In this case series, the combination of germline testing and tumor mutational assessment was used to discern the clinical relevance of variants of uncertain significance to guide immunotherapeutic decision‐making for individualized patient care.
Abstract
Lynch syndrome is characterized by germline abnormalities in mismatch repair (MMR) genes, leading to predisposition to multiple cancers [1]. A second hit to the unaffected allele is required for tumorigenesis. MMR proteins repair incorrectly paired nucleotides and prevent generation of insertions and deletions at microsatellites [2]. Aberrancies in these MMR proteins can be a result of germline mutations or somatic alterations. Defective MMR results in microsatellite instability (MSI) and a high mutational burden [3].
The clinical implications of MSI are becoming readily apparent, as presence of MSI leads to the generation of neoantigens, stimulating tumor‐associated lymphocytes [4], [5]. This has led to the use of programmed cell death protein 1 blockade for MMR‐deficient tumors [6]. The U.S. Food and Drug Administration recently approved pembrolizumab for any advanced solid tumor demonstrating MSI and nivolumab for metastatic MSI colorectal cancer. However, the clinical significance of numerous MMR gene variants remains uncertain. The International Society for Gastrointestinal Hereditary Tumors classification system categorizes 2,360 MMR variants, which can be used to gauge pathogenicity [7]. There are many variants of uncertain significance (VUS; or class 3) for which clinicians are unable to provide recommendations. In this study, we employed the combination of germline testing and tumor mutational assessment to help discern the clinical relevance of VUS and guide immunotherapeutic decisions.
Key Points.
A clinical dilemma arises when genomic testing yields variants of uncertain significance (VUS).
Germline VUS were identified in two patients with gastrointestinal malignancies, but only one patient had a second‐hit mutation in a mismatch repair gene leading to mismatch repair deficiency that conferred response to immunotherapy.
The combination of germline testing along with tumor mutational assessment can help discern the clinical relevance of VUS and can help guide therapeutic decision‐making toward individualized patient care.
Patient Story 1
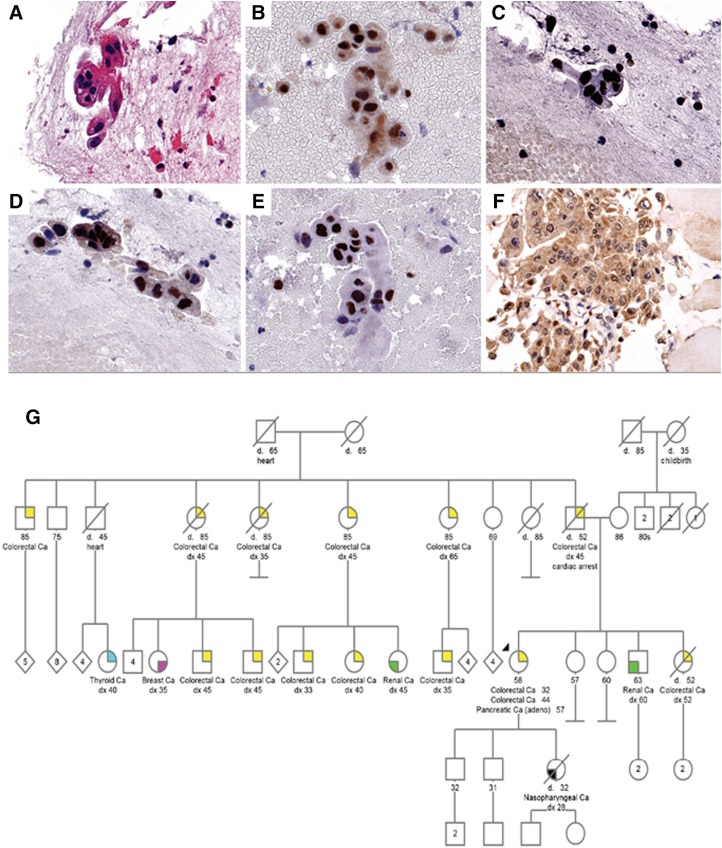
A 58‐year‐old woman was evaluated for newly diagnosed metastatic pancreatic cancer. Her history was notable for metachronous colon cancers diagnosed in the Philippines at age 32 and 44. Twelve years later, she presented with epigastric pain. Computed tomography (CT) showed a 1.9 × 1.7 cm hypoattenuating pancreatic head mass. Cancer antigen 19‐9 (CA 19‐9) was elevated at 9,121 U/mL. Endoscopic ultrasound revealed a 3.3 × 2.6 cm pancreatic mass. CT also showed lesions in the C7, T1, and T8 vertebrae. Biopsy of the T1 vertebrae showed adenocarcinoma of pancreatic primary (Fig. 1A). Immunohistochemistry showed intact expression of MLH1, MSH2, MSH6, and PMS2 (Fig. 1B–1E). A second biopsy (gluteal biopsy) was done later (Fig. 1F, please see below).
Figure 1.
Fine needle aspirate of T1 vertebrae of patient 1 demonstrating clusters of malignant glandular cells (A). Mismatch repair protein staining showed intact MLH1 (B), intact MSH2 (C), intact MSH6 (D), and intact PMS2 (E) expression. Fine needle aspirate of subsequent gluteal nodule demonstrates equivocal expression of PMS2 (F). Family pedigree of patient 1 showing family history of malignancy on paternal side (G). Proband is denoted by black arrow.
Genetic Testing
Germline testing was performed via Myriad COLARIS Plus with Myriad myRisk Hereditary Cancer testing. MyVision Classification Program showed two variants of uncertain significance (VUS) including CDH1 c.2165‐15C>A (intron) and MLH1 c.374C>A (p.Ala125Glu) in the germline. Tumor testing performed on gluteal biopsy (after progression on first‐line chemotherapy) via Foundation One showed a second MLH1 c.250A>G (p.Lys84Glu) somatic variant, classified as a likely pathogenic variant by the International Society for Gastrointestinal Hereditary Tumors (InSiGHT) as well as high tumor mutational burden of 25 mutations per megabase and microsatellite instability (MSI). The tumor specimen also showed the MLH1 c.374C>A variant, which was identified in the germline.
Family History
Family history was notable for father with colon cancer at age 45, sister with colon cancer at age 52, five paternal aunts and uncles with colon cancer, and five paternal first cousins with colon cancer diagnosed in their 30s and 40s (Fig. 1G). Family history fulfilled Amsterdam criteria.
Therapy
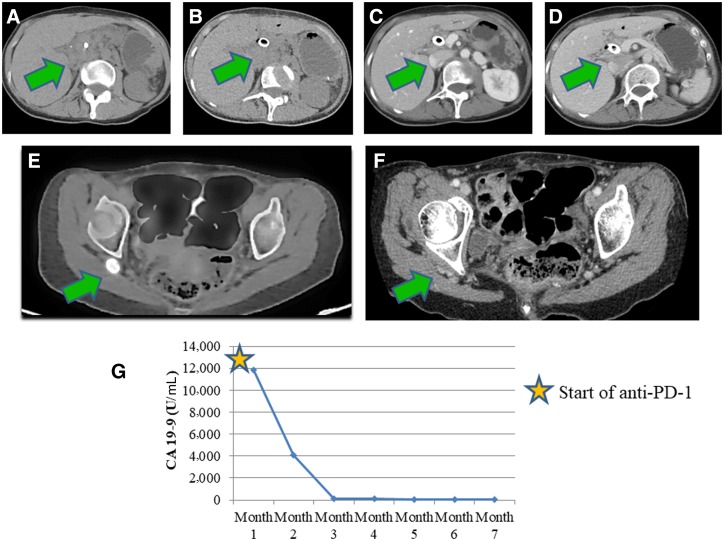
She was initially started on gemcitabine and nab‐paclitaxel. Follow‐up positron emission tomography/computed tomography (PET/CT) revealed a new 1.4 cm right gluteal soft tissue nodule, for which biopsy showed metastatic adenocarcinoma. Immunohistochemistry showed intact expression of MLH1, MSH2, and MSH6 with equivocal PMS2 testing (incomplete PMS2 expression with a predominant nuclear rimming pattern; Fig. 1F). Therapy was subsequently switched to pembrolizumab. Interval CT showed decrease in size of pancreatic mass (Fig. 2A–2D) and complete resolution of the gluteal mass (Fig. 2E, 2F). There was a decrease in CA 19‐9 after six cycles of pembrolizumab (Fig. 2G).
Figure 2.
Computed tomography (CT) of abdomen of patient 1 after treatment. (A–D): Interval resolution of pancreatic head mass. Positron emission tomography‐CT showing right gluteal mass (green arrow) with fluoro‐deoxyglucose avidity prior to treatment with immunotherapy (E) and after treatment with immunotherapy (F). (G): CA 19‐9 levels during treatment course with immunotherapy. Reference range for CA 19‐9 is <35 U/mL. Abbreviations: CA 19‐9, cancer antigen 19‐9; PD‐1, programmed cell death protein 1.
Patient Story 2
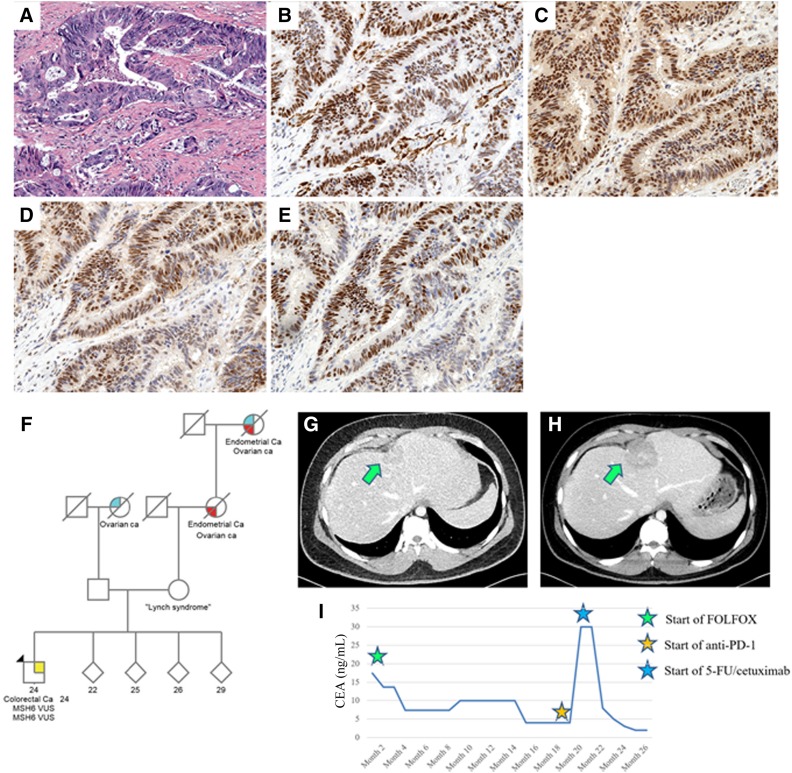
A 24‐year‐old man presented with diarrhea, hematochezia, and weight loss. Colonoscopy showed a circumferential tumor in the rectum, and biopsy revealed adenocarcinoma. CT showed mesenteric lymphadenopathy. He was started on folinic acid, 5‐fluorouracil, and oxaliplatin for 11 cycles with a partial response and subsequently underwent low anterior resection and liver resection. Eight months later, PET/CT scan showed mural thickening at the rectum and new liver lesions suspicious for recurrent disease. Immunohistochemistry of liver biopsy showed intact expression of MLH1, MSH2, MSH6, and PMS2 (Fig. 3A–3E).
Figure 3.
Liver biopsy showing metastatic colorectal carcinoma (A). Mismatch repair protein staining showed intact MLH1 (B), intact MSH2 (C), intact MSH6 (D), and intact PMS2 (E) expression. (F): Family pedigree of patient 2. Proband is denoted by black arrow. Computed tomography of abdomen of patient 2 showing hepatic lesion prior to therapy (G) and after therapy (H). (I): CEA levels during treatment course with cytotoxic chemotherapy and with immunotherapy. Reference range for CEA is <5 ng/mL. Abbreviations: 5‐FU, 5‐fluorouracil; CEA, carcinoembryonic antigen; FOLFOX, folinic acid, 5‐fluorouracil, and oxaliplatin; PD‐1, programmed cell death protein 1; VUS, variants of uncertain significance.
Genetic Testing
Germline testing via Invitae Common Hereditary Cancers Panel (42‐gene panel) showed two known VUS in the MSH6 gene, located in cis on the same chromosome inherited from his mother: [c.3260C>A(p.Pro1087His)] and [c.3312T>A(p.Phe1 104Leu)]. The c.3260C>A germline variant was thought to be disruptive per three predictive algorithms. The two variants in MSH6 were of uncertain significance, but his young age was suspicious for Lynch syndrome. Tumor testing via Foundation One was done after 11 cycles of chemotherapy (at the time of surgery) and showed microsatellite stability by polymerase chain reaction and a low tumor mutational burden of four mutations per megabase. The tumor specimen also showed the two MSH6 variants in cis that were found in the germline. No second‐hit somatic mutation was found in MSH6. Other variants found included APC p.Ile1311fs*4 and TP53 p.Glu286Ala.
Family History
Family history was notable for mother with the same MSH6 VUS but no history of cancer, maternal grandmother with uterine and ovarian cancers, maternal great grandmother with uterine and ovarian cancers, and paternal grandmother with ovarian cancer (Fig. 3F).
Therapy
He was started on pembrolizumab for two cycles. Interval CT showed enlargement of the rectal mass, liver lesions, pelvic lymphadenopathy, peritoneal involvement, and clinical deterioration with worsening pelvic pain requiring hospital admission (Fig. 3G, 3H). Carcinoembryonic antigen (CEA) rose to 30 mg/mL (Fig. 3I). He was thus restarted on cytotoxic chemotherapy with 5‐fluorouracil plus cetuximab and subsequently had a partial response (Fig. 3I).
Molecular Tumor Board
Interpretation of Genotyping and Molecular Results
In this report, we employed the combination of germline testing and tumor molecular testing to show that tumor molecular testing guides anti‐programmed cell death protein 1 (PD‐1) therapy and can provide evidence for pathogenicity of mismatch repair (MMR) variants. In patient 2, tumor testing did not reveal a second MMR hit, MSI, or a high mutational burden, and the patient had rapid clinical deterioration and disease progression on pembrolizumab, all suggesting that this cancer is not driven by a deficient MMR system. In patient 1, the initial bone biopsy specimen had intact stains but upon progression, a gluteal lesion was biopsied revealing PMS2 deficiency, and indeed the tumor testing revealed high mutational burden, MSI, and a second MLH1 mutation. Bone biopsy tissue can be difficult to interpret, and, given the suspicious family history, a second biopsy was important to establish the MMR deficiency, which led to a treatment change. Therefore, all evidence pointed to MMR deficiency driving the cancer, and the patient had a remarkable response to pembrolizumab. The coupling of tumor molecular testing with genomic data is especially pertinent to patients who are found to have a VUS. This approach is becoming more clinically meaningful as we enter an era with vast quantities of readily accessible genomic data that must be analyzed and selectively applied toward precision medicine for individual patients.
The National Comprehensive Cancer Network guidelines recommend screening all colorectal and endometrial cancers for Lynch syndrome via MMR immunohistochemistry or MSI by PCR [8]. Recent reports suggest that around 2%–4% of all advanced cancers may have MSI by next‐generation sequencing testing [9], [10]. Therefore, some investigators have advocated testing all cases of advanced cancers, particularly as this could influence the use of anti‐PD‐1 therapy as well as identify individuals with Lynch syndrome. A deeper understanding of the contribution of germline variants will be important for risk stratification efforts and for the delivery of precision medicine.
This study also emphasizes the need for understanding the functional implications for VUS, as these can play a role in treatment response and patient outcomes when combined with tumor molecular testing. Both patients were found to have variants in MMR genes, but only patient 1 had a tumor that was driven by an MMR variant and conferred responsiveness to immunotherapy. The MLH1 c.374C>A (p.Ala125Glu) variant is a missense variant. Prior studies have shown that in the case of the MLH1 gene, pathogenic missense variants can sometimes lead to intact expression of a nonfunctional MLH1 protein, with the PMS2 protein missing alone [11]. The genomics of patient 1 are consistent with these reports as an isolated PMS2 loss by immunohistochemistry was seen on the gluteal biopsy (Fig. 1F). Patient 2 had two MSH6 variants (located in cis) but neither of them contributed to MMR deficiency, as the tumor was not hypermutated, not microsatellite unstable, and negative for a second hit to MSH6 in the tumor. The patient did not respond to pembrolizumab. Importantly, he eventually responded favorably after starting cytotoxic chemotherapy with an epidermal growth factor receptor monoclonal antibody. One can argue that the favorable response was due to delayed effect of immunotherapy, but this is unlikely given that MMR defects did not drive his tumor, and he had clinical deterioration while on immunotherapy. In the case of patient 1, multiple family members were at risk for malignancies arising in the context of defective MMR, as confirmed in the pedigree.
Implications for Therapy
In summary, this case series underscores the utility of germline testing for MMR variants along with tumor molecular testing, particularly in patients with VUS or strong family history. Identification of an MMR‐deficient tumor can help guide treatment decisions in patients who have progressive or refractory disease and might influence surveillance in family members carrying this variant. The combination of MMR genomics and tumor molecular testing can help delineate the significance of a VUS. A caveat is that, although this approach might have clinical relevance for a patient, we cannot conclude that a variant should be reclassified based on this data alone. It is important for all investigators to contribute clinical and sequencing data to databases such as InSiGHT to assist in reclassification of variants of uncertain significance.
Patient Update
As of the submission of this manuscript, patient 1 continues on cycle 17 of pembrolizumab. Her recent CT showed stable disease, and CA 19‐9 have been stable. Patient 2 continues on cycle 19 of chemotherapy (initially folinic acid, 5‐fluorouracil, irinotecan plus cetuximab, then 5‐fluorouracil plus panitumumab). His recent CT showed stable metastatic disease with no new lesions. CEA has been stable.
Glossary of Genomic Terms and Nomenclature
Microsatellite instability: The presence of instability (insertions and deletions) at microsatellite loci when comparing tumor DNA to normal DNA due to impaired ability of DNA repair proteins to oversee correct replication of DNA.
Mismatch repair: The process by which DNA‐modifying enzymes, such as MLH1, MSH2, MSH6, and PMS2, repair erroneous nucleotide misincorporations in a strand‐specific manner.
Tumor mutational burden: The sum of all genetic alterations within a coding area of the genome, typically presented as the number of mutations per megabase.
Variant of uncertain significance: A genetic alteration at the genomic level (usually a missense mutation) for which the clinical relevance has not been characterized and for which the implications in human health are uncertain.
Footnotes
For Further Reading: Valerie Lee, Adrian Murphy, Dung T. Le et al. Mismatch Repair Deficiency and Response to Immune Checkpoint Blockade. The Oncologist 2016;21:1200–1211.
Implications for Practice: Mismatch repair deficiency has contributed to our understanding of carcinogenesis for the past 2 decades and now identifies a subgroup of traditionally chemotherapy‐insensitive solid tumors as sensitive to PD‐1 blockade. This article seeks to educate oncologists regarding the nature of mismatch repair deficiency, its impact in multiple tumor types, and its implications for predicting the responsiveness of solid tumors to immune checkpoint blockade.
Author Contributions
Conception/design: Sigurdis Haraldsdottir
Provision of study material or patients: Shyam A. Patel, Teri A. Longacre, Uri Ladabaum, Alexandra Lebensohn, Sigurdis Haraldsdottir
Collection and/or assembly of data: Shyam A. Patel, Teri A. Longacre, Uri Ladabaum, Alexandra Lebensohn, Albert Lin, Sigurdis Haraldsdottir
Manuscript writing: Shyam A. Patel, Sigurdis Haraldsdottir
Final approval of manuscript: Shyam A. Patel, Teri A. Longacre, Uri Ladabaum, Alexandra Lebensohn, Albert Lin, Sigurdis Haraldsdottir
Disclosures
The authors indicated no financial relationships.
References
- 1.Lynch HT, Snyder CL, Shaw TG et al. Milestones of Lynch syndrome: 1895‐2015. Nat Rev Cancer 2015;15:181–194. [DOI] [PubMed] [Google Scholar]
- 2.Dudley B, Brand RE, Thull D et al. Germline MLH1 mutations are frequently identified in Lynch syndrome patients with colorectal and endometrial carcinoma demonstrating isolated loss of PMS2 immunohistochemical expression. Am J Surg Pathol 2015;39:1114–1120. [DOI] [PubMed] [Google Scholar]
- 3.Stadler ZK, Battaglin F, Middha S et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next‐generation sequencing panels. JClin Oncol 2016;34:2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llosa NJ, Cruise M, Tam A et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter‐inhibitory checkpoints. Cancer Discov 2015;5:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dudley JC, Lin MT, Le DT et al. Microsatellite instability as a biomarker for PD‐1 blockade. Clin Cancer Res 2016;22:813–820. [DOI] [PubMed] [Google Scholar]
- 6.Le DT, Uram JN, Wang H et al. PD‐1 blockade in tumors in tumors with mismatch‐repair deficiency. N Engl J Med 2015;375:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson BA, Spurdle AB, Plazzer JP et al. Application of a 5‐tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus‐specific database. Nat Genet 2014;46:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Provenzale D, Gupta S, Ahnen DJ et al. Genetic/familial high‐risk assessment: Colorectal Version 1.2016, NCCN Clinical Practice Guidelines in Oncology. JNatl Compr Canc Netw 2016;14:1010–1030. [DOI] [PubMed] [Google Scholar]
- 9.Middha S, Zhang L, Nafa K et al. Reliable pan‐cancer microsatellite instability assessment by using targeted next‐generation sequencing data. JCO Precision Oncol 2017;DOI: 10.1200/PO.17.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonneville R, Krook MA, Kautto EA, et al. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precision Oncol 2017. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosty C, Clendenning M, Walsh MD et al. Germline mutations in PMS2 and MLH1 in individuals with solitary loss of PMS2 expression in colorectal carcinomas from the Colon Cancer Family Registry Cohort. BMJ Open 2016;6:e010293. [DOI] [PMC free article] [PubMed] [Google Scholar]