Abstract
Background
Statistically significant linkage of melanoma to chromosome 9q21 was previously reported in a Danish pedigree resource and independently confirmed in Utah high-risk pedigrees, indicating strong evidence that this region contains a melanoma predisposition gene.
Methods
Whole-exome sequencing of pairs of related melanoma case subjects from two pedigrees with evidence of 9q21 linkage was performed to identify the responsible predisposition gene. Candidate variants were tested for association with melanoma in an independent set of 454 unrelated familial melanoma case subjects and 396 unrelated cancer-free control subjects from Utah, and 1534 melanoma case subjects and 1146 noncancer control subjects from Texas (MD Anderson) via a two-sided Fisher exact test.
Results
A rare nonsynonymous variant in Golgi Membrane Protein 1 (GOLM1), rs149739829, shared in two hypothesized predisposition carriers in one linked pedigree was observed. Segregation of this variant in additional affected relatives of the index carriers was confirmed. A statistically significant excess of carriers of the variant was observed among Utah case subjects and control subjects (odds ratio [OR] = 9.81, 95% confidence interval [CI] = 8.35 to 11.26, P < .001) and statistically significantly confirmed in Texas case subjects and control subjects (OR = 2.45, 95% CI = 1.65 to 3.25, P = .02).
Conclusion
These findings support GOLM1 as a candidate melanoma predisposition gene.
The clinical and economic burden of cutaneous malignant melanoma (CMM) is substantial and increasing (1), with a lifetime risk of 1% (2). Familial aggregation studies conducted in population genealogies have provided substantial evidence for a genetic contribution (3–6), and multiple CMM predisposition genes have been identified. Variants in CDKN2A (p16) (7) explain a high percentage of familial CMM (40% of melanoma families and 2% of case subjects) (8). Other CMM predisposition genes, each explaining a small number, or percentage, of CMM pedigrees, include ACD (six families) (9), BAP1 (24 families) (10), CDK4 (17 families) (11), CDKN2B (p14ARF; 1% of families) (12), MITF (1% of CMM) (13), POT1 (12 families) (14,15), TERF2IP (five families) (8), and TERT (one family) (16); however, a large number of high-risk CMM pedigrees and case subjects remain unexplained by any of these genes.
Almost a quarter century after the study of melanoma pedigrees identified CDKN2A as the first CMM predisposition gene (7,17), it is still recognized that high-risk pedigree studies are a powerful method for identification of rare predisposition variants with large effects and should be performed when these rare resources are available (18–20).
Statistically significant evidence for linkage of CMM to chromosome 9q21 was previously reported in the Danish population (21) and confirmed in Utah pedigrees (22). To identify the predisposition genes and variants that contribute to CMM at this locus, whole-exome sequencing was performed on pedigree members that shared the hypothesized segregating predisposition haplotype in each of two Utah CMM pedigrees showing evidence of 9q21 linkage.
Methods
Utah CMM Pedigrees
Hundreds of high-risk CMM pedigrees have been identified and studied in Utah using genealogy linked to state-wide Utah Cancer Registry (UCR) data from 1966 to the present in the Utah Population Database (UPDB). CMM case subjects recorded in the UCR (National Cancer Institute [NCI] Surveillance, Epidemiology, and End Results [SEER] Registry from 1973) represent histologically confirmed, independent primary cancers. Two Utah high-risk CMM pedigrees were previously reported to show linkage to the 9q21 region (22). Linkage analysis identified hypothesized predisposition haplotype carriers at the 9q21 locus from each linked pedigree, and whole-exome sequence data were generated for two hypothesized carriers from one pedigree and three from the other. Whole-exome sequence data from 12 pairs of CMM-affected cousins from an additional 12 Utah high-risk CMM pedigrees (all screened negative for known melanoma predisposition genes CDKN2A, CDKN2B, or CDK4) were used to perform a burden test for GOLM1 with pVAAST software, version 2.2.0 (Omicia, Inc., Oakland, CA).
Utah Case and Control Subjects
Utah CMM case subjects included 923 case subjects who are members of hundreds of Utah high-risk CMM pedigrees; all CMM diagnoses came from the Utah Cancer Registry or Huntsman Cancer Institute Clinics and were histopathologically confirmed. Utah control subjects included 396 unrelated cancer-free subjects who had no first-degree relatives with cancer and no second-degree relatives with melanoma. This study was approved by the Institutional Review Board, and informed consent was obtained from all participants.
Texas Case and Control Subjects
The 1534 CMM case subjects and 1146 control subjects from Texas were participants in a previously described hospital-based case–control study at MD Anderson (23). All participants self-identified as non-Hispanic with European ancestry and were recruited between March 1998 and August 2013. Control participants were cancer-free and were recruited from friends or acquaintances of patients. The case–control study was approved by the Institutional Review Board, and informed consent was obtained from all participants.
Utah Exome Data Analysis
Whole-exome sequencing for 29 Utah CMM case subjects selected as cousin pairs (and one cousin trio) from 14 high-risk CMM pedigrees (including the two 9q-linked pedigrees) was performed at the Huntsman Cancer Institute’s Genomics Core facility. DNA libraries were prepared from 3 micrograms of DNA using the Agilent SureSelect XT Human All Exon + UTR (v5) capture kit. Samples were run on the Illumina HiSeq 2000 instrument, which generates paired-end reads up to 100 base pairs in length. Raw reads were mapped to the human genome v19 reference genome using BWA-mem (24) for alignment, and variants were called using Genome Analysis Toolkit (GATK) (25) software following Broad Institute Best Practices Guidelines. Exome capture resulted in an average of 85.7% of target bases being covered by greater than 10× coverage across the genome. Variants were called with GATK software and annotated with Annovar (26). Variants occurring outside the exon capture kit intended area of coverage were removed.
Texas Exome Data Analysis
Whole-exome sequencing for 396 Utah cancer-free control subjects was performed at the MD Anderson Sequencing and Microarray Facility. DNA libraries were prepared from 0.5 micrograms of DNA per sample using the Agilent SureSelect Clinical Research Exome capture kit. Samples were run on the Illumina HiSeq 4000 instrument, with eight samples per lane and 150 base pair paired-end reads. Raw reads were mapped, and variants were called simultaneously with the Utah melanoma case subjects. Exome capture resulted in an average of 98.2% of target bases being covered by greater than 10× coverage across the genome.
Genotyping of GOLM1 Variant
The GOLM1 candidate variant was confirmed in the Utah index carriers and their relatives with Sanger sequencing. In addition, the entire set of 923 Utah CMM case subjects, 1534 Texas CMM case subjects, and 1146 Texas control subjects were genotyped for single nucleotide polymorphism (SNP) rs149739829 from genomic DNA using the TaqMan Assay C_175102585_10 (Applied Biosystems, Carlsbad, CA) according to the manufacturer’s specifications on a Bio-Rad CFX96 (Utah samples) and CFX384 (Texas samples) real-time PCR instruments.
Statistical Analysis
Statistical analysis for case–control association used a two-sided Fisher exact test; a P value of less than .05 was considered statistically significant. Clusters of related CMM case subjects in pedigrees were characterized as high risk by comparing observed with expected numbers of CMM case subjects among members of a descending pedigree based on age-, sex-, and birthplace-specific CMM rates estimated in the UPDB (27). A test for significance of the relative risk statistic and 95% confidence intervals assume that the observed number of case subjects follows a Poisson distribution, with mean equal to the expected value (3).
The Pedigree Variant Annotation, Analysis, and Search Tool (pVAAST), version 2.2.0, was used to perform a burden test for GOLM1 in exome data from the 14 Utah pedigrees (28). pVAAST combines case–control information, functional variant prediction, and evidence of segregation for variants shared among related case subjects to evaluate the combined statistical evidence of disease-gene association in familial sequencing studies (28). Background genomes used to run pVAAST included 1000 Genomes Phase 1 samples (1K-Genomes) (29) denoted as having Northern European ancestry and the Complete Genomics Diversity Panel samples (http://www.completegenomics.com/public-data/69-genomes/). When the same individual was sequenced by both 1K-Genomes and the Complete Genomics Diversity Panel, the 1K-Genomes sample was removed, resulting in 1112 samples. Analysis excluded low-quality variants with statistically significant departure from Hardy-Weinberg equilibrium (P < .0001) or with missing genotype rates greater than 10.0% among background genomes.
Some of the GOLM1 candidate variant carriers from Utah (n = 16) had been genotyped on Illumina Human OmniExpress high-density SNP array platform. SNP genotypes were used to estimate shared haplotypes between carriers to assess the possibility of an inheritance from a common ancestor using Shared Genomic Segments analysis (SGS) (30). SGS analysis identifies the set of contiguous markers at which genotyped individuals could share alleles; long runs of such markers indicate identity-by-descent sharing from a common ancestor (30). The length of the shared haplotype in carriers was used to estimate the number of meioses between the haplotype carriers and a common ancestor. Thomas et al. (1994) showed that the extent of haplotype sharing L, measured in centi-Morgans, around a locus shared identical-by-descent by k individuals linked together in a pedigree comprising d meioses has the Gamma(2,d/100) distribution (31). Thus, dL/100 has the Gamma(2,1) distribution, from which E(dL/100) = 2 and P(0.242 ≤ dL/100 ≤ 5.572) = 0.95. In addition, Kingman (1982) showed that the time to coalescence for k haplotypes is given by
(32)
Results
Exome Sequencing of Utah CMM Case Subjects
Analysis of exome data at chromosome 9q21 for the hypothesized predisposition haplotype carriers from each of two 9q21 linked high-risk CMM pedigrees identified a rare coding variant in GOLM1 that was shared by the three hypothesized carriers in one pedigree. This GOLM1 variant (rs149739829) is a nonsynonymous variant substitution of C to T in exon 8 that codes a Serine > Leucine amino acid substitution. Available population frequency estimates (minor allele frequency/number of alleles tested) include A = 0.00398, 483/121 370 ExAC browser (33); A = 0.0010, 5/5008 1000 Genomes database v3 (29); and A = 0.00339, 44/12 962 NHLBI Exomes (GO-ESP; Exome Variant Server, NHLBI GO Exome Sequencing Project; Seattle, WA; http://evs.gs.washington.edu/EVS/).
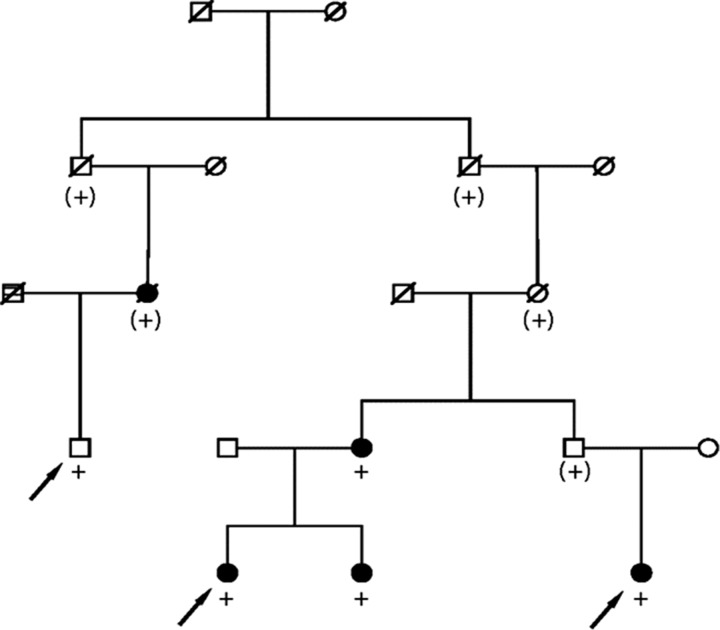
Sanger sequencing confirmed the presence of the rare rs149739829 allele in the three sequenced subjects and in two additional CMM-affected relatives in the original pedigree; the variant was inferred in an additional CMM case subject in the pedigree (the affected parent of one carrier; informed consent was obtained for each living individual represented) (Figure 1). A test of statistical excess of CMM among all 797 descendants of the pedigree founders who are included in the UPDB (not all shown) identified a total of eight CMM case subjects, 1.7 expected (two-sided P < .001). Four of the variant carriers have at least two independent melanomas, and the fifth had an additional cancer. The average age at diagnosis of the melanomas was 40 years; all melanomas were stage 1 (localized) at diagnosis per SEER staging data.
Figure 1.
High-risk cutaneous malignant melanoma (CMM) pedigree segregating a rare variant in GOLM1. Arrows indicate index predisposition haplotype carriers with whole-exome sequencing data, black fill indicates CMM diagnosis, + indicates carriage of the rare allele at rs149739829 confirmed by Sanger Sequencing, and (+) indicates inferred carriage.
Case–Control Association for rs149739829
The 923 Utah CMM case subjects consisted of 454 unrelated case subjects and 469 CMM case subjects related to them in pedigrees. The genotype data for variant rs149739829 (Taqman assay) in the 454 unrelated Utah CMM case subjects was compared with data on this variant from the 396 exomes from unrelated cancer-free Utah control subjects; 22 case subjects and two control subjects carried the variant (odds ratio [OR] = 9.81, 95% confidence interval [CI] = 8.35 to 11.26, Fisher two-sided P < .001). Variant rs149739829 was also genotyped in an independent population from MD Anderson Cancer Center. Among a total of 1534 CMM case subjects and 1146 control subjects, 26 case subjects and eight control subjects carried the variant (OR = 2.45, 95% CI = 1.65 to 3.25, Fisher two-sided P = .02).
Additional rs149739829 Variant-Carrying Utah Pedigrees
Additional carriers of the rare allele at rs149739829 were identified among the 469 Utah CMM related case subjects. We observed two other pairs of related case subjects who carried the rare variant (a pair of second cousins and a pair of first cousins). The first pedigree has 2000 descendants, including five Utah CMM case subjects (with six expected; two-sided P = .69). The second pedigree has 300 descendants, including four CMM case subjects (with 0.8 expected; two-sided P = .009).
pVAAST Burden Test
A burden test was performed for the GOLM1 gene as the only genomic feature in 29 Utah CMM exomes together with 1111 control subjects of European ancestry. pVAAST estimated a burden score of 13.563 (permutation-based 95% CI = 6.3E-4 to 1.0E-5, two-sided P < .001), demonstrating that the observed allele frequency among the case subjects is unlikely under the null hypothesis of no disease association.
Two GOLM1 variants received a positive score in the burden test, rs149739829 and another nonsynonymous variant (rs142242230) detected in two unrelated sequenced CMM case subjects. This additional GOLM1 variant rs142242230, chr9:88,648,217 (hg19), is a nonsynonymous variant substitution of C to T in exon 9 that codes a Glycine > Glutamic acid amino acid substitution with population frequency estimates (given as minor allele frequency/number of alleles tested): T = 0.0042, 506/121 186 (ExAC) (33), T = 0.0026, 13/5008 (1000 Genomes) (29), T = 0.0053, 69/12 937 (GO-ESP). The second GOLM1 variant was not statistically significantly associated with CMM in the 454 Utah CMM case subjects and 396 control subjects, where 17 case subjects and 11 control subjects were observed (OR = 1.35, 95% CI = 0.59 to 2.12, Fisher two-sided P = .44), and was not considered further here.
Identification and Age of Shared Haplotype in GOLM1 Variant Carriers
SGS analysis identified that all 16 GOLM1 candidate variant carriers with high-density SNP array data shared a 306 Kb region of chromosome 9 (88.641–88.714 Mb) containing the GOLM1 gene.
Thus, by the method of moments, an observation of L = 306 Kb, which is approximately 0.306 centi-Morgans, gives an estimate of d = 2*100/0.306 = 653.6 meioses (95% CI = 79.1 to 1820.9). Thus, for k = 16, an estimate for the time to most recent common ancestor is 184.7 generations (95% CI = 22.3 to 514.5). Assuming 20 years per generation, this is estimated to be 3693 years ago (95% CI = 447 to 10 289).
Discussion
A powerful and efficient study design for predisposition gene identification, focused on sequencing members of extended high-risk pedigrees with evidence for linkage, combined with validation in multiple populations, has identified GOLM1 as a candidate predisposition gene for CMM. Previous analysis of large CMM pedigrees led to the discovery of CDKN2A as a predisposition gene for CMM (7,17). These outcomes speak to the continued value of high-risk pedigree resources for predisposition gene discovery.
The GOLM1 gene encodes Golgi-related Golgi membrane protein 1 (GOLM1, GOLPH2, or GP73), which is a type II transmembrane protein localized predominately to the cis- and medial-Golgi cisternae in cells of epithelial lineage within most human tissue types (34). GOLM1 has a short 12–amino acid N-terminal cytoplasmic domain, a 13–amino acid transmembrane domain that regulates its Golgi localization (35), and a 376–amino acid Golgi luminal domain containing multiple N- and O-linked glycosylation sites and a coiled-coil domain that mediates GOLM1 dimerization (35). A pro-protein protease cleavage site spanning residues 52–55 allows for removal of the large luminal domain and secretion of cleaved GOLM1 into the extracellular milieu (35).
Secreted GOLM1 has been used as a biomarker for clinical diagnosis of hepatocellular carcinoma (HCC) (36) and chronic liver disease (37) as numerous reports have demonstrated that viral infection of hepatocytes by hepatitis B or C virus increases GOLM1 expression and serum levels (34,38). In patients with virus-induced HCC, elevated GOLM1 is associated with poorer patient survival (39–41). Increased GOLM1 expression has also been reported in gastric cancer (42), prostate cancer (43), and melanoma (44). Elevated GOLM1 expression is correlated with decreased disease-free survival and cancer-specific overall survival in melanoma patients (44) as its expression coincides with clinical features indicative of aggressive disease such as increased tumor thickness, ulceration, and lymph node metastasis (44).
While the native function of GOLM1 is still poorly understood, high GOLM1 expression in HCC cells promotes proliferation, migration, and invasion in vitro and ultimately metastasis of HCC xenografts in vivo (41,45,46). Loss-of-function experiments show that silencing elevated GOLM1 expression attenuates metastasis by inhibiting the epithelial-to-mesenchymal transition required for migration and invasion (47). These GOLM1-modulated cellular phenotypes enable malignant cell progression to metastasis and are mechanistically regulated through altered membrane trafficking of receptor tyrosine kinases (RTKs). Aberrant, unregulated RTK signaling is a hallmark of oncogenic transformation and cancer metastasis (48).
Using HCC cell lines, Ye et al. demonstrated that GOLM1 is recruited from the trans-Golgi network to Rab5-positive early endosomes upon epidermal growth factor stimulation of HCC cells, where it mediates the “slow recycling” of the epidermal growth factor receptor (EGFR) via re-entry through the Golgi network back to the cell surface in lieu of lysosomal degradation. GOLM1 binds EGFR or other RTKs (c-MET, VEGFR) through residues 5–12 in its short N-terminus and thus promotes RTK postendocytic recycling via a Rab11-regulated recycling compartment (41). Thus, increased levels of GOLM1 promote elevated EGFR expression on the cell surface by enhanced recycling, which results in elevated Protein Kinase B pathway activity that drives HCC metastasis, rather than mitogenic signal attenuation through lysosomal RTK degradation (41). While these and other reports demonstrate that elevated levels of GOLM1 are sufficient to drive oncogenesis and metastasis, no reports to date have demonstrated a GOLM1 mutation that alters its cellular function and thus predisposes to malignant transformation. The GOLM1 variant of interest, at amino acid position 307, is within 2 amino acids of a known phosphorylation site, S309; phosphorylation of S309 might be affected by the variant. The kinase implicated in modifying S309 is Fam20C, which is responsible for generating the vast majority of the secreted phosphoproteome, including substrates linked to tumor cell migration (49).
Given the recognized link between GOLM1 expression and melanoma progression (44) and the association of the variant with melanoma predisposition reported here, we hypothesize that this variant may increase cellular levels of the protein either through increased protein stability or decreased degradation in melanocytes. Future studies will test this hypothesis.
Melanoma incidence is rising, increasing the need to better identify persons at risk to facilitate earlier and more efficient skin screening for detection. It is likely that GOLM1 will only explain a small number of melanomas, as is the case for most of the melanoma predisposition genes identified. Nevertheless, each predisposition gene identified increases our understanding of the biology underlying melanoma. Currently, only the CDKN2A (p16) test is commercially available; each new predisposition identified can potentially identify additional individuals at risk.
There are some limitations that must be noted. CMM phenotypes were identified primarily from the Utah Cancer Registry, an NCI SEER registry registering all cancers in Utah since 1973. CMM case subjects diagnosed before 1973, outside Utah, or in individuals without linked genealogy data were censored. Utah’s population is reflective of its founding Mormon pioneers, which consisted of largely unrelated groups of northern Europeans from Scotland, Wales, Denmark, Sweden, and England. Subsequent migration, along with a more recent substantial influx of international migrants, contributes to a population mix that is thought to be representative of the broader US and northern Europe. Our focus on established Utah high-risk pedigrees (requiring genealogy data) limited the population we analyzed to original Utah pioneer pedigrees, which are exclusively of Northern European extraction. It should be recognized that extrapolations to other populations should not be made without additional evaluation.
In conclusion, we report evidence for the role of a rare GOLM1 variant (rs149739829) in CMM predisposition. The detected variant conveys moderate risk in a set of population-ascertained case subjects (Texas) and high risk among familial case subjects (Utah).
Funding
This work was supported by the National Cancer Institute (R01 CA195614 to CH and LACA, RO1 CA102422 to LACA, and P50CA093459 to CH).
This research was supported by the Utah Cancer Registry, funded by Contract No. HHSN261201000026C from the National Cancer Institute’s Surveillance, Epidemiology, and End Results Program with additional support from the Utah Department of Health and University of Utah. Partial support for the Utah Population Database was provided by Huntsman Cancer Institute, Huntsman Cancer Foundation, University of Utah, and the Huntsman Cancer Institute’s Cancer Center Support grant (P30 CA42014) from the National Cancer Institute. LACA is supported by the George E. Wahlen Department of Veterans Affairs Medical Center. SAL is supported by the Oregon Health and Science University Knight Cancer Institute and the John D. Gray Endowed Chair. JCF has support from the Utah Center for Clinical and Translational Science, funded by National Center for Advancing Translational Sciences award 1ULTR001067. Computer resources were provided by the University of Utah Center for High Performance Computing. Support for the Texas data set was provided in part by contributions to the University of Texas MD Anderson Cancer Center Moon Shots Program, the Miriam and Jim Mulva Melanoma Research Fund, and the Marit Peterson Fund for Melanoma Research.
Notes
Affiliations of authors: Genetic Epidemiology, Department of Internal Medicine (CCT, JS, JMF, AT, LACA), Huntsman Cancer Institute (SLH, MRS, KT, DG, LACA), Center for Cell and Genome Science (MB), and Department of Biology (MB), University of Utah, Salt Lake City, UT; Department of Epidemiology, University of Texas MD Anderson Cancer Center, Houston, TX (CH, YY, HZ); Department of Surgery, University of Utah Health Sciences Center, Salt Lake City, UT (SLH, MRS, KT); Department of Dermatology (DG, SRF, LM, JJZ) and Department of Biomedical Informatics (JCF), University of Utah School of Medicine, Salt Lake City, UT; Utah Department of Health, Salt Lake City, UT (JW); George E. Wahlen Department of Veterans Affairs Medical Center, Salt Lake City, UT (LM, LACA); Department of Dermatology and Knight Cancer Institute, Oregon Health and Science University, Portland, OR (SL).
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
References
- 1. Guy GP, Machlin SR, Ekwueme DU, Yabroff KR.. Prevalence and costs of skin cancer treatment in the US, 2002-2006 and 2007-2011. Am J Prev Med. 2015;482:183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Howlader N, Noone AM, Krapcho M, et al. , eds. SEER Cancer Statistics Review, 1975-2008. Bethesda, MD: National Cancer Institute; 2011. [Google Scholar]
- 3. Teerlink CC, Albright FS, Lins L, Cannon-Albright LA.. A comprehensive survey of cancer risks in extended families. Genet Med. 2012;141:107–114. [DOI] [PubMed] [Google Scholar]
- 4. Albright FS, Teerlink CC, Werner TL, Cannon-Albright LA.. Significant evidence for a heritable contribution to cancer predisposition: A review of cancer familiality by site. BMC Cancer. 2012;12:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, et al. Cancer as a complex phenotype: Pattern of cancer distribution within and beyond the nuclear family. PLoS Med. 2004;13:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frank C, Sundquist J, Yu H, Hemminki A, Hemminki K.. Concordant and discordant familial cancer: Familial risks, proportions and population impact. Int J Cancer. 2017;1407:1510–1516. [DOI] [PubMed] [Google Scholar]
- 7. Kamb A, Shattuck-Eidens D, Eeles R, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;81:23–26. [DOI] [PubMed] [Google Scholar]
- 8. Aoude LG, Wadt KA, Pritchard AL, Hayward NK.. Genetics of familial melanoma: 20 years after CDKN2A. Pigment Cell Melanoma Res. 2015;282:148–160. [DOI] [PubMed] [Google Scholar]
- 9. Aoude LG, Pritchard AL, Robles-Espinoza CD, et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J Natl Cancer Inst. 2015;1072:dju408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;4310:1018–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soufir N, Avril MF, Chompret A, et al. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet. 1998;72:209–216. [DOI] [PubMed] [Google Scholar]
- 12. Randerson-Moor JA, Harland M, William S, et al. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;101:55–62. [DOI] [PubMed] [Google Scholar]
- 13. Cronin JC, Wunderlich J, Loftus SK, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;224:435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robles-Espinoza CD, Harland M, Ramsay AJ, et al. POT1 loss-of-function variants predispose to familial melanoma. Nat Genet. 2014;465:478–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi J, Yang XR, Ballew B, et al. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;465:482–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;3396122:959–961. [DOI] [PubMed] [Google Scholar]
- 17. Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;81:15–21. [DOI] [PubMed] [Google Scholar]
- 18. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;4617265:747–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Hum Genet. 2012;13110:1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ott J, Wang J, Leal SM.. Genetic linkage analysis in the age of whole-genome sequencing. Nat Rev Genet. 2015;165:275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jönsson G, Bendahl P, Sandberg T, et al. Mapping of a novel ocular and cutaneous malignant melanoma susceptibility locus to chromosome 9q21.32. J Nat Cancer Inst. 2005;9718:1377–1382. [DOI] [PubMed] [Google Scholar]
- 22. Cannon-Albright LA, Teerlink CC, Farnham JM, et al. Linkage analysis of extended high-risk pedigrees replicates a cutaneous malignant melanoma predisposition locus on chromosome 9q21. J Invest Dermatol. 2013;1331:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amos CI, Wang LE, Lee JE, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;2024:5012–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li H, Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler Transform. Bioinformatics. 2010;265:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;209:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang K, Li M, Hakonarson H.. ANNOVAR: Functional annotation of genetic variants from high-throughput sequence data. Nucleic Acids Res. 2010;3816:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cannon-Albright LA. Utah family-based analysis: pPst, present and future. Hum Hered. 2008;654:209–220. [DOI] [PubMed] [Google Scholar]
- 28. Hu H, Huff CD, Moore B, Flygare S, Reese MG, Yandell M.. VAAST 2.0: Improved variant classification and disease-gene identification using a conservation-controlled amino acid substitution matrix. Genet Epidemiol. 2013;376:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. 1000 Genomes Project Consortium, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;4917422:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thomas A, Camp NJ, Farnham JM, Allen-Brady K, Cannon-Albright LA.. Shared genomic segment analysis. Mapping disease predisposition genes in extended pedigrees using SNP genotype assays. Ann Hum Genet. 2008;72(Pt 2):279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas A, Skolnick MH, Lewis CM.. Genomic mismatch scanning in pedigrees. IMA J Math Appl Med Biol. 1994;111:1–16. [DOI] [PubMed] [Google Scholar]
- 32. Kingman JFC. On the genealogy of large populations. J Applied Prob. 1982;19:27–43. [Google Scholar]
- 33. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;5367616:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kladney RD, Bulla GA, Guo L, et al. GP73, a novel Golgi-localized protein upregulated by viral infection. Gene. 2000;249(1–2):53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu L, Li L, Xie H, Gu Y, Peng T.. The Golgi localization of GOLPH2 (GP73/GOLM1) is determined by the transmembrane and cytoplamic sequences. PLoS One. 2011;611:e28207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsuchiya N, Sawada Y, Endo I, Saito K, Uemura Y, Nakatsura T.. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;2137:10573–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu Z, Liu L, Pan X, et al. Serum Golgi Protein 73 (GP73) is a diagnostic and prognostic marker of chronic HBV liver disease. Medicine (Baltimore). 2015;9412:e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu L, Yao W, Wang F, Rong X, Peng T.. GP73 is upregulated by hepatitis C virus (HCV) infection and enhances HCV secretion. PLoS One. 2014;93:e90553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang Y, Su Y, Zhao Y, Pan C, Chen L.. Golgi phosphoprotein3 overexpression is associated with poor survival in patients with solid tumors: A meta-analysis. Int J Clin Exp Pathol. 2015;89:10615–10624. [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y, Zou Z, Zhu B, Hu Z, Zeng P.. CXCL10 decreases GP73 expression in hepatoma cells at the early stage of hepatitis C virus (HCV) infection. Int J Mol Sci. 2013;1412:24230–24241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ye QH, Zhu WW, Zhang JB, et al. GOLM1 modulates EGFR/RTK cell-surface recycling to drive hepatocellular carcinoma metastasis. Cancer Cell. 2016;303:444–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu G, Zhang Y, He F, et al. Expression of GOLPH2 is associated with the progression of and poor prognosis in gastric cancer. Oncol Rep. 2014;325:2077–2085. [DOI] [PubMed] [Google Scholar]
- 43. Kojima S, Enokida H, Yoshino H, et al. The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet. 2014;592:78–87. [DOI] [PubMed] [Google Scholar]
- 44. Donizy P, Kaczorowski M, Biecek P, Halon A, Szkudlarek T, Matkowski R.. Golgi-related proteins GOLPH2 (GP73/GOLM1) and GOLPH3 (GOPP1/MIDAS) in cutaneous melanoma: Patterns of expression and prognostic significance. Int J Mol Sci. 2016;1710:E1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen X, Wang Y, Tao J, et al. mTORC1 up-regulates GP73 to promote proliferation and migration of hepatocellular carcinoma cells and growth of xenograft tumors in mice. Gastroenterology. 2015;1493:741–752. [DOI] [PubMed] [Google Scholar]
- 46. Jin D, Tao J, Li D, et al. Golgi protein 73 activation of MMP-13 promotes hepatocellular carcinoma cell invasion. Oncotarget. 2015;632:33523–33533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang Y, Liu Q, Zhang H, et al. Silencing of GP73 inhibits invasion and metastasis via suppression of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncol Rep. 2017;372:1182–1188. [DOI] [PubMed] [Google Scholar]
- 48. Lamorte L, Park M.. The receptor tyrosine kinases: Role in cancer progression. Surg Oncol Clin N Am. 2001;102:271–288. [PubMed] [Google Scholar]
- 49. Tagliabracci VS, Wiley SE, Guo X, et al. A single kinase generates the majority of the secreted phosphoproteome. Cell. 2015;1617:1619–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]