Abstract
Remarkably little information is available about the interaction between the gut microbiota and intestinal immunity in fish and crustaceans. In our study, we used Illumina MiSeq sequencing and real-time quantitative PCR to compare the microbial community and immunity genes expression in the foregut, midgut and hindgut of Chinese mitten crab (Eriocheir sinensis). Our results indicated that the community richness of the midgut is higher than in the foregut or the hindgut, although the bacterial diversity in the hindgut is higher. The predominant phyla were Tenericutes and Firmicutes in the foregut, Tenericutes and Proteobacteria in the midgut and Proteobacteria, Tenericutes and Bacteroidetes in the hindgut. When compared with the midgut, the expression of antimicrobial peptides (AMPs) were significantly elevated in the hindgut (P < 0.05), and the gene expression of EsRelish (IMD pathway) was higher than the Toll signaling pathway genes. Actinobacteria and Lactobacillus had negative correlationship with the expression of AMPs, although Acinetobacter, Bacteroides, Flavobacterium can up-regulate the expression of AMP genes. Collectively, our data indicate that microbiota are site-specific within the digestive tracts of crabs and the bacterial community and intestinal immunity have a close relationship in E. sinensis.
Electronic supplementary material
The online version of this article (10.1186/s13568-018-0722-0) contains supplementary material, which is available to authorized users.
Keywords: Eriocheir sinensis, Intestinal bacterial community, Gut immunity, Antimicrobial peptides, Gene expression
Introduction
The connections between gut flora and host was one of the most important impactors influence aquatic animal health (Chaiyapechara et al. 2012; Yan et al. 2012). The intestinal flora is vital to the development (Shang et al. 2017), immunity and disease resistance of gut (Cerf-Bensussan and Gaboriau-Routhiau 2010). The digestive tract is the richest part of human body in terms of number and diversity of bacterial species (Quigley 2013). The dysregulation of intestinal bacteria was closely related to chronic inflammatory and epithelial barrier dysfunction and other illnesses (Abreu 2010; Olmos et al. 2010). There is a lot of information and fast growing about this topic. Comparative analysis of the intestinal bacterial community and expression of gut immunity genes is very important.
In vertebrates and invertebrates, antimicrobial peptides (AMPs) play a crucial role in gut immunity (Destoumieux-Garzón et al. 2016; Zhang and Gallo 2016) and form effective defense mechanisms against multitudinous pathogens. Many studies on AMPs and invertebrate gut defense have been performed in insects, especially Drosophila melanogaster, because this species is the model organism of invertebrate (Broderick 2016; Loch et al. 2017; Rolff and Schmid-Hempel 2016). The key role of AMPs in gut immune response is demonstrated through oral reactive oxygen species (ROS) blockers to Drosophila mutants that are defective in the NF-κB pathway, resulting in increased mortality (Ryu et al. 2006). This mortality rate decreases when a single AMP is expressed in the intestines of these flies (Ryu et al. 2008). In addition, closely regulated AMPs in the gut of Drosophila seem to be indispensable for maintaining the homeostasis of the intestinal flora (Ryu et al. 2008). The production of AMPs was affected by the Toll and IMD pathways in Drosophila hemocytes, but mainly by the latter in gut (Garcia-Garcia et al. 2013). As the central components of Toll and IMD signaling pathways, e.g., EsTolls, EsDorsal, EsPelle, EsRelish (a novel NF-κB-like transcription factor), these genes have been proven to participate in the innate immunity of Chinese mitten crab Eriocheir sinensis (Crustacea: Decapoda: Brachyura: Varunidae) and defense against several pathogens (Li et al. 2010; Ying et al. 2015; Yu et al. 2013a, b). The above genes could function as an adapter protein in Toll and IMD signaling and regulate the production of crab AMP genes in hemocytes (Mu et al. 2011; Ying et al. 2015; Zhang et al. 2010). Then there were AMP genes in hemocytes have various biological functions in E. sinensis (Destoumieux-Garzón et al. 2016). Hence, these AMPs can help the screening of therapeutic or preventive agents for healthy breeding of crabs. However, the function of these very important genes in the crab intestine is not clear. The study of AMPs in gut immunity of crustaceans is rare and it is unclear which signaling pathways regulate the production of AMPs.
The E. sinensis is widely cultivated in China and other countries because of rich nutritional value and high economic value (Chen and Zhang 2007). The culture of E. sinensis has increased dramatically in the past decade following a rise in demand. Current production in China is 820,000 tons with an output value of more than 50 billion in 2015 (China Fishery Statistics Yearbook 2016). In recent years, frequent outbreaks of diseases have resulted the decreased production and economic losses (Ding et al. 2017). Accordingly, considerable effort has been expended on intestinal health research of E. sinensis. In the present study, we hypothesized that different regions of the digestive tract in E. sinensis have difference in the intestinal epithelium bacterial communities and that may be related to the different expression of digestive tract immune genes. The goal of our research was to explore the bacterial diversity, gut immunity genes expression and the relationship between the two in the various regions of the E. sinensis digestive tract. Our results suggest a role for the dominant bacteria in each region of the digestive tract and provide novel insight into the bacterial community and gut immunity of E. sinensis. These data will be helpful to understand the relationships between symbiotic bacteria and the host.
Materials and methods
Crab culture and crab intestine collection
Healthy female E. sinensis (9.5 ± 0.5) g were collected with permission from Panjin Guanghe Crab Industry Co., Ltd. (Panjin, Liaoning, China) in July 2016. In the laboratory, crabs were fed with formula diets (Shenyang Hefeng Aquatic Feed Co., Ltd., Shenyang, Liaoning, China) in a concrete recirculating aquaculture system with the following conditions: dissolved oxygen > 7.0 mg L−1, pH 7.5–8.3, ammonia < 0.2 mg L−1, and nitrite < 0.01 mg L−1. Following acclimation (1 week), feeding was stopped 24 h prior to handling and sampling to minimize the stress on E. sinensis. Because the tissue of the crab digestive tract is very small, 24 crabs were used for the determination of intestinal flora, the accumulation of 8 crabs is a sample and repeat three times for each sample. In addition, 6 crabs were used for the determination of intestinal immune gene expression, the accumulation of 2 crabs is a sample and repeat three times for each sample. First, washed the body surface with sterile water. Second, disinfected the outside of the body with 75% ethanol for 2 min. Third, dissected the crab in the aseptic state to remove the digestive tract and were then divided into three segments (foregut, midgut, hindgut). Samples were put into sterile EP tubes and stored at − 80 °C for analysis of gut microbiota and expression of gut immune gene.
DNA isolation, PCR amplification and Illumina MiSeq sequencing
The digestive tract samples were taken for DNA extraction (0.25–0.3 g wet weight) using the E.Z.N.A.® Soil DNA Kit (OMEGA Bio-tek, Norcross, GA, US). We used PCR to amplify the V3–V4 regions of the bacterial 16S rRNA gene. The primers used in the present study were 338F (5′-barcode-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (amplicon length: 468 bp), 20 μL of PCR mixture containing 5× FastPfu buffer (4 μL), 2.5 mM dNTPs (2 μL), 5 μM forward primer (0.8 μL), 5 μM revese primer (0.8 μL), FastPfu Polymerase (0.4 μL), and template DNA (10 ng). 16S rRNA sequencing were conducted on Illumina MiSeq platform. Paired-end sequencing 2 × 300 bp was performed to sequence all libraries using an Illumina MiSeq platform according to standard protocols.
Illumina MiSeq sequencing and data processing
We used the QIIME (version 1.17 http://qiime.org/) package to conduct sequences processing. UPARSE (version 7.1 http://drive5.com/uparse/) and UCHIME were used respectively to cluster operational taxonomic units (OTUs) with a 97% similarity cutoff and chimera identification. The diversity and richness of bacterial community were identified by the ACE, Chao1, Shannon and Simpson, which were conducted utilizing the Mothur program (version v.1.30.1 http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity). We used RDP Classifier (http://rdp.cme.msu.edu/) for sequences classification against the silva database (Amato et al. 2013). The Venn diagram and heatmap were illustrated using the R package (http://www.R-project.org/). The raw sequencing data were uploaded into the NCBI Sequence Read Archive database (SRA; http://www.ncbi.nlm.nih.gov/Traces/sra/) under accession SRP110849.
Metagenome prediction using PICRUSt
The metagenomes prediction were made with PICRUSt (Langille et al. 2013). The gene content was predicted for each sample according to KEGG and EggNOG. In the present study, the predictive function component was divided into level 3 of EggNOG (evolutionary genealogy: Non-supervised Orthologous Groups) database pathways. The richness was predicted according to the phylogenetic, and the assessed value is 0.8.
Intestinal tissue RNA extraction and RT-qPCR
Total RNA extraction of the digestive tract samples was perform using Trizol® reagent (Invitrogen, Carlsbad, CA). The RNA quality and concentration were analyzed spectrophotometrically using a BioDrop μLITE (BioDrop, Cambridge, UK). Reverse transcription of total RNA (1 μg) was conducted in a 20 μL system using PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio. Inc., Dalian, China).
The primer sequences for the Toll signaling pathway genes [EsToll2 (GenBank accession No. KC011816), EsPelle (GenBank accession No. KP795393), EsDorsal (GenBank accession No. KC900086)], IMD signaling pathway genes [EsRelish (GenBank accession No. GQ871279)], antibacterial peptides [EsALF1 (GenBank accession No. DQ793214), EsALF2 (GenBank accession No. GU014699), EsCrus1 (GenBank accession No. GQ200832), EsCrus2 (GenBank accession No. GQ200833)], and Es-β-Actin (GenBank accession No. HM053699) are described in Additional file 1: Table S1.
All of the primers were synthesized by GenScript Biotechnology Co. Ltd. (Nanjing, China). RT-qPCR was performed employing the LightCycler 480 II system (Roche Applied Science, Branford, CT, USA) in conjunction with SYBR green dye, and the following programme: 95 °C for 30 s, then 45 cycles of 95 °C for 5 s and 60 °C for 40 s, then 1 cycle of 95 °C for 5 s, 60 °C for 60 s, and 95 °C, and terminated at 50 °C for 30 s. The 20 μL reaction mixture consisted of 10 μL of 2× SYBR Premix Ex Taq II (Takara Bio. Inc., Dalian, China), 1.0 μL of each primer and 1.5 μL of cDNA. Gene expression levels were compared with those of Es-β-Actin, and the 2−∆∆Ct method was adopted to data analysis using the light cycler 480 software.
Statistical analyses
The statistical analyses were performed using the SPSS 17.0 software package (SPSS Inc., Chicago, USA). In the various regions of the E. sinensis digestive tract was regarded as the fixed effect. Values were expressed as the mean ± SEM. The diversity of digestive tract bacterial community and the relative mRNA expression of eight gut immune genes (EsToll2, EsPelle, EsDorsal, EsRelish, EsALF1,2 and EsCrus1,2) in the same regions of the E. sinensis intestinal tract were compared using a one-way analysis of variance (ANOVA), followed by Bonferroni’s multiple comparisons post hoc test. COG function classification in the intestinal microbiota and the relative mRNA expression of gut immune gene in the intestinal tract of E. sinensis were compared using Independent-Samples T test. Significance was declared at P < 0.05. Principal coordinates analysis (PCoA) based on UniFrac distances were performed to evaluate the overall differences in the gut bacterial community (Jiang et al. 2013). Student’s t test (equal variance) was used to analyse bacterial communities in the intestinal tract samples. Spearman coefficient was used for correlation detection.
Results
Bacterial composition determined by MiSeq sequencing
Microbial community of the digestive tract was determined using Illumina MiSeq. 32128 sequences were obtained by quality screening for alpha-diversity research. Rarefaction analysis was shown in Additional file 1: Fig. S1. Microbial composition was counted by the proportion of OTUs. Assessment of microbial diversity in the foregut, midgut and hindgut of crabs were completed using α-diversity indexes. The ACE and Chao values were significantly increased in the midgut (P < 0.05). Microbial composition in the hindgut were higher α-diversity indexes than those in the midgut or foregut, as revealed through the Shannon and Simpson indexes (Additional file 1: Table S2).
Microbial community compositions
At the phylum level, a total of 16 phyla were detected in the crab digestive tract, and result showed 12 phyla were present in all three groups (Additional file 1: Fig. S2A). At the genus level, there were 109, 161 and 112 taxa in the foregut, midgut and hindgut groups, respectively (Additional file 1: Fig. S2B).
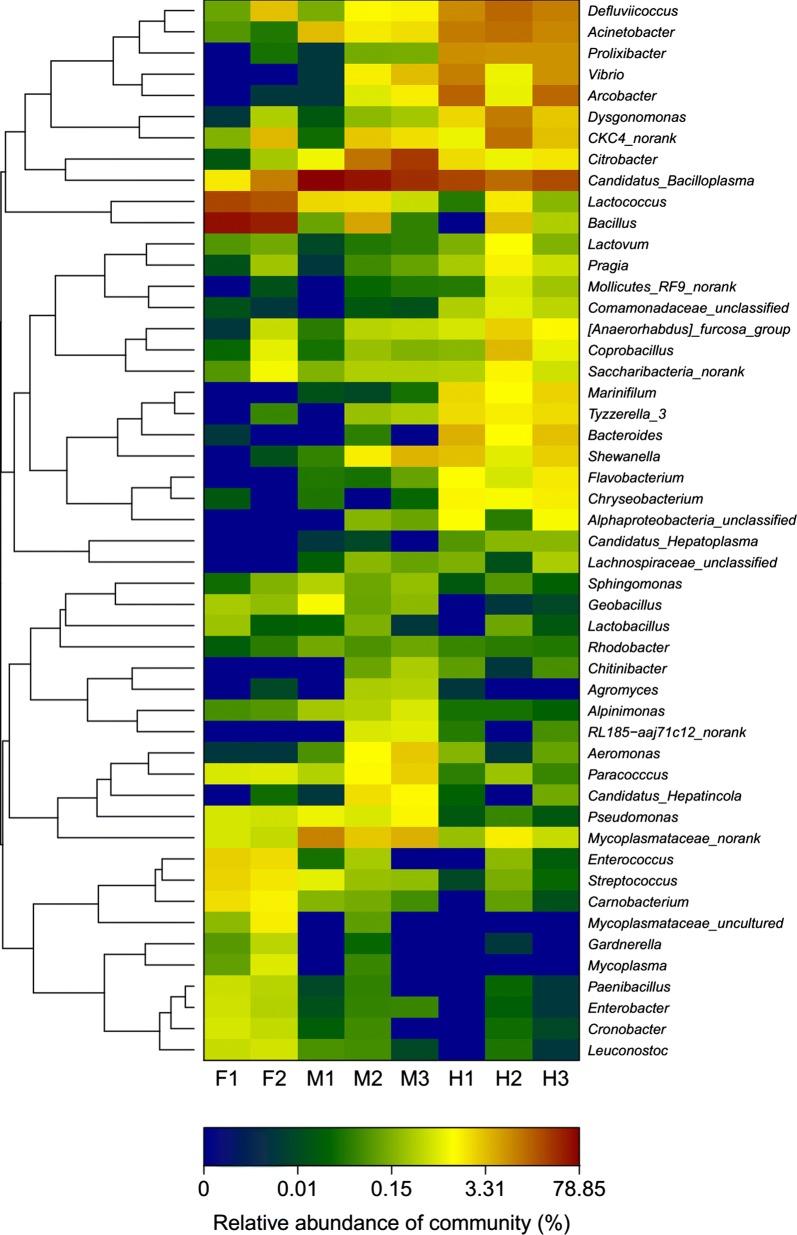
At the phylum level of the digestive tracts (F, M, H) of all the crabs, Tenericutes were the most (33.00%), then Firmicutes (29.48%), Proteobacteria (28.36%), and Bacteroidetes (7.87%). Within the intestines (M, H) of all the crabs, Tenericutes were the largest number (41.48%), then Proteobacteria (36.18%), Firmicutes (10.65%), and Bacteroidetes (10.35%) (Additional file 1: Fig. S3). At the genus level of the digestive tract (F, M, H), the dominant genera were composed of Bacillus (14.53%), Lactococcus (6.28%), Citrobacter (6.02%), Acinetobacter (4.88%), Rhodospirillaceae (4.78%), Arcobacter (4.06%), Erysipelotrichaceae (3.18%), Vibrio (2.89%), Prolixibacter (2.74%), Mycoplasmataceae (2.47%), Dysgonomonas (2.06%), Shewanella (1.55%), and Bacteroides (1.13%). Within the intestines (M, H), the dominant genera were composed of Candidatus Bacilloplasma (38.07%), Citrobacter (7.97%), Acinetobacter (6.48%), Rhodospirillaceae (5.80%), Arcobacter (5.41%), Vibrio (3.85%), Prolixibacter (3.65%), Erysipelotrichaceae (3.60%), Mycoplasmataceae (2.84%), Dysgonomonas (2.68%), Shewanella (2.06%), Bacillus (1.55%), Bacteroides (1.50%), Lactococcus (1.17%), Tyzzerella_3 (1.09%), and Marinifilum (1.01%) (Additional file 1: Fig. S4). The top fifty bacterial taxa are represented in a heat map (Fig. 1).
Fig. 1.
Heatmap of the dominant digestive flora at the genus level. F foregut, M midgut, H hindgut
Microbial community similarity among foregut, midgut and hindgut
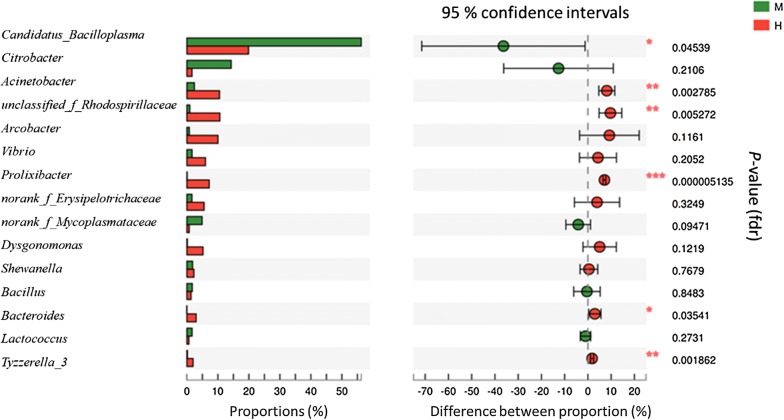
PCoA showed a clustering of the foregut, midgut and hindgut by PC1 (51.95%) and PC2 (29.6%) of the explained variance (Additional file 1: Fig. S5). At the genus level, the abundance of Candidatus Bacilloplasma was 56.24% (midgut) and 19.9% (hindgut), of which the midgut had significantly higher levels than the hindgut (P = 0.045) (Fig. 2). By contrast, there was a significantly decreased level of bacteria from the Acinetobacter (P = 0.003), unclassified_f_Rhodospirillaceae (P = 0.005), Prolixibacter (P = 0.000), Bacteroides (P = 0.035), and Tyzzerella_3 (P = 0.002) taxa in the midgut (Fig. 2).
Fig. 2.
Student’s t-test bar plot on genus level of bacterial communities in the intestinal tract samples. The ordinate indicates the name of the genus level, the abscissa represents the percentage of the abundance of the sample. The color of the bar represents different groups M: midgut (green), H: hindgut (red). 0.01 < P ≤ 0.05*, 0.001 < P ≤ 0.01**, P ≤ 0.001***
Metagenome prediction of gut microbiota
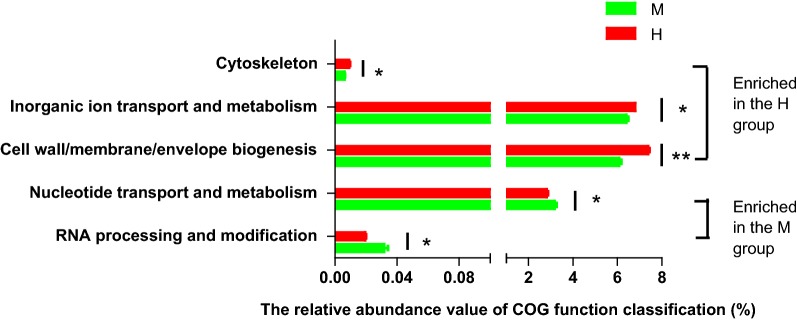
A total of 25 gene families were identified in all samples, five function classification were significantly differentially rich between the midgut and the hindgut (P < 0.05) (Fig. 3). Compared with the hindgut, the midgut had a higher relative abundance of nucleotide transport and metabolism (P = 0.034) and RNA processing and modification (P = 0.032) gene families, and a lower relative abundance of cytoskeleton (P = 0.025), inorganic ion transport and metabolism (P = 0.008), and cell wall/membrane/envelope biogenesis (P = 0.000) gene families.
Fig. 3.
COG function classification in the microbiota of the intestinal tract samples. M: midgut (green), H: hindgut (red). 0.01 < P ≤ 0.05*, 0.001 < P ≤ 0.01**
Gene expression in the intestinal tract
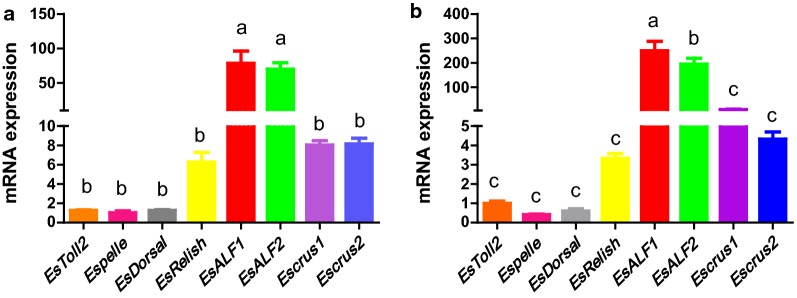
The relative mRNA expression of the Toll signaling pathway (EsToll2, EsDorsal, EsPelle), IMD signaling pathway (EsRelish) and AMPs (EsALF1, EsALF2, EsCrus1, EsCrus2) were detected by real-time quantitative polymerase chain reaction (RT-qPCR). Compared with the midgut, EsToll2 (P = 0.008), EsRelish (P = 0.005), EsALF1 (P = 0.001), EsALF2 (P = 0.006), EsCrus1 (P = 0.021), and EsCrus2 (P = 0.002) gene expression were significantly up-regulated in the hindgut (P < 0.05). However, EsDorsal (P = 0.077) and EsPelle (P = 0.102) mRNA expression levels were only slightly higher in the hindgut (Fig. 4). In addition, the RT-qPCR analysis showed that the expression changes of eight genes had similar trends in the midgut and the hindgut. The levels of EsALF1 and EsALF2 mRNA expression were significantly higher than six other genes in the intestinal tract (P < 0.05). EsRelish, EsCrus1 and EsCrus2 expression levels were higher than those of the Toll signaling pathway genes (EsToll2, EsDorsal, EsPelle), but no significant difference was detected (Fig. 5).
Fig. 4.
Comparative analysis on gut immune gene expression (relative to Es-β-Actin) in the intestinal tract of E. sinensis (mean ± SEM, n = 3). a–h: relative expression profile of 8 immune-related genes (EsToll2, EsPelle, EsDorsal, EsRelish, EsALF1, EsALF2, EsCrus1, EsCrus2) in crab midgut and the hindgut. M: midgut (red), H: hindgut (green). 0.01 < P ≤ 0.05*, P ≤ 0.01**
Fig. 5.
Comparative analysis on eight gut immune genes expression (relative to Es-β-Actin) in the same regions of the E. sinensis intestinal tract (mean ± SEM, n = 3). a Midgut, b hindgut. Different letters indicate significant differences (P < 0.05)
Correlation analysis
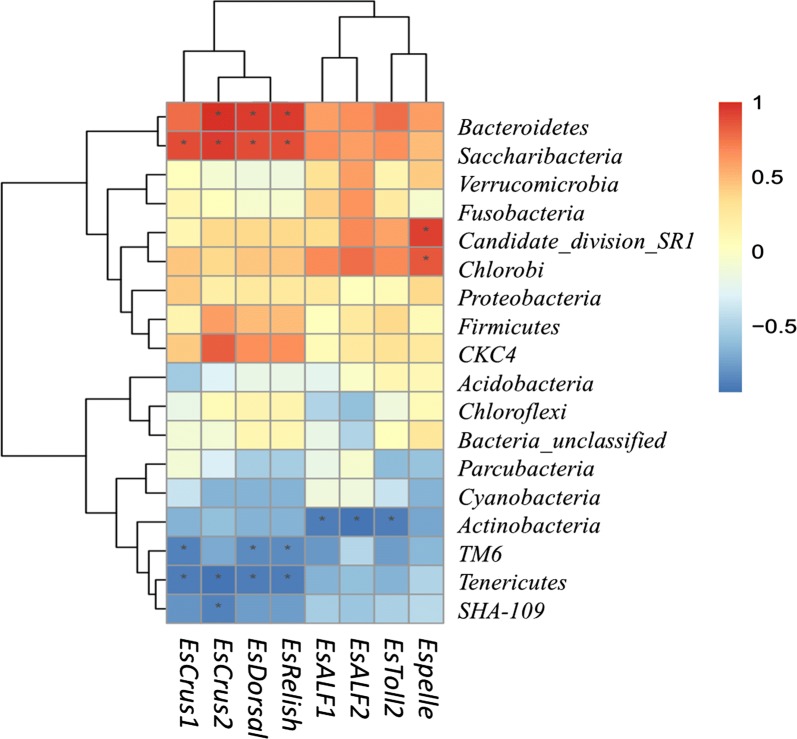
The relationships among immune gene expression and intestinal microbiota were evaluated. At the phylum level (Fig. 6), the results showed that Actinobacteria negatively correlated with EsToll2, EsALF1, and EsALF2 gene expression (P < 0.05). Candidate_division_SR1 and Chlorobi and EsPelle gene expression were positive correlations (P < 0.05). In addition, the abundance of four taxa (tow negative: TM6, Tenericutes; tow positive: Bacteroidetes, Saccharibacteria) was related to increase EsRelish, EsCrus1, EsCrus2 gene expression (P < 0.05). At the genus level (Additional file 1: Fig. S6), Acinetobacter positively correlated and Paracoccus negatively correlated with the mRNA expression of EsALF1 (P < 0.05). The abundance of nine taxa (six negative: Defluviimonas, Paracoccus, Alpinimonas, Gemmobacter, MNG7_norank, Pseudomonas; three positive: Chryseobacterium, Bacteroides, Candidatus Hepatoplasma) was correlated with increased EsALF2 gene expression (P < 0.05). The abundance of nine taxa (one negative: Candidatus Bacilloplasma; eight positive: Acinetobacter, Mollicutes_RF9_norank, Lactovum, Pragia, Saccharibacteria_norank, [Anaerorhabdus]_furcosa_group, Defluviicoccus, Dysgonomonas) was correlated with increased EsCrus1 gene expression (P < 0.05). The abundance of seventeen taxa (five negative: Candidatus Bacilloplasma, Rhodobacter, Gemmobacter, MNG7_norank, Clostridium_sensu_stricto_1; twelve positive: Candidatus Hepatoplasma, Mollicutes_RF9_norank, Lactovum, Pragia, Saccharibacteria_norank, [Anaerorhabdus]_furcosa_group, Defluviicoccus, Dysgonomonas, Comamonadaceae_unclassified, Morganella, Coprobacillus, Sphingobacterium) was related to increase EsCrus2 gene expression (P < 0.05). The abundance of eight taxa was related to increase mRNA expression of EsToll2 (P < 0.05). The abundance of fourteen taxa was correlated with increased mRNA expression of EsRelish (P < 0.05), as shown in Additional file 1: Fig. S6.
Fig. 6.
Correlation analyses between intestinal microorganism (phylum level) and gut immune genes expression. R values are correlation coefficient shown in different colors in the figure, the red color represents a positive correlation, the blue color represents a negative correlation. *P < 0.05
Discussion
In the human and mouse gut, Firmicutes and Bacteroidetes are the dominant phyla (Consortium 2012). In chickens, Firmicutes is the most prevalent, followed by Proteobacteria, Bacteroidetes and Actinobacteria (Choi et al. 2014). In ruminants, Firmicutes is the predominant phylum, followed by Bacteroidetes and Proteobacteria (Ye et al. 2016). In fish, Proteobacteria and Fusobacteria are the most dominant gut phyla (Clements et al. 2014; Tyagi and Singh 2017). In invertebrates, Proteobacteria and Firmicutes dominate the gut of Drosophila melanogaster (Broderick and Lemaitre 2012). Tenericutes and Proteobacteria are dominant in L. vannamei and E. sinensis (Zhang et al. 2014a, 2016). Proteobacteria and Bacteroidetes are the two dominant populations in E. sinensis (Li et al. 2007). Our research results showed that Firmicutes and Tenericutes were the predominant phyla in the foregut, Tenericutes and Proteobacteria were predominant in the midgut, and Proteobacteria, Tenericutes and Bacteroidetes were predominant in the hindgut of E. sinensis. In crabs, the epithelial surface structure of the midgut and the hindgut is different and therefore so is the bacterial adhesion (Chen et al. 2015). Although the dominant species are similar, the abundances are different, indicating that bacteria in the phyla Tenericutes, Bacteroidetes, Firmicutes and Proteobacteria maintain a close relationship with the host (Chen et al. 2015). Our findings were consistent with the above and suggest that there are evolutionary relationships between these animals. The intestinal flora and symbiotic microbes are very similar in crustaceans, although the functions of these symbiotic bacteria remain unknown.
We observed that the ACE and Chao values were significantly increased in the midgut. The microbial composition in the hindgut showed higher α-diversity indexes (Shannon and Simpson) comparing the midgut and the foregut. The gut flora of pond crabs had more inter-subject variation compared to wild crabs (Li et al. 2007). The unweighted UniFrac PCoA and Venn diagram revealed differences between the bacterial communities within the foregut, midgut and hindgut, indicating that the bacterial community and digestive tract function are closely related. These differences may be due to several reasons. The gastrointestinal organizational structure is different in the foregut, midgut and hindgut. Additionally, the various digestive tract regions have different functions; the foregut (stomach) is used to grind food, the midgut is responsible for digestion and absorption, and the hindgut stores stool and defends against microbes.
At the genus level, the abundance of Candidatus Bacilloplasma was significantly increased in the midgut compared to the microbes of the hindgut. By contrast, there was a significant decrease in Acinetobacter, unclassified_f_Rhodospirillaceae, Prolixibacter, Bacteroides, and Tyzzerella_3 in the midgut. Candidatus Bacilloplasma was a new lineage of bacteria specifically related to the intestinal surface of Porcellio scaber (Kostanjsek et al. 2007). They are rod-shaped bacteria that colonize the gut surface of crustacea and are affiliated with the class Mollicutes. Mollicutes is a common bacterium which is symbiotic and beneficial to the host in the intestinal tract of arthropods (Leclercq et al. 2014). Many of the Mollicutes, including Candidatus Bacilloplasma, colonize the digestive tract with food (Kostanjsek et al. 2007). Early research showed that there were rich in flora in the hindgut of Decapoda, but no absorption of nutrients, therefore, there is no competition with the hosts. Later, there has been an increasing interest on the midgut microbiota of Decapoda as suitable part for the research of microorganism–host interactions, and where nutrient absorption takes place (Meziti et al. 2010). However there is limited information comparing results between midgut and hindgut. In the present study, Candidatus Bacilloplasma were significantly more abundant in the midgut, which is where food digestion primarily occurs, indicating that the bacterial community and digestive tract function have a close relationship. Rhodospirillaceae, which are photosynthetic bacteria and contain abundant nutrients and functional factors (Qiu et al. 2008), were found to be significantly more abundant in the hindgut, suggesting that these bacteria could be potentially used as a probiotic.
The microbiota of the digestive tract are influenced by genetic background, environment and diet, etc. Early research had reported that gut microorganism are very key role in nutrients absorption and degradation (Zhang et al. 2014b), living environments (Huang et al. 2014), and immunity (Ye et al. 2016). Recently, research has aimed to determine the relationship between intestinal microbiota and the host in invertebrates (Xiong et al. 2015; You et al. 2014; Zhang et al. 2014a). Although intestinal microbes had closely related to the immune system, their interaction with gut immunity in invertebrates, especially in crustaceans, remains unknown.
Intestinal flora plays a key role in maintaining host health. In this study, the difference in predictive functional pathways between the midgut and hindgut indicated that change of microbial composition may have the ability to modify their functional capabilities. Our data showed that the genes responsible for nucleotide transport and metabolism and RNA processing and modification were up-regulated in the midgut compared with the hindgut. However, the genes were down-regulated associated with the cytoskeleton, inorganic ion transport and metabolism, and cell wall/membrane/envelope biogenesis. Because there are only a few studies concerning the gut bacterial community of crabs, the biological significance of these distinct differences remains unclear. Inorganic ion transport and metabolism includes phosphate, sulfate, and various cation transporters (Gill et al. 2006). The bacterial proteins identified were classified into COG functional categories, there is more highly represented in inorganic ion transport and metabolism of healthy children compared with obese children with non-alcoholic fatty liver disease (Michail et al. 2015). The increased abundance of cell wall/membrane/envelope biogenesis genes may suggest involvement with transmembrane transport and the exocytosis of antibiotics to resist the effects of tetracycline hydrochloride, indicating that gut microbiota can improve antibiotic resistance capability. Therefore, opportunistic bacteria can survive within the mouse gut (Yin et al. 2015).
There is a wealth of information about the intestinal immune regulation mechanism of vertebrate and its interaction with the symbiotic microbiota on the surface of intestinal mucosa (Rooks and Garrett 2016). However, there is little information concerning the gut immune mechanisms of invertebrates at the barrier epithelia (Garcia-Garcia et al. 2013). The relationship between commensal microbiota and gut immunity of crustaceans is important, although few data are available. We measured the relative mRNA expression of three genes in the Toll signaling pathway, one gene in the IMD signaling pathway and four AMP genes in the guts of crabs by RT-qPCR. We showed that the mRNA expression levels of EsToll2, EsRelish, EsALF1, EsALF2, EsCrus1, and EsCrus2 were significantly up-regulated in the hindgut compared with the midgut. However, the EsDorsal and EsPelle mRNA expression levels were only slightly higher in the hindgut. These results are consistent with previous reports (Watthanasurorot et al. 2012) and speculated that the hindgut flora has more regulation of intestinal immunity than the midgut flora, and it can better explain the fact that pathogens can enter easily the body through the midgut. In addition, the expression changes of eight genes showed similar trends in the midgut and the hindgut. EsALF1 and EsALF2 mRNA expression were significantly higher than the six other genes within the intestinal tract. Expression of EsRelish, EsCrus1 and EsCrus2 were found to be higher than the Toll signaling pathway genes (EsToll2, EsDorsal, EsPelle), although there was no significant difference. As previously reported, the intestinal immunity of amphioxus indicated that the expression of the TLRs gene is very low and the response to pathogens is small, which is contrary to vertebrates (Garcia-Garcia et al. 2013). This study revealed different expression of immune genes in the midgut and the hindgut of E. sinensis, and different sensitivity to external stimuli, which may be related to its different organizational structure. AMPs (EsALF1, EsALF2) act a pivotal part in intestinal immunity and may be regulated by the IMD pathway. In a study on the black tiger shrimp (Penaeus monodon), the digestive organ leads to quite severe injury after oral infection by Vibrio harveyi. RT-qPCR analysis showed that V. harvey induced up-regulation of AMP (ALF3, crustin and penaeidin) genes expression with infection time in the intestine of juvenile shrimp (Soonthornchai et al. 2010).
Innate immunity is the only host defense mechanism in invertebrates which lacks the high selection mechanisms of adaptive immunity. Recent reports have suggested that EsToll1 and EsToll2 are expressed in the digestive tract of E. sinensis (Yu et al. 2013a), although their gene expression level is not high. This result is consistent with our findings. Interestingly, TLRs were expressed at low levels in the gut of Drosophila melanogaster. Many reports have shown that the correlation between the composition of gut-associated bacteria and inflammatory parameters can serve as a biological indicator to evaluate the occurrence of diseases (Newsholme and de Bittencourt Jr 2016; Xiong et al. 2017; Zeng et al. 2017). However, the function and relationship of intestinal microbes and the occurrence of diseases in fish and crustacean are still poorly understood. Our results showed that the abundance of Actinobacteria negatively related to the relative mRNA expression of EsToll2, EsALF1, and EsALF2. Previous results showed that marine Actinobacteria can produce enzymes and can provide an important niche for probiotics in aquaculture with an increasing demand for both probiotics and prebiotics (Das et al. 2008). Subsequent studies showed that Actinobacteria isolated from the intestinal microorganism of two freshwater fish conferred resistance to human and fish pathogens and demonstrated its potential to produce biologically active compounds (Jami et al. 2015). Thus, an actinobacterial microarray can reflect the changes in body condition and culture environment (Wang et al. 2017). We speculate that Actinobacteria are symbiotic bacteria in the crab intestines and do not increase AMP gene expression. At the genus level, Acinetobacter, Bacteroides, Flavobacterium and 19 other species of bacteria can up-regulate the expression of AMPs to protect against pathogen invasion. Some bacteria, such as Lactobacillus, were highly abundant in the intestinal flora but did not have a positive correlation with AMP expression, suggesting that the intestinal mucosal immune system identifies these bacteria as symbiotic bacteria. The use of complex strategies to regulate the NF-κB signaling pathway and to inhibit the expression of AMPs is not clear and requires further research.
In conclusion, this study showed that the foregut, midgut and hindgut of E. sinensis have distinct microbiota. Community richness in the midgut was higher than that in the foregut or the hindgut, although the bacterial diversity in the hindgut was higher these data suggest that symbiotic microbiota are site-specific within the digestive tract of crabs. Data from this study also revealed the differences in the bacterial communities of the midgut and the hindgut. RT-PCR revealed that the mRNA expression level of AMPs was significantly up-regulated in the hindgut compared with the midgut and that the gene expression of EsRelish (IMD pathway) was higher than the Toll signaling pathway genes. AMP genes (EsALF1, EsALF2) play an important role in intestinal immunity and are presumably regulated by the IMD pathway. Correlation analysis revealed that the relationship between bacteria in the intestine of E. sinensis and the production of AMPs and provide information on the potential probiotics and biological indicators to evaluate the occurrence of disease. Overall, these results provide a comprehensive picture of the bacterial structure of the E. sinensis digestive tract and help to further elucidate disease prevention and treatment in crustaceans.
Additional file
Additional file 1. Additional figures and tables.
Authors’ contributions
LL and JD designed the study; JD and XZ conducted the study and collected the data; GW and JL analyzed the data; YZ helped with the manuscript writing; JD and RZ wrote the paper; LL had primary responsibility for the final content. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its additional files.
Consent for publication
The authors agree to publication in the journal.
Ethics approval and consent to participate
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Funding
This work was supported by the Natural Science Foundation of Liaoning Province (No. 20170540805), Overseas Training Project of Liaoning Colleges and Universities(2018LNGXGJWPY-YB017), Young Top Talents in the “Program for Promoting Liaoning Talents”, The earmarked fund for Modern Agro-industry Technology Research System (CARS-48)and Research start Plan for High level Talents of Shenyang Agricultural University (No. 880416005) and National Key Research and Development Program of China (No. 2017YFD0502200).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jing Dong, Email: dong9834@163.com.
Xiaodong Li, Email: lxd001@ceraap.com.
Ruiyang Zhang, Email: zhangruiyangsyau@163.com.
Yingying Zhao, Email: likeyingying@126.com.
Gaofeng Wu, Email: gaofengwu@126.com.
Jinling Liu, Email: 1405842573@qq.com.
Xiaochen Zhu, Email: asawinter2003@hotmail.com.
Lin Li, Phone: 86-13840169129, Email: lilin619619@163.com.
References
- Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;7:1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA. Friend, foe or food? Recognition and the role of antimicrobial peptides in gut immunity and Drosophila-microbe interactions. Phil Trans R Soc B. 2016;371:20150295. doi: 10.1098/rstb.2015.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf-Bensussan N, Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- Chaiyapechara S, Rungrassamee W, Suriyachay I, Kuncharin Y, Klanchui A, Karoonuthaisiri N, Jiravanichpaisal P. Bacterial community associated with the intestinal tract of P. monodon in commercial farms. Microb Ecol. 2012;63:938–953. doi: 10.1007/s00248-011-9936-2. [DOI] [PubMed] [Google Scholar]
- Chen DW, Zhang M. Non-volatile taste active compounds in the meat of Chinese mitten crab (Eriocheir sinensis) Food Chem. 2007;104:1200–1205. doi: 10.1016/j.foodchem.2007.01.042. [DOI] [Google Scholar]
- Chen X, Di P, Wang H, Li B, Pan Y, Yan S, Wang Y. Bacterial community associated with the intestinal tract of Chinese mitten crab (Eriocheir sinensis) farmed in Lake Tai, China. PLoS ONE. 2015;10:e0123990. doi: 10.1371/journal.pone.0123990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Kim G, Cha C. Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult Sci. 2014;93:1942–1950. doi: 10.3382/ps.2014-03974. [DOI] [PubMed] [Google Scholar]
- Clements KD, Angert ER, Montgomery WL, Choat JH. Intestinal microbiota in fishes: what’s known and what’s not. Mol Ecol. 2014;23:1891–1898. doi: 10.1111/mec.12699. [DOI] [PubMed] [Google Scholar]
- Consortium HMP Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Ward LR, Burke C. Prospects of using marine actinobacteria as probiotics in aquaculture. Appl Microbiol Biot. 2008;81:419–429. doi: 10.1007/s00253-008-1731-8. [DOI] [PubMed] [Google Scholar]
- Destoumieux-Garzón D, Rosa RD, Schmitt P, Barreto C, Vidal-Dupiol J, Mitta G, Gueguen Y, Bachère E. Antimicrobial peptides in marine invertebrate health and disease. Phil Trans R Soc B. 2016;371:20150300. doi: 10.1098/rstb.2015.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZF, Cao MJ, Zhu XS, Xu GH, Wang RL. Changes in the gut microbiome of the Chinese mitten crab (Eriocheir sinensis) in response to White spot syndrome virus (WSSV) infection. J Fish Dis. 2017;40:1561–1571. doi: 10.1111/jfd.12624. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia E, Galindo-Villegas J, Mulero V. Mucosal immunity in the gut: the non-vertebrate perspective. Dev Comp Immunol. 2013;40:278–288. doi: 10.1016/j.dci.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraserliggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Li X, Wang L, Shao Z. Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquac Res. 2014;47:1737–1746. doi: 10.1111/are.12628. [DOI] [Google Scholar]
- Jami M, Ghanbari M, Kneifel W, Domig KJ. Phylogenetic diversity and biological activity of culturable Actinobacteria isolated from freshwater fish gut microbiota. Microbiol Res. 2015;175:6–15. doi: 10.1016/j.micres.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Jiang XT, Peng X, Deng GH, Sheng HF, Wang Y, Zhou HW, Tam FY. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb Ecol. 2013;66:96–104. doi: 10.1007/s00248-013-0238-8. [DOI] [PubMed] [Google Scholar]
- Kostanjsek R, Strus J, Avgustin G. “Candidatus Bacilloplasma,” a novel lineage of Mollicutes associated with the hindgut wall of the terrestrial isopod Porcellio scaber (Crustacea: Isopoda) Appl Environ Microb. 2007;73:5566–5573. doi: 10.1128/AEM.02468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Zaneveld J, Caporaso JG, Mcdonald D, Dan K, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–823. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Dittmer J, Bouchon D, Cordaux R. Phylogenomics of “Candidatus Hepatoplasma crinochetorum”, a lineage of mollicutes associated with noninsect arthropods. Genome Biol Evol. 2014;6:407–415. doi: 10.1093/gbe/evu020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Guan W, Wei G, Liu B, Xu J, Zhao L, Zhang Y. Phylogenetic analysis of intestinal bacteria in the Chinese mitten crab (Eriocheir sinensis) J Appl Microbiol. 2007;103:675–682. doi: 10.1111/j.1365-2672.2007.03295.x. [DOI] [PubMed] [Google Scholar]
- Li F, Wang L, Zhang H, Zheng P, Zhao J, Qiu L, Zhang Y, Song L. Molecular cloning and expression of a Relish gene in Chinese mitten crab Eriocheir sinensis. Int J Immunogenet. 2010;37:499–508. doi: 10.1111/j.1744-313X.2010.00954.x. [DOI] [PubMed] [Google Scholar]
- Loch G, Zinke I, Mori T, Carrera P, Schroer J, Takeyama H, Hoch M. Antimicrobial peptides extend lifespan in Drosophila. PLoS ONE. 2017;12:e0176689. doi: 10.1371/journal.pone.0176689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziti A, Ramette A, Mente E, Kormas KA. Temporal shifts of the Norway lobster (Nephrops norvegicus) gut bacterial communities. FEMS Microbiol Ecol. 2010;74:472–484. doi: 10.1111/j.1574-6941.2010.00964.x. [DOI] [PubMed] [Google Scholar]
- Michail S, Lin M, Frey MR, Fanter R, Paliy O, Hilbush B, Reo NV. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol. 2015;91:1–9. doi: 10.1093/femsec/fiu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu C, Zheng P, Zhao J, Wang L, Qiu L, Zhang H, Gai Y, Song L. A novel type III crustin (Crus Es 2) identified from Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2011;31:142–147. doi: 10.1016/j.fsi.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Newsholme P, de Bittencourt Jr PIH. Gut associated bacteria are critical to metabolism, inflammation and health. Curr Opin Clin Nutr. 2016;19:245–249. doi: 10.1097/MCO.0000000000000293. [DOI] [PubMed] [Google Scholar]
- Olmos J, Ochoa L, Paniagua-Michel J, Contreras R. Functional feed assessment on Litopenaeus vannamei using 100% fish meal replacement by soybean meal, high levels of complex carbohydrates and Bacillus probiotic strains. Mar Drugs. 2010;9:1119–1132. doi: 10.3390/md9061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Xu S, Cai Y, Yang YZ, Xie H. Antagonistic effects of four strains of Rhodospirillaceae and factors affecting inhibitory function. Trans Chin Soc Agr Eng. 2008;24:194–198. [Google Scholar]
- Quigley EMM. Gut bacteria in health and disease. Gastroenterol Hepatol. 2013;9:560–569. [PMC free article] [PubMed] [Google Scholar]
- Rolff J, Schmid-Hempel P. Perspectives on the evolutionary ecology of arthropod antimicrobial peptides. Phil Trans R Soc B. 2016;371:20150297. doi: 10.1098/rstb.2015.0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Ha EM, Oh CT, Seol JH, Brey PT, Jin I, Dong GL, Kim J, Lee D, Lee WJ. An essential complementary role of NF-κB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 2006;25:3693–3701. doi: 10.1038/sj.emboj.7601233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Kim SH, Lee HY, Bai JY, Nam YD, Bae JW, Lee DG, Shin SC, Ha EM, Lee WJ. Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science. 2008;319:777–782. doi: 10.1126/science.1149357. [DOI] [PubMed] [Google Scholar]
- Shang Q, Song G, Zhang M, Shi J, Xu C, Hao J, Li G, Yu G. Dietary fucoidan improves metabolic syndrome in association with increased Akkermansia population in the gut microbiota of high-fat diet-fed mice. J Funct Foods. 2017;28:138–146. doi: 10.1016/j.jff.2016.11.002. [DOI] [Google Scholar]
- Soonthornchai W, Rungrassamee W, Karoonuthaisiri N, Jarayabhand P, Klinbunga S, Söderhäll K, Jiravanichpaisal P. Expression of immune-related genes in the digestive organ of shrimp, Penaeus monodon, after an oral infection by Vibrio harveyi. Dev Comp Immunol. 2010;34:19–28. doi: 10.1016/j.dci.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Tyagi A, Singh B. Microbial diversity in Rohu fish gut and inland saline aquaculture sediment and variations associated with next-generation sequencing of 16S rRNA gene. J Fish Life sci. 2017;2:1–8. [Google Scholar]
- Wang J, Zhao W, Liang Y, Han Y, Wang X, Zhang X, Ma X, Zhang D, Huang Z. Development of a prototype 16S rRNA gene-based microarray for monitoring planktonic actinobacteria in shrimp ponds. Int Aquat Res. 2017;9:71–80. doi: 10.1007/s40071-017-0158-1. [DOI] [Google Scholar]
- Watthanasurorot A, Söderhäll K, Jiravanichpaisal P. A mammalian like interleukin-1 receptor-associated kinase 4 (IRAK-4), a TIR signaling mediator in intestinal innate immunity of black tiger shrimp (Penaeus monodon) Biochem biophys Res Co. 2012;417:623–629. doi: 10.1016/j.bbrc.2011.12.019. [DOI] [PubMed] [Google Scholar]
- Xiong J, Wang K, Wu J, Qiuqian L, Yang K, Qian Y, Zhang D. Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl Microbiol Biotechnol. 2015;99:6911–6919. doi: 10.1007/s00253-015-6632-z. [DOI] [PubMed] [Google Scholar]
- Xiong J, Zhu J, Dai W, Dong C, Qiu Q, Li C. Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ Microbiol. 2017;19:1490–1501. doi: 10.1111/1462-2920.13701. [DOI] [PubMed] [Google Scholar]
- Yan Q, Gast CJVD, Yu Y. Bacterial community assembly and turnover within the intestines of developing Zebrafish. PLoS ONE. 2012;7:e30603. doi: 10.1371/journal.pone.0030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Liu J, Feng P, Zhu W, Mao S. Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci Rep. 2016;6:20329. doi: 10.1038/srep20329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Zhang XX, Wu B, Xian Q. Metagenomic insights into tetracycline effects on microbial community and antibiotic resistance of mouse gut. Ecotoxicology. 2015;24:2125–2132. doi: 10.1007/s10646-015-1540-7. [DOI] [PubMed] [Google Scholar]
- Ying H, Chen YH, Zhang YZ, Feng JL, Zhao LL, Zhu HX, Wen W, Qian R. Identification, characterization, and functional studies of a Pelle gene in the Chinese mitten crab, Eriocheir sinensis. Fish Shellfish Immunol. 2015;45:704–716. doi: 10.1016/j.fsi.2015.05.027. [DOI] [PubMed] [Google Scholar]
- You H, Lee WJ, Lee WJ. Homeostasis between gut-associated microorganisms and the immune system in Drosophila. Curr Opin Immunol. 2014;30:48–53. doi: 10.1016/j.coi.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Yu AQ, Jin XK, Li S, Guo XN, Wu MH, Li WW, Wang Q. Molecular cloning and expression analysis of a dorsal homologue from Eriocheir sinensis. Dev Comp Immunol. 2013;41:723–727. doi: 10.1016/j.dci.2013.08.013. [DOI] [PubMed] [Google Scholar]
- Yu AQ, Jin XK, Guo XN, Li S, Wu MH, Li WW, Wang Q. Two novel Toll genes (EsToll1 and EsToll2) from Eriocheir sinensis are differentially induced by lipopolysaccharide, peptidoglycan andzymosan. Fish Shellfish Immunol. 2013;35:1282–1292. doi: 10.1016/j.fsi.2013.07.044. [DOI] [PubMed] [Google Scholar]
- Zeng M, Inohara N, Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gallo RL. Antimicrobial peptides. Curr Biol. 2016;26:R14–R19. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang L, Wang L, Yang J, Gai Y, Qiu L, Song L. The second anti-lipopolysaccharide factor (EsALF-2) with antimicrobial activity from Eriocheir sinensis. Dev Comp Immunol. 2010;34:945–952. doi: 10.1016/j.dci.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Zhang M, Sun Y, Chen K, Yu N, Zhou Z, Chen L, Du Z, Li E. Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture. 2014;434:449–455. doi: 10.1016/j.aquaculture.2014.09.008. [DOI] [Google Scholar]
- Zhang M, Liu N, Qian C, Wang Q, Wang Q, Long Y, Huang Y, Zhou Z, Yan X. Phylogenetic and functional analysis of gut microbiota of a fungus-growing higher termite: bacteroidetes from higher termites are a rich source of β-glucosidase genes. Microb Ecol. 2014;68:416–425. doi: 10.1007/s00248-014-0388-3. [DOI] [PubMed] [Google Scholar]
- Zhang M, Sun Y, Chen L, Cai C, Qiao F, Du Z, Li E. Symbiotic bacteria in gills and guts of Chinese mitten crab (Eriocheir sinensis) differ from the free-living bacteria in water. PLoS ONE. 2016;11:e0148135. doi: 10.1371/journal.pone.0148135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional figures and tables.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its additional files.