Abstract
Proton assignment of nuclear magnetic resonance (NMR) spectra of homopyrimidine/homopurine tract oligonucleotides becomes extremely challenging with increasing helical length due to severe cross-peak overlap. As an alternative to the more standard practice of 15N and 13C labeling of oligonucleotides, here, we describe a method for assignment of highly redundant DNA sequences that uses single-site substitution of the thymine isostere 2,4-difluoro-5-methylbenzene (dF). The impact of this approach in facilitating the assignment of intractable spectra and analyzing oligonucleotide structure and dynamics is demonstrated using A-tract and TATA box DNA and two polypurine tract-containing RNA:DNA hybrids derived from HIV-1 and the Saccharomyces cerevisiae long-terminal repeat-containing retrotransposon Ty3. Only resonances proximal to the site of dF substitution exhibit sizable chemical shift changes, providing spectral dispersion while still allowing chemical shift mapping of resonances from unaffected residues distal to the site of modification directly back to the unmodified sequence. It is further illustrated that dF incorporation can subtly alter the conformation and dynamics of homopyrimidine/homopurine tract oligonucleotides, and how these NMR observations can be correlated, in the cases of the TATA box DNA, with modulation in the TATA box-binding protein interaction using an orthogonal gel assay.
1. INTRODUCTION
Fluorinated nucleoside analogs have been used as structural and chemical probes in several nuclear magnetic resonance (NMR) spectroscopy studies of nucleic acid structure and protein–nucleic acid interactions (Carlow, Short, & Wolfenden, 1996; Chu & Horowitz, 1991a,b; Hammann, Norman, & Lilley, 2001; Kwon, Jiang, Song, & Stivers, 2002; Liu, Chu, Liu, & Horowitz, 1997; Olejniczak et al., 2002; Rastinejad, Evilia, & Lu, 1995; Rastinejad & Lu, 1993). In these analogs, the fluorine (19F) nuclei provide NMR signals that can serve as sensitive reporters of the structural and chemical environment around the site of substitution. In general, fluorine has several advantages as a NMR active nucleus (Rastinejad et al., 1995). Like the proton (1H), fluorine (19F) has a spin of ½ and is found in 100% natural abundance. Moreover, fluorine signals display a broader chemical shift range than protons (700 ppm for 19F vs. 15 ppm for 1H), as well as greater sensitivity to local changes in chemical environment and conformational dynamics. Due to its high gyromagnetic ratio and 100% natural abundance, fluorine nuclei can be detected directly using sensitive, one-dimensional (1D) NMR methods much in the same way 1D 1H spectra are acquired. In addition, two-dimensional (2D) heteronuclear Overhauser effect spectroscopy (1H,19F HOESY) experiments (Metzler, Leighton, & Lu, 1988), which are analogous to homonuclear Overhauser effect spectroscopy (1H,1H NOESY) experiments, can be readily measured to define the local (<5 Å radius) proton environment around a 19F nucleus.
Solid-phase phosphoramidite chemistry has been the predominant approach used to synthesize oligonucleotides that are selectively substituted with fluorinated nucleoside analogs (Carlow et al., 1996; Chu and Horowitz, 1991a,b; Graber, Moroder, & Micura, 2008; Hammann et al., 2001; Kwon et al., 2002; Liu et al., 1997; Olejniczak et al., 2002; Puffer et al., 2009; Rastinejad et al., 1995; Rastinejad and Lu, 1993; Scott, Geierstanger, Williamson, & Hennig, 2004). Among the different fluorine analogues, selective incorporation of the fluorinated pyrimidines 5-fluorouracil (5-FU) and 5-fluorocytidine (5-FC) has been the most widely used to enable 19F NMR studies of both RNA and DNA. When incorporated into an oligonucleotide strand that is then hybridized with its complement to form a helical duplex, these pyrimidine analogs place a 19F NMR reporter nucleus in the major groove of the Watson–Crick helical structure, making them sensitive to detecting interactions and conformational changes associated with this groove. Alternatively, enzyme-based synthetic schemes have been developed, which generate 19F-labeled ribonucleotide triphosphates that can be incorporated into RNA molecules using in vitro transcription with T7 RNA polymerase (Hennig, Scott, Sperling, Bermel, & Williamson, 2007; Scott et al., 2004). Using the approach, 2-fluoroadenosine-containing RNA oligonucleotides have been generated and shown by NMR to be relatively nonperturbing of helical structure and to provide a useful 19F NMR reporter nucleus in the minor groove of the helical structure.
2,4-Difluoro-5-methylbenzene (dF), a nonpolar aromatic nonhydrogen bonding isostere of thymine, is another example of a fluorinated pyrimidine analog that has been used to probe the role of hydrogen bonding and steric effects in oligonucleotides (Kool & Sintim, 2006). When incorporated into an oligonucleotide strand that is then hybridized with its complement to form a helical duplex, dF places 19F NMR reporter nuclei in both the major and minor grooves of the duplex structure (Fig. 1A), making it suitable for detecting interactions and conformational changes associated with both grooves. NMR studies have shown this analog is also generally nonperturbing to the helical geometry of a DNA duplex. Kool and coworkers showed that incorporating dF into an oligonucleotide that formed a 12-mer DNA duplex with its complement resulted in no significant observable changes in the structure of the duplex when compared to the unmodified control sample (Guckian, Krugh, & Kool, 1998). In this study, changes in the electronic environment adjacent to the site of substitution were found to cause an upfield shift of proximal imino protons between dF-substituted and control duplexes. Moreover, a slight destabilization of the duplex, observed by thermal melting, was observed and attributed to a lack of hydrogen bonding between dF and its base-pairing partner at the site of insertion. dF stacking within the helix, however, is stronger than that of thymine and therefore partially compensates for this lack of hydrogen bonding (Guckian et al., 1996).
Figure 1.
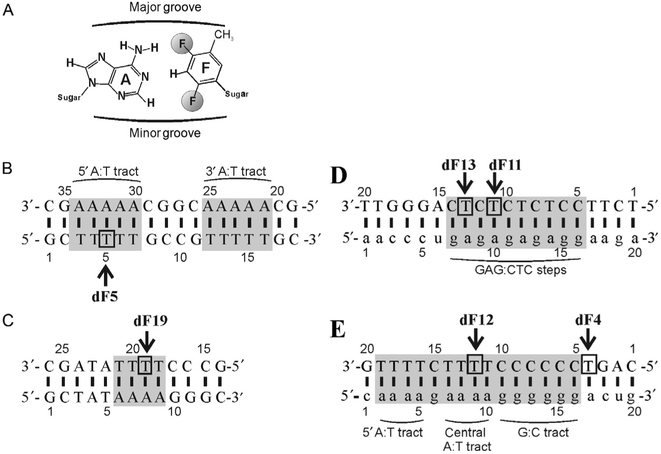
(A) dF structure and its positioning opposite an adenosine base, (B) A-tract and (C) TATA DNA duplex constructs, and the (D) Ty3 and (E) HIV-1 PPT-containing RNA/DNA constructs, respectively. DNA bases are in upper case and RNA bases in lower case. The numbering of bases is given from 5’ to 3’. The position of dF substitution is indicated by an arrow and a box. The r(GAG):d(CTC) steps in the Ty3 sequence, two (rA4):(dT4) tracts and the (rG6):(dC6) tract in the HIV-1 sequence, the two (dA5):(dT5) tracts in the A-tract sequence, and the (dA4):(dT4) tract in the TATA sequence are highlighted in gray.
Application of the dF labeling strategy for NMR studies is presented here as an tool for the analysis of homopyrimidine/homopurine-rich nucleic acid sequences: A-tract and TATA box DNA duplexes derived from Arabidopsis thaliana and two polypurine tract (PPT)-containing RNA:DNA hybrid duplexes derived from HIV-1 and the Saccharomyces cerevisiae long-terminal repeat (LTR)-retrotransposon Ty3. Using these constructs, we illustrate how dF insertion disperses overlapped NMR signals within highly redundant oligonucleotide sequences, thus facilitating proton resonance assignments of otherwise intractable spectra. We additionally show how NMR can be used to detect subtle perturbation in the conformation and dynamics of a duplex formed with a dF-substituted oligonucleotide, demonstrating how dF can also serve as a sensitive, in situ molecular probe (Rausch, Qu, Yi-Brunozzi, Kool, & Le Grice, 2003; Yi-Brunozzi et al., 2008). Lastly, it is shown how NMR observed changes in duplex conformation and dynamics can be further correlated, in the cases of the TATA box DNA, with modulation in the TATA box-binding protein interaction using an orthogonal gel-binding assay.
2. PREPARATION OF dF-SUBSTITUTED OLIGONUCLEOTIDES FOR NMR ANALYSIS
2.1. Design of Oligonucleotides
Strategic placement of dF within oligonucleotides can alleviate spectral crowding and facilitate assignment of 1H resonances. To illustrate such placement, we chose sequences that contained varying degrees of redundancy, including A-tract and TATA box DNA duplexes derived from A. thaliana (Kim & Burley, 1994) and two PPT-containing RNA:DNA hybrid duplexes derived from HIV-1 and S. cerevisiae LTR-retrotransposon Ty3 (Fig. 1). For the A-tract, TATA box, and HIV PPT constructs, dF was inserted at the second or third position of a 4- or 5-nucleotide dT tract to disrupt redundancy. In a second HIV PPT construct, dF was inserted at position 4 to attempt to relieve, in part, the signal overlap arising from the rG:dC tract. For the Ty3 sequences, two separate constructs with dF inserted at positions 11 and 13 were prepared to break the redundancy of the r(GAG):d(CTC) runs.
2.2. Reagents
5’-Dimethoxytrityl-(2,4-difluorotoluyl nucleoside), 3’-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (Glen Research Corp.).
1 × TBE (tris–borate–EDTA) buffer: 89 mM Tris base, 89 mM boric acid, 2 mM ethylenediaminetetraacetic acid (EDTA), pH 8.3. All components were purchased from Sigma-Aldrich.
1 × TBM buffer: 90 mM Tris base, 89 mM boric acid, 1 mM MgCl2 NMR buffer: 80 mM NaCl, 10 mM Na2HPO4/NaH2PO4, pH 7.0, in 90% H2O and 10% D2O. All components were purchased from Sigma-Aldrich.
2.3. Synthesis of dF-Substituted Oligonucleotides
dF-substituted and canonical DNA and RNA oligonucleotides were synthesized at a 1 μmol scale using standard phosphoramidite chemistry on a PE Biosystems Expedite 8909 nucleic acid synthesizer. No changes were needed for the synthesis of dF-substituted oligonucleotides from the standard protocols supplied by the instrument manufacturer.
2.3.1. Purification of dF-Substituted Oligonucleotides
The individual strands were purified using preparative denaturing polyacryl-amide gel electrophoresis (PAGE) using 20% acrylamide at 19:1 acrylamide: bisacrylamide. PAGE was performed at 30 W and approximately 50 °C in 1 × TBE buffer. The desired oligonucleotide product was visualized with ultraviolet (UV) irradiation and cut from the gel. The samples were then electroeluted from the gel strips at 200 V with ½× TBE buffer using an Elutrap device (Whatman Scientific). Fractions from the Elutrap were collected every hour for 6 h and analyzed via UV-vis spectroscopy to confirm the presence of oligonucleotide. All fractions containing sample were pooled.
2.3.2. Sample Conditioning for NMR Spectroscopy
Electroeluted nucleic acid was concentrated with a 3 kDa centrifugal filter and then buffer exchanged in the same filter sequentially with water (twice), 1 M NaCl (once), water (thrice), and the final NMR buffer (thrice) to a volume of 250 μl. Washing with 1 M NaCl is especially important with RNA to displace residual Tris buffer, which is found to bind to some RNA molecules. Each single-stranded oligonucleotide was microdialyzed against a 3 kDa membrane to assure full buffer exchange. Following concentration and buffer exchange, the single-stranded oligonucleotides were quantified spectrophotometrically at 260 nm, assuming a molar extinction coefficient equal to the sum of the ribo- and/or deoxyribonucleotides using the nearest-neighbor two-state model. DNA and RNA/DNA hybrid duplexes were formed by mixing equimolar amounts of the complementary strands, heating the solution to 90 °C for 3 min, and then slow cooling to room temperature. Homogeneous duplex formation was visualized and stoichiometry further confirmed using native PAGE (15% acrylamide at 19:1 acrylamide: bisacrylamide in 1 × TBM buffer run at 40 W and 4 °C). Final duplex concentrations were typically 1.0 mM in 275 μl and 90% H2O, 10% D2O in NMR buffer. All samples were loaded into a 5-mm symmetrical, susceptibility-matched NMR microtube (Shigemi, Inc.) for NMR experiments.
2.3.3. Exchange of Samples into 99.96% D2O
The following procedure was employed for NMR experiments requiring that the sample be in 99.96% D2O. The plunger was removed from the Shigemi tube and set aside. The top of the tube was plugged with parafilm to prevent accidental expulsion of sample during the freeze-drying process. Crushed dry ice was used to freeze the upper portion of the tube above the sample solution. The tube was then removed from the dry ice and gently shaken to freeze the sample along its walls. This process was performed iteratively until the entire sample was frozen along the walls of the NMR tube. The RNA sample was subsequently lyophilized, reconstituted in 99.96% D2O, frozen a second time using the same procedure, and lyophilized again. After the second lyophilization, the sample was once again reconstituted in 99.96% D2O and used for NMR experiments.
3. NMR METHODS
3.1. Proton NMR Data Acquired in 90%:10% H2O:D2O
NMR experiments were collected on a 600 MHz Bruker AVANCE I NMR spectrometer equipped with a room temperature, triple resonance (i.e., 1H, 13C, and 15N) TXI probe.
Initially, a standard 1D 1H experiment was acquired using a water flip-back Watergate for water suppression. The imino 1H region, 10.0–15.0 ppm, was inspected to assure that the duplex was properly formed. Duplex formation is ascertained from qualitatively visualizing peak shape (30±10 Hz imino linewidth) and counting the appropriate number of peaks. Imino resonances from terminal base pairs are known to exchange rapidly with solvent and are usually not observed. For duplex oligonucleo-tides, most other imino resonances from the Watson–Crick pairs should be observed.
A 2D nuclear Overhauser effect spectroscopy (NOESY) experiment also employing a water flip-back Watergate for solvent suppression was then collected at typically two different mixing times (e.g., 100 and 300 ms). This standard experiment allows the user to map sequential correlations of imino resonances along the length of the sequences. This assignment process has been reviewed extensively (Flinders & Dieckmann, 2006).
3.2. Proton and Fluorine NMR Data Acquired in 99.96% D2O
After exchange of the NMR sample into D2O, another 2D NOESY, typically at a mixing time of 250 ms, and a double quantum filtered correlation spectroscopy (DQF-COSY) experiments were collected for analysis of non-exchangeable proton resonances from the base and sugars that comprise each strand of the duplex. This analysis has been reviewed extensively (Flinders and Dieckmann, 2006).
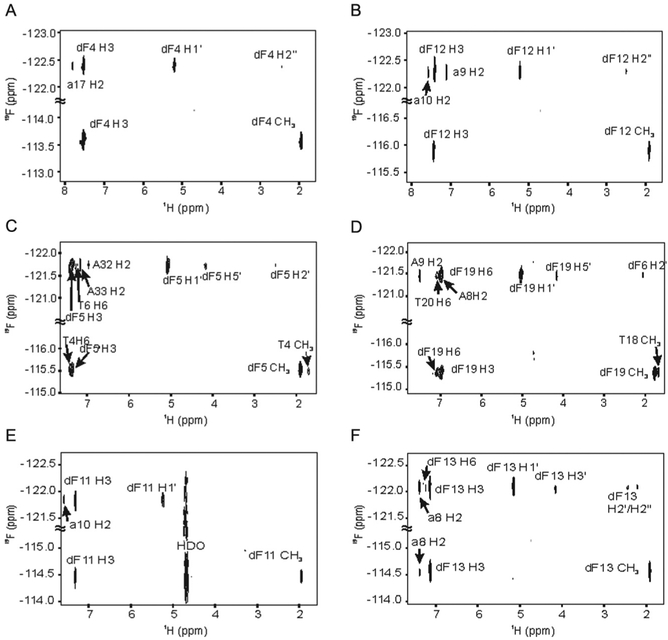
19F,1H-heteronuclear single-quantum coherence and 19F,1H-heteronuclear Overhauser effect spectroscopy (HOESY, τmix 300 ms) experiments were acquired using a dual resonance (1H,19F) HF probe. The HOESY experiment demonstrated proper helical stacking of the dF base based on the observed HF NOE patterns (Fig. 2). Specifically, the presence of cross-peaks to neighboring thymidines and cross-strand adenosines indicates that the dF-modified base is properly base stacked within the helix.
Figure 2.
1H-19F NOESY spectra of the (A) HIV dF4 duplex; (B) HIV dF12 duplex; (C) A-tract dF5 duplex; (D) TATA box dF19 duplex; (E) Ty3 dF11 duplex; (F) Ty3 dF13 duplex, all in 99.96% D2O at 30 °C. The proton carrier frequency was set to 4.708 ppm and the fluorine carrier frequency to −118 ppm. Sweep widths were set to 6009 Hz for 1H and 5646 Hz for 19F, 128 complex points in t1, 4 K complex points in t2, and 480 scans per increment.
4. dF AS AN NMR ASSIGNMENT TOOL
The DNA duplex and RNA/DNA hybrids that instigated the development of this approach exhibit severe resonance overlap that arises from sequence redundancy, making complete spectral analysis a formidable challenge. As will be shown, dF can dramatically perturb the chemical shifts of proton resonances proximal to the site of substitution and thus significantly reduce spectral overlap. Since the electronic environment of distal resonances are unaffected by dF substitution, the chemical shifts of these resonances remain in the same position as the unmodified sequences. The reduction in spectral overlap, combined with the close matching of distal resonance signals between the modified and unmodified sequences, facilitates the assignment of duplexes formed by dF-substituted oligonucleotides as well as mapping of resonance assignments back to the unmodified sequence.
4.1. Illustration of the 1H Imino Resonance Assignment Strategy Using dF Substitution
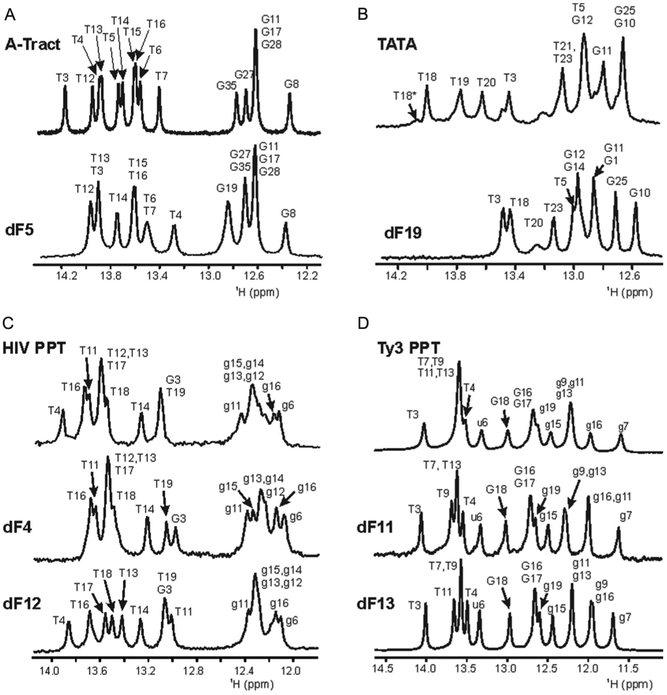
Representative 1D imino 1H spectra, the “fingerprint” region of nucleic acid duplexes, are shown in Fig. 3. While the dispersion resulting from dF substitution of imino resonances from the TATA box and A-tract duplexes was negligible, the effect of dF substitution is clearly illustrated for the imino spectra of dF-modified Ty3 and HIV PPT duplexes. Four imino resonances of the Ty3 construct were unresolved in the wild-type sequence: T7, T9, T11, and T13. Both the Ty3 dF11 and dF13 sequences dispersed the PPT duplex signals, allowing assignment of resonances distal from the site of substitution. In a similar manner, the imino spectra of the HIV wild-type sequence exhibit resonance overlap in the thymine region (~13.5 ppm) and in the G:C tract (~12.3 ppm). Again, both the HIV dF12 and dF4 substitutions dispersed resonances and facilitated assignments of imino resonances associated with the T-tracts and G:C tract. For the HIV dF12 substitution, chemical shifts of the central rA:dT tract were significantly shifted to allow assignment of the thymine resonances distal from the central rA:dT tract, which remained in approximately the same position as found in the unmodified sample. Using a similar strategy, the HIV dF4 substitution yielded sufficient resolution in the G:C tract to facilitate assignment of all rG imino resonances.
Figure 3.
Comparison of the imino regions of the 1D water flip-back Watergate 1H NMR spectra of (A) A-tract DNA; (B) TATA box DNA; (C) HIV PPT; and (D) Ty3 PPT duplexes, all at 10 °C. In all panels, the unmodified duplex is the top spectrum, and the position of dF substitution in the modified duplex is indicated in the bottom spectra. Imino proton assignments are indicated. RNA is lower case and DNA upper case. Panels (C) and (D) are modified from published data Yi-Brunozzi et al. (2008).
4.2. Illustration of the 1H Nonexchangeable Resonance Assignment Strategy Using dF Substitution
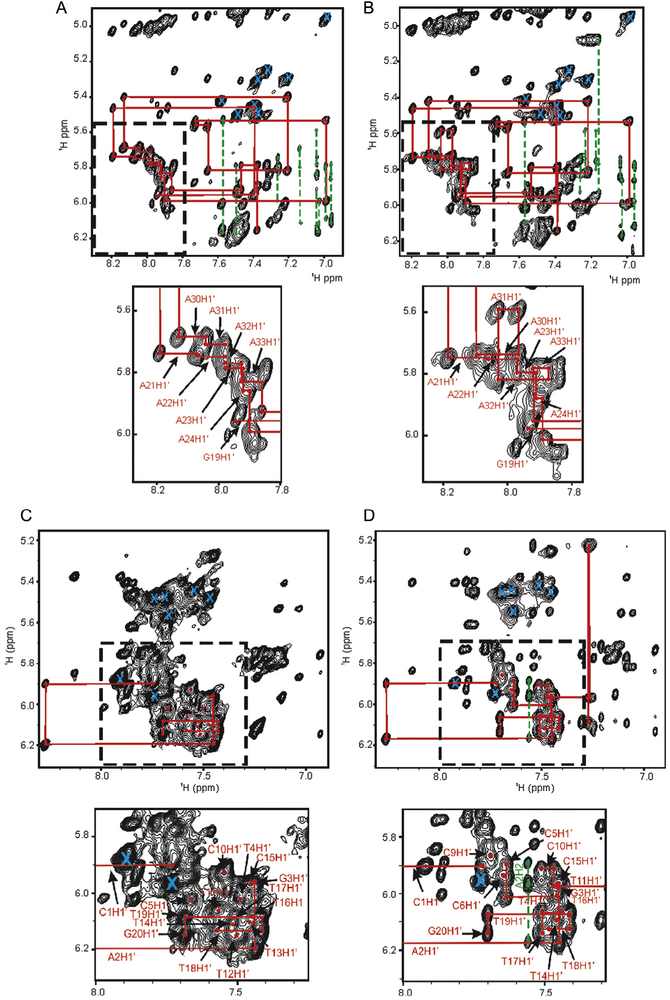
The process of assigning backbone resonances through inter/intramolecular NOEs, particularly aromatic (H6/H8/H2) and anomeric (H1’)/H5 correlations, can also be especially challenging with high sequence redundancy. The region of the 2D NOESY spectra containing these correlations is approximately from 6.8 to 8.3 ppm (H8/H6/H2) in 1D and 4.8 to 6.3 ppm (H1’/H5) in the second (Fig. 4). In order to obtain detailed information for the structure and dynamics of an oligonucleotide, as complete an assignment of these correlations in this region as is possible is desired. As was observed for the imino spectra, the utility of dF as a tool for NMR resonance assignment is clearly evident by its dispersion of overlapped signals in a 2D NOESY experiment on the dF5-substituted A-tract (Fig. 4A and B). This spectrum shows slight shifting of the H8–H1’cross-peaks of residues A31–A35. The inset clearly shows subtle shifting of dF resonances proximal its site of substitution, facilitating assignment of the analogous cross-peaks in the distal A-tract associated with residues A21–A25.
Figure 4.
Expansion of the anomeric (H1’) to aromatic region of a NOESY (τNOE =250 ms) experiment applied to a 1.0 mM sample of the (A) unmodified A-tract DNA duplex, (B) A-tract dF5 DNA duplex, (C) unmodified HIV, and (D) HIV dF12 duplex. The local dispersion of peaks due to dF substitution is emphasized by the expansions in the dotted boxes below each spectrum. The sequential assignment “NOE walk” for residues G19 through C36 is shown in red lines for the A-tract and C1 though G20 for the HIV spectra. For clarity, assignment labels were removed from the full spectrum but retained in the expansions. Dashed green lines are used to annotate correlations of adenosine H2 protons with 5’ sequential and 3’ cross-strand anomeric (H1’) protons. An “X” in cyan indicates a cytosine H5–H6 correlation.
The unmodified HIV PPT provides an even clearer example of a severely overlapped NOESY spectrum which is not possible to assign using standard proton-based approaches for resonance assignment. In addition, preparing RNA through in vitro transcription was problematic due to the run of six guanosines, so facile stable isotopic labeling with 13C or 15N was not practical. Through comparative analysis of the unmodified, dF12 and dF4 NOESY data, complete assignment could be made for all H8, H6, H2, H1’, and H5 resonances (Fig. 4C and D). In this case, the dF12 substitution resulted in dispersion of signals from the central A:T tract which facilitated assignments in the 5’ A:T tract, while dF4 substitution afforded resolution that facilitated assignments of resonances in the G:C tract.
4.3. Quantifying Chemical Shift Perturbations in the dF-Modified Constructs
4.3.1. Computation of Chemical Shift Differences
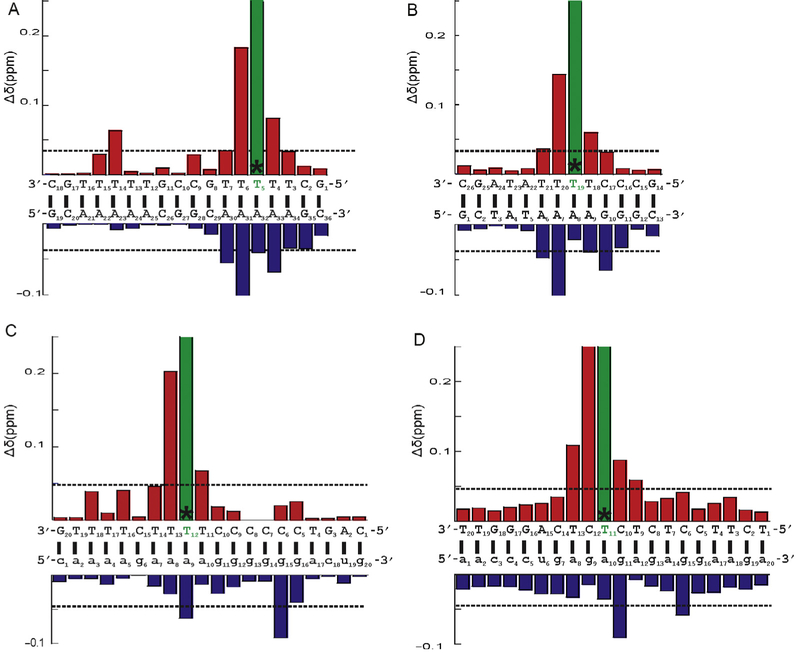
From the assignments of each unmodified and dF-substituted A-tract and TATA box DNA, HIV-1 and Ty3 RNA:DNA PPT hybrids (Yi-Brunozzi et al., 2008), a chemical shift difference histogram was computed to visualize the range in magnitude of the perturbations resulting from dF substitution for the anomeric to aromatic region (Fig. 5). The deviation of chemical shifts of all aromatic protons and H1’ were normalized for each nucleotide, and each nucleotide in a dF-substituted duplex was compared with the corresponding residue in the unmodified duplex. A similar plot could be created for any other region (or for a combined plot of all or other resonances).
Figure 5.
Proton chemical shift changes associated with (A) T5→dF substitution of the A-tract DNA duplex, (B) T19→dF substitution of the TATA box DNA duplex, (C) T12→dF substitution of the HIV-1 PPT, and (D) T11→dF substitution of the Ty3 PPT. DNA and RNA sequences of each hybrid are shown in upper case and lower case letters, respectively. Positions of dT→dF substitution are highlighted in green and an asterisk “*.” Each bar represents the average difference in normalized ppm values obtained for equivalent nucleotides within dF-substituted and unmodified duplexes. All aromatic and H1’ resonances were included in the average. The dotted gray line designates the threshold of a significant chemical shift difference. Chemical shift changes (ppm) for dF-substituted DNA strands are shown above the sequence schematics (red bars), while unmodified strands are depicted below (blue bars).
4.3.2. Visualization of Chemical Shift Differences for All Constructs
Introduction of dF clearly results in all cases in a significant local alteration of chemical shift for the dF base protons, while proximal bases were perturbed by approximately 0.2 ppm or less. These bases included T4, T6, and A30–33 for the A-tract DNA duplex; A6, A7, G10, T18, and T20 for the TATA DNA duplex; T11, T13, a9, and g15 for HIV; and T9, C10, C12, T13, g11, and g15 for Ty3. Most other resonances of nucleotides distal to the site of substitution were shifted in the spectrum from the unmodified positions by an insignificant average of approximately 0.05 ppm or less. These histograms confirm and quantify what is observed visually in the spectra; that is, perturbations resulting from dF substitution are primarily local and assignment of signals from nucleotides distal from the site of isostere substitution can readily be mapped onto the unmodified sequence.
5. STRUCTURAL AND DYNAMIC PERTURBATIONS OBSERVED BY dF SUBSTITUTION
While these examples highlight the utility of single-site dF substitutions as a means of alleviating spectral overlap for the purpose of assigning resonances, it was additionally observed that dF modifications in these sequences can function as a subtle structural probe of conformational dynamics. In these instances, a subtle change in structure afforded by the dF substitution appears to select for a sampling of different conformational states with different exchange rates that can be detected on the NMR time scale.
5.1. Altered Sugar Pucker Conformations in the PPT dF Series
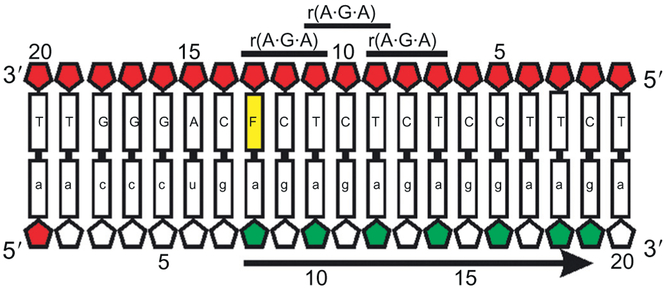
The unmodified Ty3 PPT construct was observed to have predominantly C3’-endo sugar puckers on the RNA strand, which is the conformation found in an A-form RNA helix (Yi-Brunozzi et al., 2008). In a C3’-endo sugar pucker, the torsional angle between the H1’ and H2’ protons is such that the three bond coupling constant (3JHH) is less than 1 Hz, and no cross-peak is observed between the H1’ and H2’ protons in a DQF-COSY spectrum. However, if the conformational equilibrium of the sugar shifts to a mixture of C3’- and C2’-endo puckers, the 3JHH coupling constant between H1’ and H2’ becomes measureable, and a cross-peak in the DQF-COSY becomes visible. For the Ty3 dF13 and dF11 samples, the dF substitution on the DNA strand resulted in a shift to a mixed sugar pucker conformation on the RNA strand that propagated to every other base down the length of the PPT to residue g16 (Fig. 6). This observed propagation of altered sugar puckering along the RNA strand was attributed to stacking of repeated 5’-r(A·G·A)-3’ symmetric steps in the TY3 sequence (Yi-Brunozzi et al., 2008). To confirm this was a phenomenon specific to the helical stacking in the Ty3 PPT sequence, a C10T substitution was made and shown to also produce a similar altering of sugar pucker that propagated down the RNA strand of the PPT sequence.
Figure 6.
A schematic representation of the Ty3 dF13 PPT hybrid sequence. dF13 substitution is in yellow. Wild-type ribose sugars in mixed C3’/C2’-endo conformation are shown in red. The symmetric 5’-r(A·G·A)-3’ steps are labeled. Highlighted collectively in green are ribose sugars for which a C3’-endo to C3’/C2’-endo sugar pucker switch is observed when the PPT hybrids, and arrow indicates the propagation of the sugar pucker switch to the 3’ end of the sequence. Figure was modified from published data Yi-Brunozzi et al. (2008).
In contrast to the TY3 PPT, the HIV PPT dF construct series showed negligible long-range effects on PPT conformation and dynamics. This suggested that this RNA/DNA hybrid PPT may not exhibit the same structural coupling due to an absence of repeated symmetric base stacking step that serve in the Ty3 system to propagate structural perturbation down the length of the PPT sequence.
5.2. Changes in the Dynamic Equilibrium of the TATA Box DNA Duplex
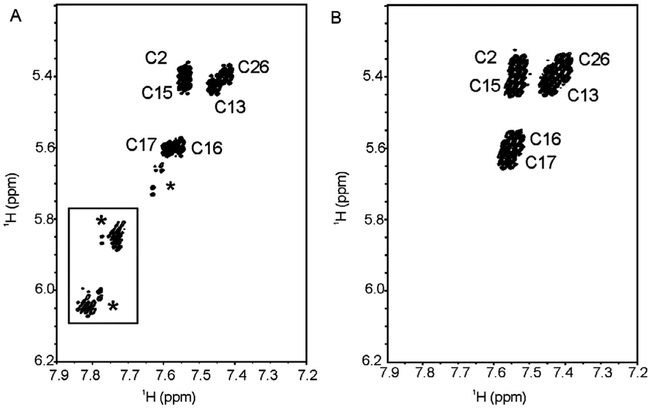
Changes in the spectral heterogeneity and line widths of the TATA DNA samples further demonstrates that dF can introduce subtle changes in the conformational dynamics of a duplex. The unmodified TATA box duplex, in general, displayed greater conformational heterogeneity (e.g., additional imino proton signals from minor species) than the dF19-substituted duplex. Indeed, the heterogeneity observed in the TATA box duplex 1D imino proton spectra was previously observed for imino resonance T18 but not discussed (Fig. 3B; Chen & Russu, 2004). An effect of dF substitution on the conformational dynamics of the TATA box duplexes is further illustrated in the 2D DQF-COSY. In this experiment, unassigned H6–H5 resonance cross-peaks from cytosines from minor conformation(s) are clearly evident for the unmodified sequence and absent in the dF19-substituted duplex (Fig. 7, boxed region). Analogous cross-peaks can also be observed in the 2D NOESY spectrum of the unmodified duplex (data not shown). In total, the NMR data suggest that dF substitution biases the conformational equilibrium of the TATA box DNA duplex to a subset of conformational states sampled by the unmodified duplex. As the TATA DNA duplex is well known to be malleable and unwound upon binding to the TATA-binding protein (TBP) (Davis, Majee, & Kahn, 1999; Juo et al., 1996; Kim and Burley, 1994), we surmised that the NMR observed effect of dF substitution on the TATA box DNA duplex might have an impact on the formation of the TBP–TATA complex. The next section (5.3) describes gel-based methods that were used to explore the differential binding of the two TATA substrates to TBP.
Figure 7.
Expansion of the H6–H5 region of a DQF-COSY of the (A) unmodified and (B) dF19-containing TATA duplexes collected in 99.96% D2O at 30 °C. An asterisk indicates an unassigned peak. The boxed region highlights cross-peaks of interest that are further discussed in the text. For clarity, positive and negative contours are black.
5.3. An Orthogonal Gel Assay Correlating dF NMR Results from TATA Box DNA with TATA Box Protein Binding
The dF-induced changes in the DNA conformational ensembles, as observed in the NMR experiments can be predictors of altered or impaired function. To test these predictions, an orthogonal functional assay can be performed using the modified and unmodified sequences to make a direct comparison. For the TATA box DNA duplex example, an electrophoretic mobility shift assay (EMSA) for binding to the TATA box-binding protein (TBP) is described that was used to compare the behavior of the unmodified and dF19-modified TATA box DNA duplexes.
5.3.1. TBP-Binding Assay
Unmodified and dF-substituted DNA oligonucleotides were 5’-end labeled using T4 polynucleotide kinase and γ–32P-ATP, purified by denaturing PAGE, and eluted overnight in 0.3 M sodium acetate, pH 5.2, 1.0 mM EDTA at 37 °C. DNA was recovered by ethanol precipitation and reconstituted in water. Labeled oligonucleotides were mixed with equimolar amounts of their unlabeled complements at 0.4 μM final concentration in 10 mM Tris, pH 7.4, 25 mM KCl and annealed in a thermal cycler by heating to 85 °C for 3 min followed by slow cooling to 4 °C at 0.1 °C/s. TBP was purified essentially as previously reported (Petri, Hsieh, & Brenowitz, 1995), and the active fraction, determined by EMSA at saturating DNA, was 13%. TBP was serially diluted in 25 mM HEPES (pH 8.0), 0.3 M KCl, 20% glycerol, 1 mM EDTA, and 1 mM DTT. Final active TBP concentrations in the binding reactions were 180, 91.8, 46.9, 23.5, 5.9, 2.9, 1.5, and 0.73 nM. Binding reactions contained 4 nM substrate, 50 mM HEPES (pH 8.0), 200 mM potassium glutamate, 1 mM CaCl2, 2.5 Mm DTT, 10 mM MgOAc, 100 μg/ml gelatin, 50 μg/ml BSA, 0.05% NP-40 (Igepal), and 10% glycerol. Negative controls contained TBP dilution buffer. Reactions were assembled at 4 °C, then placed on ice, and incubated a further 5 min. One-tenth volume of 50% glycerol containing 0.05% (w/v) bromophenol blue and xylene cyanol was added, and samples were loaded onto a 15% acrylamide gel (75:1 acrylamide:bis; prerun at 4 °C for 15 min at 230 V) containing 0.045 M Tris base, 0.045 M boric acid (pH 8.3), 1 mM Na2EDTA, 2 mM DTT, 3 mM MgCl2, and 1% glycerol. Electrophoresis was performed at 4 °C at 350 V for 3 h. The gel was dried before exposure to a phosphor screen. Screens were scanned with a Typhoon Trio Plus variable mode imager and bands quantified by ImageQuant Total Lab software (GE Healthcare).
5.3.2. Binding of TBP to Unmodified and dF-Substituted TATA Box DNA Duplexes
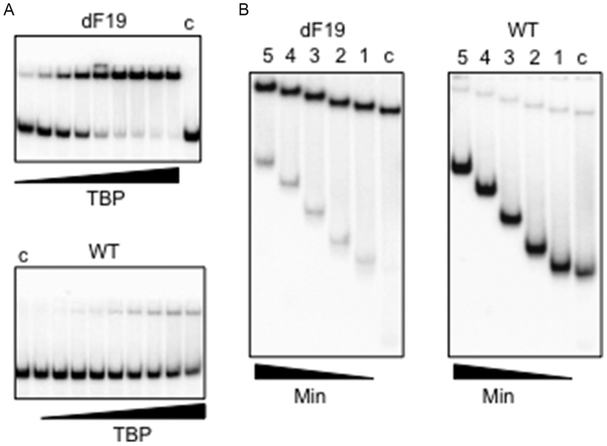
As shown in Fig. 8A, the dF-substituted duplex supported significantly more shifted product than the unmodified sequence overall TBP concentrations tested. At the highest protein:DNA concentration, 100% of the dF-substituted TATA duplex was bound, compared to only 18% with unmodified DNA. The apparent increased affinity and lifetime of protein–DNA complex formation for the dF19-substituted TATA duplex suggests an increased helical malleability with respect to the unmodified sequence that likely results from changes in the stacking geometry and stability at the site of dF substitution. In contrast, since the A-T pairs in the TATA box DNA complex remain hydrogen bonded, the lack of H-bonding for dF seems unlikely to stabilize the protein–DNA complex.
Figure 8.
TBP binding to unmodified and dF-substituted TATA box DNA duplexes. (A) Nondenaturing polyacrylamide gels showing unmodified and dF19-substituted duplexes (4 nM) binding to TBP, varying in twofold increments (left to right) from 0.73 to 180 nM, Lanes c: no protein. (B) Competition assays of TBP complexes with unmodified and dF-substituted TATA box DNA duplex. Lanes from right to left for each substrate: Lanes c, no competitor; Lane 1, 10 min; Lane 2, 30 min; Lane 3, 60 min; Lane 4, 90 min; Lane 5, 120 min. Both the disappearance of bound DNA and appearance of free DNA are noticeably slower for the dF-substituted substrate.
5.3.3. Relative Thermodynamics and Kinetics of TBP:DNA Complexes
If the enhanced affinity reflects preorganization of the dF-substituted TATA duplex, this would create a lower-barrier pathway for the normally slow formation of the TBP–DNA complex (Hoopes, LeBlanc, & Hawley, 1992). dF substitution might then be expected to increase the on-rate, but this is difficult to measure directly, and affinity changes are more frequently reflected in the off-rate, as suggested by the initial binding assay results. To further address the basis for the enhanced thermodynamic stability of the TBP complex with dF19-substituted TATA duplexes, one can instead carry out a competition experiment that measures the stability of the complex in solution as a function of time in the presence of excess competitor DNA. In each experiment, the labeled double-stranded DNA substrate (4 nM final) was incubated with TBP (92 nM active concentration) for 15 min on ice to form the complex prior to addition of a 50-fold excess of cognate cold competitor (Fig. 8B). Binding reactions were performed under the same buffer conditions and analyzed via PAGE as described above. One sample was loaded on each gel (lane “c”) before adding competitor. After addition of competitor, aliquots were removed at 10, 30, 60, 90, and 120 min. One-tenth volume of 50% glycerol containing 0.05% (w/v) bromophenol blue and xylene cyanol was added, and samples were loaded onto the running gel. The gel was dried, exposed to a phosphor screen, and analyzed as above. Based on a qualitative comparison of the rates of appearance of free DNA and disappearance of bound DNA for the two TBP–DNA complexes, the off-rate of the complex formed with the dF-modified duplex was found to be significantly slower than the off-rate of the unmodified duplex. However, the difference in solution off-rates alone does not seem to explain the dramatic difference in gel stability. The molecular determinants of stability in the nonequilibrium EMSA are not well understood, but presumably they include responses to ionic strength, mechanical interactions with the gel matrix, and differential effects on on-rates versus off-rates. If the on-rate for the TBP–dF oligonucleotide is fast, reassociation in the gel matrix cage may be the main source of stabilization.
The observed slower rate of dissociation, the implied increased rate of association, and the enhanced affinity observed between the dF-modified construct and the TBP supports the idea that the DNA may be preorganized to bind TBP and that affinity depends on the energetic cost of the DNA deformation needed for TBP complex formation, in accord with earlier work (Arndt, Wobbe, Ricupero-Hovasse, Struhl, & Winston, 1994; Davis et al., 1999; Strahs, Barash, Qian, & Schlick, 2003). The dF substitution also appears to affect less well-understood mechanisms that control kinetic stability in the EMSA.
6. CONCLUSIONS
In this chapter, single-site dF substitutions in A-tact and TATA box DNA and two RNA/DNA PPT-containing hybrid duplexes were used to demonstrate the utility of this base analog as a tool for spectral assignment and as a probe of structure/dynamics in highly redundant, homopurine-rich DNA sequences. dF substitution offers an alternative and cost-effective means of assigning spectra that are otherwise intractable. In all cases, insertion within pyrimidine tracts results in local perturbation of resonances of adjacent base pairs due to the proximity of the electronegative 19F but leaves the chemical shift of all other nucleotides relatively unaffected. dF substitution significantly facilitated assignment of proton resonances, particularly in the imino, aromatic, and H1’ regions of the spectrum. In addition, no significant change in the relaxation behavior of resonances associated with residues more than one base pair from the site of dF insertion nor deterioration of spectral quality was observed, as is often seen with other labeling (e.g., 13C) approaches. Taken together, data from the DNA duplexes demonstrate that dF can be used as a tool to facilitate proton resonance assignments and could be easily extended to other pyrimidine/purine-rich sequences found in regulatory DNAs of other biological systems.
In addition to its use as a tool for assignment of 1H NMR signals, incorporation of dF into an oligonucleotide may be exploited as a subtle structural probe. dF substitutions of the DNA strand of the Ty3 PPT constructs induced long-range sugar pucker switches in the RNA complement, suggesting an inherent PPT flexibility that may induce recognition by the cognate reverse transcriptase. We additionally used an orthogonal assay of TBP–TATA box DNA binding to demonstrate that the altered conformational dynamics observed by NMR had a measurable impact on the kinetics and thermodynamics of the protein–DNA interaction. Indeed, it has become apparent that while dF introduces minimal changes in ground-state structure, it does decrease the overall thermal stability of the modified duplex (Kool and Sintim, 2006). In addition, in sequences like TATA or HIV-1, which adopt alternative low energy structures, the lack of H-bonding or changes in the stacking interactions at the site of dF incorporation appears to lower the energetic barriers to interconversion between structures or to “relax” the structure to one dominant conformation.
In conclusion, our results from applications to purine-rich sequence elements show that single-site substitution of dF: (1) facilitates assignment of otherwise intractable NMR spectra of highly redundant, A:T tract nucleic acid duplexes and (2) serves as a probe of subtle differences in the structure and dynamics of nucleic acid duplexes.
ACKNOWLEDGMENTS
This work was supported in part by the NIH (GM 59107) to J.P.M. S.F.J.L.G. is supported by the intramural research program of the National Cancer Institute, National Institutes of Health. NMR instrumentation at IBBR was purchased in part with support from the W.M. Keck Foundation, the NIH/NCRR and NIST. The authors additionally acknowledge support of the Hollings Marine Laboratory NMR facility.
Note: Certain commercial equipment, instruments, and materials are identified in this paper in order to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the material or equipment identified is necessarily the best available for the purpose.
REFERENCES
- Arndt KM, Wobbe CR, Ricupero-Hovasse S, Struhl K, & Winston F (1994). Equivalent mutations in the two repeats of yeast TATA-binding protein confer distinct TATA recognition specificities. Molecular and Cellular Biology, 14, 3719–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlow DC, Short SA, & Wolfenden R (1996). Role of glutamate-104 in generating a transition state analogue inhibitor at the active site of cytidine deaminase. Biochemistry, 35, 948–954. [DOI] [PubMed] [Google Scholar]
- Chen C, & Russu IM (2004). Sequence-dependence of the energetics of opening of at basepairs in DNA. Biophysical Journal, 87, 2545–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WC, & Horowitz J (1991a). Fluorine-19 NMR studies of the thermal unfolding of 5-fluorouracil-substituted Escherichia coli valine transfer RNA. FEBS Letters, 295, 159–162. [DOI] [PubMed] [Google Scholar]
- Chu WC, & Horowitz J (1991b). Recognition of Escherichia coli valine transfer RNA by its cognate synthetase: A fluorine-19 NMR study. Biochemistry, 30, 1655–1663. [DOI] [PubMed] [Google Scholar]
- Davis NA, Majee SS, & Kahn JD (1999). TATA box DNA deformation with and without the TATA box-binding protein. Journal of Molecular Biology, 291, 249–265. [DOI] [PubMed] [Google Scholar]
- Flinders J, & Dieckmann T (2006). NMR spectroscopy of ribonucleic acids. Progress in Nuclear Magnetic Resonance Spectroscopy, 48, 137–159. [Google Scholar]
- Graber D, Moroder H, & Micura R (2008). 19F NMR spectroscopy for the analysis of RNA secondary structure populations. Journal of the American Chemical Society, 130, 17230–17231. [DOI] [PubMed] [Google Scholar]
- Guckian KM, Krugh TR, & Kool ET (1998). Solution structure of a DNA duplex containing a replicable difluorotoluene-adenine pair. Nature Structural Biology, 5, 954–959. [DOI] [PubMed] [Google Scholar]
- Guckian KM, Schweitzer BA, Ren RX, Sheils CJ, Paris PL, Tahmassebi DC, et al. (1996). Experimental measurement of aromatic stacking affinities in the context of duplex DNA. Journal of the American Chemical Society, 118, 8182–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammann C, Norman DG, & Lilley DM (2001). Dissection of the ion-induced folding of the hammerhead ribozyme using 19F NMR. Proceedings of the National Academy of Sciences of the United States of America, 98, 5503–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig M, Scott LG, Sperling E, Bermel W, & Williamson JR (2007). Synthesis of 5-fluoropyrimidine nucleotides as sensitive NMR probes of RNA structure. Journal of the American Chemical Society, 129, 14911–14921. [DOI] [PubMed] [Google Scholar]
- Hoopes BC, LeBlanc JF, & Hawley DK (1992). Kinetic analysis of yeast TFIIDTATA box complex formation suggests a multi-step pathway. The Journal of Biological Chemistry, 267, 11539–11547. [PubMed] [Google Scholar]
- Juo ZS, Chiu TK, Leiberman PM, Baikalov I, Berk AJ, & Dickerson RE (1996). How proteins recognize the TATA box. Journal of Molecular Biology, 261, 239–254. [DOI] [PubMed] [Google Scholar]
- Kim JL, & Burley SK (1994). 1.9 A resolution refined structure of TBP recognizing the minor groove of TATAAAAG. Nature Structural Biology, 1, 638–653. [DOI] [PubMed] [Google Scholar]
- Kool ET, & Sintim HO (2006). The difluorotoluene debate—A decade later. Chemical Communications, 35, 3665–3675. [DOI] [PubMed] [Google Scholar]
- Kwon K, Jiang YL, Song F, & Stivers JT (2002). 19F NMR studies of vaccinia type IB topoisomerase. Conformational dynamics of the bound DNA substrate. The Journal of Biological Chemistry, 277, 353–358. [DOI] [PubMed] [Google Scholar]
- Liu M, Chu WC, Liu JC, & Horowitz J (1997). Role of acceptor stem conformation in tRNAVal recognition by its cognate synthetase. Nucleic Acids Research, 25, 4883–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler WJ, Leighton P, & Lu P (1988). Two-dimensional heteronuclear NOE spectroscopy; application to 19F-labeled oligodeoxyribonucleotides. Journal of Magnetic Resonance, 76, 534–539. [Google Scholar]
- Olejniczak M, Gdaniec Z, Fischer A, Grabarkiewicz T, Bielecki L, & Adamiak RW (2002). The bulge region of HIV-1 TAR RNA binds metal ions in solution. Nucleic Acids Research, 30, 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri V, Hsieh M, & Brenowitz M (1995). Thermodynamic and kinetic characterization of the binding of the TATA binding protein to the adenovirus E4 promoter. Biochemistry, 34, 9977–9984. [DOI] [PubMed] [Google Scholar]
- Puffer B, Kreutz C, Rieder U, Ebert MO, Konrat R, & Micura R (2009). 5-Fluoro pyrimidines: Labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Research, 37, 7728–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F, Evilia C, & Lu P (1995). Studies of nucleic acids and their protein interactions by 19F NMR. Methods in Enzymology, 261, 560–575. [DOI] [PubMed] [Google Scholar]
- Rastinejad F, & Lu P (1993). Bacteriophage T7 RNA polymerase. 19F-nuclear magnetic resonance observations at 5-fluorouracil-substituted promoter DNA and RNA transcript. Journal of Molecular Biology, 232, 105–122. [DOI] [PubMed] [Google Scholar]
- Rausch JW, Qu J, Yi-Brunozzi HY, Kool ET, & Le Grice SF (2003). Hydrolysis of RNA/DNA hybrids containing nonpolar pyrimidine isosteres defines regions essential for HIV type 1 polypurine tract selection. Proceedings of the National Academy of Sciences of the United States of America, 100, 11279–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LG, Geierstanger BH, Williamson JR, & Hennig M (2004). Enzymatic synthesis and 19F NMR studies of 2-fluoroadenine-substituted RNA. Journal of the American Chemical Society, 126, 11776–11777. [DOI] [PubMed] [Google Scholar]
- Strahs D, Barash D, Qian X, & Schlick T (2003). Sequence-dependent solution structure and motions of 13 TATA/TBP (TATA-box binding protein) complexes. Biopolymers, 69, 216–243. [DOI] [PubMed] [Google Scholar]
- Yi-Brunozzi HY, Brinson RG, Brabazon DM, Lener D, Le Grice SF, & Marino JP (2008). High-resolution NMR analysis of the conformations of native and base analog substituted retroviral and LTR-retrotransposon PPT primers. Chemistry & Biology, 15, 254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]