Bacterial–fungal interactions and oral diseases
The oral cavity contains up to 700 different species of microorganisms, including both bacteria and fungi [1]. The interactions of these communities of different organisms has become of increasing interest, particularly with respect to cross-kingdom interactions involving fungi and bacteria, which have been associated with severity of dental caries (tooth decay) and mucosal infections. Here, we provide a short review of the significance and mechanisms for the interactions between C. albicans and streptococci, the most common fungal and bacterial organisms in the oral cavity [2–5].
Candida albicans and oral streptococci coinfections are associated with enhanced virulence of dental caries and more severe oropharyngeal diseases (Fig 1) [6,7]. Specifically, C. albicans partners with Streptococcus gordonii, S. oralis, and S. sanguinis to enhance bacterial colonization and biofilm formation. In addition, C. albicans becomes more invasive, exacerbating mucosal tissue infection and destruction [8,9]. Mixed C. albicans–bacterial infections are also associated with denture stomatitis, the inflammation of the oral mucosa under dentures. Furthermore, C. albicans–bacterial communities have been clinically found in other oral niches, including periodontal pockets and endodontic canals [6].
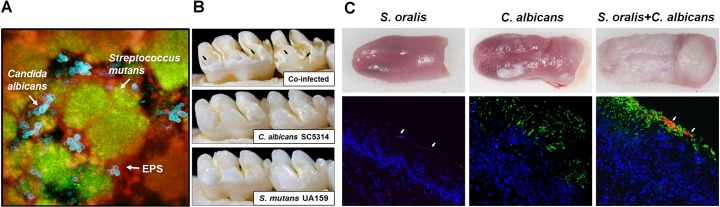
Fig 1. Candida–streptococcal interactions and oral diseases.
A. Confocal fluorescence microscopy images of C. albicans–S. mutans mixed biofilms, illustrating the spatial relationship between C. albicans (blue), S. mutans (green), and exopolysaccharides (red). B. Images of teeth from rats infected with S. mutans, C. albicans, or coinfected. Black arrows indicate severe carious lesions of coinfections in which enamel is missing, which exposes underlying dentin. Such rampant caries was absent in the animals infected by S. mutans or C. albicans alone. C. Fluorescence microscopy images of harvested mouse tongues infected with S. oralis (red, see arrows), C. albicans (green), or both. Coinfection substantially increased bacterial–fungal biofilm accumulation, soft tissue invasion, and inflammatory response. Original images provided by Dr. Anna Dongari-Bagtzoglou; adapted from Sobue T. and colleagues, Methods Mol Biol. 1356:137–52, 2016, with permission. EPS, exopolysaccharides.
C. albicans–streptococcal biofilms are an important contributor to the development of early childhood caries that affects toddler-age children [10–12]. Severe childhood caries is a particularly virulent form of caries that causes extensive and painful tooth destruction, induced by protracted consumption of sucrose containing foods and beverages [11]. Typically, C. albicans is usually absent on teeth of healthy, caries-free children [10]. Furthermore, C. albicans does not interact strongly with S. mutans (a caries-causing pathogen), nor is it an efficient colonizer of mineralized tooth enamel by itself. However, the high level of sucrose in the oral cavity increases the physical coadhesion between the C. albicans and S. mutans as well as tooth surface colonization and drastically enhances the microbial burden, aciduricity, and production of extracellular matrix. Ultimately, the extensive mixed-kingdom and acidogenic biofilm leads to severe tooth decay in a process that can be recapitulated in a rodent model under sugar-rich diets [12].
Fungal and bacterial cell surface adhesins mediate C. albicans interactions with mitis group streptococci on mucosal surfaces
C. albicans physically interacts with mitis group streptococci (MGS) species such as S. gordonii, S. sanguinis, and S. oralis through well-characterized cell wall surface proteins/receptors on both organisms [6,13]. Streptococcal cell surface adhesins SspA and SspB (from the antigen I/II polypeptide family) interact with the C. albicans surface, while ALS and HWP1 adhesins on the fungal cell wall appear to mediate binding to MGS (Fig 2). Specifically, SspB and Als3 directly bind C. albicans and S. gordonii together through the N-terminal domain of Als3 [14]. These interactions may also involve O-mannosyl residues in Als adhesins and other cell wall proteins, such as Sap9 [15,16].
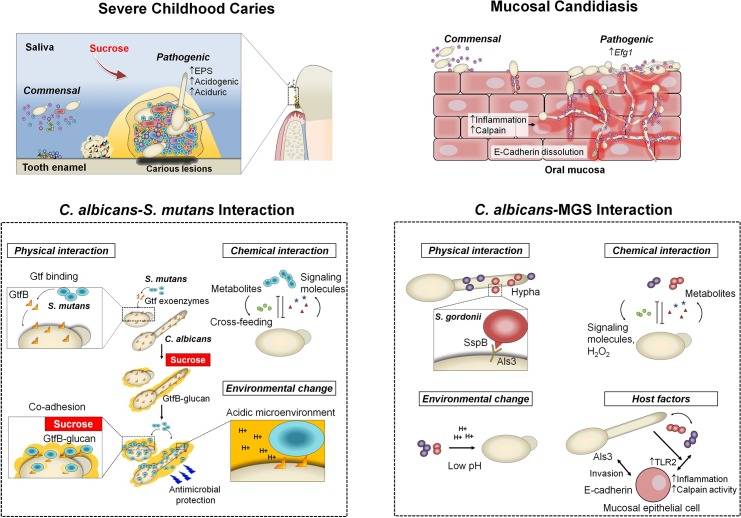
Fig 2. Pathogenic mechanisms of C. albicans–oral streptococcal cross-kingdom interactions.
Complex physical and chemical interactions (including cross-feeding and metabolites exchange) as well as environmental and host factors govern the development of pathogenic bacterial–fungal biofilms, including spatial organization, virulence, and drug protection/resistance. These interactions can be cooperative or competitive to mediate symbiotic, antagonistic, or synergistic relationships, often modulated by host and environmental factors to promote the onset and amplify the severity of the disease. Host diet (dietary sugars, particularly sucrose) promote the interactions between C. albicans and S. mutans by providing a substrate for EPS α-glucans production by streptococcal Gtfs that enhances coadhesion and bacterial–fungal tooth colonization, stimulating cross-kingdom biofilms. This interaction enhances the carriage of the cariogenic pathogen and acid production, while the presence of C. albicans increases EPS matrix production (via Gtf induction and fungal-derived EPS) and biofilm aciduricity, resulting in cariogenic conditions on tooth surface. Likewise, the pathogenic impact of C. albicans interactions with MGS on mucosal surfaces is also influenced by host factors. The interactions of S. oralis with C. albicans on mucosal surfaces cause exacerbated inflammatory responses and increased neutrophilic activity. C. albicans increase the biomass of S. oralis and this leads to increased mucosal TLR2 expression, activating proinflammatory signaling. C. albicans and S. oralis also synergistically increase epithelial μ-calpain activity, a proteolytic enzyme that targets E-cadherin from epithelial junctions. The bacteria influence fungal physiology by promoting hyphal formation via the Efg1 filamentation pathway and expression of secreted aspartyl proteases, which further induces proteolytic degradation of E-cadherin, facilitating invasion and tissue destruction. Efg1; EPS, exopolysaccharides; Gtf, glucosyltransferase; MGS, mitis group streptococci; TLR2.
The consequences of C. albicans–streptococcal interactions have been demonstrated in vivo. C. albicans and S. oralis coinfection results in increased tissue invasion and heightened mucosal inflammatory responses compared with infection by either organism alone [9]. This latter feature appears to be due to increased induction of multiple neutrophil-activating cytokines and up-regulation of TLR2-dependent inflammatory genes as well as enhanced epithelial μ-calpain activity [9,17] (Fig 2).
C. albicans interacts with S. mutans exoenzymes (glucosyltransferases) to promote interspecies biofilm formation on tooth surface
In contrast to MGS, C. albicans does not directly bind to the cariogenic pathogen S. mutans. Instead, glucosyltransferases (Gtfs) secreted by S. mutans promote the generation of an extensive extracellular matrix in the presence of C. albicans, leading to virulent mixed biofilms under sugar-rich conditions of severe childhood caries [18,19]. S. mutans-derived GtfB binds avidly to the C. albicans cell surface and converts sucrose to large amounts of extracellular polysaccharides (EPS) α-glucans on the fungal surface (Fig 2). The EPS provides bacterial binding sites for S. mutans and concurrently allows C. albicans to bind to and colonize teeth [12]. Consequently, the interaction between S. mutans and C. albicans is mediated by both secreted Gtfs and their glucan product. This mechanism is distinct from the more typical cell–cell binding interactions observed between MGS, staphylococci, or bacillus and C. albicans, although the role of Gtfs in MGS species has not been extensively studied [13].
To further understand the mechanistic basis of this “biochemical interaction,” we identified the C. albicans surface molecules to which GtfB binds. C. albicans mutants lacking either N- or O-linked mannans (located on the outer most layer of the fungal cell wall) showed severely reduced GtfB binding. As a result, these mannoprotein-deficient mutants developed poor mixed-species biofilms with S. mutans, showed reduced EPS α-glucans content, and reduced microbial carriage on teeth in vivo [18]. Likewise, S. mutans defective in GtfB does not yield mixed-species biofilms with C. albicans.
In a rodent model, the sucrose-dependent partnership between C. albicans and S. mutans synergistically enhanced bacterial–fungal carriage within plaque biofilm, leading to aggressive onset of tooth decay with rampant carious lesions similar to those found in severe childhood caries [12]. The potential mechanisms for severe caries have been an active subject of research, which entails at least, in part, enhanced microbial carriage, cross-feeding metabolic interactions, and the accumulation of adherent acidic biofilms on teeth facilitated by an EPS matrix surrounding acidogenic–aciduric organisms, as reviewed recently [5,10] (Fig 2).
The role of the C. albicans master regulator Efg1 is required for MGS mixed biofilms but not for S. mutans mixed biofilms
C. albicans Efg1 regulates key hyphae-associated biofilm effector molecules, and homozygous efg1 deletion mutants form only rudimentary biofilms [20]. The nature of the biofilm formed between S. mutans and C. albicans differs from single species C. albicans biofilms because deletion of two transcription factors that are essential for C. albicans biofilms, Efg1 and Bcr1, does not affect the amount of fungal cells in the mixed biofilm. This is likely due to the fact that GtfB binds these mutants with similar afinity compared to wild types and generates robust extracellular α-glucans matrix that allows C. albicans to coadhere and form biofilm with S. mutans [18].
In contrast, efg1ΔΔ mutants are unable to form mixed biofilms with S. oralis [21]. Interestingly, overexpression of the Efg1-regulated adhesin ALS1 partially restores C. albicans–S. oralis biofilm formation to efg1ΔΔ, suggesting that Als1 is a key mediator of this mixed biofilm [21]. Consistent with this notion, C. albicans strains lacking either ALS1 or ALS3 also are deficient for S. oralis mixed biofilm formation [21]. Als3 is also crucial for C. albicans–S. gordonii mixed biofilms through a mechanism involving an interaction between Als3 and SspB [22]. However, C. albicans strains lacking ALS3 are able to form mixed biofilms with S. mutans under sucrose-rich conditions, showing similar levels of fungal cells as those formed with wild-type strains [18]. Thus, the interactions of C. albicans with oral streptococci vary significantly with the specific species of bacteria. Additional studies will be needed to understand how these differences affect the colonization and disease severity at distinct oral niches.
C. albicans–bacterial biofilm relationship is critically dependent on EPS matrix and chemical interactions
The EPS matrix critically influences the relationship between C. albicans and oral streptococci within the biofilm [23]. The matrix provides a scaffold for both surface adhesion and cell-to-cell cohesion while at the same time establishing chemical and nutrient gradients by modulating diffusion [5]. Like most microbes, the matrix of Candida species is comprised of the protein, carbohydrate, nucleic acid, and lipids. In particular, a complex containing mannan and β-glucan constituents sequesters antifungal drugs to protect Candida cells from their effects [23]. Nearly a dozen C. albicans proteins involved in polysaccharide synthesis and modification (e.g., Phr1, Bgl2, Alg11, and Mnn11) are indispensable for production of the matrix [23]. In mixed biofilms, the fungal derived biofilm matrix also protects some prokaryotic pathogens (e.g., Staphylococcus aureus and Escherichia coli) against antibacterial drugs [24]. Similarly, S. mutans-derived α-glucans surrounding fungal cells form an additional “drug-trapping matrix” that prevents uptake of the antifungal fluconazole, reducing killing efficacy [25].
Complex signaling, cross-feeding, and metabolic interactions within the biofilm shape its microenvironment and lead to pathogenic synergies that modulate the onset and severity of oral diseases (Fig 2). A range of signaling/quorum sensing (QS) molecules and other factors appear to facilitate these synergies, including AI-2, peptidoglycan fragments, exoenzymes, and hydrogen peroxide (H2O2) [2–4,13,26]. For example, nutrient byproducts as well as AI-2 signaling and H2O2 from S. gordonii stimulate C. albicans hyphal development within the biofilm [26], while S. oralis presence also activates expression of fungal aspartyl proteases [13]. Conversely, C. albicans can promote streptococcal proliferation by providing growth-stimulating factors and reducing oxygen tension [13,26]. The impact of C. albicans and MGS synergism on the host–pathogen interaction has been demonstrated in vivo whereby mixed biofilm (with S. oralis) growth enhances neutrophil infiltration, leading to increased severity of soft tissue lesions [9,17] (Fig 2). This is distinct from single-species C. albicans biofilms, which are notable for inhibition of neutrophil influx and subsequent function.
A further example of the consequences of the EPS matrix and chemical interactions has been observed between C. albicans and S. mutans. S. mutans converts sucrose to glucose that can be more readily metabolized by C. albicans [27,28, 29]. Importantly, C. albicans activates S. mutans competence [28], virulence genes, and GtfB production via QS molecules such as farnesol [27]. Furthermore, C. albicans secretes its own matrix products such as β-glucan and creates an EPS-producing loop within the S. mutans mixed biofilm [12]. As a result, the organisms enhance the carriage of cariogenic pathogens, biofilm accumulation, and acid production, promoting a localized and persistent acidogenic–aciduric microenvironment that potentiates demineralization of tooth enamel and may explain the synergistic enhancement of caries severity.
Although the cross-kingdom synergies are involved in the pathogenesis of both mucosal and dental diseases, the interactions can also repress functions of the member species to modulate population growth, biofilm structure, community changes, and spatial organization [5] (Fig 2). For example, S. mutans-derived metabolites such as mutanobactin A and fatty acid signaling trans-2-decenoic acid inhibit C. albicans hyphal formation [30,31]. These effects, in addition to the generation of a hyphae-inhibiting, acidic environment, can explain why yeast forms are associated with S. mutans clusters in the deeper layers of mixed biofilms [12]. Furthermore, competence-stimulating peptides released by S. mutans [32] and S. gordonii [33] also disrupt hyphal formation in C. albicans cells. These hyphae-inhibiting effects are consistent with the fact that the Efg1 filamentation pathway is not required for mixed C. albicans–S. mutans biofilm growth and cariogenicity as noted above, suggesting that in contrast to mucosal candidiasis, filamentation may not be a virulence-promoting phenotype in mineralized tissue infections such as dental caries. Paradoxically, farnesol produced by C. albicans, which stimulates S. mutans growth and gtfB expression at low concentrations (25–50 μM), disrupts bacterial growth at high concentrations (>100 μM) [27]. Therefore, a tightly regulated cooperative and antagonistic balance through stimulus-inhibition mechanisms appears to mediate bacterial–fungal coexistence and survival within biofilms, which can become synergistic when conditions are conducive for disease (Fig 2).
In summary, the polymicrobial nature of biofilm-associated oral diseases has been increasingly recognized. Clinical data, together with in vivo studies, provide compelling evidence of the importance of cross-kingdom interactions in the severity of mucosal diseases and dental caries. Complex cell–cell and cell–EPS matrix interactions, spatial organization, and chemical/metabolic factors modulate biofilm development and virulence. These fungal–bacterial interactions are facilitated by host factors (immunity, diet, and salivary function) to modify the local microenvironment and promote oral diseases. Elucidating how bacterial–fungal interactions occur spatiotemporally (cooperative, competitive, or both simultaneously) to mediate symbiotic, antagonistic, or synergistic states may shed new light into the pathogenic mechanisms and identify more effective therapeutic targets. Since cross-kingdom biofilms exist throughout the gastrointestinal tract, principles and molecules that emerge from these studies may lead to novel approaches to prevent and eradicate other intractable polymicrobial biofilms at various clinical niches.
Acknowledgments
The authors are grateful to Dr. Anna Dongari-Bagtzoglou for critically reading the manuscript and providing the original images in Fig 1. We are also thankful to Dr. Dongyeop Kim for the preparation of the figures, particularly the diagram in Fig 2.
Funding Statement
This work was supported by the following grants from the National Institutes of Health: 1R01DE018023 (HK), 1R01DE025220 (HK), R01AI073289 (DRA), and 1R01AI133409 (DJK). The funders had no role in study design, data collections and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marsh PD, Zaura E. Dental biofilm: ecological interactions in health and disease. J Clin Periodontol. 2017;44: S12–S22. 10.1111/jcpe.12679 [DOI] [PubMed] [Google Scholar]
- 2.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial–fungal interactions. Nat Rev Microbiol. 2010;8: 340–349. 10.1038/nrmicro2313 [DOI] [PubMed] [Google Scholar]
- 3.Harriott MM, Noverr MC. Importance of Candida–bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19: 557–563. 10.1016/j.tim.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohse MB, Gulati M, Johnson AD, Nobile CJ. Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol. 2018;16: 19–31. 10.1038/nrmicro.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen WH, Burne RA, Wu H, Koo H. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol. 2018;26: 229–242. 10.1016/j.tim.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donnell LE, Millhouse E, Sherry L, Kean R, Malcolm J, Nile CJ, et al. Polymicrobial Candida biofilms: friends and foe in the oral cavity. FEMS Yeast Res. 2015;15: 1–14. 10.1093/femsyr/fov077 [DOI] [PubMed] [Google Scholar]
- 7.Allison DL, Willems HME, Jayatilake JAMS, Bruno VM, Peters BM, Shirtliff ME. Candida–Bacteria Interactions: Their Impact on Human Disease. Microbiol Spectr. 2016;4:VMBF-0030-2016. 10.1128/microbiolspec.VMBF-0030-2016 [DOI] [PubMed] [Google Scholar]
- 8.Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80: 620–32. 10.1128/IAI.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Sobue T, Bertolini M, Thompson A, Dongari-Bagtzoglou A. Streptococcus oralis and Candida albicans Synergistically Activate μ-Calpain to Degrade E-cadherin From Oral Epithelial Junctions. J Infect Dis. 2016;214: 925–34. 10.1093/infdis/jiw201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao J, Huang X, Alkhers N, Alzamil H, Alzoubi S, Wu TT, et al. Candida albicans and Early Childhood Caries: A Systematic Review and Meta-Analysis. Caries Res. 2018;52: 102–112. 10.1159/000481833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajishengallis E, Parsaei Y, Klein MI, Koo H. Advances in the microbial etiology and pathogenesis of early childhood caries. Mol Oral Microbiol. 2017;32: 24–34. 10.1111/omi.12152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai C-H, et al. Symbiotic Relationship between Streptococcus mutans and Candida albicans Synergizes Virulence of Plaque Biofilms In Vivo. Infect Immun. 2014;82: 1968–1981. 10.1128/IAI.00087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29: 99–116. 10.1111/omi.12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bamford C V, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 2015;161: 18–29. 10.1099/mic.0.083378-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutton LC, Nobbs AH, Jepson K, Jepson MA, Vickerman MM, Aqeel Alawfi S, et al. O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. MBio. 2014;5: e00911 10.1128/mBio.00911-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutton LC, Jenkinson HF, Lamont RJ, Nobbs AH. Role of Candida albicans secreted aspartyl protease Sap9 in interkingdom biofilm formation. Pathog Dis. 2016;74: ftw005. 10.1093/femspd/ftw005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, et al. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 2014;16: 214–231. 10.1111/cmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017;13: e1006407 10.1371/journal.ppat.1006407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, et al. Role of Glucosyltransferase B in Interactions of Candida albicans with Streptococcus mutans and with an Experimental Pellicle on Hydroxyapatite Surfaces. Appl Environ Microbiol. 2011;77: 6357–6367. 10.1128/AEM.05203-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A Recently Evolved Transcriptional Network Controls Biofilm Development in Candida albicans. Cell. 2012;148: 126–138. 10.1016/j.cell.2011.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Sobue T, Bertolini M, Thompson A, Vickerman M, Nobile CJ, et al. S. oralis activates the Efg1 filamentation pathway in C. albicans to promote cross-kingdom interactions and mucosal biofilms. Virulence. 2017;8: 1602–1617. 10.1080/21505594.2017.1326438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Interaction of Candida albicans Cell Wall Als3 Protein with Streptococcus gordonii SspB Adhesin Promotes Development of Mixed-Species Communities. Infect Immun. 2010;78: 4644–4652. 10.1128/IAI.00685-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell KF, Zarnowski R, Sanchez H, Edward JA, Reinicke EL, Nett JE, et al. Community participation in biofilm matrix assembly and function. Proc Natl Acad Sci. 2015;112: 4092–4097. 10.1073/pnas.1421437112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong EF, Tsui C, Kucharíková S, Andes D, Van Dijck P, Jabra-Rizk MA. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. MBio. 2016;7: e01365–16. 10.1128/mBio.01365-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Liu Y, Benhamou RI, Sanchez H, Hwang G, Fridman M, et al. Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 2018; 1427–1442. 10.1038/s41396-018-0113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nobbs AH, Jenkinson HF. Interkingdom networking within the oral microbiome. Microbes Infect. 2015;17: 484–492. 10.1016/j.micinf.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim D, Sengupta A, Niepa THR, Lee BH, Weljie A, Freitas-Blanco VS, et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep. 2017;7: 1–14. 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sztajer H, Szafranski SP, Tomasch J, Reck M, Nimtz M, Rohde M, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014;8: 2256–2271. 10.1038/ismej.2014.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi DCP, Gleason JE, Sanchez H, Schatzman SS, Culbertson EM, Johnson CJ, et al. Candida albicans FRE8 encodes a member of the NADPH oxidase family that produces a burst of ROS during fungal morphogenesis. PLoS Pathog. 2017;13: e1006763 10.1371/journal.ppat.1006763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyner PM, Liu J, Zhang Z, Merritt J, Qi F, Cichewicz RH. Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org Biomol Chem. 2010;8: 5486 10.1039/c0ob00579g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vílchez R, Lemme A, Ballhausen B, Thiel V, Schulz S, Jansen R, et al. Streptococcus mutans Inhibits Candida albicans Hyphal Formation by the Fatty Acid Signaling Molecule trans-2-Decenoic Acid (SDSF). ChemBioChem. 2010;11: 1552–1562. 10.1002/cbic.201000086 [DOI] [PubMed] [Google Scholar]
- 32.Jarosz LM, Deng DM, Van Der Mei HC, Crielaard W, Krom BP. Streptococcus mutans competence-stimulating peptide inhibits candida albicans hypha formation. Eukaryot Cell. 2009;8: 1658–1664. 10.1128/EC.00070-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jack AA, Daniels DE, Jenkinson HF, Vickerman MM, Lamont RJ, Jepson MA, et al. Streptococcus gordonii comCDE (competence) operon modulates biofilm formation with Candida albicans. Microbiology. 2015;161: 411–421. 10.1099/mic.0.000010 [DOI] [PMC free article] [PubMed] [Google Scholar]