodule comprising TAF5, TAF6 and TAF9. Presumably, CCT facilitates folding of the WD40 repeat domain and stabilizes TAF5 in a conformation compatible with forming the multitude of interactions with TAF6-TAF9 via the TAF5 NTD as observed in the crystal structure.
Our results evidence that TAF6-TAF9 binding is required to release TAF5 from CCT and convey that CCT rebinding may be prevented by docking of α-helix 4 of TAF9 on the lateral surface of the TAF5 WD40 domain (Fig. 2A, Supplementary Video). This handover is critical for forming holo-TFIID complex containing a full complement of TAFs and TBP. Additionally, we identify TAF5-TAF6-TAF9 as a discrete cytoplasmic TFIID subcomplex poised to engage other TAFs including the TAF4-TAF12 heterodimer, thus substantiating the concept of holo-TFIID assembly from preformed submodules and pinpointing a chaperonin as a key factor in this process.
Our finding that not only TAF5, but also its paralogue TAF5L interact with CCT, implies that the handover mechanism described here is not confined to TFIID and similar mechanisms may also govern early stages of SAGA assembly. We anticipate that our observations will have general implications for the molecular mechanisms regulating assembly of the many multiprotein machines at work in cells.
Methods
TAF5-TAF6-TAF9 complex production
TAF5-TAF6-TAF9 full-length complex was cloned, expressed and purified using a polyprotein strategy as described previously3. For all other TAF5-TAF6-TAF9 complexes, coding sequences of individual subunits and truncations were cloned into acceptor (pFL) and donor (pIDC and pIDK) vectors of the MultiBac system by sequence and ligation independent cloning (SLIC) and fused by in vitro Cre-LoxP recombination to yield a single plasmid with multiple expression cassettes33. Constructs used are listed in Supplementary Table 1. A decahistidine tag followed by a Tobacco Etch Virus (TEV) NIa protease cleavage site was placed at the N-terminus of the TAF5 subunit. Recombinant baculovirus was produced as previously described34 and used to infect SF21 insect cells (Invitrogen) at a cell density of 1.0x106/mL in SF4 medium. Cells were harvested 72-96 h after proliferation arrest by centrifugation at 4,000 g for 15 min. Cell pellets were resuspended in lysis buffer (25 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM Imidazole, 1 mM β-mercaptoethanol, 0.1 % NP-40), incubated on ice for 30 min and the lysate was cleared by centrifugation at 25,000 rpm in a JA-25.50 rotor (Beckman) for 90 min. The supernatant was mixed with Talon resin (Clontech), equilibrated in Talon A buffer (25 mM Tris-HCl pH 7.5, 300 mM NaCl, 5 mM Imidazole) and incubated for 1 h at 4° C. The resin was washed with 15 column volumes (CV) of Talon A buffer, 10 CV of Talon HS buffer (25 mM Tris-HCl pH 7.5, 1 M NaCl, 7.5 mM Imidazole), and 15 CV of Talon A buffer before eluting the bound protein complex with 5 CV of Talon B buffer (25 mM Tris-HCl pH 7.5, 200 mM NaCl, 300 mM Imidazole). 6xHis-TEV protease (prepared in house) was added at a 1:100 w/w ratio and the protein was dialyzed against a 100-fold excess of dialysis buffer (25 mM Tris-HCl pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol). The dialyzed protein was passed over Ni-NTA resin (Qiagen) equilibrated in dialysis buffer and the flow through was collected. It was then subjected to a MonoQ 5/50GL column (GE Healthcare). The protein complex was found in the flow through and separated on a Superdex S200 16/60 column (GE Healthcare) equilibrated in SEC buffer (10 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 1 mM DTT). Peak fractions were pooled and concentrated to 10 mg/mL. Small aliquots were frozen in liquid nitrogen and stored at -80 °C until further use. In TAF5ΔWD40-TAF6-TAF9 pull-down experiments with full-length TAF6 and TAF9 co-expressed with a TAF5 truncation (TAF51-343) lacking the WD40 domain (Fig. 4e), samples were separated by SDS-PAGE, blotted and probed by specific antibody (Tora laboratory) according to standard immunoblot procedures.
Crystallization, data collection and structure determination
The TAF5194-800-TAF61-92RLRRRAH-TAF91-120 complex was crystallized in 0.1 M Tris-HCl pH 6.8, 0.8 M Na-citrate and 0-0.15 M NaCl using the hanging drop vapor diffusion technique by mixing 2 μL of protein solution at 5.4-7.5 mg/mL with 1 μL of reservoir solution and equilibrated against 500 μL of reservoir solution at 4C. Optionally, crystallization drops were seeded with 0.5 μL of a 1:10.000 to 1:1.000.000 diluted micro-seed stock solution, which was prepared by crushing a single crystal in 10 μL reservoir solution. Crystals of pyramidal shape first appeared after four days and reached their maximal size of 300 x 300 x 300 μm after ten days.
For data collection at cryogenic temperatures, crystals were transferred sequentially into 2 μL drops containing 0.1 M Tris-HCl pH 6.8, 0.15 M NaCl and increasing concentrations of Na-citrate from 1.0 M to 1.6 M before flash freezing in liquid nitrogen. For experimental phasing, crystals of the complex were transferred to a 2 μL drop containing a stabilizing solution of 0.1 M Tris-HCl pH 6.8, 1.0 M Na-citrate and 0.15 M NaCl. Solid Ta6Br12 cluster was directly added to the crystallization drop containing the pre-equilibrated crystals. After 16 h, the Ta6Br12 soaked crystals were sequentially transferred into 2 μL drops containing 0.1 M Tris-HCl pH 6.8 with 0.15 M NaCl and increasing concentrations of Na-citrate from 1.2 M to 1.6 M before flash freezing in liquid nitrogen. Native datasets were collected at beamline ID14-4 (ESRF, Grenoble, France) at 100 K (1.0044 Å wavelength). Datasets of Ta6Br12 soaked TAF5-TAF6-TAF9 complex crystals were collected at beamline ProximaI (SOLEIL, Gif-sur-Yvette, France) following an optimized inverse beam SAD data collection strategy at the peak wavelength for Ta (1.25439 Å). The Ta6Br12 soaked crystals were non-isomorphous to native TAF5-TAF6-TAF9 complex crystals.
All diffraction data were processed with the XDS software package35,36. The structure of the TAF5-TAF6-TAF9 complex was solved using the Ta6Br12 SAD dataset. Three Ta6Br12 cluster sites were identified by HySS as implemented in PHENIX software suite37. The heavy atom substructure was further refined and phases were calculated with PHASER38. The initial electron density map improved significantly by density modification in PARROT39, allowing placement of four truncated molecules of the human TAF5-NTD domain (PDB-ID 2NXP) and four molecules of the D. melanogaster TAF6-TAF9 HFD pair (PDB-ID 1TAF) manually into the electron density map. Subsequently, the experimental electron density map was further improved by NCS averaging in RESOLVE40,41. The improved map allowed placement of four molecules of a 7-bladed WD40 repeat model obtained from the PHYRE server42 (based on the TAF5 protein sequence) by molecular replacement in MOLREP43. This initial model was used to phase the native dataset by molecular replacement in MOLREP. After automated model building in ARP/wARP44 and BUCCANEER39, the model was manually adjusted in repetitive rounds of refinement in PHENIX and model building in COOT. TLS refinement was used in the final rounds of refinement with 61 individual TLS groups as determined by PHENIX.
Pull-down assay of TAF5NTD-TAF6-TAF9
Cell pellet was resuspended in buffer (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM Imidazole, EDTA free complete protease inhibitor (Roche), lysed by 2 cycles of freeze thaw in liquid nitrogen and the lysate was cleared by centrifugation at 19,000 g in a Thermo Scientific™ Fiberlite™ F21-8 x 50y fixed-angle rotor for 90 min. The supernatant was mixed with Talon resin (Clontech), equilibrated in buffer A (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM Imidazole) and incubated for 1 hour at 4° C. The resin was washed with 15 column volumes (CV), 10 CV of buffer HS (25 mM Tris-HCl pH 8.0, 1000 mM NaCl, 5 mM Imidazole), and 15 CV of buffer A before eluting the bound protein complex with 5 CV of buffer B (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 200 mM Imidazole). Different samples were analysed by SDS-PAGE analysis and western blot analysis using HRP conjugated anti-His antibody (Sigma-Aldrich) for TAF5NTD and anti-TAF6 (primary, received from Laszlo Tora) followed by HRP conjugated anti-mouse (secondary, Sigma-Aldrich) for TAF6.
GFP-TAF cell line generation
Human cDNA was amplified by PCR with gene-specific primers fused to attB recombination sequences for GATEWAY cloning. The amplified sequence was recombined into the pDON201 donor vector according to manufacturer’s protocol (ThermoFisher). The cloned sequence was verified with Sanger sequencing and was transferred to the pcDNA5_FRT_TO_N_GFP26 Gateway destination vector by LR recombination according to manufacturer’s protocol (ThermoFisher). TAF9 and TAF5 mutant cDNAs were purchased from GenScript (Piscataway, NJ, USA). Both TAF5m1+2 and TAF5m2+3 were obtained in a single round of mutagenesis PCR. All destination vectors were co-transfected with pOG44 plasmid encoding for the Flp recombinase into HeLa Flp-In/T-REx cells26 using polyethyleneimine (PEI) transfection to generate stable Dox-inducible expression cell lines.
Cell culture methods
HeLa Flp-In/T-REx cells, containing the Flp Recombination Target site and expressing the Tet Repressor, were grown in Dulbecco's Modified Eagle's Medium (DMEM), 4.5 g/L glucose, supplemented with 10% v/v fetal bovine serum, 10 mM L-glutamine and 100 U/mL penicillin/streptomycin (all purchased from Lonza), together with 5 μg/ml blasticidin S (InvivoGen, San Diego, CA) and 200 μg/ml zeocin (Invitrogen, Carlsbad, CA) as selection drugs for FRT and Tet repressor, respectively. All cell lines used were mycoplasma-negative. Recombined cells were selected by replacing zeocin by 250 μg/mL hygromycin B (Roche Diagnostics, Mannheim, Germany) 48 h after PEI transfection. Expression of GFP-tagged protein was induced by addition of 1 μg/mL doxycycline, for 16 to 18 h. HEK293T cells used for transient expression experiments were grown in Dulbecco's Modified Eagle's Medium (DMEM), 4.5 g/L glucose, supplemented with 10% v/v fetal bovine serum, 10 mM L-glutamine and 100 U/mL penicillin/streptomycin (all purchased from Lonza).
Immunoblotting procedures
Cell were seeded in 6-well dishes at 30,000 cell/well and induced with doxycycline for to 18 h prior to harvesting. Cell lysates were prepared in 1x sample buffer (160 mM Tris-HCl pH 6.8, 4% SDS, 20% glycerol, 0.05% bromophenol blue). Equal amounts of cell lysates were separated by a 10% SDS-PAGE and transferred onto PVDF membrane. GFP purification samples for immunoblotting were collected by elution with sample buffer. The membrane was developed with the appropriate antibodies and ECL. Immunoblots were analyzed using ChemiDoc imaging system (BioRad). The images were subjected to linear contrast/brightness enhancement in Photoshop (CS6, 13.0.6 x64, extended) when needed for data representation purposes. Antibodies used in the study include: GFP (JL-8, Clontech), TCP1 (91A, ThermoFisher), CCT2 (kind gift from Willianne Vonk/Judith Frydman), GAPDH (clone 6C5, mAb374, Millipore), Penta His (Qiagen, Hilden, Germany), TAF5 (in-house, 25TA 2G7) and TAF6 (in-house, 1TA-1C2).
Extract preparation and GFP co-immunoprecipitation (co-IP)
Cells were seeded in 15-cm dishes (Greiner Cellstar, Sigma-Aldrich) and grown to 70-80% confluence prior to doxycycline induction. GFP-protein expression was verified using EVOS fluorescence microscopy (Thermo Fischer). Next, induced cells were harvested and nuclear and cytoplasmic extracts were obtained using a modified version of the Dignam and Roeder procedure45. Protein concentrations were determined by Bradford assay (BioRad). 1 mg of nuclear or 3 mg of cytoplasmic extract was used for GFP-affinity purification as described46. In short, protein lysates were incubated in binding buffer (20 mM Hepes-KOH pH 7.9, 300 mM NaCl, 20% glycerol, 2 mM MgCl2, 0.2 mM EDTA, 0.1% NP-40, 0.5 mM DTT and 1x Roche protease inhibitor cocktail) on a rotating wheel for 1 h at 4° C in triplicates with GBP-coated agarose beads (Chromotek) or control agarose beads (Chromotek). The beads were washed two times with binding buffer containing 0.5% NP-40, two times with PBS containing 0.5% NP-40, and two times with PBS. On-bead digestion of bound proteins was performed overnight in elution buffer (100 mM Tris-HCl pH 7.5, 2 M urea, 10 mM DTT) with 0.1 μg/ml of trypsin at RT and eluted tryptic peptides were bound to C18 stagetips prior to mass spectrometry analysis.
Whole cell extracts were collected in lysis buffer (50 mM Tris-HCl pH 7.9, 10% Glycerol, 100 mM NaCl, 10 mM MgCl2, 0.5 mM DTT, 0.1% NP40 and 1x complete protease inhibitor cocktail (Roche)), by incubating the cells with the lysis buffer on ice for 10 min. Next the protein samples were collected by manual harvesting and centrifuged at 14,000 rpm C for 20 min at 4° C. Supernatant was snap-frozen and stored at -80° C.
Mass spectrometry and data analysis
Tryptic peptides were eluted from the C18 stagetips in H2O:acetonitril (50:50) with 0.1% formic acid and dried prior to resuspension in 10% formic acid. A third of this elution was injected into Q-Exactive (Thermo Fischer) in the MS/MS mode with 90 min total analysis time. Blank samples consisting of 10% formic acid were run for 45 min between GFP and non-GFP samples, to avoid carry-over between runs. The raw data files were analyzed with MaxQuant software (version 1.5.3.30) using Uniprot human FASTA database46,47. Label-free quantification values (LFQ) and match between run options were selected. Intensity based absolute quantification (iBAQ) algorithm was also activated for subsequent relative protein abundance estimation27. The obtained protein files were analyzed by Perseus software (MQ package, version 1.5.4.0), in which contaminants and reverse hits were filtered out. Protein identification based on non-unique peptides as well as proteins identified by only one peptide in the different triplicates were excluded to increase protein prediction accuracy.
For identification of the bait interactors LFQ intensity-based values were transformed on the logarithmic scale (log2) to generate Gaussian distribution of the data. This allows for imputation of missing values based on the normal distribution of the overall data (in Perseus, width = 0.3; shift = 1.8). The normalized LFQ intensities were compared between grouped GFP triplicates and non-GFP triplicates, using 1% permutation-based false discovery rate (FDR) in a two-tailed t-test. The threshold for significance (S0), based on the FDR and the ratio between GFP and non-GFP samples, was kept at the constant value of 1 for comparison purposes. Relative abundance plots were obtained by comparison of the iBAQ values of GFP interactors. The values of the non-GFP iBAQ values were subtracted from the corresponding proteins in the GFP pull-down and were next normalized on a chosen co-purifying protein for scaling and data representation purposes27.
siRNA mediated knockdown experiments
Cells were transfected using HiPerfect and reverse transfection protocol based on the manufacturer’s instructions (Qiagen, Valencia, CA). After 72 h cells were collected for immunoblotting, mRNA analysis or immunofluorescent confocal microscopy. Protein lysates were collected in sample buffer. RNA was extracted using RNeasy kit following the manufacturer’s protocol including a DNase treatment step (Qiagen, Valencia, CA). For immunofluorescent analysis cells grown on coverslips were fixed in 4% Paraformaldehyde-PBS for 20 min, RT, and stored in 0.4% PF-PBS at 4° C prior to analysis.
RT-qPCR analysis
cDNA for RT-qPCR analysis was synthesized from 500 ng of the total RNA using Superscript II (Invitrogen, Carlsbad, CA) with the supplied random primers mix. RT-qPCR was performed in a 25 μl final volume using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). qPCR analysis was performed in CFX Connect Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and extracted data analyzed in Excel following the ΔΔCq method.
Confocal microscopy
Paraformaldehyde-fixed cells were subjected to short cell membrane permeabilization with 0.5% Triton X-100 in PBS for 5 min. After quenching the cross-linking with 50 mM glycine, the samples were blocked for 30 min at RT with 5% natural goat serum (NGS) and subsequently incubated with primary antibodies for 2 hours at RT. After three sequential washing steps with PBS, secondary antibodies conjugated to a fluorophore were added, together Phalloidin (Abcam) for staining of actin for 45 min at RT. For nuclear staining, the samples were incubated with DAPI dye (2 mg/L, Sigma-Aldrich, St. Louis, MO) for 5 min prior to mounting the coverslips on microscopy slides (ThermoFischer Scientific, Waltham, MA) with Immu-Mount (Invitrogen, Carlsbad, CA).
Images were obtained using SP8-X confocal microscope (Leica microsystems, Germany) using a HC PL APO 63x/1.40 Oil CS2 objective. Gain and offset settings were adjusted according to the fluorescence signal, but they were kept constant in comparative experimental designs such as doxycycline-induction tests. Exported files were next subjected to linear contrast and brightness processing in Photoshop (CS6, 13.0.6 x64, extended) for image representation purposes.
Pulse-chase experiments
Forward setup (tracing newly-synthesized proteins): cells were seeded in 10 cm culture dishes and grown to approximately 90% confluence. Next, protein translation was blocked by addition of 50 μg/ml cycloheximide (CHX; Sigma-Aldrich). 30 min after CHX block, cells were induced with 1 μg/ml Dox for 6 hours to accumulate GFP-protein mRNA. Prior to sample collection, cells were washed with PBS two times for complete removal of CHX and medium without Dox was refreshed for the duration of the chosen time points. Whole cell extracts were collected for each of the time points and used for GFP affinity purification. Enriched proteins were separated with SDS-PAGE and detected by immunoblotting.
Reverse setup (blocking new protein synthesis): cells were grown in 10 cm culture dishes to 70% confluence prior Dox-induction. 50 μg/ml of CHX was added after 16 hours of induction to block new protein synthesis and to allow gradual disassociation of the newly-synthesized proteins from their respective folding partners. Cells were harvested at different time points after translational block induction and used for GFP affinity purification in order to analyze associated proteins. Samples were analyzed by SDS-PAGE and immunoblotting.
Supplementary Material
One Sentence Summary.
Proteomics and structure analyses reveal assembly mechanisms of the transcription factor TFIID identifying chaperonin CCT as a checkpoint in the process.
Acknowledgments
We thank S. Trowitzsch (Goethe University Frankfurt), P. Legrand and A. Thompson (Synchrotron SOLEIL, Gif-sur-Yvette), W. Vonk (Princess Maxima Centre Utrecht), R. Baas (Netherlands Cancer Institute Amsterdam) and M. Vermeulen (Radboud University Nijmegen) for assistance and reagents. We greatly appreciate discussion with R. Sawakar (MPI for Immunobiology and Epigenetics, Freiburg). This research was supported by Netherlands Organization for Scientific Research (NWO) grants 022.004.019 (S.V.A), ALW820.02.013 (H.T.M.T), and 184.032.201 Proteins@Work (E.C, T.Y.L., H.R.V., A.J.R.H), a Kékulé fellowship from the Fonds der Chemischen Industrie (M.H.), a European Research Council Advanced grant ERC-2013-340551 (L.T.), Agence Nationale de Recherche research grants ANR-10-IDEX-0002-02 and ANR-10-LABX-0030-INRT (L.T.) and a Wellcome Trust Senior Investigator Award 106115/Z/14/Z (I.B.). This work used the platforms of the Grenoble Instruct-ERIC Center (ISBG UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (ANR-10-INBS-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). This research received support from BrisSynBio, a BBSRC/EPSRC Research Centre for synthetic biology at the University of Bristol (BB/L01386X/1).
Footnotes
Accession codes
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 6F3T. All mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under the dataset identifier PXD011293.
Data availability Statement
Atomic coordinates and crystallographic structure factors have been deposited in the Protein Data Base under accession code PDB 6F3T. Proteomics data has been deposited in the PRIDE database under accession code PXD011293. All other data supporting findings in this study are available from the corresponding authors upon reasonable request.
Author Contributions:
H.T.M.T. and I.B. conceived the study with input from S.V.A., M.H., L.T. and A.J.R.H.. S.V.A. carried out all cell biology experiments, assisted by M.M. and E.S.. The majority of the quantitative proteomics analyses were carried out by E.C. and T.Y.L., assisted by S.V.A.. R.M.E. and H.R.V. carried out proteomics analyses of the pulse-chase experiment. M.H. carried out all recombinant protein work, crystallization and structural analysis, assisted by L.S., K.G. and with input from I.B.. S.V.A., M.H., L.T., A.J.R.H., H.T.M.T. and I.B. designed experiments, interpreted data and wrote the manuscript together with input from all authors.
Competing Interest Statement
The authors declare no competing interests.
References
- 1.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniossek C, et al. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- 4.Louder RK, et al. Structure of promoter bound TFIID and model of human pree-initiation complex assembly. Nature. 2016;531:604–609. doi: 10.1038/nature17394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller F, Zaucker A, Tora L. Developmental regulation of transcription initiation: more than just changing the actors. Curr Opin Genet Dev. 2010;20:533–540. doi: 10.1016/j.gde.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Helmlinger D, Tora L. Sharing the SAGA. Trends Biochem Sci. 2017;42:850–861. doi: 10.1016/j.tibs.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baptista T, et al. SAGA is a General Cofactor for RNA Polymerase II Transcription. Mol Cell. 2017;68:130–143.e135. doi: 10.1016/j.molcel.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warfield L, et al. Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID. Mol Cell. 2017;68:118–129.e115. doi: 10.1016/j.molcel.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spedale G, Timmers HT, Pijnappel WW. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 11.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 12.Ellis RJ. Assembly chaperones: a perspective. Philos Trans R Soc Lond B Biol Sci. 2013;368 doi: 10.1098/rstb.2011.0398. 20110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 14.Ramos PC, Dohmen RJ. PACemakers of proteasome core particle assembly. Structure. 2008;16:1296–1304. doi: 10.1016/j.str.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 16.Lopez T, Dalton K, Frydman J. The Mechanism and Function of Group II Chaperonins. J Mol Biol. 2015;427:2919–2930. doi: 10.1016/j.jmb.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pines A, et al. TRiC controls transcription resumption after UV damage by regulating Cockayne syndrome protein A. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03484-6. 1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz IG, et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol. 2011;18:14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- 19.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 20.Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nat Rev Mol Cell Biol. 2010;11:789–801. doi: 10.1038/nrm2973. [DOI] [PubMed] [Google Scholar]
- 21.Wright KJ, Marr MT, 2nd, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci U S A. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trowitzsch S, et al. Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat Commun. 2015;6 doi: 10.1038/ncomms7011. 6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta K, et al. Architecture of TAF11/TAF13/TBP complex suggests novel regulation properties of general transcription factor TFIID. Elife. 2017;6 doi: 10.7554/eLife.30395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheer E, Delbac F, Tora L, Moras D, Romier C. TFIID TAF6-TAF9 complex formation involves the HEAT repeat-containing C-terminal domain of TAF6 and is modulated by TAF5 protein. J Biol Chem. 2012;287:27580–27592. doi: 10.1074/jbc.M112.379206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 26.van Nuland R, et al. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol. 2013;33:2067–2077. doi: 10.1128/mcb.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya S, Takada S, Jacobson RH. Structural analysis and dimerization potential of the human TAF5 subunit of TFIID. Proc Natl Acad Sci U S A. 2007;104:1189–1194. doi: 10.1073/pnas.0610297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie X, et al. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 30.Miyata Y, et al. The molecular chaperone TRiC/CCT binds to the Trp-Asp 40 (WD40) repeat protein WDR68 and promotes its folding, protein kinase DYRK1A binding, and nuclear accumulation. J Biol Chem. 2014;289:33320–32. doi: 10.1074/jbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuellar J, et al. The structure of CCT-Hsc70 NBD suggests a mechanism for Hsp70 delivery of substrates to the chaperonin. Nat Struct Mol Biol. 2008;8:858–64. doi: 10.1038/nsmb.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han Z, et al. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. doi:10.1016. [DOI] [PubMed] [Google Scholar]
- 33.Haffke M, Viola C, Nie Y, Berger I. Tandem recombineering by SLIC cloning and Cre-LoxP fusion to generate multigene expression constructs for protein complex research. Methods Mol Biol. 2013;1073:131–140. doi: 10.1007/978-1-62703-625-2_11. [DOI] [PubMed] [Google Scholar]
- 34.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3:1021–32. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/s0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/s0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/s0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/s0907444906022116. [DOI] [PubMed] [Google Scholar]
- 40.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchr Rad. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 41.Terwilliger TC. Automated structure solution, density modification and model building. Acta Crystallogr D Biol Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- 42.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 43.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/s0907444909042589. [DOI] [PubMed] [Google Scholar]
- 44.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protocols. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carey MF, Peterson CL, Smale ST. Dignam and Roeder nuclear extract preparation. CSH Protocols. 2009;2009 doi: 10.1101/pdb.prot5330. pdb.prot5330. [DOI] [PubMed] [Google Scholar]
- 46.Spruijt CG, Baymaz HI, Vermeulen M. Identifying specific protein-DNA interactions using SILAC-based quantitative proteomics. Methods Mol Biol. 2013;977:137–157. doi: 10.1007/978-1-62703-284-1_11. [DOI] [PubMed] [Google Scholar]
- 47.Low TY, et al. Quantitative and qualitative proteome characteristics extracted from in-depth integrated genomics and proteomics analysis. Cell Rep. 2013;5:1469–1478. doi: 10.1016/j.celrep.2013.10.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.