Summary
Proper morphogenesis of dendrites plays a fundamental role in the establishment of neural circuits. The molecular mechanism by which dendrites grow highly complex branches is not well understood. Here, using the C. elegans PVD neuron, we demonstrate that high-order dendritic branching requires actin polymerization driven by coordinated interactions between two membrane proteins, DMA-1 and HPO-30, and with their cytoplasmic interactors, the RacGEF TIAM-1 and the actin nucleation promotion factor WAVE Regulatory Complex (WRC). The dendrite branching receptor DMA-1 directly binds to the PDZ domain of TIAM-1, while the claudin-like protein HPO-30 directly interacts with the WRC. On dendrites, DMA-1 and HPO-30 form a receptor-associated signaling complex to bring TIAM-1 and the WRC to close proximity, leading to elevated assembly of F-actin needed to drive high-order dendrite branching. The synergistic activation of F-actin assembly by scaffolding distinct actin regulators might represent a general mechanism in promoting complex dendrite arborization.
Keywords: dendrite branching, actin assembly, synergy
Introduction
Dendrite morphogenesis, especially the formation of highly-branched dendritic arbors, is essential for the establishment of neural circuits throughout the nervous system. Genetic and cell biological studies showed that dendrite arborization requires both extrinsic and intrinsic mechanisms, including ligand-receptor signaling, transcription regulation, cytoskeleton remodeling and membrane trafficking (Dong et al., 2015; Jan and Jan, 2010). Several extrinsic cues and cell surface receptors have been identified as important guidance molecules for dendrite formation. For example, the secreted protein semaphorin 3A (Sema3A) attracts the apical dendrites of pyramidal neurons to grow towards the pial surface in the mammalian cortex (Polleux et al., 2000). The cell-adhesion receptors DSCAM and protocadherin act as hemophilic repulsive cues to mediate the self-avoidance of dendrites in Drosophila and mammalian neurons (Hughes et al., 2007; Lefebvre et al., 2012; Matthews et al., 2007; Soba et al., 2007).
Within dendrites, the actin cytoskeleton acts as a major structural component to drive morphogenesis (Jan and Jan, 2010). The Rho family GTPase Rac plays a critical role in regulating actin reorganization needed for dendrite branching (Andersen et al., 2005; Emoto et al., 2004; Luo, 2000). Activated Rac binds to the actin nucleation promotion factor WAVE Regulatory Complex (WRC), which, in turn, stimulates the Arp2/3 complex to polymerize actin (Chen et al., 2017; Chen et al., 2010; Eden et al., 2002; Ismail et al., 2009). The activity of Rac is controlled by various Rac-specific Guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). Several RacGEFs (e.g., Trio and Tiam1) have been shown to be important for dendrite morphogenesis (Iyer et al., 2012; Tolias et al., 2005). A large body of research on neuron morphogenesis supports the idea that links between membrane receptors and GEFs and GAPs serve as major means to relay receptor signaling to the actin cytoskeleton in axon guidance and dendrite formation (Dent et al., 2011; Huber et al., 2003). For example, UNC-40/DCC and NMDA receptor are both linked to Tiam1 to regulate actin remodeling in axon guidance and development of dendritic arbors and spines (Demarco et al., 2012; Tolias et al., 2005). Furthermore, the WRC could potentially connect many receptors to actin through its direct interaction with a short peptide motif, named the WRC Interacting Receptor Sequence (WIRS), found in the intracellular domains (ICDs) of a large variety of membrane proteins (Chen et al., 2014a). This interaction was shown to be important for the cell adhesion receptor SYG-1 to control actin assembly at the presynaptic terminals of the C. elegans HSN neuron to drive axon branching and synapse formation (Chia et al., 2014). However, whether dendrites use similar mechanisms to control the remodeling of actin is largely unknown.
Recently, several groups, including ours, have started to use the C. elegans multi-dendritic PVD neurons as a model system to dissect the molecular mechanisms of dendrite development. During larval development, PVD elaborates complex and stereotyped dendritic arbors by sequentially adding primary (1o), secondary (2o), tertiary (3o) and quaternary (4o) dendrites (Figures 1A–1B) (Smith et al., 2010). The 3o branches grow precisely along the border of the outer body wall muscles, while the 4o branches innervate the narrow space between the epidermis and the body wall muscles, suggesting that the epidermal and muscle tissues provide guidance cues to regulate branch formation (Albeg et al., 2011). Indeed, a multi-protein ligand-receptor complex has been identified as an extrinsic cue in guiding PVD dendrite morphogenesis (Diaz-Balzac et al., 2016; Dong et al., 2013; Salzberg et al., 2013; Zou et al., 2016). This complex consists of the cell adhesion molecules SAX-7/L1CAM and MNR-1/Menorin, which are enriched in epidermis, the chemotaxin LECT-2/LECT2, which is secreted by the body wall muscle cells, and the cell adhesion receptor DMA-1, which is specifically expressed in PVD dendrites. SAX-7 forms a striped pattern on epidermis that correlates with the location of the 3o and 4o branches (Dong et al., 2013; Liang et al., 2015; Salzberg et al., 2013), whereas the epidermis-specific expression of MNR-1 specifies the attachment points for PVD dendrites (Dong et al., 2013; Salzberg et al., 2013). Both genetic and biochemical experiments support a model where the three ligand proteins from the epidermis/muscle cells, including SAX-7, MNR-1 and LECT-2, simultaneously bind to the extracellular domain of the dendrite-specific receptor DMA-1, leading to the formation of an inter-cellular complex which specifies the precise location of dendritic arborization (Diaz-Balzac et al., 2016; Liu and Shen, 2012; Zou et al., 2016). However, it remains unknown how this extrinsic cue is transmitted through DMA-1 to instruct intracellular signaling in the PVD neuron and, subsequently, drive dendrite branching.
Figure 1. act-4, tiam-1 and gex-3 are required for dendrite branching in PVD neurons.
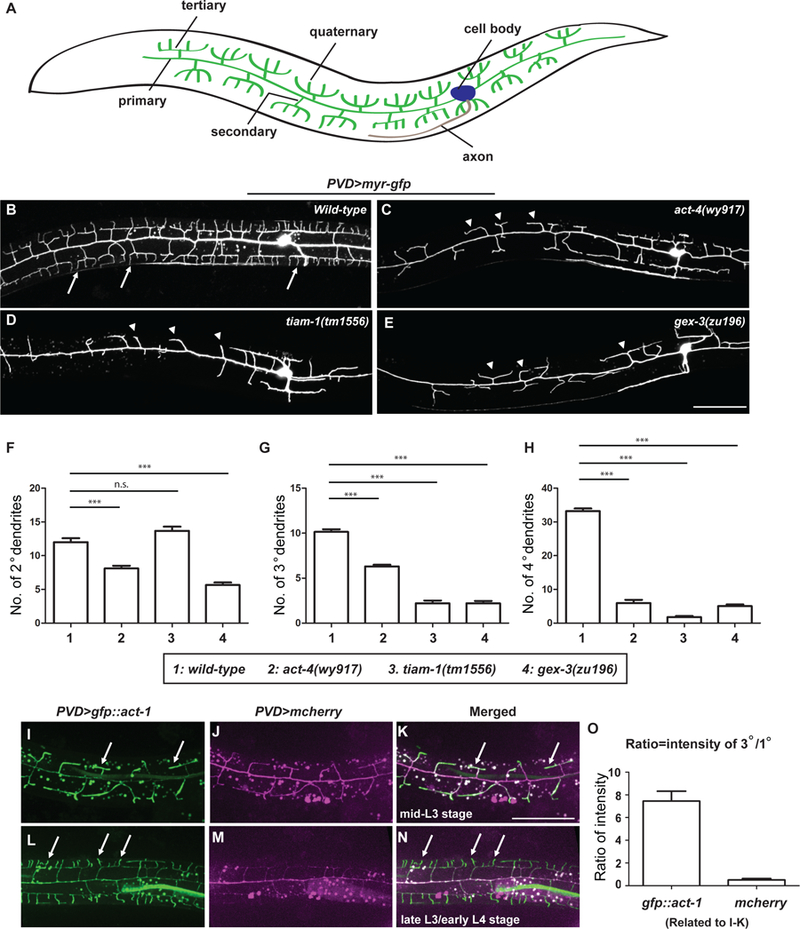
(A) Schematic showing the morphology of PVD dendritic arbors. Lateral view with anterior on left and dorsal up is shown. The same orientation is used for all fluorescent images in this work. (B-E) Representative confocal images showing the morphologies of PVD dendritic arbors labeled by a transgene ser2prom3>myr-gfp, in (B) wild-type, (C) act-4(wy917), (D) tiam-1(tm1556), and (E) gex-3(zu196) animals at late L4 stage. The most anterior and posterior parts are not shown. All PVD neuron fluorescence images in this work are labeled and presented by the same means unless otherwise noted. Arrows: 4o branches in the wild-type animals. Arrow heads: 2o branches that failed to form 4o or even 3o branches. Scale bar = 50 μm. (F-H) Quantifications of the number of 2o, 3o and 4o branches in a region 100 μm anterior to the PVD cell body. Data were obtained using 20 animals for each genotype and are presented as mean ± SEM. One-sided ANOVA with Dunnett’s test was used for statistical analysis. ***: p< 0.0001. n.s.: not significant. (I-N) Representative fluorescent images showing expression of GFP::actin during PVD dendrite branching and growth at indicated developmental stages. PVD was labeled by a transgene ser2prom3>mcherry. Arrows: dendrites with enriched GFP::actin. Scale bar = 50 μm. (O) Quantifications of the ratio of GFP::ACT-1 and mCherry intensity between 3o and 1o branches. Thirty-five 3o branches and 7 primary dendrites from 7 independent L3 stage animals were measured and the results are presented as mean ± SEM. See also Figure S1 and S2.
In addition to DMA-1, a dendrite-specific, claudin-like transmembrane protein HPO-30, was also identified as an essential regulator of PVD dendritic branching (Smith et al., 2013). Loss of hpo-30 results in a severe defect in dendrite morphogenesis similar to the dma-1 mutants (Smith et al., 2013). Claudins are transmembrane proteins important for the establishment and function of tight-junctions in mammals, through both cis- and trans- interactions (Krause et al., 2008). In addition to their major roles in tight-junction formation, claudins may play a role in mediating signaling. For example, Claudin-1 has been shown to act as a co-receptor for the hepatitis C virus during a late step of viral entry (Evans et al., 2007). It is not known how HPO-30 mediates signaling to control dendrite branching.
Here, we address the above questions of how extrinsic cues are connected to intracellular signaling to control actin remodeling and dendrite branching. Our genetic and biochemical results demonstrate that dendritic branching requires actin assembly driven by cooperative interactions between dendritic membrane proteins DMA-1 and HPO-30 and the actin regulators RacGEF TIAM-1 and the WRC. The intracellular domains of DMA-1 and HPO-30 directly bind to TIAM-1 and the WRC, respectively. DMA-1 and HPO-30 interact with each other to form a receptor-associated signaling complex, leading to co-recruitment of TIAM-1 and the WRC to the dendritic membranes and synergistic enhancement of F-actin assembly to facilitate formation of highly branched dendritic arbors.
Results
Formation of high-order dendrites requires the assembly of filamentous actin and actin regulators TIAM-1 and the WRC
To understand the molecular mechanisms that control the formation of highly branched dendrites during development, we used the multi-dendritic PVD sensory neurons in C. elegans (Figure 1A) as a model and performed both unbiased forward genetic screens and candidate-based screens. Our forward genetic screens identified wy917 as a mutant that affected the dendritic branching of PVD neurons. Compared to the wild-type controls (Figure 1B), the wy917 mutant animals showed simplified arbors with normal 1o dendrites but significantly reduced numbers of the 2o, 3o and 4o dendrites (Figures 1C, 1F–1H). Using standard genetic mapping and cloning methods, we identified the causative mutation in wy917 as a single base pair change, which resulted in a G151E mutation in the ACT-4A protein. The act-4 gene encodes an isoform of actin, and the G151 amino acid residue is completely conserved throughout evolution (not shown). A mutation in the equivalent position (G152A) in the human skeletal muscle -actin gene (ACTA1) was identified to cause a severe congenital myopathy in an infant patient (Ravenscroft et al., 2011). This mutation gave rise to abnormal assembly and mislocalization of actin rods both in vivo and in vitro (Ravenscroft et al., 2011), suggesting that the G151E mutation identified here in act-4 may analogously impair actin assembly. The defect of dendrite formation could be fully rescued using a PVD specific promoter (ser2prom3) to drive expression of the wild type act-4 in PVD neurons, suggesting that act-4 acts in a cell-autonomous manner (Figures S1A, S1D–1F).
Paralleling our unbiased genetic screening, our candidate-based screen identified two additional genes important to PVD dendritic branching. These include tiam-1, a conserved RacGEF, and gex-3, which encodes the homolog of mammalian Nap1, a component of the WRC. Similar to the act-4 mutation, loss of either tiam-1 or gex-3 dramatically reduced high-order dendrite branches, with tiam-1 primarily affecting the 3o and 4o dendrites and gex-3 affecting all of the 2o, 3o and 4o dendrites (Figures 1D–1H) (Smith et al., 2010). In contrast, a mutation in wsp-1, the ortholog of another actin regulator WASP, did not lead to any PVD dendrite morphology defects, indicating that the WRC is selectively involved in PVD dendrite morphogenesis (data not shown). Both tiam-1 and gex-3 act cell-autonomously in PVD neurons, since the observed defects could be similarly rescued by PVD-specific re-expression of tiam-1 and gex-3, respectively (Figures S1B–1F). Together, these results suggest that formation of highly branched dendrites in PVD neurons depends on proper actin assembly and requires the function of both TIAM-1 and the WRC.
To directly visualize the distribution of the actin cytoskeleton during dendrite formation, we generated a ser2prom3>gfp::act-1 transgene. At the mid-L3 stage, when the 3o dendrites started branching off the 2o dendrites, GFP::actin was enriched in the 2o and 3o dendrites compared to the 1o dendrite (Figures 1I–1K, 1O), whereas similar measurements of a cytosolic PVD>mcherry transgene showed no enrichment in the 3o dendrites (Figures 1I–1K, 1O). Similarly, at late-L3 and early-L4 stage when 4o branches started to form, GFP::actin was enriched in the 3o and 4o dendrites compared to the 1o and 2o dendrites (Figures 1L–1N). We also analyzed a transgene PVD>lifeact::gfp, which specifically labels the filamentous actin (F-actin) in the PVD neurons (Riedl et al., 2008). Similar to the GFP::actin strategy, Lifeact::GFP was also transiently enriched in newly formed dendrites (Figure S2), suggesting that the observed enrichment of GFP::actin was mostly F-actin. These results corroborate the notion that actin assembly acts as a driving force during PVD dendrite formation.
The intracellular domain of DMA-1 interacts with TIAM-1 and is required for dendritic branching
As a receptor for extrinsic dendritic branching signals, DMA-1 forms an inter-cellular complex by simultaneously binding to two membrane proteins on epidermis cells, SAX-7 and MNR-1, and the chemotactic molecule LECT-2 secreted from muscle cells (Diaz-Balzac et al., 2016; Dong et al., 2013; Salzberg et al., 2013; Zou et al., 2016). DMA-1 is a single-transmembrane protein. It contains a large extracellular domain consisting of 15 leucine-rich repeat (LRR) units, which directly interacts with SAX-7, MNR-1 and LECT-2 (Liu and Shen, 2012; Zou et al., 2016). DMA-1 also contains a short intracellular domain (ICD) of 73 amino acid residues with unknown functions. To understand whether the DMA-1 ICD is important for dendritic arbor development, we tested if expression of a DMA-1 construct that lacked the ICD (DMA-1 △ICD) could rescue the dendrite branching phenotype in dma-1 null mutant animals. In contrast to the expression of full length DMA-1, which fully rescued the defects in high-order dendrite formation (Figures 2A–2C, 2G–2I), transgenic animals expressing DMA-1 △ICD showed a dramatic reduction of the 4o dendrites and a milder, albeit significant, reduction of the 3o branches (Figures 2D, 2G–2I). Interestingly, expression of a full-length DMA-1 with a GFP tag fused to the C-terminus of the ICD (DMA-1::GFP) similarly failed to restore the formation of the 3o and 4o branches in dma-1 null mutants (Figures 2E, 2G–2I ). Note that both DMA-1 △ ICD::GFP and DMA-1::GFP were detected on the dendritic membranes, suggesting that the observed defects from deleting the intracellular domain or fusing a GFP to the C-terminus of DMA-1 were not due to protein folding or trafficking (Figures S3A–3B). This result was corroborated by our CRISPR/Cas9 knock-in animals, in which we removed the ICD or fused a C-terminal ZF1::YFP in the endogenous locus of dma-1 (DMA-1 △ICDendo and DMA-1::YFPendo, respectively). These two strains exhibited a similar reduction of the 4o dendrites compared to the transgenic strains (Figures S4A–4E). Together, these results suggest that DMA-1 ICD is required for the formation of the 3o and 4o dendrites and that the addition of GFP or YFP at the C terminus perturbs its function.
Figure 2. Interaction between the cytoplasmic tail of DMA-1 and TIAM-1 is required for dendrite formation.

(A-F) Representative confocal images showing the morphologies of PVD dendritic arbors in animals of indicated genotypes. Arrows: 4o branches. Arrow heads: 2o branches that failed to form 4o or even 3o branches. Four distinct constructs were used to express full-length DMA-1 (C), DMA-1 without the last 68 amino acids in the intracellular domain (D), full-length DMA-1 with a C-terminal GFP fusion (E) and DMA-1 without the last 4 amino acids in the intracellular domain (F). Scale bar = 50 μm. (G-I) Quantification of the number of 2o, 3o and 4o branches in a region 100 μm anterior to the PVD cell body. Data were obtained using 20 animals for each genotype and are presented as mean ± SEM. One-sided ANOVA with Tukey test was used for statistical analysis. *: p<0.05. ***: p< 0.0001. n.s.: not significant. (J) Coomassie-blue-stained SDS-PAGE gel showing pull-down assay using GST or GST-TIAM-1 PDZ domains as bait and MBP-tagged DMA-1 ICDs as prey. The input lanes were loaded with 8 pmol of the prey proteins. Open arrow head indicates retained prey proteins. (K-L) Representative confocal images showing the labeling of TIAM-1::GFP in animals of indicated genotypes. Arrows: 3o /4o branches labeled by TIAM-1::GFP. Scale bar = 50 μm. (M-N) Representative confocal images showing the morphologies of PVD dendritic arbors in animals of indicated genotypes. Arrows: 4o branches. Scale bar = 50 μm. (O) Quantifications of the number of 4o branches in a region 100 μm anterior to the PVD cell body. Data were obtained using 20, 20, 15, 13 and 20 animals for each genotype and are presented as mean ± SEM. One-sided ANOVA with Tukey test was used for statistical analysis. **: p<0.001. ***: p< 0.0001. n.s.: not significant. See also Figure S3, S4 and S5.
What is the function of the DMA-1 ICD? Does it interact with a downstream effector to control dendritic branching? We first noticed that the DMA-1 ICD contained a putative WRC Interacting Receptor Sequence (WIRS) motif, which could potentially allow DMA-1 to recruit the WRC to membranes through a direct WIRS/WRC interaction (Chen et al 2014). However, our in vitro GST pull-down assay failed to detect a significant interaction between the DMA-1 ICD and the WRC (data not shown). Furthermore, mutating the WIRS motif in animals did not affect dendritic branching (Figures S5A–5E), suggesting that the WIRS motif is not important to the function of the DMA-1 ICD. Further analysis of the DMA-1 ICD identified its last four amino acid residues as a putative type II PDZ binding motif (Lee and Zheng, 2010). To test whether this motif is important for DMA-1’s function, we expressed a mutant form of DMA-1 that lacked the last four residues (DMA-1 △4AA) in dma-1 null mutants. We found that the animals also exhibited significantly reduced numbers of the 4o dendrites, to a level similar to those observed in the DMA-1 △ICDendo and the DMA-1::YFPendo animals (Figures 2F–2I, S4C–4E). Thus, DMA-1 likely transduces signals through its PDZ binding motif to a downstream PDZ domain-containing protein(s). This prompted us to test whether the ICD of DMA-1 could interact with TIAM-1, which contains a PDZ-like domain (hereafter referred to as PDZ domain) (Demarco et al., 2012). For this, we performed GST pull-down experiments by using recombinantly purified maltose-binding protein (MBP)-tagged DMA-1 ICD and GST-tagged TIAM-1 PDZ. We found that the immobilized GST-TIAM-1 PDZ, but not GST, was able to retain MBP-DMA-1 ICD (Figure 2J). Furthermore, deleting the putative PDZ-binding motif from DMA-1 ICD (MBP-DMA-1 ICD △4AA) or mutating two conserved amino acids predicted to be located at the binding pocket of the TIAM-1 PDZ domain (PDZ KL/AA) specifically abolished the binding (Figure 2J).
To directly test if DMA-1 recruits TIAM-1 to dendritic branching sites in vivo, we expressed TIAM-1::GFP in PVD and compared its localization between wild-type and dma-1 △ ICD mutants (Figures 2K–2L). During the formation of 3o and 4o branches, TIAM-1::GFP exhibits punctate localization along the 1o dendrite, is almost undetectable on the 2o dendrite, and shows strong localization on the 3o and 4o dendrites. The striking contrast between the 2o and 3o/4o branches indicates that TIAM-1 is indeed enriched in the newly formed 3o/4o branches in the WT animals, with 20 out of 23 WT animals showing TIAM-1::GFP positive 3o/4o branches. This enrichment is largely gone in the dma-1 (wy908) mutants, where only 3 out of 27 dma-1 (wy908) animals showed TIAM-1::GFP positive 3o/4o branches, indicating that the intracellular domain of DMA-1 recruits TIAM-1 (Figures 2K–2L). Furthermore, the TIAM-1::GFP puncta along the primary dendrite were not affected by the dma-1 mutation, suggesting that the dma-1 mutation specifically affected the enrichment of TIAM-1 on newly formed branches but not overall protein synthesis and folding (Figures 2K–2L).
To further confirm if the intracellular domain of DMA-1 primarily acts by recruiting TIAM-1, we tested whether directly tethering TIAM-1 to DMA-1 could substitute for the function of the DMA-1 ICD. For this, we created a fusion construct, PVD>dma-1ΔICD::tiam-1, in which we deleted the entire cytoplasmic domain of DMA-1 and replaced it with the full length TIAM-1. In control experiments, where TIAM-1 was not tethered to DMA-1 when expressed in dma-1 null mutants, no rescue could be observed (Figures 2M, 2O). In contrast, when TIAM-1 was tethered to DMA-1 △ ICD, it significantly rescued 3o/4o branches (Figures 2N, 2O), suggesting that tethering TIAM-1 to DMA-1 is largely sufficient to carry out the signaling function of DMA-1 ICD in PVD dendrite morphogenesis. It is worth noting that the tethered TIAM-1 also resulted in many ectopic branches, which might be due to the constitutive association between TIAM-1 and DMA-1 in this construct (Figure 2N). Based on this result and together with the above results of the subcellular localization of TIAM-1, the direct interaction between TIAM-1 and DMA-1 and the similar phenotypes between tiam-1 and dma-1 △ICDendo, we conclude that the DMA-1 ICD acts by recruiting the TIAM-1 RacGEF through a PDZ domain interaction, which may increase local concentrations of active Rac to promote actin polymerization.
HPO-30 associates with DMA-1
While dma-1 △ICDendo mutants showed strong defects in the 4o dendrite formation, they retained most of the 3o branches and a small number of 4o branches (Figures S4A, 4C–4E). This was different from the dma-1 null allele, which completely lacked the 3o and 4o dendrites (Figures 2B, 2G–2I), suggesting that DMA-1 might signal to other protein(s) independent of its intracellular domain. We hypothesized that DMA-1 may act together with another dendritic receptor. A previous study by Smith et. al. showed that HPO-30, a dendritic claudin-like protein, regulates dendrite branching and stabilization (Smith et al., 2013). Like DMA-1, HPO-30 also functions cell-autonomously in PVD neurons. Loss of hpo-30 similarly abolished the formation of 3o and 4o branches (Figures 3A–3B, 3E–3G). Moreover, the dma-1; hpo-30 double mutants did not further enhance or suppress the dendritic branching defects observed in single mutant animals (Figures 3B–3G), consistent with the notion that dma-1 and hpo-30 might act in the same genetic pathway for dendrite formation.
Figure 3. HPO-30 interacts with DMA-1 both in vivo and in vitro.
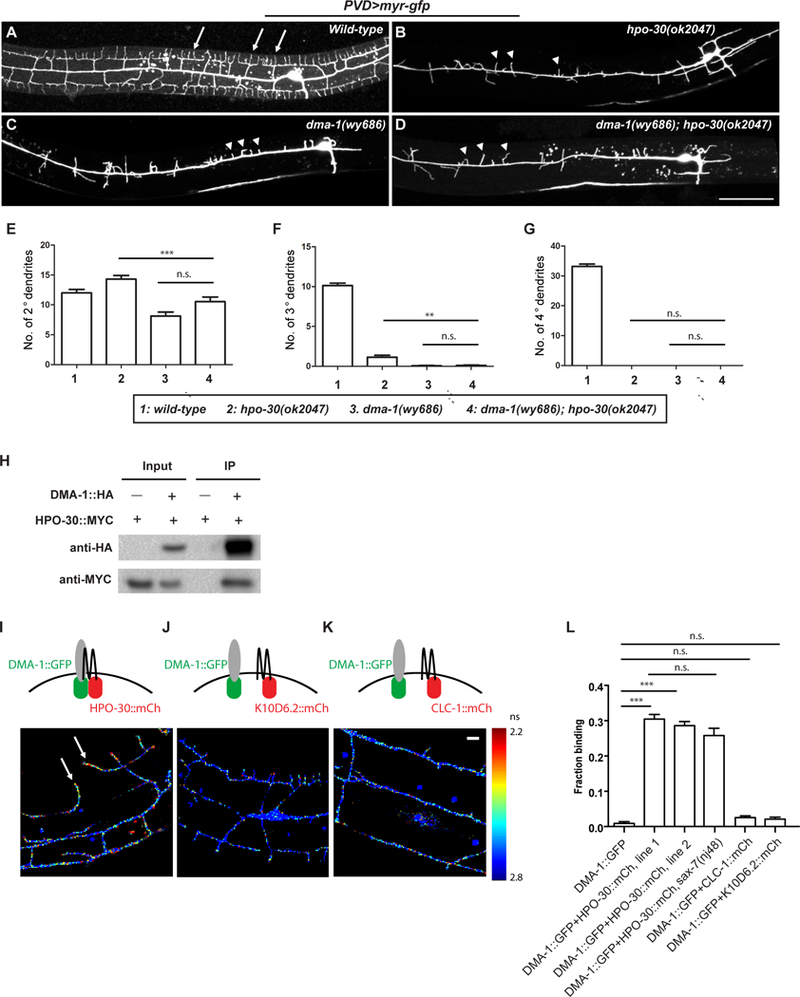
(A-D) Representative confocal images showing the morphologies of PVD dendritic arbors in animals of indicated genotypes. Arrows: 4o branches in the wild-type animals. Arrow heads: 2o branches that failed to form 4o or even 3o branches. Scale bar = 50 μm. (E-G) Quantification of the number of 2o, 3o and 4o branches in a region 100 μm anterior to the PVD cell body. Data were obtained using 20 animals for each genotype and are presented as mean ± SEM. One-sided ANOVA with Tukey test was used for statistical analysis. **: p<0.001. ***: p< 0.0001. n.s.: not significant. (H) Western blot of co-IP experiments. DMA-1-HA was used as the bait. (I-K) Fluorescence lifetime images of PVD neurons expressing DMA-1::GFPnovo2 with HPO-30::mCherry or with negative controls K10D6.2::mCherry or CLC-1::mCherry. Heat map represents measured fluorescence lifetime of DMA-1::GFPnovo2. Shorter life time (red signal) corresponds to higher FRET efficiency and stronger interaction. Arrows: DMA-1::GFPnovo2 co-expressed with HPO-30::mCherry exhibit short lifetime in 4o branches of wild-type PVD neurons. Scale bar = 10 μm. (L) Quantification of the images in (I-K), showing the fraction of DMA-1::GFPnovo2 bound to mCherry-tagged proteins. Data are represented as mean ± SEM, with 20 animals quantified for each genotype. ***: p< 0.001 by Student’s t-test. n.s.: not significant.
We next sought to determine whether HPO-30 and DMA-1 physically interact with each other to form a receptor complex. We first used a co-immunoprecipitation assay, which showed that MYC-tagged HPO-30 co-precipitated with HA-tagged DMA-1 from Drosophila S2 cells (Figure 3H). To further validate the interaction in vivo, we quantitatively measured fluorescence resonance energy transfer (FRET) between DMA-1::GFPnovo2 and HPO-30::mCherry molecules using two photon fluorescence lifetime imaging microscopy (2pFLIM) in live animals (Lee et al., 2009; Yasuda et al., 2006). When DMA-1::GFPnovo2 was co-expressed with HPO-30::mCherry in the PVD neuron, the lifetime of DMA-1::GFPnovo2 was significantly reduced (Figure 3I), suggesting that the fluorophores underwent strong FRET and that the two receptors were in close proximity. In contrast, co-expression of DMA-1::GFPnovo2 with two other claudin-like proteins, K10D6.2::mCherry or CLC-1::mCherry, did not affect the lifetime of DMA-1::GFPnovo2 (Figures 3J–3K), suggesting the observed FRET between DMA-1::GFPnovo2 and HPO-30::mCherry was specific. Quantification of the fluorescence lifetime images suggested that a large fraction of DMA-1::GFPnovo2 (~30%) bound to HPO-30::mCherry in two independent transgenes, whereas the control proteins exhibited no binding to DMA-1::GFPnovo2 (Figure 3L). We note that the interaction between DMA-1 and HPO-30 was not affected in sax-7 null mutants (Figure 3L), suggesting that the interaction is likely independent of the binding of extracellular ligands. Combining the in vitro and in vivo analysis, we conclude that HPO-30 acts as a signaling co-organizer of DMA-1 to drive dendrite morphogenesis.
HPO-30 is not required for the binding between DMA-1 receptor and the co-ligand complex
Previously we have shown that DMA-1 uses its extracellular domain to assemble a co-ligand complex through direct interactions with three epidermis/muscle proteins, SAX-7, MNR-1 and LECT-2 (Zou et al., 2016). Knowing that HPO-30 acts as a signaling co-organizer of DMA-1, we sought to determine whether HPO-30 is also a component of this multi-protein receptor-ligand complex and whether HPO-30 affects the assembly of the complex. For this, we performed single molecule pull-down experiments using proteins expressed either in Drosophila S2 cells or in C. elegans (Jain et al., 2011; Jain et al., 2012; Zou et al., 2016). In these experiments, HA- or FLAG-tagged proteins were immobilized by biotinylated anti-HA or anti-FLAG antibodies bound to a quartz cover slide coated with NeutrAvidin, where the HA- or FLAG-tagged proteins acted as bait proteins. Fluorescent protein-tagged prey proteins that were retained by the immobilized bait proteins were determined by quantifying the intensity and co-localization of fluorescent signals using single-molecule total internal reflection (TIRF) microscopy (Figure 4A) (Jain et al., 2011; Jain et al., 2012).
Figure 4. HPO-30 is dispensable for the formation of receptor-ligand complex.
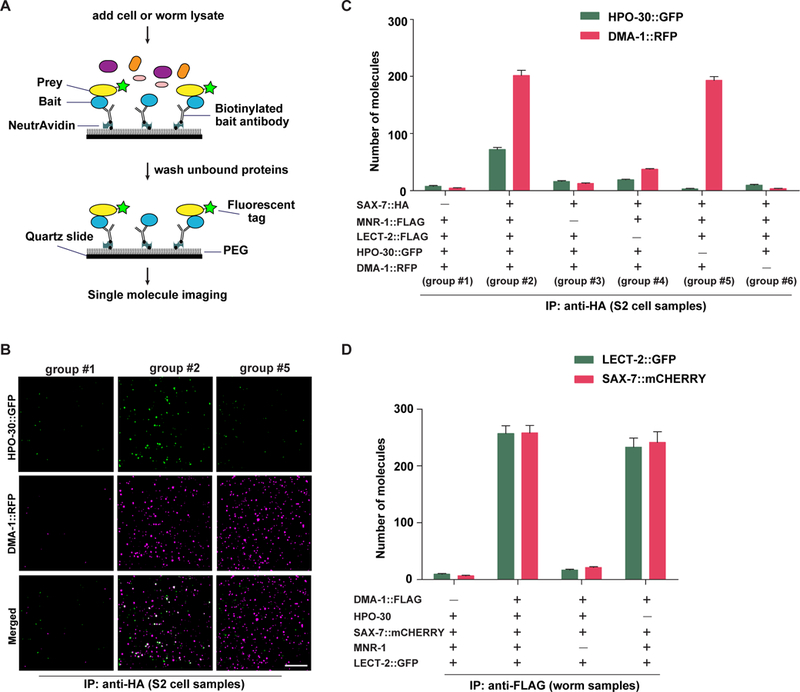
(A) Schematic showing the experimental design of the single molecule pull-down (SiMPull) assay. (B) Representative TIRF images showing single molecules labeled with GFP or RFP retained on the cover slides. Scale bar = 10 μm. Composition of indicated group IDs is shown in (C). (C) Quantification of the SiMPull experiments using proteins expressed from S2 cells. (D) Quantification of the SiMPull experiments using proteins extracted from worm lysates.
In the first set of experiments, the potential prey proteins included different combinations of SAX-7::HA, MNR-1::FLAG, LECT-2::FLAG, DMA-1::RFP and HPO-30::GFP expressed from Drosophila S2 cells. When all five proteins were present, DMA-1::RFP and HPO-30::GFP were efficiently retained by SAX-7::HA (group #2 in Figures 4B–4C). Removing the bait, SAX-7::HA, from the reaction (group #1 in Figures 4B–4C) revealed little to no background binding of DMA-1::RFP and HPO-30::GFP to the cover slide. When we examined the colocalization of the two channels, we found that 56.1% of DMA-1::RFP speckles co-localized with HPO-30::GFP, consistent with the notion that HPO-30 and DMA-1 form a signaling co-organizer complex.
When MNR-1 or LECT-2 was excluded from the reaction (group #3 and #4 in Figure 4C, respectively), SAX-7 no longer retained either DMA-1 or HPO-30 efficiently, consistent with our previous results (Zou et al., 2016) and suggesting that formation of a SAX-7-DMA-1-HPO-30 complex requires both MNR-1 and LECT-2. Furthermore, when HPO-30::GFP was excluded from the reaction, the pull-down efficiency for DMA-1::RFP was not affected (group #2 vs group #5 in Figures 4B–4C), suggesting that HPO-30 was not required for DMA-1 to complex with SAX-7. In contrast, when DMA-1::RFP was removed from the reaction, HPO-30::GFP could no longer be retained (group #6 in Figure 4C), indicating that HPO-30 itself did not bind to SAX-7 and likely formed a SAX-7-DMA-1-HPO-30 complex through an interaction with DMA-1.
In a second, orthogonal set of experiments, we repeated the single molecule immunoprecipitation assays by using proteins expressed from C. elegans to validate their interactions in vivo. In these experiments, DMA-1::FLAG was used as bait. Similar numbers of LECT-2::GFP and SAX-7::mCherry were retained by DMA-1::FLAG in both hpo-30(+) and hpo-30(−) strains (Figure 4D), consistent with the above results that HPO-30 was not required for DMA-1 to form the extracellular receptor-ligand complex. As a control, the receptor-ligand complex was disrupted in mnr-1 mutants (Figure 4D), consistent with our previous observation that formation of a stable receptor-ligand complex requires the presence of all four proteins (DMA-1, SAX-7, MNR-1 and LECT-2). Together, our results suggest that HPO-30 does not directly contribute to the formation of the receptor-ligand complex. Instead, HPO-30 may function as a signaling co-organizer through a direct interaction with DMA-1.
The intracellular domain of HPO-30 binds to the WRC
The results above suggest that although HPO-30 is required for the DMA-1-mediated dendrite branching, it does not act by facilitating the formation of the extracellular receptor-ligand complex. Therefore, we hypothesized that HPO-30 may function through intracellular signaling. Like claudin proteins, HPO-30 has four transmembrane helices and a short C-terminal ICD (51 amino acids for HPO-30). To test our hypothesis, we asked if the ICD of HPO-30 directly interacts with TIAM-1 or the WRC, the two actin regulators required for PVD dendrite morphogenesis (Figures 1D–1H).
We first asked if HPO-30 binds to TIAM-1, based on the observation that the HPO-30 ICD also contained a putative PDZ binding motif at the last four amino acids, and that although the DMA-1 ICD interacted with TIAM-1, the dma-1 △ICDendo mutants showed less severe defects in dendritic branching than the tiam-1 mutants. However, we did not detect an interaction between HPO-30 ICD and the TIAM-1 PDZ domain in a GST pull-down assay (Figure S6A). Furthermore, unlike DMA-1, HPO-30 lacking the putative PDZ binding motif (HPO-30 △5AA) was still able to fully restore the branching defect of the hpo-30 null mutants (Figures S6B–6F). Therefore, the putative PDZ binding motif in the HPO-30 ICD is not important for dendritic branching. HPO-30 likely interacts with a different binding partner to mediate branch formation.
We next asked if HPO-30 ICD interacted with the WRC using our GST pull-down approach. Indeed, the immobilized GST-HPO-30 ICD specifically retained a recombinantly purified, MBP-tagged C. elegans WRC ((MBP)3-ceWRC), but not the (MBP)2 tag or a subcomplex containing only WAVE, ABI and HSPC300 ((MBP)3-ceSubcomplex) (Figure 5A). These results suggest that the HPO-30 ICD may recruit the intact WRC through a direct interaction. The HPO-30 ICD does not appear to contain a WIRS peptide motif, suggesting it may use a novel mechanism to bind the WRC (Chen et al., 2014a; Chia et al., 2014). The HPO-30 ICD similarly interacted with the recombinantly purified human WRC (hWRC), suggesting that the interaction is conserved (Figure 5B). To map the binding region, we performed an alanine scan experiment by systematically mutating residues throughout the HPO-30 ICD to alanine, and then examined how the mutations affected the binding in pull-down assays. Due to technical difficulties in obtaining an adequate amount of ceWRC, for the assays below we used hWRC, which we could obtain in larger quantities. Our analysis suggested that many regions of the HPO-30 ICD contributed to the binding. In Figure 5B, by replacing strings of five amino acids with alanines, we found that mutating residues in the C-terminal half of the ICD appreciably reduced binding between the HPO-30 ICD and the WRC, including residues 21–47 but not the last four residues (putative PDZ motif). In contrast, mutating residues in the N-terminal half of the ICD did not affect the binding (except for residues 11–15, which when mutated, did reduce binding), suggesting that the C-terminal half of the HPO-30 ICD makes a major contribution to the interaction.
Figure 5. HPO-30 interacts with the WRC.
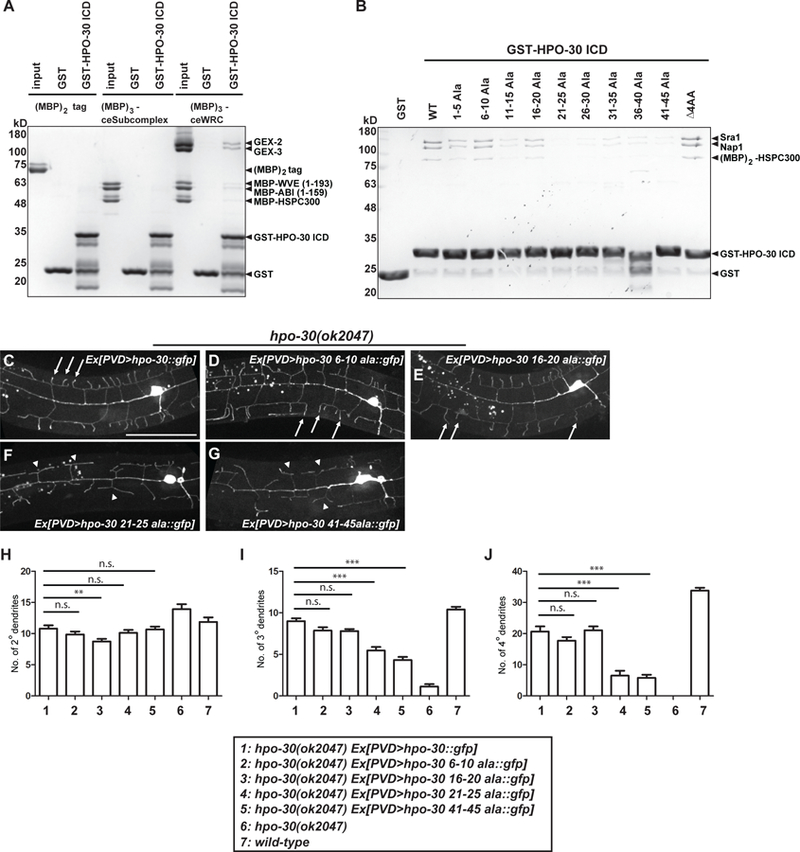
(A-B) Coomassie-blue-stained SDS-PAGE gel showing GST-HPO-30 ICD pull-down of C. elegans WRC (A) and human WRC (B). In (A), the input lanes were loaded with 3 pmol of the MBP-tagged proteins. In (B), GST-HPO-30 ICDs with indicated alanine scan mutations were compared. The numbers in the name of each construct indicate the position of the mutated amino acids in the cytosolic part of HPO-30 (the last 51 amino acids). △4AA means deletion of the last 4 amino acids of HPO-30. (C-G) Representative confocal images showing the morphologies of PVD dendritic arbors. PVD was labeled by GFP fused to the re-expressed wild-type or mutant forms of HPO-30 in hpo-30 null mutant animals. Arrows: 4o branches in fully rescued animals. Arrow heads: 2o branches that failed to form 4o branches. Scale bar = 50 μm. (H-J) Quantification of the number of 2o, 3o and 4o branches in a region 100 μm anterior to the PVD cell body. Data are represented as mean ± SEM, with 15 animals quantified for each genotype. One-sided ANOVA with Dunnett’s test was used for statistical analysis. **: p<0.001. ***: p< 0.0001. n.s.: not significant. See also Figure S3 and S6.
To test if the direct interaction between HPO-30 and the WRC is important for dendrite branch formation in vivo, we expressed four of the HPO-30 alanine scan mutants in hpo-30 null strains, and evaluated their ability to restore branch formation. Consistent with the above in vitro binding results, mutating the N-terminal residues of the HPO-30 ICD (residues 6–10 or 16–20) rescued the dendritic branching defect to an extent comparable to the wild type HPO-30 rescue, whereas mutating the C-terminal residues (residues 21–25 or 41–45) failed to fully rescue the dendritic defects (Figures 5C–5J). Combining both the in vitro and in vivo results, we conclude that the signaling co-organizer HPO-30 acts though the WRC to promote branch formation.
The intracellular domains of HPO-30 and DMA-1 act synergistically
The above evidence raises an intriguing model whereby the DMA-1-HPO-30 signaling co-organizer complex cooperates by recruiting TIAM-1 and the WRC to close proximity to orchestrate actin polymerization, which in turn drives exuberant dendritic branching. To test the synergy between DMA-1 and HPO-30, we compared the effects of individually perturbing each interaction versus simultaneously disrupting both binding events in vivo.
We first perturbed the HPO-30-WRC interaction by expressing an HPO-30 lacking the last 10 amino acids of the ICD, tagged with GFP on its C terminus (HPO-30△10AA::GFP). This construct was expressed under a PVD neuron-specific promoter in hpo-30 null mutants. Compared to expression of a full-length HPO-30 tagged with GFP (HPO-30::GFP), HPO-30△ 10AA::GFP constructs was expressed at similar levels and properly localized to dendritic surface (Figures S3C–3D), suggesting that deletion of the last 10 amino acids did not affect protein folding or trafficking. Compared to the rescuing ability of the full length HPO-30::GFP, HPO-30 △10AA::GFP was able to rescue 20.4% of the 4o branches and 53.6% of the 3o branches (Figures 6A–6D, 6G–6I). This result corroborates the above alanine scan experiments which showed that the C-terminal half of the HPO-30 ICD is critical for binding the WRC. Moreover, these data indicate that by disrupting the HPO-30-mediated signaling, the DMA-1-TIAM-1 signaling alone can support 3o branching to some degree, but was not sufficient for 4o branch formation.
Figure 6. Synergy between HPO-30 and DMA-1 is required for dendrite formation.

(A-F) Representative confocal images showing the morphologies of PVD dendritic arbors in animals of indicated genotypes. △ 10 AA means deletion of the last 10 amino acids of HPO-30. Arrows: 4o branches in the wild-type worms and hpo-30 mutant animals carrying a functional transgene. Arrow heads: 2o branches that failed to form 4o or even 3o branches. Scale bar = 50 μm. (G-I) Quantification of the number of 2o, 3o and 4o branches in a region 100 μm anterior to the PVD cell body. Data are represented as mean ± SEM with at least 15 animals quantified for each genotype. One-sided ANOVA with Tukey test was used for statistical analysis. *p<0.05. **: p<0.001. ***: p< 0.0001. n.s.: not significant.
Similarly, we perturbed the DMA-1-TIAM-1 interaction by inserting a ZF1-YFP cassette at the C terminus of DMA-1 in the endogenous dma-1 locus (DMA-1::ZF1::YFP). Compared to the wild-type strains, the knock-in animals reduced the 3o dendritic branches to 50.2% and the 4o branches to 17.6% (Figures S4B–4E). This result indicates that by disrupting the DMA-1-TIAM-1 interaction, the HPO-30-WRC interaction alone can largely support 3o branching but was not sufficient for growing the 4o branches.
Finally, we simultaneously disrupted both interactions by combining these two genetic manipulations. For this, we created a strain that contained both the hpo-30 null and DMA-1::ZF1::YFP knock-in backgrounds, and expressed HPO-30△10AA::GFP driven by a PVD neuron-specific promoter. In contrast to the milder effects seen in animals with a single interaction disruption, the doubly disrupted animals completely failed to make 3o and 4o branches (Figures 6F–6I), demonstrating a synergistic effect of losing both the HPO-30-WRC and DMA-1-TIAM-1 interactions. Together, these results suggest that HPO-30 and DMA-1 act synergistically to co-localize two distinct actin regulators to promote F-actin assembly at the dendritic branching loci. This synergy is required for the establishment of branched arbors and is particularly critical for the higher order dendritic branches.
Discussion
HPO-30 functions as a signaling co-organizer of DMA-1
Several lines of evidence support the notion that HPO-30 is a signaling co-organizer of DMA-1. First, both the hpo-30 and dma-1 null mutants showed similar phenotypes. In both mutants, the 3o and 4o dendrites were completely missing. Among all the mutants that we have isolated with defective PVD dendrite morphology, only these two mutants showed nearly identical phenotypes. Second, both hpo-30 and dma-1 functioned cell autonomously in PVD to regulate arbor morphology. Third, hpo-30; dma-1 double mutants showed similar phenotypes as the single mutants, suggesting that they likely function in the same genetic pathway. Fourth, HPO-30 and DMA-1 physically interact with each other both in vivo and in vitro.
Interestingly, HPO-30 did not appear to participate in the formation of the inter-cellular ligand-receptor complex, because it did not enhance the binding between DMA-1 and other components of the complex, and was dispensable for the complex formation. However, these in vitro assays might not provide sufficient sensitivity to detect the contribution of HPO-30 to ligand binding. HPO-30 has four transmembrane domains and two extracellular loops, which contain the signature cysteine motif characteristic of the claudin-family proteins. Claudins form tight junctions through homotypic interactions both in trans and in cis (Krause et al., 2008). The crystal structure of a claudin showed that the cis interactions were responsible for concentrating claudins into clusters while the trans interactions were important for juxtaposition of the opposing membranes in close proximity (Suzuki et al., 2014). It is conceivable that the HPO-30-DMA-1 interaction on PVD membrane may increase the local concentration of DMA-1 to promote ligand binding. Consistent with this notion, the deletion variant of HPO-30 lacking the ICD that binds to the WRC showed a milder phenotype compared with the hpo-30 null mutants, suggestive of additional functions of HPO-30.
Actin regulators act downstream of DMA-1 and HPO-30 to form dendritic branches
It is well established that actin plays a critical role in neuron morphogenesis. Therefore, it is not surprising that mutations in major actin regulators such as WRC, Rac family of small GTPases (unpublished result, Zou W and Shen K) or TIAM-1 would perturb PVD morphology (Aguirre-Chen et al., 2011; Smith et al., 2010). Nevertheless, it is interesting to compare the loss-of-function phenotypes between the actin regulators and those of dma-1 and hpo-30. The actin regulator mutants displayed dramatically simplified dendritic arbors with reduced branch numbers, but the overall menorah shapes were maintained. In dma-1, hpo-30 or sax-7 mutants, the menorah shapes were lost due to the complete absence or disruption of the 3o and 4o branches. This is consistent with the notion that the DMA-1-HPO-30 receptor complex and their extracellular ligands serve as the topmost signaling cue to initiate the morphogenesis events. DMA-1 and HPO-30 subsequently recruit and direct the actin regulators to the desired subcellular loci, with DMA-1 binding to the RacGEF TIAM1 and HPO-30 binding to the WRC, respectively. These interactions appear to be important for dendrite formation because perturbing these interactions with mutations in dma-1 or hpo-30 resulted in reduction of the 3o and 4o branches. The reduction of 4o branches is particularly dramatic, hinting that a higher level of signaling might be required to form the 4o branches.
Our genetic experiments demonstrated that the synergy between the WRC and TIAM-1 recruitments was essential for high-order branches. Missing one of the two interactions only led to a mild reduction of the 3o branches and a strong, but incomplete, reduction of the 4o branches. In contrast, missing both interactions resulted in a complete loss of both 3o and 4o branches. The simultaneous recruitment of the WRC and TIAM-1 to the receptor associated signaling complex can potentially not only increase the amplitude of actin assembly by increasing the local concentration of active Rac and the WRC, but also enhance the spatial precision of signaling. Another interesting question is whether DMA-1 or HPO-30 simply recruits and organizes actin regulators, or whether the binding interactions further activate the actin regulators. Future experiments to directly measure the activity of actin regulators will shed light on this question.
Analogous signaling strategies are likely used to achieve spatial specificity of the F-actin-based morphogenesis
It is well established that the Rac-WRC-Arp2/3 signaling axis plays important roles in controlling actin polymerization and membrane protrusion to drive cell morphogenesis, especially in neurons (Dent et al., 2011; Lowery and Vactor, 2009). It is not well understood how these molecules achieve high spatial and temporal specificity in directing F-actin assembly during a morphogenesis event. Many studies have shown that axon guidance receptors can directly interact with Rac-specific GEFs, which can elevate localized activation of Rac at membranes of action. For example, the ROBO receptor in Drosophila was shown to recruit the SOS RacGEF through the adaptor protein Dock (Yang and Bashaw, 2006). Similarly, the DCC receptor binds to the DOCK180 RacGEF using its intracellular domain (Li et al., 2008) and the Frazzled receptor (Drosophila homolog of DCC) can recruit a distinct RacGEF, TRIO (Forsthoefel et al., 2005). These studies suggest that while the exact molecular components may be different, the interactions between receptors and GEFs may be generally utilized to provide specificity of Rac activation.
On the other hand, membrane receptors could also achieve spatial specificity by directly interacting with the Rac effector WRC. Our biochemical, structural, and genetic work showed that numerous receptors, including DCC, ROBO and Frazzled, can potentially recruit the WRC to membranes using a short WIRS peptide motif found in their intracellular domains (Chen et al., 2014a; Chia et al., 2014). It is plausible that the aforementioned WIRS receptors could further enhance the spatial specificity of signaling by scaffolding the RacGEFs and the WRC to close proximity. Our work provides direct evidence supporting this hypothesis by showing that the WRC and RacGEF TIAM-1 can be synergistically recruited to promote dendrite branching (Figure 7). The fact that in this specific case, the colocalization of the WRC and GEF is facilitated by a receptor associated signaling complex, and that HPO-30 uses a novel mechanism to bind the WRC, further suggests that such synergistic recruitments might be more prevalent in cells and might be afforded by a larger spectrum of interaction mechanisms than currently realized.
Figure 7. DMA-1 and HPO-30 link extrinsic signals to intracellular regulation of the actin cytoskeleton to control dendrite branching.
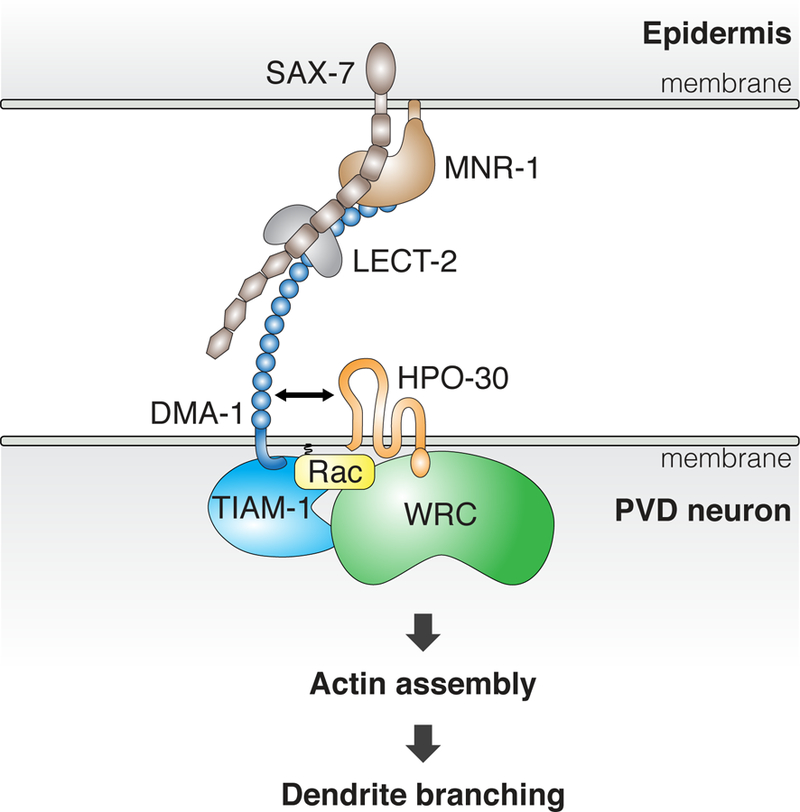
Cartoon representation showing our current understanding of how the DMA-1-HPO-30 protein complex promotes dendrite formation.
STAR Methods text
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-HA | Sigma | Cat#: H3663; RRID: AB_262051 |
| Rabbit anti-MYC | Santa Cruz Biotechnology |
Cat# sc-789; RRID:AB_631274 |
| Mouse anti-FLAG | Sigma | Cat# F2555; RRID:AB_796202 |
| Bacterial and Virus Strains | ||
| OP50 | Caenorhabditis Genetics Center |
https://cgc.umn.edu/strain/OP50 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Glutathione Sepharose 4B beads | GE Healthcare | Cat# 17075605 |
| Amylose beads | New England Biolabs | Cat# E8021L |
| Source 15Q beads | GE Healthcare | Cat# 17094705 |
| Source 15S beads | GE Healthcare | Cat# 17094405 |
| Anti-HA affinity gel | Sigma-Aldrich | Cat# E6779 |
| GST-TIAM-1 PDZ (a.a. QRKRE...VNKKT) | This paper | N/A |
| GST-TIAM-1 PDZ KL/AA (with a.a. VRKTNGRLGL mutated to VRATNGRAGL) |
This paper | N/A |
| GST-DMA-1 ICD (a.a. RERQY…SYFGI) | This paper | N/A |
| GST-HPO-30 ICD and various Alanine scan mutants (a.a. TSKHA…DDSSM) |
This paper | N/A |
| MBP-DMA-1 ICD (a.a. RERQY…SYFGI) | This paper | N/A |
| MBP-DMA-1 ICD △4AA (a.a. RERQY…KPGSS) | This paper | N/A |
| MBP-HPO-30 ICD (a.a. TSKHA…DDSSM) | This paper | N/A |
| MBP-HPO-30 ICD △4AA (a.a. TSKHA…VVIDD) | This paper | N/A |
| Recombinant CeWRC, including His6-Gex2 FL, His6- Gex3 FL, MBP-WVE (1–193), MBP-Abi (1–159), and MBP-HSPC300 FL |
This paper | N/A |
| Recombinant hWRC, including His6-hSra1 FL, His6- hNap1 FL, MBP-hWAVE1 (1–178), MBP-hAbi2 (1–158), and (MBP)2-HSPC300 FL |
Chen et al., 2014a | N/A |
| Critical Commercial Assays | ||
| Phusion High-Fidelity DNA polymerase | New England Biolabs | Cat # M0530S |
| Experimental Models: Cell Lines | ||
| Drosophila S2 cells | Invitrogen | Cat # R69007; Cell Line: Schneider 2 |
| Experimental Models: Organisms/Strains | ||
| C. elegans strains, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kang Shen (kangshen@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were grown on nematode growth medium (NGM) plates seeded with OP50 E. coli. Worm strains were maintained at 20 °C or room temperature (22.5 °C). N2 Bristol was used as the wild-type strain. A list of C. elegans strains used in this study is shown in Table S1.
METHOD DETAILS
DNA Manipulations and Transgenes
Most of the plasmid constructs were generated in pSMdelta vector backbone (a derivative of pPD49.26). Transgenes expressed from extrachromosomal arrays were generated using standard gonad transformation by injection (Mello and Fire, 1995). Pmyo-2>mcherry, Pmyo-2>gfp, Pmyo-3>mcherry, Podr-1>gfp, Podr-1>rfp, Punc-122>rfp or unc-119(+) plasmid was injected at 2–40 ng/µl as co-injection markers.
CRISPR/Cas9-mediated Genome Editing
To generate mutations to perturb the function of the intracellular domain of DMA-1, Peft-3>cas9 (50 ng/µl) (Dickinson et al., 2013), U6>dma-1-sg#1 (50 ng/µl, target sequence: 5’-GTGTGTTTGCCATCGTGAG-3’) and Pmyo-3>mcherry (5 ng/µl) were injected into wyIs592 (PVD>myr-gfp) strain. F1 worms with abnormal PVD dendritic morphology were rescued and F2 worms with 100% dendrite morphogenesis defects were kept. The genomic DNA of dma-1 locus were amplified by PCR and sequenced. wy908 contained an 8 base pairs deletion that caused a frame shift leading to premature stop codons in the cytosolic domain of dma-1.
To insert a zf1::yfp (with floxed Cbr-unc-119 in one of the introns of yfp) cassette into the endogenous locus of dma-1, zf1::yfp(+floxed Cbr-unc-119) was amplified from pJN601 and served as the repair template (Armenti et al., 2014). Two micro-homology arms (40bp and 46 bp, respectively) were added onto the PCR primers (Paix et al., 2014). Peft-3>cas9 (50 ng/µl), U6>dma-1-sg#2 (20 ng/µl, target sequence: 5’-G ATTCCACCAGCACCTCCAAAACC-3’), U6>dma-1-sg#3 (20 ng/µl, target sequence: 5’-G CCAAAATAGGATGATCCAGGTTT −3’), U6>dma-1-sg#4 (20 ng/µl, target sequence: 5’-G GATGATCCAGGTTTTGG-3’), repair template (10 ng/µl), Pmyo-2>mcherry (2 ng/µl), Pmyo-3>mcherry (5 ng/µl) and Podr-1>rfp (20 ng/µl) were injected into unc-119(ed4) strain. dma-1(wy996) was isolated based on 100% non-unc and lack of red fluorescence protein expression. wy1000 was generated by injecting Peft-3>Cre into wy996 strain to remove the floxed Cbr-119 (Dickinson et al., 2013). Genomic DNA of dma-1::zf1::yfp was amplified by PCR and sequenced. No extra mutation was identified.
Isolation, mapping and cloning of wy917 mutant
dma-1(wy908); wyIs592 worms at L4 stage were mutagenized with 50 mM ethyl methane sulfonate (EMS). Five F1 animals were picked into each plate and F2 animals were screened under a fluorescent compound microscope for enhanced PVD dendrite morphogenesis defects. wy917 was isolated from a screen of 3000 haploid genomes. Standard SNP-SNIP mapping and fosmid transformation experiments were used to map and clone the gene affected by wy917 (Davis et al., 2005). act-4 genomic DNA was amplified by PCR and sequenced to identify the causative mutation.
Confocal Imaging of C. elegans
Hermaphroditic C. elegans were anesthetized using 10 mM levamisole in M9 buffer, mounted on 2% agar pads and imaged using a Zeiss LSM710 confocal microscope (Carl Zeiss) with a Plan-Apochromat 40×/1.3NA objective (for most images showed in this study) or a spinning disk confocal microscope with a 63×/1.4NA objective (for images showed in Figures 1I–1N, 5C–5G, S2 and S3). 15–30 Z-stacks (0.5 or 0.75 μm/step) were generated to cover the entire PVD dendritic arbors. Maximum-intensity projections were generated using ZEN2009 software or ImageJ.
Co-immunoprecipitation (Co-IP)
To express proteins of interest for Co-IP experiments, Drosophila S2 cells were cultured in Schneider’s insect medium (Sigma) according to the manufacturer’s description and transfected using Effectene (Qiagen). Three days after transfection, S2 cells were harvested to prepare cell lysates. Anti-HA affinity gel (Sigma) was used to incubate with the cell lysates for 2 hours at 4 ℃ with rotation. The gel was washed with cell lysis buffer (1xPBS, 1% Triton X-100 and 1% protease inhibitor cocktail (Sigma)) three times. Proteins were eluted at 65℃ using 2% SDS elution buffer and detected using Western blot analysis with mouse antibody to HA (1:1000, Sigma), rabbit antibody to Myc (1:2000, Santa Cruz Biotechnology), and HRP-conjugated goat antibodies to mouse (1:20,000, Jackson Immuno Research).
Protein purification and GST pull-down
Both the ceWRC and hWRC were recombinantly expressed and purified essentially as previously described (Chen et al., 2014b). GST-tagged HPO-30 ICD and TIAM-1 PDZ were expressed in BL21 (DE3) T1R cells at 18 °C overnight and purified using glutathione sepharose beads (GE Healthcare). MBP-tagged HPO-30 ICD and DMA-1 ICD were similarly expressed and purified using amylose beads (New England Biolabs). The tagged ICDs and PDZ domain were further purified using Source 15Q and/or Source 15S ion exchange columns (GE Healthcare) before use.
GST pull-down was performed as previously described (Chen et al., 2014a). Briefly, 130 to 300 pmol of GST-tagged bait and 130 to 300 pmol of prey proteins were mixed with 20 µL of glutathione sepharose beads (GE Healthcare) in 1 mL of pulldown buffer (20 mM HEPES pH 7, 50 mM NaCl, 5% (w/v) glycerol and 5 mM β-mercaptoethanol) at 4 °C for 30 min. After three washes using 1 mL of the pulldown buffer, bound proteins were eluted with GST elution buffer (100 mM Tris-HCl pH 8.5, 50 mM NaCl, 5% (w/v) glycerol, 5 mM β-mercaptoethanol and 30 mM reduced glutathione) and examined by SDS-PAGE.
Two-photon fluorescence lifetime imaging microscopy (2pFLIM)
FRET imaging using a custom-built two-photon fluorescence lifetime imaging microscope was conducted in Ryohei Yasuda’s laboratory as previously described (Murakoshi et al., 2008; Murakoshi et al., 2011; Yasuda, 2006). Briefly, worms expressing an integrated PVD>dma-1::GFPnovo2 marker (with the GFPnovo2 inserted before the cytosolic domain of DMA-1 to avoid hindering the PDZ binding motif) (Arakawa et al., 2008) were injected with extrachromosomal arrays carrying PVD>hpo-30::mcherry, PVD>K10D6.2::mcherry or PVD>clc-1::mcherry. Expression and membrane localization of the proteins were verified using regular fluorescent microscopy. During the 2pFLIM experiment, GFPnovo2 and mCherry were simultaneously excited with a Ti:sapphire laser (Maitai, Spectraphysics) tuned at a wavelength of 920 nm. Fluorescent lifetime images were obtained using a time-correlated single photon counting board (SPC-140; Becker and Hickl) controlled with custom software (Yasuda et al., 2006). The fluorescence lifetime of GFPnovo2 was obtained as described before (Lee et al., 2009; Yasuda, 2006; Yasuda et al., 2006). The value is then set as the fixed lifetime value for the free DMA-1::GFPnovo2 donors. PVD neurons co-expressing DMA-1::GFPnovo2 and HPO-30::mCherry or one of the negative controls were then imaged using the same setting and FRET between GFPnovo2 and mCherry were measured using 2pFLIM. The fraction of DMA-1::GFPnovo2 bound to HPO-30::mCherry or controls was calculated by fitting the fluorescence lifetime curve with a double exponential function (Lee et al., 2009; Yasuda, 2006; Yasuda et al., 2006).
Single Molecule Pull-down Assay (SiMPull)
SiMPull assay was performed as previously described (Zou et al., 2016). Briefly, Drosophila S2 cells over-expressing proteins of interested were pelleted and lysed in lysis buffer (50 mM HEPES pH 7.7, 150 mM NaCl, 2 mM MgCl2, 1 mM EDTA pH 8.0, 1% Triton X-100 with protease inhibitors) at 4°C for 1 hr. After centrifugation 16000g 15min, supernatants were collected and total protein concentration for each sample was measured by BCA assay (Thermo Fisher Scientific). C. elegans grown on twenty 6-cm NGM plates were harvested to form “worm pearls” using a liquid nitrogen-based method. Worm pearls (300mg wet weight) were thawed in 150 µl of lysis buffer (50 mM HEPES pH 7.7, 50 mM KCl, 2 mM MgCl2, 250 mM sucrose, 1 mM EDTA pH 8.0, with protease inhibitors). After sonication on ice (5’ pulse with 59” pause, 5 cycles) to break cuticle, 100 mM NaCl and 1% Triton X-100 were added into solution and samples were rotated at 4°C for 1 hr. After centrifugation, supernatants were collected and total protein concentration for each sample was measured by BCA assay. Worm and S2 cell lysates were adjusted by lysis buffer to desired concentrations to achieve optimum density of fluorescent proteins on the surface of imaging slides (100–400 molecules in a 2000 μm2 imaging area). Proteins immobilized on the imaging slides were visualized by a TIRF microscope equipped with excitation laser 488 nm (GFP) and 561 nm (mCherry or RFP), and DV2 dichroic 565dcxr dual-view emission filters (520/30 nm and 630/50 nm). 5–13 different regions of the imaging surface were imaged and analyzed.
QUANTIFICATION AND STATISTICAL ANALYSIS
For C. elegans confocal imaging experiments, a 100 μm region anterior to the PVD cell body was selected to quantify the number of the 2o, 3o and 4o branches in each genotype. For SiMPull experiments, single-molecule colocalization between GFP and mCherry was calculated as previously described (Jain et al., 2011). Co-localization was defined if two molecules (labeled by mCherry and GFP, respectively) were within a 2-pixel distance (~300 nm). Co-localization index equals to the number of molecules where colocalization occurred divided by the total number of mCherry molecules. Statistical comparisons were conducted using Student’s t-test (to test for differences between two groups) or one-sided ANOVA with the Tukey correction or Dunnett’s test (to test for differences between three or more groups).
Supplementary Material
Acknowledgements
This work was supported by the Howard Hughes Medical Institute and the National Institute of Neurological Disorders and Stroke (1R01NS082208), National Natural Science Foundation of China (31571061) and the CAS/SAFEA International Partnership Program for Creative Research Teams to K.S., National Natural Science Foundation of China (31741056) to W.Z., the National Institute of Neurological Disorders and Stroke (1R01NS079611) to D.M., and start-up funds to B. C. from the Iowa State University and the Roy J. Carver Charitable Trust. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and the MITANI Lab through the National Bio-Resource Project of the MEXT, Japan. We thank Drs. Suhong Xu, Zhiping Wang, Erik Lundquist, Jeremy Nance, Jordan Ward, Bob Goldstein, Erik Jorgensen, Yuji Kohara and Liqun Luo for kindly sharing equipment and/or reagents, and Cen Gao for technique assistance.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Aguirre-Chen C, Bulow HE, and Kaprielian Z (2011). C. elegans bicd-1, homolog of the Drosophila dynein accessory factor Bicaudal D, regulates the branching of PVD sensory neuron dendrites. Development 138, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albeg A, Smith CJ, Chatzigeorgiou M, Feitelson DG, Hall DH, Schafer WR, Miller DM 3rd, and Treinin M (2011). C. elegans multi-dendritic sensory neurons: morphology and function. Mol Cell Neurosci 46, 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen R, Li Y, Resseguie M, and Brenman JE (2005). Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J Neurosci 25, 8878–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H, Kudo H, Batrak V, Caldwell RB, Rieger MA, Ellwart JW, and Buerstedde J-M (2008). Protein evolution by hypermutation and selection in the B cell line DT40. Nucleic Acids Research 36, e1-e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenti ST, Lohmer LL, Sherwood DR, and Nance J (2014). Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development 141, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, and Rosen MK (2014a). The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Chou HT, Brautigam CA, Xing W, Yang S, Henry L, Doolittle LK, Walz T, and Rosen MK (2017). Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Padrick SB, Henry L, and Rosen MK (2014b). Biochemical reconstitution of the WAVE regulatory complex. Methods Enzymol 540, 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, and Rosen MK (2010). Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PH, Chen B, Li P, Rosen MK, and Shen K (2014). Local F-actin network links synapse formation and axon branching. Cell 156, 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, and Jorgensen EM (2005). Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarco RS, Struckhoff EC, and Lundquist EA (2012). The Rac GTP exchange factor TIAM-1 acts with CDC-42 and the guidance receptor UNC-40/DCC in neuronal protrusion and axon guidance. PLoS Genet 8, e1002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Gupton SL, and Gertler FB (2011). The Growth Cone Cytoskeleton in Axon Outgrowth and Guidance. Cold Spring Harbor perspectives in biology 3, a001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Balzac CA, Rahman M, Lazaro-Pena MI, Martin Hernandez LA, Salzberg Y, Aguirre-Chen C, Kaprielian Z, and Bulow HE (2016). Muscle- and Skin-Derived Cues Jointly Orchestrate Patterning of Somatosensory Dendrites. Curr Biol 26, 2397. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Liu OW, Howell AS, and Shen K (2013). An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell 155, 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Shen K, and Bulow HE (2015). Intrinsic and extrinsic mechanisms of dendritic morphogenesis. Annu Rev Physiol 77, 271–300. [DOI] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, and Kirschner MW (2002). Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793. [DOI] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, and Jan YN (2004). Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell 119, 245–256. [DOI] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, and Rice CM (2007). Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, Liebl EC, Kolodziej PA, and Seeger MA (2005). The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila. Development 132, 1983–1994. [DOI] [PubMed] [Google Scholar]
- Huber AB, Kolodkin AL, Ginty DD, and Cloutier JF (2003). Signaling at the growth cone: ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci 26, 509–563. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, and Schmucker D (2007). Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron 54, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, and Rosen MK (2009). The WAVE regulatory complex is inhibited. Nat Struct Mol Biol 16, 561–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SC, Wang D, Iyer EP, Trunnell SA, Meduri R, Shinwari R, Sulkowski MJ, and Cox DN (2012). The RhoGEF trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLoS One 7, e33634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, and Ha T (2011). Probing cellular protein complexes using single-molecule pull-down. Nature 473, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Xiang YK, and Ha T (2012). Single-molecule pull-down for studying protein interactions. Nat Protoc 7, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, and Jan LY (2010). Branching out: mechanisms of dendritic arborization. Nat Rev Neurosci 11, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, and Blasig IE (2008). Structure and function of claudins. Biochim Biophys Acta 1778, 631–645. [DOI] [PubMed] [Google Scholar]
- Lee HJ, and Zheng JJ (2010). PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-JR, Escobedo-Lozoya Y, Szatmari EM, and Yasuda R (2009). Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 458, 299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, and Sanes JR (2012). PROTOCADHERINS MEDIATE DENDRITIC SELF-AVOIDANCE IN THE MAMMALIAN NERVOUS SYSTEM. Nature 488, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao X, Liu G, Xiong W, Wu J, and Rao Y (2008). Netrin signal transduction and the guanine nucleotide exchange factor DOCK180 in attractive signaling. Nat Neurosci 11, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Dong X, Moerman DG, Shen K, and Wang X (2015). Sarcomeres Pattern Proprioceptive Sensory Dendritic Endings through UNC-52/Perlecan in C. elegans. Dev Cell 33, 388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu OW, and Shen K (2012). The transmembrane LRR protein DMA-1 promotes dendrite branching and growth in C. elegans. Nat Neurosci 15, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, and Vactor DV (2009). The trip of the tip: understanding the growth cone machinery. Nat Rev Mol Cell Biol 10, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L (2000). Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci 1, 173–180. [DOI] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, and Grueber WB (2007). Dendrite self-avoidance is controlled by Dscam. Cell 129, 593–604. [DOI] [PubMed] [Google Scholar]
- Mello C, and Fire A (1995). DNA transformation. Methods Cell Biol 48, 451–482. [PubMed] [Google Scholar]
- Murakoshi H, Lee S-J, and Yasuda R (2008). Highly sensitive and quantitative FRET–FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biology 36, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Wang H, and Yasuda R (2011). Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature 472, 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Wang Y, Smith HE, Lee CY, Calidas D, Lu T, Smith J, Schmidt H, Krause MW, and Seydoux G (2014). Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198, 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Morrow T, and Ghosh A (2000). Semaphorin 3A is a chemoattractant for cortical apical dendrites. Nature 404, 567–573. [DOI] [PubMed] [Google Scholar]
- Ravenscroft G, Wilmshurst JM, Pillay K, Sivadorai P, Wallefeld W, Nowak KJ, and Laing NG (2011). A novel ACTA1 mutation resulting in a severe congenital myopathy with nemaline bodies, intranuclear rods and type I fibre predominance. Neuromuscul Disord 21, 31–36. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg Y, Diaz-Balzac CA, Ramirez-Suarez NJ, Attreed M, Tecle E, Desbois M, Kaprielian Z, and Bulow HE (2013). Skin-derived cues control arborization of sensory dendrites in Caenorhabditis elegans. Cell 155, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, O’Brien T, Chatzigeorgiou M, Spencer WC, Feingold-Link E, Husson SJ, Hori S, Mitani S, Gottschalk A, Schafer WR, et al. (2013). Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron 79, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Watson JD, Spencer WC, O’Brien T, Cha B, Albeg A, Treinin M, and Miller DM 3rd (2010). Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev Biol 345, 18–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, and Jan YN (2007). Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 54, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, and Fujiyoshi Y (2014). Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344, 304–307. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, and Greenberg ME (2005). The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron 45, 525–538. [DOI] [PubMed] [Google Scholar]
- Yang L, and Bashaw GJ (2006). Son of sevenless directly links the Robo receptor to rac activation to control axon repulsion at the midline. Neuron 52, 595–607. [DOI] [PubMed] [Google Scholar]
- Yasuda R (2006). Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Current Opinion in Neurobiology 16, 551–561. [DOI] [PubMed] [Google Scholar]
- Yasuda R, Harvey CD, Zhong H, Sobczyk A, van Aelst L, and Svoboda K (2006). Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci 9, 283–291. [DOI] [PubMed] [Google Scholar]
- Zou W, Shen A, Dong X, Tugizova M, Xiang YK, and Shen K (2016). A multi-protein receptor-ligand complex underlies combinatorial dendrite guidance choices in C. elegans. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


