Abstract
Kinetochores are multicomponent complexes responsible for coordinating the attachment of centromeric DNA to mitotic-spindle microtubules. The point centromeres of budding yeast are organized into three centromeric determining elements (CDEs), and are associated with the centromere-specific nucleosome Cse4. Deposition of Cse4 at CEN loci is dependent on the CBF3 complex that engages CDEIII to direct Cse4 nucleosomes to CDEII. To understand how CBF3 recognizes CDEIII and positions Cse4, we determined a cryo-EM structure of a CBF3−CEN complex. CBF3 interacts with CEN DNA as a head-to-head dimer that includes the whole of CDEIII and immediate 3’ regions. Specific CEN-binding of CBF3 is mediated by a Cep3 subunit of one of the CBF3 protomers that forms major groove interactions with the conserved and essential CCG and TGT motifs of CDEIII. We propose a model for a CBF3−Cse4−CEN complex with implications for understanding CBF3-directed deposition of the Cse4 nucleosome at CEN loci.
Centromeres mediate the attachment of chromosomes to the mitotic spindle by coordinating the assembly of kinetochores, large proteinaceous complexes that bind microtubules 1–3. In mitosis, kinetochores couple chromosome motion to the energy released by microtubule depolymerisation, which powers chromosome segregation. Incorrectly attached kinetochores trigger a mitotic checkpoint pathway that inhibits the anaphase-promoting complex to prevent chromosome mis-segregation 4–6. The regional centromeres of higher eukaryotes comprise megabases of chromosomal DNA and engage multiple microtubules. In contrast, the point centromeres of budding yeast chromosomes 7 are defined by conserved ~125 nucleotide segments 8,9 that are sufficient to direct kinetochore-mediated attachment of each chromosome to a single microtubule 10–12. Budding yeast point centromeres are organized into three elements: CDEI, CDEII and CDEIII 8 (Fig. 1i). The short CDEI and CDEIII segments are highly conserved across all 16 S. cerevisiae chromosomes, whereas CDEII elements, although lacking sequence similarity, share an A-T rich region of 80-90 nucleotides. Disruption of the invariant CCG motif of CDEIII inactivates the centromere 13–15.
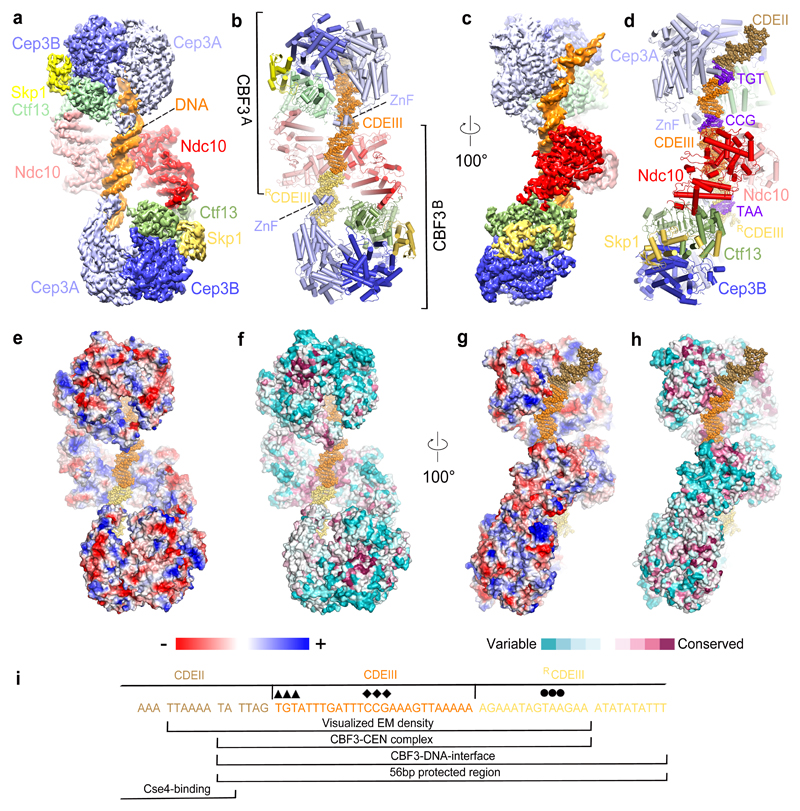
Figure 1. Overall structure of the dimeric−CEN3 complex.
(a) and (c) Two orthogonal views of the EM density map of the complex, (b) and (d) Corresponding cartoon representations. The TGT and CCG motifs of CDEIII are colored blue. (e) and (g) Electrostatic surface representation of the dimeric CBF3−CEN3 complex. Color code is displayed below. (f) and (h) Surface conservation of the CBF3−CEN3 dimer. Conservation score is indicated below. (i) Region of CEN3 sequence and 3’ flanking region recognized by the dimeric CBF3-CEN3 complex. Sequences indicated with diamonds and circles are the CCG and TAA motifs recognized by the Zn2Cys6 cluster of CBF3A and CBF3B, respectively. The TGT motif contacted by αMN helix of Cep3A is indicated with arrowheads.
Centromeres of all species are marked by centromere-specific nucleosomes in which the canonical histone H3 is replaced by a specific histone (CENP-A); CENP-A in animals and Cse4 in S. cerevisiae 16. CENP-A nucleosomes coordinate kinetochore assembly by recruiting inner kinetochore scaffolding subunits such as CENP-C (Mif2) and CENP-N (Chl4). CENP-A nucleosome deposition at human centromeres is epigenetically specified by pre-existing CENP-A nucleosomes. In S. cerevisiae, the multi-protein CBF3 complex assembles on CDEIII of CEN loci 17 to direct Cse4 nucleosome deposition centered onto the adjacent CDEII 18–20, creating a nuclease resistant region of ~200 bp 21–24. The four proteins that comprise CBF3 (Cep3, the F-box protein Ctf13, Skp1 and Ndc10) are essential for yeast viability with Cep3, Ctf13 and Skp1 forming CBF3core. The Cep3 homodimer incorporates two DNA-binding Gal4-like Zn2-Cys6 clusters 25, one of which provides CEN sequence specificity by engaging the essential CCG motif of CDEIII 14,15,26–28. Ctf13 and Ndc10 contribute to CBF3-CEN interactions by binding to DNA without sequence specificity 26,27. To understand how CBF3 interacts with S. cerevisiae centromeric DNA and mediates Cse4 nucleosome deposition to CEN loci 19,20, we reconstituted CBF3-CEN DNA complexes for single particle cryo-EM analysis.
Results
Overall architecture of CBF3
To generate a CBF3−CEN complex, all four CBF3 proteins were co-expressed, the CBF3 complex was purified and then assembled onto the 147 bp CEN3 DNA duplex 29 (Supplementary Fig. 1a). Cryo-electron micrographs of the CBF3−CEN3 complex revealed a heterogeneous mixture of particles (Supplementary Fig. 2a-d and Table 1 and Supplementary Table 1). Approximately a third of particles consisted of CBF3core (Cep3 dimer, Ctf13, Skp1) without DNA. However, other particles had larger sizes, with one notable class featuring an extended structure with two prominent lobes joined by a smaller central lobe (Supplementary Fig. 2a,b – boxed particles). 3D reconstruction and classification indicated that these particles correspond to a dimeric CBF3−CEN3 complex in which two CBF3core complexes, bridged by a central Ndc10 dimer, are bound to a single CEN3 DNA duplex. This creates a structure with pseudo dyad symmetry. The entire CBF3−CEN3 complex was reconstructed to an overall resolution of 4.4 Å. Using multi-body refinement in RELION 30, segmented rigid domains of CBF3−CEN3 were determined at higher resolution (Fig. 1 and Supplementary Fig. 2d-g,I). We also observed monomeric CBF3−CEN3 complexes (Supplementary Fig. 2d and Supplementary Table 1). To obtain a higher-resolution reconstruction of CBF3 for atomic-model building, we masked the EM density corresponding to a single CBF3 protomer (CBF3msk) in the 3D classes of both dimeric and monomeric CBF3−CEN3 complexes and used focused 3D refinement. The resultant CBF3msk reconstruction was determined to 3 Å, sufficient for de novo modeling with confidence (Table 1 and Supplementary Fig. 2d,h,j).
Table 1. Cryo-EM data collection, refinement and validation statistics.
| CBF3core−Ndc10DBD (EMD-0095, PDB 6GYP) |
CBF3−CEN3 (EMD-0096, PDB 6GYS) |
CBF3msk (EMD-0099, PDB 6GYU) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 132,075 | 132,075 | 132,075 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | ~28 | ~28 | ~28 |
| Defocus range (μm) | 2.4-3.5 | 2.4-3.5 | 2.4-3.5 |
| Pixel size (Å) | 1.06 | 1.06 | 1.06 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 757,433 | 987,484 | 987,484 |
| Final particle images (no.) | 73,894 | 22,668 | 198,010 |
| Map resolution (Å) | 3.6 | 4.4 | 3.0 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 3.3-4.5 | 3.7-8.3 | 2.5-3.7 |
| Refinement | |||
| Initial model used (PDB code) | 2QUQ,4ACO,1FQV,2HAP | 2QUQ,4ACO,1FQV,2HAP | 2QUQ,1FQV,2HAP |
| Model resolution (Å) | 3.6 | 4.4 | 3.0 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | |||
| Map sharpening B factor (Å2) | -120 | -110 | -120 |
| Model composition | |||
| Non-hydrogen atoms | 17,811 | 38,882 | 18,157 |
| Protein residues | 2,800 | 5,688 | 2,800 |
| Ligands | 0 | 8 | 2 |
| B factors (Å2) | |||
| Protein | 174.4 | 304.1 | 148.9 |
| Ligand | - | 395.4 | 181.3 |
| R.m.s. deviations | 0.006 | 0.007 | 0.005 |
| Bond lengths (Å) | 0.895 | 1.018 | 0.872 |
| Bond angles (°) | 0.693 | 0.811 | 0.780 |
| Validation | |||
| MolProbity score | 1.71 | 1.77 | 1.72 |
| Clashscore | 4.86 | 6.07 | 5.00 |
| Poor rotamers (%) | 0.50 | 0.29 | 0.74 |
| Ramachandran plot | |||
| Favored (%) | 92.83 | 93.36 | 93.58 |
| Allowed (%) | 7.08 | 6.48 | 6.28 |
| Disallowed (%) | 0.09 | 0.16 | 0.14 |
In a related complex, (CBF3core−Ndc10DBD−CEN3), we replaced full-length Ndc10 with Ndc10DBD (residues 1-540) that includes only its DNA-binding domain (DBD) and lacks the C-terminal residues (Ndc10C) shown to mediate Ndc10 dimerization 31,32 (Supplementary Fig. 1b). EM analysis of CBF3core−Ndc10DBD−CEN3 showed mainly a mixture of DNA-free CBF3core and CBF3core-Ndc10DBD particles suggesting that Ndc10C enhances CBF3−CEN3 DNA binding (Supplementary Fig. 3a). We determined 3D reconstructions of DNA-free CBF3core and CBF3core−Ndc10DBD at 3.9 Å and 3.6 Å, respectively (Supplementary Fig. 3 and Table 1 and Supplementary Table 1).
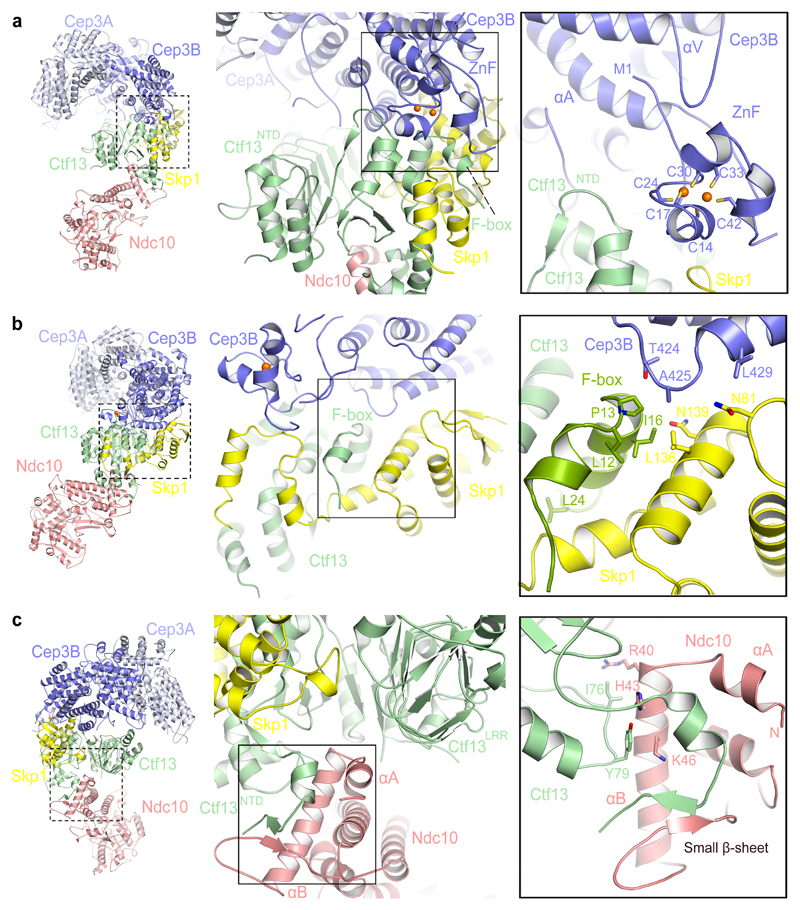
The high quality of the 3 Å EM density map of CBF3msk derived from the CBF3−CEN3 cryo-EM data allowed atomic modeling and refinement of its complete structure (Supplementary Fig. 2h,j,k and Table 1). This was then used to interpret the 3.6 Å map of CBF3core−Ndc10DBD (without DNA) (Supplementary Fig. 3f and Table 1) and the CEN3-bound CBF3 dimer (Fig. 1 and Supplementary Fig. 2e-g and Table 1). CBF3core forms a rigid assembly through extensive contacts involving its component subunits; a Cep3 homodimer and Ctf13-Skp1 heterodimer (Fig. 1b,d). Our CBF3core structure resembles that of the recently published CBF3core 33, although at higher resolution, allowing the building of Ctf13. The Cep3B subunit of the Cep3 homodimer contacts both subunits of the Ctf13-Skp1 heterodimer with its N-terminal Zn2-Cys6 (ZnF) cluster becoming ordered by a composite interface formed of Ctf13 (including the F-box) and Skp1 (Fig. 2a,b). Skp1 interacts with Cep3B and cullin subunits through the same interface, explaining how Cep3 protects Ctf13 from ubiquitin-dependent degradation 34 28. The Zn2-Cys6 cluster of the Cep3A subunit of CBF3core−Ndc10DBD (without DNA) is disordered, similar to that in crystal structures of free Cep3 35,36. However in both Cep3 subunits, Met1 and Phe2 are anchored within a hydrophobic pocket formed by the αA and αV helices. For Cep3A, this would restrict the conformational variability of its Zn2-Cys6 cluster. When bound to CEN3, the Cep3A Zn2-Cys6 cluster engages the CCG motif of CDEIII 28 (Fig. 1a and Fig. 3a,g,i). In contrast to the large interfaces shared between subunits of CBF3core, Ndc10 associates only with Ctf13 and this is through a relatively small contact surface area (1354 Å2) situated opposite the Ctf13-Cep3B interface (Fig. 1a and Fig. 2c). Due to this small interface Ndc10 appears flexible in the DNA-free complex (Supplementary Fig. 4b).
Figure 2. CBF3 inter-subunit interfaces.
(a) Interactions of the Cep3B Zn2Cys6 cluster at the Skp1 and Ctf13 interface. (b) Contacts of the Ctf13 F-box with Skp1 and Cep3B. L12, P13 and L24 are essential for Ctf13−Skp1 interactions. (c) The N-terminus of Ndc10 contacts Ctf13. Mutating I76 and Y79 of Ctf13 disrupts Ctf13-Ndc10 interactions (Supplementary Fig. 1c).
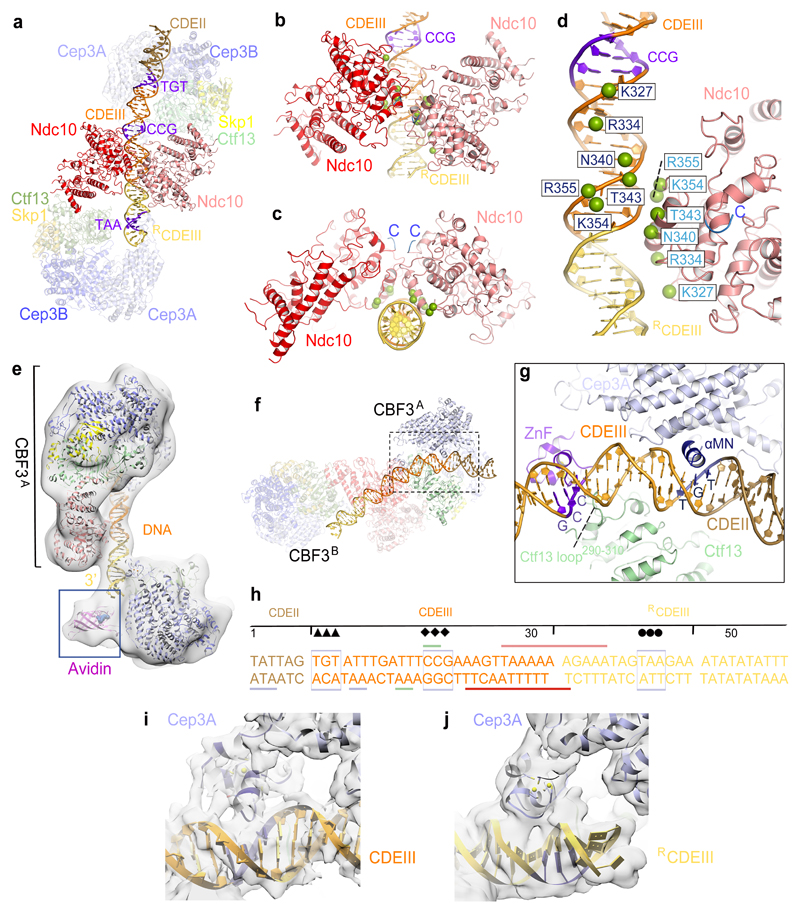
Figure 3. Basis for CBF3−CEN3 recognition.
(a) Overview of the dimeric CBF3−CEN3 complex. (b) and (c) Two views of the Ndc10DBD-DNA interface. DNA induces dimerization of Ndc10DBD. (d) Details of Ndc10DBD−DNA contacts. Residues in contact with DNA are shown as green spheres. Cartoon representation of only one Ndc10 subunit is shown. (e) EM density map of the CBF3−CEN3-biotin avidin complex orients and positions the DNA duplex. (f) Overview of Cep3B-Ctf13 contacts with DNA in CBF3A. (g) Details of Cep3A−Ctf13−CDEIII interface, showing Zn2Cys6 cluster contacting the CCG motif and αMN helix contacting the TGT motif. Ctf13 contacts the minor groove midway between the CCG and TGT motifs. (h) Schematic of CBF3-DNA interactions. Sequence of the nuclease resistant 56 bp segment protected by CBF3 is shown 17. Arrow, diamond and circle symbols as in Fig. 1i. Colored horizontal lines indicate contacts to CBF3 subunits. (i) and (j) EM density of the Zn2Cys6 clusters interacting with CEN3-DNA for (i) Cep3A of CBF3A and (j) Cep3A of CBF3B.
In the 3 Å map of CBF3msk well defined EM density for Ctf13 allowed complete tracing of the protein sequence (Table 1). As proposed from biochemical studies 28, Ctf13 is at the center of the complex (Fig. 1a and Fig. 2a). The N terminal F-box of Ctf13 (residues 1-28) forms hydrophobic interactions with both Skp1 and Cep3B (Fig. 2a,b). Relative to other Skp1-F-box interactions, Ctf13 forms extensive contacts to Skp1 involving its mainly α-helical N-terminal domain (Ctf13NTD). This domain also contacts the Zn2-Cys6 cluster and Ndc10DBD (Fig. 2a,b). Interactions of the conserved Leu12, Pro13 and Leu24 residues of the F-box with Skp1 are consistent with their biochemically defined function of stabilizing Ctf13−Skp1 interactions 28 (Fig. 2b). The C-terminal region of the LRR domain of Ctf13 (Ctf13LRR) contacts both Skp1 and Cep3B, the latter contact augmented by the α-helical insert of Ctf13LRR.
Ndc10 interacts with CBF3core through an N-terminal α-helical hairpin that inserts into a crevice between Ctf13NTD and Ctf13LRR (Fig. 2c). Helix αA of the hairpin, not visible in free Ndc10 31,32, becomes ordered through interactions with Ctf13. The Ctf13−Ndc10 interface is augmented by a small anti-parallel β-sheet formed of the N-terminal β-strands of both proteins (Fig. 2c). Ile76 and Tyr79 of Ctf13NTD interact with Arg40 and Lys46 of the Ndc10 αB helix respectively. Replacing Ile76 and Tyr79 with either Ala or Arg abolished Ctf13 binding to Ndc10 (Supplementary Fig. 1c), validating the importance of this interaction in stabilizing CBF3. A mitotic stability assay, assessing the efficiency of mini-chromosome segregation, confirmed the importance of this interface in vivo (Supplementary Fig. 5). In yeast strains harboring the Ctf13 mutants, the efficiency of mini-chromosome segregation was severely reduced, equivalent to that caused by disruption of the essential CCG motif of CDEIII. Combining the Ctf13 and CCG mutants did not further impair mini-chromosome segregation, showing that the Ctf13 and CCG mutations disrupt an inter-dependent function.
CBF3 interacts with CEN3 DNA as a dimer
We fitted the refined CBF3msk coordinates and a 47 bp DNA duplex into the EM density map of the CBF3−CEN3 dimer (Fig. 1a-d). Two CBF3 protomers (CBF3A and CBF3B), bound to a gently bent DNA duplex, associate through the central dimerized Ndc10 (Fig. 1a,b and Fig. 3a and Supplementary video 1). EM density for Ndc10 subunits in CBF3−CEN3 is similar to that in CBF3core−Ndc10DBD (without DNA) with no visible density for Ndc10C in the majority of defined 3D classes (Supplementary Fig. 2d). This indicates significant flexibility of Ndc10C. In the low-pass filtered maps, we visualized the fragmented densities of two Ndc10DBD C-termini situated in close proximity, suggesting the possibility that two Ndc10 subunits, self-associated through Ndc10DBD on DNA, may also self-associate through their Ndc10C (Fig. 3c and Supplementary Fig. 6a). Ndc10C-dimerization would likely stabilize the DNA-bound complex, an idea supported by the very small proportion of dimeric CBF3−CEN3 complexes in the CBF3core−Ndc10DBD−CEN3 cryo-EM dataset.
At 4.3 Å, the resolution of the CEN3 EM density map did not allow definitive identification of DNA bases (Fig. 1a,c and Supplementary Fig. 2g). Thus, to determine the orientation and register of CEN3 DNA, we prepared CEN3 DNA biotinylated at the CDEIII end for avidin labeling. The labeled DNA maintained affinity for CBF3. A cryo-EM reconstruction of the CBF3−CEN3−avidin complex at 12.5 Å revealed clear EM density for the avidin label only at one DNA end (Fig. 3e). We then fitted a DNA duplex that is consistent with CBF3 CEN-DNA crosslinking data 27, and that positions the Cep3A Zn2-Cys6 cluster at the CDEIII CCG motif 28.
The CBF3 dimer binds to CEN3 DNA in a roughly two-fold symmetric manner, although because the protein complex extends beyond the 3’end of CEN3 DNA, the DNA duplex is positioned asymmetrically with respect to the CBF3 dimer (Fig. 1b,d). CBF3A contacts CDEIII and 6 bp of CDEII. Strikingly, this corresponds to the 5’ end of the 56 bp region protected from DNA cleavage in the CBF3−CEN3 DNA complex 17 (Fig. 1i). CBF3B contacts the entire DNA duplex (14 bp) 3’ of CDEIII (Fig. 1i). Only 12 bp of CDEII are visible, with CEN3 DNA 5’ of this segment being disordered. Both CBF3A and CBF3B engage the DNA duplex through a channel created by the Cep3 dimer and Ctf13 (Fig. 1a and Fig. 3a). The 3’end of the CEN3 DNA exiting the CBF3A DNA-binding channel then contacts Ndc10 of CBF3A some 10 bp 3’ of the CCG motif (Fig. 3a-d). Ndc10−DNA interactions in the CBF3−CEN3 complex involve non-specific DNA sugar-phosphate interactions and are identical to those of the K. lactis Ndc10−DNA complex 31, and consistent with a mutagenesis study of S. cerevisiae Ndc10 32 (Fig. 3b-d). In the CBF3−CEN3 complex however, a dimer of Ndc10DBD is arranged with dyad symmetry on the DNA duplex. The two Ndc10DBD self-associate through a small interface of 451 Å2 surface area that involves the α-helix responsible for mediating Ndc10-DNA interactions. As Ndc10DBD is not known to dimerize without DNA 32, this protein-protein interface is presumably of low intrinsic affinity.
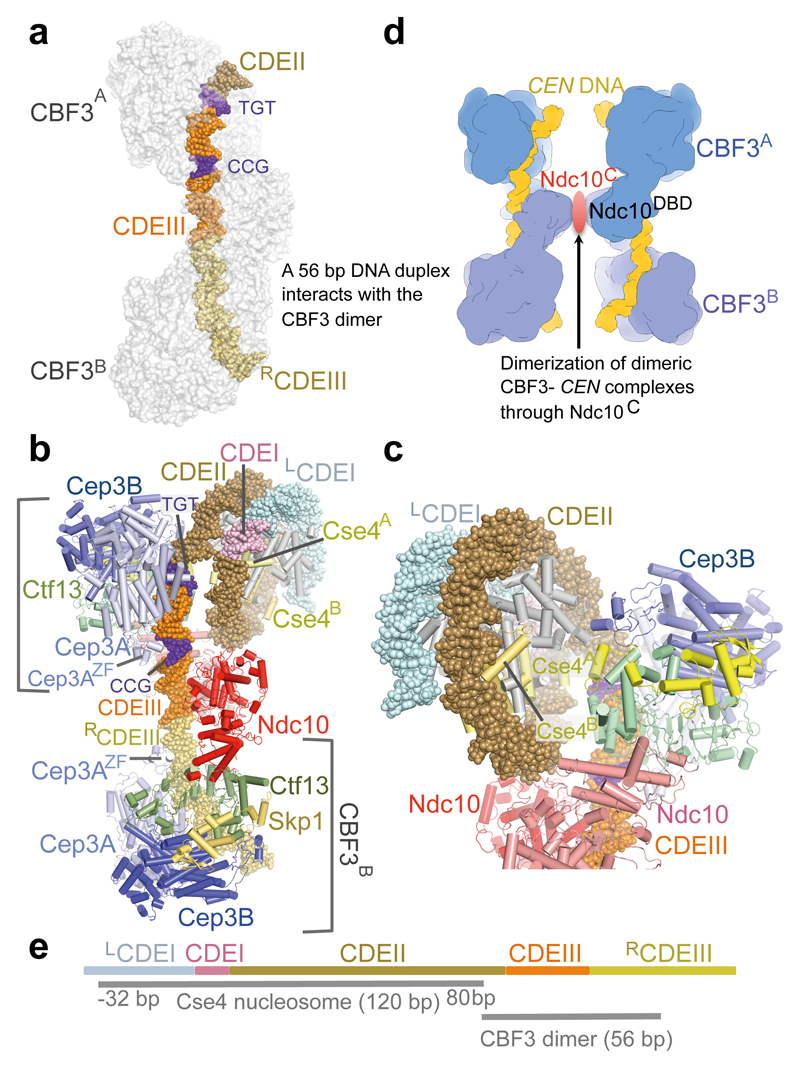
CBF3B extends beyond the DNA duplex present in the CBF3−CEN3 complex. We therefore modeled an extended DNA duplex downstream of CBF3B guided by the DNA contacts with CBF3A (Fig. 4a). This indicated that the DNA in contact with the CBF3 dimer comprises CDEIII together with 6 bp of CDEII and 24 bp to the right of CDEIII (RCDEIII) (Fig. 1i and Fig. 4a). The modeled complex exactly matches the 56 bp DNase resistant core protected by CBF3 17.
Figure 4. Model of CBF3−Cse4 nucleosome complex.
(a). Model of dimeric CBF3 with nuclease resistant 56 bp DNA segment. CBF3 is represented with a transparent molecular surface. (b) CBF3−Cse4−CEN structure modeled on the cryo-EM CBF3−CEN complex and the CENP-A crystal structure 40 (PDB 3AN2). The two CENP-A subunits (labeled as Cse4A and Cse4B) are highlighted. (c) Close up of the view of the CBF3−Cse4−CEN model showing close proximity of the N-terminus of Cse4A and Ctf13. (d) Cartoon showing possible pairing of CBF3−CEN complexes mediated by dimerization of Ndc10C domains. (e) Schematic of the proposed CBF3−Cse4 nucleosome complex at the CEN3 locus. CDEI, CDEII and CDEIII, and the DNA contact regions of Cse4 and CBF3 are drawn to scale. For Cse4, the start and end of the binding site are shown, -32 bp: 32 bp to the left of the LCDEI-CDEI junction, 80 bp: 80 bp to the right of the CDEI-CDEII junction.
Cep3A of CBF3A forms two important sequence-specific contacts to CDEIII. The Zn2-Cys6 cluster engages the major groove of DNA contacting the essential CCG motif 28 (Fig. 3f-i). Modeling of this interaction was based on the Hap1−DNA structure 37. The second sequence-specific contact is created by residues 318-328 of Cep3A that in the presence of CEN3 fold into an α-helix (αMN) that inserts into the major groove of DNA 1.25 turns 5’ to the CCG motif. The αMN helix contacts the conserved TGT motif that is essential for optimal CBF3 binding 28 (Fig. 3g,h and Supplementary Fig. 6b). In between these two Cep3A sequence-specific contact sites, the Ctf13 loop290-310 inserts into the minor groove and interacts with the DNA phosphate backbone (Fig. 3g). In the CBF3B promoter of the CBF3 dimer, the Zn2Cys6 cluster of Cep3A inserts into the major groove of RCDEIII some 20 bp 3’ of the CCG motif (Fig. 1b and Fig. 3a,h,j). Our structural results agree with DNA-protein crosslinking data that Cep3 interacts with the CCG and TGT motifs of CDEIII with Ctf13 contacting a region of CDEIII midway between these motifs 27,28. The positively charged DNA binding interfaces of CBF3 are evolutionarily conserved among budding yeasts (Fig. 1e-h). Our cryo-EM model of the dimeric CBF3−CEN3 complex shares a marked resemblance to atomic force microscopy images of a CBF3−CEN complex 38. These AFM images revealed a bilobal globular domain from which extend two DNA arms subtended by ~55°. Similar to our structure, CBF3 was located asymmetrically to the right of CDEIII.
While our paper was under review the cryo-EM structure of CBF3core with monomeric Ndc10 was reported 39. This is essential similar to our DNA-free monomeric CBF3core-Ndc10DBD reconstruction (Supplementary Fig. 3f).
Implications for CBF3-Cse4 nucleosome interactions
In human CENP-A nucleosomes, 120 bp of α-satellite DNA generate 1.4 gyres to surround the histone octamer 40. Budding yeast Cse4 nucleosomes deposited on CEN DNA are centered on CDEII and protect a micrococcal nuclease-resistant kernel of 125-135 bp 19. In our CBF3−CEN cryo-EM structure the curvature of the 12 bp of CDEII matches that of the CENP-A nucleosome DNA 40. We modeled a CBF3-Cse4-CEN3 complex, assuming the Cse4 nucleosome is an octamer and wraps 115-120 bp of DNA 29,41. We placed the entry point of the nucleosome four bp 5’ of the CDEII-CDEIII junction (Fig. 1i and Fig. 4b,c). The modeled CBF3-Cse4-CEN3 complex is devoid of steric clashes, superimposes the major and minor grooves of the CBF3-bound and nucleosome DNA, and allows Ndc10 to interact with CDEII and possibly with Cbf1 engaged at CDEI, in agreement with a previous study 42. The N-terminus of Cse4 approaches Ctf13 (Fig. 4c), suggesting a mechanism for specific recognition of Cse4 nucleosomes by CEN-bound CBF3 43. Our model is compatible with both Cse4−H4−H2A−H2B hemisomes 44 and octameric nucleosomes 45. Whereas a left-handed supercoiled DNA can be readily accommodated, consistent with previous reports 29,41, our modeling suggests that a nucleosome with right-handed chirality would collide with CBF3A (Supplementary Fig. 6c,d). The 120 bp wrapped by the Cse4 nucleosome together with 56 bp of the CBF3 dimer would generate a protein-DNA interface of ~180 base pairs. This matches well to the single-base pair resolution mapping of Cse4 nucleosomes 20,46,47 and the size of nuclease resistant centromere cores 18,21,24. Due to the symmetry of the CBF3 dimer bound to the CEN3 DNA, an interesting feature of this model is the possibility that the CBF3 dimer interacts with two Cse4 nucleosomes. Budding yeast centromeres containing more than one Cse4 nucleosome have been proposed 48.
In the CBF3−CEN3 structure, Ndc10DBD is dimerized on DNA (Fig. 3a-c). Using SEC-MALS we analyzed the molecular mass of CBF3 in the presence and absence of CEN3 DNA. This indicated that CEN3 DNA enhanced the dimerization of a CBF3core−Ndc10 dimer to generate a CBF3−CEN3 complex incorporating two Ndc10 dimers (Supplementary Fig. 7), consistent with Ndc10 forming higher order multimers 28. Extended 89 bp DNA segments (33 nucleotides to the right of the core 56 bp segment) bind an additional CBF3 complex that is stimulated by Ndc10 27. One possibility is that a dimer of Ndc10 dimers mediates bridging of two DNA-bound CBF3 dimers facilitating either DNA looping or pairing of sister chromatids (Fig. 4d). Intriguingly, AFM images of CBF3−CEN DNA complexes revealed paired complexes from which project three to four DNA arms 38.
Discussion
The structure of a CBF3−CEN3 complex provides the first insights into the architecture of point centromeres, revealing a head-to-head CBF3 dimer that explains the CBF3 nuclease protected region of CEN DNA 17 and CBF3−CEN crosslinking data 27. Three of the CBF3 subunits (Cep3, Ctf13 and Ndc10) interact with DNA. Two zinc fingers of the CBF3 dimer, one from each Cep3 dimer of the two CBF3 protomers directly engage the major groove of the DNA. The DNA-binding zinc finger of Cep3A of CBF3A interacts with the conserved CCG of the CDEIII motif of CEN3, explaining the crucial role and conservation of the CCG motif 13–15. The second sequence-specific contact within CDEIII involves the αMN helix of Cep3B of CBF3A that contacts the conserved TGT motif that is essential for optimal CBF3 binding 28. Both the zinc finger of Cep3A and αMN helix of Cep3B assume order on binding CDEIII. We present a model for the CBF3−Cse4−CEN3 complex that supports the idea that CENP-A nucleosomes wrap DNA as a left-handed helix. The close proximity of the CENP-A specific Cse4 protein to CBF3 suggests a mechanism for how CBF3 would recruit Cse4 nucleosomes to CEN loci thereby creating a platform for the assembly of the constitutive centromere-associated network (CCAN) of the inner kinetochore by engaging CENP-C and CENP-N. This study provides a framework for understanding the structure of the centromere-inner kinetochore interface.
Online Methods
Cloning, expression and purification of recombinant CBF3−CEN3 complex
Coding fragments of Cep3, Ndc10, Ctf13 and Skp1 were amplified by PCR from Saccharomyces cerevisiae genomic DNA and cloned into a pU1 plasmid using a modified Multibac expression system 49 as described 50. A double StrepII tag together with a TEV cleavage site was attached to the C-terminus of Cep3. All four genes were further assembled into a pU2 plasmid for CBF3 complex expression. We define CBF3 as Cep3, Ctf13, Skp1 and Ndc10. CBF3core as Cep3, Ctf13 and Skp1. CBF3 is equivalent to CBF3core-Ndc10. Ndc101-540 (Ndc10DBD) was combined with Cep3, Ctf13 and Skp1 into pU2 for CBF3core-Ndc10DBD expression. The C-termini of Ndc10 and Ndc10DBD were modified with a TEV-His tag for Ndc10 and Ndc10DBD purification. The Ctf13I76A-Y79A and Ctf13I76R-Y79R mutations were made and combined with Cep3, Ndc10 and Skp1 in pU2 for the mutant CBF3 complexes. All complexes were expressed in the insect cell-baculovirus system using Hi5 cells as described 50.
To prepare CEN3 DNA, 24 copies of the 147 base pairs of CEN3 29 (sequence: ATC AAA TAG TAC AAA TAA GTC ACA TGA TGA TAT TTG ATT TTA TTA TAT TTT TAA AAA AAG TAA AAA ATA AAA AGT AGT TTA TTT TTA AAA AAT AAA ATT TAA AAT ATT AGT GTA TTT GAT TTC CGA AAG TTA AAA AAG AAA TAG TAA GAA GAT: red indicates the CEN3 sequence, CDEI and CDEIII, underlined) were assembly into a pUC18 plasmid, separated by EcoRV sites. The plasmid was isolated from E. coli using a Plasmid Giga Kit (Qiagen). CEN3 was purified with resource Q anion exchange chromatography (GE Healthcare) after digestion with EcoRV-HF (NEB) at 37 °C overnight. For CEN3 CDEIII-end biotin-avidin labeling, two PCR primers were synthesized (Sigma) with a 5’-biotin modification on the reverse primer. The PCR product of CEN3biotin was incubated in PBS with avidin protein (Sigma) on ice for 30 min, resulting in CEN3biotin-avidin.
Ndc10, Cep3, Ctf13 and Skp1 genes were co-expressed in Hi5 cells. Forty-eight hours after infection Hi5 cells were harvested and the lysate was loaded onto a Strep-Tactin ® Column (QIAGEN) and the CBF3 complex was eluted with 2.5 mM desthiobiotin (Sigma). The tag was cleaved using TEV protease overnight at 4 °C. The complex was further purified on Resource Q anion exchange and Superose 6 size exclusion chromatography. To compensate for sub-stoichiometric levels of Ndc10 (which expressed at lower levels than Cep3, Ctf13 and Skp1) in the CBF3 complex, Ndc10 was expressed in Hi5 cells and purified by HisTrap™HP column (GE Healthcare) following Resource Q anion exchange and S200 size exclusion chromatography.
CBF3core−Ndc10DBD complex was purified by HisTrap™HP column (GE Healthcare) and the His6-tag was cleaved by TEV protease overnight at 4 °C. After a second HisTrap™HP column the complex was further purified by Resource Q anion exchange and Superose 6 size exclusion chromatography.
To prepare the CBF3−CEN3 complex, CBF3 and CEN3 were incubated in a 1:1 molar ratio with addition of additional Ndc10 at 22 °C for 20 min. The complex was purified by Superose 6 size exclusion chromatography. Similar procedures were applied for the CBF3core−Ndc10DBD−CEN3 and CBF3−CEN3biotin-avidin complexes.
Multi-Angle Light Scattering (MALS)
SEC-MALS was performed using a Wyatt MALS system. CBF3 and CBF3−CEN3 complexes were injected onto an analytical Superose 6 gel filtration column pre-equilibrated in 20 mM Hepes (pH 8.0), 200 mM NaCl, 0.5 mM DTT. The light scattering and protein concentration at each point across the peaks in the chromatograph were used to determine the absolute molecular mass from the intercept of the Debye plot using Zimm’s model as implemented in the ASTRA v5.3.4.20 software (Wyatt Technologies Corp.). In order to determine the inter-detector delay volumes, band broadening constants and the detector intensity normalization constants for the instrument, we used BSA as a standard prior to sample measurement.
Electron microscopy data collection
3.0 μl of the CBF3core−Ndc10DBD, CBF3core−Ndc10DBD−CEN3, CBF3−CEN3 and CBF3−CEN3biotin-avidin complexes at concentrations of ~0.4 mg/ml were applied to glow-discharged copper 300 mesh Quantifoil R1.2/1.3 holey carbon grids (Quantifoil Micro Tools GmbH) (no carbon support). The grids were flash plunged into liquid ethane using an FEI Vitrobot Mark IV (waiting time, 20 s, blotting time, 2 s). A total of four data sets were collected: (i) CBF3core−Ndc10DBD; (ii) CBF3core−Ndc10DBD−CEN3; (iii) CBF3−CEN3 and (iv) CBF3−CEN3biotin-avidin. EM image stacks were collected with a Falcon III camera in counting mode on a FEI Titan Krios electron microscope at a nominal magnification of 75 K (yielding a pixel size of 1.06 Å). The images were recorded at a dose rate of 0.5 electrons per pixel per second and the total exposure time was 60 s (75 frames) with the FEI automated low-dose data-collection program EPU. Defocus varied from -2.2 to -3.4 µm with an interval of 0.2 µm.
Image processing
Movie frames were first aligned using MotionCor2 51. CTF parameters were estimated with Gctf 52. The initial template-free particle picking was performed with Gautomatch (developed by Kai Zhang, http://www.mrc-lmb.cam.ac.uk/kzhang/Gautomatch/). Subsequent image processing was carried out in RELION 2.1 package 53. To determine a reconstruction of CBF3core−Ndc10DBD (Supplementary Fig. 3 and Table 1) from the CBF3core−Ndc10DBD data set, a subset of 420 micrographs (of 724) was used for Gautomatch template-free particle picking, and the resulting 124,092 coordinates were imported into RELION 2.1 for particle extraction and reference-free 2D classification. Selected averages from 2D classification were used for an initial model reconstruction with SIMPLE-PRIME 54. This was used for template-based particle auto-picking in RELION from both the CBF3core−Ndc10DBD and CBF3core−Ndc10DBD−CEN3 data sets. The extracted particles were subject to two rounds of reference-free 2D classifications resulting in a dataset of 625,194 particles from the combined total of 1,895 micrographs. A tandem cascade of 3D classification against the model built with SIMPLE-PRIME was performed, and initial iterations were performed without angular search restriction for each round of classification. 145,747 particles were assigned to CBF3core−Ndc10DBD, whereas 184,356 were assigned to the CBF3core. After multi-reference based 3D classifications, 87,156 particles of CBF3core−Ndc10DBD were selected for the final refinement. For CBF3core, different conformations of the zinc finger motifs were characterized after a multi-reference 3D classification. The dataset including all the particles generated the highest resolution reconstruction. The final resolutions for CBF3core−Ndc10DBD and CBF3core are 3.58 Å and 3.91 Å, respectively, based on the gold-standard FSC=0.143 criterion 55 (Supplementary Fig. 3g).
For the CBF3−CEN3 dataset (Supplementary Fig. 2 and Table 1), a similar workflow was applied to the image processing, and a low-pass filtered structure of CBF3core−Ndc10DBD was used as a reference in the 3D classification. A dimeric CBF3−CEN3 complex and a monomeric CBF3−CEN3 complex were characterized. A total of 99,089 particles were assigned to the dimeric CBF3−CEN3 complex and 22,668 (class 5) of these were seen bound to a longer DNA duplex. This class was used for reference-based refinement to generate a 4.4 Å map (Supplementary Fig. 2d). 197,322 particles were assigned to the monomeric CBF3−CEN3 complex. We applied multi-body refinement in RELION 30 to segment three domains of the dimeric CBF3−CEN3 complex to improve their local resolutions: (i) the upper CBF3A-core; (ii) CEN3 DNA with associated Ndc10DBD and (iii) the lower CBF3B-core, resulting in reconstructions at 3.89 Å, 4.33 Å and 4.16 Å, respectively (Supplementary Fig. 2e-g). The flexible orientation between the CBF3core and Ndc10DBD is stabilized by the CEN DNA (Supplementary Fig. 4). A local mask encompassing the upper CBF3 complex only (CBF3A) (with Zn cluster of Cep3B) was employed for both a 3D classification and the subsequent density improvement in the refinement. All the particles assigned to either the dimeric CBF3−CEN3 or the monomeric CBF3−CEN3 complexes were subject to 3D classification and the particles in the best quality classes were saved. A total of 198,010 particles were used for the final reconstruction, and a 3.05 Å resolution map was obtained for CBF3msk (Supplementary Fig. 2h,j,k).
Before visualization, a negative B factor determined with RELION was applied to the density map for sharpening. The modulation transfer function (MTF) of the detector was corrected in the post-processing step with RELION. The local resolution was estimated with ResMap 56 integrated in RELION 2.1 53.
Model building and structure refinement
Three maps are defined as: (i) 3.6 Å map of CBF3core−Ndc10DBD (no CEN3 DNA) (Supplementary Fig. 3f), (ii) 4.4 Å map of CBF3−CEN3 DNA (Supplementary Fig. 2d) and (iii) 3.0 Å map of CBF3msk (Supplementary Fig. 2h), (Tables 1 and 2).
The crystal structures of the S. cerevisiae Cep3 dimer 35 (PDB:2QUQ) and Ndc10 32 (PDB:4ACO) (residues 44-537) and the model of S. cerevisiae Skp1 were fitted into monomeric CBF3core−Ndc10DBD−CEN3. The S. cerevisiae Skp1 model was modeled on human Skp1 57 (PDB 1FQV) with I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER). Based on the excellent quality of the EM density map, an atomic model of the Ctf13 was build de novo using COOT 58. The secondary structure and disordered regions of the Ctf13 sequence were analyzed with PHYRE2 59 and PSIPred 60. Amino acids were assigned based mainly on the predicted secondary structures and clearly defined bulky residue densities. The final model of Ctf13 lacks the N-terminal two residues and two unstructured loop regions (50-54 and 205-252). The zinc finger motif (1-53) in the Cep3 N-terminal was built guided by the Hap1 structure (PDB: 2HAP) 37. The extreme N-terminus of Ndc10 except the missing first 26 amino acids shows clear side chain densities that facilitated the model building. This CBF3 model was fit into the cryo-EM map of the dimeric CBF3−CEN3 complex following minor adjustments. Based on the position of avidin in the CBF3−CEN3biotin-avidin cryo-EM map and information from published CBF3−CEN3 crosslinking data 27, we built the CEN DNA model. The model was optimized by several rounds of real-space refinement in PHENIX (phenix.real_space_refine) 61. Standard stereochemical and secondary-structural constraints were applied during the real-space refinement. The final models were evaluated with MolProbity (http://molprobity.biochem.duke.edu/) 62.
Modeling of the CBF3−Cse4−CEN3 complex
To model the CBF3−Cse4−CEN3 complex we made the following assumptions:
CBF3: The position of CBF3 on CEN3 DNA remains unchanged and that CBF3 undergoes no conformational changes. Relative to its DNA-free state, CBF3 does not undergo major conformational change in the CBF3−CEN3 complex.
Cse4: The Cse4 nucleosome was modeled based on the CENP-A crystal structure 40. In this structure the CENP-A nucleosome is formed from a 147 bp palindromic DNA derived from a human α-satellite DNA sequence. 120 bp of α-satellite DNA generate 1.4 gyres to surround the histone octamer. This contrasts to 146 bp for a canonical H3 nucleosome 63 (with an α-satellite sequence element) and also with cryo-EM structures of CENP-A nucleosomes reconstituted using the Widom 601 sequences in which 146 bp of wrapped DNA is wrapped around the octamer 64,65. Moreover, a nucleosome core particle containing a Poly(dA.dT) sequence element wraps 147 bp of DNA 66.
Cse4 nucleosomes provide less resistance to micrococcal nuclease digestion, in which only 115 to 125 bp of DNA are protected 29,41. These studies, together with single molecule DNA unzipping data and small angle X-ray scattering results 29 indicate that in Cse4-nucleosomes, the DNA is less tightly bound at the entry and exit regions compared with canonical nucleosomes. From these data we assume that the CENP-A-α-satellite nucleosome would represent a reasonable model for a Cse4−CEN3 nucleosome structure (120 bp forming the nucleosome). However, we note that in the Cse4 nucleosome reconstitution studies 29,41, either Widom 601 DNA or pBR322 plasmid DNA was used instead of CEN DNA
In our predicted model we assume that the Cse4 nucleosome is an octamer rather than a Cse4−H4−H2A−H2B hemisome. A Cse4−H4−H2A−H2B hemisome would be expected to have a similar radius as a Cse4 octamer nucleosome. The difference between the two would therefore relate to the length of CEN DNA wrapped around an octamer compared with a hemisome, with possibly 120 bp for the former and 80 bp for the latter.
In the CBF3−CEN3 structure, the ordered region of CDEII (12 bp adjacent to CDEIII) adopts a structure with the same curvature as the DNA wrapped around the CENP-A nucleosome 40. We superimposed this structure onto the ‘entry’ DNA of the CENP-A nucleosome 40, keeping the major and minor grooves of the CBF3−CEN3 DNA and the CENP-A DNA in register. This means that the ‘entry’ point for the modeled Cse4 nucleosome is 5 bp to the left of the CDEII-CDEIII junction.
Because 120 bp of DNA wrap around the CENP-A nucleosome, placing the entry point 5 bp left of the CDEII-CDEIII junction means that the remaining 80 bp, all of CDEI (assuming CDEII is 84 bp and CDEI is 8 bp) and 32 bp to the left of CDEI, are included in the Cse4 nucleosome. This positions the dyad axis of the Cse4 octamer 18 bp to the right of the CDEII-CDEI junction.
- Variations of the model include:
-
(i)The position of the nucleosome entry point relative to the CDEII-CDEIII junction. Changing the entry point relative to the CDEII-CDEIII junction would cause the nucleosome to rotate relative to CBF3. This would alter the position of Cse4 relative to Ctf13, Cep3 and Ndc10. However, placing the entry point closer to the CDEII-CDEIII junction causes steric clashes between the nucleosome and Ndc10, whereas placing the entry point further from the CDEII-CDEIII junction causes steric clashes between the nucleosome and Cep3B and Ctf13.
-
(ii)The length of DNA wrapped around the Cse4 octamer (i.e. a more loosely wrapped nucleosome than 120 bp).
-
(iii)An 80 bp hemisome, in which case the wrapped DNA would extend only to the CDEII-CDEI junction – assuming the entry point at the CDEII-CDEIII junction is as modeled for the octamer.
-
(i)
Mitotic stability assays
A modified S. cerevisiae strain (BJ2168Ctf13+) expressing an additional S. cerevisiae Ctf13 gene with its native promoter was generated by transforming BJ2168 with a 2µ origin plasmid pYes2 incorporating the S. cerevisiae Ctf13 allele and the URA3 selection marker. The transformed cells were selected on synthetic media lacking uracil, and the presence of the plasmid-encoded Ctf13 was verified by PCR using a primer pair over-spanning the Ctf13 and URA3 genes. Based on BJ2168Ctf13+, a genomic Ctf13 knockout strain (BJ2168ΔCtf130) was created by replacing the chromosomal Ctf13 with a PCR fragment of the KanMX6 gene flanked by two 50-base pair sequences complementary to both ends of the promoter and terminator regions of the chromosomal Cft13 by homologous recombination, selected with Kanamycin (G418, Melford Laboratories Ltd.). The BJ2168ΔCtf130 strain, viable due to the functional Ctf13 in the pYes2-URA3 plasmid, was confirmed by sequencing the recombination joins of the PCR products amplified by primer pairs over-spanning the KanMX6 gene and the promoter or terminator regions outside of the complementary sequences used for PCR fragment for gene replacement experiment of chromosomal Ctf13. A Ctf13wt-LEU2 construct was created by cloning Ctf13wt with its own native promoter into a modified pYes2 plasmid with LEU2 replacing URA3 as a selection marker. Subsequently, Ctf13I76A-Y79A and Ctf13I76R-Y79R mutations were made based on the Cft13wt-LEU2 construct. Ctf13wt and its mutants were transformed into the BJ2168Δctf13 strain, selected on the synthetic media lacking uracil and leucine, and with Kanamycin G418. The positive colonies were verified by PCR using primer pair over-spanning the Ctf13 and Leu2 genes. The resultant strains with no chromosomal Ctf13 contain URA3-Ctf13 and Leu2-Ctf13wt or its mutants. The URA3-Ctf13 was then driven out by 5-FOA-URA3 counter selection. The selected colonies were verified by PCR using a primer pair over-spanning Ctf13 and URA3 genes.
A fragment of ARS1::TRP1::CEN3 flanked by two EcoRI sites was cloned in pUC18 plasmid. Based on this plasmid, a CEN3CdeIIICCG-AGC mutation was made. Mini-chromosomes (1442 bp) of CEN3WT and CEN3CdeIIICCG-AGC were made by cyclised ligation fragment ARS1::TRP1::CEN3 released from the pUC18 plasmids by EcoRI. The mini-chromosomes were transformed into yeast strains harboring Ctf13 wild type or its mutants, selected on synthetic media lacking leucine, tryptophan and surplus G418.
Single colonies of yeast strains carrying wild type and mutant Ctf13, in combination with a mini-chromosome with either wild type of mutant CEN3 from synthetic complete (Sc)-Leu-Trp+G418 selective plates were picked to inoculate 3 ml of Sc-Leu+G418 medium, a nonselective growth medium for mini-chromosome. After 48 h growth at 30 °C, cells were plated for single colonies onto Sc-Leu+G418 plates. After 3 days incubation at 30 °C, colonies were plated onto Sc-Leu-Trp+G418 selective plates. These plates were incubated for 3 days at 30 °C, and the percentage of grown population bearing mini-chromosome was determined.
Supplementary Material
Acknowledgments
This work was funded by MRC grant (MC_UP_1201/6) and CR-UK grant (C576/A14109) to D.B. We thank S. Chen, G. Cannone and G. McMullan for help with EM data collection, J. Grimmett and T. Darling for computing and J. Shi for help with the insect cell expression.
Footnotes
Data availability and accession codes. EM maps are deposited with EMDB (EMD-0095, EMD-0096, EMB-0097). Protein coordinates are deposited with RCSB (6GYP, 6GYS, 6GYU). Other data are available upon reasonable request.
Author contributions. D.B. directed the project. Z.Z. cloned all constructs. J.Y and Z.Z purified complexes. Z.Z. performed the mitotic stability assay. J.Y. and S.M. performed SEC-MALS experiments. K.Y. prepared EM grids, collected and analyzed EM data. K.Y. and D.B. wrote the manuscript.
Competing Interests. The authors declare that there are no competing interests.
References
- 1.Bloom K, Costanzo V. Centromere Structure and Function. Prog Mol Subcell Biol. 2017;56:515–539. doi: 10.1007/978-3-319-58592-5_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musacchio A, Desai A. A Molecular View of Kinetochore Assembly and Function. Biology (Basel) 2017;6 doi: 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenni S, Dimitrova YN, Valverde R, Hinshaw SM, Harrison SC. Molecular Structures of Yeast Kinetochore Subcomplexes and Their Roles in Chromosome Segregation. Cold Spring Harb Symp Quant Biol. 2017 doi: 10.1101/sqb.2017.82.033738. [DOI] [PubMed] [Google Scholar]
- 4.Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr Biol. 2015;25:R1002–18. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 5.Joglekar AP. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology (Basel) 2016;5 doi: 10.3390/biology5040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfieri C, Zhang S, Barford D. Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C) Open Biol. 2017;7 doi: 10.1098/rsob.170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–9. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–44. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 9.Hieter P, et al. Functional selection and analysis of yeast centromeric DNA. Cell. 1985;42:913–21. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 10.Peterson JB, Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976;22:219–42. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- 11.Cottarel G, Shero JH, Hieter P, Hegemann JH. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3342–9. doi: 10.1128/mcb.9.8.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kingsbury J, Koshland D. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell. 1991;66:483–95. doi: 10.1016/0092-8674(81)90012-x. [DOI] [PubMed] [Google Scholar]
- 13.McGrew J, Diehl B, Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:530–8. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng R, Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4522–34. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegemann JH, Shero JH, Cottarel G, Philippsen P, Hieter P. Mutational analysis of centromere DNA from chromosome VI of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2523–35. doi: 10.1128/mcb.8.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–13. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 17.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–25. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 18.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci U S A. 2007;104:14706–11. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci U S A. 2011;108:12687–92. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krassovsky K, Henikoff JG, Henikoff S. Tripartite organization of centromeric chromatin in budding yeast. Proc Natl Acad Sci U S A. 2012;109:243–8. doi: 10.1073/pnas.1118898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–17. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 22.Bloom KS, et al. Chromatin conformation of yeast centromeres. J Cell Biol. 1984;99:1559–68. doi: 10.1083/jcb.99.5.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders M, Fitzgerald-Hayes M, Bloom K. Chromatin structure of altered yeast centromeres. Proc Natl Acad Sci U S A. 1988;85:175–9. doi: 10.1073/pnas.85.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Funk M, Hegemann JH, Philippsen P. Chromatin digestion with restriction endonucleases reveals 150-160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol Gen Genet. 1989;219:153–60. doi: 10.1007/BF00261171. [DOI] [PubMed] [Google Scholar]
- 25.Lechner J. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 1994;13:5203–11. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorger PK, et al. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc Natl Acad Sci U S A. 1995;92:12026–30. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–96. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell ID, Grancell AS, Sorger PK. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J Cell Biol. 1999;145:933–50. doi: 10.1083/jcb.145.5.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dechassa ML, et al. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakane T, Kimanius D, Lindahl E, Scheres SH. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife. 2018;7 doi: 10.7554/eLife.36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho US, Harrison SC. Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nat Struct Mol Biol. 2011;19:48–55. doi: 10.1038/nsmb.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perriches T, Singleton MR. Structure of yeast kinetochore Ndc10 DNA-binding domain reveals unexpected evolutionary relationship to tyrosine recombinases. J Biol Chem. 2012;287:5173–9. doi: 10.1074/jbc.C111.318501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leber V, Nans A, Singleton MR. Structural basis for assembly of the CBF3 kinetochore complex. EMBO J. 2018;37:269–281. doi: 10.15252/embj.201798134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 35.Bellizzi JJ, 3rd, Sorger PK, Harrison SC. Crystal structure of the yeast inner kinetochore subunit Cep3p. Structure. 2007;15:1422–30. doi: 10.1016/j.str.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purvis A, Singleton MR. Insights into kinetochore-DNA interactions from the structure of Cep3Delta. EMBO Rep. 2008;9:56–62. doi: 10.1038/sj.embor.7401139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King DA, Zhang L, Guarente L, Marmorstein R. Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation. Nat Struct Biol. 1999;6:22–7. doi: 10.1038/4893. [DOI] [PubMed] [Google Scholar]
- 38.Pietrasanta LI, et al. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc Natl Acad Sci U S A. 1999;96:3757–62. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Lukoynova N, Miah S, Lucas J, Vaughan CK. Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10. Cell Rep. 2018;24:744–754. doi: 10.1016/j.celrep.2018.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tachiwana H, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–5. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 41.Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2011;286:4021–6. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol Biol Cell. 2003;14:4557–68. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morey L, Barnes K, Chen Y, Fitzgerald-Hayes M, Baker RE. The histone fold domain of Cse4 is sufficient for CEN targeting and propagation of active centromeres in budding yeast. Eukaryot Cell. 2004;3:1533–43. doi: 10.1128/EC.3.6.1533-1543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henikoff S, et al. The budding yeast Centromere DNA Element II wraps a stable Cse4 hemisome in either orientation in vivo. Elife. 2014;3:e01861. doi: 10.7554/eLife.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nechemia-Arbely Y, et al. Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J Cell Biol. 2017;216:607–621. doi: 10.1083/jcb.201608083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henikoff S, Henikoff JG. “Point” centromeres of Saccharomyces harbor single centromere-specific nucleosomes. Genetics. 2012;190:1575–7. doi: 10.1534/genetics.111.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife. 2017;6 doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–82. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitzgerald DJ, et al. Multiprotein expression strategy for structural biology of eukaryotic complexes. Structure. 2007;15:275–9. doi: 10.1016/j.str.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Z, Yang J, Barford D. Recombinant expression and reconstitution of multiprotein complexes by the USER cloning method in the insect cell-baculovirus expression system. Methods. 2016;95:13–25. doi: 10.1016/j.ymeth.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Leiro R, Scheres SHW. A pipeline approach to single-particle processing in RELION. Acta Crystallogr D Struct Biol. 2017;73:496–502. doi: 10.1107/S2059798316019276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elmlund H, Elmlund D, Bengio S. PRIME: probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure. 2013;21:1299–306. doi: 10.1016/j.str.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schulman BA, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–6. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 64.Chittori S, et al. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science. 2018;359:339–343. doi: 10.1126/science.aar2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pentakota S, et al. Decoding the centromeric nucleosome through CENP-N. Elife. 2017;6 doi: 10.7554/eLife.33442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bao Y, White CL, Luger K. Nucleosome core particles containing a poly(dA.dT) sequence element exhibit a locally distorted DNA structure. J Mol Biol. 2006;361:617–24. doi: 10.1016/j.jmb.2006.06.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.