Abstract
Domestication converts perennial and photoperiodic ancestral cotton to day-neutral cotton varieties, and the selection of short-season cotton varieties is one of the major objectives of cotton breeding. However, little is known about the mechanism of flowering time in cotton. Here, we report a cotton HD-ZIP I-class transcription factor (GhHB12) specifically expressed in axillary buds, which antagonisticlly interacts with GhSPL10/13 to repress the expression of GhFT, GhFUL, and GhSOC1, resulting in bushy architecture and delayed flowering under long-day conditions. We found that GhHB12-mediated ancestral upland cotton phenotypes (bushy architecture and delayed flowering) could be rescued under short-day conditions. We showed that overexpressing of GhrSPL10 partially rescues the bushy architecture and delayed flowering phenotypes, while overexpression of GhmiR157 reinforced these phenotypes in GhHB12-overexpressing plants. This study defines a regulatory module which regulates cotton architecture, phase transition and could be applied in the breeding of early maturing cotton varieties.
Subject terms: Agricultural genetics, Flowering, Patterning, Plant domestication
Xin He et al. present a characterization of GhHB12, a HD-ZIP family transcription factor expressed in upland cotton axillary buds. They show that GhHB12 regulates flowering time, plant architecture and phase transition via a regulatory module that could be harnessed to improve cotton for mechanical harvesting.
Introduction
Domestication is a selection process conducted by humans to adapt plants and animals to human needs. Despite the geographically diverse distribution of domestication centers, a remarkably similar set of traits (domestication syndrome traits) have been selected in a wide range of crops, including the architecture of plants, flowering time, distribution of reproductive structures, seed size, and dormancy1. The domestication of many crops resulted in more determinate architectures with compact growth habits, favorable and synchronous flowering times, and higher yields2,3. Over the past decade, researchers have identified some specific genes (domestication genes) that control the most important morphological changes associated with domestication.
Squamosa promoter-binding-like protein (SPL) is a plant-specific transcription factor family that regulates plant development and stress response, such as branch development, inflorescence development, timing of vegetative and reproductive phase change and response to stresses4. It is targeted by MicroRNA 156/157(miR156/157) for cleavage and/or translational repression, and both SPL and MicroRNA 156/157 are key genes in crop domestication5,6. In rice, OsSPL13 and OsSPL16 (SBP-Like) control grain size, shape, and quality via regulating cell proliferation7,8, and OsSPL14 controls panicle and shoot branching9–11. Moreover, novel functions were continuously recorded for SPL, such as the formation of the casing that surrounds the kernels of teosinte, fruit ripening of tomato, and tuberization of potato12–14. Overexpression of miR156/157 usually results in a bushy architecture and delays the flowering of plants via repressing SPL7–11,15,16. A common effect observed in domesticated crops is that the onset of flowering becomes less dependent on day length and vernalization, resulting in shorter life cycles, and the key domestication genes are the CENTRORADIALIS/TFL1/SP (CETS) family genes FLOWERING LOCUS T (FT) and TERMINAL FLOWER1 (TFL1) and MADS-box genes FLOWERING LOCUS C (FLC), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and FRUITFULL (FUL)17–27.
Upland cotton (Gossypium hirsutum) is the most important natural textile fiber source worldwide. The main stem of cotton is monopodial (meaning that the axis of growth continues from a single point) and remains indeterminate throughout development, producing lateral branches from the leaf axils indefinitely. Then, the lateral branches develop and differentiate into vegetative branches or fruiting branches. The vegetative branch often develops monopodially (meaning that the apical meristem is terminated and growth is continued by one or more lateral meristems, which repeat the process) from the lower leaf axils in the same way as the growth of the main stem, while the fruiting branch develops sympodially from the upper leaf axils, with each sympodial unit consisting of a terminal flower, a subtending leaf and an axillary bud to continue the branch28.
Ancestral upland cotton is mostly short-day photoperiodic, with exclusive monopodial (vegetative) branches and late-flowering under non-inductive long days. In contrast, domesticated cotton is day-neutral, with sympodial fruiting branches initiating as early as at the fifth to seventh node, and produces 1–3 vegetative branches below the node of the first fruiting branch (NFFB). Thus, fruit branching and NFFB are major events in the development of cotton architecture and important productivity-related agronomic traits considered in domestication programs29,30. Unfortunately, the regulatory mechanism of the transition from vegetative growth to reproductive development in cotton remains unclear.
Previously, an expressed sequence tag (EST), DW511649, was isolated as a candidate gene responsive to environmental stresses in cotton using data-mining and expression pattern analysis31. The full length of gene was obtained according to this EST, which putatively encodes a HD-ZIP I-class transcription factor, and was designated as GhHB12. Here, we demonstrate that this HD-ZIP transcription factor is specifically expressed in axillary buds, and can interact with GhSPL10 and GhSPL13. As a result, the expression of GhFT, GhFUL, and GhSOC1 is repressed, which promotes cotton vegetative branch outgrowth and delays fruiting branch development under long-day conditions. In addition, the GhHB12-mediated ancestral upland cotton phenotypes (bushy architecture and delayed flowering) in cotton can be rescued under short-day conditions. Further analysis revealed that up-regulation of GhrSPL10 can also partially rescue the bushy architecture and delayed flowering phenotypes, while overexpression of GhmiR157 reinforced these phenotypes in GhHB12-overexpressing plants. This study defines a novel regulatory module that regulates cotton architecture, phase transition and photoperiod sensitivity, which may help the breeding of early mature cotton varieties suitable for mechanical harvest.
Results
GhHB12 promotes bushy architecture and delayed flowering
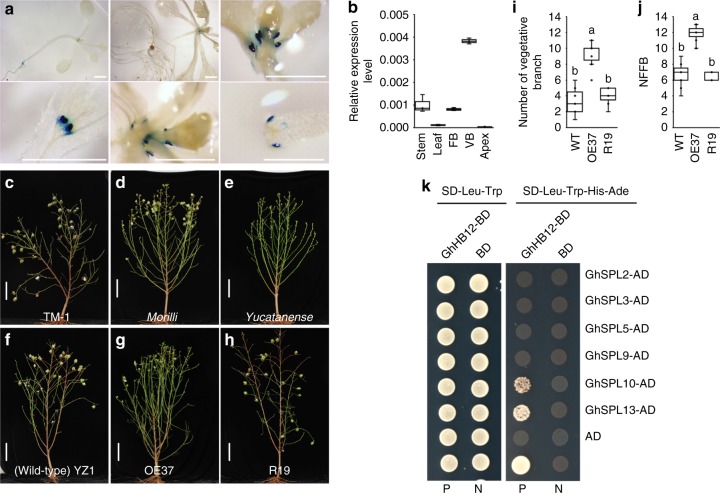
In our previous study, the cotton EST DW511649 was isolated as a putative abiotic-stress-responsive gene, which could be induced by salt and ABA treatments31. To further elucidate the function of DW511649, we cloned a putative full length cDNA sequence from domesticated upland cotton (G. hirsutum) variety YZ1 using rapid-amplification of cDNA ends (RACE) according to the sequence of DW511649. Alignment and phylogenetic analysis showed that this gene encoded a cotton HD-ZIP I-class transcription factor and had the highest similarity to Arabidopsis ATHB12, and thus was named as GhHB12 (Supplementary Figure 1a–1c). Spatial–temporal expression analysis showed that GhHB12 was highly expressed in the buds of cotton vegetative branches (Fig. 1b). To further elucidate the expression profile of GhHB12, a 996-bp promoter region of GhHB12 was isolated from YZ1, and several putative cis-acting regulatory elements involved in light-response, circadian control, stress-response and plant hormone-response were predicted (Supplementary Figure 1d and 1e), implying that GhHB12 may function in the responses to various environmental cues. The 905-bp promoter region of GhHB12 was fused to the GUS reporter gene and transformed into Arabidopsis. The proGhHB12::GUS construct in transgenic Arabidopsis was predominantly expressed in axillary buds (Fig. 1a).
Fig. 1.
Identification of GhHB12. a Histochemical localization of GUS activity in proGhHB12:GUS Arabidopsis plants. Scale bars = 2 mm. b Detection of the expression of GhHB12 in stem, leaf, fruit branch bud (FB), vegetative branch bud (VB), and apex of domesticated upland cotton (Gossypium hirsutum L. YZ1) by qRT-PCR. The GhUBQ7 gene was used as the endogenous reference gene. The data represent the mean ± SD of three technical replicates. c–h Photographs of domesticated upland cotton TM-1 (c), YZ1 (f), OE37 (g, GhHB12 overexpression in the YZ1 background), and R19 (h, GhHB12 RNAi in the YZ1 background), ancestral upland cotton morilli (d) and yucatanense (e) plants. Scale bars = 20 cm. i and j Comparison of the number of vegetative branches (i) and the node of the first vegetative branch (NFFB) (j) among YZ1, OE37, and R19 plants. Error bars indicate the standard deviation of 12–15 plants, and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test). k Physical interaction of GhHB12 with GhSPL10 and GhSPL13 validated using the GAL-4-based Y2H assay. SD synthetic dropout, BD binding domain, AD activation domain, P positive control, N negative control
To evaluate the functions of GhHB12 in axillary bud and branch development, we constructed 35S::GhHB12 and GhHB12-RNAi vectors, and GhHB12-overexpression (OE) and RNA interference (RNAi)-transformed plants were obtained with YZ1 through an A. tumefaciens-mediated method (see the experimental procedures). Three overexpressing lines (OE37, OE39, and OE42) and three RNAi lines (R7, R17, and R19) were selected for further analysis (Supplementary Figure 2a and 2b). Moreover, GhHB12-overexpressing Arabidopsis (monopodial growth pattern plant) and tomato (sympodial growth pattern plant) were obtained (Supplementary Figure 3). Among them, the GhHB12-overexpressing cotton lines showed phenotypes of more vegetative branches and delayed flowering time in Wuhan (China) under long day conditions. The growth of GhHB12-RNAi lines was similar to that of the wild type (YZ1), and the number of vegetative branches of GhHB12-RNAi lines was slightly smaller than that of the wild type (Fig. 1c–j and Supplementary Figure 2c–2k). Interestingly, the number of branches and flowering time showed no differences between GhHB12-overexpressing Arabidopsis lines and the wild type Arabidopsis (Col-0) (Supplementary Figure 3a), while overexpression of GhHB12 in tomato led to late flowering and leafy inflorescences (Supplementary Figure 3b). These results indicated that GhHB12 is a crucial regulator of the sympodial growth pattern of plant branch development and phase transition.
GhHB12 interacts with GhSPL10 and GhSPL13
SPL is a transcription factor family that regulates plant development, such as branch development, inflorescence development, and phase transition, and is regulated by MicroRNA 156/157(miR156/157)16. Overexpression of miR156/157 usually results in bushy architecture and delays flowering in plants7–11,15,16. The phenotypes of bushy architecture and inhibited flowering in GhHB12-overexpressing cotton plants led to the hypothesis that GhHB12 may interact with GhSPL. To test this hypothesis, we cloned six cotton miR156/157-targeted GhSPLs (GhSPL2, GhSPL3, GhSPL5, GhSPL9, GhSPL10, and GhSPL13), and detected the interactions between them and GhHB12 by the yeast two-hybrid system (Y2H) assay. As shown in Fig. 1k, the strains co-expressing GhHB12-BD (BD: GAL4 DNA-binding domain) and GhSPL10/13-AD (AD: activation domain) were able to grow on SD-Trp-Leu-His-Ade (medium lacking tryptophan, leucine, histidine, and adenine), whereas the clones expressing GhHB12-BD and GhSPL2/3/5/9-AD only grew on medium supplemented with histidine and adenine, indicating that GhHB12 interact with GhSPL10 and GhSPL13, but not with GhSPL2, GhSPL3, GhSPL5, or GhSPL9.
GhHB12 and GhmiR157 coordinate in vegetative development
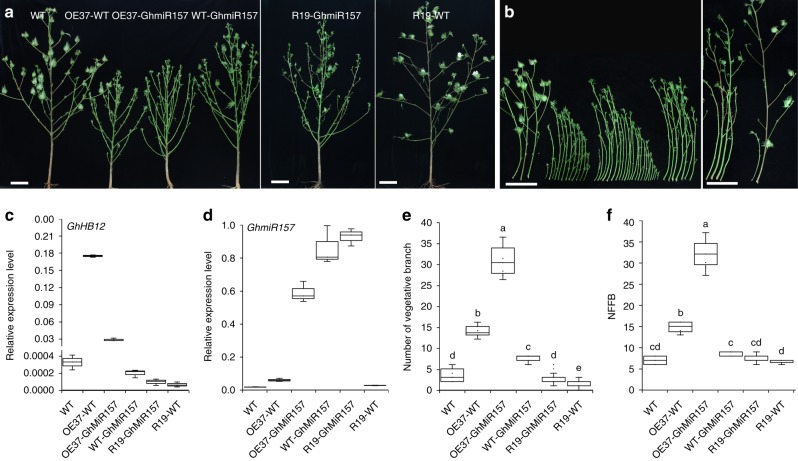
To further dissect the functions of cotton miR156/157 and SPLs, GhmiR157, MIMI157 (GhmiR157 target mimicry vector generated by replacing the miR399 complementary motif of the 522-bp genomic sequence of Arabidopsis IPS1 with cotton GhmiR157 complementary motif through PCR according to the previous study32), and GhrSPL10 (a GhmiR157-resistant version without the GhmiR157 response element generated by introducing seven synonymous codon mutations into the predicted GhmiR157-binding site of GhSPL10 using recombinant PCR) were overexpressed in cotton as previously described33–35 (Supplementary Figure 4). Overexpression of GhmiR157 greatly increased the number of vegetative branches34 and slightly delayed the flowering time in cotton (Fig. 2a and Supplementary Figure 4b). The growth of the MIMI157-overexpressing cotton line (designated as MIMI157-8) and GhrSPL10-overexpressing cotton line (designated as GhrSPL10-7) was similar to that of the wild-type YZ1 (Supplementary Figure 4c and 4d).
Fig. 2.
GhHB12 and GhmiR157 synergistically promote cotton vegetative branches and delay reproductive development. a–f Photographs of architectures (a) and vegetative branches (b), qRT-PCR analysis of the transcript levels of GhHB12 (c), and GhmiR157 (d), number of vegetative branches (e) and NFFB (f) of WT (wild-type), OE37-WT (hybrid plants of GhHB12-overexpression line and WT), OE37-GhmiR157 (hybrid plants of GhHB12-overexpression and GhmiR157-overexpression lines), WT-GhmiR157 (hybrid plants of WT and GhmiR157-overexpression line), R19-GhmiR157 (hybrid plants of GhHB12-RNAi and GhmiR157-overexpression lines), and R19-WT (hybrid plants of GhHB12-RNAi line and WT). The GhUBQ7 gene was used as the endogenous reference gene. The data represent the mean ± SD of three technical replicates in c and d. Error bars indicate the standard deviation of 7–17 plants, and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test) in e and f
To investigate the possible genetic interactions between GhHB12 and GhmiR157, WT (wild-type YZ1) and GhmiR157-38 (GhmiR157-overexpressing cotton line) were crossed with the OE37 (GhHB12-overexpression line) and R19 (GhHB12-RNAi line), respectively, and the branching phenotype of the resulting F1 generation hybrid plants was examined. The vegetative branch number and NFFB of hybrid plants of GhHB12-overexpression and GhmiR157-overexpression lines were dramatically increased compared with those of hybrid plants of GhHB12-overexpression line and WT, and hybrid plants of GhmiR157-overexpression line and WT, whereas the vegetative branch number of hybrid plants of GhHB12-RNAi and GhmiR157-overexpression lines was significantly smaller than that of hybrid plants of GhmiR157-overexpression line and WT (Fig. 2), suggesting that GhHB12 and GhmiR157 synergistically promote the outgrowth of cotton vegetative branches and delay reproductive development.
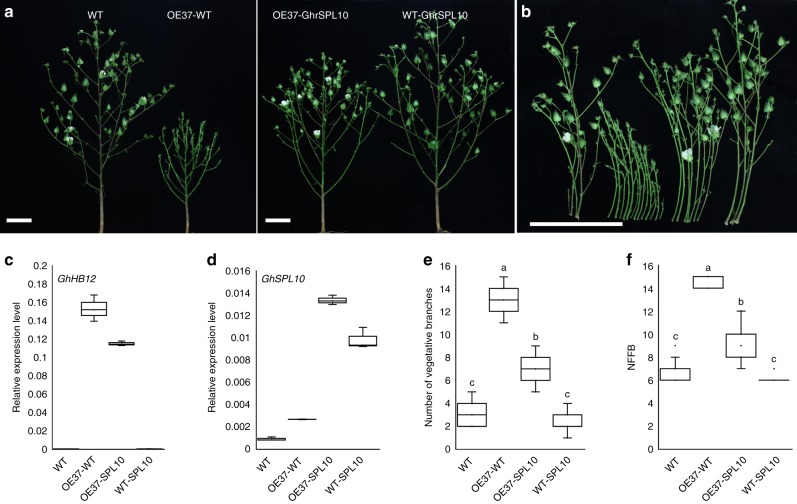
To further investigate the genetic interactions between GhHB12 and GhSPL10, WT and GhrSPL10-7 plants were crossed with the OE37 and R19 plants, respectively, and the branching phenotype of the resulting F1 generation hybrid plants was examined. The vegetative branch number and NFFB of hybrid plants of GhHB12-overexpression and GhrSPL10-overexpression lines were significantly fewer than those of hybrid plants of GhHB12-overexpression line and WT (Fig. 3), suggesting that GhHB12 regulates cotton vegetative branch development and flowering time through antagonistically interacting with GhSPL10. Taken together, the results suggested that GhHB12 and GhmiR157-GhSPLs coordinate to regulate cotton architecture and phase transition.
Fig. 3.
GhHB12 and GhSPL10 regulate vegetative branches and reproductive development in an antagonistic manner. a–f Photographs of architectures (a) and vegetative branches (b), qRT-PCR analysis of the transcript levels of GhHB12 (c), and GhSPL10 (d), number of vegetative branches (e) and NFFB (f) of WT (wild-type), OE37-WT (hybrid plants of GhHB12-overexpression line and WT), OE37-GhrSPL10 (hybrid plants of GhHB12-overexpression and GhrSPL10-overexpression lines), and WT-GhrSPL10 (hybrid plants of WT and GhrSPL10-overexpression line). The GhUBQ7 gene was used as the endogenous reference gene. The data represent the mean ± SD of three technical replicates in c and d. Error bars in e and f indicate the standard deviation of 9–17 plants, and different letters indicate significant differences at P < 0.05 (Duncan’s multiple range test)
GhHB12 inhibits the expression of genes in phase transition
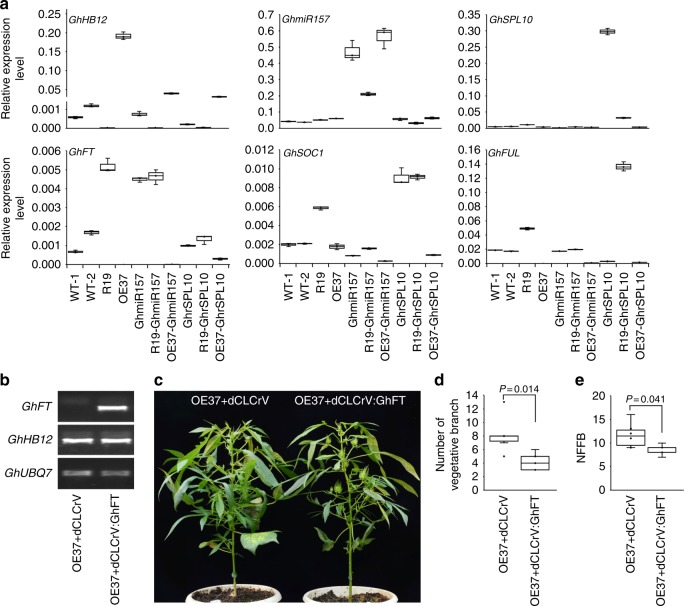
It has been reported that some MADS transcription factors are the direct target genes of SPLs, such as SOC1, FUL, and AP1, which play important roles in plant phase transition and floral development16,36. To explore the downstream genes regulated by the GhHB12 and GhmiR157-GhSPLs pathways in cotton branch development and phase transition, we examined the expression levels of genes homologous to SOC1, FUL, and AP1 in the leaves and shoot apices of five-leaf stage cotton seedlings. The results showed that GhFUL was dramatically repressed in GhHB12-overexpressing plants; GhSOC1 was down-regulated in GhmiR157-overexpressing plants and was further repressed in OE37-GhmiR157 plants, whereas GhSOC1, GhFUL, and GhAP1 were up-regulated in hybrid plants of GhHB12-RNAi and GhrSPL10-overexpression lines compared with in R19 and GhrSPL10 plants (Fig. 4a and Supplementary Figure 5a). In addition, the expression levels of some cotton GhSPLs decreased in GhmiR157-overexpressing plants and GhHB12-overexpressing plants, but were up-regulated in GhHB12-RNAi plants (Supplementary Figure 5a). These results demonstrated that GhHB12 suppresses the genes related to flowering time partly through the GhmiR157-GhSPLs pathway.
Fig. 4.
GhHB12 regulates cotton vegetative branches and reproductive development by suppressing GhFT and GhFUL transcription. a qRT-PCR analysis of the transcript levels of GhHB12, GhmiR157, GhSPL10, GhFT, GhSOC1, and GhFUL in WT and transgenic plants. The GhUBQ7 gene was used as the endogenous reference gene. The data represent the mean ± SD of three technical replicates. b RT-PCR analysis of the transcript levels of GhHB12 and GhFT in GhHB12-overexpressing plants inoculated with dCLCrV or dCLCrV:GhFT. c Photographs of GhHB12-overexpressing plants inoculated with dCLCrV or dCLCrV:GhFT. Number of vegetative branches (d) and NFFB (e) of GhHB12-overexpressing plants inoculated with dCLCrV or dCLCrV:GhFT. Error bars in d and e indicate the standard deviation of 5–6 plants. *P < 0.05 indicate significant differences between two groups (Student’s t-test)
GhHB12-mediated phenotypes are partially dependent on GhFT
CENRORADIALIS/TFL1/SP (CETS) family gene FLOWERING LOCUS T (FT) and its antagonist TERMINAL FLOWER1 (TFL1) play important roles in determinate growth and indeterminate growth of barley, sunflower, tomato, and strawberry17,19,20,24,26,37, and FT is directly regulated by SPL3 in Arabidopsis38. The variations of GhFT and GhTFL1 activities would affect cotton architecture and photoperiod sensitivity. Overexpressing GhSFT/GhFT (Gh_D08G2407 and Gh_A08G2015) in ancestral and photoperiodic cotton resulted in flowering under non-inductive long days, and silencing GhFT in domesticated and day-neutral cotton led to the phenotypes of more vegetative branches and delayed flowering time21. Therefore, we determined the expression of GhFT in the leaves and shoot apex of five-leaf stage seedlings and found that its expression was dramatically suppressed in GhHB12-overexpressing plants, while overexpression of GhrSPL10 partially attenuated this suppression (Fig. 4a), implying that GhHB12 and GhSPL10 antagonistically regulate GhFT. Additionally, there was an inversely proportional relationship in the expression levels between GhHB12 and GhFT in cotton vegetative branches and fruit branches (Fig. 1b and Supplementary Figure 5b), and the expression of either GhHB12 or GhFT was rhythmic and regulated by photoperiod (Supplementary Figure 5c), which was consistent with the enrichment of light responsive cis-acting elements in the promoter of GhHB12 (Supplementary Figure 1d). These results implied that GhHB12 represses the expression of GhFT, which may subsequently change the photoperiod sensitivity and architecture of cotton.
Interestingly, the phenotypes of bushy architecture and delayed flowering in GhHB12-overexpressing plants were completely rescued when grown in the Hainan (China) winter nursery (natural short-day). The growth of GhHB12-overexpressing plants was similar to that of the wild-type (YZ1) (Supplementary Figure 5d–5f). These results indicated that the photoperiodic effects of GhHB12 on the phenotype may depend on the expression of GhFT. To confirm this hypothesis, we transiently expressed GhFT in GhHB12-overexpressing plants via the dCLCrV system, a disarmed geminivirus Cotton leaf crumple virus was used for virus-induced flowering21. GhHB12-overexpressing plants inoculated with dCLCrV:GhFT flowered earlier than dCLCrV-infected GhHB12-overexpressing plants (Fig. 4b–e). Taken together, these results demonstrated that the regulatory effect of GhHB12 on cotton architecture, reproductive development, and photoperiod sensitivity is partially dependent on GhFT.
Discussion
The main stem of cotton is monopodial and remains indeterminate throughout development, producing lateral branches (monopodial vegetative branches or sympodial fruiting branches) from the leaf axils indefinitely28. Cotton domestication converts perennial and photoperiodic ancestral cotton to day-neutral cotton varieties, and the selection of short-season cotton varieties suitable for mechanical harvest is a major objective of cotton breeding39. Although numerous QTLs regulating cotton branch development and flowering time have been identified40–44, little is known about the regulatory mechanism of cotton branch development and transition from vegetative growth to reproductive development.
Genes involved in axillary bud initiation control both vegetative and reproductive branching, whereas genes controlling axillary bud outgrowth have specific roles only at certain stages. Mir156/157-SPL plays vital roles in vegetative and reproductive phase changes, and overexpression of miR156/157 usually results in a bushy and dwarf architecture and delays flowering in plants4–6. Similar to miR156/157, GhmiR157 could increase the outgrowth of vegetative branches in cotton. However, overexpression of GhmiR157 slightly delayed the flowering time and increased plant height in cotton34 (Fig. 2 and Supplementary Figure 4b). These results demonstrate that the homologous genes and pathways among different species may have functional variations, and the branch development, phase transition, and plant height are controlled by a complex regulatory network in cotton.
In this study, we confirmed that a cotton HD-zip I class transcription factor GhHB12, which is specifically expressed in axillary buds (Fig. 1a, b) and directly interacts with miR156/157-targeted GhSPL10 and GhSPL13 (Fig. 1k), coordinates with the GhMir157-GhSPLs pathway to regulate the expression of genes related to cotton flowering time (e.g. GhFT, GhSOC1, and GhFUL), and subsequently modulates cotton architecture and phase transition (Figs. 2–4 and Supplementary Figure 5). It is noteworthy that overexpressing GhHB12 in tomato (sympodial growth pattern plant) led to late flowering and leafy inflorescences, while it showed no effects on Arabidopsis (monopodial growth pattern plant) (Supplementary Figure 3). These results indicate that the monopodial branch and sympodial branch developments of different species are controlled in various ways, and GhHB12 is a crucial suppressor of the sympodial growth pattern of plant branch development and phase transition.
It has been reported that genes related to plant flowering time (SOC1, FUL, and FT) and the SPL genes could regulate each other in response to photoperiod signals to promote flowering16,36,38,45. soc1 and ful mutants showed delayed flowering time in Arabidopsis46,47. Overexpression of GhSOC1 promoted flowering in Arabidopsis and caused dwarfing of plant height in cotton48. ft mutant showed delayed flowering time in many plants, but exhibited decreased branch number in Arabidopsis49,50. Silencing of GhFT in domesticated and day-neutral cotton resulted in more vegetative branches and delayed flowering time21, which are similar to the phenotypes of GhHB12-overexpression line. As expected, up-regulation of GhrSPL10 partially rescued the phenotypes of bushy architecture and delayed flowering of GhHB12-overexpressing plants (Fig. 3); the expression of GhFT, GhFUL, and GhAP1 was supressed in GhHB12-overexpressing cotton plants, while overexpression of GhrSPL10 partially attenuated this suppression (Fig. 4a). Furthermore, transient expression of GhFT in GhHB12-overexpressing plants also partially rescued the phenotypes of bushy architecture and delayed flowering (Fig. 4b–e). Unexpectedly, GhrSPL10 could up-regulate GhSOC1 and GhAP1, but not GhFT and GhFUL (Fig. 4a and Supplementary Figure 5a), presumably because GhSOC1 was the direct target of GhrSPL10, while GhFT and GhFUL were the direct targets of GhHB12, and GhAP1 was the direct target of both GhrSPL10 and GhHB12. When GhHB12 and GhrSPL10 were co-overexpressed in cotton, they inhibited each other and subsequently regulated the expression of GhSOC1, GhFT, GhFUL, and GhAP1 antagonistically. Additionally, the variations in the expression of GhSPL2 and GhSPL3 under the regulation of GhHB12, GhrSPL10, and GhmiR157 were similar to those of GhFT and GhFUL (Supplementary Figure 5a). All results indicated that there is a complex regulatory network composed of GhHB12, GhmiR157-GhSPLs, and GhFT/GhSOC1/GhFUL/GhAP1 that regulates the photoperiodic flowering of cotton during domestication. Therefore, further research is needed to clarify which of them were selected for during cotton domestication and how the genes related to cotton flowering time are regulated by the interactions between GhHB12 and GhSPLs or other proteins.
Previous studies have shown that GhFT is regulated by daily oscillations in photoperiod and contributes to cotton photoperiod sensitivity21,51. We found that both GhHB12 and GhFT are regulated by the circadian clock and photoperiod (Supplementary Figure 5c), and the GhHB12-mediated phenotypes were rescued under short-day conditions (Supplementary Figure 5d–f). It is consistent with the enrichment of light responsive cis-acting elements in the promoter of GhHB12 (Supplementary Figure 1d). Additionally, in our previous study, the expression of GhHB12 was elevated by ABA and salt treatment31. Most HD-Zip I class transcription factors were reported to be involved in developmental reprogramming in response to changes of environmental conditions52,53 (e.g. drought, salt, and light). Although the molecular basis underlying the rescue of the GhHB12-mediated phenotypes under short-day conditions has not been clarified in this report, our results suggested that GhHB12 is a node of convergence between cotton development and response to various environmental changes. Hence, further investigation of the possible post-translational modification and protein modification of GhHB12 will expand our understanding of the functions of GhHB12 in cotton growth and development under different conditions.
In conclusion, the functions of the GhHB12 and GhMir157-GhSPLs pathways revealed in this study have significant implications for understanding the regulation of cotton architecture and phase transition, as well as for the breeding of early mature cotton varieties suitable for mechanical harvest.
Methods
Plant materials and growth conditions
Domesticated upland cotton (Gossypium hirsutum) YZ1 and TM-1 and ancestral upland cotton latifolium, palmeri, punctatum, yucatanense, marie-galante, morilli, and richmonolii were used in the experiments. The wild-type (YZ1), transgenic cotton plants, and ancestral upland cotton were grown under natural long-day conditions in the experimental field of Huazhong Agricultural University (Wuhan, China, 30.4°N, 114.2°E) during summer and were grown under natural short-day conditions in the field of Sanya (Hainan, China, 18.2°N, 109.5°E) during winter with normal daily management.
Construct generation and transformation
The cDNA of GhHB12 was cloned into the pGWB418 plasmid; a genomic sequence containing GhmiR157 precursor and MIMI157 (mimicry GhmiR157) was cloned into the pGWB402 plasmid33,34. GhrSPL10 was generated by two rounds of mutagenic PCR and then cloned into the pGWB408 plasmid to produce overexpression plants35. The vector pHellesgate4 was used for GhHB12-RNAi. A 969-bp promoter sequence of GhHB12 was cloned into the vector pGWB433. Agrobacterium-mediated transformation was carried out to generate transgenic cotton and tomato plants, and YZ1 (cotton) and A57 (tomato) were used as wild-type and transgenic receptors.
GUS (β-glucuronidase) staining
proGhHB12:GUS transgenic Arabidopsis seedlings were incubated in GUS staining solution for 12 h at 37 °C and then washed with 75% ethanol. The GUS staining solution was composed of 0.9 g l–1 5-bromo-4-chloro-3-indolylglucuronide, 50 mM sodium phosphate buffer (pH 7.0), 20% (v/v) methanol, and 100 mg l–1 chloromycetin. The samples were examined and photographed with a stereomicroscope (Leica Microsystems, Germany).
Southern blotting and qRT-PCR analysis
Genomic DNA was extracted from the young leaves of wild-type (YZ1) and transgenic cotton plants using a DP305 plant genomic DNA kit (Tiangen Biotech, Beijing), digested with Hind III and resolved in 0.8% agarose gel, then transferred to Hybond N+ nylon membranes (Amersham, UK). Fragment of NPTII was used as probes and labeled with 32P (Promega Labeling Kit, Promega). Hybridization was performed at 65 °C for 16–18 h and was followed by washing two times in 2 × SSC, for 5 min each, and then two times in 0.1 × SSC, for 10 min each, to remove unbound probes. Then the washed membranes were scanned using FLA-5000 Plate/Fluorescent Image Analyzer (Fuji Poto Film, Tokyo, Japan). qRT-PCR was performed using the 7500 Real-Time PCR System (ABI, Foster City, USA) with SYBR Green (Bio-Rad, USA). The GhUBQ7 (GenBank: DQ116441) gene from G. hirsutum was used as the endogenous reference gene. The relative transcript level was determined and normalized using the reference level and averaged over the three technical replicates. The primers used in this study are listed in Supplementary Data 1. The raw gel image of the RT-PCR results is shown in Supplementary Figure 6.
Yeast two-hybrid assays
The full-length sequences were cloned into pGADT7. pGBKT7 fused with GhHB12 was used to transform Y2H, and pGADT7 fused with GhSPL2, GhSPL3, GhSPL5, GhSPL9, GhSPL10, and GhSPL13 was transferred into Y187 yeast using the Transformation System (Clontech, Cat. no. 630489). Interactions between these transcription factors after mating were determined by growth on SD medium with the -Ade/-His/-Trp/-Leu assay as described by the manual (Clontech, Cat. no. 630489).
Virus inoculations
dCLCrV a disarmed geminivirus Cotton leaf crumple virus was used for virus-induced flowering21. CLCrVB, dCLCrV, and dCLCrV: GhFT vectors were generated previously21, and were introduced into A. tumefaciens strain GV3101. A. tumefaciens containing CLCrVB and A. tumefaciens containing dCLCrV or dCLCrV: GhFT were mixed in equal amounts and infiltrated into the cotyledons of GhHB12-overexpressing seedlings at 5 days post germination by syringe infiltration. Inoculated seedlings were covered overnight at 25 °C, and maintained in a growth chamber at 25 °C under long-day conditions (16 h light/8 h dark) for 3 weeks, and then were transplanted into the glasshouse at 26–30 °C for long-day conditions (14 h light/10 h dark).
Supplementary Information
Description of Additional Supplementary Files
Acknowledgements
We thank Professor Brian G. Ayre for providing the dCLCrV and dCLCrV: GhFT, Professor Ye, Zhibiao for providing the A57 tomato seeds. This work was supported by National Key R&D Program of China (Grant no. 2018YFD0100403) and China Agricultural Research System (Grant no. CARS-18-09).
Author contributions
X.H. performed most of the experiments and analyzed the data; T.W. and Z.X. generated some of the hybrid plants; T.W. performed the transient expression of dCLCrV and dCLCrV: GhFT; N.L. and L.W. generated the GhMir157, MIMI157, and GhrSPL10 transgenic plants; Q.H. and X.L. performed the Y2H; L.Z. and X.Z. supervised the project; L.Z., X.Z., and X.H. designed the research. X.H. wrote the manuscript. X.Z. and L.Z. revised the manuscript.
Data availability
The authors declare that all the data generated during this study are available in the manuscript, figures and Supplementary Information Files. Source data underlying the graphs and charts presented in the figures are available in Supplementary Data 2.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s42003-018-0234-0.
References
- 1.Hammer K. The domestication syndrome. Kulturpflanze. 1984;32:11–34. doi: 10.1007/BF02098682. [DOI] [Google Scholar]
- 2.Lenser T, Theissen G. Molecular mechanisms involved in convergent crop domestication. Trends Plant Sci. 2013;18:704–714. doi: 10.1016/j.tplants.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Meyer RS, DuVal AE, Jensen HR. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 2012;196:29–48. doi: 10.1111/j.1469-8137.2012.04253.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, et al. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 2010;52:946–951. doi: 10.1111/j.1744-7909.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- 5.Teotia S, Tang GL. To bloom or not to bloom: role of MicroRNAs in plant flowering. Mol. Plant. 2015;8:359–377. doi: 10.1016/j.molp.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Wang HY. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant. 2015;8:677–688. doi: 10.1016/j.molp.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Si LZ, et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 8.Wang SK, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 9.Jiao YQ, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010;42:541–U536. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 10.Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010;42:545–U102. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, et al. Coordinated regulation of vegetative and reproductive branching in rice. Proc. Natl Acad. Sci. USA. 2015;112:15504–15509. doi: 10.1073/pnas.1521949112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 13.Martin A, et al. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development. 2009;136:2873–2881. doi: 10.1242/dev.031658. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, et al. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Xu M, et al. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006263. doi: 10.1371/journal.pgen.1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackman BK, Strasburg JL, Raduski AR, Michaels SD, Rieseberg LH. The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. 2010;20:629–635. doi: 10.1016/j.cub.2010.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, et al. A 2.833-kb insertion in BnFLC.A2 and its homeologous exchange with BnFLC.C2 during breeding selection generated early-flowering rapeseed. Mol. Plant. 2018;11:222–225. doi: 10.1016/j.molp.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Comadran J, et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012;44:1388–1392. doi: 10.1038/ng.2447. [DOI] [PubMed] [Google Scholar]
- 20.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010;42:459–U138. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 21.McGarry RC, et al. Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytol. 2016;212:244–258. doi: 10.1111/nph.14037. [DOI] [PubMed] [Google Scholar]
- 22.Mouhu K, et al. The Fragaria vesca homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 represses flowering and promotes vegetative growth. Plant Cell. 2013;25:3296–3310. doi: 10.1105/tpc.113.115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natalia PM, Ambrose BA, Litt A. Poppy APETALA1/FRUITFULL orthologs control flowering time, branching, perianth identity, and fruit development. Plant Physiol. 2012;158:1685–1704. doi: 10.1104/pp.111.192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soyk S, et al. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017;49:162–168. doi: 10.1038/ng.3733. [DOI] [PubMed] [Google Scholar]
- 25.Tian ZX, et al. Artificial selection for determinate growth habit in soybean. Proc. Natl Acad. Sci. USA. 2010;107:8563–8568. doi: 10.1073/pnas.1000088107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wigge PA, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, et al. A naturally occurring InDel variation in BraA.FLC.b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 2012;12:151. doi: 10.1186/1471-2229-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gore UR. Morphogenetic studies on the inflorescence of cotton. Bot. Gaz. 1935;97:118–138. doi: 10.1086/334540. [DOI] [Google Scholar]
- 29.Guo YF, McCarty JC, Jenkins JN, Saha S. QTLs for node of first fruiting branch in a cross of an upland cotton, Gossypium hirsutum L., cultivar with primitive accession Texas 701. Euphytica. 2008;163:113–122. doi: 10.1007/s10681-007-9613-1. [DOI] [Google Scholar]
- 30.Li CQ, Zhang JB, Hu GH, Fu YZ, Wang QL. Association mapping and favorable allele mining for node of first fruiting/sympodial branch and its height in Upland cotton (Gossypium hirsutum L.) Euphytica. 2016;210:57–68. doi: 10.1007/s10681-016-1697-z. [DOI] [Google Scholar]
- 31.Zhu L, et al. Genome-wide identification of genes responsive to ABA and cold/salt stresses in Gossypium hirsutum by data-mining and expression pattern analysis. Agric. Sci. China. 2011;10:499–508. doi: 10.1016/S1671-2927(11)60030-8. [DOI] [Google Scholar]
- 32.Franco-Zorrilla JM, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 33.Liu N, et al. Small RNA and degradome profiling reveals a role for miRNAs and their targets in the developing fibers of Gossypium barbadense. Plant J. 2014;80:331–344. doi: 10.1111/tpj.12636. [DOI] [PubMed] [Google Scholar]
- 34.Liu N, et al. MicroRNA 157-targeted SPL genes regulate floral organ size and ovule production in cotton. BMC Plant Biol. 2017;17:7. doi: 10.1186/s12870-016-0969-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, et al. The GhmiR157a/GhSPL10 regulatory module controls initial cellular dedifferentiation and callus proliferation in cotton by modulating ethylene-mediated flavonoid biosynthesis. J. Exp. Bot. 2018;69:1081–1093. doi: 10.1093/jxb/erx475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi A, et al. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell. 2009;17:268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koskela EA, et al. Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol. 2012;159:1043–1054. doi: 10.1104/pp.112.196659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JJ, et al. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012;159:461–478. doi: 10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu S, Huang Z. Inheritance analysis on earliness components of short season cotton varieties in G. hirsutum. Sci. Agric. Sin. 1990;23:48–54. [Google Scholar]
- 40.Fang L, et al. Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 2017;49:1089–1098. doi: 10.1038/ng.3887. [DOI] [PubMed] [Google Scholar]
- 41.Huang C, et al. Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol. J. 2017;15:1374–1386. doi: 10.1111/pbi.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song Q, Zhang T, Stelly DM, Chen ZJ. Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 2017;18:99. doi: 10.1186/s13059-017-1229-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su J, et al. Genome-wide association study identified genetic variations and candidate genes for plant architecture component traits in Chinese upland cotton. Theor. Appl. Genet. 2018;131:1299–1314. doi: 10.1007/s00122-018-3079-5. [DOI] [PubMed] [Google Scholar]
- 44.Wang M, et al. Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 2017;49:579–587. doi: 10.1038/ng.3807. [DOI] [PubMed] [Google Scholar]
- 45.Jung JH, Ju Y, Seo PJ, Lee JH, Park CM. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012;69:577–588. doi: 10.1111/j.1365-313X.2011.04813.x. [DOI] [PubMed] [Google Scholar]
- 46.Borner R, et al. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- 47.Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- 48.Zhang XH, et al. Functional characterization of GhSOC1 and GhMADS42 homologs from upland cotton (Gossypium hirsutum L.) Plant Sci. 2016;242:178–186. doi: 10.1016/j.plantsci.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Hiraoka K, Yamaguchi A, Abe M, Araki T. The florigen genes FT and TSF modulate lateral shoot outgrowth in Arabidopsis thaliana. Plant Cell Physiol. 2013;54:352–368. doi: 10.1093/pcp/pcs168. [DOI] [PubMed] [Google Scholar]
- 50.Pin PA, Nilsson O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 2012;35:1742–1755. doi: 10.1111/j.1365-3040.2012.02558.x. [DOI] [PubMed] [Google Scholar]
- 51.Guo D, et al. Molecular cloning and functional analysis of the FLOWERING LOCUS T (FT) homolog GhFT1 from Gossypium hirsutum. J. Integr. Plant Biol. 2015;57:522–533. doi: 10.1111/jipb.12316. [DOI] [PubMed] [Google Scholar]
- 52.Ariel FD, Manavella PA, Dezar CA, Chan RL. The true story of the HD-Zip family. Trends Plant Sci. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 53.Harris JC, Hrmova M, Lopato S, Langridge P. Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol. 2011;190:823–837. doi: 10.1111/j.1469-8137.2011.03733.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The authors declare that all the data generated during this study are available in the manuscript, figures and Supplementary Information Files. Source data underlying the graphs and charts presented in the figures are available in Supplementary Data 2.