Abstract
Background
Tobacco smoking creates DNA damage, inducing mutations and potentially altering the tumor immune microenvironment. These types of genetic and immune microenvironment alterations are critical factors known to affect tumor response to immunotherapy. Here we analyze the association between the mutational signature of tobacco smoking, tumor mutational load, and metrics of immune activity in squamous cell carcinomas arising in the head and neck and lung.
Methods
Using RNA and DNA sequencing data from The Cancer Genome Atlas head and neck (HNSC; n = 287) and lung (LUSC; n = 130) squamous cell carcinoma data sets and two independent gene expression data sets (HNSC, n = 136; LUSC, n = 75), we examined associations between the mutational smoking signature, mutation count, immune cell infiltration, cytolytic activity, and interferon-γ signaling.
Results
An increasing mutational smoking signature was associated with statistically significantly increased overall mutational load in both HNSC (ρ = .33, P = 1.01 × 10−7) and LUSC (ρ = .49, P = 2.80 × 10−9). In HNSC, a higher mutational smoking signature was associated with lower levels of immune infiltration (ρ = –.37, P = 1.29 × 10−10), cytolytic activity (ρ = –.28, P = 4.07 × 10−6), and interferon-γ pathway signaling (ρ = .39, P = 3.20 × 10−11). In LUSC, these associations were reversed (ρ = .19, P = .03; ρ = .20, P = .02; and ρ = .18, P = .047, respectively). Differentially expressed genes between smoking-high and smoking-low tumors revealed broad tobacco-induced immunosuppression in HNSC, in contrast to a tumor-inflamed microenvironment in smokers with LUSC.
Conclusions
In squamous cell carcinomas, the genetic smoking signature is associated with higher mutational load, but variable effects on tumor immunity can occur, depending on anatomic site. In HNSC, smoking is predominantly immunosuppressive; in LUSC, more pro-inflammatory. Both tumor mutation load and immune microenvironment affect clinical response to immunotherapy. Thus, the mutational smoking signature is likely to have relevance for immunotherapeutic investigation in smoking-associated cancers.
Tobacco smoking is a risk factor for the development of cancers of the lung, head and neck, and other sites. Tobacco carcinogens create a specific mutational signature in tumors and are associated with increased overall mutation count (1). It is also likely that immunologic sequelae of smoking may contribute to the development of cancer, with both pro-inflammatory (2) and immunosuppressive (3) effects having been described. Both of these factors—mutational load and the tumor immune microenvironment—have been associated with tumor response to immunotherapy (4,5). Clinical data indicate that smokers with lung cancer have higher response rates to immune checkpoint inhibitor therapy (6,7), while smokers with head and neck cancer tend to have lower response rates (8).
Because only a minority (10%–15%) of patients with head and neck and non–small cell lung cancer experience clinical benefit with immune checkpoint inhibitor therapy, there is a need to identify factors that determine clinical response. Identifying predictive biomarkers can maximize clinical benefit for patients by guiding choice of therapy, and potentially illuminate novel biologic mechanisms.
In order to better understand why the majority of smoking-related tumors do not respond to immune checkpoint inhibitor therapy, we analyzed the mutational signature of smoking and its association with tumor mutational load and the immune microenvironment—parameters that modulate immunotherapy response—in squamous cell carcinomas arising in the head and neck (HNSC) and lung (LUSC).
Methods
Data Sources
The gene-level RSEM-normalized RNA-sequencing (RNAseq) data for The Cancer Genome Atlas (TCGA) head and neck (HNSC, n = 287) and lung (LUSC, n = 130) squamous cell carcinoma cohorts (9,10), and other cancer types were obtained from the UC Santa Cruz cancer genomics browser (https://genome-cancer.ucsc.edu) (11). We calculated the cytolytic (CYT) score (12), enrichment in interferon-γ (IFNg) signaling (via gene set enrichment analysis using the REACTOME IFNg geneset; http://www.broadinstitute.org/gsea/msigdb/cards/REACTOME_INTERFERON_GAMMA_SIGNALING), and ESTIMATE Immune Score (13) for each sample. Additional data sets for HNSC and LUSC, including raw count RNAseq level 1 files, exome sequencing data in level 3 curated MAF files, clinical data, and Affymetrix SNP6 copy number data, were also downloaded from the Broad Institute Firehose pipeline (https://gdac.broadinstitute.org). Sample purity was inferred from SNP6 copy number array data in ASCAT (14). For the mutational smoking signature, we used the published deconstructSigs mutational signature 4 data (15), representing the mutational pattern associated with tobacco carcinogens such as benzo[a]pyrene, characterized mainly by C>A mutations (16). The numerical signature weight represents the contribution of the process to the mutations in a given tumor sample.
Additional published gene expression microarray data sets were analyzed. The expression matrix for the head and neck cancer data set (n = 138, of which 136 had smoking history) of Walter et al. (17) was downloaded from the Gene Expression Omnibus (data set GSE39366) and comprised background-corrected and LOESS-normalized data from customized Agilent 4x44K arrays. The expression matrix for the lung squamous cell cancer data set (n = 75) of Lee et al. (data set GSE8894) (18) comprised GC-content robust multiarray averaging-normalized data from Affymetrix HG U133 Plus 2.0 arrays. Probes were filtered by selecting the probe with the median absolute deviation for the gene, and normalized values were used for calculation of the ESTIMATE Immune Score. Because these data were obtained on different platforms and normalized independently, the results of immune metrics are not comparable across data sets.
Bioinformatic and Statistical Analyses
The primary hypothesis to be tested was whether there was an association between the mutational smoking signature and immune metrics. Correlations with immune metrics were performed with a nonparametric Spearman coefficient, and multivariable least-squares regression was used to adjust for purity. Power calculations indicated that the HNSC cohort was powered to identify a Spearman ρ of .165, and the LUSC cohort ρ value was .245, at an α of .05 and 1-β of .80. For dichotomous comparisons, HNSC and LUSC samples were ranked together by signature 4 and classified as “smoking-high” or “smoking-low” based on highest and lowest quartiles, respectively, such that the same numerical thresholds were used for both HNSC and LUSC. Comparisons of independent sample means of non–normally distributed data such as the tobacco signature were performed with the two-tailed Wilcoxon rank-sum test, and comparisons of normally distributed immune metrics were performed with the two-tailed t test. Multivariable analyses in HNSC that included tumor subsite or human papillomavirus (HPV) status were limited to 215 cases with nonoverlapping single anatomic subsites and known HPV status, as delineated in Supplementary Tables 1 and 2 (available online). Analysis of correlations between clinical smoking and immunologic metrics in independent expression data sets (17,18), clinical response rates in patients, and the interaction term in the multivariable survival model were directional hypotheses evaluated with a single-tailed t test. Survival analyses of overall survival, where events are defined as death from any cause, utilized univariate or multivariable Cox regression. The proportional hazards assumption was assessed using log(-log(survival)) plots for different strata.
Differentially expressed genes (DEGs) were identified using RNAseq raw counts in DESeq2(19) with a false discovery rate (FDR)–adjusted P value of less than .10. For HNSC, the calculation of DEGs incorporated anatomic subsite as a covariate and included HPV-negative cases only. Pathway enrichment was then assessed with input of DEGs with an FDR P value of less than .10 into Ingenuity Pathway Analysis. An orthogonal pathway analysis was also carried out with gene set enrichment analysis (GSEA), inputting gene expression data for smoking-high and smoking-low LUSC and HNSC (HPV-negative only) tumors and plotting results by normalized enrichment score and –log (FDR-adjusted P value).
Scatterplot regression lines were generated with LOESS regression using Epanechikov kernel. Analyses were performed in R 3.3.3 (R Project for Statistical Computing, Vienna, Austria) and IBM SPSS 24 (IBM Corporation, Armonk, NY). Complete immunologic, genomic, and clinical data are provided in Supplementary Tables 2 and 3 (available online). Clinical analyses were approved by the Institutional Review Board at Memorial Sloan Kettering Cancer Center.
Results
We analyzed the mutational smoking signature and tumor-infiltrating immune microenvironment in several cancer types sequenced by the TCGA. The smoking signature represents the contribution of the tobacco-associated mutational process to the mutations in a sample and is most prevalent in head and neck and lung cancers. Further analyses were performed in HNSC and LUSC (9,10). A nonzero smoking signature was more prevalent in LUSC than in HNSC (89.2% vs 76.7%, P = .003) (Supplementary Figure 1 and Supplementary Table 1, available online), correlating with clinical smoking history (pack-years) in HNSC (Spearman ρ = .52, P < .001, n = 232 with complete data) but not in LUSC (ρ = –.02, P = .89, n = 127) (Supplementary Figure 2, available online), consistent with prior data and the low numbers of nonsmokers in LUSC (<5% of LUSC patients were never-smokers) (1,6). HNSC and LUSC tumors were ranked together by smoking signature and classified in highest (“smoking-high” > .394) and lowest (“smoking-low” < .099) quartiles (Supplementary Tables 2 and 3, available online).
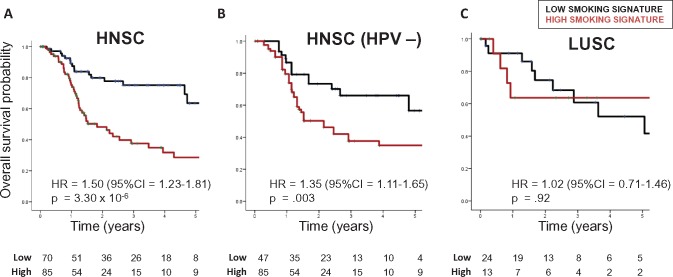
In HNSC, patients with smoking-high tumors had poorer overall survival (hazard ratio [HR] = 1.50, 95% confidence interval [CI] = 1.23 to 1.81, P = 3.30×10−6) (Figure 1A;Supplementary Table 4, available online). The negative prognostic impact of smoking was independent of HPV status (Figure 1B;Supplementary Table 4, available online) and remained statistically significant when considering adjuvant therapy in multivariable Cox regression (Supplementary Table 4, available online). In contrast, the smoking signature was not prognostic in LUSC (HR = 1.02, 95% CI = 0.71 to 1.46, P = .92) (Figure 1C) or when controlling for adjuvant therapy (Supplementary Table 4, available online). A statistically significant interaction between cancer type and smoking signature (P = .03) (Supplementary Table 4, available online) indicated that the prognostic impact of smoking differs between HNSC and LUSC.
Figure 1.
Overall survival in (A) head and neck squamous cell carcinoma (HNSC; median follow-up = 536 days), (B) human papillomavirus–negative HNSC (499 days), and (C) lung squamous cell carcinoma (686 days), by genetic smoking signature status. Smoking signature was defined as highest or lowest quartile, and hazard ratios determined using Cox regression. CI = confidence interval; HNSC = head and neck squamous cell carcinoma; HPV = human papillomavirus; HR = hazard ratio; LUSC = lung squamous cell carcinoma.
To examine the effects of smoking on the tumor immune microenvironment, we interrogated levels of immune activation and infiltration in RNAseq data, analyzing three immune metrics: cytolytic activity (CYT score, incorporating cytolytic effectors of CD8+ T cells: GZMA, and PRF1), IFNg signaling, and ESTIMATE Immune Score. Across eight TCGA cancer types, these three orthogonal measures correlated closely, suggesting that they each reflect aspects of tumor immunity (Supplementary Figure 3, available online).
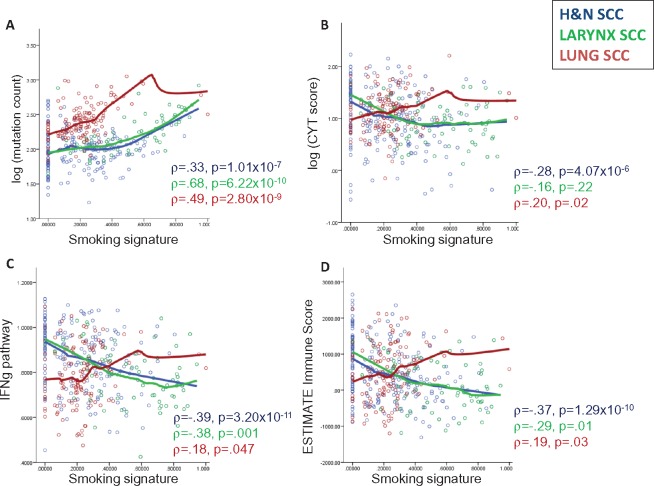
In HNSC, most (47/73, 64.4%) smoking-high tumors arose in the larynx, consistent with the strong etiologic link between smoking and laryngeal cancers. We performed further analyses comparing HNSC in all subsites, HNSC in the larynx only, and LUSC. Smoking was associated with increasing mutation count in HNSC (ρ = .33, P = 1.01×10−7), larynx (ρ = .68, P = 6.22×10−10), and LUSC (ρ = .49, P = 2.80×10−9) tumors (Figure 2A). Mutational load was not consistently associated with trends in tumor immune infiltration in HNSC or LUSC (Supplementary Figures 3 and 4, available online), consistent with prior data in LUSC (21).
Figure 2.
Scatterplots of associations between genetic smoking signature and (A) tumor mutational load (log-transformed), (B) cytolytic score (log-transformed), (C) enrichment in interferon-γ signaling, and (D) overall immune infiltration, as measured by ESTIMATE Immune Score. Correlations for head and neck squamous cell carcinoma (HNSC; all sites), HNSC (larynx only), and lung squamous cell carcinoma were determined using Spearman correlation (ρ with P value) and curve fitting with LOESS regression. CYT = cytolytic; H&N = head and neck; SCC = squamous cell carcinoma.
In HNSC (all subsites), a higher smoking signature was consistently associated with lower levels of immune infiltration (ρ = –.37, P = 1.29×10−10), IFNg signaling (ρ = –.39, P = 3.20×10−11), and cytolytic activity (ρ = –.28, P = 4.07×10−6). Similar associations were observed in laryngeal tumors. However, these trends were all reversed in LUSC: ρ = .19, P = .03; ρ = .18, P = .047; ρ = .20, P = .02 (Figure 2B–D;Supplementary Figure 5 and Supplementary Table 5, available online).
Associations remained statistically significant when adjusting for human papillomavirus (HPV) status in HNSC tumors and tumor purity in all tumors (Supplementary Table 5, available online). In HNSC, the immunosuppressive effects of smoking were observed when analysis was limited to HPV-negative tumors (Supplementary Figure 6, available online) or stratified by subsite (Supplementary Figure 7, available online) or prior radiation therapy (Supplementary Figure 8, available online).
We confirmed these associations in independent HNSC and LUSC expression data sets (17,18). The mutational smoking signature was not discernible in these samples without exome sequencing, so immune metrics were correlated with clinical smoking history. In the HNSC data set of Walter et al., smoking history was associated with lower immune infiltration (n = 136, Immune Score 2268 vs 2590, P = .02) (Supplementary Figure 9, available online). This trend was reversed in the LUSC data set of Lee et al. (n = 75, 1341 vs 670, P = .11) (Supplementary Figure 10, available online).
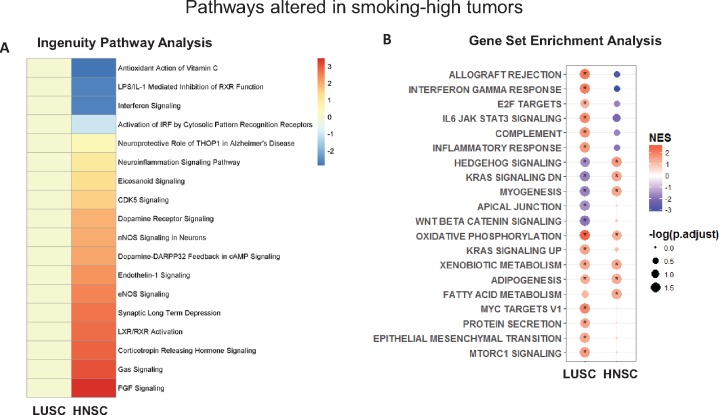
In the TCGA HNSC (HPV-negative) and LUSC data sets, differentially expressed genes (DEGs) were identified by comparing smoking-high to smoking-low tumors (with HNSC subsite as a covariate) (Supplementary Tables 6 and 7 and Supplementary Figure 11, available online). These DEGs were enriched for genes involved in nicotine degradation, xenobiotic metabolism, and glutathione-mediated detoxification pathways. Using Ingenuity Pathway Analysis to determine directionally altered pathways, smoking was associated with immunosuppressive pathways in HNSC, but not in LUSC: repression of interferon signaling (P = .01), type I interferon response via interferon regulatory factor (P = .03), leukocyte (P = 2.63×10−4) and granulocyte (P = 5.25×10−4) function (Figure 3A;Supplementary Tables 8 and 9, available online). Using an orthogonal pathway analysis technique (Gene Set Enrichment Analysis), a similar distinction was seen: there was a strong pro-inflammatory phenotype in LUSC and an immunosuppressive phenotype in HNSC (Figure 3B).
Figure 3.
A) Heatmap of Ingenuity Pathway Analysis–defined canonical pathways altered in either head and neck squamous cell carcinoma (HNSC) or lung squamous cell carcinoma (LUSC), showing an immunosuppressive phenotype in smoking-high HNSC, with the color legend representing directional z-score (Supplementary Tables 8 and 9, available online). B) Dot plot of results from Gene Set Enrichment Analysis (GSEA) of pathways enriched in smoking-high tumors, showing a pro-inflammatory phenotype in smoking-high LUSC, but an immunosuppressive phenotype in smoking-high HNSC. Complete expression data of smoking-high and smoking-low LUSC and HNSC (human papillomavirus–negative only) were analyzed for enrichment of GSEA hallmark gene sets, and results are plotted by color (normalized enrichment score; legend on the upper right) and size (–log false discovery rate [FDR]–adjusted P value; legend on the lower right, with * indicating FDR-adjusted P < .05). HNSC = head and neck squamous cell carcinoma; LUSC = lung squamous cell carcinoma; NES = normalized enrichment score.
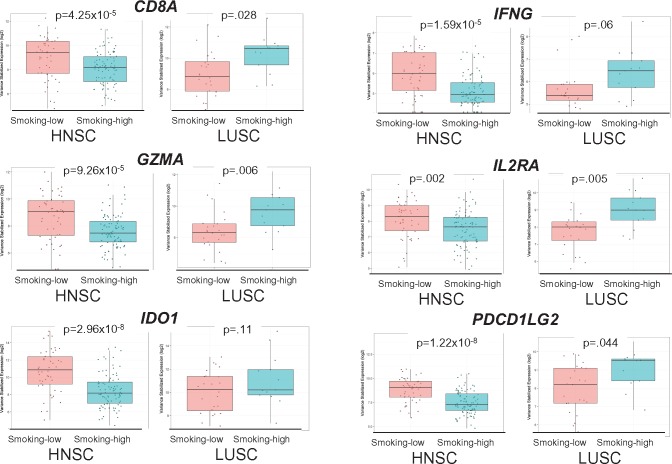
Many immune-related genes were downregulated in smoking-high HNSC but upregulated in LUSC, including T-cell receptors (CD8A, PDCD1 [PD-1], CTLA4), immunoregulatory molecules (CD274 [PD-L1], PDCD1LG2 [PD-L2], IDO1, IDO2), cytotoxic effectors (GZMA, GZMB, GZMH, GZMK, PRF1), cytokines (CXCL10, CXCL11, IL2RA, IL13RA2, IL15, IL20, IL27), and MHCII molecules (HLA-DRA, HLA-DQA1) (Figure 4; Supplementary Table 10 and Supplementary Figure 12, available online). These immunologic DEGs represented four of the six genes in the previously described “IFNg signature,” which predicts response to anti-PD1 treatment (Supplementary Table 11, available online) (5), supporting the likely relevance of these findings to immunotherapy response.
Figure 4.
Scatter box plots of differentially expressed immunologic genes in smoking-high vs smoking-low tumors, where dots represent samples; horizontal lines represent median values; boxes represent interquartile range; whiskers represent minimum/maximum values; outlier values beyond 1.5× the interquartile range. CD8A = cluster of differentiation 8a (encoding the CD8 alpha chain); GZMA = granzyme A; HNSC = head and neck squamous cell carcinoma; IDO1 = indoleamine 2,3-dioxygenase 1; IFNG = interferon gamma; IL2RA = interleukin 2 receptor subunit alpha; LUSC = lung squamous cell carcinoma; PDCD1LG2 = programmed cell death 1 ligand 2 (PD-L2).
Heavy smoking has been associated with TP53 mutations, both in lung and head and neck cancers (22,23). Indeed, the smoking signature was higher in TP53-mutated vs TP53 wild-type tumors, both in HNSC (0.35 vs 0.25, P = .002) and LUSC (0.23 vs 0.13, P = .13). The trend was not statistically significant in LUSC, likely due to small numbers of TP53 wild-type tumors (n = 6). TP53-mutated HNSC tumors had statistically significantly lower levels of immune infiltration (Immune Score 337 vs 624, P = .003), while TP53-mutated LUSC tumors demonstrated an opposite trend (Immune Score 626 vs 360, P = .38). The other commonly mutated genes in HNSC and LUSC, which have not been linked with tobacco exposure, were not associated with immune infiltration (Supplementary Table 5, available online). The only gene statistically significantly associated with higher levels of immunity was CASP8 (HNSC Immune Score 820 vs 443, P = .04; LUSC Immune Score 1574 vs 594, P = .04). This is consistent with prior findings that CASP8 was the most enriched mutation in tumors with high cytolytic activity (12).
Taken together, these data are consistent with reported response rates to immune checkpoint inhibitor therapies, which are higher among smokers with lung cancer but lower among smokers with HNSC (6–8). Because published HNSC response data stratified by HPV and smoking status are not available, we analyzed the clinical response data of 81 HNSC patients treated at our institution with anti-PD-1/PD-L1 drugs. We found that the rate of clinical benefit (complete/partial response or stable disease) was statistically significantly higher in never-smokers compared with current/former smokers (57.1% vs 33.3%, P = .03). This trend remained statistically significant when analysis was limited to HPV-negative tumors (P = .02) (Supplementary Table 12, available online).
Discussion
The mutagenic effects of cigarette smoking operate similarly in lung and head and neck squamous cell carcinomas. In both cancer types, tumors with an increasing proportion of mutations attributable to smoking have higher overall mutational loads. However, the clinical and immunologic correlates of smoking differ in these two squamous cell carcinoma types. In HNSC, the mutational smoking signature is associated with poorer survival and strong immunosuppressive effects, which are seen even adjusting for subsite and HPV status. In contrast, in LUSC, smoking is associated with a more inflamed tumor microenvironment.
These findings are consistent with clinical data indicating that smokers with lung cancer tend to have higher response rates to immune checkpoint inhibitors. For example, Rizvi et al. reported that in non–small cell lung cancer patients (both squamous cell and adenocarcinoma) treated with anti-PD-1 therapy, the mutational smoking signature was a more precise determinant of response than clinical smoking history. Objective response rates to therapy were substantially higher among smoking signature–high tumors, compared with smoking signature–low tumors (56% vs 17%, P = .03) (6). Although clinical smoking history seems to be less predictive than the mutational smoking signature, in the phase III trial of firstline nivolumab in stage IV non–small cell lung cancer, never-smokers had a statistically significantly higher likelihood of progression or death with nivolumab therapy (HR = 2.51, 95% CI = 1.31 to 4.83) vs chemotherapy, compared with former or current smokers (HR = 1.03–1.14) (7). Data in HNSC are more limited: the only available data of HNSC patients treated with immune checkpoint blockade with clinical smoking history are from the phase III trial of nivolumab in patients with recurrent HNSC. Among these patients, the survival benefit associated with nivolumab was numerically lower in current/former smokers (HR = 0.71, 95% CI = 0.52 to 0.99) compared with never-smokers (HR = 0.58, 95% CI = 0.32 to 1.06) (8). Although these data were not stratified by HPV status, response rates were similar in HPV+ and HPV- tumors (15.9% vs 14.5%) (24). Our analysis of 81 HNSC patients treated at our institution provides additional support for the higher response rates seen in never-smokers. Future genomic analyses that incorporate the mutational smoking signature will be able to define the effects of smoking in both disease sites more precisely.
Interestingly, the association between patient-reported smoking history and the smoking signature was stronger in HNSC than in LUSC. This may be due to the very high prevalence of tobacco exposure in LUSC patients (∼97%) compared with HNSC patients (81%), attenuating the power of this comparison. The vast majority (80%) of LUSC patients were actually former/reformed smokers, a population in which the associations between clinical smoking history and the smoking signature may be obscured by patient recollection and other mutational processes occurring over intervening years (1).
Smoking has direct suppressive and pro-apoptotic effects on T cells, linked to reactive oxygen and nitrogen species (3). The immunosuppressive effects of tobacco smoke on T cells may be counterbalanced in the lung context by strongly pro-inflammatory effects, which have been well described in chronic obstructive pulmonary disease (2,25). In the context of lung cancer (adenocarcinomas), smoking-associated transversions have been found to be associated with higher levels of immune markers (such as tumor PD-L1 and granzyme B staining) (26).
Comparable chronic airway inflammation is not common in the head and neck, where immunosuppressive mechanisms may instead dominate. These pro- and anti-immunity effects of smoking coexist with the enrichment in immunogenic neoantigens that would be anticipated with smoking mutagenicity, and which is seen in both HNSC and LUSC. It is likely that, in LUSC, the combined effects of increased neoantigen load and an inflamed microenvironment account for the markedly higher response rates to immunotherapy observed in smokers. In contrast, in HNSC, the increase in immunogenic neoantigen load associated with smoking may be countered by a more immunosuppressive microenvironment.
These findings raise the possibility that tumor mutational burden, which is highly predictive of immunotherapy response in lung cancer, may have lower predictive value in head and neck cancer. Preliminary data from pembrolizumab-treated HNSC patients suggest that gene expression–based measures of tumor inflammation are more predictive of response than mutational load (27). These hypotheses will require further investigation.
We found that smoking signature was associated with TP53 mutations, but not other driver mutations, in HNSC and LUSC (22,23). Among statistically significantly mutated genes, CASP8 was the only gene in which mutations were associated with the status of the immune microenvironment. In both HNSC and LUSC, CASP8 mutations were associated with statistically significantly higher immune infiltration, consistent with prior pan-cancer analyses of cytolytic activity (12). CASP8-mutated tumor cells, which are deficient in the extrinsic apoptosis pathway, may be able to evade cytotoxic T-cell killing (28) and thereby undergo positive selection in highly immune-enriched environments.
There are important limitations to this analysis. These associations cannot implicate causality, for example, whether effects are mediated by T cells, neoantigen presentation, or other mechanisms. While consistent with initial clinical observations, further prospective data will be needed to definitively connect the smoking signature with immunotherapy response. The mutational smoking signature is likely to be more strongly correlated with immunotherapy response than clinical smoking history, as has been seen in lung cancer (6).
This study demonstrates the importance of the mutational smoking signature as an indicator of both tumor mutational burden and smoking-associated modulation of the immune microenvironment. These associations vary by cancer site. Future analyses will help to define the additional value of the smoking signature, combined with other factors such as tumor mutational load, PD-L1 expression, and measures of the T-cell inflamed microenvironment, in patients with smoking-associated cancers who are treated with immunotherapy. Our data indicate that it will be important to assess the effects of smoking separately in different cancer types.
The etiology of smoking-associated cancers is attributable to both mutagenic and immunomodulatory mechanisms. These data illuminate the profound yet context-dependent effects of smoking on the tumor immune microenvironment of squamous cell cancers. Smoking can exert either immunosuppressive or pro-inflammatory effects on the microenvironment, which appear to vary depending on anatomic site. Because both mutational load and immune infiltration affect the likelihood of clinical response to immune checkpoint inhibitors, these data will have relevance for future clinical investigation of immunotherapy in smoking-related cancers, particularly among the increasing number of tumors now being studied with genomic profiling.
Funding
This work was supported by National Institutes of Health DE024774 and Cancer Center Support Grant P30 CA008748, Cycle for Survival, the Damon Runyon Cancer Research Foundation, the Frederick Adler Fund, and the Jayme and Peter Flowers Fund.
Notes
Affiliations of authors: Immunogenomics and Precision Oncology Platform (AD, FK, DC, KWL, NR, TAC, LGTM), Human Oncology and Pathogenesis Program, Memorial (AD, DC, KWL, TAC, LGTM), Department of Radiation Oncology (NR, TAC), and Department of Surgery (RJW, LGTM), Memorial Sloan Kettering Cancer Center, New York, NY.
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
We thank Drs. Neil Hayes, Vonn Walter, Matthew Wilkerson, Jhingook Kim, and Soo Jung for providing clinical data for the external expression data sets. The authors wish to acknowledge the patients and investigators who contributed to this work as part of The Cancer Genome Atlas (TCGA). The results published here are, in part, based upon data generated by the TCGA. Further information about the TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov.
TAC is a co-founder of Gritstone Oncology. LGTM reports personal fees from Merck outside of the submitted work. All other authors declare no completing interests.
Supplementary Material
References
- 1. Alexandrov LB, Ju YS, Haase K, et al. Mutational signatures associated with tobacco smoking in human cancer. Science. 2016;3546312:618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hodge G, Nairn J, Holmes M, et al. Increased intracellular T helper 1 proinflammatory cytokine production in peripheral blood, bronchoalveolar lavage and intraepithelial T cells of COPD subjects. Clin Exp Immunol. 2007;1501:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hernandez CP, Morrow K, Velasco C, et al. Effects of cigarette smoke extract on primary activated T cells. Cell Immunol. 2013;2821:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;5157528:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ayers M, Lunceford J, Nebozhyn M, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;1278:2930–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;3486230:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage iv or recurrent non-small-cell lung cancer. N Engl J Med. 2017;37625:2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferris RL, Blumenschein G Jr, Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;37519:1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;5177536:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;4897417:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen R, Seshan V. FACETS: Fraction and allele-specific copy number estimates from tumor sequencing. Memorial Sloan-Kettering Cancer Center, Dept. of Epidemiology & Biostatistics, Collection of Biostatistics Research Archive. Working Paper Series 29. 2015. http://biostats.bepress.com/mskccbiostat/paper29/. Accessed July 4, 2017.
- 14. Van Loo P, Nordgard SH, Lingjaerde OC, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;10739:16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosenthal R, McGranahan N, Herrero J, et al. DeconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;5007463:415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walter V, Yin X, Wilkerson MD, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;82:e56823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee ES, Son DS, Kim SH, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;1422:7397–7404. [DOI] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;1512:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005;9716:1180–1184. [DOI] [PubMed] [Google Scholar]
- 21. Choi M, Kadara H, Zhang J, et al. Mutation profiles in early-stage lung squamous cell carcinoma with clinical follow-up and correlation with markers of immune function. Ann Oncol. 2017;281:83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suzuki H, Takahashi T, Kuroishi T, et al. p53 mutations in non-small cell lung cancer in Japan: Association between mutations and smoking. Cancer Res. 1992;523:734–736. [PubMed] [Google Scholar]
- 23. Brennan JA, Boyle JO, Koch WM, et al. Association between cigarette smoking and mutation of the p53 gene in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995;33211:712–717. [DOI] [PubMed] [Google Scholar]
- 24. Gillison ML. Nivolumab versus investigator's choice therapy among patients with human papillomavirus (HPV)-associated squamous cell carcinoma of the head and neck (SCCHN): Updated results from CheckMate 141. Proceedings of the American Head and Neck Society Annual Meeting; April 2017; San Diego, CA. http://ahns.jnabstracts.com/Detail?ID=81644:S061. Accessed January 3, 2018.
- 25. Jackute J, Zemaitis M, Pranys D, et al. Distribution of CD4(+) and CD8(+) T cells in tumor islets and stroma from patients with non-small cell lung cancer in association with COPD and smoking. Medicina (Kaunas). 2015;515:263–271. [DOI] [PubMed] [Google Scholar]
- 26. Kadara H, Choi M, Zhang J, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol. 2017;281:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haddad RI. Genomic determinants of response to pembrolizumab in head and neck squamous cell carcinoma (HNSCC). J Clin Oncol. 2017;35(Suppl):Abstr 6009. [Google Scholar]
- 28. Medema JP, de Jong J, van Hall T, et al. Immune escape of tumors in vivo by expression of cellular FLICE-inhibitory protein. J Exp Med. 1999;1907:1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.