Summary
Innate immune factors may restrict hematopoietic stem cell (HSC) genetic engineering and contribute to broad individual variability in gene therapy outcomes. Here, we show that HSCs harbor an early, constitutively active innate immune block to lentiviral transduction that can be efficiently overcome by cyclosporine H (CsH). CsH potently enhances gene transfer and editing in human long-term repopulating HSCs by inhibiting interferon-induced transmembrane protein 3 (IFITM3), which potently restricts VSV glycoprotein-mediated vector entry. Importantly, individual variability in endogenous IFITM3 levels correlated with permissiveness of HSCs to lentiviral transduction, suggesting that CsH treatment will be useful for improving ex vivo gene therapy and standardizing HSC transduction across patients. Overall, our work unravels the involvement of innate pathogen recognition molecules in immune blocks to gene correction in primary human HSCs and highlights how these roadblocks can be overcome to develop innovative cell and gene therapies.
Keywords: hematopoietic stem cells, gene therapy, gene editing, innate immunity, lentiviral vectors, IFITM3, transduction enhancers, cyclosporine
Graphical Abstract
Highlights
-
•
The innate immune factor IFITM3 blocks VSV-G-mediated lentiviral (LV) entry into HSCs
-
•
CsH treatment causes rapid and transient IFITM3 degradation
-
•
CsH potently enhances LV gene transfer and editing in human long-term repopulating HSCs
-
•
CsH negates IFITM3-mediated variability in LV transduction efficiency across donors
Petrillo et al. demonstrate that the innate antiviral factor IFITM3 constitutively inhibits entry of lentiviral vectors into hematopoietic stem cells. CsH efficiently overcomes this block by degrading IFITM3, significantly improving HSC gene transfer and editing efficiency. These findings will improve lentiviral gene therapy in IFITM3-positive stem cells.
Introduction
The limited efficiency of gene manipulation in human hematopoietic stem and progenitor cells (HSPC) remains a major hurdle for effective clinical application of genetic therapies for a wide range of disorders. Indeed, high vector doses and prolonged ex vivo culture are still required for effective gene transfer, even with the most established lentiviral vector (LV)-based delivery platforms. Various transduction enhancers have been identified (Heffner et al., 2018, Petrillo et al., 2015, Wang et al., 2014, Zonari et al., 2017), which impact the LV life cycle at different stages from vector entry to integration. This highlights the existence of multiple barriers to gene transfer in HSPC.
We previously observed that cyclosporine A (CsA) enhances transduction in HSPC, contrasting with its well-known inhibitory activity against lentiviruses (Petrillo et al., 2015, Rasaiyaah et al., 2013). In differentiated cells, CsA inhibits lentiviral infection through interfering with the interaction of the HIV-1 capsid with the host cofactor cyclophilin A (CypA), which is important for optimal DNA synthesis, capsid uncoating and nuclear import of the viral pre-integration complex (PIC) (Hilditch and Towers, 2014). It has been unclear how CsA enhances LV transduction in HSPC.
There is increasing evidence that HSPC are responsive to type-I interferon (IFN)-mediated innate immune signaling (Essers et al., 2009, Haas et al., 2015, Hirche et al., 2017, Nagai et al., 2006). Although we have demonstrated that LV transduction does not trigger type I IFN signaling in HSPC (Piras et al., 2017), it has recently been shown that stem cells constitutively express genes that are typically IFN-inducible. This protects HSPC from viral infections (Wu et al., 2018). Although many of these antiviral host factors are known to potently restrict retroviral infections in mammalian cells (Towers and Noursadeghi, 2014), their potential impact on LV gene transfer in HSPC remains poorly characterized (Kajaste-Rudnitski and Naldini, 2015).
Here, we identify a potent steady-state restriction of LV-mediated gene transfer in human HSPC. We demonstrate that this barrier can be efficiently overcome by the non-immunosuppressive cyclosporine H (CsH), leading to significantly enhanced transduction and gene editing efficiencies in human HSPC.
Results
A CypA-Independent Cyclosporine Reveals an Early Block to LV Transduction in HSPCs
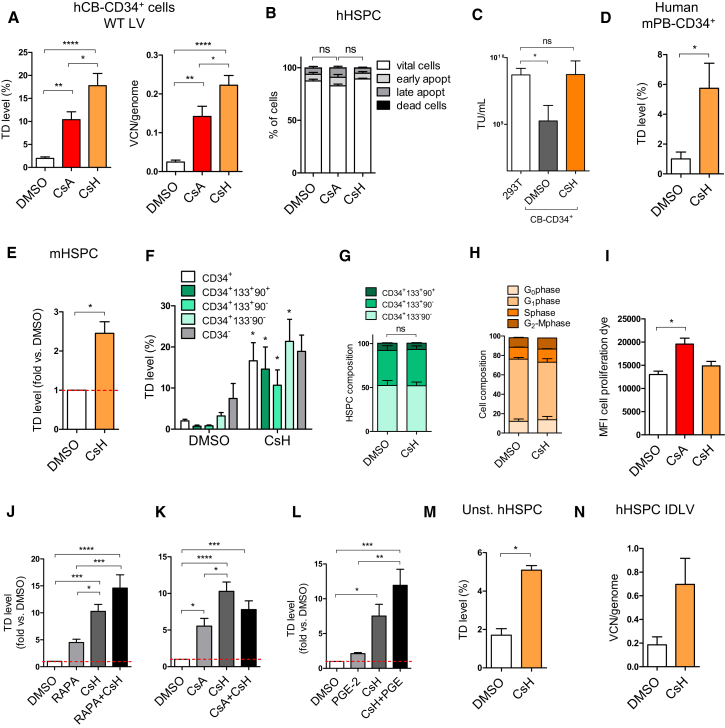
The reduction of LV infection in differentiated cells by CsA is due to inhibition of CypA recruitment to the incoming HIV-1 capsid (CA) (Sokolskaja and Luban, 2006, Towers, 2007, Towers et al., 2003). In agreement with a cofactor role for CypA during LV transduction, depletion of CypA led to lower transduction of human HSPC (Figures S1A–S1D). This implies that the capacity of CsA to increase LV transduction in HSPC is likely suboptimal, given that it will also interfere with this positive CypA-vector interaction. Based on these results, and our previous observation that the immunosuppressive arm of CsA is not involved in enhancing LV transduction in HSPC (Petrillo et al., 2015), we tested a naturally occurring cyclosporine, cyclosporine H (CsH), which does not bind CypA and is not immunosuppressive (Figure S1E) (Jeffery, 1991). Remarkably, CsH was more potent than CsA at the same 8μM dose and increased LV transduction up to 10-fold in human cord blood (CB)-derived HSPC (Figure 1A). Higher doses of CsH further increased transduction (Figure S1F) but were toxic (Figures 1B and S1G). CsH improved transduction as early as 2 hr post-exposure but optimal efficacy was achieved after overnight (16 hr) exposure (Figures S1H and S1I). The enhancement was lower if CsH was removed prior to transduction but could be restored by blocking de novo protein synthesis during the 6 hr of vector exposure (Figure S1J). Remarkably, CsH rendered HSPC as permissive as the highly transducible 293T cell line (Figure 1C). Importantly, CsH was effective in the clinically relevant human mobilized peripheral blood (mPB)-derived CD34+ cells, in murine HSPC (Figures 1D and 1E) and in all CD34+ subpopulations, including in the more primitive CD34+CD133+CD90+ fraction (Figure 1F), without altering the subpopulation composition nor the cell-cycle status (Figures 1G and 1H). Unlike CsA, no proliferation delay was observed with CsH, in line with CsH not being immunosuppressive (Figure 1I). CsH was more potent and additive with transduction enhancers Rapamycin (Rapa) (Petrillo et al., 2015, Wang et al., 2014) and PGE2 (Heffner et al., 2018, Zonari et al., 2017) but not with CsA (Figures 1J–1L). CsH also increased LV transduction in unstimulated HSPC (Figure 1M), but did not alter transduction in primary human monocyte-derived macrophages (MDM) (Figure S1K). This was irrespective of SAMHD1-mediated LV restriction, which can be overcome by incorporating the simian immunodeficiency virus macaque (SIVmac) accessory protein Vpx into the LV particle (Bobadilla et al., 2013). Conversely, we saw a statistically significant increase in the percentage of transduced (GFP+) activated T cells with CsH (Figure S1L). These data are in contrast with the inhibitory effect of CsA that we and others have observed in this cell type (Donahue et al., 2017, Lahaye et al., 2016, Petrillo et al., 2015). This further underscores how the CypA-dependent CsA inhibition of LV infection likely masks its enhancement of transduction. Indeed, CsA increased transduction of the CypA-independent P90A LV capsid mutant, which does not recruit CypA, to similar extent as CsH in HSPC (Figure S1M). CsH also increased transduction of the CPSF6-independent N74D capsid mutant (Lee et al., 2010) (Figure S1N), ruling out CPSF6 in the CsH enhancement mechanism. Of note, transduction by an SIVmac-derived vector, which does not interact with human CypA (Luban et al., 1993), was also increased by both cyclosporines in human CD34+ cells (Figure S1O). To further dissect the step of the viral life cycle affected by CsH, we measured viral DNA replication intermediates early after transduction. Consistent with the increase in infection, CsH led to an increase in late-reverse transcription (RT) products and 2LTR circles, the latter a marker of LV nuclear import (Follenzi et al., 2000) (Figures S1P and S1Q). Of note, no changes in the 2LTR/late-RT ratio were observed upon CsH treatment (Figure S1R), indicating that CsH relieves an early block to transduction prior to DNA synthesis without affecting nuclear import. Concordantly, CsH also enhanced transduction of an integrase-defective LV (IDLV), as measured by an increase in vector DNA per cell (Figure 1N). CsA was also able to increase IDLV transduction when packaged with the CypA-independent A88T mutant capsid (Busnadiego et al., 2014) (Figure S1S). These results indicate that both cyclosporines relieve an early block to LV transduction in human HSPC and identify CsH as the most effective HSPC transduction enhancer described thus far.
Figure 1.
CsH Is a Potent and Non-toxic Enhancer of Lentiviral Vector Transduction in Hematopoietic Stem Cells
(A) Human cord-blood (CB) cells were transduced with a VSV-g lentiviral (LV) at a multiplicity of infection (MOI) of 1 transducing unit (TU)/293T cell ± 8 μM CsA or CsH. Percentages of transduced cells and vector copy numbers/human genome (VCN/genome) were assessed at 5 or 14 days post-transduction, respectively (mean ± SEM; n = 20; one-way ANOVA with Bonferroni’s multiple comparison, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001).
(B) Impact of CsA and CsH on apoptosis was assessed in hCB-CD34+ cells 48 hr post-transduction (mean ± SEM; n = 6; Dunn’s adjusted Kruskal-Wallis test; ns, not significant).
(C) VSV-g pseudotyped LV was titered in parallel in 293T cells or human CB-CD34+ cells ± 8 μM CsH (mean ± SEM; n = 4; one-way ANOVA with Bonferroni’s multiple comparison; ns, not significant, ∗p ≤ 0.05).
(D and E) Transduction efficiencies (MOI = 1) in human mobilized peripheral blood (mPB)-CD34+ cells (mean ± SEM, n = 4, Mann-Whitney test, ∗p ≤ 0.05) (D) or murine hematopoietic stem and progenitor cells (mHSPC) (mean ± SEM, n = 8, Wilcoxon signed rank test, ∗p = 0.0078) (E).
(F) Transduction efficiencies (MOI = 1) in the different subpopulations of human mPB-CD34+ cells (mean ± SEM, n = 4, Mann-Whitney test versus each DMSO, ∗p ≤ 0.05).
(G and H) The composition (G) and cell-cycle status (H) of human mPB- or CB-CD34+ cells, respectively, were evaluated 48 hr post-transduction.
(I) Impact of CsA and CsH on cell proliferation was assessed in hCB-CD34+ cells 48 hr post-transduction (mean ± SEM; n = 4; Dunn’s adjusted Kruskal-Wallis test, ∗p ≤ 0.05).
(J–L) hCB- (J and K) and mPB- (L) derived HSPC were transduced in the presence of different drug combinations with LV at an MOI of 1 or 10 (mean ± SEM; n = 7 for I and J, and n = 4 for K; one-way ANOVA with Bonferroni’s multiple comparison, ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001).
(M) Transduction efficiencies ± 8 μM CsH (MOI = 10) in unstimulated hCB-CD34+ cells (unst. hHSPC) (mean ± SEM, n = 4, Mann-Whitney, ∗p ≤ 0.05).
(N) hCB-CD34+ cells were transduced with an integrase defective LV (IDLV) vector at MOI = 50 ± 8 μM CsH.
CsH Increases LV Transduction and Gene Editing Efficiency in SCID-Repopulating HSPCs
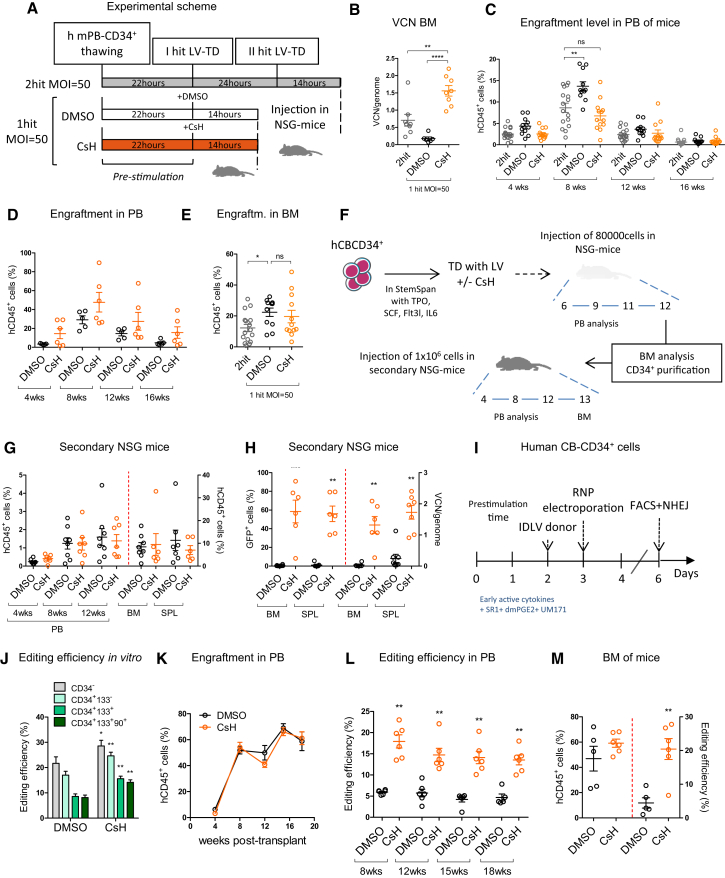
To assess CsH-enhanced transduction in a more clinically relevant setting, we transplanted human mPB-CD34+ cells transduced with clinical grade LV expressing the alpha-L-iduronidase (IDUA) transgene (IDUA-LV), designed to treat patients affected by type I mucopolysaccharidoses (MPS-I) (Visigalli et al., 2010, Visigalli et al., 2016), into the xenograft NSG mouse model of human hematopoiesis and followed engraftment and transduction efficiency in vivo (Figure 2A). As a control, cells were transduced twice in the absence of CsH, as per the current standard protocol. No differences in colony-forming capacity were observed between control and CsH-treated cells. The two-hit transduction protocol showed a slightly increased colony output, likely reflecting the higher percentage of progenitor cells due to the longer ex vivo culture period (Figure S2A). Remarkably, one single LV dose in the presence of CsH was enough to yield significantly higher gene marking in vitro (Figures S2B and S2C), as well as in long-term repopulating HSC and derived progeny in vivo (Figures 2B, S2D, and S2E). This surpassed the reference protocol by 2-fold and achieved an almost 10-fold increase in long-term gene marking in vivo compared to the single transduction control group. CsH exposure did not alter the short-term engraftment capacity of HSPC compared to the two-hit protocol but remained slightly below levels achieved with the single-hit control (Figures 2C and S2F). This could be related to the enhanced level of transduction as observed previously (Piras et al., 2017). Indeed, exposure of mPB-CD34+ cells to CsH alone did not impact early HSPC engraftment (Figures 2D and S2G). Importantly, we observed higher long-term engraftment, associated with shorter ex vivo culture, in the BM and spleens for CsH-treated transduced cells (Figures 2E and S2H–S2L). CsH also significantly enhanced LV transduction in bona fide long-term HSC repopulating secondary recipients without affecting their engraftment capacity (Figures 2F–2H and S2M–S2P).
Figure 2.
CsH Increases Gene Transfer and Editing in Long-Term SCID-Repopulating Human HSPCs
(A) Experimental scheme of the different transduction protocols using a clinical-grade LV and human mPB-derived CD34+ cells.
(B) VCN/genome were measured in the bone marrow (BM) at 18 weeks (mean ± SEM; n = 8; one-way ANOVA with Bonferroni’s multiple comparison, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001).
(C and D) Engraftment levels in the peripheral blood of mice from different treatment groups transduced (C) or not (D) with IDUA-LV, respectively (mean ± SEM; n ≥ 11; Dunn’s adjusted Kruskal-Wallis; ns, not significant, ∗∗p ≤ 0.01).
(E) Engraftment levels in the BM at 18 weeks post-transplant (mean ± SEM; n ≥ 11; Dunn’s adjusted Kruskal-Wallis; ns, not significant, ∗p ≤ 0.05).
(F) Experimental design of the secondary transplantation experiment using a purified PGK-GFP LV and human CB-CD34+ cells.
(G) Engraftment levels in the PB, BM, and SPL of secondary mice 13 weeks after transplantation.
(H) Transduction efficiencies (left: GFP+ cells; right: VCN/human genome) in BM and SPL of mice 13 weeks post-injection (mean ± SEM; n = 7, 8 mice per group, Mann-Whitney test versus DMSO control, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
(I) Scheme of the gene editing protocol for human CB-derived CD34+ cells.
(J) Percentage of edited cells at AAVS1 locus measured within the indicated subpopulations 3 days after editing (mean ± SEM; n = 7, Mann-Whitney test, ∗p ≤ 0.05, ∗∗p ≤ 0.01).
(K) Human CD45+ cell engraftment in PB at indicated times after transplantation of IL2RG edited CB-CD34+ cells (n = 5).
(L) Percentage of gene editing by homology-directed repair (HDR) measured within human cells in mice from (K).
(M) Percentage of human gene edited cells and editing efficiency measured in the BM of mice 19 weeks post-transplantation.
As CsH enhanced IDLV transduction in human HSPC, we tested its effect on the efficiency of IDLV-mediated HSPC gene editing (Figure 2I). Remarkably, delivery of the donor IDLV in the presence of CsH increased gene editing efficiency in human HSPC (Figure 2J) without altering the relative composition of the hematopoietic subpopulations (Figure S2Q). This increase was even more pronounced in the primitive CD34+CD133+CD90+ fraction (Figure 2J) and significantly higher targeting efficiency was maintained long-term in vivo, reaching an almost 4-fold increase in gene editing over controls, without affecting engraftment (Figures 2K–2M, S2R, and S2S). Further molecular analysis confirmed that CsH specifically increased targeted integration by homology-directed repair (HDR) with the overall fraction of edited alleles remaining unchanged (Figure S2T). CsH did not affect AAV6-based gene editing in HSPC (Figure S2U), consistent with it not improving AAV6-mediated donor delivery (Figure S2V). Together, these observations clearly illustrate the value of CsH as an enhancer of LV gene transfer and editing and strongly support its use in therapeutic HSPC gene engineering.
Cyclosporines Counteract an IFN-Inducible Block to VSV Glycoprotein-Dependent Lentiviral Vector Entry
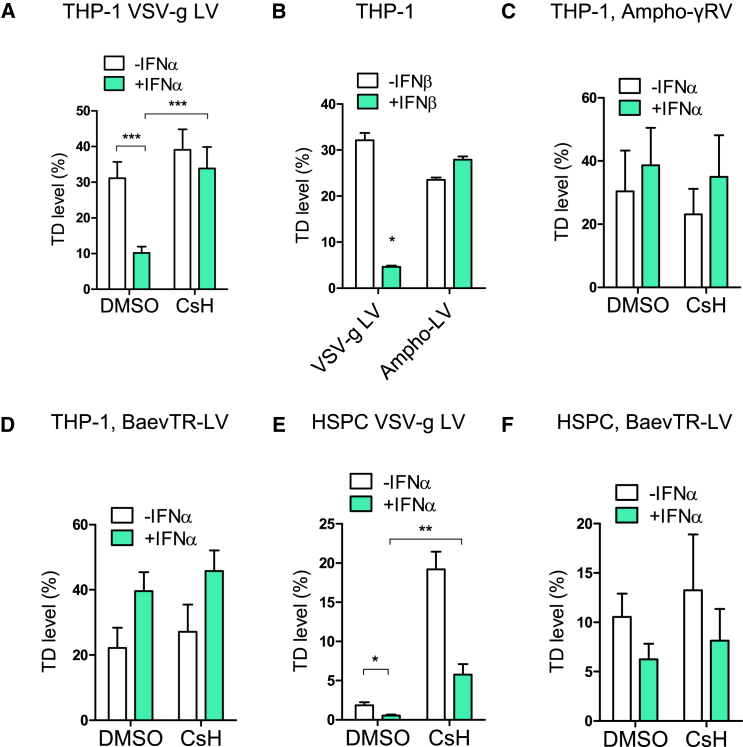
To understand how CsH enhances gene transfer/editing in HSPC, we considered how CsA improves VSV glycoprotein (VSV-G) pseudotyped LV transduction of IFNα-stimulated monocytic THP-1 cells (Bulli et al., 2016). We found that CsH also has this effect (Figures 3A and S3A). Neither calcineurin-inhibitor FK506 nor depletion of calcineurin, a known target of CsA, rescued transduction in this way (Figures S3B and S3C). The best characterized CsH target, formyl peptide receptor 1 (FPR1) (de Paulis et al., 1996, Prevete et al., 2015), is also not involved as IFN continued to suppress LV transduction in FPR1-depleted THP-1 cells (Figure S3D).
Figure 3.
CsH Counteracts an IFN-Inducible Block to VSV-G-Mediated Vector Entry
(A) THP-1 cells were pre-stimulated or not with 1,000 IU/mL of human IFNα for 24 hr followed by transduction with a VSV-g pseudotyped LV (MOI = 1) +/− 8 μM CsH (mean ± SEM; n = 10; Mann-Whitney test, ∗∗∗p ≤ 0.001).
(B) THP-1 cells ± 1 ng/mL of human IFNβ were transduced with VSV-g or amphotropic Molony murine leukemia virus (MLV) pseudotyped LVs (mean ± SEM, n = 3, Mann-Whitney test, ∗p = 0.05).
(C) THP-1 cells were pre-stimulated with human IFNα and transduced with Ampho-MLV pseudotyped RV ± CsH (mean ± SEM, n = 4).
(D) THP-1 ± IFNα were transduced with a BaEV-TR LV (MOI = 0.5–1) +/− CsH (mean ± SEM, n = 4).
(E) Human CB-derived CD34+ cells ± human IFNα were transduced with a VSV-g pseudotyped LV (MOI = 1) +/− 8 μM CsH (mean ± SEM; n = 6; Mann-Whitney test, ∗p ≤ 0.05, ∗∗p ≤ 0.01).
(F) Human CB-derived CD34+ cells ± IFNα were transduced with BaEV-TR LV (MOI = 0.5-1) +/− CsH (mean ± SEM, n ≥ 4).
See also Figure S3.
Similar to the CsH-sensitive transduction block in HSPC (Figures S1P–S1R), the type I IFN-mediated block in THP-1 is evident by viral DNA synthesis (Figure S3E). Inhibition by type I IFN requires de novo protein synthesis as cyclohexamide (CHX) rescued transduction in IFNβ-treated cells (Figure S3F). HIV-1 capsid mutants known to be altered for interactions with host cofactors CypA and CPSF6/Nup153 (Lee et al., 2010, Matreyek et al., 2013, Schaller et al., 2011) remained sensitive to IFN (Figure S3G). Similar IFN-induced inhibition and CsA-mediated rescue were observed with diverse VSV-G pseudotyped retroviral vectors (Figure S3H). However, vectors pseudotyped with the MLV-derived amphotropic envelope glycoprotein remained insensitive to both type I IFN-mediated inhibition and CsH (Figures 3B and 3C), indicating that restriction and CsH sensitivity is influenced by the viral envelope. In agreement with an envelope-dependent mechanism, LV pseudotyped with the modified Baboon endogenous retroviral envelope glycoprotein (BaEV-TR) (Girard-Gagnepain et al., 2014) remained insensitive to both type I IFN and CsH in THP-1 cells (Figure 3D). Importantly, in IFNα-treated HSPC, CsH also rescued VSV-G pseudotyped LV transduction (Figures 3E and S3I) but not BaEV-TR LV, which remained less sensitive to IFNα (Figure 3F). Together, these results are consistent with cyclosporine-mediated rescue of a type I IFN-inducible block to VSV-G-mediated LV entry in HSPC/THP-1.
CsH Counteracts IFITM3 Antiviral Activity in HSPCs
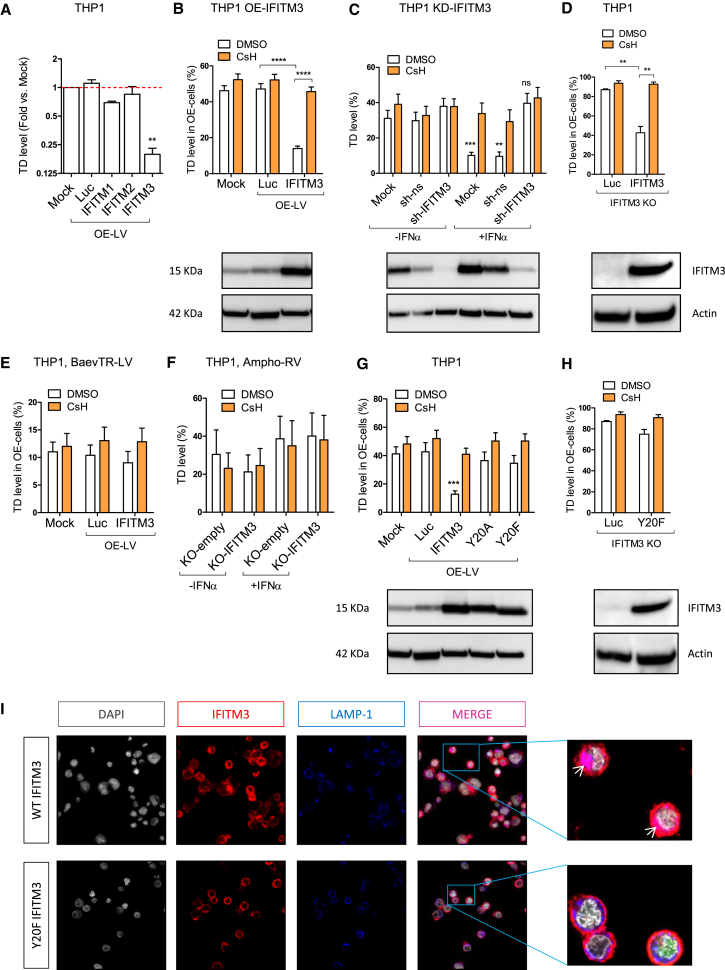
Members of the family of interferon induced transmembrane proteins (IFITMs), in particular IFITM3, have broad antiviral activity against several viruses including VSV and retroviruses such as HIV-1, but not MLV (Brass et al., 2009, Perreira et al., 2013). IFITM3 acts at the level of viral entry into target cells (Amini-Bavil-Olyaee et al., 2013, Li et al., 2013). To test whether IFITMs are involved in the effect of CsH, we first overexpressed the antiviral IFITM 1, 2, or 3 in THP-1 cells, followed by transduction with a reporter LV to evaluate IFITM impact on transduction efficiency (Figure S4A). Only overexpression of IFITM3 resulted in a significant decrease in LV transduction (Figures 4A and S4B), and importantly, inhibited transduction was fully rescued by CsH (Figure 4B). Furthermore, depletion or deletion of IFITM3 led to a full rescue of IFNα-induced LV restriction in THP-1 cells and abrogated CsH-mediated enhancement (Figures 4C, S4C, and S5A–S5F). We specifically overexpressed IFITM3 in THP-1 cells deleted for the endogenous IFITM2/3 and further confirmed its role in LV restriction and cyclosporine enhancement (Figure 4D). Similar to CsH-mediated effects, IFITM3-dependent restriction was independent of interactions of the LV capsid with host factors including CypA and CPSF6/Nup153 (Figure S4D). IFITM3 also impaired IDLV transduction (Figure S4E). As the CsH-mediated increase in LV transduction is specific to VSV-G-mediated entry, which occurs by particle endocytosis (Sun et al., 2005), we tested the capacity of IFITM3 to block BaEV-TR or Ampho-pseudotyped vectors that both fuse at the plasma membrane (Girard-Gagnepain et al., 2014, Ragheb et al., 1995). Consistent with their endocytosis-independent mechanism of entry, neither pseudotype was inhibited in THP-1 cells expressing IFITM3 (Figures 4E and 4F). On the other hand, AAV6 remained insensitive to the action of type I IFN and IFITM3 (Figure S4F), in agreement with its insensitivity to CsH in HSPC (Figure S2V). Phosphorylation of the tyrosine residue in position 20 (Y20) of IFITM3 has been shown to be required for its activity against VSV-G pseudotyped RV (Jia et al., 2012). Indeed, overexpressed IFITM3 Y20 mutants failed to restrict VSV-G pseudotyped LV transduction in THP-1 cells (Figures 4G, 4H, S4G, and S4H) and remained mostly at the plasma membrane (Figure 4I), potentially explaining the lack of restriction.
Figure 4.
CsH Counteracts IFITM3 Antiviral Activity
(A) THP-1 cells were transduced with an LV co-expressing GFP and IFITM proteins or Luciferase (Luc) as a control (OE-LV) and then re-challenged with a second reporter LV. Transduction efficiencies are as fold versus mock (mean ± SEM, n ≥ 6, Wilcoxon signed rank test versus mock = 1, ∗∗p = 0.0078).
(B) THP-1 cells overexpressing Luc or IFITM3 were transduced with a reporter LV ± 8 μM CsH (mean ± SEM, n ≥ 20, Mann-Whitney test, ∗∗∗∗p ≤ 0.0001).
(C) THP-1 cells depleted for IFITM3 (sh-IFITM3) or expressing a non-silencing small hairpin RNA (shRNA) control (sh-ns) were re-transduced ± IFNα pre-treatment and ± CsH (mean ± SEM, n = 10, Mann-Whitney test versus each control without hIFNα; ns, not significant, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001).
(D) THP-1 deleted for IFITM3 were transduced with control Luc or IFITM3 expressing LV and re-challenged with a reporter LV ± 8 μM CsH (mean ± SEM, n = 5, Mann-Whitney test, ∗∗p ≤ 0.01).
(E) THP-1 cells expressing exogenous Luc or IFITM3 were transduced with a BaEV-TR LV ± CsH (mean ± SEM, n = 12).
(F) THP-1 cells deleted for IFITM3 (KO-IFITM3) or control (KO-empty) were transduced with an Ampho pseudotyped RV ± CsH (mean ± SEM, n = 4). IFITM3 protein levels were evaluated at the time of transduction.
(G and H) THP-1 overexpressing the WT or mutated forms of IFITM3 (mean ± SEM, n = 12, Mann-Whitney test versus Luc control, ∗∗∗p ≤ 0.001) (G) or THP-1 deleted for endogenous IFITM3 overexpressing the Luc control or the Y20F form of IFITM3 were re-exposed to a reporter LV ± 8 μM CsH (mean ± SEM, n = 5) (H).
(I) Co-localization (purple areas evidence by white arrows) of IFITM3 protein (in red) with the lysosome associated membrane protein 1 (LAMP1) marker (in blue) was evaluated by immunofluorescence in THP-1 cells deleted for the endogenous IFITM3 overexpressing the WT or Y20F form of IFITM3.
See also Figure S4.
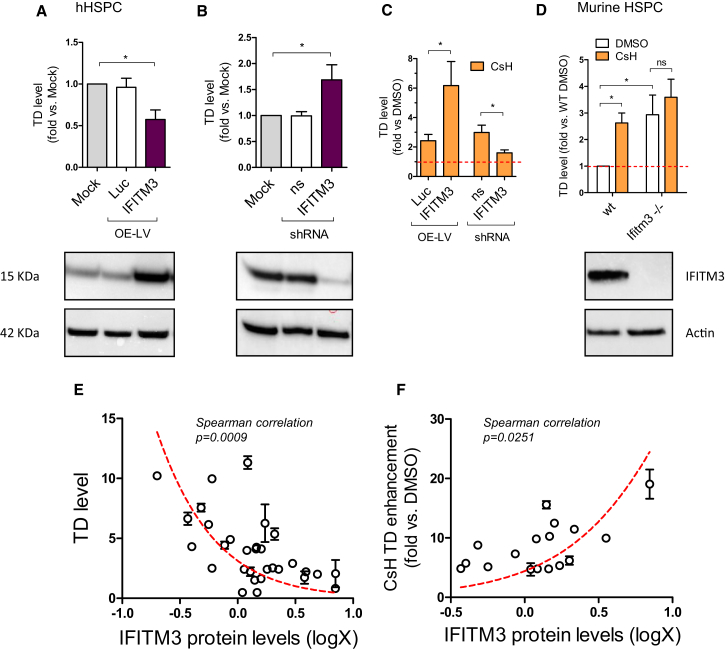
Importantly, overexpression of IFITM3 in HSPC resulted in a significant decrease in LV transduction (Figure 5A). Conversely, depletion of IFITM3 resulted in an enhancement of transduction (Figure 5B). Moreover, CsH-mediated enhancement of transduction was stronger in IFITM3-overexpressing HSPC and was almost completely lost with IFITM3 depletion (Figure 5C). These data were confirmed in vivo as Lin− HSPC isolated from the bone marrow of Ifitm3−/− mice (Lange et al., 2008) were more permissive to LV transduction and were largely insensitive to CsH-mediated transduction enhancement as compared to Lin− HSPC isolated from wild-type counterparts (Figure 5D).
Figure 5.
CsH Rescues IFITM3-Dependent Early Impairment of LV Transduction in HSPCs
(A and B) IFITM3 was expressed (A) or depleted (B) in human CB-CD34+ cells (A: mean ± SEM, n = 7, Wilcoxon signed rank test versus mock = 1, ∗p = 0.0156; B: mean ± SEM, n = 8, Wilcoxon signed rank test versus mock = 1, ∗p = 0.0313).
(C) IFITM3 protein levels were evaluated by western blot 3–5 days post overexpression and cells were re-challenged with an LV ± 8 μM CsH (Mann-Whitney test, ∗p ≤ 0.05).
(D) WT or Ifitm3 knock-out (Ifitm3−/−) murine Lin− HSPC were transduced with LV at MOI 1 ± CsH (mean ± SEM, n = 3 experiments in duplicate each from 7 WT/Ifitm3−/− mice, Wilcoxon signed rank test versus DMSO = 1, ∗p = 0.0313; Mann-Whitney test; ns, not significant).
(E) Human CD34+ cells from individual donors were transduced at MOI 1 in duplicate and IFITM3 protein levels were evaluated at the time of transduction by WB, quantified by densitometry using ImageJ software, normalized on Actin and then log-transformed (x axis). Transduction efficiencies were measured by fluorescence-activated cell sorting (FACS) 5 days post-transduction and reported on y axis (mean ± SEM, n = 31 HSPC donors, Spearman correlation, ∗∗∗p ≤ 0.001, r = −0.5671).
(F) Human CD34+ cells from individual donors were transduced at MOI 1 in duplicate ± 8 μM CsH and IFITM3 protein levels were evaluated and quantified at the time of transduction as above (x axis). Transduction efficiencies are expressed in terms of fold-increase in presence of CsH versus DMSO (y axis) (mean ± SEM, n = 19 HSPC donors, Spearman correlation, ∗p ≤ 0.05, r = 0.5405).
See also Figure S5.
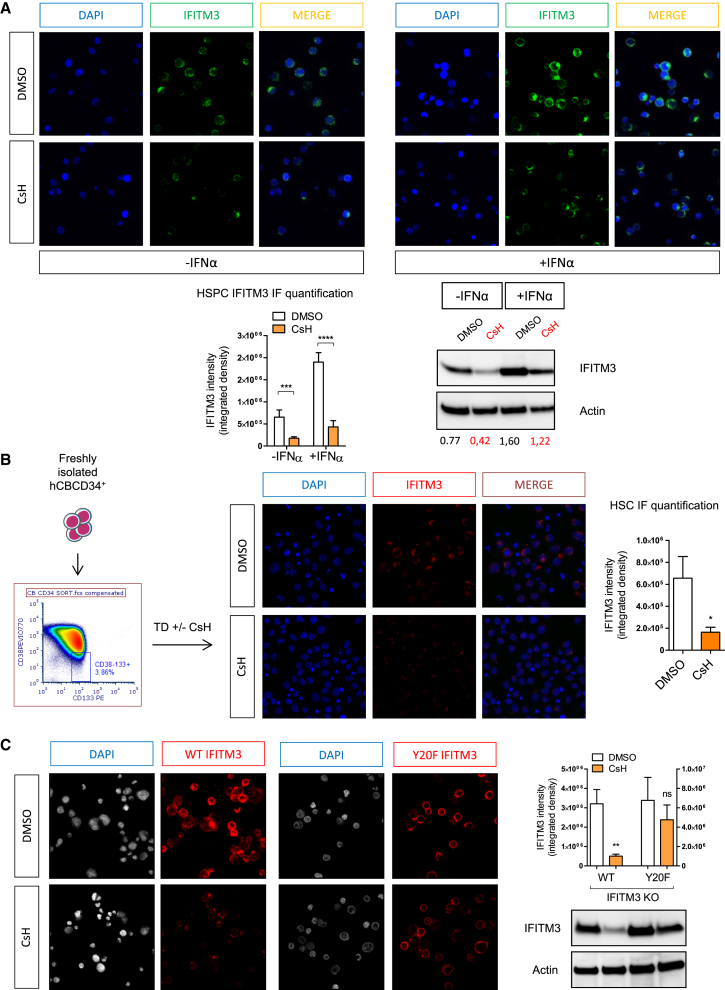
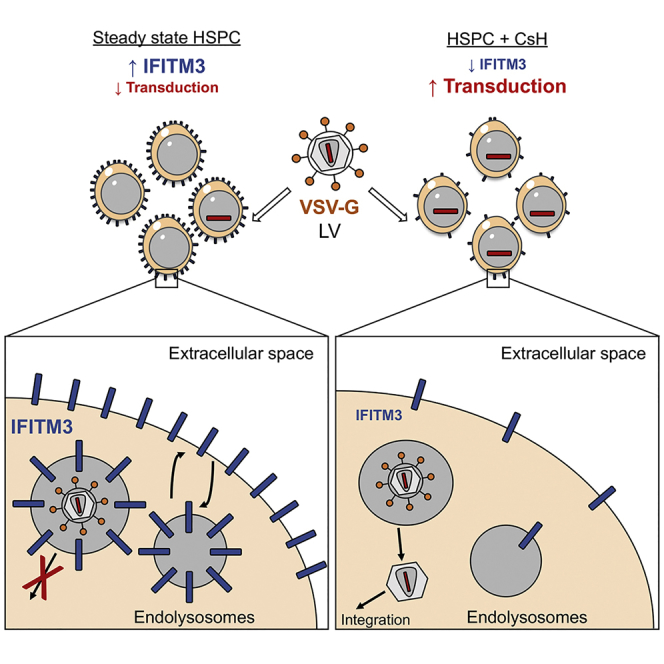
Intriguingly, individual human HSPC donors expressed different levels of IFITM3 protein, which negatively correlated with LV transduction efficiency (Figure 5E). As expected, CsH enhanced transduction most effectively in samples with higher IFITM3 expression (Figure 5F). Finally, we investigated the impact of CsH on IFITM3 expression in bulk or sorted HSPC or THP-1 stimulated with type I IFN. Significantly lower IFITM3 protein, but not mRNA, was detected after CsH treatment in both cell types (Figures 6A, 6B, and S6A–S6D). Interestingly, the inactive IFITM3 Y20F mutant was insensitive to CsH-induced loss in THP-1 cells (Figure 6C). Of note, removal of CsH prior to transduction resulted in partial restoration of IFITM3 protein levels within the 6 hr transduction window. This was prevented by blocking de novo protein synthesis during vector exposure in HSPC (Figure S6E). These results are in line with CsH being most effective when added with the vector (Figure S1J). The transient effect of CsH on IFITM3 protein levels was confirmed in THP-1 cells overexpressing IFITM3, where IFITM3 protein levels were restored within 6 hr of CsH removal (Figure S6F). Overall, these results illustrate how CsH potently enhances HSPC gene manipulation efficiencies by overcoming an IFITM3-mediated block to VSV-G-pseudotyped LV infection.
Figure 6.
CsH Degrades IFITM3
(A) IFITM3 protein levels were evaluated in human bulk CB-CD34+ cells ± IFNα by immunofluorescence and quantified as integrated density with ImageJ software after an overnight exposure to CsH (mean ± SEM of three independent experiments in duplicate, n = 18 images; Mann-Whitney test, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001) or by western blot as above (one representative blot out of two is shown).
(B) Freshly isolated FACS-sorted CD34+38−133+ HSC were transduced ± CsH and IFITM3 protein levels were evaluated by immunofluorescence as above (mean ± SEM, n = 10 images; Mann-Whitney test versus DMSO, ∗p ≤ 0.05).
(C) IFITM3 protein levels, evaluated by immunofluorescence or by WB as above, in THP-1 deleted for endogenous IFITM3 overexpressing the WT or the Y20F form of IFITM3 were then exposed ± CsH (mean ± SEM of three independent experiments, n = 14 images; Mann-Whitney test, ∗∗p ≤ 0.01).
See also Figure S6.
Discussion
Because of concerns of toxicity related to the immunosuppressive function of CsA, a number of non-immunosuppressive cyclosporine derivatives have been developed and tested for several applications (Peel and Scribner, 2013). Nevertheless, most of these still inhibit the host factor CypA. Here, we have shown that CypA is an important cofactor for efficient lentiviral transduction of HSPC. Therefore, we sought a non-immunosuppressive cyclosporine that did not inhibit CypA, identifying CsH as, to our knowledge, the most potent enhancer of HSPC gene transfer (Heffner et al., 2018, Lewis et al., 2018, Wang et al., 2014, Zonari et al., 2017, Petrillo et al., 2015). Our side-by-side comparisons illustrate CsH treatment outperforms even the standard double-hit protocol. Our findings are of particular relevance in gene therapy settings in which high levels of gene marking are required but difficult to achieve, such as for hemoglobinopathies, where the large and complex human β-globin gene expression cassette limits clinical-scale LV production (Baldwin et al., 2015). CsH utility may extend to other stem cells populations (Geis et al., 2017, Noser et al., 2006). Moreover, CsH helps the IDLV-based editing platforms compete with other non-integrating methods based on AAV or oligonucleotide-mediated donor DNA delivery (De Ravin et al., 2017, Dever et al., 2016, Schiroli et al., 2017, Wang et al., 2015), in particular for patients harboring pre-existing adaptive immunity against some AAV serotypes that may lead to immune recognition of vector and transduced cells (Boutin et al., 2010). Of note, the inability of CsH to increase AAV6-based donor delivery and gene editing, together with the unaltered overall DNA repair observed in CsH exposed HSPC, indicates that CsH improves gene editing specifically through increasing IDLV donor delivery rather than other effects on the efficiency of DNA break repair. Interestingly, CsH seemed to increase targeting efficiencies in particular in the more primitive HSC, previously shown to be more sensitive than committed progenitors to the cytotoxicity of the gene targeting procedure and less proficient at performing HDR (Genovese et al., 2014).
Our work reveals that CsH overcomes IFITM3-mediated antiviral restriction in HSPC at steady-state. Concordantly, stem cells, including HSPC, have recently been shown to be naturally resistant to RNA viral infections due to intrinsic expression of subsets of antiviral ISGs including IFITM proteins (Wu et al., 2018). This further supports the idea that constitutively expressed innate immune factors contribute to poor sensitivity of HSPC to gene transfer and editing. The antiviral role of IFITM proteins has been extensively described for a wide range of viruses (Lu et al., 2011, Perreira et al., 2013, Qian et al., 2015), although the mechanisms of action remain unclear. IFITM sensitivity of HIV-1 strains is determined by the co-receptor usage of the viral envelope glycoproteins as well as IFITM subcellular localization within the target cell (Foster et al., 2016). In LV designed for gene therapy the HIV-1 envelope glycoprotein has been replaced with the VSV-G envelope glycoprotein. In agreement with our observations, IFITM proteins have been recently shown to limit VSV-G pseudotyped lentivirus-based vector gene transfer to airway epithelia (Hornick et al., 2016). Interestingly, pseudotyping LV with the BaEV-TR glycoprotein has been suggested to significantly improve transduction efficiencies in human CD34+ cells compared to VSV-G pseudotyped vectors, including in unstimulated HSPC (Girard-Gagnepain et al., 2014), which is potentially explained by their capacity to bypass IFITM3 restriction. Rapamycin and PGE2 are also thought to enhance transduction at the level of VSV-G-mediated viral entry, (Wang et al., 2014, Zonari et al., 2017), therefore the involvement of IFITM3 in their effect is worth investigating. Consistent with previous reports suggesting that IFITM3 acts at the level of endosomal membrane fusion (Chesarino et al., 2014, Foster et al., 2016, Jia et al., 2012), AAV6-mediated donor DNA delivery did not benefit from CsH. This vector does not have an envelope and therefore does not fuse with endosomal membranes to enter the cytoplasm (Nonnenmacher and Weber, 2012). Interestingly, CsH seems to abrogate IFITM3 by inducing its degradation in HSPC and THP-1 cells but fails to impact the non-restrictive Y20F mutant, potentially due to its misplaced cellular localization mainly at the plasma membrane or because Y20 phosphorylation has been suggested to prevent its ubiquitination by NEDD4 E3 ligase (Chesarino et al., 2014, Chesarino et al., 2015). Effects of CsH on IFITM3 were transient as protein levels were restored within 6 hr from CsH removal, likely explaining the requirement for the drug during LV exposure for maximal effects.
Importantly, we observed a significant negative correlation between IFITM3 protein levels and HSPC permissivity to LV transduction between donors. The least permissive donors expressed the highest IFITM3 levels and benefitted most from CsH enhancement of transduction. These data suggest that CsH will also abrogate donor variability during ex vivo gene therapy, a major benefit for the design of better controlled clinical trials and, eventually, standardized medicinal products based on modification of HSC. Indeed, donor variability remains a significant issue for the field as underscored also by the high variability in gene marking recently observed for β-Thalassemia gene therapy patients (Thompson et al., 2018). Variations in expression levels of the antiretroviral host factor TRIM5α have also been suggested to correlate with human CD34+ cell permissiveness to lentiviral transduction (Evans et al., 2014) although human TRIM5α inhibition of HIV-1 is demonstrably weak (Keckesova et al., 2006). It is possible that, differences in TRIM5α levels, and other ISGs, reflect variable IFITM3 levels rather than direct TRIM5α effects on transduction.
Overall, we have described how manipulating a single host factor can have a dramatic impact on gene transfer efficiencies in HSPC. Our findings are an important step toward the development of maximally effective HSPC gene engineering protocols and uncover hitherto unknown molecular mechanisms of innate immune defense in human hematopoietic stem cells. We expect exploitation in innovative cell and gene therapies and in broadly effective antiviral strategies.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human FCR Blocking | Miltenyi Biotec | Cat# 130-059-901 |
| Purified Rat Anti-Mouse CD16/CD32 | BD Biosciences | Cat# 553141; RRID:AB_394656 |
| Anti-human CD45 APC-eFluor 780 | eBioscience | Cat# 47-0459-42; RRID:AB_1944368; clone HI30 |
| Anti-human CD19 PE | BD Biosciences | Cat# 345789; clone SJ25C1 |
| Anti-human CD33 BV | BD Biosciences | Cat# 562854; RRID:AB_2737405; clone WM53 |
| Anti-human CD3 APC | BD Biosciences | Cat# 555335; RRID:AB_398591; clone UCHT1 |
| Anti-human CD13 BV | BD Biosciences | Cat# 562596; RRID:AB_2737672; clone WM15 |
| Anti-human CD34 PeCy7 | BD Biosciences | Cat# 348811; clone 8G12 |
| Anti-human CD34 VB | Miltenyi Biotec | Cat# 130-095-393; RRID:AB_10827793 |
| Anti human CD38 perCP/Cy5.5 | BioLegend | Cat# 356614; RRID:AB_2562183; clone HB-7 |
| Anti-human CD38 PeVio770 | Miltenyi Biotec | Cat# 130-099-151; RRID:AB_2660384 |
| Anti-human CD90 APC | BD Biosciences | Cat# 559869; RRID:AB_398677; clone 5E10 |
| Anti-human CD133/2 PE | Miltenyi Biotec | Cat# 130-090-853; RRID:AB_244346; clone 293C3 |
| Anti-human CD45RA PE | Miltenyi Biotec | Cat# 130-092-248; RRID:AB_615094 |
| Anti-human CD45RA FITC | BD Biosciences | Cat# 335039; clone HI100 |
| Mouse IgG1 isotype control | BD PharMingen | Cat# 51-35405X; clone MOPC-21 |
| Mouse polyclonal anti-CypA | Santa-Cruz Biotechnology | Cat# sc-134310; RRID:AB_2169131 |
| Mouse monoclonal anti-CypB | Santa-Cruz Biotechnology | Cat# sc-130626; RRID:AB_2169421 |
| Rabbit polyclonal anti-IFITM3 | Proteintech | Cat# 11714-1-AP; RRID:AB_2295684 |
| Rabbit polyclonal anti-IFITM2 | Proteintech | Cat# 12769-1-AP; RRID:AB_2122089 |
| Mouse monoclonal anti-IFITM1 | Proteintech | Cat# 60074-1-Ig; RRID:AB_2233405 |
| Mouse monoclonal anti-β-Actin | Sigma-Aldrich | Cat# A2228; RRID:AB_476697; clone AC-74 |
| Mouse monoclonal anti-LAMP1 | Abcam | Cat# ab25630; RRID:AB_470708 |
| Rabbit IgG HRP Linked Whole Ab | GE Healthcare | Cat# NA934; RRID:AB_772206 |
| Mouse IgG, HRP-Linked Whole Ab | GE Healthcare | Cat# NA931; RRID:AB_772210 |
| Donkey anti-Rabbit IgG, AlexaFluor 488 | Thermo Fisher Scientific | Cat# A-21206; RRID:AB_141708 |
| Donkey anti-Rabbit IgG, AlexaFluor 555 | Thermo Fisher Scientific | Cat# A-31572; RRID:AB_162543 |
| Donkey anti-Rabbit IgG, AlexaFluor 647 | Thermo Fisher Scientific | Cat# A-31573; RRID:AB_2536183 |
| Biological Samples | ||
| Umbilical cord blood | Ospedale San Raffaele (TIGET01/09) | N/A |
| Peripheral blood | Ospedale San Raffaele (TIGET01/09) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cyclosporine A | Sigma-Aldrich | Cat# C1832-5MG |
| Cyclosporine H | Sigma-Aldrich | Cat# SML1575-5MG |
| Rapamycin | Sigma-Aldrich | Cat# R8781 |
| FK-506 | Sigma-Aldrich | Cat# 109581-93-3 |
| Cyclohexamide | Sigma-Aldrich | Cat# 66-81-9 |
| Prostaglandin E2 | Cayman | Cat# 14750 |
| Human IFNα | pbl assay science | Cat# 11105-1 |
| Puromycin | Sigma-Aldrich | Cat# P8833-25MG |
| DAPI (4′,6-diamidino-2-phenylindole) | Roche | Cat# 10236276001 |
| Hoechst 33342 | Invitrogen | Cat# H3570 |
| StemSpan SFEM | VODEN | Cat# 09650 |
| GMP Serum-free Stem Cell Growth | Cellgenix | Cat# 20802-0500 |
| Recombinant human stem cell factor | Peprotech | Cat# 300-07 |
| Recombinant human thrombopoietin | Peprotech | Cat# 300-18 |
| Recombinant human Flt3 ligand | Peprotech | Cat# 300-19 |
| Recombinant human IL6 | Peprotech | Cat# 200-06 |
| Recombinant human IL3 | Peprotech | Cat# 200-03 |
| StemRegenin 1 (SR1) | BioVision | Cat# 1967 |
| UM171 | STEMCell Technologies | Cat# 72912 |
| 3TC | Sigma-Aldrich | Cat# 134678-17-4 |
| RetroNectin (r-Fibronectin CH-296) | Takara Bio | Cat# T100A |
| Bolt 4%–12% Bis-Tris Plus Gels | Thermo Fisher Scientific | Cat# NW04120BOX |
| 20X Bolt MES SDS Running Buffer | Invitrogen | Cat# B0002 |
| 10X Bolt Sample Reducing Agent | Thermo Fisher Scientific | Cat# B0009 |
| 4X Bolt LDS Sample Buffer | Thermo Fisher Scientific | Cat# B0007 |
| CD34 MicroBead Kit, human | Miltenyi Biotec | Cat# 130-046-702 |
| Lineage cell depletion kit | Miltenyi Biotec | Cat# 130-090-858 |
| CytofixFixation Buffer | BD Biosciences | Cat# 554655 |
| Permeabilizing Solution 2 | BD Biosciences | Cat# 347692 |
| CD4+ T Cell Isolation Kit, human | Miltenyi Biotec | Cat# 130-096-533 |
| Pan Monocyte Isolation Kit, human | Miltenyi Biotec | Cat# 130-096-537 |
| Critical Commercial Assays | ||
| Annexin V Apoptosis Detection Kit I | BioLegend | Cat# 640918; RRID:AB_1279044 |
| Cell Proliferation Dye eFluor® 670 | Thermo Fisher Scientific | Cat# 65-0840-85 |
| PE Mouse Anti-Ki-67 Set | BD Biosciences | Cat# 556027; RRID:AB_2266296 |
| ReliaPrep RNA Cell Miniprep System | Promega | Cat# Z6011 |
| QIAamp DNA Micro Kit | QIAGEN | Cat# 56304 |
| RNAeasy micro | QIAGEN | Cat# 74004 |
| RNAeasy Plus micro | QIAGEN | Cat# 74034 |
| SuperScript Vilo kit | Invitrogen | Cat# 11754250 |
| P3 Primary Cell 4D-Nucleofector X Kit | Lonza | Cat# V4XP-3032 |
| T7 Endonuclease I | New England Biolabs | Cat# M0302L |
| LV GFP gene (Mr03989638_mr) | Thermo Fisher Scientific | Cat# 4331182 |
| Human PPIA gene (Hs99999904_m1) | Thermo Fisher Scientific | Cat# 4331182 |
| Human PPIB gene (Hs00168719_m1) | Thermo Fisher Scientific | Cat# 4331182 |
| Human IRF7 gene (Hs01014809_g1) | Thermo Fisher Scientific | Cat# 4331182 |
| Human ISG15 gene (Hs01921425_s1) | Thermo Fisher Scientific | Cat# 4331182 |
| Human IFITM1 gene (Hs00705137_s1) | Thermo Fisher Scientific | Cat# 4331182 |
| Human IFITM2 gene (Hs00829485_sH) | Thermo Fisher Scientific | Cat# 4331182 |
| Human IFITM3 gene (Hs03057129_s1) | Thermo Fisher Scientific | Cat# 4331182 |
| Human HPRT1 gene (Hs01003267_m1) | Thermo Fisher Scientific | Cat# 4331182 |
| Experimental Models: Cell Lines | ||
| 293T cells (HEK293T) | ATCC | Cat# CRL-11268; RRID:CVCL_1926 |
| THP-1 | ATCC | Cat# TIB-202; RRID:CVCL_0006 |
| CB-CD34 | Lonza | Cat# 2C-101 |
| BM-CD34 | Lonza | Cat# 2M-101C |
| mPB-CD34 | All Cells | Cat# mPB015F/mPB016F |
| Experimental Models: Organisms/Strains | ||
| NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJArc Mus musculus | Jackson laboratory (IACUC 784 and 749) | Cat# ARC:NSG, RRID:IMSR_ARC:NSG |
| Ifitm3−/− mice | Wellcome Trust Sanger Institute; Lange et al., 2008 | N/A |
| Oligonucleotides | ||
| See Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pCCLsin.cPPT.hPGK.eGFP.Wpre transfer | Petrillo et al., 2015 | N/A |
| pMDLg/Prre packaging | Petrillo et al., 2015 | N/A |
| pCMV-Rev | Petrillo et al., 2015 | N/A |
| pMD2.VSV-G | Petrillo et al., 2015 | N/A |
| pMD.Lg/pRRE.D64VInt | Petrillo et al., 2015 | N/A |
| pMDLg/pRRE-N74D | GenScript Inc | N/A |
| pMDLg/pRRE-P90A | GenScript Inc | N/A |
| BaEV-TR | Girard-Gagnepain et al., 2014 | N/A |
| RVrkat43.2MLV GFP | Montini et al., 2006 | N/A |
| pCM-gag-pol | Montini et al., 2006 | N/A |
| AmphoRV | Ragheb et al., 1995 | N/A |
| SIVmac-GFP | Mangeot et al., 2002 | N/A |
| SIV3+ | Mangeot et al., 2002 | N/A |
| pSVRΔNBDM | Griffin et al., 2001 | N/A |
| pSVRΔNBGFPΔH | Griffin et al., 2001 | N/A |
| pONY3 | Mitrophanous et al., 1999 | N/A |
| pONY8 | Mitrophanous et al., 1999 | N/A |
| FP93 | Poeschla et al., 1998 | N/A |
| pGINSIN | Poeschla et al., 1998 | N/A |
| plko shRNA CypA | Open Biosystems | TRCN0000049232 |
| plko shRNA IFITM3 | Sigma-Aldrich | TCRN0000118022 |
| Software and Algorithms | ||
| FACSDIVA software | BD Biosciences | N/A |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| FCS Express Flow | De Novo Software | https://www.denovosoftware.com/site/DownloadResearch.shtml |
| Vector NTI | Invitrogen | N/A |
| Image Lab | Biorad | N/A |
| QuantaSoft | Biorad | N/A |
| QuantStudio Real-Time PCR software | Applied Biosystems | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact, Anna Kajaste-Rudnitski (kajaste.anna@hsr.it).
Experimental Model and Subject Details
Mice
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG, RRID:IMSR_ARC:NSG) Mus musculus were purchased from Jackson laboratory. All animal procedures were performed according to protocols approved by the Animal Care and Use Committee of the Ospedale San Raffaele (IACUC 784 and 749) and communicated to the Ministry of Health and local authorities according to the Italian law. Female 8-10 week old mice were used in all studies to allow better engraftment of human HSPC cells upon tail vein transplantation (Notta et al., 2010). Mice were observed carefully by laboratory staff and veterinarian personnel for health and activity. Animals were monitored to ensure that food and fluid intake meets their nutritional needs. Mice were housed in cages with microisolator tops on ventilated or static racks in a specific pathogen-free facility. All caging materials and bedding are autoclaved. All mice were randomized into different HSC transplantation groups. On the basis of a standard backward sample size calculation, we transplanted at least three to ten mice per condition and performed at least two independent experiments to obtain a sufficient number of mice to perform statistical analysis. Human cell engraftment was blindly assessed by serial bleeding or bone marrow as well as spleen analysis at sacrifice. At the end of the experiments mice were euthanized by CO2 and organs were harvested and analyzed as reported in the Method Details section.
Cells
Primary cells were isolated from umbilical cord blood or from mononuclear cells collected upon informed consent from healthy volunteers according to the Institutional Ethical Committee approved protocol (TIGET01/09). Otherwise, cord blood (CB), bone marrow (BM) or G-CSF mobilized peripheral blood (mPB) CD34+ cells were directly purchased from Lonza or Hemacare. For the majority of the experiments HSPC from male and female donors were pooled together to have sufficient number of cells for each experiment. Regarding the correlation study between LV transduction and IFITM3 protein levels we collected HSPC from the cord blood of single donors without indications regarding the sex.
Total bone marrow from n = 7 background-matched 8-10 week old wild-type and Ifitm3−/− mice (Wellcome Trust Sanger Institute) (Lange et al., 2008) were collected and murine Lin− cells were isolated as described in Method Details.
Method Details
Vectors
Third generation LV stocks were prepared, concentrated and titered as previously described (Dull et al., 1998, Follenzi et al., 2000). Briefly, self-inactivating (SIN) LV vectors were produced using the transfer vector pCCLsin.cPPT.hPGK.eGFP.Wpre, the packaging plasmid pMDLg/pRRE, Rev-expressing pCMV-Rev and the VSV-G envelop-encoding pMD2.VSV-G plasmids. IDLV was produced as previously described (Lombardo et al., 2007) substituting the packaging plasmid pMDLg/pRRE with pMD.Lg/pRRE.D64VInt. For SINLV capsid mutants, vectors were produced as described above, except that the wild-type pMDLg/pRRE was replaced with a packaging plasmid harboring a specific point-mutation in the p24 coding region as follows: pMDLg/pRRE-N74D; pMDLg/pRRE-P90A. All modified packaging plasmids were purchased from GenScript Inc. For pseudotyping LVs with the mutant baboon retrovirus envelope, pMD2.VSV-G was replaced by the BaEV-TR during vector production as previously described (Girard-Gagnepain et al., 2014). Vpx-containing lentiviral vector stocks were produced as previously described (Bobadilla et al., 2013). The SIN-RV was produced as previously described (Montini et al., 2006) using as transfer vector RVrkat43.2MLV GFP, the packaging plasmid pCM-gag-pol and the VSV-G envelop-encoding pMD2.VSV-G plasmid or pseudotyped with the amphotropic envelope glycoprotein (AmphoRV). Simian immunodeficiency virus macaque- (SIVmac) based vectors were produced as previously described (Mangeot et al., 2002) using an GFP encoding genome SIVmac-GFP, SIVmac packaging plasmid SIV3+ kindly provided by Francois Loic Cosset and VSV-G pseudotyped. HIV-2 was prepared using HIV-2 packaging construct pSVRΔNBDM (also known as HIV-2 pack) and HIV-2 genome encoding GFP pSVRΔNBGFPΔH (also known as HIV-2 GFP) kindly provided by Andrew Lever (Griffin et al., 2001). SIV sooty mangabey (SIVsm) GFP encoding vector was made using an SIVmac delta Env construct encoding SIVsm Gag-Pol and with GFP in place of nef kindly provided by Welkin Johnson. Equine Infectious Anaemia Virus (EIAV) GFP was prepared using EIAV packaging construct pONY3 and GFP encoding EIAV genome pONY8 kindly provided by Kyriacos Mitrophanous (Mitrophanous et al., 1999). Feline immunodeficiency virus (FIF GFP) was made using FIVpetaluma packaging construct FP93 and FIV GFP encoding genome pGINSIN kindly provided by Eric Poeschla (Poeschla et al., 1998). Moloney MLV GFP was produced using MLV packaging construct CMVintron and MLV genome encoding GFP CNCG kindly provided by Francois Loic Cosset. AAV6 donor templates for homology-directed repair (HDR) were generated from a construct containing AAV2 inverted terminal repeats, produced by triple-transfection method and purified by ultracentrifugation on a cesium chloride gradient as previously described (Wang et al., 2015). Clinical-grade arylsulfatase A (ARSA) and α-l-iduronidase (IDUA) LVs were produced by MolMed (Milan, Italy) using a large scale validated process as previously reported (Biffi et al., 2013). The bidirectional LVs (Amendola et al., 2013) were used to overexpress the coding sequence (CDS) of candidate human genes under the control of the human phosphoglycerate kinase (PGK) promoter and the eGFP and the minimal cytomegalovirus (mCMV) promoter forming the antisense expression unit. Knock-down (KD) experiments were performed using plko or vectors encoding shRNA against human CypA (from Open Biosystems, TRCN0000049232), IFITM3 (from Sigma, TCRN0000118022) or non-silencing (ns) as controls, or pcSIREN vectors expressing shRNA against CNA, all under the human U6 promoter. The plko ns was designed to express an unrelated shRNA with no predicted target as control. Knock-out (KO) experiments were performed using LVs co-expressing a mammalian codon-optimized Cas9 nuclease along with a single guide RNA (sgRNA) (Shalem et al., 2014) against the gene of interest (sgRNA IFITM3: GGGGGCTGGCCACTGTTGACAGG). As control LV KO-empty with no sgRNA was used.
Cells
Cell lines
The human embryonic kidney 293T cells (HEK293T, RRID:CVCL_1926) were maintained in Iscove’s modified Dulbecco’s medium (IMDM; Sigma), instead human THP-1 cells (RRID:CVCL_0006) were maintained in Roswell Park Memorial Institute medium (RPMI; Lonza). Both medium were supplemented with 10% fetal bovine serum (FBS; GIBCO), penicillin (100 IU/ml), streptomycin (100 μg/ml) and 2% glutamine.
Primary cells
Human CD34+ HSPC, CD4+ and CD14+ monocytes were isolated through positive magnetic bead selection according to manufacturer’s instructions (Miltenyi) from umbilical cord blood or from mononuclear cells collected upon informed consent from healthy volunteers according to the Institutional Ethical Committee approved protocol (TIGET01/09). Otherwise, cord blood (CB), bone marrow (BM) or G-CSF mobilized peripheral blood (mPB) CD34+ cells were directly purchased from Lonza or Hemacare. CD4+ T cells were activated in RPMI, supplemented with 10% FBS, penicillin (100 IU/ml), streptomycin (100 μg/ml), 2% glutamine, phytohaemagglutanin (PHA) (2 μg/ml, Roche) and IL-2 (300IU/mL, Novartis) for three days and maintained in RPMI, supplemented with 10% FBS, penicillin (100 IU/ml), streptomycin (100 μg/ml), 2% glutamine and IL-2 (300 UI/ml). Monocyte-derived macrophages (MDM) were differentiated from isolated CD14+ monocytes in DMEM supplemented with 10% FBS, penicillin (100 IU/ml), streptomycin (100 μg/ml), 2% glutamine and 5% human serum AB (Lonza) for seven days. Primary T lymphocytes from healthy donors’ PB mononuclear cells were isolated and activated using magnetic beads conjugated to anti-human CD3 and CD28 antibodies (Dynabeads human T-activator CD3/CD28; Invitrogen) in IMDM medium (GIBCO-BRL) supplemented with penicillin, streptomycin, glutamine, 10% FBS, and 5 ng/ml of IL-7 and IL-15 (PeproTech) for 2 days as described (Provasi et al., 2012). BM cells were retrieved from femurs, tibias, and homer of 8-10 week old wild-type and Ifitm3−/− mice (Wellcome Trust Sanger Institute, (Lange et al., 2008). HSPCs were purified by Lin- selection using the mouse Lineage Cell Depletion Kit (Miltenyi Biotec) according to the manufacturer’s instructions.
All cells were maintained in a 5% CO2 humidified atmosphere at 37°C.
Compounds
Cyclosporine A, Cyclosporine H, Rapamycin, FK-506 and cyclohexamide (CHX) (all from Sigma-Aldrich) were resuspended and stored following the manufacturer’ s instructions. They were added to the transduction medium at the indicated concentration (8 μM for Cyclosporines and 10 μg/ml for Rapa) and washed out with the vector 16-20 hours later. For the FK-506 experiment, THP1 cells were pre-treated with IFNb (1000U) for 24 h and FK-506 (0.5-1000nM) was added at the time of transduction and left in the media until cells were harvested 48 h post transduction. CHX (1 μg/ml) was added with IFNb treatment (1000U) 24 h prior to transduction in THP-1 cells. CHX (10 μg/ml) was added to the medium one hour before CsH wash and re-added during LV transduction for 6 hours in human HSPC. Where described, Prostaglandin E2 (Dinoprostone from Yonsung) was added at the final concentration of 10 μM two hours before LV transduction. Human IFNα pre-stimulation was performed for 24 hours together with human cytokines cocktail at the indicated concentration.
Transduction
If not otherwise specified, all vectors used in this work are vesicular stomatitis virus glycoprotein (VSV-g)-pseudotyped. All cells were transduced at the indicated multiplicity of infection (MOI) as calculated by titration of vector stocks on 293T cells and expressed as transducing units (TU)/293T cell. For transduction, human CB-derived hematopoietic stem/progenitor cells (HSPC) were cultured in serum-free StemSpan medium (StemCell Technologies) supplemented with penicillin (100 IU/ml), streptomycin (100 μg/ml), 100 ng/ml recombinant human stem cell factor (rhSCF), 20 ng/ml recombinant human thrombopoietin (rhTPO), 100 ng/ml recombinant human Flt3 ligand (rhFlt3), and 20 ng/ml recombinant human IL6 (rhIL6) (all from Peprotech) 16 to 24 hours prior to transduction. HSPC were then transduced at a concentration of 1 × 106 cells per milliliter with a given vector for 16 hours at the indicated multiplicity of infection (MOI), expressed as transducing units (TU)/293T cell, in the same medium. For CHX experiments in HSPC, after 24 hours of prestimulation cells were exposed or not to CsH for 16 hours and then transduced for 6 hours in different conditions: vector alone called “Reference protocol”; exposed to the vector after cell wash in absence “Wash protocol” or in presence “CHX protocol” of CHX. Bone marrow and G-CSF mobilized peripheral blood CD34+ cells were placed in culture on retronectin-coated non tissue culture-treated wells (T100A Takara) in CellGro medium (Cell Genix) containing a cocktail of cytokines: 60 ng/ml IL-3, 100 ng/ml TPO, 300 ng/ml SCF, and 300 ng/ml FLT-3L (all from Cell Peprotech). Cells were then transduced with the indicated dose of vectors for 14-15 hours in the same cytokine-containing medium. After transduction with a single hit reporter LV cells were washed and maintained in serum-free medium supplemented with cytokines as above until the reading of the percentage of positive cells by FACS, after which they were maintained in IMDM supplemented with 10% FBS, 25 ng/ml rhSCF, 5 ng/ml rhIL6-3, 25 ng/ml rhFlt3 and 5 ng/ml rhTPO for another seven days before analysis of vector copy numbers. In the protocol that foresees two rounds of transduction, selected for clinical application, cells were washed for 10 hours in CellGro SCGM medium supplemented with cytokines and underwent a second hit of transduction in the same conditions as the first, as reported previously (Biffi et al., 2013). At the end of transduction, cells were counted and collected for clonogenic assays, flow cytometry, and in vivo studies. Remaining cells were plated in IMDM 10% fetal bovine serum (FBS) with cytokines (IL-3, 60 ng/ml; IL-6, 60 ng/ml; SCF, 300 ng/ml) and cultured for a total of 14 days. Thereafter, cells were collected for molecular and biochemical studies. Unstimulated HSPC were transduced freshly isolated in StemSpan medium supplemented with penicillin (100 IU/ml), streptomycin (100 μg/ml) for 16-24hours and then maintained in presence of human cytokines and 10μM of the reverse-transcriptase inhibitor 3TC (from SIGMA) to avoid subsequent transduction due to cytokines stimulation.
Murine Lin- HSPC were cultured in serum-free StemSpan medium (StemCell Technologies) containing penicillin, streptomycin, glutamine and a combination of mouse cytokines (20 ng/ml IL-3, 100 ng/ml SCF, 100 ng/ml Flt-3L, 50 ng/ml TPO all from Peprotech), at a concentration of 106 cells/ml. After three hours of pre-stimulation, cells were transduced with an LV for 16-20 hours in the same medium. Cells were then washed and maintained in serum-free medium supplemented with cytokines as above until the reading of the percentage of positive cells by FACS, after which they were diluted in IMDM supplemented with 10% FBS.
MDM were transduced 7 days after differentiation. T lymphocytes were transduced at a concentration of 106 cells/ml, after 2-3 days of stimulation. The cells were exposed to the vector for 16-20 hours.
Gene editing of human cells
LV donor templates for HDR were generated using HIV-derived, third-generation self-inactivating transfer constructs. IDLV stocks were prepared and titered as previously described (Lombardo et al., 2007). AAV6 donor templates for HDR were generated from a construct containing AAV2 inverted terminal repeats, produced by triple-transfection method and purified by ultracentrifugation on a cesium chloride gradient as previously described (Wang et al., 2015). For gene editing experiments in human HSPC, 106 CD34+ cells/ml were stimulated in serum-free StemSpan medium (StemCell Technologies) supplemented with penicillin, streptomycin, glutamine, 1 μM SR-1 (Biovision), 50 μM UM171 (STEMCell Technologies), 10 μM PGE2 added only at the beginning of the culture (Cayman), and human early-acting cytokines (SCF 100 ng/ml, Flt3-L 100 ng/ml, TPO 20 ng/ml, and IL-6 20 ng/ml; all purchased from Peprotech) (Schiroli et al., 2017). Transduction with IDLV was performed at MOI 100, in presence or not of CsH, after 2 days of prestimulation. CD34+ cells were transduced with AAV6 at 10ˆ4 vg/cell 15’ after electroporation. IDLV or AAV6 donor templates with homologies for AAVS1 locus (encoding for a PGK.GFP reporter cassette; (Genovese et al., 2014) or targeting the intron 1 of IL2RG (encoding for IL2RG corrective cDNA; (Schiroli et al., 2017) were utilized as indicated. After 24 hours from IDLV transduction, cells were washed with PBS and electroporated (P3 Primary Cell 4D-Nucleofector X Kit, program EO-100; Lonza) with 1.25 μM of ribonucleoproteins (RNP). RNPs were assembled by incubating at 1:1.5 molar ratio s.p.Cas9 protein (Integrated DNA Technologies) with synthetic cr:tracrRNA (Integrated DNA Technologies) for 10’ at 25°C. Electroporation enhancer (Integrated DNA Technologies) was added prior to electroporation according to manufacturer’s instructions. Genomic sequences recognized by the gRNAs are the following: TCACCAATCCTGTCCCTAGtgg for AAVS1 locus and ACTGGCCATTACAATCATGTggg for intron 1 IL2RG. Gene editing efficiency was measured from cultured cells in vitro 3 days after electroporation. For AAVS1 edited cells, editing by homology-directed repair (HDR) was quantified by flow cytometry measuring the percentage of cells expressing the GFP marker. For IL2RG edited cells, HDR was quantified by digital droplet PCR analysis designing primers and probe on the junction between the vector sequence and the targeted locus and on control sequences utilized as normalizer (human TTC5 genes) as previously described (Schiroli et al., 2017). NHEJ was was measured by mismatch-sensitive endonuclease assay by PCR-based amplification of the targeted locus followed by digestion with T7 Endonuclease I (NEB) according to the manufacturer’s instructions.
Transplantation of human HSPC in NSG mice
Human mPB-CD34+ cells were pre-stimulated and transduced as described before with IDUA-LV at an MOI of 50 in presence or not of DMSO/CsH. After transduction 2-5x105 cells were infused into the tail vein of sublethally irradiated 8-10 week-old NSG mice (radiation dose: 200 cGy for mice weighting 18-25 g and of 220 cGy for mice above 25 g of weight). Transduced and untransduced cells were also cultured in vitro for 14 days for further analysis. In vitro cultured cells, BM and spleen cells isolated from transplanted mice at time of sacrifice were then used to quantify the VCN by qPCR. For secondary transplantation experiments 8 × 104 stimulated human CB-derived CD34+ cells were injected into the tail vein of primary NSG mice after an over-night transduction with a PGK-GFP Molmed purified vector (DSP05). Peripheral blood was sampled at indicated times post-transplant and analyzed. At sacrifice, the cells from the spleen and BM isolated from the primary recipients were analyzed at flow cytometry and the CD34+ cells were purified from the BM through positive magnetic bead selection on LD and MS columns (Miltenyi) according to the manufacturer’s instructions. Purity was verified by FACS prior to pooling by condition and injection into secondary recipients. Between 9 × 105 and 1 × 106 CD34+ cells isolated from the primary hosts were injected into the tail vein of sublethally irradiated secondary NSG mice (8–10 weeks old). Peripheral blood was sampled at indicated times post-transplant. 13 weeks post-transplantation, all mice were sacrificed by CO2 to analyze the BM and the spleen of secondary mice as described above (Piras et al., 2017). For transplantation of gene edited cells, 3x105 CD34+ cells treated for editing at day 5 of culture were injected intravenously into NSG mice after sub-lethal irradiation (180-200 cGy). Human CD45+ cell engraftment and the presence of gene-edited cells were monitored by serial collection of blood from the mouse tail and, at the end of the experiment (19 weeks after transplantation), BM was harvested and analyzed.
Colony-forming cell assay
Colony-forming cell assays were performed by plating 8x102 human HSPC transduced in presence of the different compounds in a methylcellulose-based medium (Methocult GF4434; Stem Cell Technologies). Fifteen days later colonies were scored by light microscopy for colony numbers and morphology as erythroid or myeloid. Moreover, they were collected as a pool and as a single colony, and lysed for molecular analysis to evaluate transduction efficiencies with clinical grade LV.
Flow cytometry and cell sorting
All cytometric analyses were performed using the FACS Canto III instrument and LSRFortessa instruments (BD Biosciences) and analyzed with the FACS Express software (De Novo Software). Sorting procedures were performed on a MoFlo XDP sorter (Beckman Coulter).
FACS sorting
Fresh human cord blood-derived CD34+ cells were FACS-sorted according to CD133 and CD38 expression as shown in Figure 6C. The sorted subpopulations were exposed or not to CsH for 16 hours and then analyzed by IF for IFITM3 protein expression.
Transduced cells
GFP or BFP expression in transduced cells was measured 5-7 days post-transduction and expressed as transduction level (%). Adherent MDM were detached by scraping in 5mM PBS-EDTA, washed and resuspended in PBS containing 2% fetal bovine serum (FBS). Cells grown in suspension were washed and resuspended in PBS containing 2% FBS. To measure HSPC subpopulation composition cells were harvested 16 or 72 hours post-TD, incubated with anti-human receptor blocking antibodies for 15 min at 4°C and then stained for 20 min at 4°C with anti-human CD34 (RRID:AB_10827793), CD38 (RRID:AB_2562183), CD45RA (RRID:AB_615094), CD90 (RRID:AB_398677) or with anti-human CD34, CD133 (RRID:AB_244346), CD90 antibodies (for antibodies see Key Resources Table). To exclude dead cells from the analysis, 10 ng/ml 7-aminoactinomycin D (7-AAD) was added.
Peripheral blood from mice
For each mouse, 250 μL of peripheral blood were added to 15 μL of PBS containing 45 mg/mL EDTA. For immunostaining a known volume of whole blood (100 μl) was first incubated with anti-human FcR Blocking Reagent and anti-mouse FcγIII/II receptor (Cd16/Cd32) blocking antibodies for 15 min at 4°C and then incubated in the presence of anti-human CD45 (RRID:AB_1944368), CD19, CD13, CD3 (RRID:AB_398591) (for antibodies see Key Resources Table) for 20 min at 4°C. Erythrocytes were removed by lyses with the TQ-Prep workstation (Beckman-Coulter) in the presence of an equal volume of FBS (100 μl) to protect white blood cells.
Bone marrow
BM cells were obtained by flushing the femurs in PBS 2% FBS solution. Cells (1x106 cells) were washed, resuspended in 100 μL of PBS containing 2% FBS, and incubated with anti-human receptor (Cd16/Cd32) blocking antibodies for 15 min at 4°C. Staining was then performed with anti-human CD45, CD19, CD33 (for antibodies see Key Resources Table) for 20 min at 4°C. Cells were washed and finally resuspended in PBS containing 2% FBS.
Spleen
Spleens were first smashed and the resulting cell suspension was passed through 40 μm nylon filter and washed in cold phosphate buffered saline (PBS) containing 2mM EDTA and 0.5% bovine serum albumine (BSA). Cells were incubated with anti-human receptor (Cd16/Cd32) blocking antibodies for 15 min at 4°C and then stained with anti-human CD45, CD19, CD13, CD3 (for antibodies see Key Resources Table) for 20 min at 4°C. Cells were finally washed and resuspended in PBS containing 2% FBS.
Ki67-Hoechst and Annexin V flow cytometry
Cells were washed and fixed using BD Cytofix buffer (Cat. #554655), washed and permeabilized with BD Perm 2 (Cat. # 347692), washed and stained with PE-conjugated Ki67 antibody (BD, RRID:AB_2266296) and finally resuspended in BD Cytofix buffer with Hoechst at 1 μg/mL. The cells were then analyzed on a BD LSRII machine with UV laser. The apoptosis assays were performed with the Annexin V Apoptosis Detection Kit I (BD Pharmigen, RRID:AB_1279044) according to the manufacturer’s instructions and 48 hours after transduction.
Cell proliferation assay
Cells were stained with Cell Proliferation Dye eFluor® 670 (Affimetrix, eBioscience) after 24 hours of cytokines pre-stimulation and before cell transduction. This fluorescent dye binds to any cellular protein containing primary amines, and as cells divide, the dye is distributed equally between daughter cells that can be measured as successive halving of the fluorescence intensity of the dye. At different time points after TD, cells were harvested and analyzed at flow cytometry. Cell Proliferation Dye eFluor® 670 has a peak excitation of 647 nm and can be excited by the red (633 nm) laser line. It has a peak emission of 670 nm and can be detected with a 660/20 band pass filter (equivanet to APC, Alexa Fluor® 647, or eFluor® 660).
RNA extraction, qPCR and gene expression analysis
RNA extraction from cells was performed using the RNeasy micro Kit or RNeasy Plus micro Kit (QIAGEN) or ReliaPrep RNA Cell Miniprep System (Promega). Briefly, cells were lysed in Buffer RLT plus, supplemented with beta-mercaptoethanol. RNA was then extracted according to manufacturer’s instructions. The extracted mRNAs were reverse transcribed (RT) using the SuperScript Vilo kit (11754250; Invitrogen). RT-qPCR analyses were performed using TaqMan probes from Applied Biosystems to detect endogenous mRNA levels. Otherwise, we designed specific primers to quantify the overexpression of the coding sequence of human IFITM2 and IFITM3 genes (see below). Q-PCR was run for 40 cycles using the Viia 7 instrument while the Viia 7 software was then used to extract the raw data (Ct). To determine gene expression, the difference (ΔCt) between the threshold cycle (Ct) of each gene and that of the reference gene was calculated by applying an equal threshold. Relative quantification values were calculated as the fold-change expression of the gene of interest over its expression in the reference sample, by the formula 2–ΔΔCt. The expression was normalized using the housekeeping gene HPRT1. Human Taqman probes from Applied Biosystems were used and reported in Key Resources Table.
Replication intermediates
CB-derived CD34+ cells were transduced at an MOI of 100, in presence or absence of CsA or CsH. To analyze viral replication intermediates, transduced cells were washed and resuspended in Monini lysis buffer (0.1% polyoxyethylene 10 lauryl ether (Sigma), 0.1 mg/mL proteinase K (Promega) (25 μl/ 1X105 cells), incubated at 65°C for 2 h and heat inactivated at 94°C for 15 min (Monini et al., 1999). Lysis of the cells to retrieve Late-RT and 2-LTR intermediates was performed at 6 or 24 hours post-transduction respectively. Late-RT and 2LTR circles were measured by quantitative droplet digital-PCR (dd-PCR) assay with primers described below and normalized using the human TERT gene.
To measure RT products in transduced THP-1 cells, DNA was harvested at 6 h post-transduction using the QIAGEN DNeasy Blood and Tissue kit, as per manufacturer’s instructions. qPCR was performed using 2x TaqMan Gene Expression Master Mix (ThermoFisher) and primers and probe for GFP present in the LV genome (LV GFP gene). All the primers used were reported in Key Resources Table.
Genomic DNA extraction and qPCR
DNA from human CD34+ liquid culture, hematopoietic colony pool samples as well as from murine tissues was extracted using a Maxwell 16 instrument (Promega) or Blood & Cell Culture DNA micro kit (QIAGEN). DNA from single colonies was lysed in Monini buffer as previously described (Cassetta et al., 2013). Vector copies per diploid genome (vector copy number, VCN) of the integrated lentiviral vectors were quantified by quantitative droplet digital-PCR (dd-PCR) using the primers against the primer binding site region of LVs. VCN quantification of the total lentiviral DNA (integrated and non-integrated) was performed as previously described (Mátrai et al., 2011) at three days post-transduction. Copy numbers of the reverse transcribed retroviral vector genome (both integrated and non-integrated) was performed by quantitative droplet digital-PCR (dd-PCR) discriminating it from plasmid carried over from the transient transfection using the primers called ΔU3 sense and PBS antisense and reported in Key Resources Table. Vector copy numbers and replication intermediates were normalized to genomic DNA content, which was assessed using the human TERT gene. VCN analysis by ddPCR involved quantification of target and reference loci through the use of duplex target and reference assays. In QuantaSoftsoftware copy number was determined by calculating the ratio of the target molecule concentration to the reference molecule concentration, times the number of copies of reference species in the genome (usually 2). Transduction efficiencies were evaluated by ddPCR on individual colonies from CFC assay performed on the HSCs transduced with clinical grade LVs and expressed as percentage (%) of LV+ colonies on total tested as previously reported (Biffi et al., 2013). PCR reaction for ddPCR is:
| Cycling step | Temperature ° C | Time | Number of Cycles | |
|---|---|---|---|---|
| Enzyme Activation | 95 | 5 min | 1 | ddPCR 200 or 300nM |
| Denaturation | 95 | 30 s | 40 | |
| Annealing/Extension | 63 | 1 min | 40 | |
| Signal Stabilization | 4 | 5 min | 1 | |
| 90 | 5 min | 1 | ||
| Hold (Optional) | 4 | Infinite | 1 |
Western Blot
Whole cell extracts were prepared as previously described (Kajaste-Rudnitski et al., 2006, Kajaste-Rudnitski et al., 2011). Samples were subjected to SDS-PAGE using Bolt 4%–12% Bis-Tris Plus Gels (Thermo Fisher Scientific), transferred to PVDF membrane by electroblotting, and blotted with mouse polyclonal antibody (Ab) raised against CypA (1:500 dilution, RRID:AB_2169131); mouse anti-CypB monoclonal Ab (1:500 dilution, RRID:AB_2169421); rabbit anti-IFITM3 polyclonal Ab (1:1000 dilution, RRID:AB_2295684); rabbit anti-IFITM2 polyclonal Ab (1:2500, RRID:AB_2122089); mouse anti-IFITM1 monoclonal Ab (1:5000, RRID:AB_2233405). A mouse monoclonal anti-beta-actin Ab (1;1000 dilution, RRID:AB_476697) was used as a normalizer. After the incubation with primary antibodies, PVDF membranes were washed three times with tris-buffered saline (TBS) 0.1% tween 20 for 5 minutes and then incubated for one hour with rabbit or mouse IgG secondary antibodies (1:10000; RRID:AB_772206, RRID:AB_772210).
Immunofluorescence microscopy
3x104 to 5x104 THP-1 or CBCD34+ cells were seeded in Multitest slide glass 10-well 8mm (from MP Biomedicals) precoated with poly-L-lysine solution (Sigma-Aldrich) for 20 minutes. Cells were fixed with 4% paraformaldehyde (in 1X PBS) for 20 minutes at room temperature and permeabilized with 0.1% Triton X-100 for 20 minutes at room temperature. For blocking non-specific sites cells were incubated 30 minutes in PBG (5% BSA, 2% gelatin from cold water fish skin, from Sigma-Aldrich) and then stained for 2 till 16 hours with rabbit anti-IFITM3 polyclonal antibody (1:200 dilution) and or with a mouse monoclonal anti-LAMP1 (1:100, RRID:AB_470708). After 3 washes with 1X phosphate-buffered saline, cells were incubated with donkey anti-Rabbit IgG, Alexa Fluor 488 (RRID:AB_141708) or 555 (RRID:AB_162543) or 647 (RRID:AB_2536183) (1:500 dilution from Thermo Fisher Scientific) for 2 hours at room temperature. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) for 10 minutes at room temperature. Images were recorded using the TCS SP5 Leica confocal microscope, 60x with oil and quantified as integrated density with ImageJ software.
Quantification and Statistical Analysis
All statistical analyses were conducted with GraphPad Prism 5.04 version. In all studies, values are expressed as mean ± standard error of the mean (SEM) and all n numbers represent biological repeats. Statistical analyses were performed by Mann Whitney or Wilcoxon Signed Rank test between means of two groups and by ANOVA or Dunn’s adjusted Kruskal–Wallis for multiple comparisons, as indicated in the Figure legends. For WB quantifications, signal was quantified by densitometry using the ImageJ software and normalized to actin, then log10 transformed for the purposes of the correlation statistics and graphical representation when needed. For correlation studies Spearman’s rank correlation coefficient was calculate. Experimental models were randomly assigned to treatment groups.
Differences were considered statistically significant at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, “ns” represents non significance.
Acknowledgments
We thank Cesare Covino from Alembic for help with the confocal imaging acquisition and analysis; Alessandro Aiuti, Alessandra Biffi, and MolMed Spa for the clinical-grade LV used in this study; Erika Zonari for the help with FACS sorting procedures; François-Loic Cosset for the BaEV-TR envelope expression plasmid; and Nathaniel Landau for the Vpx-incorporating packaging construct. F.P., G.U., A.M.S.G., and C.P. conducted this study as partial fulfillment of their Ph.D.s in Molecular Medicine, Program in Cellular and Molecular Biology, International Ph.D. School, Vita-Salute San Raffaele University, Milan, Italy. This work was supported by the Italian Telethon Foundation (A.K.-R.). G.J.T. is funded by a Wellcome Trust Senior Biomedical Research Fellowship (108183), the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC (339223), and the National Institute for Health Research University College London Hospitals Biomedical Research Centre. L.T. is funded by a Wellcome Trust Sir Henry Wellcome Post-Doctoral Fellowship (106079). S.C. is funded by the Wellcome Trust (206194). M.N. is funded by a Wellcome Trust Investigator Award.
Author Contributions
C.P., L.G.T., and G.U. conducted experiments and analyzed data. A.M.S.G. designed and tested CRISPR/Cas9 knockout reagents. F.P. participated in in vivo transplantation experiments. G.S. and P.G. performed gene editing experiments. S.J.P., F.A., and M.N. contributed to generating and interpreting IFITM3 data in THP-1. S.C. provided the IFITM3−/− mice bone marrows. I.C. provided technical assistance. B.G. and L.N. provided reagents and intellectual input. C.P., L.G.T., G.J.T., and A.K.-R. designed the research studies, analyzed data, and wrote the manuscript.
Declaration of Interests
A.K.-R., C.P., F.P., G.S., P.G., B.G., and L.N. are inventors on pending and issued patents on lentiviral vector technology and gene transfer filed by the Salk Institute, Cell Genesys, or Telethon Foundation and San Raffaele Scientific Institute.
Published: November 8, 2018
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.stem.2018.10.008.
Supplemental Information
References
- Amendola M., Giustacchini A., Gentner B., Naldini L. A double-switch vector system positively regulates transgene expression by endogenous microRNA expression (miR-ON vector) Mol. Ther. 2013;21:934–946. doi: 10.1038/mt.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini-Bavil-Olyaee S., Choi Y.J., Lee J.H., Shi M., Huang I.C., Farzan M., Jung J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin K., Urbinati F., Romero Z., Campo-Fernandez B., Kaufman M.L., Cooper A.R., Masiuk K., Hollis R.P., Kohn D.B. Enrichment of human hematopoietic stem/progenitor cells facilitates transduction for stem cell gene therapy. Stem Cells. 2015;33:1532–1542. doi: 10.1002/stem.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- Bobadilla S., Sunseri N., Landau N.R. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther. 2013;20:514–520. doi: 10.1038/gt.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- Brass A.L., Huang I.C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulli L., Apolonia L., Kutzner J., Pollpeter D., Goujon C., Herold N., Schwarz S.M., Giernat Y., Keppler O.T., Malim M.H., Schaller T. Complex interplay between HIV-1 capsid and MX2-independent alpha interferon-induced antiviral factors. J. Virol. 2016;90:7469–7480. doi: 10.1128/JVI.00458-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busnadiego I., Kane M., Rihn S.J., Preugschas H.F., Hughes J., Blanco-Melo D., Strouvelle V.P., Zang T.M., Willett B.J., Boutell C. Host and viral determinants of Mx2 antiretroviral activity. J. Virol. 2014;88:7738–7752. doi: 10.1128/JVI.00214-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassetta L., Kajaste-Rudnitski A., Coradin T., Saba E., Della Chiara G., Barbagallo M., Graziano F., Alfano M., Cassol E., Vicenzi E., Poli G. M1 polarization of human monocyte-derived macrophages restricts pre and postintegration steps of HIV-1 replication. AIDS. 2013;27:1847–1856. doi: 10.1097/QAD.0b013e328361d059. [DOI] [PubMed] [Google Scholar]
- Chesarino N.M., McMichael T.M., Hach J.C., Yount J.S. Phosphorylation of the antiviral protein interferon-inducible transmembrane protein 3 (IFITM3) dually regulates its endocytosis and ubiquitination. J. Biol. Chem. 2014;289:11986–11992. doi: 10.1074/jbc.M114.557694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino N.M., McMichael T.M., Yount J.S. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog. 2015;11:e1005095. doi: 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paulis A., Ciccarelli A., de Crescenzo G., Cirillo R., Patella V., Marone G. Cyclosporin H is a potent and selective competitive antagonist of human basophil activation by N-formyl-methionyl-leucyl-phenylalanine. J. Allergy Clin. Immunol. 1996;98:152–164. doi: 10.1016/s0091-6749(96)70237-3. [DOI] [PubMed] [Google Scholar]
- De Ravin S.S., Li L., Wu X., Choi U., Allen C., Koontz S., Lee J., Theobald-Whiting N., Chu J., Garofalo M. CRISPR-Cas9 gene repair of hematopoietic stem cells from patients with X-linked chronic granulomatous disease. Sci. Transl. Med. 2017;9:eaah3480. doi: 10.1126/scitranslmed.aah3480. [DOI] [PubMed] [Google Scholar]
- Dever D.P., Bak R.O., Reinisch A., Camarena J., Washington G., Nicolas C.E., Pavel-Dinu M., Saxena N., Wilkens A.B., Mantri S. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue D.A., Porrot F., Couespel N., Schwartz O. SUN2 silencing impairs CD4 T cell proliferation and alters sensitivity to HIV-1 infection independently of cyclophilin A. J. Virol. 2017;91:e02303-16. doi: 10.1128/JVI.02303-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D., Naldini L. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M.A., Offner S., Blanco-Bose W.E., Waibler Z., Kalinke U., Duchosal M.A., Trumpp A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- Evans M.E., Kumkhaek C., Hsieh M.M., Donahue R.E., Tisdale J.F., Uchida N. TRIM5alpha variations influence transduction efficiency with lentiviral vectors in both human and Rhesus CD34 cells in vitro and in vivo. Mol. Ther. 2014;22:348–358. doi: 10.1038/mt.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follenzi A., Ailles L.E., Bakovic S., Geuna M., Naldini L. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 2000;25:217–222. doi: 10.1038/76095. [DOI] [PubMed] [Google Scholar]
- Foster T.L., Wilson H., Iyer S.S., Coss K., Doores K., Smith S., Kellam P., Finzi A., Borrow P., Hahn B.H., Neil S.J.D. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 2016;20:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geis F.K., Galla M., Hoffmann D., Kuehle J., Zychlinski D., Maetzig T., Schott J.W., Schwarzer A., Goffinet C., Goff S.P., Schambach A. Potent and reversible lentiviral vector restriction in murine induced pluripotent stem cells. Retrovirology. 2017;14:34. doi: 10.1186/s12977-017-0358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard-Gagnepain A., Amirache F., Costa C., Lévy C., Frecha C., Fusil F., Nègre D., Lavillette D., Cosset F.L., Verhoeyen E. Baboon envelope pseudotyped LVs outperform VSV-G-LVs for gene transfer into early-cytokine-stimulated and resting HSCs. Blood. 2014;124:1221–1231. doi: 10.1182/blood-2014-02-558163. [DOI] [PubMed] [Google Scholar]
- Griffin S.D., Allen J.F., Lever A.M. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001;75:12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas S., Hansson J., Klimmeck D., Loeffler D., Velten L., Uckelmann H., Wurzer S., Prendergast A.M., Schnell A., Hexel K. Inflammation-induced emergency megakaryopoiesis driven by hematopoietic stem cell-like megakaryocyte progenitors. Cell Stem Cell. 2015;17:422–434. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Heffner G.C., Bonner M., Christiansen L., Pierciey F.J., Campbell D., Smurnyy Y., Zhang W., Hamel A., Shaw S., Lewis G. Prostaglandin E2 increases lentiviral vector transduction efficiency of adult human hematopoietic stem and progenitor cells. Mol. Ther. 2018;26:320–328. doi: 10.1016/j.ymthe.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilditch L., Towers G.J. A model for cofactor use during HIV-1 reverse transcription and nuclear entry. Curr. Opin. Virol. 2014;4:32–36. doi: 10.1016/j.coviro.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirche C., Frenz T., Haas S.F., Döring M., Borst K., Tegtmeyer P.K., Brizic I., Jordan S., Keyser K., Chhatbar C. Systemic virus infections differentially modulate cell cycle state and functionality of long-term hematopoietic stem cells in vivo. Cell Rep. 2017;19:2345–2356. doi: 10.1016/j.celrep.2017.05.063. [DOI] [PubMed] [Google Scholar]
- Hornick A.L., Li N., Oakland M., McCray P.B., Jr., Sinn P.L. Human, pig, and mouse interferon-induced transmembrane proteins partially restrict pseudotyped lentiviral vectors. Hum. Gene Ther. 2016;27:354–362. doi: 10.1089/hum.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery J.R. Cyclosporine analogues. Clin. Biochem. 1991;24:15–21. doi: 10.1016/0009-9120(91)90105-n. [DOI] [PubMed] [Google Scholar]
- Jia R., Pan Q., Ding S., Rong L., Liu S.L., Geng Y., Qiao W., Liang C. The N-terminal region of IFITM3 modulates its antiviral activity by regulating IFITM3 cellular localization. J. Virol. 2012;86:13697–13707. doi: 10.1128/JVI.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A., Naldini L. Cellular innate immunity and restriction of viral infection: implications for lentiviral gene therapy in human hematopoietic cells. Hum. Gene Ther. 2015;26:201–209. doi: 10.1089/hum.2015.036. [DOI] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A., Mashimo T., Frenkiel M.P., Guénet J.L., Lucas M., Desprès P. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J. Biol. Chem. 2006;281:4624–4637. doi: 10.1074/jbc.M508649200. [DOI] [PubMed] [Google Scholar]
- Kajaste-Rudnitski A., Marelli S.S., Pultrone C., Pertel T., Uchil P.D., Mechti N., Mothes W., Poli G., Luban J., Vicenzi E. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-kappaB-responsive long terminal repeat elements. J. Virol. 2011;85:5183–5196. doi: 10.1128/JVI.02302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z., Ylinen L.M., Towers G.J. Cyclophilin A renders human immunodeficiency virus type 1 sensitive to Old World monkey but not human TRIM5 alpha antiviral activity. J. Virol. 2006;80:4683–4690. doi: 10.1128/JVI.80.10.4683-4690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye X., Satoh T., Gentili M., Cerboni S., Silvin A., Conrad C., Ahmed-Belkacem A., Rodriguez E.C., Guichou J.F., Bosquet N. Nuclear envelope protein SUN2 promotes cyclophilin-A-dependent steps of HIV replication. Cell Rep. 2016;15:879–892. doi: 10.1016/j.celrep.2016.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange U.C., Adams D.J., Lee C., Barton S., Schneider R., Bradley A., Surani M.A. Normal germ line establishment in mice carrying a deletion of the Ifitm/Fragilis gene family cluster. Mol. Cell. Biol. 2008;28:4688–4696. doi: 10.1128/MCB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Ambrose Z., Martin T.D., Oztop I., Mulky A., Julias J.G., Vandegraaff N., Baumann J.G., Wang R., Yuen W. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe. 2010;7:221–233. doi: 10.1016/j.chom.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G., Christiansen L., McKenzie J., Luo M., Pasackow E., Smurnyy Y., Harrington S., Gregory P., Veres G., Negre O., Bonner M. Staurosporine increases lentiviral vector transduction efficiency of human hematopoietic stem and progenitor cells. Mol. Ther. Methods Clin. Dev. 2018;9:313–322. doi: 10.1016/j.omtm.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A., Genovese P., Beausejour C.M., Colleoni S., Lee Y.L., Kim K.A., Ando D., Urnov F.D., Galli C., Gregory P.D. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Lu J., Pan Q., Rong L., He W., Liu S.L., Liang C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luban J., Bossolt K.L., Franke E.K., Kalpana G.V., Goff S.P. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Mangeot P.E., Duperrier K., Nègre D., Boson B., Rigal D., Cosset F.L., Darlix J.L. High levels of transduction of human dendritic cells with optimized SIV vectors. Mol. Ther. 2002;5:283–290. doi: 10.1006/mthe.2002.0541. [DOI] [PubMed] [Google Scholar]
- Mátrai J., Cantore A., Bartholomae C.C., Annoni A., Wang W., Acosta-Sanchez A., Samara-Kuko E., De Waele L., Ma L., Genovese P. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matreyek K.A., Yücel S.S., Li X., Engelman A. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog. 2013;9:e1003693. doi: 10.1371/journal.ppat.1003693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanous K., Yoon S., Rohll J., Patil D., Wilkes F., Kim V., Kingsman S., Kingsman A., Mazarakis N. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 1999;6:1808–1818. doi: 10.1038/sj.gt.3301023. [DOI] [PubMed] [Google Scholar]
- Monini P., Colombini S., Stürzl M., Goletti D., Cafaro A., Sgadari C., Buttò S., Franco M., Leone P., Fais S. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi’s sarcoma. Blood. 1999;93:4044–4058. [PubMed] [Google Scholar]
- Montini E., Cesana D., Schmidt M., Sanvito F., Ponzoni M., Bartholomae C., Sergi Sergi L., Benedicenti F., Ambrosi A., Di Serio C. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Nagai Y., Garrett K.P., Ohta S., Bahrun U., Kouro T., Akira S., Takatsu K., Kincade P.W. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher M., Weber T. Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther. 2012;19:649–658. doi: 10.1038/gt.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noser J.A., Towers G.J., Sakuma R., Dumont J.M., Collins M.K., Ikeda Y. Cyclosporine increases human immunodeficiency virus type 1 vector transduction of primary mouse cells. J. Virol. 2006;80:7769–7774. doi: 10.1128/JVI.02427-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F., Doulatov S., Dick J.E. Engraftment of human hematopoietic stem cells is more efficient in female NOD/SCID/IL-2Rgc-null recipients. Blood. 2010;115:3704–3707. doi: 10.1182/blood-2009-10-249326. [DOI] [PubMed] [Google Scholar]
- Peel M., Scribner A. Cyclophilin inhibitors as antiviral agents. Bioorg. Med. Chem. Lett. 2013;23:4485–4492. doi: 10.1016/j.bmcl.2013.05.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreira J.M., Chin C.R., Feeley E.M., Brass A.L. IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo C., Cesana D., Piras F., Bartolaccini S., Naldini L., Montini E., Kajaste-Rudnitski A. Cyclosporin a and rapamycin relieve distinct lentiviral restriction blocks in hematopoietic stem and progenitor cells. Mol. Ther. 2015;23:352–362. doi: 10.1038/mt.2014.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras F., Riba M., Petrillo C., Lazarevic D., Cuccovillo I., Bartolaccini S., Stupka E., Gentner B., Cittaro D., Naldini L., Kajaste-Rudnitski A. Lentiviral vectors escape innate sensing but trigger p53 in human hematopoietic stem and progenitor cells. EMBO Mol. Med. 2017;9:1198–1211. doi: 10.15252/emmm.201707922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeschla E.M., Wong-Staal F., Looney D.J. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- Prevete N., Liotti F., Marone G., Melillo R.M., de Paulis A. Formyl peptide receptors at the interface of inflammation, angiogenesis and tumor growth. Pharmacol. Res. 2015;102:184–191. doi: 10.1016/j.phrs.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Provasi E., Genovese P., Lombardo A., Magnani Z., Liu P.Q., Reik A., Chu V., Paschon D.E., Zhang L., Kuball J. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 2012;18:807–815. doi: 10.1038/nm.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Le Duff Y., Wang Y., Pan Q., Ding S., Zheng Y.M., Liu S.L., Liang C. Primate lentiviruses are differentially inhibited by interferon-induced transmembrane proteins. Virology. 2015;474:10–18. doi: 10.1016/j.virol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragheb J.A., Yu H., Hofmann T., Anderson W.F. The amphotropic and ecotropic murine leukemia virus envelope TM subunits are equivalent mediators of direct membrane fusion: implications for the role of the ecotropic envelope and receptor in syncytium formation and viral entry. J. Virol. 1995;69:7205–7215. doi: 10.1128/jvi.69.11.7205-7215.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasaiyaah J., Tan C.P., Fletcher A.J., Price A.J., Blondeau C., Hilditch L., Jacques D.A., Selwood D.L., James L.C., Noursadeghi M., Towers G.J. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T., Ocwieja K.E., Rasaiyaah J., Price A.J., Brady T.L., Roth S.L., Hué S., Fletcher A.J., Lee K., KewalRamani V.N. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiroli G., Ferrari S., Conway A., Jacob A., Capo V., Albano L., Plati T., Castiello M.C., Sanvito F., Gennery A.R. Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Sci. Transl. Med. 2017;9:eaan0820. doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolskaja E., Luban J. Cyclophilin, TRIM5, and innate immunity to HIV-1. Curr. Opin. Microbiol. 2006;9:404–408. doi: 10.1016/j.mib.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Sun X., Yau V.K., Briggs B.J., Whittaker G.R. Role of clathrin-mediated endocytosis during vesicular stomatitis virus entry into host cells. Virology. 2005;338:53–60. doi: 10.1016/j.virol.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Thompson A.A., Walters M.C., Kwiatkowski J., Rasko J.E.J., Ribeil J.A., Hongeng S., Magrin E., Schiller G.J., Payen E., Semeraro M. Gene therapy in patients with transfusion-dependent β-thalassemia. N. Engl. J. Med. 2018;378:1479–1493. doi: 10.1056/NEJMoa1705342. [DOI] [PubMed] [Google Scholar]
- Towers G.J. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers G.J., Noursadeghi M. Interactions between HIV-1 and the cell-autonomous innate immune system. Cell Host Microbe. 2014;16:10–18. doi: 10.1016/j.chom.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers G.J., Hatziioannou T., Cowan S., Goff S.P., Luban J., Bieniasz P.D. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- Visigalli I., Delai S., Politi L.S., Di Domenico C., Cerri F., Mrak E., D’Isa R., Ungaro D., Stok M., Sanvito F. Gene therapy augments the efficacy of hematopoietic cell transplantation and fully corrects mucopolysaccharidosis type I phenotype in the mouse model. Blood. 2010;116:5130–5139. doi: 10.1182/blood-2010-04-278234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visigalli I., Delai S., Ferro F., Cecere F., Vezzoli M., Sanvito F., Chanut F., Benedicenti F., Spinozzi G., Wynn R. Preclinical testing of the safety and tolerability of lentiviral vector-mediated above-normal alpha-L-iduronidase expression in murine and human hematopoietic cells using toxicology and biodistribution good laboratory practice studies. Hum. Gene Ther. 2016;27:813–829. doi: 10.1089/hum.2016.068. [DOI] [PubMed] [Google Scholar]
- Wang C.X., Sather B.D., Wang X., Adair J., Khan I., Singh S., Lang S., Adams A., Curinga G., Kiem H.P. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124:913–923. doi: 10.1182/blood-2013-12-546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Exline C.M., DeClercq J.J., Llewellyn G.N., Hayward S.B., Li P.W., Shivak D.A., Surosky R.T., Gregory P.D., Holmes M.C., Cannon P.M. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015;33:1256–1263. doi: 10.1038/nbt.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Dao Thi V.L., Huang Y., Billerbeck E., Saha D., Hoffmann H.H., Wang Y., Silva L.A.V., Sarbanes S., Sun T. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172:423–438. doi: 10.1016/j.cell.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonari E., Desantis G., Petrillo C., Boccalatte F.E., Lidonnici M.R., Kajaste-Rudnitski A., Aiuti A., Ferrari G., Naldini L., Gentner B. Efficient ex vivo engineering and expansion of highly purified human hematopoietic stem and progenitor cell populations for gene therapy. Stem Cell Reports. 2017;8:977–990. doi: 10.1016/j.stemcr.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.