Abstract
Retinitis pigmentosa (RP) is an inherited degenerative disease causing severe retinal dystrophy and visual impairment mainly with onset in infancy or adolescence. Targeted next-generation sequencing (NGS) has become an efficient tool to encounter the enormous genetic heterogeneity of diverse retinal dystrophies, including RP. To identify disease-causing mutations in unselected, consecutive RP patients, we conducted Sanger sequencing of genes commonly involved in the suspected genetic RP subtype, followed by targeted large-panel NGS if no mutation was identified, or NGS as primary analysis. A high (70%) detection rate of disease-causing mutations was achieved in a large cohort of 116 unrelated patients. About half (48%) of the solved RP cases were explained by mutations in four genes: RPGR, EYS, PRPF31 and USH2A. Overall, 110 different mutations distributed across 30 different genes were detected, and 46 of these mutations were novel. A molecular diagnosis was achieved in the majority (82–100%) of patients if the family history was suggestive for a particular mode of inheritance, but only in 60% in cases of sporadic RP. The diagnostic potential of extensive molecular analysis in a routine setting is also illustrated by the identification of unexpected genotype-phenotype correlations for RP patients with mutations in CRX, CEP290, RPGRIP1, MFSD8. Furthermore, we identified numerous mutations in autosomal dominant (PRPF31, PRPH2, CRX) and X-linked (RPGR) RP genes in patients with sporadic RP. Variants in RP2 and RPGR were also found in female RP patients with apparently sporadic or dominant disease. In summary, this study demonstrates that massively parallel sequencing of all known retinal dystrophy genes is a valuable diagnostic approach for RP patients.
Introduction
Retinitis pigmentosa (RP) is one of the most common forms of inherited retinal degenerations, affecting about 1:3.000 individuals.[1, 2] It is characterized by a primary rod photoreceptor degeneration and consecutive cone photoreceptor death, leading to night blindness and subsequent progressive vision loss. Legal blindness at working age is common in RP patients, with devastating professional and social implications for the patients.[1, 2] Although typical characteristics of RP have been described, there is broad phenotypic variability, regarding both clinical presentation and age of onset. RP is genetically heterogeneous, with more than 60 disease-causing RP genes reported so far (RetNet, https://sph.uth.edu/retnet/). With novel upcoming therapeutic options such as gene therapy, the identification of the disease-causing mutations has gained importance.[3]
Diagnostic genotyping in RP has become very efficient with the implementation of next-generation sequencing (NGS),[4–7] enabling parallel sequencing of all known RP (and, if needed, many more) genes which improves our understanding of the disorder’s pathogenesis.
Here, we report unexpected molecular findings and novel genotype-phenotype correlations in a cohort of 116 consecutive and unrelated patients seen in a retinal dystrophy clinic of a German tertiary referral center. In addition, the application of targeted NGS of all known RP genes provides insight into the mutational spectrum of this representative cohort.
Methods
Patients
This retrospective single-center cross-sectional study included 116 consecutive and unrelated patients who were investigated at the Department of Ophthalmology, University of Bonn, Germany. After clinical diagnosis of RP, molecular screening was performed. The study was in adherence with the declaration of Helsinki. Institutional review board approval (Ethics Committee, Medical Faculty, University of Bonn, Germany) and patients' informed consent were obtained. All patients underwent genetic counseling, which included family history assessment. Asymptomatic relatives of index patients received genetic counselling prior to mutation testing.
Image acquisition and functional testing
Clinical assessment included standardized anterior segment and dilated fundus examination, best corrected visual acuity (BCVA), and visual field testing (30–2 threshold program; HFA II; Carl Zeiss Meditec, Dublin, CA, USA), and, in selected cases, electroretinography (ERG). Retinal imaging consisted of spectral domain optical coherence tomography (OCT), fundus autofluorescence (AF) imaging (both, Spectralis HRA+OCT, Heidelberg Engineering, Heidelberg, Germany), fundus photography (Zeiss, Visucam, Oberkochen, Germany) and wide-field fundus imaging (Optos PLC, Dunfermline, United Kingdom).
Molecular genetic analysis
Genomic DNA was extracted from blood lymphocytes by a standard protocol. A two-tier procedure was implemented for patients suggestive for autosomal recessive or sporadic RP. A step-wise molecular screening was chosen, with initial Sanger sequencing of genes commonly involved in the pathogenesis of the suspected genetic RP subtype (EYS and RP1 for autosomal recessive RP, PRPF31 for autosomal dominant RP).[8] If no mutation was identified in this initial sequencing of singular genes, targeted NGS on an Illumina Hiseq1500 system was carried out for the RP genes known at the respective time of analysis (see S1 Table) after enrichment using NimbleGen sequence capture technology (as described previously[8]). Importantly, the NGS panels also contained the genes for clinically overlapping conditions such as cone/cone-rod dystrophies, Leber’s congenital amaurosis (LCA) and syndromes with retinal dystrophies, allowing for an extended genetic assessment if no mutation was found in genes previously associated with RP, without need for additional experimental efforts. To detect X-linked RP-causing mutations, we added NGS of amplicons comprising RPGRORF15 to panel-NGS of the remaining exons of RPGR and RP2. Based on our step-wise molecular screening, Sanger sequencing and targeted NGS identified disease-causing mutations in 20 and 61 cases, respectively. Verification of mutations identified in NGS and segregation analyses were carried out by PCR and subsequent Sanger sequencing.
To determine the most likely inheritance mode, pedigrees were generated based on the patients’ family history including at least three generations. Autosomal recessive inheritance was assumed in case of parental consanguinity and/or if only siblings were affected. Autosomal dominant inheritance was assumed if there was a positive family history for at least three successive generations, and likely autosomal dominant if different but not successive generations were affected in absence of known consanguinity (e.g., assuming reduced penetrance, or because a linking individual died early or lost contact). X-linked inheritance was assumed in families with only males being severely affected and at least considered if male-to-male transmission was lacking in families with RP patients in different generations. RP was categorized as sporadic in case of negative family history. If no family history was available, e.g. because the patient was adopted and without contact with his biological parents, no candidate inheritance mode was defined.
Variants were filtered against dbNSFP v2.0, dbSNP v137, gnomAD (exomes) and the Human Gene Mutation Database (HGMD Professional 2017.3). The cut-off for the maximum minor allele frequency (MAF) was set to 1%.[9] Nonsense, frameshift, large deletions and canonical splice site variants were regarded pathogenic. Rare non-synonymous single nucleotide variations were considered likely pathogenic when at least half of the algorithms of used in silico prediction software tools predicted that the variant is probably damaging and when it was predicted as conserved with conservation prediction algorithms. Functional predictions were carried out using SIFT, PolyPhen2, MutationTaster, MutationAssessor, FATHMM, LRT, VEST, CADD, PROVEAN and DANN. Splice sites were predicted with AdaBoost and RF. Assessment of conservation was done by PhyloP, GERP++, PhastCons, SiPhy, Grantham Distance and BLOSUM62.
Results and discussion
Approximately 99% of all coding exons were covered at least 20-fold. Disease-causing mutations were identified in 81 (70%) out of the 116 analyzed patients, a high diagnostic yield compared to previous reports using targeted NGS in RP patients (S2 and S3 Tables).[4–8, 10–20]. The wide range of reported diagnostic yields in cohorts >50 patients (25%-80%)[4, 7, 8, 10–17] may be due to differences in cohort sizes, populations, inclusion criteria, differences in clinical assessment, NGS platforms, and bioinformatic pipelines. Cases without mutations in the genes analysed herein can be due to causative variants in non-coding regions of these genes or in genes not yet known to underlie retinal degeneration, or–less likely–due to mutations in regions below optimal coverage. Overall, 110 different mutations distributed across 30 different genes were detected in this study, and 46 of these mutations were novel at the time of molecular diagnosis (Fig 1, S2 and S4 Tables).
Fig 1. Distribution of mutations in consecutive, unrelated RP patients.
Genes mutated in patients who were reversely phenotyped (based on the genetic findings) are indicated by asterisk. The number of patients with mutations in the different genes is displayed on the right. Black, autosomal recessive. Blue, autosomal dominant. Green, X-linked.
The clinical classification was revised in four patients who had variants in genes usually associated with an early macular degeneration (CERKL, PROM1 and ABCA4). They presented with very late disease stages, preventing a clear classification of the initial phenotype as cone-rod or rod-cone dystrophy. However, re-evaluations of these patients’ histories were compatible with retinal dystrophies that initially affected the central retina and/or the cone system. Classification may be particularly difficult in patients with CERKL mutations, as their initial symptoms may manifest in dim light conditions, although their retinal phenotype may indicate a dystrophy primarily affecting the central retina.[21] Thus, a combination of comprehensive clinical phenotyping and molecular testing improves the accuracy of diagnoses in genetically and clinically heterogeneous diseases.
About half (48%) of the remaining 77 solved RP cases were explained by mutations in four genes: RPGR (n = 12, 16%), EYS (n = 10, 13%), PRPF31 (n = 8, 10%) and USH2A (n = 7, 9%). Based on the genetic findings, inheritance turned out to be autosomal recessive in 56% (n = 45), autosomal dominant in 26% (n = 21), and X-linked in 19% (n = 15) of patients. If the family history was highly suggestive for a particular mode of inheritance (n = 45), a molecular diagnosis was achieved in 82–100%, then confirming the assumed inheritance (Table 1). The mutation detection rate was lower (60%; 42 out of 70 patients) in cases of sporadic RP (see below), similar to a previous reported cohort of patients with macular and cone/cone-rod dystrophies.[22]
Table 1. More of inheritance based on family history, on genetic results, and mutation detection rate for each group.
| Assumed inheritance based on family history | n | Inheritance based on mutation | n | Mutation detection rate % |
|---|---|---|---|---|
| Autosomal recessive | 24 | arRP | 20 | 83% |
| Autosomal dominant (n = 15) or likely autosomal dominant (n = 2) | 17 | adRP | 13 | 82% |
| XLRP | 1 | |||
| X-linked | 4 | XLRP | 4 | 100% |
| Unclassified | 1 | adRP | 1 | 100% |
| Sporadic | 70 | arRP | 25 | 60% |
| adRP | 7 | |||
| XLRP | 10 |
In the following, unexpected findings identified in 21 (27%) out of the 77 solved RP patients are described in detail (variants displayed in Table 2). These include a high proportion of autosomal dominant or X-linked mutations in patients with sporadic RP, novel or uncommon genotype-phenotype correlations (with no mutations in genes knowingly associated with the respective phenotype), and X-linked mutations in females with apparently sporadic or dominant RP.
Table 2. Molecular findings in patients with unexpected genotyping results.
| ID (#) | Panel | Gender (m/f) | Gene | Zygosity | Exon/Intron (IVS) | Nucleotide | Protein | Segregation analysis |
Reference |
|---|---|---|---|---|---|---|---|---|---|
| 36 | II | f | CEP290 | Heterozygous | Exon 12 Intron 26 |
c.982C>T c.2991+1655A>G |
p.Gln328* splice site/p.Cys998* |
yes | novel [8, 23] |
| 37 | II | m | MFSD8 | Homozygous | Exon 13 | c.1445G>C | p.Arg482Pro | yes | novel |
| 39 | II | m | RPGRIP1 | Homozygous | Exon 19 | c.3100_3238del139 | p.Gln1034Thrfs*23 | no | novel |
| 46 | I | f | PRPF31 | Heterozygous | Exon 8 | c.839T>G | p.Val280Gly | yes | novel |
| 47 | II | m | PRPF31 | Heterozygous | Exon 1–3 | deletion of exons 1–3 | deletion of exons 1–3 | yes | [8, 24] |
| 48 | III | f | PRPF31 | Heterozygous | Intron 7 | c.698-1G>A | splice | no | [25] |
| 49 | I | f | PRPF31 | Heterozygous | Exon 1–5 | deletion of exons 1–5 | deletion of exons 1–5 | yes | [8, 24] |
| 50 | Sanger | m | PRPF31 | Heterozygous | Exon 8 | c.816_830delCTACATCTACCACAG | p.Tyr273_Ser277del | no | novel |
| 63 | III | m | CRX | Heterozygous | Exon 3 | c.122G>A | p.Arg41Gln | yes | [26] |
| 66 | IV | f | PRPH2 | Heterozygous | Exon 1 | c.422A>G | p.Tyr141Cys | no | [27] |
| 68 | III | f | RPGR | Heterozygous | ORF15 | c.2442_2445del AGAG | p.Gly817Lysfs*2 | no | [28] |
| 69 | ORF15-Amplicon | m | RPGR | Hemizygous | ORF15 | c.2452G>T | p.Glu818* | no | novel |
| 71 | IV | m | RPGR | Hemizygous | Exon 8 | c.917A>C | p.His306Pro | no | novel |
| 72 | ORF15-Amplicon | m | RPGR | Hemizygous | ORF15 | c.2630delA | p.Glu877Glyfs*212 | yes | novel |
| 73 | IV | m | RPGR | Hemizygous | Exon 9 | c.1006A>T | p.Asn336Tyr | no | novel |
| 74 | ORF15-Amplicon | m | RPGR | Hemizygous | ORF15 | c.2426_2427delAG | p.Glu809Glyfs*25 | no | [28] |
| 75 | III | f | RPGR | Heterozygous | ORF15 | c.2405_2406delAG | p.Glu802Glyfs*32 | no | [28] |
| 76 | IV | m | RPGR | Hemizygous | ORF15 | c.3034delG | p.Glu1012Lysfs*77 | no | novel |
| 77 | V | m | RPGR | Hemizygous | Exon 3 | c.194G>T | p.Gly65Val | no | [29–31] |
| 79 | III | f | RP2 | Heterozygous | Exon 3 | c.829dupG | p.Ala277Glyfs*11 | no | novel |
| 80 | I | m | RP2 | Hemizygous (47,XXY karyotype, skewed X inactivation) |
Exon 2 | c.630_633delTCGT | p.Arg211Phefs*26 | no | novel |
High proportion of autosomal dominant and X-linked inheritance in sporadic RP
In the majority of patients with sporadic RP, the retinopathy is autosomal recessively inherited, usually with a small recurrence risk in the offspring. Although this was confirmed by our findings, it is noteworthy that X-linked (n = 10) and autosomal dominant (n = 7) mutations accounted for disease in 24% of the sporadic cases (Table 1).
Similar rates of X-linked RP have been reported for overall sporadic (4–6%,[10, 32]) or for male sporadic cases (15%-30%[33, 34]). Of note, two sporadic female patients with X-linked RP were identified in this study (see below). Most autosomal dominant mutations in sporadic RP patients were identified in PRPF31 (n = 5, #46–50 in S2 Table). Only one patient carried a CRX mutation (see below, patient #63), and one patient had a previously described assumingly pathogenic variant in PRPH2 (c.422A>G, p.Tyr141Cys; patient #67; variant not carried by her mother, no further family members were available for segregation analysis). Previously it has been shown that mutations in PRPH2 cause highly variable and mild RP phenotypes, and index patients may be erroneously classified as simplex cases because of undiagnosed family members with mild disease.[35] Incomplete penetrance of PRPF31-associated RP is a well-documented phenomenon.[36] Our study underlines the high rate of non-penetrance in PRPF31 mutations and/or that the family history of relatively small pedigrees is less reliable in identifying such low- or non-penetrant dominant mutations.[36, 37] Segregation analysis for PRPF31 mutations was possible in three families, revealing a de novo mutation in one patient (#47, S2 Table). Healthy individuals with PRPF31 mutations were fully examined and showed no morphological fundus changes (including peripheral AF recordings). Of note, they had slightly reduced or borderline low responses on scotopic ERG testing, indicating that PRPF31 mutation carriers may exhibit reduced retinal function at subclinical level.[38, 39]
RP caused by a CRX mutation previously associated with cone-rod dystrophy
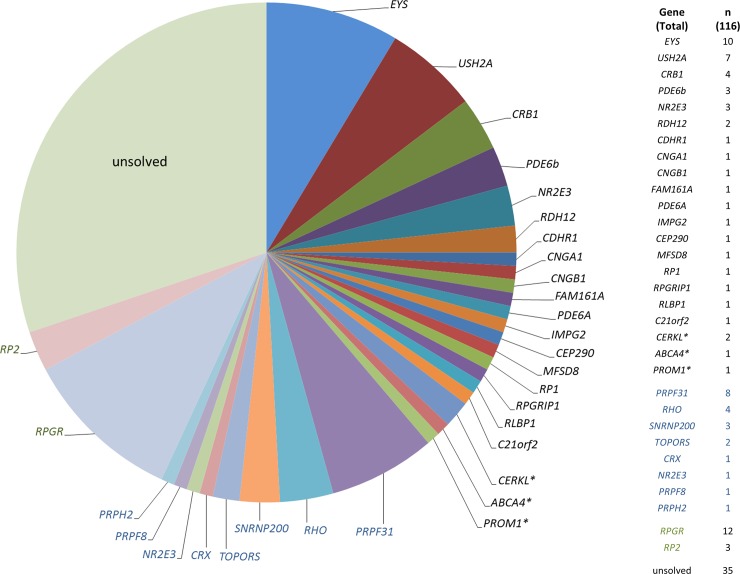
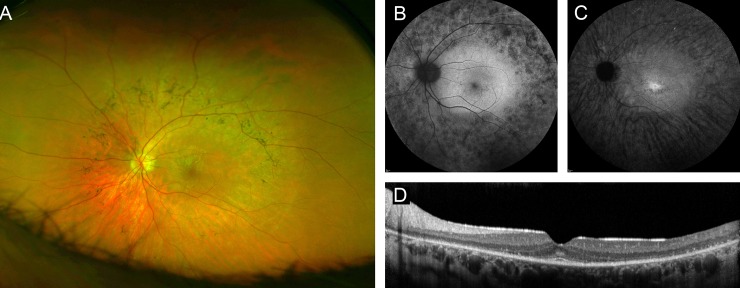
An 83-year old patient (#63, S2 Table). without visual limitations throughout his professional life reported impaired dark adaption, nyctalopia and glare as first symptoms in his early 8th decade of life. Funduscopy showed typical RP fundus changes (Fig 2). He also noted progressive loss of visual acuity over the past 7 years, and visual acuity was now 20/200 in the right eye and hand movements on the left eye. The visual field was severely constricted, and ERG responses were not detectable. Although there was no known affected family member, exclusion of a dominant inheritance of this late onset RP was not possible because most of his relatives died at relatively young age. Genetic testing showed a heterozygous missense mutation (c.122G>A, p.Arg41Gln) in exon 3 of the CRX gene. The same mutation had previously been described in families with autosomal dominant late-onset cone-rod dystrophy,[26] indicating a novel genotype-phenotype correlation for this particular mutation.
Fig 2. Right eye of a patient with RP due to an CRX mutation.
(A) widefield false-color image, fundus AF with (B) 488 nm and (C) 787 nm excitation light, and (D) spectral-domain optical coherence tomography. Only one eye is shown due to high symmetry between eyes.
Non-syndromic RP caused by mutations in CEP290
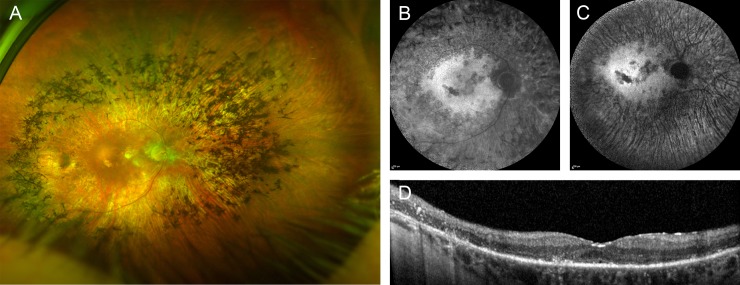
A female patient (#36, S2 Table) with reduced night vision and dark adaption since childhood had graduated from university without visual aids. Starting in her mid to late twenties, she had noticed progressive vision loss and was diagnosed with RP at the age of 27. Now 70 years-old, visual acuity was reduced to light perception in both eyes. Clinical examination showed widespread atrophy of the outer retina and bone-spicule pigmentations (Fig 3). Targeted NGS revealed heterozygosity for a nonsense mutation (c.982C>T, p.Gln328*) in exon 12 and a deep-intronic mutation (c.2991+1655A>G) in intron 26 that creates a strong splice-donor site in CEP290 and a premature stop codon, p.Cys998*. Compound-heterozygosity was deduced because the patient’s son carried the nonsense, but not the splice site mutation.
Fig 3. Left fundus of a 70-year-old women with RP caused by compound-heterozygous CEP290 mutations.
(A) widefield fundus imaging, (B, C) near infrared reflectance imaging, (D) spectral-domain optical coherence tomography. Only one eye is shown due to high symmetry between eyes.
Biallelic mutations in CEP290 cause several syndromic ciliopathies and non-syndromic LCA.[13, 23, 40–42] Of note, our patient had no abnormalities in motor or cognitive development, renal cysts or other morphological abnormalities indicating a CEP290-associated syndrome. Although an association of CEP290 mutations with relatively milder non-syndromic retinal phenotypes has been described in a few previous reports, vision loss in those patients either occurred in the first decade of life or the history of vision loss was not described in detail.[4, 7, 8, 16, 43, 44] Only one recent comparable study reported a patient with an apparently similar disease course.[45] The latter and our case also show that retinal disease due to CEP290 mutations is not necessarily associated with poor cone function,[46] since both patients had normal visual acuity apart from reduced night vision for at least the first two decades of life. It is likely that the phenotypic continuum of CEP290-related non-syndromic retinal dystrophies can be attributed to modifying retina-relevant variants elsewhere in the genome. The identification of CEP290-related RP, a second non-syndromic phenotype associated with mutations in this gene, further supports the categorization of Joubert syndrome (JBTS) genes as strong candidates for isolated retinopathies. OFD1 and, very recently, AHI1, have also been shown to cause both JBTS and isolated retinal degeneration.[47, 48]
Non-syndromic RP caused by RPGRIP1 mutations
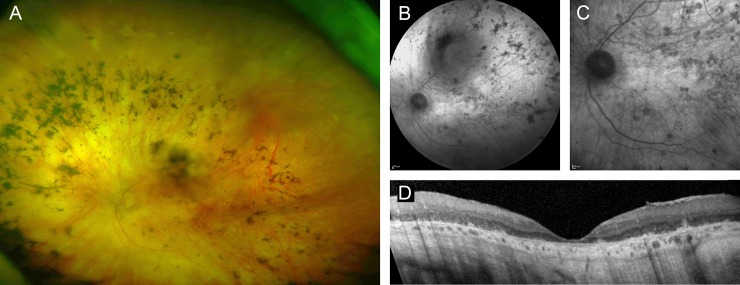
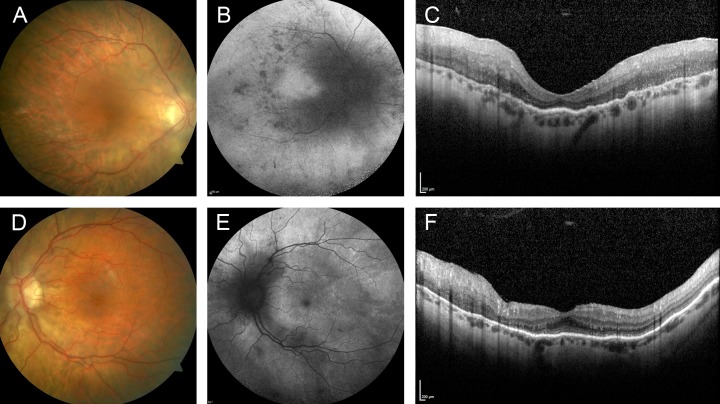
A 42-year-old male patient (#39, S2 Table) was diagnosed with sporadic RP at the age of 13 years. He reported nyctalopia, dark adaption problems and glare since childhood. At the age of 37 years, he started using visual aids. He underwent cataract surgery, and visual acuity now was 20/50 in the right and 20/63 in the left eye. Visual fields were severely constricted and full-field ERG showed no detectable responses. Funduscopy showed alterations characteristic for RP, with widespread retinal pigment epithelial and photoreceptor atrophy, and bone spicule pigmentations (Fig 4). Targeted NGS revealed a novel homozygous deletion (c.3100_3238del139, p.Gln1034Thrfs*23) of exon 19 of RPGRIP1, resulting in a frameshift predicted to result in unstable mRNA or protein truncation.
Fig 4. Right eye of a patient with RP and a homozygous RPGRIP1 mutation.
Fundus color image (A), fundus AF with (B) 488 nm and (C) 787 nm excitation light, (D, E) spectral-domain optical coherence tomography. Only one eye is shown due to high symmetry between eyes.
Mutations in RPGRIP are a known cause of LCA [42, 49, 50] and juvenile RP.[51–53] Besides its uncommon benign disease course, the presented case is exceptional because visual acuity was good in early life and remained relatively stable until an age beyond 40 years. This contrasts with previous observations which had suggested severe cone functional loss despite relative sparing of the fovea anatomy on OCT images.[53]
Non-syndromic RP caused by mutations in MFSD8
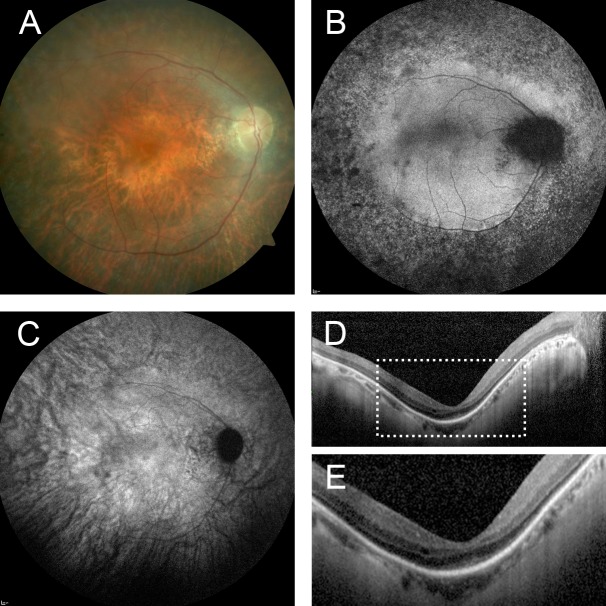
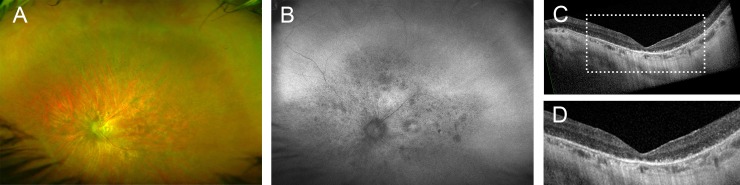
A 35-year old patient (#37, S2 Table) who initially noticed nyctalopia, dark adaption problems and glare at the age of 31 years showed fundus changes typical for RP (Fig 5). Best corrected visual acuity was 20/40 and 20/25, and there was severe concentric constriction of the visual fields. ERG showed no detectable responses. Genetic testing identified a homozygous variant (c.1445G>C, p.Arg482Pro) in exon 13 in the MFSD8 gene. In silico assessment with various programs strongly supports the categorization as disease-causing mutation, as did the results from segregation analysis: The index patient’s likewise affected brother and a second brother who reported difficulties seeing in dim light (not examined) carried the MFSD8 missense mutation in homozygous state, whereas a sister with normal vision carried the wild-type allele.
Fig 5. Left eye of a patient with RP and a homozygous MFSD8 mutation.
(A) widefield false-color image, fundus AF with (B) 488 nm and (C) 787 nm excitation light, (D) spectral-domain optical coherence tomography. Only one eye is shown because there was high similarity of both eyes.
MFSD8 mutations are known to cause variant late-infantile neuronal ceroid lipofuscinosis (vLINCL; CLN7), an early-onset severe lysosomal storage disorder with intralysosomal accumulation of autofluorescent lipopigments, presenting with seizures, mental regression and retinopathy, which was either not further specified [54–58] or suggestive for RP.[59] Recently, MFSD8 mutations have been described in families with non-syndromic autosomal recessive macular dystrophy with central cone involvement, isolated maculopathy and generalized retinopathy.[60, 61] Similarly, the patient in our study showed no neurologic features typical for vLINCL, confirming MFSD8 as a non-syndromic retinopathy gene. The MFSD8 genotype-phenotype correlation proposed earlier,[61] with vLINCL resulting if both mutations are severe (in particular truncating mutations), whereas milder mutations (such as hypomorphic missense mutations) on at least one gene copy result in non-syndromic retinal degeneration, likely also applies in the family reported herein.
RP2 and RPGR mutations in female patients with apparently sporadic or dominant RP
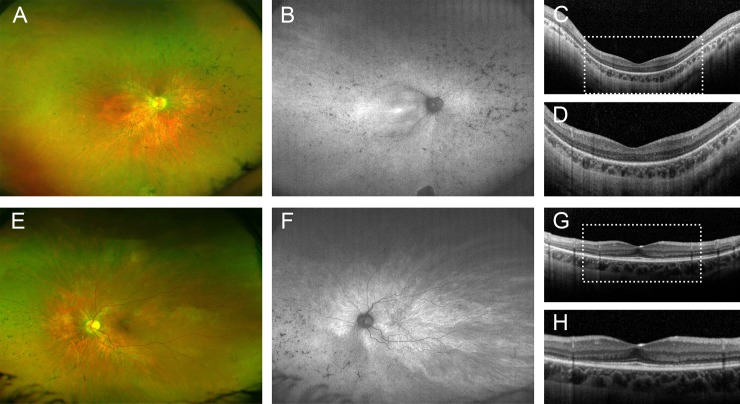
Patient A (RP2): In this 16-years-old young woman (#80, S2 Table), unilateral reduced vision was first recognized at the age of four years, but no further examinations had been initiated at that time. When examined at the age of 12 years, she reported impaired central vision, but no nyctalopia. Visual acuity in the right and left eye was 20/63 and 20/20, respectively. The right eye was emmetropic and the left eye was myopic (spherical equivalent -3,5 dpt). The visual fields were severely constricted in the right eye, and there was a nasal superior visual field loss in the left eye. The ERG showed extinct rod responses in her right eye, while responses in her left eye were severely reduced. When she was examined at the age of 16, visual acuity had deteriorated only in the right eye (now 20/100). Fundus examination revealed narrowed vessels, outer retinal atrophy and bone spicule pigmentations, all much more pronounced in the right eye. In addition, the left eye showed a tapetal-like reflex (Fig 6). Fundus AF confirmed the asymmetry and revealed a pattern of radial lines extending into the fundus periphery in the left eye, which is a characteristic finding in carriers of X-linked RP. NGS analysis identified a one base-pair duplication (c.829dupG, p.Ala277Glyfs*11) in exon 3 in the RP2 gene. No retinal disease was known in other family members, assessment of the parental retinal phenotype was not possible, and samples for segregation analysis were not available.
Fig 6. Phenotype of an RP patient with an RP2 mutation.
A-D right eye, E-H left eye Right eye: (A, E) widefield fundus imaging and (B, F) widefield fundus autofluorescence, (C, D, G, H) spectral-domain optical coherence tomography.
Patient B (RPGR): This 39-years-old myopic (spherical equivalent -5 dpt in the right eye and -6,75 dpt in the left eye) woman (#69, S2 Table) with nyctalopia since childhood reported decreasing visual acuity and visual fields since her twenties. Visual acuity was 20/400 in the right eye and 20/63 in the left eye. ERG examination was not tolerated by the patient. Funduscopy revealed changes characteristic for RP including bone spicule pigmentation and attenuated retinal vessels. Fundus AF showed areas of increased and decreased AF in the right eye and a fine pattern of radial lines radiating peripherally from the fovea in the left eye. On OCT imaging, there was widespread thinning of the photoreceptor layer in both eyes with foveal sparing in the left eye. Furthermore, OCT imaging revealed thickening of the inner retina mainly around the optic disc, which was more obvious in the right than in the left eye (Fig 7). We identified a heterozygous four-base-pair deletion (c.2442_2445del, p.Gly817Lysfs*2) in ORF15 of RPGR. None of the patient’s parents had a history suggestive for retinal disease, and examination of the mother including AF-recordings showed no characteristics for a carrier state of X-linked RP. Genetic analysis of the mother revealed wild-type RPGR alleles, indicating a de novo RPGR mutation in the index patient.
Fig 7. Phenotype of an RP patient with an RPGR mutation.
A-E right eye, F-J left eye: Fundus color imaging (A, D), fundus autofluorescence with 488 nm excitation light (B, E) and (C, F) spectral-domain optical coherence tomography.
Patient C (RPGR): This 52-years-old myopic (spherical equivalent approximately -4 dpt in both eyes) woman (#76, S2 Table) with difficulty in seeing in the dark since childhood reported a progressive reduction of visual acuity over the past 10–20 years. Visual acuity was above 20/50 in adolescence, 20/400-20/200 around the age of 40 years and now 20/800. ERG examination showed no detectable responses. Funduscopy revealed changes characteristic for RP in both eyes. There was symmetric and widespread thinning of the photoreceptor layer on OCT imaging (Fig 8). Although fundus AF imaging showed no sign for an X-linked carrier state, we identified a heterozygous two-base-pair deletion (c.2405_2406delAG, p.Glu802Glyfs*32) in ORF15 of RPGR. The patient’s maternal great-uncle was visually impaired, and her maternal great-grandfather was blind, compatible with autosomal dominant inheritance with reduced penetrance. The mother of the patient died at the age of 50 years and had no visual problems.
Fig 8. Phenotype of an RP patient with an RPGR mutation.
(A) widefield fundus AF, (B) widefield false-color image and (C, D) spectral-domain optical coherence tomography. Only one eye is shown due to high symmetry between eyes.
Female carriers of X-linked RP consistently have peripheral retinal pigment epithelial atrophy.[62] Most carriers may experience mild or moderate reduction of visual function, with a minority becoming legally blind.[63] Although rare, severe RP may occur in female carriers of X-linked RP and simulate autosomal dominant inheritance[15, 31, 62, 64–66], as in Patient C. Comprehensive genetic testing has been shown to detect mutations in RPGR or RP2 in cohorts assumed to have autosomal dominant RP, leading to a genetic re-classification of those families.[15, 31] However, to the best of our knowledge, a sporadic female RP patient diagnosed with X-linked RP has only been reported once.[5] Of note, parental testing for the RPGR mutation of patient B indicated that it occurred de novo.
Herein, severe manifestations of X-linked RP were found in two sporadic female carriers and in one patient with a family history suggesting autosomal dominant inheritance with variable expressivity. Two of these three patients revealed very subtle signs for X-linked RP which may easily be missed by standard clinical examinations. This includes asymmetry between eyes which has been reported as a typical sign in X-linked RP-carriers,[67] although marked asymmetry was specifically mentioned in only one out of 61 carriers.[62] Another peculiar finding on fundus AF imaging in female carriers of X-linked RP is a radial pattern extending peripherally from the fovea,[68–70] as observed in a more or less subtle form in the less affected eye of Patients A and B. Moreover, peripapillary thickening of inner retinal layers as observed in patient 2 has been reported in males with X-linked RP.[71, 72] Thus, a subtle X-linked RP carrier phenotype was present in the less severely affected eye in two patients, but not in the third patient who presented with a progressed bilateral disease stage. Unbiased molecular genetic testing eventually revealed the correct diagnosis in these female patients with sporadic RP or with a family history suggestive for autosomal dominant inheritance with variable expressivity.
Conclusion
In summary, this study demonstrates the enormous genetic heterogeneity of RP and a high detection rate of disease-causing mutations in RP patients using targeted NGS. Given the continuous decline of NGS costs, initial Sanger sequencing of likely disease-causing genes does not appear necessary anymore. Novel genotype-phenotype correlations (CRX, RPGRIP1) were uncovered, and recently reported novel correlations were confirmed (CEP290, MFSD8). PRPF31 and RPGR mutations revealed unexpected inheritance modes in a subset of patients. Massively parallel sequencing of all known retinal dystrophy genes is a valuable diagnostic approach in RP patients.
Supporting information
RP genes included in the NGS panels at the respective time of analysis. Importantly, the NGS panels also contained genes for clinically overlapping conditions such as cone/cone-rod dystrophies, Leber’s congenital amaurosis and syndromes with retinal dystrophies, allowing for an extended genetic assessment if no mutation was found in genes previously associated with RP. +: added genes, -: removed genes (compared to the respective previous panel version). arRP, X-linked RP: I (8/10), 32 genes; II (8/13), 63 genes; III (4/15), 73 genes; IV (8/15), 74 genes; V (1/16), 85 genes. adRP: I (8/10), 23 genes; II (8/13), 21 genes; III (4/15), 25 genes; IV (1/16), 31 genes.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Bioscientia, a publicly traded diagnostic company, provided support in the form of salaries for C.N., S.L., D.Z., C.B., T.E. and H.J.B., but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section. This work was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC); ProRetina Foundation, Germany and the German Research Foundation (MG, grant #GL920/1-1; PLM grant # MU4279/1-1). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. No sponsor or funding agency had any involvement in the design, collection, analysis and interpretation of the data, manuscript writing or the decision to submit the manuscript for publication.
References
- 1.Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40 Epub 2006/10/13. 10.1186/1750-1172-1-40 ; PubMed Central PMCID: PMC1621055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–809. Epub 2006/11/23. 10.1016/S0140-6736(06)69740-7 . [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Fernandez De La Camara C, Nanda A, Salvetti AP, Fischer MD, MacLaren RE. Gene therapy for the treatment of X-linked retinitis pigmentosa. Expert Opin Orphan Drugs. 2018;6(3):167–77. Epub 2018/07/31. 10.1080/21678707.2018.1444476 ; PubMed Central PMCID: PMC6059358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neveling K, Collin RW, Gilissen C, van Huet RA, Visser L, Kwint MP, et al. Next-generation genetic testing for retinitis pigmentosa. Hum Mutat. 2012;33(6):963–72. Epub 2012/02/16. 10.1002/humu.22045 ; PubMed Central PMCID: PMC3490376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Sullivan J, Mullaney BG, Bhaskar SS, Dickerson JE, Hall G, O'Grady A, et al. A paradigm shift in the delivery of services for diagnosis of inherited retinal disease. J Med Genet. 2012;49(5):322–6. Epub 2012/05/15. 10.1136/jmedgenet-2012-100847 . [DOI] [PubMed] [Google Scholar]
- 6.Shanks ME, Downes SM, Copley RR, Lise S, Broxholme J, Hudspith KA, et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet. 2013;21(3):274–80. Epub 2012/09/13. 10.1038/ejhg.2012.172 ; PubMed Central PMCID: PMC3573204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glöckle N, Kohl S, Mohr J, Scheurenbrand T, Sprecher A, Weisschuh N, et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur J Hum Genet. 2014;22(1):99–104. Epub 2013/04/18. 10.1038/ejhg.2013.72 ; PubMed Central PMCID: PMC3865404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberger T, Neuhaus C, Khan AO, Decker C, Preising MN, Friedburg C, et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS One. 2013;8(11):e78496 Epub 2013/11/23. 10.1371/journal.pone.0078496 ; PubMed Central PMCID: PMC3827063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–55. Epub 2011/09/29. 10.1038/nrg3031 . [DOI] [PubMed] [Google Scholar]
- 10.Zhao L, Wang F, Wang H, Li Y, Alexander S, Wang K, et al. Next-generation sequencing-based molecular diagnosis of 82 retinitis pigmentosa probands from Northern Ireland. Hum Genet. 2015;134(2):217–30. Epub 2014/12/05. 10.1007/s00439-014-1512-7 ; PubMed Central PMCID: PMC4347882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, Guan L, Shen T, Zhang J, Xiao X, Jiang H, et al. Mutations of 60 known causative genes in 157 families with retinitis pigmentosa based on exome sequencing. Hum Genet. 2014;133(10):1255–71. Epub 2014/06/19. 10.1007/s00439-014-1460-2 . [DOI] [PubMed] [Google Scholar]
- 12.Wang F, Wang H, Tuan HF, Nguyen DH, Sun V, Keser V, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014;133(3):331–45. Epub 2013/10/25. 10.1007/s00439-013-1381-5 ; PubMed Central PMCID: PMC3945441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Wang H, Sun V, Tuan HF, Keser V, Wang K, et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J Med Genet. 2013;50(10):674–88. Epub 2013/07/13. 10.1136/jmedgenet-2013-101558 ; PubMed Central PMCID: PMC3932025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oishi M, Oishi A, Gotoh N, Ogino K, Higasa K, Iida K, et al. Comprehensive molecular diagnosis of a large cohort of Japanese retinitis pigmentosa and Usher syndrome patients by next-generation sequencing. Invest Ophthalmol Vis Sci. 2014;55(11):7369–75. Epub 2014/10/18. 10.1167/iovs.14-15458 . [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-San Jose P, Corton M, Blanco-Kelly F, Avila-Fernandez A, Lopez-Martinez MA, Sanchez-Navarro I, et al. Targeted Next-Generation Sequencing Improves the Diagnosis of Autosomal Dominant Retinitis Pigmentosa in Spanish Patients. Invest Ophthalmol Vis Sci. 2015;56(4):2173–82. Epub 2015/02/24. 10.1167/iovs.14-16178 . [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Zhang VW, Feng Y, Tian X, Li FY, Truong C, et al. Dependable and efficient clinical utility of target capture-based deep sequencing in molecular diagnosis of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014;55(10):6213–23. Epub 2014/08/07. 10.1167/iovs.14-14936 . [DOI] [PubMed] [Google Scholar]
- 17.Bravo-Gil N, Gonzalez-Del Pozo M, Martin-Sanchez M, Mendez-Vidal C, Rodriguez-de la Rua E, Borrego S, et al. Unravelling the genetic basis of simplex Retinitis Pigmentosa cases. Sci Rep. 2017;7:41937 Epub 2017/02/06. 10.1038/srep41937 ; PubMed Central PMCID: PMC5291209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riera M, Navarro R, Ruiz-Nogales S, Mendez P, Bures-Jelstrup A, Corcostegui B, et al. Whole exome sequencing using Ion Proton system enables reliable genetic diagnosis of inherited retinal dystrophies. Sci Rep. 2017;7:42078 Epub 2017/02/10. 10.1038/srep42078 ; PubMed Central PMCID: PMC5299602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Xu M, Verriotto JD, Li Y, Wang H, Gan L, et al. Next-generation sequencing-based molecular diagnosis of 35 Hispanic retinitis pigmentosa probands. Sci Rep. 2016;6:32792 Epub 2016/09/07. 10.1038/srep32792 ; PubMed Central PMCID: PMC5011706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Iorio V, Karali M, Brunetti-Pierri R, Filippelli M, Di Fruscio G, Pizzo M, et al. Clinical and Genetic Evaluation of a Cohort of Pediatric Patients with Severe Inherited Retinal Dystrophies. Genes (Basel). 2017;8(10). Epub 2017/10/21. 10.3390/genes8100280 ; PubMed Central PMCID: PMC5664130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aleman TS, Soumittra N, Cideciyan AV, Sumaroka AM, Ramprasad VL, Herrera W, et al. CERKL mutations cause an autosomal recessive cone-rod dystrophy with inner retinopathy. Invest Ophthalmol Vis Sci. 2009;50(12):5944–54. Epub 2009/07/07. 10.1167/iovs.09-3982 . [DOI] [PubMed] [Google Scholar]
- 22.Birtel J, Eisenberger T, Gliem M, Muller PL, Herrmann P, Betz C, et al. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci Rep. 2018;8(1):4824 Epub 2018/03/21. 10.1038/s41598-018-22096-0 ; PubMed Central PMCID: PMC5859282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79(3):556–61. Epub 2006/08/16. 10.1086/507318 ; PubMed Central PMCID: PMC1559533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almoguera B, Li J, Fernandez-San Jose P, Liu Y, March M, Pellegrino R, et al. Application of Whole Exome Sequencing in Six Families with an Initial Diagnosis of Autosomal Dominant Retinitis Pigmentosa: Lessons Learned. PLoS One. 2015;10(7):e0133624 Epub 2015/07/22. 10.1371/journal.pone.0133624 ; PubMed Central PMCID: PMC4509755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts L, Ratnapriya R, du Plessis M, Chaitankar V, Ramesar RS, Swaroop A. Molecular Diagnosis of Inherited Retinal Diseases in Indigenous African Populations by Whole-Exome Sequencing. Invest Ophthalmol Vis Sci. 2016;57(14):6374–81. Epub 2016/11/30. 10.1167/iovs.16-19785 ; PubMed Central PMCID: PMC5132076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19(6):1329–36. Epub 1998/01/14. S0896-6273(00)80423-7. . [DOI] [PubMed] [Google Scholar]
- 27.Sohocki MM, Daiger SP, Bowne SJ, Rodriquez JA, Northrup H, Heckenlively JR, et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001;17(1):42–51. Epub 2001/01/04. 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K ; PubMed Central PMCID: PMC2585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vervoort R, Lennon A, Bird AC, Tulloch B, Axton R, Miano MG, et al. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000;25(4):462–6. Epub 2000/08/10. 10.1038/78182 . [DOI] [PubMed] [Google Scholar]
- 29.Fahim AT, Bowne SJ, Sullivan LS, Webb KD, Williams JT, Wheaton DK, et al. Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS One. 2011;6(8):e23021 Epub 2011/08/23. 10.1371/journal.pone.0023021 ; PubMed Central PMCID: PMC3155520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowne SJ, Sullivan LS, Koboldt DC, Ding L, Fulton R, Abbott RM, et al. Identification of disease-causing mutations in autosomal dominant retinitis pigmentosa (adRP) using next-generation DNA sequencing. Invest Ophthalmol Vis Sci. 2011;52(1):494–503. Epub 2010/09/24. 10.1167/iovs.10-6180 ; PubMed Central PMCID: PMC3053293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Churchill JD, Bowne SJ, Sullivan LS, Lewis RA, Wheaton DK, Birch DG, et al. Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2013;54(2):1411–6. Epub 2013/02/02. 10.1167/iovs.12-11541 ; PubMed Central PMCID: PMC3597192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge Z, Bowles K, Goetz K, Scholl HP, Wang F, Wang X, et al. NGS-based Molecular diagnosis of 105 eyeGENE probands with Retinitis Pigmentosa. Sci Rep. 2015;5:18287 Epub 2015/12/17. 10.1038/srep18287 ; PubMed Central PMCID: PMC4678898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branham K, Othman M, Brumm M, Karoukis AJ, Atmaca-Sonmez P, Yashar BM, et al. Mutations in RPGR and RP2 account for 15% of males with simplex retinal degenerative disease. Invest Ophthalmol Vis Sci. 2012;53(13):8232–7. Epub 2012/11/15. 10.1167/iovs.12-11025 ; PubMed Central PMCID: PMC3522443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelletier V, Jambou M, Delphin N, Zinovieva E, Stum M, Gigarel N, et al. Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: genotype-phenotype correlations and impact on genetic counseling. Hum Mutat. 2007;28(1):81–91. Epub 2006/09/14. 10.1002/humu.20417 . [DOI] [PubMed] [Google Scholar]
- 35.Manes G, Guillaumie T, Vos WL, Devos A, Audo I, Zeitz C, et al. High prevalence of PRPH2 in autosomal dominant retinitis pigmentosa in france and characterization of biochemical and clinical features. Am J Ophthalmol. 2015;159(2):302–14. Epub 2014/12/03. 10.1016/j.ajo.2014.10.033 . [DOI] [PubMed] [Google Scholar]
- 36.Rose AM, Shah AZ, Venturini G, Krishna A, Chakravarti A, Rivolta C, et al. Transcriptional regulation of PRPF31 gene expression by MSR1 repeat elements causes incomplete penetrance in retinitis pigmentosa. Sci Rep. 2016;6:19450 Epub 2016/01/20. 10.1038/srep19450 ; PubMed Central PMCID: PMC4725990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inglehearn CF, Tarttelin EE, Plant C, Peacock RE, al-Maghtheh M, Vithana E, et al. A linkage survey of 20 dominant retinitis pigmentosa families: frequencies of the nine known loci and evidence for further heterogeneity. J Med Genet. 1998;35(1):1–5. Epub 1998/02/25. ; PubMed Central PMCID: PMC1051177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans K, al-Maghtheh M, Fitzke FW, Moore AT, Jay M, Inglehearn CF, et al. Bimodal expressivity in dominant retinitis pigmentosa genetically linked to chromosome 19q. Br J Ophthalmol. 1995;79(9):841–6. Epub 1995/09/01. ; PubMed Central PMCID: PMC505271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore AT, Fitzke F, Jay M, Arden GB, Inglehearn CF, Keen TJ, et al. Autosomal dominant retinitis pigmentosa with apparent incomplete penetrance: a clinical, electrophysiological, psychophysical, and molecular genetic study. Br J Ophthalmol. 1993;77(8):473–9. Epub 1993/08/01. 10.1136/bjo.77.8.473 ; PubMed Central PMCID: PMC504578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppieters F, Lefever S, Leroy BP, De Baere E. CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum Mutat. 2010;31(10):1097–108. Epub 2010/08/07. 10.1002/humu.21337 . [DOI] [PubMed] [Google Scholar]
- 41.Perrault I, Delphin N, Hanein S, Gerber S, Dufier JL, Roche O, et al. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum Mutat. 2007;28(4):416 Epub 2007/03/09. 10.1002/humu.9485 . [DOI] [PubMed] [Google Scholar]
- 42.den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27(4):391–419. Epub 2008/07/18. 10.1016/j.preteyeres.2008.05.003 . [DOI] [PubMed] [Google Scholar]
- 43.Shen T, Guan L, Li S, Zhang J, Xiao X, Jiang H, et al. Mutation analysis of Leber congenital amaurosisassociated genes in patients with retinitis pigmentosa. Mol Med Rep. 2015;11(3):1827–32. Epub 2014/11/08. 10.3892/mmr.2014.2894 . [DOI] [PubMed] [Google Scholar]
- 44.Littink KW, Pott JW, Collin RW, Kroes HY, Verheij JB, Blokland EA, et al. A novel nonsense mutation in CEP290 induces exon skipping and leads to a relatively mild retinal phenotype. Invest Ophthalmol Vis Sci. 2010;51(7):3646–52. Epub 2010/02/05. 10.1167/iovs.09-5074 . [DOI] [PubMed] [Google Scholar]
- 45.Ge Z, Bowles K, Goetz K, Scholl HP, Wang F, Wang X, et al. NGS-based Molecular diagnosis of 105 eyeGENE probands with Retinitis Pigmentosa. Scientific reports. 2015;5:18287 Epub 2015/12/17. 10.1038/srep18287 ; PubMed Central PMCID: PMC4678898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cideciyan AV, Aleman TS, Jacobson SG, Khanna H, Sumaroka A, Aguirre GK, et al. Centrosomal-ciliary gene CEP290/NPHP6 mutations result in blindness with unexpected sparing of photoreceptors and visual brain: implications for therapy of Leber congenital amaurosis. Hum Mutat. 2007;28(11):1074–83. Epub 2007/06/08. 10.1002/humu.20565 . [DOI] [PubMed] [Google Scholar]
- 47.Webb TR, Parfitt DA, Gardner JC, Martinez A, Bevilacqua D, Davidson AE, et al. Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa (RP23). Hum Mol Genet. 2012;21(16):3647–54. Epub 2012/05/24. 10.1093/hmg/dds194 ; PubMed Central PMCID: PMC3406759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen TT, Hull S, Roepman R, van den Born LI, Oud MM, de Vrieze E, et al. Missense mutations in the WD40 domain of AHI1 cause non-syndromic retinitis pigmentosa. J Med Genet. 2017. Epub 2017/04/27. 10.1136/jmedgenet-2016-104200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerber S, Perrault I, Hanein S, Barbet F, Ducroq D, Ghazi I, et al. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur J Hum Genet. 2001;9(8):561–71. Epub 2001/08/31. 10.1038/sj.ejhg.5200689 . [DOI] [PubMed] [Google Scholar]
- 50.Dryja TP, Adams SM, Grimsby JL, McGee TL, Hong DH, Li T, et al. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet. 2001;68(5):1295–8. Epub 2001/04/03. 10.1086/320113 ; PubMed Central PMCID: PMC1226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Booij JC, Florijn RJ, ten Brink JB, Loves W, Meire F, van Schooneveld MJ, et al. Identification of mutations in the AIPL1, CRB1, GUCY2D, RPE65, and RPGRIP1 genes in patients with juvenile retinitis pigmentosa. J Med Genet. 2005;42(11):e67 Epub 2005/11/08. 10.1136/jmg.2005.035121 ; PubMed Central PMCID: PMC1735944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walia S, Fishman GA, Jacobson SG, Aleman TS, Koenekoop RK, Traboulsi EI, et al. Visual acuity in patients with Leber's congenital amaurosis and early childhood-onset retinitis pigmentosa. Ophthalmology. 2010;117(6):1190–8. Epub 2010/01/19. 10.1016/j.ophtha.2009.09.056 . [DOI] [PubMed] [Google Scholar]
- 53.Jacobson SG, Cideciyan AV, Aleman TS, Sumaroka A, Schwartz SB, Roman AJ, et al. Leber congenital amaurosis caused by an RPGRIP1 mutation shows treatment potential. Ophthalmology. 2007;114(5):895–8. Epub 2007/02/20. 10.1016/j.ophtha.2006.10.028 . [DOI] [PubMed] [Google Scholar]
- 54.Aiello C, Terracciano A, Simonati A, Discepoli G, Cannelli N, Claps D, et al. Mutations in MFSD8/CLN7 are a frequent cause of variant-late infantile neuronal ceroid lipofuscinosis. Hum Mutat. 2009;30(3):E530–40. Epub 2009/01/30. 10.1002/humu.20975 . [DOI] [PubMed] [Google Scholar]
- 55.Siintola E, Topcu M, Aula N, Lohi H, Minassian BA, Paterson AD, et al. The novel neuronal ceroid lipofuscinosis gene MFSD8 encodes a putative lysosomal transporter. Am J Hum Genet. 2007;81(1):136–46. Epub 2007/06/15. 10.1086/518902 ; PubMed Central PMCID: PMC1950917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahajnah M, Zelnik N. Phenotypic heterogeneity in consanguineous patients with a common CLN8 mutation. Pediatr Neurol. 2012;47(4):303–5. Epub 2012/09/12. 10.1016/j.pediatrneurol.2012.05.016 . [DOI] [PubMed] [Google Scholar]
- 57.Mandel H, Cohen Katsanelson K, Khayat M, Chervinsky I, Vladovski E, Iancu TC, et al. Clinico-pathological manifestations of variant late infantile neuronal ceroid lipofuscinosis (vLINCL) caused by a novel mutation in MFSD8 gene. Eur J Med Genet. 2014;57(11–12):607–12. Epub 2014/10/02. 10.1016/j.ejmg.2014.09.004 . [DOI] [PubMed] [Google Scholar]
- 58.Kousi M, Siintola E, Dvorakova L, Vlaskova H, Turnbull J, Topcu M, et al. Mutations in CLN7/MFSD8 are a common cause of variant late-infantile neuronal ceroid lipofuscinosis. Brain. 2009;132(Pt 3):810–9. Epub 2009/02/10. 10.1093/brain/awn366 . [DOI] [PubMed] [Google Scholar]
- 59.Craiu D, Dragostin O, Dica A, Hoffman-Zacharska D, Gos M, Bastian AE, et al. Rett-like onset in late-infantile neuronal ceroid lipofuscinosis (CLN7) caused by compound heterozygous mutation in the MFSD8 gene and review of the literature data on clinical onset signs. Eur J Paediatr Neurol. 2015;19(1):78–86. Epub 2014/12/03. 10.1016/j.ejpn.2014.07.008 . [DOI] [PubMed] [Google Scholar]
- 60.Khan KN, El-Asrag ME, Ku CA, Holder GE, McKibbin M, Arno G, et al. Specific Alleles of CLN7/MFSD8, a Protein That Localizes to Photoreceptor Synaptic Terminals, Cause a Spectrum of Nonsyndromic Retinal Dystrophy. Invest Ophthalmol Vis Sci. 2017;58(7):2906–14. Epub 2017/06/07. 10.1167/iovs.16-20608 . [DOI] [PubMed] [Google Scholar]
- 61.Roosing S, van den Born LI, Sangermano R, Banfi S, Koenekoop RK, Zonneveld-Vrieling MN, et al. Mutations in MFSD8, encoding a lysosomal membrane protein, are associated with nonsyndromic autosomal recessive macular dystrophy. Ophthalmology. 2015;122(1):170–9. Epub 2014/09/18. 10.1016/j.ophtha.2014.07.040 . [DOI] [PubMed] [Google Scholar]
- 62.Bird AC. X-linked retinitis pigmentosa. Br J Ophthalmol. 1975;59(4):177–99. Epub 1975/04/01. ; PubMed Central PMCID: PMC1042592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Comander J, Weigel-DiFranco C, Sandberg MA, Berson EL. Visual Function in Carriers of X-Linked Retinitis Pigmentosa. Ophthalmology. 2015;122(9):1899–906. Epub 2015/07/06. 10.1016/j.ophtha.2015.05.039 PubMed Central PMCID: PMC4562908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Souied E, Segues B, Ghazi I, Rozet JM, Chatelin S, Gerber S, et al. Severe manifestations in carrier females in X linked retinitis pigmentosa. J Med Genet. 1997;34(10):793–7. Epub 1997/11/14. ; PubMed Central PMCID: PMC1051083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pomares E, Riera M, Castro-Navarro J, Andres-Gutierrez A, Gonzalez-Duarte R, Marfany G. Identification of an intronic single-point mutation in RP2 as the cause of semidominant X-linked retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2009;50(11):5107–14. Epub 2009/06/12. 10.1167/iovs.08-3208 . [DOI] [PubMed] [Google Scholar]
- 66.Rozet JM, Perrault I, Gigarel N, Souied E, Ghazi I, Gerber S, et al. Dominant X linked retinitis pigmentosa is frequently accounted for by truncating mutations in exon ORF15 of the RPGR gene. J Med Genet. 2002;39(4):284–5. Epub 2002/04/16. 10.1136/jmg.39.4.284 ; PubMed Central PMCID: PMC1735080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobson SG, Yagasaki K, Feuer WJ, Roman AJ. Interocular asymmetry of visual function in heterozygotes of X-linked retinitis pigmentosa. Exp Eye Res. 1989;48(5):679–91. Epub 1989/05/01. 10.1016/0014-4835(89)90009-2 . [DOI] [PubMed] [Google Scholar]
- 68.Wegscheider E, Preising MN, Lorenz B. Fundus autofluorescence in carriers of X-linked recessive retinitis pigmentosa associated with mutations in RPGR, and correlation with electrophysiological and psychophysical data. Graefes Arch Clin Exp Ophthalmol. 2004;242(6):501–11. Epub 2004/06/03. 10.1007/s00417-004-0891-1 . [DOI] [PubMed] [Google Scholar]
- 69.Acton JH, Greenberg JP, Greenstein VC, Marsiglia M, Tabacaru M, Theodore Smith R, et al. Evaluation of multimodal imaging in carriers of X-linked retinitis pigmentosa. Exp Eye Res. 2013;113:41–8. Epub 2013/05/15. 10.1016/j.exer.2013.05.003 ; PubMed Central PMCID: PMC3974115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ogino K, Oishi M, Oishi A, Morooka S, Sugahara M, Gotoh N, et al. Radial fundus autofluorescence in the periphery in patients with X-linked retinitis pigmentosa. Clin Ophthalmol. 2015;9:1467–74. Epub 2015/09/01. 10.2147/OPTH.S89371 ; PubMed Central PMCID: PMC4544811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aleman TS, Cideciyan AV, Sumaroka A, Schwartz SB, Roman AJ, Windsor EA, et al. Inner retinal abnormalities in X-linked retinitis pigmentosa with RPGR mutations. Invest Ophthalmol Vis Sci. 2007;48(10):4759–65. Epub 2007/09/28. 10.1167/iovs.07-0453 ; PubMed Central PMCID: PMC3178894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hood DC, Lin CE, Lazow MA, Locke KG, Zhang X, Birch DG. Thickness of receptor and post-receptor retinal layers in patients with retinitis pigmentosa measured with frequency-domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2009;50(5):2328–36. Epub 2008/11/18. 10.1167/iovs.08-2936 ; PubMed Central PMCID: PMC2835526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RP genes included in the NGS panels at the respective time of analysis. Importantly, the NGS panels also contained genes for clinically overlapping conditions such as cone/cone-rod dystrophies, Leber’s congenital amaurosis and syndromes with retinal dystrophies, allowing for an extended genetic assessment if no mutation was found in genes previously associated with RP. +: added genes, -: removed genes (compared to the respective previous panel version). arRP, X-linked RP: I (8/10), 32 genes; II (8/13), 63 genes; III (4/15), 73 genes; IV (8/15), 74 genes; V (1/16), 85 genes. adRP: I (8/10), 23 genes; II (8/13), 21 genes; III (4/15), 25 genes; IV (1/16), 31 genes.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.