Abstract
Exosomes are nanosized vesicles that are abundant in biological fluids. In recent years, exosomes have attracted increasing attention as their cargo may provide promising biomarkers for the early diagnosis of and therapy for many diseases, such as cancer. In addition to ultracentrifugation (UC), many alternative methods including size-exclusion chromatography (SEC) have been developed for isolating exosomes. It has been reported that the SEC method provided improved performance relative to the UC method in isolating exosomes from plasma, where the former contained less residual blood protein contamination. We have compared the SEC method with an optimized UC method in isolating exosomes from human serum. This was based on dilution of the serum to reduce the viscosity and a prolonged cycle of UC, followed by another four cycles. We found that >95% of serum proteins were removed without a significant loss of exosome proteins relative to SEC. We also combined one cycle of UC with SEC and found that this method provided improved results relative to the SEC method, although the serum protein contamination was several times higher than that of our optimized UC method. The TEM showed that the size distribution of exosomes isolated from each of the three methods was similar.
Keywords: exosomes, SEC, serum, UC
Graphical Abstract
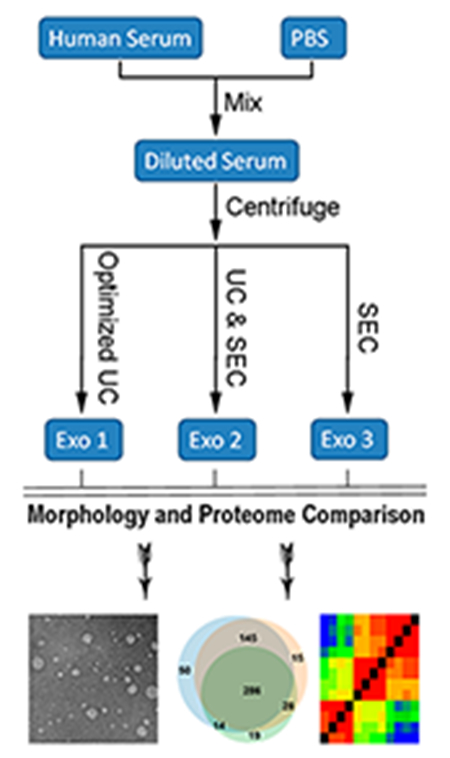
INTRODUCTION
Exosomes are 30–120 nm sized membrane-bound vesicles1,2 originating from the luminal membrane of multivesicular bodies (MVBs) in all types of normal and tumor cells.3–6 They are believed to participate in cell–cell communication by loading cargo proteins, metabolites, and nucleic acids (mRNA, miRNA) from their original cells,2,5,7,8 which can be released into the extracellular space upon fusion with the plasma membrane.9–11 There are many cell surface membrane proteins that can be detected in exosomes, some of which may be used for the early detection, diagnosis, and prognosis of cancer.12–18.
The commonly used method for isolation of exosomes is ultracentrifugation (UC). This method can be time-consuming and requires access to an ultracentrifuge. Several other methods, such as immunoaffinity,19 ultrafiltration,20 polymeric precipitation,21 and size-exclusion chromatography (SEC), have been recently developed as alternatives.22,23 However, the polymeric precipitation method has little specificity and usually captures unwanted extracellular vesicles other than exosomes. The immunoaffinity methods require large amounts of antibodies, which are costly and may extract larger vesicles other than exosomes. Ultrafiltration generally has low exosome yield because of the adhesion of exosomes to filters. Several types of microfluidic chip separations based on immunoaffinity or SEC have also been developed to isolate exosomes.24,25 The throughput of these methods is generally low and not suitable for preanalytical processes, such as discovery.24
SEC has the ideal performance in separating extracellular vesicles from blood proteins.26,27 Although it was reported to have the limitation of low exosome yield, the SEC method has been applied for the analysis of clinical samples.28,29 A recent study used a commercial qEV SEC column (Izon Science, Christchurch, New Zealand) and compared it with the exoEasy kit.30 The authors found that the SEC method resulted in better protein yield and size distributions for the isolation of exosomes from human plasma compared with the exoEasy kit. Another study compared the SEC method with the UC method in isolating exosomes from plasma and found that the former provided improved performance when there was less serum protein contamination.31
In the current study, we compare the commercial qEV SEC column with our optimized UC method for exosome isolation from human serum. We studied human serum because this will be essential for clinical applications. In brief, our optimized UC method was based on dilution of the serum with PBS to reduce the viscosity and a prolonged first cycle of UC, followed by another four cycles of UC. We have also studied the use of one cycle of UC coupled to the qEV SEC column for isolating exosomes for further analysis. NanoSight and transmission electron microscopy (TEM) were used to compare the yield and size distribution of exosomes. We have performed LC–MS/MS-based proteome analysis to compare the protein categories, exosome protein markers, and degree of exosome purification from blood proteins for the three methods.
EXPERIMENTAL SECTION
Human Serum Sample
The commercially available serum (Innovative Research, Novi, MI) was pooled from a cohort of 1500 to 3000 healthy normal humans. Each donor unit was tested and found to be negative for HIV-1 and −2, HCV, HBsAg, and PRP by FDA-approved methods. The serum was stored at −80 °C until use. The initial volume of serum was 18 mL. It was diluted with an equal volume of PBS (AppliChem, St. Louis, MO) to decrease the viscosity. The diluted serum was centrifuged at 2000g for 10 min and 10 000g for 30 min at 4 °C to remove cell debris and large extracellular vesicles. The supernatant of serum was divided into three equal aliquots, which were designated as serum sample 1 (SS1), serum sample 2 (SS2), and serum sample 3 (SS3). Each sample contained 12 mL of diluted serum.
Five Cycles of Ultracentrifugation
The SS1 was further divided into six equal aliquots (SS1-1, SS1-2, SS1-3, SS1-4, SS1-5, SS1-6), where each aliquot contained 2 mL of diluted serum. Each aliquot of the SS1 was transferred to Ultra-Clear tubes (Beckman Coulter, Indianapolis, IN), diluted by PBS into 4 mL, and centrifuged at 100 000g using a Beckman Optima XL-70 ultracentrifuge for 120 min at 4 °C. The supernatant was carefully removed by pipet. To avoid the loss of exosome pellets, 2 mm of supernatant was left in the tubes. Compared with exosome protein, the presence of serum protein was dominant. Thus a couple of cycles of UC were not sufficient to remove the serum protein for the analysis of the exosome proteome. In our previous work, we showed that five cycles of UC were necessary to efficiently remove the serum protein contamination.32 The sedimentary pellets in each tube were suspended in 4 mL of PBS to dilute the supernatant and centrifuged at 100 000g for 70 min at 4 °C, followed by the removal of the supernatant. In total, five cycles of UC were performed to purify exosomes to eliminate serum protein contamination. Samples SS1-1 and SS1-2 were analyzed by NanoSight and TEM. Samples SS1-3, SS1-4, SS1-5, and SS1-6 were each analyzed by mass spectrometry separately to provide replicate runs.
SEC Columns
The SS2 was further divided into six equal aliquots (SS2-1, SS2-2, SS2-3, SS2-4, SS2-5, SS2-6), where each aliquot contained 2 mL of diluted serum. The qEV SEC 10 mL columns (iZON Science, Christchurch, New Zealand) were used here to isolate exosomes. After rinsing the columns with 15 mL of PBS for equilibration, each aliquot of SS2 was transferred on the SEC columns. It should be noted that the loading volume of sample was 0.5 mL each time. PBS was continually added to the columns so that they did not run dry. The first 3.5 mL of elute was discarded because it was the void volume. The next 0.5 mL of elute was collected for the exosome analysis. The columns were then flushed using 30 mL of PBS and cleaned using 10 mL of 0.5 M NaOH. The columns were rinsed with 50 mL of PBS for equilibration, which were then ready for the next loading. To prepare each sample, two columns were used, where each column was used twice. Samples SS2-1 and SS2-2 were analyzed by NanoSight and TEM. Samples SS2-3, SS2-4, SS2-5, and SS2-6 were each separately analyzed by mass spectrometry.
One Cycle of Ultracentrifugation and SEC Columns
The SS3 was further divided into six equal aliquots (SS3-1, SS3-2, SS3-3, SS3-4, SS3-5, SS3-6), where each aliquot contained 2 mL of diluted serum. Each aliquot of SS3 was transferred to Ultra-Clear tubes (Beckman Coulter, Indianapolis, IN), diluted by PBS into 4 mL, and centrifuged at 100 000g using a Beckman Optima XL-70 ultracentrifuge for 120 min at 4 °C. After the supernatant was removed, the pellets were suspended and diluted by PBS to a final volume of 0.5 mL. Then, the sample was transferred to a pre-equilibrated qEV SEC 10 mL column. PBS was continually added to the columns. The first 3.5 mL of eluent was discarded, and the next 0.5 mL of eluent was collected for the exosome analysis. To prepare each sample, only one column was used. Samples SS3-1 and SS3-2 were analyzed by NanoSight and TEM. Samples SS3-3, SS3-4, SS3-5, and SS3-6 were each analyzed by mass spectrometry separately.
NanoSight Analysis
The recoveries of exosomes by the above three methods were evaluated using the NanoSight NS300 (Malvern, Worcestershire, U.K.). Each exosome sample was diluted by PBS to 1 mL and continuously infused into the NanoSight by an automatic syringe pump at a flow rate of 30 μL/min. The focus was adjusted, and the temperature was set to 25 °C. The exosome movement was captured five times, where each time was 60 s. The concentration of exosomes was then calculated by a built-in application.
Transmission Electron Microscopy
TEM was used to measure the size of the exosomes. First, glow discharge was performed to make the surface of the carbon film (Hatfield, PA) hydrophilic. Each exosome sample was diluted by PBS to 1 mL, where 5 μL was dropped on the carbon film and incubated for 2 min. Next, 5 μL of 2.5% w/v glutaldehyde was used to fix the exosomes for 5 min. The film was negatively stained using 5 μL of 1% uranyl acetate for 1 min. After each of the above steps, the liquid was removed by a small piece of filter paper. Exosomes on carbon films were then imaged in a Philips CM-100 TEM instrument.
NanoLC–MS/MS
The tryptic digests of exosome samples were desalted by homemade C18 tips33 and then separated on an EASY–nLC 1000 liquid chromatograph system (Thermo Fisher Scientific, San Jose, CA) with a 250 mm reverse-phase (RP) C18 column. The samples were resolved under a 120 min linear gradient from 2 to 35% acetonitrile in 0.1% formic acid at a constant flow rate of 300 nL/min.
The samples were analyzed on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) operated in positive ion mode. The capillary temperature and the spray voltage were set as 200 °C and 2.5 kV. The data were acquired in a data-dependent mode, where up to 20 of the strongest MS1 peaks were selected for subsequent MS2 analysis. For every selected peak, collision-induced dissociation (CID) was performed. The MS1 spectra (m/z 350–1650) and the MS2 spectra were acquired in the Orbitrap and LTQ, respectively.
Data Analysis
All raw data files were processed by the MaxQuant computational proteomics platform (version 1.6.1.0).34 The parameters were set as follows: database, human UniProt; enzyme, trypsin; fixed modification, carbamidomethyl (C); variable modifications, oxidation (M) and protein N-terminal acetylation; up to two missed cleavages allowed. The MS1 mass tolerance was set as 20 and 6 ppm for the first search and main search, respectively; the MS2 mass tolerance was set as 0.5 Da. The false discovery rates (FDRs) for peptides and proteins were both set as 1%.
Western Blot
Exosomes proteins from each method were separated on a 4–15% SDS-PAGE gradient gel (Bio-Rad, Berkeley, CA) and transferred to a PVDF membrane (Bio-Rad, Berkeley, CA). After blocking, the membrane was incubated overnight with anti-CD63, anti-ALBU, or anti-Calnexin antibody (Sigma-Aldrich, St. Louis, MO), followed by incubation with HRP-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA) and was visualized using a chemiluminescence method kit (Merck Millipore, Billerica, MA).
RESULTS AND DISCUSSION
Comparison of Exosome Recoveries
In this work, we have compared three methods for isolating exosomes. These include using multiple cycles of ultracentrifugation (UC method), a commercial qEV SEC column (SEC method), and a combination of these two methods, which first uses one cycle of UC, followed by further purification with the qEV SEC column (UC&SEC method). The workflow of the three methods for isolating exosomes from human serum is shown in Figure 1. It has been reported that the efficiency of the UC method for isolating exosomes from highly viscous biofluid is very low.35 Reducing the viscosity of the serum and increasing the time of UC was found to significantly increase the exosome recovery.36 Herein we have mixed the serum with an equal volume of PBS to reduce the viscosity of serum before using each of the isolation methods. We set 120 min for the first cycle of UC and 70 min for each of the next four cycles because the viscosity of PBS was much lower.
Figure 1.
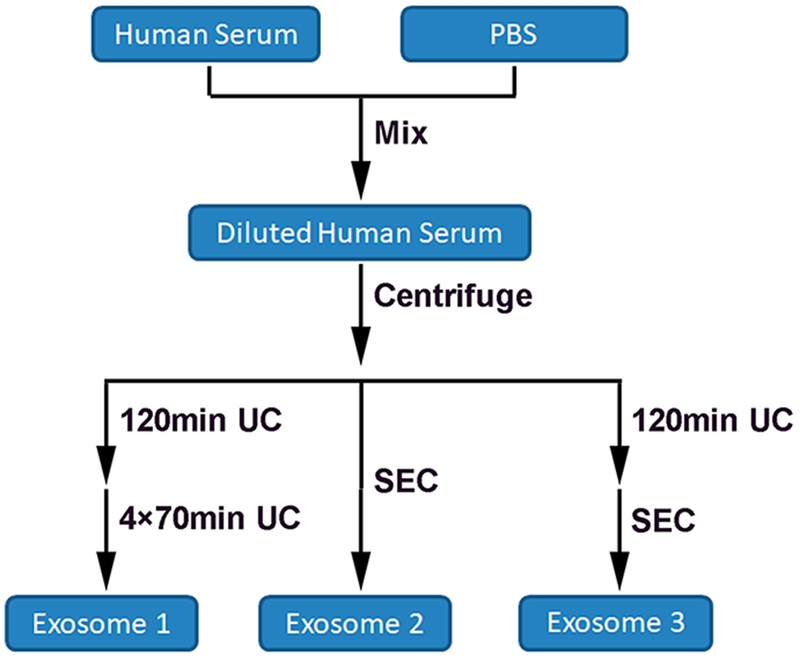
Workflow of exosomes isolated by three different methods from human serum. Human serum was diluted with an equal volume of PBS to reduce the viscosity, followed by the centrifugation to remove cell debris and large extracellular vesicles. Next, exosomes were isolated using multiple cycles of ultracentrifugation (UC method), a commercial qEV SEC 10 mL column (SEC method), and a combination of these two methods, which first uses one cycle of UC, followed by further purification with the qEV SEC column (UC&SEC method).
We used the NanoSight method to measure the number of exosomes isolated. Before the use of UC or SEC column isolation, the concentration of serum protein was too high for this light scattering method, so that we could not read any information from NanoSight. After the first and the fifth cycles of UC, we found that the number of exosomes based on 1 mL of starting material for serum was around (1.5 and 1.0) × 109, respectively. The results showed that the loss of exosomes in PBS by UC was not significant even after multiple cycles of UC. To evaluate the recovery of exosomes in serum from the first cycle of UC, we collected the supernatant and performed UC again to isolate exosomes. The NanoSight result showed that only ~2.0 × 108 exosomes were acquired from the collected supernatant, which was much less than the number of exosomes from the serum by the first cycle of UC. It seems that the prolonged UC time and reduced serum viscosity by PBS efficiently isolated exosomes. Also, the precipitated exosomes might not stick to the tube bottom firmly. To make sure these exosomes were not lost, ~2 mm of supernatant above the bottom was left when we slowly removed the supernatant after each cycle of UC.
In comparison, we investigated the use of the qEV SEC 10 mL column for isolating exosomes from serum. The instructions of the commercial SEC column show that the eighth, ninth, and tenth fractions (each fraction = 0.5 mL) of the eluent contain exosomes, whereas the eighth fraction of eluent contains very little serum protein. We also measured the protein amount of each of the three fractions using the BCA assay and found that the ninth and tenth fractions had a much higher protein amount compared with the eighth (Figure 2). To eliminate the serum protein contamination, we only collected the eighth 0.5 mL of eluent. After the isolation of exosomes by the SEC column, the number of exosomes from 1 mL of serum was ~1.3 × 109. Compared with one cycle and five cycles of UC, the SEC column method resulted in the relative isolation of 85 and 130% of exosomes, respectively. We also investigated the combined use of one cycle of UC, followed by the qEV SEC column. The number of exosomes isolated by the UC&SEC method from 1 mL of serum was ~1.2 × 109. Although its recovery was lower than that by the SEC column method, it was still 20% higher than that by five cycles of UC. It should be noted that potential protein aggregates in the SEC samples could possibly add concentration to the signal, although the serum underwent centrifugation at 10 000g for 30 min beforehand.
Figure 2.
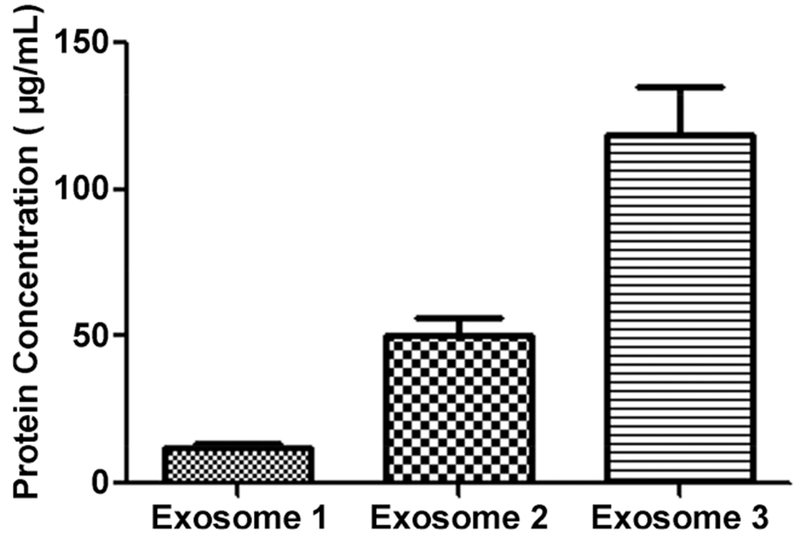
Protein concentration of fractions 8, 9, and 10 eluted from SEC column. In the SEC method, 0.5 mL of diluted serum was loaded onto the SEC column. The eluent was collected by fractions, where each fraction was 0.5 mL. The protein amount of each fraction was then measured using the BCA assay three times, and it was found that the ninth and tenth fractions had a much higher protein amount compared with the eighth.
Before the isolation by the three methods, low-speed centrifugations were performed to remove cell debris and large extracellular vesicles. The TEM images showed that the exosomes isolated from the human serum by the three different methods had similar morphology and size distribution (72 ± 5 nm), where the mean diameters of exosomes from the optimized UC, UC&SEC, and SEC methods were 67, 75, and 77 nm, respectively (Figure 3). In addition, these exosomes had intact membrane structures.
Figure 3.
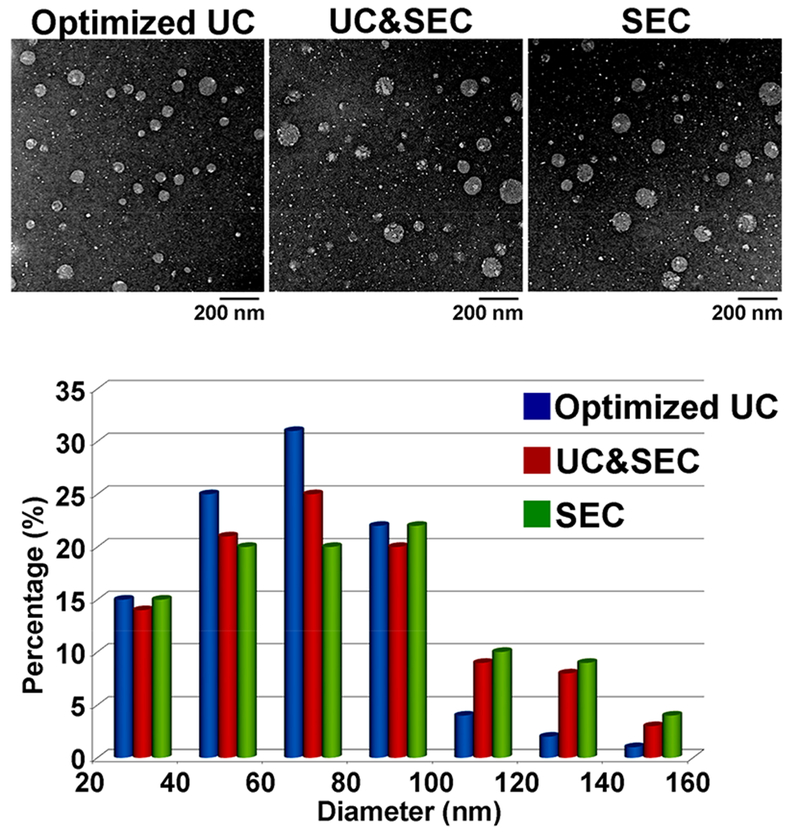
TEM images and the size distribution of the exosomes. TEM images showed that the exosomes isolated from the human serum by the three different methods had similar morphology and size distribution. The mean diameters of exosomes from optimized UC, UC&SEC, and SEC method were 67, 75, and 77 nm, respectively.
Recovery and Purity Comparison of Exosome Protein by LC–MS/MS
We processed and digested exosome proteins using the filter-aided sample preparation (FASP) method,37,38 followed by desalting and LC–MS/MS analysis. Herein we measured the protein concentration of the sample from each method by BCA assay and found that the sample from the SEC method had a much higher protein amount. We loaded each sample prepared from the same volume (1 mL) of serum for LC–MS/MS analysis. However, the protein amount in the sample prepared by the SEC method was too high to load on the mass spectrometer. We finally loaded 10% of sample from the SEC method for LC–MS/MS analysis. Usually, the loading protein amounts are statistically normalized among different samples using the intensities of total proteins or sometimes the highest abundance protein. However, such normalization is not suitable in this case. The highest abundance proteins in SEC samples were the serum proteins, which were efficiently removed in the UC and UC&SEC methods. Meanwhile, the protein number and the abundance of each protein in samples from the three methods were significantly different.
The experiment was performed four times for each sample of the three different methods. Thus a total of 12 samples were analyzed by mass spectrometry. We identified in total 578 proteins (Supplemental Table S-1) using the MaxQuant 1.6.1.0 computational proteomics platform, where 495, 474, and 347 proteins were from UC, UC&SEC, and SEC methods, respectively. The number of common proteins identified in all four runs for each of the methods was 242 for UC, 216 for UC&SEC, and 162 for SEC, respectively. The Venn diagram shows the overlap of the identified proteins from the three isolation methods (Figure 4a). The samples from the SEC method had the highest protein amount but identified the fewest proteins, whereas the samples from the optimized UC method identified the most proteins. We believe that the optimized UC method is the optimal method for the identification of the exosome proteome. The SEC method alone cannot avoid excessive contamination of serum proteins. Although it retains more exosome proteins and should be ideal for RNA analysis, the SEC has obvious limitations in the discovery of the exosome proteome. It should be noted that the SEC would be improved using well-known modifications in the liquid chromatography (LC) method to reduce shoulder effects, whereby the exosomes could be better enriched with less serum protein contamination. The heat map of the Pearson correlation coefficient (PCC) of the mass spectrometry results was used to reflect the reproducibility of each method and the differences among the methods (Figure 4b). The PCC of four replicates in each method was ~0.95, whereas the PCC of two runs in different methods was generally between 0.67 and 0.90. Because a smaller PCC meant a larger difference between two samples, the heat map showed that the proteomes of different methods had a larger variation than that of four replicates of the same method. The reproducibility of each method appeared overall to be very good. The proteomes observed using the SEC method were much more different than those of the other two methods. We speculate that this might be caused by the larger amount of blood proteins in the SEC samples.
Figure 4.
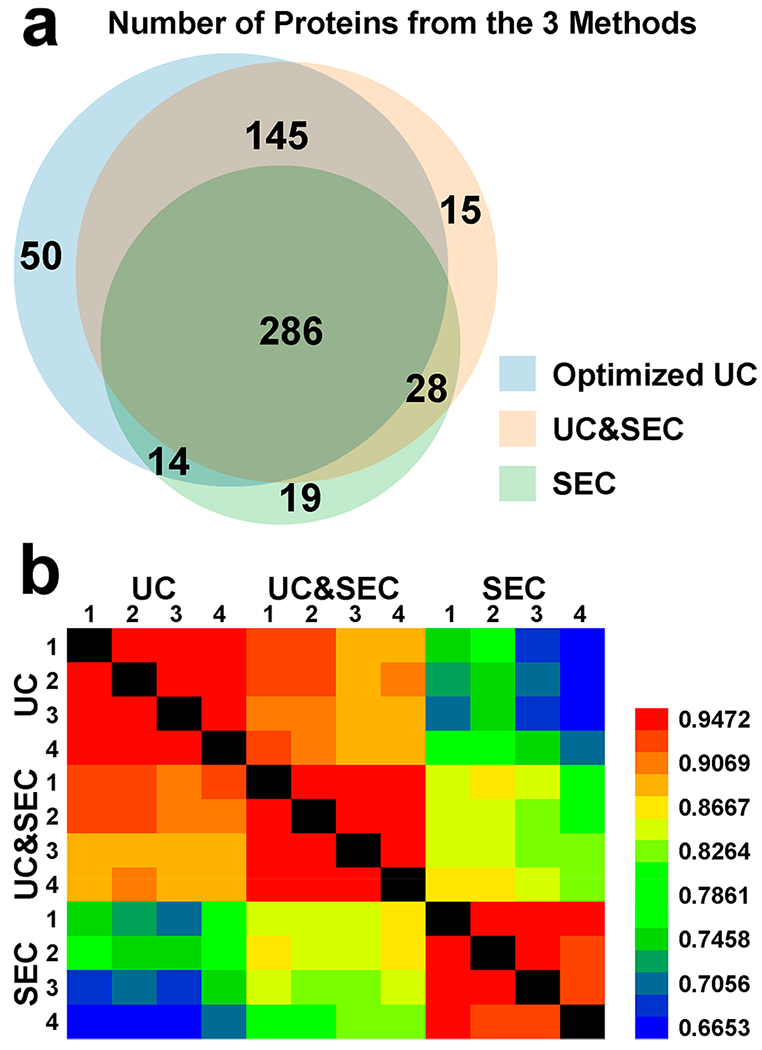
Proteomics analysis of exosomal proteins isolated from the three isolation methods. (a) Venn diagram showed the overlap of the identified proteins from the three isolation methods. (b) Heat map of Pearson correlation coefficient shows that the proteomes of different methods had a larger variation than that of four replicates of the same method.
We also studied various exosome markers that were detected by mass spectrometry analysis. Among these proteins, we did not detect the negative exosome marker Calnexin. We performed Western blot assay and verified the absence of Calnexin. We compared the recovery of exosome protein by the label-free method for quantifying the exosome markers CD9, CD63, and CD81 among the three methods because these marker proteins were identified in samples from all three methods. We also detected another positive exosome marker TSG101 in samples from UC and UC&SEC methods but not in samples from the SEC method, possibly due to the low ratio of markers to serum proteins.
Because the loading amount of the SEC sample was one tenth of that prepared by the other two methods, we amplified by 10 times the peak intensity of the SEC data when compared with the peak intensities of the other two methods. The protein amounts of three exosome markers CD9, CD63, and CD81 in samples from the UC&SEC column method are 1.3, 1.4, and 2.7 times that from the optimized UC method, respectively (Figure 5a). NanoSight results showed that the SEC method had more exosomes than the UC&SEC method, but the mass spectrometry data suggested that the amount of CD63 prepared by the UC&SEC method was larger. We speculate that there may be a large amount of serum protein contamination that suppresses the signal from exosome proteins in the SEC sample in the mass spectrometer. We then performed Western blot assay of CD63 and found that its abundance prepared by the optimized UC was 80% of that from the other two methods (Supplemental Figure S1).
Figure 5.
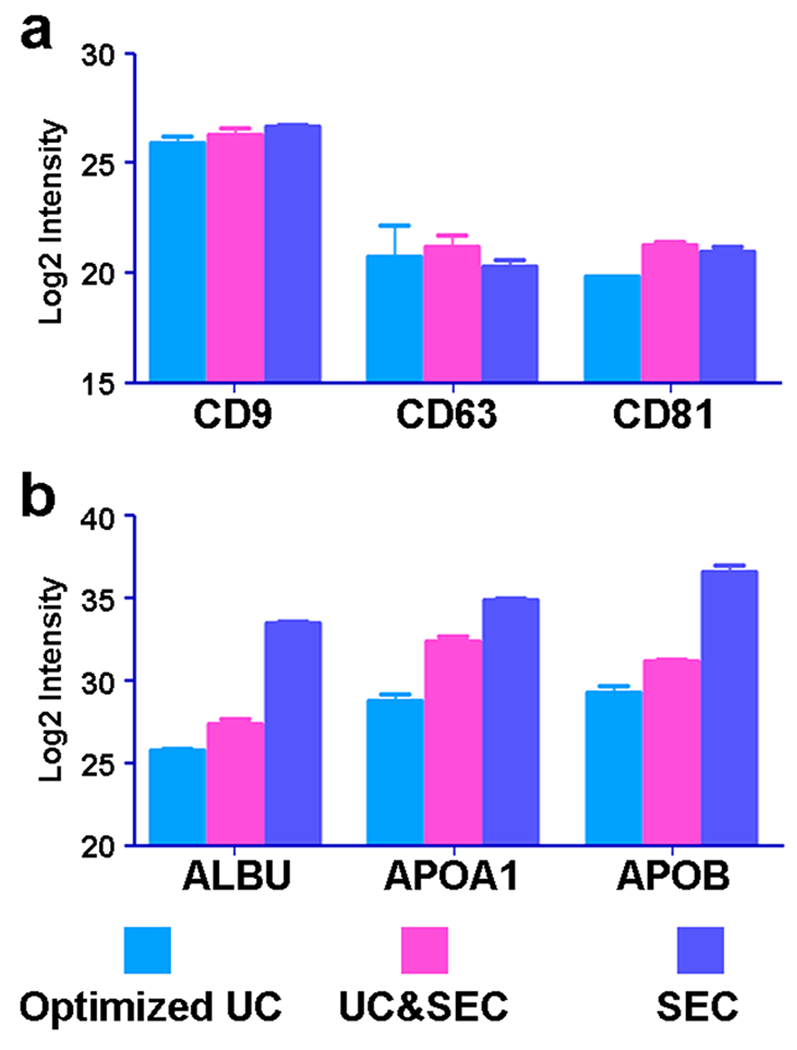
Relative abundance of exosome markers and serum contamination protein in the samples from the three methods. (a) Relative abundance of three exosome markers (CD9, CD63, and CD81) identified in the samples from the three methods was similar. The protein amounts of CD9, CD63, and CD81 in samples from the UC&SEC column method are 1.3, 1.4, and 2.7 times that of the optimized UC method, respectively. (b) Protein amounts of ALBU, APOA1, and APOB in samples from the SEC column method are 149.2, 38.6, and 127.5 times of that of the optimized UC method, respectively.
In the samples from the SEC method, the amounts of exosome protein and serum protein were the highest relative to those from the other two methods, but the amount of serum protein had a much larger ratio. According to the intensities of proteins in the mass spectrometer, APOB was the most abundant serum protein in samples from the SEC method. The protein amounts of ALBU, APOA1, and APOB in samples from the SEC column method are 149.2, 38.6, and 127.5 times that from the optimized UC method, respectively (Figure 5b). Western blot results verified that the abundances of ALBU from samples prepared by the optimized UC and SEC methods were the least and the most abundant, respectively (Supplemental Figure S2). Although it had a slightly lower exosome recovery than that from other two methods, the sample from our optimized UC method had the highest purity of exosome protein, which is critical for further proteomic analysis. Although the amount of various IgG in samples from the SEC method was slightly less than that from other two methods, the SEC method alone was limited for proteome analysis because serum proteins and various lipoproteins were dominant.
Theoretically, multiple cycles of the SEC column method could also probably remove serum protein more efficiently than one cycle with the accompanying loss. The speed of the flow through a used column becomes slower than a new one even after thorough washing, suggesting that the content of fractions might have a tiny shift, although the protein amounts in various fractions were not significantly changed between new columns and those used once according to our experiment. The use of UC before the SEC column was found to remove the serum protein contamination relative to the SEC method. As a result, the speed of the flow through a used column did not become significantly slower. Thus the UC&SEC method saves the lifetime of the qEV SEC column and increases the purity of exosomes for proteomics, although the qEV SEC columns were only used once in the UC&SEC method.
CONCLUSIONS
We optimized the UC method by diluting the serum with PBS to reduce the viscosity, prolonging the first cycle of UC and using another four cycles of UC. Our results suggest that serum proteins were efficiently removed without a significant loss of exosomes by our optimized UC method relative to the SEC method. We also combined one cycle of UC with SEC and found that this method provided improved results relative to the SEC method, although the blood protein contamination was slightly higher than that of our optimized UC method. The use of each these methods will depend on the application where the UC method would be best for proteomic discovery, whereas the UC&SEC method would be better for targeted proteomics. The SEC method has obvious limitations in the discovery of the exosome proteome due to large amounts of serum protein contamination, but it retains more exosome proteins and should be ideal for RNA analysis.
Supplementary Material
Figure S1: Western blot results of CD63 from the samples prepared by the optimized UC, UC&SEC, and SEC methods.
Figure S2: Western blot results of ALBU from the samples prepared by the optimized UC, UC&SEC, and SEC methods. (PDF)
Supplemental Table S1: Total proteins identified and quantified in the three methods. (XLSX)
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health under grant R01GM49500 (D.M.L.) and the National Cancer Institute under grant R21CA189775 (D.M.L.). We acknowledge the assistance of the Wayne State University Proteomics Core that is supported through National Institutes of Health grants P30 ES020957, P30 CA 022453, and S10 OD010700. We acknowledge Dr. Anoop Pal from iZON Science, Ltd., who provided us qEV 10 mL SEC columns.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00479.
The authors declare no competing financial interest.
REFERENCES
- (1).Kleeff J; Costello E; Jackson R; Halloran C; Greenhalf W; Ghaneh P; Lamb RF; Lerch MM; Mayerle J; Palmer D; Cox T; Rawcliffe CL; Strobel O; Buchler MW; Neoptolemos JP The impact of diabetes mellitus on survival following resection and adjuvant chemotherapy for pancreatic cancer. Br. J. Cancer 2016, 115, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yang C; Robbins PD The roles of tumor-derived exosomes in cancer pathogenesis. Clin. Dev. Immunol 2011, 2011, 842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Vlassov AV; Magdaleno S; Setterquist R; Conrad R Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim. Biophys. Acta, Gen. Subj 2012, 1820, 940–948. [DOI] [PubMed] [Google Scholar]
- (4).Hood JL; San RS; Wickline SA Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 2011, 71, 3792–3801. [DOI] [PubMed] [Google Scholar]
- (5).Huang SZ; Huang Y; Chen MJ; Chen W; Huang Z; Li H; Li JC; Ren ZR; Zeng YT A study of transgenic IFV cattle integrated with human serum albumin gene. Yichuan Xuebao 2000, 27, 573–579. [PubMed] [Google Scholar]
- (6).Whiteside TL Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv. Clin. Chem 2016, 74, 103–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Al-Hajj M; Wicha MS; Benito-Hernandez A; Morrison SJ; Clarke MF Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).van den Boorn JG; Dassler J; Coch C; Schlee M; Hartmann G Exosomes as nucleic acid nanocarriers. Adv. Drug Delivery Rev 2013, 65, 331–335. [DOI] [PubMed] [Google Scholar]
- (9).Henderson MC; Azorsa DO The genomic and proteomic content of cancer cell-derived exosomes. Front. Oncol 2012, 2, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pant S; Hilton H; Burczynski ME The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem. Pharmacol 2012, 83, 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Principe S; Jones EE; Kim Y; Sinha A; Nyalwidhe JO; Brooks J; Semmes OJ; Troyer DA; Lance RS; Kislinger T; Drake RR In-depth proteomic analyses of exosomes isolated from expressed prostatic secretions in urine. Proteomics 2013, 13, 1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Shao H; Chung J; Balaj L; Charest A; Bigner DD; Carter BS; Hochberg FH; Breakefield XO; Weissleder R; Lee H Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat. Med 2012, 18, 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Simpson RJ; Lim JW; Moritz RL; Mathivanan S Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 2009, 6, 267–283. [DOI] [PubMed] [Google Scholar]
- (14).Gonzales PA; Pisitkun T; Hoffert JD; Tchapyjnikov D; Star RA; Kleta R; Wang NS; Knepper MA Large-scale proteomics and phosphoproteomics of urinary exosomes. J. Am. Soc. Nephrol 2009, 20, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Chen Y; Xie Y; Xu L; Zhan S; Xiao Y; Gao Y; Wu B; Ge W Protein content and functional characteristics of serumpurified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int. J. Cancer 2017, 140, 900–913. [DOI] [PubMed] [Google Scholar]
- (16).An M; Lohse I; Tan Z; Zhu J; Wu J; Kurapati H; Morgan MA; Lawrence TS; Cuneo KC; Lubman DM Quantitative Proteomic Analysis of Serum Exosomes from Patients with Locally Advanced Pancreatic Cancer Undergoing Chemoradiotherapy. J. Proteome Res 2017, 16, 1763–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).An M; Zhu J; Wu J; Cuneo KC; Lubman DM Circulating Microvesicles from Pancreatic Cancer Accelerate the Migration and Proliferation of PANC-1 Cells. J. Proteome Res 2018, 17, 1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chen IH; Xue L; Hsu CC; Paez JS; Pan L; Andaluz H; Wendt MK; Iliuk AB; Zhu JK; Tao WA Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 3175–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Shao H; Chung J; Lee K; Balaj L; Min C; Carter BS; Hochberg FH; Breakefield XO; Lee H; Weissleder R Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun 2015, 6, 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Musante L; Tataruch D; Gu D; Benito-Martin A; Calzaferri G; Aherne S; Holthofer H A simplified method to recover urinary vesicles for clinical applications, and sample banking. Sci. Rep 2015, 4, 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Shin H; Han C; Labuz JM; Kim J; Kim J; Cho S; Gho YS; Takayama S; Park J High-yield isolation of extracellular vesicles using aqueous two-phase system. Sci. Rep 2015, 5, 13103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li P; Kaslan M; Lee SH; Yao J; Gao Z Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Szatanek R; Baj-Krzyworzeka M; Zimoch J; Lekka M; Siedlar M; Baran J The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci 2017, 18, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chen C; Skog J; Hsu CH; Lessard RT; Balaj L; Wurdinger T; Carter BS; Breakefield XO; Toner M; Irimia D Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 2010, 10, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wu M; Ouyang Y; Wang Z; Zhang R; Huang PH; Chen C; Li H; Li P; Quinn D; Dao M; Suresh S; Sadovsky Y; Huang TJ Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Lobb RJ; Becker M; Wen Wen S; Wong CS; Wiegmans AP; Leimgruber A; Möller A Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Welton JL; Webber JP; Botos LA; Jones M; Clayton A Ready-made chromatography columns for extracellular vesicle isolation from plasma. J. Extracell. Vesicles 2015, 4, 27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Boing AN; van der Pol E; Grootemaat AE; Coumans FA; Sturk A; Nieuwland R Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).van Eijndhoven MA; Zijlstra JM; Groenewegen NJ; Drees EE; van Niele S; Baglio SR; Koppers-Lalic D; van der Voorn H; Libregts SF; Wauben MH; de Menezes RX; van Weering JR; Nieuwland R; Visser L; van den Berg A; de Jong D; Pegtel DM Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight. 2016, 1, e89631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Stranska R; Gysbrechts L; Wouters J; Vermeersch P; Bloch K; Dierickx D; Andrei G; Snoeck R Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med 2018, 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Baranyai T; Herczeg K; Onodi Z; Voszka I; Modos K; Marton N; Nagy G; Mager I; Wood MJ; El Andaloussi S; Palinkas Z; Kumar V; Nagy P; Kittel A; Buzas EI; Ferdinandy P; Giricz Z Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS One 2015, 10, e0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kim J; Tan Z; Lubman DM Exosome enrichment of human serum using multiple cycles of centrifugation. Electrophoresis 2015, 36, 2017–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).An M; Zou X; Wang Q; Zhao X; Wu J; Xu LM; Shen HY; Xiao X; He D; Ji J High-confidence de novo peptide sequencing using positive charge derivatization and tandem MS spectra merging. Anal. Chem 2013, 85, 4530–4537. [DOI] [PubMed] [Google Scholar]
- (34).Cox J; Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 2008, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- (35).Momen-Heravi F; Balaj L; Alian S; Trachtenberg AJ; Hochberg FH; Skog J; Kuo WP Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol 2012, 3, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Shelke GV; Lasser C; Gho YS; Lotvall J Importance of exosome depletion protocols to eliminate functional and RNAcontaining extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 2014, 3, 24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wisniewski JR; Zougman A; Nagaraj N; Mann M Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [DOI] [PubMed] [Google Scholar]
- (38).An M; Shen H; Cao J; Pei X; Chang Y; Ma S; Bao J; Zhang X; Bai X; Ma Y The alteration of H4-K16ac and H3-K27met influences the differentiation of neural stem cells. Anal. Biochem 2016, 509, 92–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Western blot results of CD63 from the samples prepared by the optimized UC, UC&SEC, and SEC methods.
Figure S2: Western blot results of ALBU from the samples prepared by the optimized UC, UC&SEC, and SEC methods. (PDF)
Supplemental Table S1: Total proteins identified and quantified in the three methods. (XLSX)


