This article discusses genomic copy number variations (CNV) with clinical importance in the setting anaplastic oligodendroglioma. Analysis was based on a grade III glioma patient cohort established by the French POLA network, which allowed for the identification of correlations between CNV events with clinical‐pathological variables and prognosis.
Keywords: Glioma, Oligodendroglioma, Genomics, Machine learning, Survival
Abstract
Background.
1p/19q‐codeleted anaplastic gliomas have variable clinical behavior. We have recently shown that the common 9p21.3 allelic loss is an independent prognostic factor in this tumor type. The aim of this study is to identify less frequent genomic copy number variations (CNVs) with clinical importance that may shed light on molecular oncogenesis of this tumor type.
Materials and Methods.
A cohort of 197 patients with anaplastic oligodendroglioma was collected as part of the French POLA network. Clinical, pathological, and molecular information was recorded. CNV analysis was performed using single‐nucleotide polymorphism arrays. Computational biology and feature selection based on the random forests method were used to identify CNV events associated with overall survival and other clinical‐pathological variables.
Results.
Recurrent chromosomal events were identified in chromosomes 4, 9, and 11. Forty‐six focal amplification events and 22 focal deletion events were identified. Twenty‐four focal CNV areas were associated with survival, and five of them were significantly associated with survival after multivariable analysis. Nine out of 24 CNV events were validated using an external cohort of The Cancer Genome Atlas. Five of the validated events contain a cancer‐related gene or microRNA: CDKN2A deletion, SS18L1 amplification, RHOA/MIR191 copy‐neutral loss of heterozygosity, FGFR3 amplification, and ARNT amplification. The CNV profile contributes to better survival prediction compared with clinical‐based risk assessment.
Conclusion.
Several recurrent CNV events, detected in anaplastic oligodendroglioma, enable better survival prediction. More importantly, they help in identifying potential genes for understanding oncogenesis and for personalized therapy.
Implications for Practice.
Genomic analysis of 197 anaplastic oligodendroglioma tumors reveals recurrent somatic copy number variation areas that may help in understanding oncogenesis and target identification for precision medicine. A machine learning multivariable model built using this genomic information enables better survival prediction.
Introduction
Gliomas represent approximately 80% of primary malignant brain tumors [1]. Grade 2 and 3 gliomas, also termed lower‐grade gliomas (LowerGG), have variable clinical behavior [2]. Recently, several studies uncovered three robust histomolecular subgroups of LowerGG that differ clinically: (a) gliomas with 1p/19q‐codeletion and isocitrate dehydrogenase (IDH) mutations are defined as oligodendroglioma, have better prognosis and response to therapies, and harbor somatic mutations in CIC, FUBP1, NOTCH1, and the TERT promoter; (ii) gliomas without 1p/19q‐codeletion and with IDH mutations have intermediate prognosis and had frequent mutations in TP53 and ATRX; and (iii) gliomas with neither 1p/19q‐codeletion nor IDH mutations have comparable prognosis and molecular profile to glioblastoma [2], [3], [4]. These two pivotal genetic alterations (1p/19q‐codeletion and mutations in IDH genes) have been integrated in the new classification of brain tumors published by World Health Organization (WHO) [5].
Somatic copy number variations (CNVs) play a central role in oncogenesis [6] and are implicated in survival prediction and response to therapies [7], [8]. Recently, several studies reported on the CNV landscape in LowerGG. Brat et al. showed in an analysis of The Cancer Genome Atlas (TCGA) cohort that LowerGG with 1p/19q‐codeletion and IDH mutation subtypes had low number of recurring CNV events (4q12 amplification and focal 1p deletion, including CDKN2C in a few cases). This cohort contained 82 tumors with 1p/19q‐codeletion, among which only 37 were grade 3 [2]. Suzuki et al. analyzed the CNV landscape of 715 LowerGG tumors in TCGA cohort and a Japanese cohort. This combined cohort contained 256 1p/19q‐codeleted tumors, of which 113 were grade 3 [3]. This study pooled all types of grade 2 and 3 gliomas into the CNV recurrence analysis, and this approach might mask relatively rare CNV associated with specific histologic subgroups.
We present here the CNV landscape of anaplastic oligodendroglioma, based on the largest nationwide prospective cohort of patients with grade 3 glioma, established by the Prise en charge des oligodendrogliomes anaplasiques (POLA) network [9]. This cohort enables the examination of correlation of CNV events with clinical‐pathological variables and prognosis.
Materials and Methods
Patients
A cohort of 197 patients with primary anaplastic oligodendroglioma was collected as part of the POLA network [9]. The patients’ characteristics are given in Table 1. The samples were accrued over 6 years between 2008 and 2014. Clinical‐pathological information and molecular analysis were undertaken with (a) the patient's informed consent, (b) the regulatory‐requested ethical board approval, and (c) the tenets of the Declaration of Helsinki. The patients were followed up clinically, and this was recorded by the electronic Case Report Form (eCRF) system. 1p/19q‐codeletion was defined as the loss of at least 90% of the probes mapped on 1p and 19q with concurrent chromosomes 1 and 19 centromeric breakpoints. The histological review was performed by the central pathological review team. The definition of severe nuclear atypia was marked nuclear pleomorphism with enlarged nucleoli occasionally associated with multinucleated giant cells. Tissue invasion was defined by the presence of isolated tumoral cells intercalated among axons on routine staining (hematoxylin and eosin). The clinical‐pathological variables used in the study are given in supplemental online Table 1. Four patients with zero follow‐up were excluded from the survival analysis.
Table 1. Patients’ epidemiological and clinical characteristics.
Abbreviations: PCV procarbazine, lomustine, and vincristine; SD, standard deviation; RT, radiotherapy; TMZ, temozolomide.
DNA Preparation
Cell pellets were digested with proteinase K; DNA was extracted using the QIAamp DNA mini kit (Qiagen, Hilden, Germany), and DNA concentration was measured by NanoDrop (Thermo Fisher Scientific, Waltham, MA). The quality of DNA was controlled on gel of agarose 1%.
CNV Analysis
CNV analysis was performed using the iSelect Infinium Human OmniExpress version 1.0 chip platform (n = 46; 730,397 probes; Illumina, San Diego, CA), Quad370 (n = 43; 373,397 probes; Illumina), Quad610 (n = 54; 620,711 probes; Illumina), and hcore (n = 54; 298,930 probes; Illumina). Normalized intensity signals were generated from the Illumina GenomeStudio software and then further normalized by Quantile Normalization modified with a threshold (tQN) [10]. The array‐based single‐nucleotide polymorphism (SNP) data can be accessed through ArrayExpress under accession number E‐MTAB‐5669. The normalized data was analyzed for CNV and loss of heterozygosity using the Global Parameter Hidden Markov Model (GPHMM) algorithm [11]. DNA index was dichotomized to diploid/tetraploid state, in which DNA index of ≥3 was interpreted as tetraploidy. The cutoffs for gain, high level amplification, loss, and homozygous deletion were determined by GISTIC software (Broad Institute, Cambridge, MA), as described below.
Recurrent CNV Analysis
In order to identify recurrent CNV events, GISTIC version 2.0 [12] was used. The GISTIC module identifies regions of the genome that are significantly amplified or deleted across a set of samples. Each aberration is assigned a G‐score that considers the amplitude of the aberration as well as the frequency of its occurrence across samples. False discovery rate q‐values are then calculated for the aberrant regions, using a background score distribution generated by random permutation of the marker locations in each sample. Regions with q‐values below a defined threshold are considered significant. The CNV segmentation files produced by GPHMM were used, and the copy number values were converted to log R ratio (LRR), as required by the GISTIC 2.0 software. In order to produce a unified marker file for the different platforms, the segmentation files were binned at 1,000 base pairs, similar to a protocol carried out when using GISTIC with sequencing data (GISTIC support, personal communication). The parameters used to analyze the recurrent amplifications and deletions were ‐smallmem 0, ‐broad 1, ‐brlen 0.5, ‐conf 0.95, ‐armpeel 1, ‐savegene 1, ‐gcm mean, ‐twosides, 1 ‐rx 1, ‐ta 0.1, ‐td 0.1, ‐cap 2, ‐js 4, and ‐maxseg 2000. Cutoffs for CNV state were determined by GISTIC with the following default parameters: normal: −0.1 < LRR < 0.1; loss: −1.3 < LRR < −0.1; homozygous deletion: LRR <−1.3; gain: 0.1 < LRR < 0.9, high amplification: LRR >0.9. The normal variation CNV file was taken from the Database of Genomic Variants (release date October 16, 2014) [13] in order to exclude normal CNV areas from the analysis. Copy‐neutral loss of heterozygosity (CN‐LOH) was defined as loss of heterozygosity and copy number state that equals the estimated sample ploidy. The parameters used for the analysis of recurrent CN‐LOH were ‐smallmem 0, ‐broad 1, ‐brlen 0.5, ‐conf 0.95, ‐armpeel 1, ‐savegene 1, ‐gcm mean, ‐twosides 1, ‐rx 1, ‐ta 0.1, ‐td 0.1, ‐cap 1.5, and ‐fname LOH.
Survival Analysis
Survival analysis was carried out using R (R Foundation, Vienna, Austria). For the survival analysis, we used the univariate Wald test on the Cox model (survival package [14]). We used the random forests approach for multivariable analysis of the survival data (randomForestSRC package [15]). The importance of each variable in the multivariable model was measured by (a) the variable importance (VIMP) parameter and (b) the depth of the variable in the trees [15]. The threshold for VIMP was defined as 0.01, and the threshold for depth was defined by the software. Variables were selected as informative if they were positive for at least one of these criteria and visualized using the ggRandomForests package. C‐index [16] was used to estimate the predictability of the multivariable models (survcomp package [17]).
External Validation
We computed position overlaps with cancer‐related oncogenic CNV areas based on curated CNV areas from 12 types of cancer [6]. The overlap analysis was performed using the genomic ranges package for R [18]. External validation of CNV peak survival analysis was performed using CBioPortal [19] with TCGA lowerGG provisional cohort (n = 530). The queries were made using Cosmic census genes [20] for CNV peaks containing such genes and otherwise using several genes spanning each tested CNV peak. Amplification peaks and deletion peaks were compared with corresponding peaks, and CN‐LOH peaks were compared to deletion peaks.
Results
Patients
A cohort of 197 patients with anaplastic oligodendroglioma were included in the current study. The median age at diagnosis was 50.0 years, and the sex ratio (male‐female) was 1.23. Mean follow‐up time was 3 years (range, 0–6.4 years; Table 1).
CNV Landscape
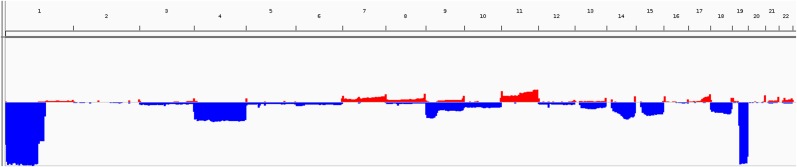
In the cohort, 117 of 197 (59%) patients were estimated to have diploid genomes, and 80 (41%) were estimated to have tetraploid genomes. Median tumor purity was 0.76 (range, 0.2–0.94). To identify and characterize the landscape of CNV in this tumor type, GISTIC analysis was carried out (Fig. 1). Recurrent chromosome arm losses (aside from 1p and 19q) were found in chromosome arms 4p, 4q, 9p, 9q, 13q, 14q, 15q, 18p, and 18q. The most frequent chromosome arms were 4p and 4q (30%) and 9p (23%). Recurrent chromosomal gains were found in chromosome arms 7q, 11p, and 11q, and the most frequent were 11q (19%) and 11p (15%). Recurrent CN‐LOH chromosomal events were identified in chromosome arms 9p, 9q, 11p, 11q, 16q, 21p, and 21q. The most frequent were 9p (15%) and 21p (8%; Fig. 2, supplemental online Tables 2 and 3).
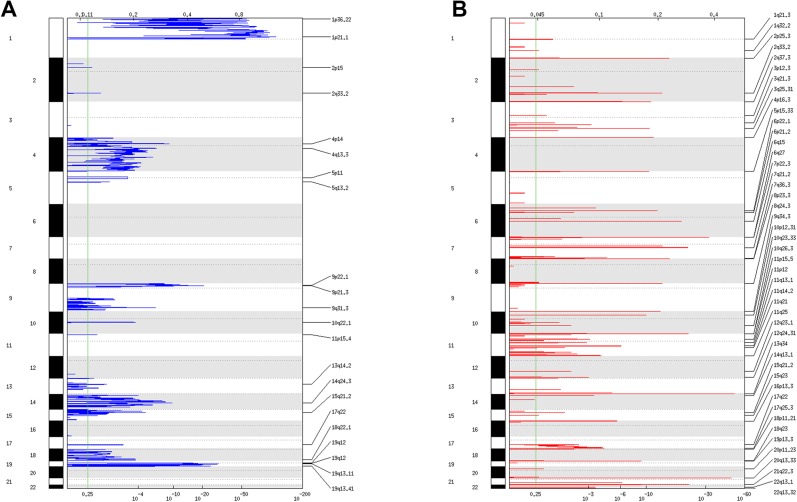
Figure 1.
Copy number variation landscape. Landscape of recurrent deletions (A) and amplifications (B) identified by GISTIC.
Figure 2.
Chromosomal view of the copy number variation landscape.
As shown in Figure 1A and B and in supplemental online Table 4, there were 46 focal amplification events and 22 focal deletion events. In addition, 113 focal CN‐LOH events were identified. The mean frequency (in the cohort) of focal amplification events was 17% (range, 3%–37%); of focal deletion events, 17% (range, 2%–31%); and of CN‐LOH events, 7% (range, 3%–17%).
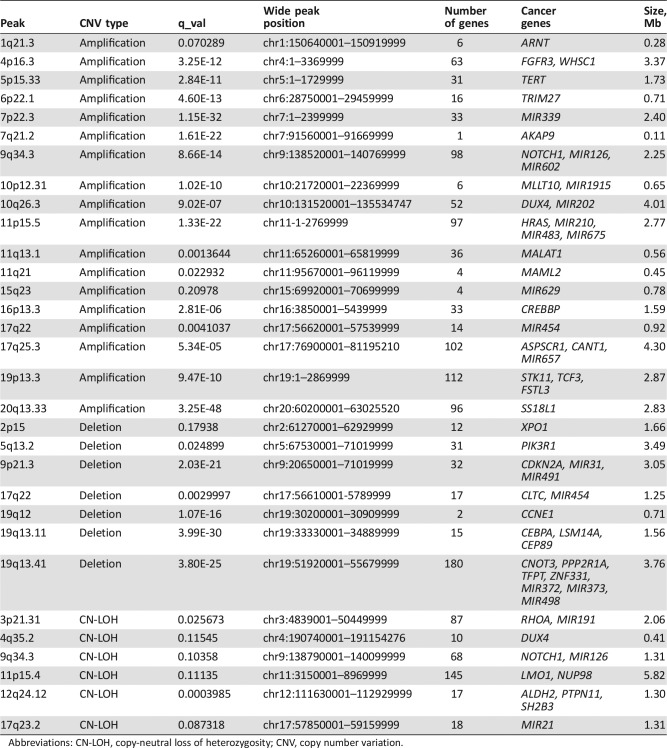
The cosmic census genes [20] and oncogenic microRNAs [21] located within the focal CNV peaks are given in Table 2. Some of these genes are already known to be involved in gliomas: FGFR3 [22], TERT, CDKN2A, NOTCH1, and PIK3R1 [3]. Importantly, nine of the cancer genes located in the focal CNV peaks are potentially druggable (FGFR3, NOTCH1, XPO1, CDKN2A, CCNE1, PPP2R1A, RHOA, NUP98, and PTPN11) [23].
Table 2. CNV focal peak containing a cancer gene and/or cancer‐related microRNA.
Abbreviations: CN‐LOH, copy‐neutral loss of heterozygosity; CNV, copy number variation.
CNV Focal Areas Overlap with Cancer‐Related CNV Areas
The CNV focal peaks identified in this study were compared to a pan‐cancer landscape of CNV peaks identified and curated from 12 tumor types [6]. As shown in supplemental online Table 5, 9 amplification peaks, 3 deletion peaks, and 15 CN‐LOH peaks overlap known cancer‐related CNV peaks (median overlap of 23%).
Correlation Between Genomic Characteristics (CNV, Ploidy) and Clinical‐Pathological Variables
There was an association between high Karnofsky score at diagnosis and the presence of amplification peak 6p22.1 (p = .04, chi square test, false discovery rate [FDR] correction) and oligoastrocytic histological aspect (p = .007, chi square test, FDR correction) and also between low Karnofsky score at diagnosis and the presence of amplification peak 2q33.2 (p = .003, chi square test, FDR correction). In addition, CN‐LOH 9p was associated with a previous history of cancer and with less tissue invasion. An additional six associations are given in supplemental online Table 6.
We have previously shown that anaplastic glioma with 1p19q codeletion can be stratified in three pathological groups predictive of prognosis according to microvascular proliferation and necrosis [24], [25]. CNV events associated with the grouping variable are given in supplemental online Table 7. Our data also predicted a worse prognosis for Group 3, which was positive for both necrosis and microvascular proliferation. Samples of this group showed significantly higher number of chromosomal alterations (p = 3.34e−05). For the complete cohort of all three groups, the total number of chromosomal CNVs equal to or greater than three predicted worse survival (p = .016).
Overall Survival Analysis
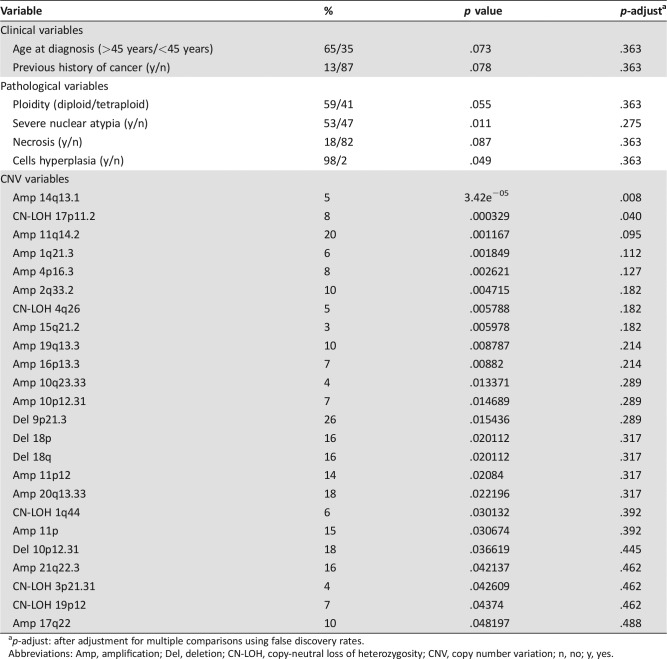
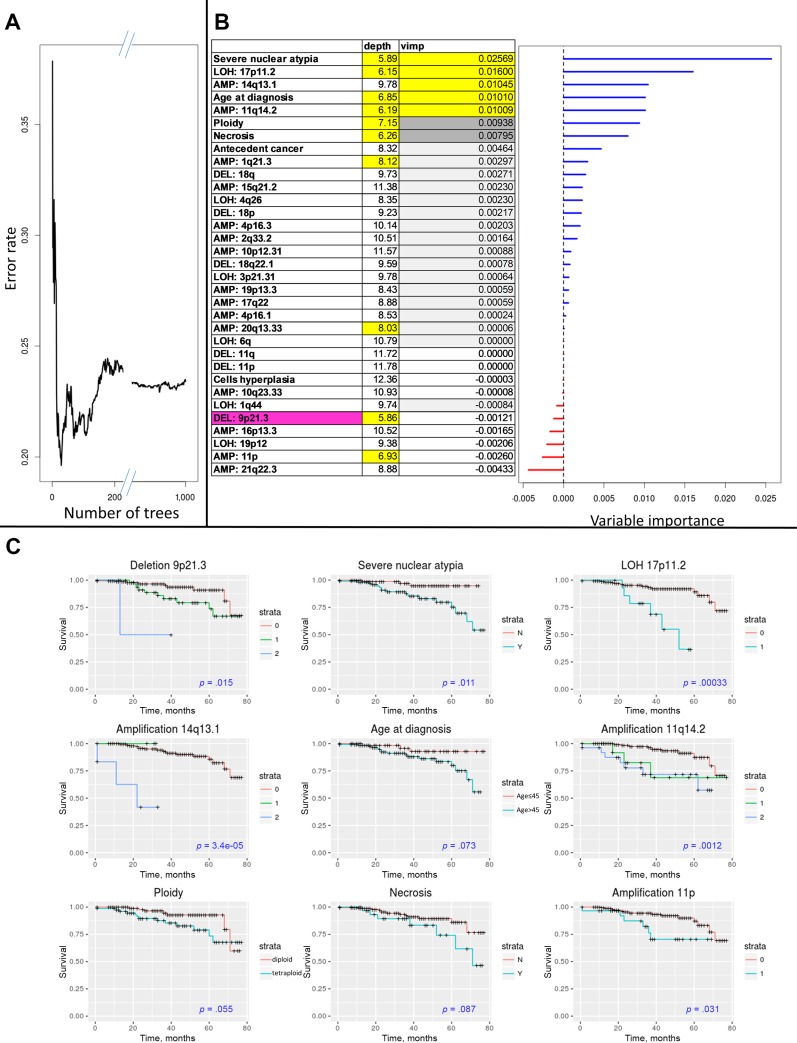
Table 3 shows the clinical variables and CNV variables associated with OS at the .05 level. Amplification peak 14q13.1 and CN‐LOH 17p11.2 were significant at the .05 level after FDR correction. Next, multivariable analysis including CNV variables (significant at the .05 level) and the clinical‐pathological variables (significant at the .1 level) was performed using a random forests method. We generated 1,000 trees, and the classification error rate was 0.23. As shown in Figure 3, the clinical‐pathological variables that contributed significantly to the model were severe nuclear atypia, necrosis, age at diagnosis, and ploidy. Images of severe and mild nuclear atypia are given in Figure 4. The CNV variables that contributed to the model were deletion of peak 9p21.3; amplification of peaks 14q13.1, 11q14.2, 20q13.33, and 1q21.3; CN‐LOH peak 17p11.2; and gain of 11p. Each of these predictors showed marked group differences in survival rates. For example, survival rates at 6 years were 54% and 95% for patients with and without severe nuclear atypia, respectively. Survival rates at 4 years were 55% and 92% for patients with and without LOH 17q11.2. The maximal differences in survival rates for the informative variables are given in supplemental online Table 8.
Table 3. Clinical, pathological, and CNV variables significantly associated with survival.
p‐adjust: after adjustment for multiple comparisons using false discovery rates.
Abbreviations: Amp, amplification; Del, deletion; CN‐LOH, copy‐neutral loss of heterozygosity; CNV, copy number variation; n, no; y, yes.
Figure 3.
Multivariable random forest analysis. (A): Classification error as function of number of trees. (B): Relative importance measures of the variables associated with overall survival: VIMP (the reduction of prediction power by omitting a variable from the analysis) and average depth of the variables in the decision trees (smaller numbers represent variables closer to the tree root and thus greater influence on the classification). (C): Survival plots for nine of the variables with VIMP >0.005. Copy number variation coding is as follows. For amplifications: 0, normal; 1, gain; 2, high amplification. For deletions: 0, normal; 1, heterozygous deletion; 2, homozygous deletion. For copy‐neutral LOH: 0, normal; 1, LOH.
Abbreviations: AMP, amplification; DEL, deletion; LOH, loss of heterozygosity; VIMP, variable importance.
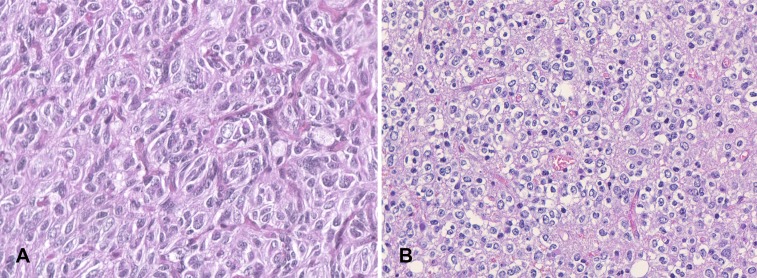
Figure 4.
Nuclear atypia. (A): A case showing severe nuclear atypia with increased nuclear‐cytoplasmic ratio and enlarged and pleomorphic nuclei. (B): A case showing slightly irregular nuclei. (Hematoxylin and eosin, ×40.)
To evaluate the relative predictive power of the clinical and the CNV variables and to rule out model overfitting, C‐index was calculated using cross validation. Briefly, C‐index is an estimate of the probability for any two individuals (chosen from the sample) that an individual with predicted longer survival indeed survived more time than his or her counterpart. C‐index of 0.5 reflects model prediction that is equivalent to random comparisons. Eighty percent of the samples were selected randomly and were used to build a random forest‐based model. The C‐index was estimated from the other 20% of the samples. This process was repeated randomly 1,000 times. The median C‐index was 0.62 for CNV variables and 0.65 for the clinical variables. The median C‐index for the combined model was 0.69. The model based on clinical variables performed better than the model based on CNV variables (p = .0002). The combined model performed best, compared with the models based on either CNV (11.3% added model accuracy; p < 2e−16) or clinical variables (6.2% added model accuracy; p = 2e−9). Accordingly, it appears that the CNV variables are informative for survival prediction and moreover complement the clinical‐pathological variables in this regard.
We then focused on the variables that had predictive power for patients’ overall survival (OS). Older age (>45 years), tetraploidy [26], severe nuclear atypia, and necrosis predict worse OS. Deletion peak 9p21.3 predicted shorter OS even for heterozygous deletion state. This peak contains CDKN2A, which has an effect on survival in several cancer types [27]. The presence of the other CNV areas described above also predicted poor OS (Fig. 3).
Progression‐Free Survival
supplemental online Table 9 gives the variables associated with progression‐free survival (PFS; p < .05). Severe nuclear atypia was also significant after FDR correction. Multivariable analysis was carried out using the random forests method for all clinical variables (p < .1) and for CNV variables (p < .05). The analysis yielded classification error of 0.4. The variables that were informative were severe nuclear atypia, CN‐LOH peak 7q31.1, CN‐LOH peak 17p11.2, and amplification peak 15q21.2. The C‐index was 0.49 for CNV variables and 0.6 for the clinical variables. For the combined model, the C‐index was 0.53. Accordingly, it appears that the CNV variables are not informative for a multivariable model of PFS prediction.
Validation of CNV Peaks Associated with Survival Using TCGA Data as an External Validation
TCGA LowerGG cohort consists of 530 patients with grade 2 and 3 gliomas and among them only 75 patients with grade 3 oligodendrogliomas; such a subcohort is underpowered for survival analysis for this tumor type and for relatively uncommon CNVs. However, we assumed that findings of CNV areas that are associated with survival in our cohort of grade 3 oligodendroglioma and that are also associated with survival in an additional larger cohort of more heterogeneous LowerGG would have higher level of confidence. Accordingly, we used the complete LowerGG cohort for validation. We examined all CNV peaks that were associated with OS or PFS in our samples (p < .05). Overall, as shown in supplemental online Table 11, 9 of 24 focal areas were significantly associated with OS and/or PFS. The presence of all the validated peaks were associated with shorter OS and/or PFS in both our sample and in TCGA dataset. Five of the validated peaks involve the following known cancer‐related genes or microRNAs, with their prevalence given in parentheses: CDKN2A deletion (26%), SS18L1 amplification (18%), RHOA/MIR191 CN‐LOH (4%), FGFR3 amplification (8%), and ARNT amplification (6%). The most prevalent CNV peaks that were validated were CDKN2A deletion (26%), SS18L1 amplification (18%), CN‐LOH 9p13.3 (16%), and 2q33.2 amplification (11%).
We also examined CNV peaks that contained a cancer‐related gene or microRNA and that were not associated with survival in our cohort but were, nevertheless, associated with survival in TCGA LowerGG cohort. This may point to genes that are involved in cancer pathogenesis but not associated with survival specifically in anaplastic oligodendroglioma tumors. The cancer genes that were positively associated with OS and/or PFS are given in supplemental online Table 11.
Of note, one of the CNV peaks most significantly associated with survival, 11q14.2 amplification, which contain TMEM135 gene and is amplified in 20% of our cohort, was not associated with OS and/or PFS in TCGA LowerGG cohort.
Discussion
Anaplastic oligodendroglioma is now defined as a distinct histomolecular entity in the WHO classification 2016 [5]. However, this tumor group remains heterogeneous. Recently, we have shown that the most common genomic alteration (i.e., 9p CN‐LOH) is associated with worse prognosis [27]. Moreover, a recently published work identified three subtypes of anaplastic oligodendrogliomas with distinct genomic alteration profiles associated with differences in prognosis [28]. Therefore, comprehensive prognostic evaluation integrating clinical, pathological, and genomic biomarkers in a large cohort of patients with grade 3 gliomas may help achieve a sharper stratification of patients.
The CNV landscape of a large cohort of patients with anaplastic oligodendroglioma is reported here. Forty‐one percent of the tumors were estimated to be hyperploid, which is in the higher spectrum of tumor hyperploidy frequency [29]. Several recurrent chromosomal CNV events were identified, most notably losses in chromosomal arms 4p, 4q, and 9p; gains in chromosomal arms 11p and 11q; and CN‐LOH in chromosomal arms 9p and 21p. In addition, 22 focal deletions, 46 focal amplifications, and 113 focal CN‐LOH were identified.
Twenty‐seven of the focal CNV areas were shown to have overlap with known pan‐cancer oncogenic CNV events. It should be noted that this overlap might be important even for those with little overlap, and the differences can be attributed to the following facts: (a) different SNP arrays platforms were used for the reference data set and for the current data set; (b) the reference databases represent a pan‐cancer sample combination, whereas our data set consisted of a very homogenous tumor subtype. Thirty‐one of the focal events contained at least one Cosmic Census gene and/or an oncogenic microRNA. Some of these genes are known to be involved in glioma biology, such as FGFR3, TERT, and CDKN2A; some are attractive putative candidates for such involvement, such as DUX4 and CCNE, known to be involved in other cancer types [30], [31]. Importantly, some of these genes are potentially druggable, such as FGFR3, NOTCH1, and CCNE1.
Interesting associations of several CNV areas with clinical‐pathological variables were identified. OS analysis was performed in two parts. First, clinical‐pathological variables and recurrent CNV areas that are associated with OS were identified. From these variables, after adjustment for multiple comparisons, amplification peak 14q13.1 and CN‐LOH 17p11.2 remained as variables strongly correlated with OS by themselves. This stringent analysis is conservative and prone to reject important variables because of the high number of statistical tests performed. In addition, this analysis does not permit the evaluation of the prognostic value of the combined set of variables. Accordingly, a random forests algorithm was applied for all the variables that were associated with OS. The variables that were most important for the model were severe nuclear atypia; necrosis; age at diagnosis and ploidy; deletion peak 9p21.3 (containing CDKN2A); amplification peaks 11q14.2, 20q13.33 (containing SS18L1), 14q13.1, and 1q21.3 (containing ARNT); and CN‐LOH peak 17p11.2. These variables included the three variables that passed the conservative single‐variable survival analysis.
We were interested to examine the relative contributions of the model and the model components for prognostic prediction. C‐index is a nonparametric measure to quantify the discriminatory power of a predictive model. A previous study showed that median C‐index for genomic CNV variables (depending on tumor type) may be in the range of 0.5 (for glioblastoma) to 0.58 (renal clear cell carcinoma) [7]. In the current analysis for anaplastic oligodendroglioma, the C‐index for genomic CNV variables was relatively high (i.e., 0.62), for the clinical‐pathological variables it was 0.65, and for both sets of variables in a combined model, the C‐index was 0.69. These results show that the CNV measures not only have predictive information but also complement the predictive information of the clinical variables, and the combined model is better than each model alone.
Analysis of PFS was associated with severe nuclear atypia after multiple comparison adjustment, and no CNV variables were significant at this level. The multivariable model performed worse compared with the analysis of OS; the classification error was 0.4. The median C‐index as measured by cross validation and 1,000 repeats was 0.49 for the CNV variables, 0.62 for the clinical variables, and 0.52 for the combined model. Hence, we conclude that the CNV variables are not predictive for PFS. The negative results for PFS compared with the positive results for OS can be attributed to lower accuracy of PFS definition compared with OS.
There are several limitations of this study. One limitation is the use of four different CNV platforms. However, the difference in densities is less than one level of magnitude (on average one probe every 4–10 kb). A second limitation is that the validation cohort of TCGA is heterogeneous (i.e., grade 2, grade 3, astrocytic, and oligodendroglial tumors). TCGA cohort included only 75 patients with grade 3 oligodendroglioma and is underpowered for survival analysis for this tumor type. However, we assumed that findings of CNV areas that are associated with survival in our cohort of grade 3 oligodendroglia and that are also associated with survival in an additional larger cohort of more heterogenous LowerGG have a higher level of confidence. In addition, the outcome analysis is limited by the apparently heterogeneous treatment given to the cohort population (Table 1).
Conclusion
This study describes the CNV landscape of anaplastic oligodendroglioma using the largest reported homogenous cohort. In addition, using robust clinical follow‐up and databases, we created a multivariable predictive model for OS using a combination of CNV genomic information and clinical‐pathological variables. Although this model cannot stand by itself for individual clinical survival prediction, it can help in the identification of high‐risk patients harboring CNV associated with shorter OS (using the model or even single markers; Fig. 3C). Moreover, the model helped in the identification of a small set of informative CNV areas that should lead to further research into driver genes for anaplastic oligodendroglioma.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
The authors thank the Association pour la Recherche sur les Tumeurs Cérébrales; Hadassah France; the program Investissements d'avenir; the national program Cartes d'Identité des Tumeurs (CIT), funded and developed by the Ligue Nationale contre le cancer; the Institut Universitaire de Cancérologie; and OncoNeuroTek. The authors also thank Professor Ao Li for advice in CNV analysis. The research leading to these results has received funding from the program “Investissements d'avenir” (ANR‐10‐IAIHU‐06).
POLA Network affiliations: Clovis Adam, Hôpital Bicêtre, Pathology Department, Le Kremlin‐Bicêtre, France; Marie Andraud, CHU Saint‐Pierre de la Réunion, Pathology Department, Saint‐Pierre de la Réunion, France; Marie‐Hélène Aubriot‐Lorton, CHU Dijon, Pathology Department, Dijon, France; Luc Bauchet, CHU de Montpellier, Neurosurgery Department, Montpellier, France; Patrick Beauchesne, CHU Nancy, Neuro‐oncology Department, Nancy, France; Lien Bekaert, CHU de Caen, Neurology Department, Caen, France; Claire Blechet, CHR Orléans, Pathology Department, Orléans, France; Mario Campone, Centre René Gauducheau, Medical Oncology Department, Saint‐Herblain, France; Antoine Carpentier, Hôpital Avicenne, Neurology Department, Bobigny, France; Ioana Carpiuc, Clinique des Cèdres, Medical Oncology Department, Cornebarrieu, France; Dominique Cazals‐Hatem, Hôpital Beaujon, Neurosurgery Department, Clichy, France; Benoit Lhermitte, CHU Strasbourg, Pathology Department, Strasbourg, France; Danchristian Chiforeanu, CHU Rennes, Pathology Department, Rennes, France; Olivier Chinot, Hôpital de la Timone, Assistance Publique ‐ Hôpitaux de Marseille, Neuro‐oncology Department, Marseille, France; Elisabeth Cohen‐Moyal, Institut Claudius Regaud, Radiotherapy Department, Toulouse, France; Philippe Colin, Clinique de Courlancy, Radiotherapy Department, Reims, France; Thierry Cruel, Mont‐Blanc Pathology, Annecy, France; Phong Dam‐Hieu, Hôpital de la cavale blanche, CHU Brest, Neurosurgery Department, Brest, France; Christine Desenclos, Hôpital Nord, CHU Amiens, Neurosurgery Department, Amiens, France; Nicolas Desse, HIA Sainte‐Anne, Neurosurgery Department, Toulon, France; Frederic Dhermain, Institut Gustave Roussy, Radiotherapy Department, Villejuif, France; Marie‐Danièle Diebold, CHU Reims, Pathology Department, Reims, France; Sandrine Eimer, CHU de Bordeaux‐GH Pellegrin, Pathology Department, Bordeaux, France; Thierry Faillot, Hôpital Beaujon, Neurosurgery Department, Clichy, France; Mélanie Fesneau, CHR Orléans, Radiotherapy Department, Orléans, France; Denys Fontaine, CHU Nice, Neurosurgery Department, Nice, France; Stéphane Gaillard, Hôpital Foch, Neurosurgery Department, Suresnes, France; Fabien Forest, Hôpital Nord, CHU Saint‐Étienne, Pathology Department, Saint‐Priest en Jarez, France; Guillaume Gauchotte, CHU Nancy, Pathology Department, Nancy, France; Claude Gaultier, CH Colmar, Neurology Department, Colmar, France; François Ghiringhelli, Centre Georges‐François Leclerc, Medical Oncology, Dijon, France; Catherine Godfraind, CHU Clermont‐Ferrand, Pathology Department, Clermont‐Ferrand, France; Edouard Marcel Gueye, Hôpital Dupuytren, CHU de Limoges, Neurosurgery Department, Limoges, France; Selma Elouadhani‐Hamdi, Hôpital Lariboisière, Neurosurgery Department, Paris, France; Jerome Honnorat, Hospices Civils de Lyon, Hôpital Neurologique, Neuro‐oncology Department, Bron, France; Toufik Khallil, CHU Clermont‐Ferrand, Neurosurgery Department, Clermont‐Ferrand, France; Francois Labrousse, Hôpital Dupuytren, CHU de Limoges, Pathology Department, Limoges, France; Willem Lahiani, Hôpital Henri Mondor, Neurosurgery Department, Henri Mondor, France; Olivier Langlois, CHU Charles Nicolle, Neurosurgery Department, Rouen, France; Annie Laquerriere, CHU Charles Nicolle, Pathology Department, Rouen, France; Delphine Larrieu‐Ciron, CHU Poitiers, Neurology Department, Poitiers, France; Emmanuelle Lechapt‐Zalcman, CHU de Caen, Pathology Department, Caen, France; Hugues Loiseau, CHU de Bordeaux‐GH Pellegrin, Neurosurgery Department, Bordeaux, France; Stéphane Lopez, CH Annecy, Oncology Department, Annecy, France; Delphine Loussouarn, CHU Nantes, Pathology Department, Nantes, France; Claude‐Alain Maurage, CHU de Lille, Pathology Department, Lille, France; Philippe Menei, CHU Angers, Neurosurgery Department, Angers, France; Marcela Ionella Mihai, Hôpital Jean Minjoz, CHU Besançon, Pathology Department, Besançon, France; Serge Milin, CHU Poitiers, Neurosurgery Department, Poitiers, France; Marie Janette Motsuo Fotso, Hôpital Nord, CHU Saint‐Étienne, Neurosurgery Department, Saint‐Priest en Jarez, France; Georges Noel, Centre Paul Strauss, Radiotherapy Department, Strasbourg, France; Fabrice Parker, Hôpital Bicêtre, Neurosurgery Department, Le Kremlin‐Bicêtre, France; Antoine Petit, Hôpital Jean Minjoz, CHU Besançon, Neurosurgery Department, Besançon, France; Isabelle Quintin‐Roué, Hôpital de la cavale blanche, CHU Brest, Pathology Department, Brest, France; Carole Ramirez, CHU de Lille, Neurosurgery Department, Lille, France; Audrey Rousseau, CHU Angers, Pathology Department, Angers, France; Cécilia Rousselot‐Denis, CHU Bretonneau, Pathology Department Tours, France; Damien Ricard, HIA du Val de Grâce, Neurology Department, Paris, France; Pomone Richard, Clinique des Cèdres, Pathology Department, Cornebarrieu, France; Valérie Rigau, CHU de Montpellier, Pathology Department, Montpellier, France; Gwenaelle Runavot, CHU Saint‐Pierre de la Réunion, Neurology Department, Saint‐Pierre de la Réunion, France; Henri Sevestre, Hôpital Nord, CHU Amiens, Pathology Department, Amiens, France; Marie Christine Tortel, Hôpital Colmar, Pathology Department, Colmar, France; Fanny Vandenbos, CHU Nice, Pathology Department, Nice, France; Elodie Vauleon, Centre Eugène Marquis, Medical Oncology, Rennes, France; Chiara Villa, Hôpital Foch, Pathology Department, Suresnes, France; Ilyess Zemmoura, CHU Bretonneau, Neurosurgery Department Tours, France
POLA Network hospitals: Amiens (C. Desenclos, H. Sevestre), Angers (P. Menei, A. Rousseau), Annecy (T. Cruel, S. Lopez), Besançon (M‐I Mihai, A. Petit), Bicêtre (C. Adam, F. Parker), Bobigny (A. Carpentier), Brest (P. Dam‐Hieu, I. Quintin‐Roué), Bordeaux (S. Eimer, H. Loiseau), Caen (L. Bekaert, E. Lechapt‐Zalcman), Clermont‐Ferrand (C. Godfraind, T. Khallil), Clichy (D. Cazals‐Hatem, T. Faillot), Colmar (C. Gaultier, MC. Tortel), Cornebarrieu (I. Carpiuc, P. Richard), Créteil (W. Lahiani), Dijon (H. Aubriot‐Lorton, F. Ghiringhelli), Lille (CA. Maurage, C. Ramirez), Limoges (EM. Gueye, F. Labrousse), Lyon (F. Ducray, A. Jouvet), Marseille (D. Figarella‐Branger, O Chinot), Montpellier (L. Bauchet, V. Rigau), Nancy (P. Beauchesne, G. Gauchotte), Nantes (M. Campone, D. Loussouarn), Nice (D. Fontaine, F. Vandenbos‐Burel), Orléans (C. Blechet, M. Fesneau), Paris (C. Dehais, JY Delattre, S. Elouadhani‐Hamdi, D. Ricard), Poitiers (D. Larrieu‐Ciron,S. Milin), Reims (P. Colin, MD. Diebold), Rennes (D. Chiforeanu, E. Vauleon), Rouen (O. Langlois, A. Laquerriere), Saint‐Etienne (F. Forest, MJ. Motso‐Fotso), Saint‐Pierre de la Réunion (M. Andraud, G. Runavot), Strasbourg (B. Lhermitte, G. Noel), Suresnes (S. Gaillard, C. Villa), Toulon (N. Desse), Toulouse (E. Cohen‐Moyal, E. Uro‐Coste), Villejuif (F. Dhermain)
Footnotes
For Further Reading: Marianne Labussière, Amithys Rahimian, Marine Giry et al. Chromosome 17p Homodisomy Is Associated With Better Outcome in 1p19q Non‐Codeleted and IDH‐Mutated Gliomas. The Oncologist 2016;21:1131–1135.
Implications for Practice: Homodisomy of chromosome 17p (CNLOH 17p) is a frequent feature in IDH‐mutated 1p19q non‐codeleted gliomas (group 2). It is constantly associated with TP53 mutation. It was found, within this specific molecular group of gliomas (corresponding to molecular astrocytomas), that CNLOH 17p is associated with a much better outcome and may therefore represent an additional prognostic marker to refine the prognostic classification of gliomas.
Contributor Information
Shai Rosenberg, Email: shair@hadassah.org.il.
Ahmed Idbaih, Email: ahmed.idbaih@aphp.fr.
Collaborators: POLA Network, C. Desenclos, H. Sevestre, P. Menei, A. Rousseau, T. Cruel, S. Lopez, M‐I Mihai, A. Petit, C. Adam, F. Parker, A. Carpentier, P. Dam‐Hieu, I. Quintin‐Roué, S. Eimer, H. Loiseau, L. Bekaert, E. Lechapt‐Zalcman, C. Godfraind, T. Khallil, D. Cazals‐Hatem, T. Faillot, C. Gaultier, MC. Tortel, I. Carpiuc, P. Richard, W. Lahiani, H. Aubriot‐Lorton, F. Ghiringhelli, CA. Maurage, C. Ramirez, EM. Gueye, F. Labrousse, F. Ducray, A. Jouvet, D. Figarella‐Branger, O Chinot, L. Bauchet, V. Rigau, P. Beauchesne, G. Gauchotte, M. Campone, D. Loussouarn, D. Fontaine, F. Vandenbos‐Burel, C. Blechet, M. Fesneau, C. Dehais, JY Delattre, S. Elouadhani‐Hamdi, D. Ricard, D. Larrieu‐Ciron, S. Milin, P. Colin, MD. Diebold, D. Chiforeanu, E. Vauleon, O. Langlois, A. Laquerriere, F. Forest, MJ. Motso‐Fotso, M. Andraud, G. Runavot, B. Lhermitte, G. Noel, S. Gaillard, C. Villa, N. Desse, E. Cohen‐Moyal, E. Uro‐Coste, and F. Dhermain
Author Contributions
Conception/design: Shai Rosenberg, Francois Ducray, Jean‐Yves Delattre, Ahmed Idbaih
Provision of study material or patients: POLA Network
Collection and/or assembly of data: Caroline Dehais, Nabila Elarouci, Yannick Marie, Karima Mokhtari, Dominique Figarella‐Branger
Data analysis and interpretation: Shai Rosenberg, Francois Ducray, Agusti Alentorn, Caroline Dehais, Nabila Elarouci, Aurelie Kamoun, Yannick Marie, Marie‐Laure Tanguy, Aurélien De Reynies, Karima Mokhtari, Dominique Figarella‐Branger
Manuscript writing: Shai Rosenberg, Ahmed Idbaih
Final approval of manuscript: Shai Rosenberg, Francois Ducray, Agusti Alentorn, Caroline Dehais, Nabila Elarouci, Aurelie Kamoun, Yannick Marie, Marie‐Laure Tanguy, Aurélien De Reynies, Karima Mokhtari, Dominique Figarella‐Branger, Jean‐Yves Delattre, Ahmed Idbaih
Disclosures
The authors indicated no financial relationships.
References
- 1. Ostrom QT, Bauchet L, Davis FG et al. The epidemiology of glioma in adults: A “state of the science” review. Neuro Oncol 2014;16:896–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network ; Brat DJ, Verhaak RG, Aldape KD et al. Comprehensive, integrative genomic analysis of diffuse lower‐grade gliomas. N Engl J Med 2015;372:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suzuki H, Aoki K, Chiba K et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 2015;47:458–468. [DOI] [PubMed] [Google Scholar]
- 4. Ceccarelli M, Barthel FP, Malta TM et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016;164:550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Louis DN, Perry A, Reifenberger G et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol 2016;131:803–820. [DOI] [PubMed] [Google Scholar]
- 6. Ciriello G, Miller ML, Aksoy BA et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 2013;45:1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuan Y, Van Allen EM, Omberg L et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnol 2014;32:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature 2015;526:343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Idbaih A, Ducray F, Dehais C et al.; POLA Network SNP array analysis reveals novel genomic abnormalities including copy neutral loss of heterozygosity in anaplastic oligodendrogliomas. PloS One 2012;7:e45950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Staaf J, Vallon‐Christersson J, Lindgren D et al. Normalization of Illumina Infinium whole‐genome SNP data improves copy number estimates and allelic intensity ratios. BMC Bioinformatics 2008;9:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li A, Liu Z, Lezon‐Geyda K et al. GPHMM: An integrated hidden Markov model for identification of copy number alteration and loss of heterozygosity in complex tumor samples using whole genome SNP arrays. Nucleic Acids Res 2011;39:4928–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mermel CH, Schumacher SE, Hill B et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy‐number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacDonald JR, Ziman R, Yuen RK et al. The Database of Genomic Variants: A curated collection of structural variation in the human genome. Nucleic Acids Res 2014;42:D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 15. Ishwaran H, Kogalur UB, Blackstone EH et al. Random survival forests. Ann Appl Stat 2008;2:841–860. [Google Scholar]
- 16. Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: Model specific population value and confidence interval estimation. Stat Med 2004;23:2109–2123. [DOI] [PubMed] [Google Scholar]
- 17. Schröder MS, Culhane AC, Quackenbush J et al. survcomp: An R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 2001;27:3206–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lawrence M, Huber W, Pagès H et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cerami E, Gao J, Dogrusoz U et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Forbes SA, Beare D, Gunasekaran P et al. COSMIC: Exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015;43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang D, Gu J, Wang T et al. OncomiRDB: A database for the experimentally verified oncogenic and tumor‐suppressive microRNAs. Bioinformatics 2014;30:2237–2238. [DOI] [PubMed] [Google Scholar]
- 22. Lasorella A, Sanson M, Iavarone A. FGFR‐TACC gene fusions in human glioma. Neuro Oncol 2017;19:475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rubio‐Perez C, Tamborero D, Schroeder MP et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 2015;27:382–396. [DOI] [PubMed] [Google Scholar]
- 24. Figarella‐Branger D, Mokhtari K, Dehais C et al.; POLA Network . Mitotic index, microvascular proliferation, and necrosis define 3 groups of 1p/19q codeleted anaplastic oligodendrogliomas associated with different genomic alterations. Neuro Oncol 2014;16:1244–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Figarella‐Branger D, Lechapt‐Zalcman E, Tabouret E et al. Supratentorial clear cell ependymomas with branching capillaries demonstrate characteristic clinicopathological features and pathological activation of nuclear factor‐kappaB signaling. Neuro Oncol 2016;18:919–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuznetsova AY, Seget K, Moeller GK et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle 2015;14:2810–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alentorn A, Dehais C, Ducray F et al.; POLA Network. Allelic loss of 9p21.3 is a prognostic factor in 1p/19q codeleted anaplastic gliomas. Neurology 2015;85:1325–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kamoun A, Idbaih A, Dehais C et al.; POLA Network. Integrated multi‐omics analysis of oligodendroglial tumours identifies three subgroups of 1p/19q co‐deleted gliomas. Nat Commun 2016;7:11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carter SL, Cibulskis K, Helman E et al. Absolute quantification of somatic DNA alterations in human cancer. Nature Biotechnol 2012;30:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J, McCastlain K, Yoshihara H et al.; St. Jude Children's Research Hospital‐Washington University Pediatric Cancer Genome Project . Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nature Genetics 2016;48:1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karakas C, Biernacka A, Bui T et al. Cytoplasmic cyclin E and phospho‐cyclin‐dependent kinase 2 are biomarkers of aggressive breast cancer. Am J Pathol 2016;186:1900–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]