Abstract
Cystic fibrosis (CF) is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Next to progressive airway disease, CF is also associated with intestinal inflammation and dysbiosis. Ivacaftor, a CFTR potentiator, has improved pulmonary and nutritional status but its effects on the intestinal microbiota and inflammation are unclear. Hence, we assessed the changes on the intestinal microbial communities (16S rRNA variable 3 gene region) and inflammatory markers (calprotectin and M2-pyruvate kinase [M2-PK]) in 16 CF individuals (8 children and 8 adults) before and after (median 6.1 months) ivacaftor. Stool calprotectin significantly decreased following ivacaftor (median [IQR]: 154.4 [102.1–284.2] vs. 87.5 [19.5–190.2] mg/kg, P = 0.03). There was a significant increase in Akkermansia with ivacaftor. Increased abundance of Akkermansia was associated with normal stool M2-PK concentrations, and decreased abundances of Enterobacteriaceae correlated with decreased stool calprotectin concentrations. In summary, changes in the gut microbiome and decrease in intestinal inflammation was associated with Ivacaftor treatment among individuals with CF carrying at least one gating CFTR mutation. Thus, CFTR-modifying therapy may adequately improve the aberrant pathophysiology and milieu of the CF gut to favor a more healthy microbiota, which in turn reduces intestinal inflammation.
Introduction
Cystic fibrosis (CF) is a life-shortening autosomal recessive disorder associated with mutations in the gene coding for the cystic fibrosis transmembrane conductance regulator (CFTR) protein1. The CFTR protein plays an important role in epithelial fluid secretion and intra-luminal hydration due to its function as an anion-selective ion channel (mainly chloride and bicarbonate). Defective CFTR leads to accumulation and plugging of inspissated, slow-to-clear mucus on the apical surfaces of epithelial cells in the airway and intestine2. From a microbial perspective, this change in the luminal milieu results in ideal conditions for colonization with opportunistic pathogens. Progressive pulmonary failure secondary to microbial colonization, infection and destructive inflammatory response are hallmarks of CF lung disease2. Similarly, the gastrointestinal tract is affected by significant alterations in the diversity and composition of microbiota in CF patients when compared to healthy controls3–5. This may, in turn, further impact on the host as the gut microbiome plays an important role in host immunology and metabolic capacity6,7. Changes in the CF gut microbiome have therefore been speculated to contribute to the development of intestinal inflammation, gastrointestinal malignancy, liver cirrhosis as well as airway colonization in CF3,8–17. In regards to intestinal inflammation, both stool calprotectin and M2-PK have been previously reported to be elevated in patients with CF compared to healthy controls4,9,13,15,16,18.
Therapeutics in CF have entered an exciting era with the availability of personalized small molecule therapies that target the basic defect(s) in the CFTR protein19. Ivacaftor, a CFTR potentiator for CF patients carrying gating mutations such as G551D, has demonstrated impressive pulmonary outcomes, including reduction in pulmonary exacerbations, and improved lung function20–22. In the gastrointestinal tract, ivacaftor has been reported to improve the abnormal small intestinal pH and histopathologic changes of inspissated mucin within small intestinal crypts seen in CF23,24. In addition, ivacaftor was associated with substantial weight increase, which has been speculated to be related to improvements in gastrointestinal physiology rather than solely by improved pulmonary function10,18,23–25. However, the impact of such therapies on the gut microbiome and intestinal inflammation as biological markers of improved intestinal physiology in CF remains unclear.
We hypothesize that improvements in gastrointestinal physiology following commencement of ivacaftor therapy, in patients with CF carrying at least one gating CFTR mutation, is associated with changes in the gut microbiome and inflammation. In this study, we examined the gut microbial communities and gut inflammation (using two biomarkers of inflammation – calprotectin and M2-pyruvate kinase [M2-PK]) in children and adults with CF before and after commencement of ivacaftor. As secondary aim, we evaluated the relationships between microbial profiles and gut inflammatory markers calprotectin and M2-PK.
Results
Subject characteristics
Stool samples were collected from 16 patients (8 children and 8 adults; 10 females and 6 males) with CF at baseline and after commencement of ivacaftor therapy. There were 14 pancreatic insufficient and 2 pancreatic sufficient patients. The genotype and exocrine pancreatic phenotype of these CF patients are summarized in Table 1.
Table 1.
Subject demographics and clinical characteristics, including changes in pulmonary function, sweat chloride, faecal calprotectin and M2-PK levels following use of ivacaftor.
| Age | Sex | Race/Ethnic | Site | Allele 1 | Allele 2 | EPF | Δ FEV1pp | Δ SC (mmol/L) | Δ Calprotectin (mg/kg) | Δ M2-PK (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | F | White/Hispanic | S | G551D | DF508 | PI | 24 | −46 | −25 | −0.5 |
| 5 | F | White | S | G551D | DF508 | PI | 12 | −5 | 212 | 196.5 |
| 6 | F | White | T | G551D | DF508 | PI | 1 | −38 | −32 | −2.4 |
| 8 | F | White | S | G551D | Q220X | PI | 23 | −76 | −46 | −4.2 |
| 8 | M | White | S | G551D | DF508 | PI | N/A | N/A | −104 | −5 |
| 9 | M | White | T | G551D | DF508 | PI | −2 | −30 | −205 | −886.2 |
| 12 | F | White | T | G551D | DF508 | PI | 10 | −46 | −333 | −2.6 |
| 15 | F | White | S | G551D | DF508 | PI | 0 | −61 | −149 | 8.1 |
| 19 | M | White | T | G551D | DF508 | PI | 32 | −31 | −33 | 196.5 |
| 22 | F | White | T | G178R | DF508 | PI | 24 | −51 | −367 | −2.4 |
| 23 | M | White | T | G551D | DF508 | PI | N/A | −35 | −631 | −13.9 |
| 32 | M | White | T | G551D | E585X | PI | 19 | −4 | 208 | −4.2 |
| 33 | F | White | T | G551D | DF508 | PI | 24 | −66 | −92 | −5 |
| 40 | M | White | T | G551D | Unknown | PS | 9 | −33 | −16 | −886.2 |
| 44 | F | White | T | G551D | DF508 | PI | 16 | −49 | 1 | −2.6 |
| 50 | F | White | T | G551D | Unknown | PS | 10 | −31 | −168 | 8.1 |
EPF = exocrine pancreatic function; F = females; M = males; N/A = not available; PI = pancreatic insufficient; PS = pancreatic sufficient; S = Sydney, Australia; T = Toronto, Canada; Δ Calprotectin = change in stool calprotectin levels following ivacaftor; Δ FEV1pp = change in percent predicted forced expiratory volume in 1 second following ivacaftor; Δ M2-PK = change in stool M2-pyruvate kinase levels following ivacaftor; Δ SC = change in sweat chloride following ivacaftor.
Overall, the median [IQR] age of participants was 17.1 [7.8–32.7] years. Median [IQR] age at time of baseline sample collection of pediatric patients was 7.8 [6.2–11.5] years and median [IQR] age at first sample of adult patients was 32.3 [21.8–42.9] years. Stools post-commencement of ivacaftor treatment were collected at a median [IQR] of 6.1 [5.6–8.2] months.
Clinical characteristics
The pediatric patients showed an increase in weight z-score from baseline to the time of repeat stool collection while on ivacaftor therapy (median [IQR]: 0.32 [−1.89–0.84] vs. 0.66 [−0.87–1.43]; P = 0.012). There was no significant change in height z-scores in the pediatric population (median [IQR]: 0.39 [−1.53–0.80] vs. 0.11 [−1.58–0.89; P = 0.89].
In adult patients, there was a significant increase in body mass index (median [IQR]: 22.4 [21.2–28.2] vs. 23.9 [22.2–28.9] kg/m;2 P = 0.028) following commencement of ivacaftor. Furthermore, there was a significant increase in weight in these patients (median [IQR]: 64.1 [54.4–86.7] vs. 65.9 [59.0–88.0] kg; P = 0.028).
There was a significant elevation in the percent predicted forced expiratory volume in 1 second (FEV1pp) from baseline to time of repeat stool collection while on ivacaftor (median [IQR]: 76.0 [56.3–96.0] vs. 90.5 [72.5–108.5]; P = 0.002). Sweat chloride concentrations significantly reduced after starting ivacaftor therapy (median [IQR]: 86 [77–91] vs. 39 [36–53]; P = 0.001).
Stool calprotectin and M2-PK
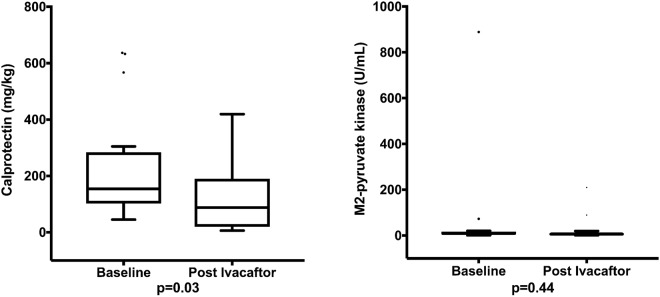
There was a significant reduction in stool calprotectin levels after commencement of ivacaftor (median [IQR]: 154.4 [102.1–284.2] vs. 87.5 [19.5–190.2] mg/kg, P = 0.03) (Fig. 1A). In contrast, there was no significant change in stool M2-PK levels from before to after commencement of treatment (median [IQR]: 8.4 [4.8–17.1] vs. 6.2 [2.8–13.3] U/mL, P = 0.44) (Fig. 1B).
Figure 1.
Comparison of fecal (A) calprotectin and (B) M2-PK before and after ivacaftor. Data are summarized as whisker-box plots presenting median (with interquartile range and range).
Bacterial microbiome before and after CFTR modulation
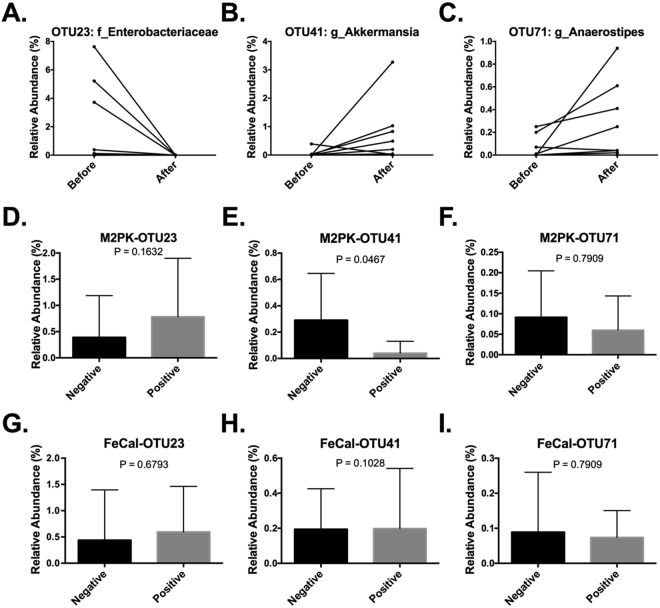
Stool samples from before and after treatment with ivacaftor were processed for microbiome profiling using 16S rRNA variable 3 (v3) gene region amplicon sequencing. To test whether any members of the gastrointestinal microbiota were correlated with improved gut health, we conducted Analysis of Composition of Microbiomes26. Following treatment, one operational taxonomic unit (OTU) of the family Enterobacteriaceae decreased while two OTUs of the genera Akkermansia and Anaerostipes increased (Fig. 2A–C). After adjusting for the false discovery rate, only Akkermansia was significantly increased following commencement of therapy (Fig. 2B).
Figure 2.
Altered levels of Enterobacteriaceae (OTU23), Akkermansia (OTU41), and Anaerostipes (OTU71) following ivacaftor therapy. (A–C) All operational taxonomic units (OTUs) with greater than 10 reads were analyzed for significant differences following ivacaftor therapy using ANCOM. (D–I) Relative abundances of relevant OTUs were compared according to categorically negative (normal) or positive (abnormal) stool levels of M2PK and calprotectin for all stool samples (combined before and after ivacaftor). Normal values for faecal calprotectin and M2-PK were based on cut-offs of ≤50 mg/kg and ≤9U/ml. Means and 95% confidence intervals of each OTU’s relative abundance are shown.
Stool bacterial microbiome were not significantly different in β-diversity and α-diversity after CFTR modulation, or by sex (Supplemental Fig. S1). However, they were statistically different (R2 = 0.09945, P = 0.00148) by location of the clinical centers (Supplemental Fig. S2).
Bacterial microbiome – correlation analyses in all stool samples
Bacterial relative abundances were compared according to categorically negative (normal) or positive (abnormal) stool levels of M2PK and calprotectin for all stool samples. A higher abundance of Akkermansia was significantly associated with negative stool M2-PK levels (M2-PK <9.1U/ml) (Fig. 2E). There were no associations between the relative abundances of Enterobacteriaceae and Anaerostipes with M2-PK levels (Fig. 2D,F). There were also no association between relative bacterial abundances and negative or positive calprotectin levels (Fig. 2G–I). However, on linear regression analysis, there was a significant association between stool calprotectin levels and the family Enterobacteriaceae; decreased relative abundances of Enterobacteriaceae was associated with reduced levels of stool calprotectin (Fig. 3A). There was no association between stool calprotectin levels and Akkermansia and Anaerostipes (Fig. 3B,C).
Figure 3.
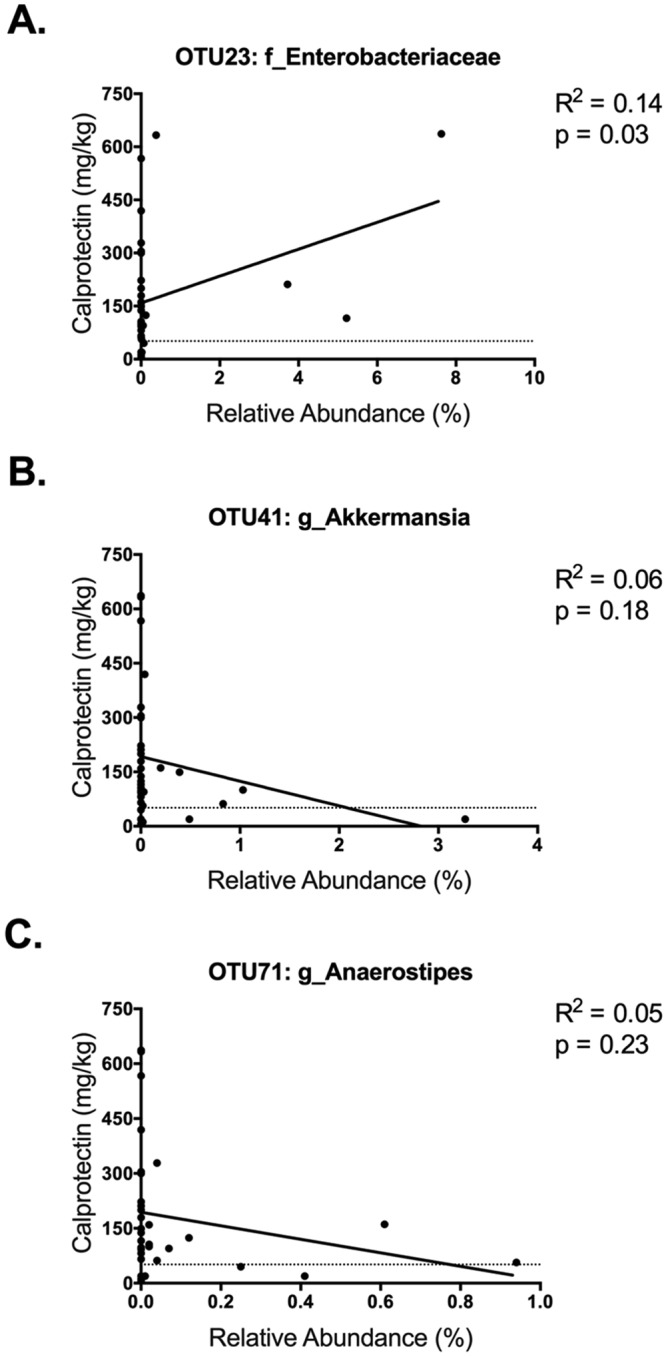
Correlations between stool calprotectin and relative abundances of (A) Enterobacteriaceae (OTU23), (B) Akkermansia (OTU41), and (C) Anaerostipes (OTU71).
Discussion
To our knowledge, this is the first study to demonstrate that treatment with ivacaftor improves intestinal inflammation in individuals with CF carrying at least one gating CFTR mutation. In this prospective observational study, we also found significant correlation between stool calprotectin levels and Enterobacteriaceae abundance, and that treatment with ivacaftor is associated with an increase in the relative abundance of Akkermansia.
Many studies have previously reported the presence of intestinal inflammation in CF patients, particularly with significant elevations in stool calprotectin and M2-PK levels in patients with CF compared to healthy controls4,9,13,15,16,18. The underlying pathogenesis of intestinal inflammation in CF remain unclear but speculated to be related to physico-chemical changes in the intestinal milieu, which is primarily due to the dysfunction in CFTR3,11,12,27, that may in turn favor alterations in the gut microbial community. In this observational study, ivacaftor therapy led to a reduction in stool calprotectin suggesting that the intestinal inflammation may be improved or reversible in patients with CF upon restoration of the intestinal millieu.
With the substantial increase in CF patients surviving into adulthood, the number of patients being diagnosed with gastrointestinal malignancies has increased17. The underlying pathogenic mechanisms remain unclear but chronic gastrointestinal inflammation in CF, which begins in early childhood16, has been shown to promote oncogenesis28,29. The impact of novel CFTR modulator therapies on the incidence of gastrointestinal cancers in patients with CF if of great interest and will require future studies.
The altered microbiome in the CF gut is characterized by a high abundance of Enterobacteriaceae, particularly Escherichia coli, which has been shown to be 10 times higher in CF compared to healthy controls8. Similarly, the Enterobacteriaceae family have also been reported to dominate the gut microbial profile in CFTR−/− mice30. Ferrets models of CF demonstrated intestinal overgrowth of E. coli at 2.5–7.5 times higher compared to controls31. Following treatment with ivacaftor, we have demonstrated a weak effect of decreased abundance of the Enterobacteriaceae family. We speculate that a statistically significant effect was not observed following false discovery rate adjustment as the Enterobacteriaceae family not only includes the familiar and well-established Gram-negative pathogens such as E. coli but also many harmless symbionts. We nevertheless propose that there was selective reduction in abundances of pathogenic members of the Enterobacteriaceae family (and thus sparing the non-pathogenic symbionts) following ivacaftor therapy due to a positive correlation between stool calprotectin levels and the abundances of Enterobacteriaceae in our cohort. This finding is consistent with a previous study reporting significant association between stool calprotectin and E. coli abundance8. This observation further supports the close link between alterations in the gut bacterial microbiome and the development of intestinal inflammation in patients with CF. Lastly, we demonstrate that modulation of intestinal CFTR function with ivacaftor and thus modification of the intestinal luminal milieu23, reverses some of the dysbiosis and inflammation in CF patients.
In our study cohort, members of the bacterial genus Akkermansia were significantly increased during therapy. Akkermansia is a Gram-negative, mucin-degrading bacteria that resides in the intestinal mucus layer32. There has been emerging interest in Akkermansia due to the association between its abundance with a healthy gut mucosa. In previous studies, the abundance of Akkermansia has been inversely correlated with intestinal inflammatory conditions, such as inflammatory bowel disease, microscopic colitis and appendicitis33–36. While its protective mechanisms remain to be fully elucidated, Akkermansia reportedly stimulates host mucosal anti-inflammatory pathways, and improves epithelial barrier integrity37–41. In addition, Akkermansia increases the expression of RegIII, an anti-microbial peptide with direct bactericidal activity against Gram-positive bacteria in the intestine40. In our study the increased abundance of Akkermansia, which is a biomarker of gut health, was significantly correlated with normal levels of stool M2-PK42. The underlying mechanism(s) for increased abundance of Akkermansia in CF following ivacaftor therapy is unknown but we speculate that this may be related to the improvement of the characteristics of the naïve CF intestinal mucus. More specifically, improved bicarbonate secretion by CFTR following ivacaftor use may have created a more amenable environment for mucin degraders such as Akkermansia, due to improved unfolding and maturation of intestinal mucins43,44.
It is notable that the overall β-diversity and α-diversity were not significantly different following CFTR modulation with ivacaftor. This suggests that the alterations in gut microbiome in CF may also be affected by non-CFTR related factors, such as the high-calorie “CF diet”. While the life-prolonging benefits of the CF diet is irrefutable, a recent study has reported that many patients with CF are achieving their dietary targets through increased consumption of ‘junk foods’ high in energy and saturated fat45. Differences in bacterial profiles according to geographic location in our cohort supports the theory that ‘environmental’ factors, such as variations in CF care, and potential variations in diet and attitudes toward diet between Australia and Canada, influence the CF gut microbiome.
Our study has several limitations. The sample size was relatively small despite a multicenter approach and an extended follow-up period of a median of 6 months. This limits our study’s power and therefore our ability to do more extensive analysis and correlations with further clinical metadata. Furthermore, we may have missed any further alteration of the microbiome, which may occur with longer treatment time. Future validation studies based on a larger cohort and multiple sampling with dietary data over a longer period are needed. We also cannot determine from our current analysis whether any direct mechanisms of interaction exist or if our observations are due to indirect effects. To achieve such mechanistic insights, future work will require conducting experiments in in vitro CF models to evaluate the interactions between host (genetic, immune and mucosal) factors and microbial communities46.
In conclusion, ivacaftor therapy positively affects the gut microbiome as well as improves intestinal inflammation in individuals with CF carrying at least one gating CFTR mutation.
Methods
Patient cohort
Patients with CF carrying at least one gating CFTR mutation who were commencing treatment with ivacaftor at The Hospital for Sick Children (Toronto, CA), St Michael’s Hospital (Toronto, CA) and Sydney Children’s Hospital Randwick (Sydney, AU) were included in this study. Patients were excluded if they had a pre-existing gut disease.
Stool and clinical data collection
Paired stool samples were collected from all patients before and after commencing ivacaftor therapy. Sample collection was were deferred/rejected if they had gastroenteritis or administered corticosteroids or non-steroidal ant-inflammatory drugs within two weeks of collection. All samples were stored at −20 °C until transfer to the laboratory, where they were then stored at −80 °C. All samples were processed together for microbial analysis. Clinical data including anthropometrics and FEV1pp were collected at time of stool collection. Sweat chloride prior and after commencement of ivacaftor were also recorded.
Stool inflammatory markers
Calprotectin was extracted and measured from frozen stool samples using the PhiCal kit (Calpro, San Diego, CA, US), according to manufacturer instructions. Fecal M2-PK was extracted and measured from stool samples using the ScheBo Tumour M2-PK test and protocol (ScheBo Biotech, Giessen, Germany). The lower limits of detection for the assays were calprotectin 5 mg/kg and M2-PK 1 U/mL. Normal values for faecal calprotectin and M2-PK were based on cut-offs of ≤50 mg/kg and ≤9U/ml respectively47,48.
DNA extraction and genomic sequencing
DNA was extracted using the Thermo Fisher Scientific MagMAX Sample Preparation System (Waltham, MA). The 16S rRNA variable 3 (v3) gene region was amplified as previously described49,50. Amplification protocols were adapted from Whelan et al.51. Each sample reaction mixture contained 5pmol of each primer, 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, and 1U Taq polymerase (Life Technologies, Carlsbad, CA). The PCR protocol consisted of an initial denaturation step at 95 °C for 5 minutes, 30 cycles, each step for 30 seconds, of 95 °C, 50 °C, and 72 °C, and a final extension step at 72 °C for 7 minutes. Previously described 341 F and 518 R 16s rRNA primers, modified for the Illumina platform (San Diego, CA) with 6-base pair unique barcode additions to the reverse primer, were utilized in this protocol50. Amplification of the v3 gene region was verified by electrophoresis on a 2% agarose gel. Amplicons were then sequenced using the Illumina MiSeq platform.
Microbial composition and statistical analysis
Resulting paired-end sequences from Illumina sequencing are available on the Sequence Read Archive (SRP#162699). Sequences were processed using a custom in-house pipeline51. Briefly, this pipeline uses Cutadapt52 to trim any reads surpassing the length of the v3 region, PANDAseq53. to align the paired-end sequences, Sickle to stringently quality-filter sequences (minimum average quality of 30)54, QIIME to size filter for sequences between 100–250 base pairs and to check for and remove chimeras55, AbundantOTU+ (clustering threshold of 97%)56 to pick operational taxonomic units (OTUs), and Ribosomal Database Project classifier to assign taxonomy using Greengenes reference database (2011 release)57,58.
After removing singleton OTUs (those with only 1 read across all samples) and any OTU that did not have a bacterial taxonomic assignment, 3,426,702 total reads (average: 95,186 reads/sample; range: 43,688- 132,742) with 1683 total OTUs (average: 263 OTUs/sample; range: 131–443) remained. To measure β-diversity, samples were normalized using proportional abundance59 and the Bray-Curtis metric was used. To measure α-diversity, samples were rarefied to 40,000 reads. Differences in β-diversity were assessed with the adonis function in the vegan package. Identification of taxa changes associated treatment was conducted using the Analysis of Composition of Microbiomes (ANCOM)26,after filtering our dataset to remove OTUs with less than 10 reads. Remaining analysis was conducted on phyloseq60, and vegan61 packages on R62 and plotted using GraphPad Prism version 6.0 for Mac OS X (La Jolla, CA). Differences in α-diversity before and after therapy were assessed using paired t-tests.
Ethics approval and consent to participate
The research ethics boards of the participating academic institutions approved the study (SickKids #1000036224, St. Michael’s Hospital #13-089, South Eastern Sydney Area Health Service #10/240), which was in compliance with the ethical principles outlined in the declaration of Helsinki. All participants, parents and/or legal guardian signed an informed consent prior to study enrolment.
Electronic supplementary material
Acknowledgements
The authors are highly grateful to all the participants in the study. We thank the following individuals at the CF Clinics at The Hospital for Sick Children (Dr. Felix Ratjen), St Michael’s Hospital (Katherine Griffin, Dr. Elizabeth Tullis) and Sydney Children’s Hospital Randwick (Rhonda Bell, Amanda Thomsen and Drs. Yvonne Belessis, Penny Field, Adam Jaffe and John Widger). We would like to thank Michelle Shah for her tireless effort in managing and processing the stool samples utilized in this study and the McMaster Genomics Facility for sequencing the samples.
Author Contributions
All authors meet requirements for authorship having contributed significantly to the project. C.Y.O. and T.G. designed the study. C.Y.O., T.G., J.A. collected patient samples. S.A.S., L.R., M.G.S. performed works related to stool microbial analysis, statistics and/or interpretation. K.Y., M.G., B.N. performed works related to stool biomarker analysis. M.G. performed statistical analysis. C.Y.O. and S.A.S. drafted the manuscript. All authors contributed to editing, and have reviewed the final version of the manuscript and consent to its publication.
Data Availability
Sequencing data are available on the Sequence Read Archive (SRP#162699).
Competing Interests
C.Y.O. is a consultant for Vertex Pharmaceuticals (unrelated to the current manuscript). The remaining authors declare no conflict of interest. T.G. received funding from Vertex Pharmaceuticals to perform stool DNA extraction, genomic sequencing and microbial composition analysis. Vertex Pharmaceuticals had no involvement in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chee Y. Ooi and Saad A. Syed contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36364-6.
References
- 1.Riordan JR, et al. Identification of the cystic fibrosis gene: Cloning and characterization of complementary DNA. Science. 1989;245:1066. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.O’Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. 2009;373(9678):1891–904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 3.Manor O, et al. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci Rep. 2016;6:22493. doi: 10.1038/srep22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruzzese E, et al. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS One. 2014;9:e87796. doi: 10.1371/journal.pone.0087796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen S, et al. Disrupted progression of the intestinal microbiota with age in children with cystic fibrosis. Sci Rep. 2016;6:24857. doi: 10.1038/srep24857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–9. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman LR, et al. Escherichia coli Dysbiosis Correlates With Gastrointestinal Dysfunction in Children With Cystic Fibrosis. Clin Infect Dis. 2014;58:396–399. doi: 10.1093/cid/cit715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pang T, et al. Elevated fecal M2-pyruvate kinase in children with cystic fibrosis: A clue to the increased risk of intestinal malignancy in adulthood? J Gastroenterol and Hepatol. 2015;30:866–71. doi: 10.1111/jgh.12842. [DOI] [PubMed] [Google Scholar]
- 10.Garg M, Ooi CY. The Enigmatic Gut in Cystic Fibrosis: Linking Inflammation, Dysbiosis, and the Increased Risk of Malignancy. Curr Gastroenterol Rep. 2017;19(2):6. doi: 10.1007/s11894-017-0546-0. [DOI] [PubMed] [Google Scholar]
- 11.Ooi CY, Durie PR. Cystic fibrosis from the gastroenterologist’s perspective. Nat Rev Gastroenterol Hepatol. 2016;13:175–85. doi: 10.1038/nrgastro.2015.226. [DOI] [PubMed] [Google Scholar]
- 12.Debyser G, et al. Faecal proteomics: A tool to investigate dysbiosis and inflammation in patients with cystic fibrosis. J Cyst Fibros. 2016;15:242–50. doi: 10.1016/j.jcf.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Flass T, et al. Intestinal lesions are associated with altered intestinal microbiome and are more frequent in children and young adults with cystic fibrosis and cirrhosis. PLoS One. 2015;10(2):e0116967. doi: 10.1371/journal.pone.0116967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madan JC, et al. Serial Analysis of the Gut and Respiratory Microbiome in Cystic Fibrosis in Infancy: Interaction between Intestinal and Respiratory Tracts and Impact of Nutritional Exposures. mBio. 2012;3:e00251–12. doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garg M, et al. Age-dependent variation of fecal calprotectin in cystic fibrosis and healthy children. J Cyst Fibros. 2017;16(5):631–636. doi: 10.1016/j.jcf.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Garg M, et al. Age-related levels of fecal M2-pyruvate kinase in children with cystic fibrosis and healthy children 0 to 10years old. J Cyst Fibros. 2018;17(1):109–113. doi: 10.1016/j.jcf.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Yamada A, et al. Risk of gastrointestinal cancers in patients with cystic fibrosis: a systematic review and meta-analysis. Lancet Oncol. 2018;19(6):758–767. doi: 10.1016/S1470-2045(18)30188-8. [DOI] [PubMed] [Google Scholar]
- 18.Dhaliwal J, et al. Intestinal inflammation and impact on growth in children with cystic fibrosis. J Pediatr Gastroenterol Nutr. 2015;60:521–6. doi: 10.1097/MPG.0000000000000683. [DOI] [PubMed] [Google Scholar]
- 19.Boyle MP, De Boeck K. A new era in the treatment of cystic fibrosis: correction of the underlying CFTR defect. Lancet Respir Med. 2013;1(2):158–63. doi: 10.1016/S2213-2600(12)70057-7. [DOI] [PubMed] [Google Scholar]
- 20.Ramsey BW, et al. A CFTR Potentiator in Patients with Cystic Fibrosis and the G551D Mutation. N Eng. J Med. 2011;365:1663–72. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies JC, et al. Efficacy and safety of ivacaftor in patients aged 6 to 11 years with cystic fibrosis with a G551D mutation. Am J Respir Crit Care Med. 2013;187(11):1219–25. doi: 10.1164/rccm.201301-0153OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Accurso FJ, et al. Effect of VX-770 in persons with cystic fibrosis and the G551D-CFTR mutation. N Engl J Med. 2010;363(21):1991–2003. doi: 10.1056/NEJMoa0909825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rowe SM, et al. Clinical Mechanism of the Cystic Fibrosis Transmembrane Conductance Regulator Potentiator Ivacaftor in G551D-mediated Cystic Fibrosis. Am J Respir Crit Care Med. 2014;190:175–84. doi: 10.1164/rccm.201404-0703OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safe M, Gifford AJ, Jaffe A, Ooi CY. Resolution of Intestinal Histopathology Changes in Cystic Fibrosis after Treatment with Ivacaftor. Ann Am Thorac Soc. 2016;13:297–8. doi: 10.1513/AnnalsATS.201509-634KV. [DOI] [PubMed] [Google Scholar]
- 25.Wainwright CE, et al. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N Engl J Med. 2015;373(3):220–231. doi: 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandal S, et al. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schippa S, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) allelic variants relate to shifts in faecal microbiota of cystic fibrosis patients. PLoS One. 2013;8(4):e61176. doi: 10.1371/journal.pone.0061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Than BLN, et al. CFTR is a tumor suppressor gene in murine and human intestinal cancer. Oncogene. 2016;35:4179–4187. doi: 10.1038/onc.2015.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arthur JC, et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science. 2012;338:120–3. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norkina O, Burnett TG, De Lisle RC. Bacterial Overgrowth in the Cystic Fibrosis Transmembrane Conductance Regulator Null Mouse Small Intestine. Infect Immun. 2004;72:6040–9. doi: 10.1128/IAI.72.10.6040-6049.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun X, et al. Gastrointestinal pathology in juvenile and adult CFTR-knockout ferrets. Am J Pathol. 2014;184:1309–22. doi: 10.1016/j.ajpath.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(Pt 5):1469–76. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 33.Kugathasan S, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet. 2017;389(10080):1710–1718. doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Png CW, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105(11):2420–8. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 35.Fischer H, et al. Altered microbiota in microscopic colitis. Gut. 2015;64(7):1185–6. doi: 10.1136/gutjnl-2014-308956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swidsinski A, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60(1):34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 37.Shin NR, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63(5):727–35. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 38.Cantarel BL, et al. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med. 2015;63(5):729–34. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneeberger M, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Everard A, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110(22):9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pang T, Leach ST, Katz T, Day AS, Ooi CY. Fecal biomarkers of intestinal health and disease in children. Front Pediatr. 2014;2:6. doi: 10.3389/fped.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schütte A, et al. Microbial-induced meprin β cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc Natl Acad Sci USA. 2014;111(34):12396–401. doi: 10.1073/pnas.1407597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambort D, et al. Calcium and pH-dependent packing and release of the gel-forming MUC2 mucin. Proc Natl Acad Sci USA. 2012;109(15):5645–50. doi: 10.1073/pnas.1120269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutherland R et al. Dietary intake of energy-dense, nutrient-poor and nutrient-dense food sources in children with cystic fibrosis. J Cyst Fibros. pii: S1569–1993(18)30083-3 (2018). [DOI] [PubMed]
- 46.Roeselers G, Ponomarenko M, Lukovac S, Wortelboer HM. Ex vivo systems to study host-microbiota interactions in the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2013;27(1):101–13. doi: 10.1016/j.bpg.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 47.NICE guidelines. Faecal calprotectin diagnostic tests for inflammatory diseases of the bowel; DAP 12 (2013). https://www.nice.org.uk/guidance/dg11.
- 48.Joshi S, Lewis SJ, Creanor S, Ayling RM. Age-related faecal calprotectin, lactoferrin and tumour M2-PK concentrations in healthy volunteers. Ann Clin Biochem. 2009;47:259–263. doi: 10.1258/acb.2009.009061. [DOI] [PubMed] [Google Scholar]
- 49.Syed SA, et al. Reemergence of Lower-Airway Microbiota in Lung Transplant Patients with Cystic Fibrosis. Ann Am Thorac Soc. 2016;13(12):2132–2142. doi: 10.1513/AnnalsATS.201606-431OC. [DOI] [PubMed] [Google Scholar]
- 50.Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl Environ Microbiol. 2011;77(11):3846–52. doi: 10.1128/AEM.02772-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whelan FJ, Surette MG. A comprehensive evaluation of the sl1p pipeline for 16S rRNA gene sequencing analysis. Microbiome. 2017;5(1):100. doi: 10.1186/s40168-017-0314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 53.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joshi N., Fass J. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) (2011).
- 55.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ye, Y. Identification and Quantification of Abundant Species from Pyrosequences of 16S rRNA by Consensus Alignment. Proceedings (IEEE Int Conf Bioinformatics Biomed). 2010, 153–157 (2011). [DOI] [PMC free article] [PubMed]
- 57.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10(4):e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McMurdie PJ, Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oksanen J. Multivariate Analysis of Ecological Communities in R: vegan tutorial. 83, 922, http://cc.oulu.fi/jarioksa/opetus/metodi/vegantutor.pdf (2011).
- 62.R Development Core Team (2011). R: A language and environment for statistical computing. The R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available on the Sequence Read Archive (SRP#162699).