Abstract
Heliothis zea nudivirus-1 (HzNV-1) is an insect virus that can induce both lytic and latent infections in various insect cell lines. During latent infection, several microRNAs (miRNAs) are produced from persistency-associated gene 1 (pag1) as the only detectable HzNV-1 transcript. Previous studies have shown that the pag1 gene suppresses the immediate-early gene hhi1 and promotes host switching into a latent infection via miRNAs derived from pag1. Although other functions of the miRNAs derived from pag1 have not yet been elucidated, several studies have suggested that miRNAs encoded from latency-associated genes can regulate histone-associated enzymes. Because pag1 is a noncoding transcript, it potentially regulates host chromatin structure through miRNAs upon infection. Nevertheless, the exact mechanism by which pag1 alters viral infections remains unknown. In this study, we found that the pag1-encoded miRNA miR-420 suppresses expression of the histone modification-associated enzyme su(var)3–9. Therefore, this miRNA causes histone modification to promote HzNV-1 infection. These results suggest that HzNV-1 may directly influence epigenetic regulation in host cells through interactions with pag1 miRNAs to promote lytic infection. This study provides us with a better understanding of both the HzNV-1 infection pathway and the relationship between viral miRNAs and epigenetic regulation.
Introduction
Heliothis zea nudivirus-1 (HzNV-1), an insect virus with a broad host range1, has a circular double-stranded DNA genome that encodes approximately 154 open reading frames2. This virus was previously classified in the Baculoviridae family based on similarity with baculovirus in regard to structure and replication mode. However, HzNV-1 was later placed in the family Nudiviridae due to its lack of occlusion bodies and higher genomic homology with nudiviruses than baculovirus3. We previously reported that HzNV-1 produces more than 100 transcripts in productive infection, with the immediate-early gene hhi1 generating high levels of a 6.2-kb transcript4. It is known that hhi1 can serve as a transcriptional activator and that it is involved in viral reactivation from latently infected cells5. In contrast to the expression pattern of most immediate-early (IE) genes of baculovirus, expression of hhi1 was recently shown to require the assistance of viral factors5. Only one gene transcript, named persistency-associated transcript (PAT1), is detectable from persistency-associated gene 1 (pag1) during latent viral infection, and this unique noncoding RNA was found to be responsible for, or at least involved in, the establishment of latent viral infection4,6.
The pag1 transcript comprises a series of microRNAs (miRNAs), with no sequences translated into protein. Wu et al. observed that several miRNAs encoded by pag1 inhibit hhi1 expression, resulting in a higher proportion of latent than lytic infection7. However, the functions of other miRNAs have not been elucidated to date. Small RNA molecules (approximately 22 nucleotides) that form stem-loop structures and function as silencers, miRNAs downregulate gene expression, in some cases by targeting the promoter of messenger RNAs (mRNAs)8,9. Many miRNA functions have been identified in insects10. For instance, miRNAs have been discovered to be involved in metabolism11,12, social behavior13 and interactions between viruses and hosts14,15. Viruses, including Heliothis virescens ascovirus (HvAV)16 and West Nile virus (WNV), have also been observed to employ their own miRNAs to regulate viral replication17.
Epigenetic regulation involves histone modification18, DNA methylation/acetylation19, and miRNA expression20. Recently, Kaposi’s sarcoma-associated herpesvirus (KSHV) has been shown to regulate its lytic/latent cycle through histone modification21, and active or repressive histone markers are reportedly distributed across the entire latent KSHV genome, suggesting that this phenomenon is dynamic22,23. Histone H3-lysine24 trimethylation (H3K27me3), the dominant repressive marker, covers the majority of the KSHV genome and suppresses lytic gene expression. In addition to KSHV, almost all human herpesviruses (HSV, KHSV and HCMV) encode viral miRNAs that inhibit IE gene expression to regulate lytic and latent infection25,26. Methylation and acetylation of histone tails are carried out by DNA methyltransferases (DNMTs) and histone acetyltransferases (HATs), respectively24,27. There are two forms of chromatin: heterochromatin and euchromatin. The former is defined as condensed chromatin in which transcription is blocked, whereas euchromatin is defined as lightly packed, unstainable chromatin that is often present during active transcription18,28. Histone-lysine N-methyltransferase Su(var) function is related to heterochromatin with position-effect variegation (PEV)29,30, and one member of the su(var) group, su(var)3–9, that causes histone H3-lysine9 (H3K9) methylation has been extensively studied in Drosophila and mammals31,32. Furthermore, su(var) is present in Spodoptera frugiperda and two additional Spodoptera species and is associated with the silencing of viral gene expression through interactions with epigenetic factors33. Another gene, tip60, encoding a HAT is also associated with pathogenic infection in Lepidoptera insects34.
HSVs have been found to modify host histone tails during virus infection. As these reports primarily focused on the promoters of lytic and latent genes and assessed histone methylation associated with gene suppression8, sites with higher levels of methylation during the latent stage have been well studied8,35. In contrast, studies of epigenetic regulation in viruses are still limited. Therefore, we used the pag1 gene, which produces the only detectable gene transcript during latent HzNV-1 infection, as a model system to study whether pag1-encoded miRNAs directly affect host histone modification. The results of this study suggest that this is not the case. We chose HzNV-1, which has features similar to those of HSVs, as our model system. We first observed a decline in H3K9me3 levels, in contrast to other reports, whereas acetylation levels were significantly enhanced by the presence of the pag1 gene. We next identified two histone transferases that might be responsible for these phenomena, su(var)3–9 and tip60. We demonstrated that su(var)3–9 was downregulated by a miRNA encoded from pag1: miR-420. We found evidence supporting that HzNV-1 uses its latent infection-associated miRNAs to control host histone modification and promote lytic infection. We propose a model that accounts for the latent infection of insect viruses and can also be applied to mammalian viruses.
Results
Histone modification detected at different time points after pag1 transfection
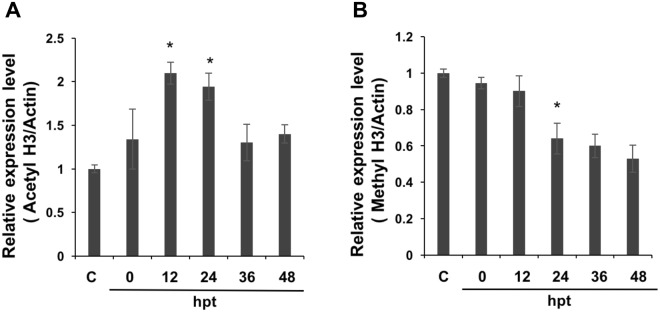
As previous studies have shown that pag1 regulated viral gene expression and influenced the type of infection5, the association between the pag1 gene and host histone status was therefore examined. Since the amino-terminal tail of histone 3 has been studied thoroughly in recent years, the histone 3 modification pattern was chosen as a reference for examining changes in histone modifications caused by pag1 using acetylation H3 and trimethylated H3K9 antibodies (Fig. 1). After pag1 was transfected into cells, strong acetylation was observed up to 36 hour post-transfection (hpt). The acetylation levels of pag1 samples were higher than those of the control group throughout the experiment and peaked at 24 hours post-transfection (Fig. 1A); conversely, methylation in host cells decreased after transfection (Fig. 1B). These data suggest that the pag1 gene may encode factor(s), which would upregulate gene expression. Although previous studies have shown that methylation levels of lytic gene transcripts may increase during latent infection, the distribution of epigenetic markers is suggested to be dynamic across the entire host genome during the latent stage.
Figure 1.
Western blot detection of histone 3 acetylation/methylation levels after pag1 transfection. (A) Acetylation levels of H3 were detected with an anti-acetyl-H3 antibody. (B) Methylation levels were detected with an anti-trimethyl-H3K9 antibody. Samples were extracted using RIPA buffer. The band areas were quantified to generate the upper bar charts. The samples were normalized to actin, and the control group was set as 1. Mean and standard deviation (SD) values are shown, and P values were calculated using Student’s t-test (*P < 0.05). All experiments were performed with three replicates.
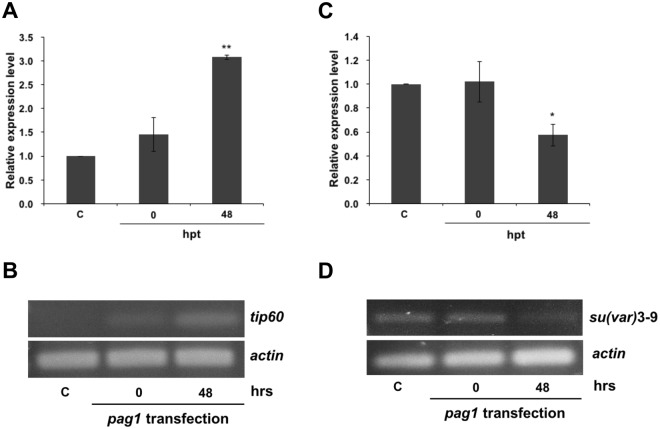
To further confirm that pag1 was able to alter host histone modifications, two histone-associated enzymes, HATs tip60 and su(var)3–9, were chosen to explore the role of the pag1 gene in the histone modification pathway. The expression levels of HAT tip60 and su(var)3–9 at 0 to 48 hours post pag1 transfection were measured. Previous studies have revealed that HAT tip60 is associated with H3 acetylation, and the result here showed that tip60 expression was increased upon pag1 transfection (Fig. 2B), which was in agreement with the western blot results (Fig. 1B) and previous findings, suggesting that acetylation increased after pag1 transfection. The acetylation levels after pag1 transfection were 3-fold higher than those in the control group at 48 hours post-transfection (Fig. 2A,B), and the presence of pag1 suppressed su(var)3–9 expression by 50% (Fig. 2C,D). It is worth noting that pag1 does not encode a protein, but instead miRNAs are produced. Several studies have suggested that the miRNAs produced from latent-associated genes are mostly involved in regulating histone-associated enzymes36. Su(var)3–9 trimethylates H3K9, and it has been identified in different animals33, though similar mechanisms in insects have not yet been identified. Therefore, miRNAs may be crucial for the mechanism by which HzNV-1 affects these two histone transferases.
Figure 2.
Real-time PCR and RT-PCR detection of tip60 and su(var)3–9 expression levels. (A) HAT tip60 gene expression levels were detected at 0 and 48 hours after pag1 transfection. (B) RT-PCR analysis of tip60 expression levels at 48 hpt. (C) Su(var)3–9 gene expression levels were detected at 0 and 48 hours after pag1 transfection by RT-qPCR. (D) RT-PCR analysis of su(var)3–9 expression levels at 48 hpt. All results were normalized to those for actin. At least three repetitions were conducted for each group. P values were calculated using Student’s t-test (*P < 0.05; **P < 0.005).
Predicted and identified miRNAs produced by the pag1 transcript
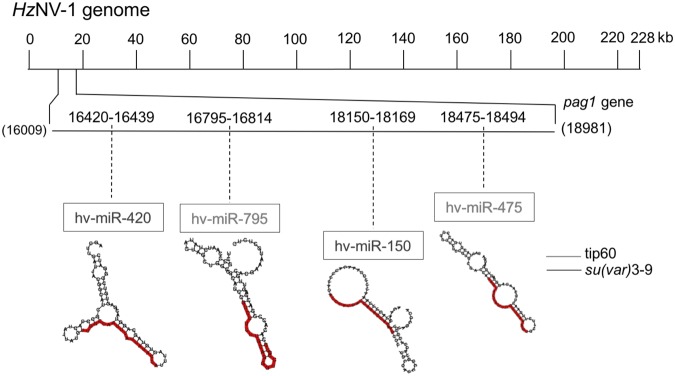
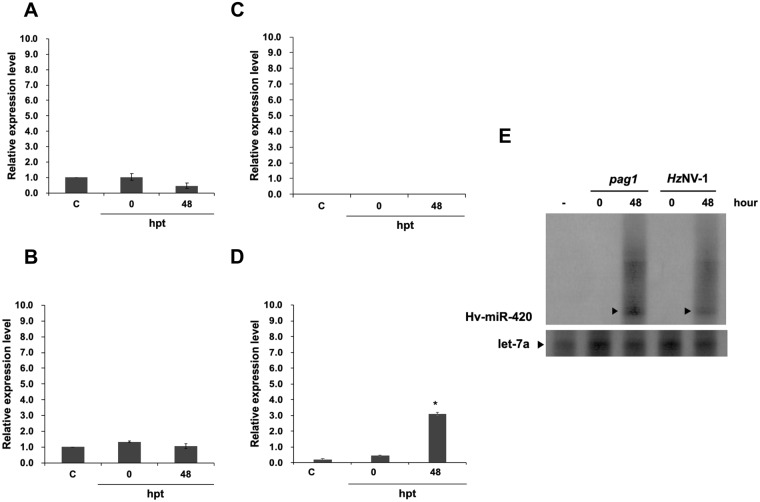
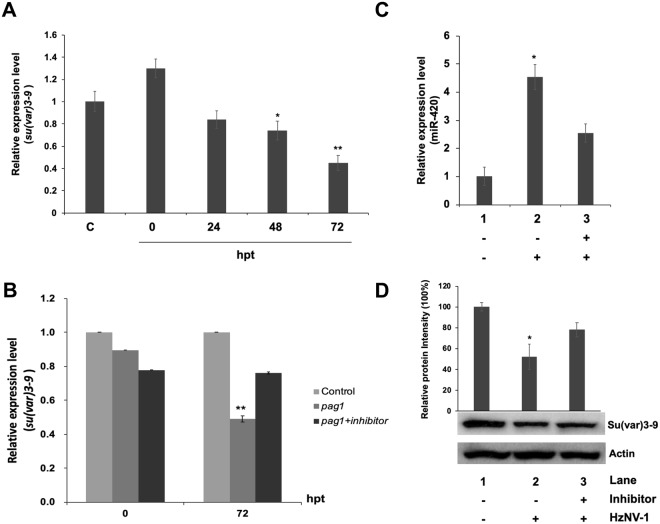
According to the NCBI database, the complete genome of HzNV-1 is approximately 228 kb, and the pag1 transcript is approximately 4 kb. To identify miRNAs that target the histone transferases encoded by tip60 and su(var)3–9, pag1-encoded miRNAs were mapped onto these genes. Prediction was performed by Vir-Mir and based on mapping scores and structures. Four candidate miRNAs (Hv-miR-420, Hv-miR-795, Hv-miR-150, Hv-miR-475) were selected for further evaluation (Fig. 3). The sequences of the miRNAs are listed in Table 1. To verify that the predicted miRNAs are actually encoded and generated from pag1, stem-loop RT-qPCR was used to measure these candidate miRNAs at different time points after pag1 transfection in Sf-21 cells. Among these miRNAs, Hv-miR-795 (Fig. 4A) and Hv-miR-475 (Fig. 4B) were detected in both pag1-transfected cells and in untransfected cells, suggesting that host cells also expressed these 2 miRNAs. Hv-miR-150 was undetectable in both groups, indicating that Hv-miR-150 is a mock miRNA (Fig. 4C). Hv-miR-420 was the only candidate miRNA whose expressed was significantly increased after pag1 transfection (Fig. 4D). To further confirm the results of stem-loop PCR, northern blot analysis was performed, showing detection of Hv-miR-420 in both pag1-transfected and HzNV-1-infected cells (Fig. 4E). These results showed that the expression level of Hv-miR-420 was significantly increased in pag1-transfected and HzNV-1-infected cells.
Figure 3.
Mapping of pag1 miRNAs that target tip60 or su(var) 3–9 in the HzNV-1 genome. The complete sequence of the HzNV-1 genome is shown in the upper portion. The pag1 gene is located from nt 16,009 to 18,981. The gray lines indicate miRNAs targeting the tip60 gene. The black lines represent miRNAs targeting the su(var)3–9 gene. The predicted structures of the miRNAs are displayed in Table 1.
Table 1.
The predicted miRNAs produced by the pag1 gene.
| Target | Name | Location | Sequence |
|---|---|---|---|
| Su(var)3–9 | Hv-miR-420 | 16420–16439 | 5′-UUGGCCAAUUUAUUAUACCA-3′ |
| Hv-miR-150 | 18150–18169 | 5′-AUCCAUGAUUAAAGUACAAG-3′ | |
| tip60 | Hv-miR-795 | 16795–16814 | 5′-GUCUUUUUGCAGGGUCCGUG-3′ |
| Hv-miR-475 | 18475–18494 | 5′-CACCACCAUUCGUUUAAAGU-3′ |
Figure 4.
Stem-loop RT-qPCR confirmation of miRNA expression after pag1 transfection. (A) Hv-miR-795 levels decreased after pag1 transfection. (B) Hv-miR-475 was not affected by pag1 in Sf-21 cells. (C) Hv-miR-150 was not detected at any time point. (D) Hv-miR-420 was identified after pag1 transfection. (E) Small RNAs harvested from pag1-transfected cells and HzNV-1 productively infected cells were analyzed by northern blotting using probes against predicted HzNV-1 miR-420 (top panels) or let-7a miRNA as a positive control (bottom panels). At least three repetitions were conducted for each group. P values were calculated using Student’s t-test (*P < 0.05).
Hv-miR-420 suppresses su(var)3–9 expression
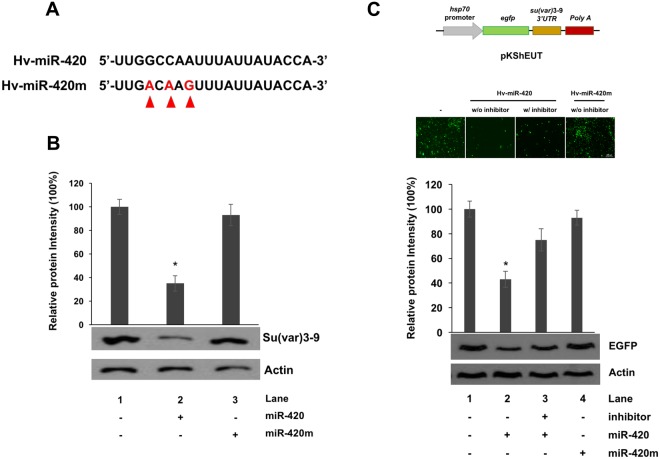
Although the functions of several pag1 miRNAs have been identified, the remaining miRNAs predicted from the pag1 transcript need to be further studied6. To address this, the miRNAs predicted from pag1 were mapped onto the tip60 and su(var)3–9 sequences (Fig. 3) and, based on the stem-loop PCR assay and the mode of action of miRNAs in general, Hv-miR-420 was selected for further experiments (Fig. 4). To evaluate the effect of miRNA-420 on su(var)3–9, Hv-miR-420 mimic was transfected into Sf-21 cells and it was found that 60% of su(var)3–9 expression was suppressed 72 hours after transfection (Fig. 5A). To prove a direct relationship between Hv-miR-420 and suppression of su(var)3–9, Hv-miR-420 inhibitor was transfected into pag1-transfected cells (pag1 transfected 24 hours prior to inhibitor transfection). Notably, the levels of su(var)3–9 expression recovered to approximately 72% of the untreated control in contrast to 50% of the expression level in the pag1-transfected cells without inhibitor treatment (Fig. 5B). Hv-miR-420 expression levels in HzNV-1-infected cells was measured and it was found to significantly increased after viral infection (Fig. 5C), where su(var)3–9 protein levels was significantly decreased (Fig. 5D). Moreover, transfection of the Hv-miR-420 inhibitor increased Su(var)3–9 protein levels compared to HzNV-1-infected cells without inhibitor (Fig. 5D). To confirm direct interaction between miR-420 and su(var)3–9, a mutant miR-420 which contained base mutations in its seed region was generated (designated miR-420m, Fig. 6A). Western blotting analysis (Fig. 6B) showed that the mutant miR420 did not affect the levels of Su(var)3–9 expression, suggesting that intact seed sequence was crucial for functioning. To assess whether the observed regulation was direct, the 3′ UTR of su(var)3–9 was fused to the 3′-end of the egfp gene to generate an EGFP reporter plasmid. Co-transfection of this EGFP reporter plasmid with Hv-miR-420 significantly decreased EGFP levels, whereas co-transfection with the Hv-miR-420m did not affect EGFP expression (Fig. 6C). These results demonstrate that su(var)3–9 expression was suppressed by increased Hv-miR-420 levels in both pag1-transfected and virus-infected cells.
Figure 5.
miR-420 regulates su(var)3–9 expression levels in Sf-21 cells. (A) Su(var)3–9 expression was downregulated by miR-420 and 60% inhibited at 72 hours post-transfection. (B) The miR-420 inhibitor rescued su(var)3–9 expression at 72 hours. (C) Stem-loop RT-qPCR confirmed miR-420 expression after HzNV-1 infection with or without the miR-420 inhibitor. (D) Su(var)3–9 expression was detected at 48 hours after HzNV-1 infection with or without the miR-420 inhibitor by western blotting. All results were normalized to the CT value of actin, and at least three repetitions were performed. The control group expression level was set to 1 or 100%. P values were calculated using Student’s t-test (*P < 0.05; **P < 0.005).
Figure 6.
Base mutations in the seed region of hv-miR-420 is unable to suppress Su(var)3–9 expression. (A) Sequences of hv-miR-420 was shown, and miRNAs with mutations in the seed region were denoted as hv-miR-420m. The mutational substituted bases are indicated by arrows. (B) Expression levels of Su(var)3–9 in various treatments were analyzed by western blotting and the expression level of Actin were used as a loading control. (C) Reporter plasmids (pKShEUT) in which the 3′-UTR of su(var)3–9 was fused to the 3′-end of the EGFP coding region, as indicated (top panels), were transfected into Sf-21 cells with miR-420 only, miR-420 plus miR-420 inhibitor or miR-420m. EGFP expression was detected at 0 or 48 hours after reporter plasmids (pKShEUT) transfection by fluorescent microscopy or western blotting. The samples were normalized to Actin, and the control group was set as 100%. At least three independent experiments were performed. P values were calculated using Student’s t-test (*P < 0.05).
Hv-miR-420 is involved in regulating HzNV-1 lytic infection
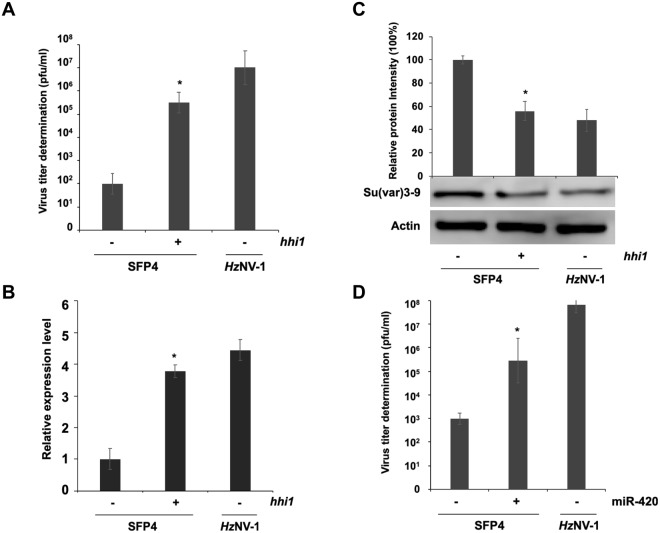
Previous studies have found that the pag1 transcript is the only HzNV-1 viral transcript expressed during both lytic and latent infections5. In this study, changes in Hv-miR-420 expression at different infection stages were examined to evaluate whether Hv-miR-420 affected histone modification and altered infection stages. Virus titers in both latent infection cell lines (SFP4) and HzNV-1-infected Sf-21 cells were analyzed following reactivation from latent stage induced by transfection of hhi1 expression plasmid (Fig. 7A). Transfection of hhi1 into HzNV-1-latently-infected SFP4 cells significantly increased Hv-miR-420 expression (Fig. 7B) but the expression of su(var)3–9 was inhibited (Fig. 7C), indicating that Hv-miR-420 played essential roles in altering histone modification by latent viruses after reactivation. To assess whether Hv-miR-420 was able to stimulate latent viruses to enter lytic infection stage, Hv-miR-420 mimic was transfected into SFP4 cells and analyzed viral reactivation. The results showed significant cell lysis within 72 hours and increased virus titers after Hv-miR-420 transfection (Fig. 7D). These results demonstrated that Hv-miR-420 was able to induce latent viruses to enter lytic infection stage, possibly by contributing to activation of viral gene expression through inhibition of histone methylation.
Figure 7.
Induction of latent viral reactivation by miR-420. (A) Viral titers estimated in productively and latently infected cells. Latently infected cells were transfected with the plasmid pKShH1. (B) Stem-loop RT-qPCR confirmed miR-420 expression in hhi1-transfected SFP4 cells or HzNV-1-infected Sf-21 cells. (C) Su(var)3–9 expression was downregulated in hhi1-transfected SFP4 cells. (D) Viral titers were estimated in latently infected cells. Latently infected cells were transfected with miR-420 mimic and control miRNA, and viral titers were determined at 72 hours post-transfection. Sf-21 cells were infected with the wild-type virus as a control. At least three repetitions were conducted for each group. P values were calculated using Student’s t-test (*P < 0.05).
Discussion
The characteristics of HzNV-1 are universal among mammalian viruses, especially HSVs, which share several common features with HzNV-1. The noncoding latency-associated transcript (LAT) gene is highly expressed during HSV or HCMV latency and encodes a series of miRNAs15,25,26,37,38. Previous studies have focused on the interaction between viral miRNAs and viral genes, and it has been reported that LAT-encoded miRNAs not only inhibit the HSV IE genes ICP0 and ICP4 but also regulate histone modification8,25,38,39. However, these studies are still limited in insect viruses.
In this study, we discovered that the latent gene of HzNV-1, pag1, was able to affect two insect histone transferases, tip60 and su(var)3–9, at the beginning phase of infection (Fig. 2). tip60 is involved in a variety of processes, such as DNA repair40, development34, and human virus association41. su(var)3–9, on the other hand, is a conserved histone transferase that also possesses multiple roles in animals and is related to the heterochromatin-associated HP1 protein together with H3K9 trimethylation31,42. We examined the expression of histone proteins containing modification in regions that are targets of the abovementioned histone transferases. Western blot analysis showed that trimethylation of H3K9 was significantly downregulated and that acetylation of H3 was upregulated after pag1 transfection (Fig. 1). A possible explanation is that when methyltransferase expression decreases, resulting in reduced trimethylation of histones (H3K9), more histones are available for acetyl modification, leading to an increase in acetylated H3.
Acetylation of histones relaxes the structure of chromatin, typically resulting in gene activation. In contrast, methylation is reported to be associated with heterochromatin and gene silencing43. For this reason, most studies are inclined to relate methylation and deacetylation with latent infection. However, the ICP0 gene of HSV was found to be acetylated during quiescence44. Additionally, three early genes of KSHV are reported to have distinct histone modifications after KSHV primary infection. Indeed, relieving HDAC repression leads to activation of lytic genes in both HCMV and EBV45,46. This study indicates that repressive and active markers are simultaneously distributed across chromatin during latency47.
Similar to the LAT gene of HSV, pag1 plays an important role during HzNV-1 latent infection. Previous studies have shown that the pag1 suppresses apoptosis induced by the lytic gene hhi148 and that pag1-encoded miRNAs cause a switch from lytic infection to a latent infection7. Although the functions of several pag1-encoded miRNAs have been demonstrated, the functions of other predicted miRNAs still need to be confirmed6. To address this issue, miRNAs predicted from pag1 were mapped onto the sequences of the tip60 and su(var)3–9 coding regions, and four candidates were chosen using universal reference miRNAs (Fig. 3). Based on stem-loop PCR and northern blot results, Hv-miR-420 was chosen for further experiments (Fig. 4). Two miRNAs (miR-795 and miR-475) (Fig. 4A,B) were expressed in untransfected cells, and there was no clear increase in their expression level upon pag1 infection, suggesting that insect cells normally express low levels of these two miRNAs and they were not likely derived from pag1 transcript. To determine the effects of Hv-miR-420 on su(var)3–9, we first transfected the miR-420 mimic into Sf-21 cells and found 60% suppression (Fig. 5A). To study opposing impacts, we then transfected miR-420 inhibitors, and nearly 30% of su(var)3–9 expression was recovered after 72 hours (Fig. 5B). Taken together, these results show that the presence of miR-420 can regulate the expression levels of su(var)3–9. It has been reported that pag1 is the only transcript expressed during both lytic and latent infections6. According to a previous report, the pag1 transcript yields several miRNAs, some of which were shown to maintain latency7. The ability of miR-420 to stimulate a virus toward lytic infection may be due to differential expression of pag1-derived miRNAs during lytic and latent stages. Our preliminary results show relatively low miR-420 expression in latently infected cells, suggesting that the generation of miR-420 was downregulated. Investigation of how the expression of pag1-derived microRNAs are regulated is currently underway.
The difficulty in establishing a stable HSV-latently-infected cell line has hampered the progress of HSV latent infection research. Nonetheless, a permanent cell line of HzNV-1 latent infection, namely, SFP4 cells, is available49 and the results of pag1 transfection could be validated in the context of HzNV-1 latent infection. Indeed, we performed experiments with pag1 to eliminate other variables correlated with HzNV-1. Our discovery of an epigenetic pathway that had been modified by Hv-miR-420 indicates that some variables regarding Hv-miR-420 and HMT su(var)3–9 in the dual stages of HzNV-1 may need to be taken into consideration in the future. According to an earlier study, pag1 is detectable in both lytic and latent stages7. Under these circumstances, we may be able to utilize a reactivating virus from SFP4 cells to compare different expression levels of Hv-miR-420, su(var)3–9, and trimethylation H3K9, which can help in determining whether this microRNA has a crucial role in reactivating HzNV-1.
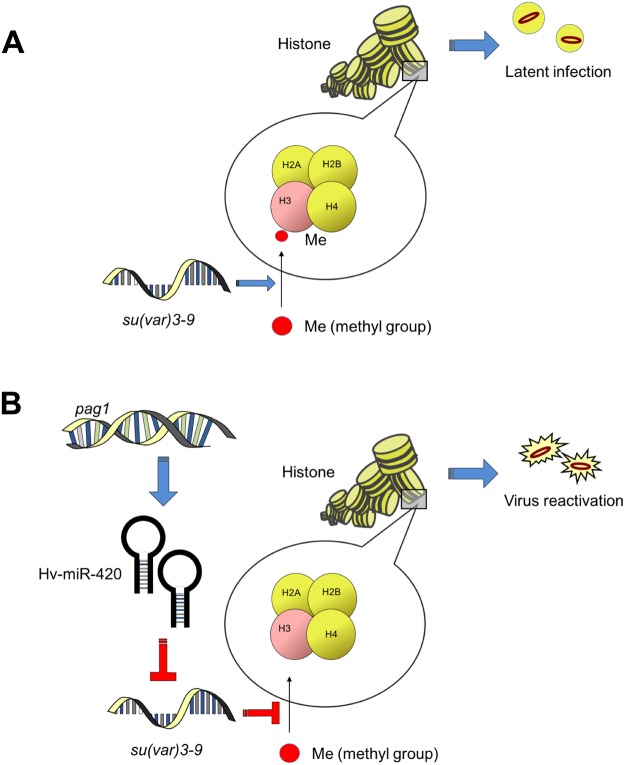
Previous studies on human latent viruses, e.g., HSV50, KSHV21, HBV51 and HCMV52, found that host HATs are affected by these viruses. In contrast, few studies have investigated interaction between viral miRNAs and histone modification. Our research presented herein addressed this interaction and revealed that Hv-miR420, a miRNA produced from the pag1 gene, regulates host HMT su(var)3–9 to suppress H3K9 trimethylation. Surprisingly, a tentative explanation is that the decrease in H3K9 trimethylation during latency is a sign of reactivation (Fig. 8). The physiological significance and in vitro tests need to be pursued to further support and verify our findings. This work on the functions of latent miRNAs from an epigenetic perspective provides a better understanding of insect latent viruses.
Figure 8.
A model of the establishment of lytic infection via miR-420-mediated suppression of histone methylation. (A) Without miR-420. (B) With miR-420.
Materials and Methods
Cell culture and virus
S. frugiperda IPLB-Sf-21 cells were cultured in TC-100 insect cell medium with 10% fetal bovine serum (FBS) at 26 °C (Gibco BRL)53. The SFP4 cell line was derived from latently infected Sf-21 cells4,5. HzNV-1 titers were calculated using TCID5054 and quantitative PCR (qPCR)55.
Plasmid DNA construction and transfection
The coding regions of hhi1 and pag1 were previously inserted into the vector pKSh to construct pKShH1 and pKShP17. To clone the 3′-UTR of su(var)3–9 (primer: 3–9 3UTR-F: ACGCACGCTCATTCTGACACACGC; 3–9 3UTR-R: TTTACAATCTTATTACATTTAC) into an EGFP reporter vector (pKShE)5, the su(var)3–9-UTR fragment was inserted into the EcoRI site after the stop codon of the EGFP gene (pKShEUT). A total of 2 × 105 Sf-21 cells were seeded into a 24-well culture plate (Corning, Action, MA) and transfected with 0.5 μg of plasmid DNA using Cellfectin (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol (Gibco BRL). EGFP expression was observed using fluorescent microscope at 2 days-post transfection. Samples were collected at 48 hours post-transfection in triplicate.
Western blotting
Proteins were isolated with 40 μl of RIPA buffer (Thermo Fisher) per well in a 24-well culture plate, and the samples were immediately stored at −80 °C until use. Western blotting was carried out using rabbit polyclonal antibodies detecting acetyl-H3 (Millipore, 1:4500), trimethyl-H3K9 (Millipore, 1:500), su(var)3–9 (Abcam, 1:4000) and EGFP (Millipore, 1:5000). Normalization was performed via detection of actin levels using a mouse polyclonal antibody (Millipore, 1:2500). All bands were analyzed and quantified using AlphaView SA.
RT-PCR and qPCR
RNA samples were collected using 100 μl of GENEzolTM reagent per cm2 of culture dish with a GENEzolTM TriRNA Pure Kit (Geneaid) and stored at −20 °C. The eluted RNA was reverse transcribed using a PrimeScriptTM RT-PCR Kit (TaKaRa), and cDNA was synthesized from 800 ng of RNA for each sample. Quantification of gene expression was carried out by adding 2 μl of cDNA to 2 × SeniFAST SYBR® Hi-ROX Mix (BIOLINE) using a 56 °C annealing temperature. PCR was carried out using the following primers:
Actin F: TCAACCCCAAGGCCAACAGAGA
Actin R: GAGGCTCCTGGGAATTTTTC
Tip60 F: CGCGAAATGGTAACAAACAG
Tip60 R: TGGAGAGCCACATAACAACTG
Su(var)3–9F: CGCCTGTCGGACTCAGTTAT
Su(var)3–9R: GAGGCTCCTGGGAATTTTTC
Computational prediction of viral miRNAs
pag1 miRNA prediction was implemented according to the full-length gene sequence of pag1 (NC_004156.1), which was obtained from the National Center for Biotechnology Information. The prediction was conducted using the miRNA prediction database (Vir-Mir) (http://alk.ibms.sinica.edu.tw). The miRNAs with high match scores chosen for analysis are listed in Table 1.
Stem-loop real-time PCR and northern blotting
Sf-21 cells transfected with pKShP1 at 0 and 48 hpt were used for extraction of total RNA using TRIzol® reagent (Invitrogen) following the manufacturer’s instructions. The method for detecting mature miRNAs with specific primers was described in a previous study56,57. The miRNA primers used for RT-PCR and real-time qPCR were as follows:
Hv-miR-420 RT: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTG
GATACGACTGGTAT-3′
Hv-miR-420 F: 5′- CACGCATTGGCCAATTTA-3′
Hv-miR-150 RT: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTG
GATACGACCTTGTA-3′
Hv-miR-150 F: 5′- CACGCAATCCATGATTAA-3′
Hv-miR-795 RT: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTG
GATACGACCACGGA-3′
Hv-miR-795 F: 5′- CACGCAGTCTTTTTGCAG-3′
Hv-miR-475 RT: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCAC
TGGATACGACACTTTA-3′
Hv-miR-475 F: 5′- CACGCACACCACCATTCG-3′
Reverse primer: 5′-CCAGTGCAGGGTCCGAGGTA-3′
Total small RNA samples from pag1-transfected and HzNV-1-infected cells were isolated using a mirVanaTM miRNA isolation kit (Ambion) according to the manufacturer’s instructions. For northern blot analysis of small RNAs, 1 µg of small RNA sample and radio-labeled Decade Marker (Ambion) were fractionated by 15% denaturing polyacrylamide gel electrophoresis (PAGE) (acrylamide∶bis ratio, 19∶1) containing 8 M urea in 0.5 × TBE buffer. RNAs were transferred by electroblotting to a Hybond-N + nylon transfer membrane (GE Healthcare) and UV cross-linked. RNA oligonucleotides (Integrated DNA Technologies) carrying the reverse complementary sequence for candidate miRNAs or let-7a were end-labeled with DIG (MP Biomedicals, Irvine, CA) to high specific activity. Hybridizations and washes were carried out using DIG hybridization buffer according to the manufacturer’s directions (Roche).
Mimic and inhibitor miRNA transfection
The Hv-miR-420 mimic was synthesized by the MDBio Incorporation, and inhibitor was produced by GenePharma Incorporation. The mimic (50 nM) or inhibitor was transfected into Sf-21 cells using 1.5 μl of LipofectamineTM RNAiMAX following the manufacturer’s protocol (Invitrogen). The sequences of the Hv-miR-420 mimic, Hv-miR-420m and inhibitor are 5′-UUGGCCAAUUUAUUAUACCA-3′; 5′-UUGACAAGUUUAUUAUACCA-3′ and 5′-UGGUAUAAUAAAUUGGCCAA-3′, respectively.
Statistical analysis
The qPCR Ct values for samples treated with plasmids were normalized to those of actin with the comparative Ct method58. Each group of experimental data was selected for comparison with the control groups using a single tail and type 1 t-test. The statistics used in this data analysis were performed with the T-TEST function in Microsoft Excel. P values are indicated in figures, with (*) representing the level of significance.
Acknowledgements
We thank Dr. Eric C. Wu for kindly revising the manuscript. This research was funded by the grants MOST107-2311-B-002-024-MY3 from the Ministry of Science and Technology, Taiwan.
Author Contributions
Guarantors of integrity of entire study, study concepts, and manuscript preparation: P.C.W., Y.H.L., S.T.L. and Y.L.W. Study design, data acquisition/analysis, literature research and manuscript preparation: P.C.W., Y.H.L., T.C.W., Y.C. and Y.L.W. Data acquisition/analysis, manuscript editing, and revision: P.C.W., C.P.W. and Y.L.W. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ralston AL, Huang YS, Kawanishi CY. Cell culture studies with the IMC-Hz-1 nonoccluded virus. Virology. 1981;115:33–44. doi: 10.1016/0042-6822(81)90086-6. [DOI] [PubMed] [Google Scholar]
- 2.Huang YS, Hedberg M, Kawanishi CY. Characterization of the DNA of a Nonoccluded Baculovirus, Hz-1V. J Virol. 1982;43:174–181. doi: 10.1128/jvi.43.1.174-181.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Kleespies RG, Huger AM, Jehle JA. The genome of Gryllus bimaculatus nudivirus indicates an ancient diversification of baculovirus-related nonoccluded nudiviruses of insects. J Virol. 2007;81:5395–5406. doi: 10.1128/JVI.02781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao YC, et al. Differential expression of Hz-1 baculovirus genes during productive and persistent viral infections. J Virol. 1992;66:1442–1448. doi: 10.1128/jvi.66.3.1442-1448.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu YL, et al. The early gene hhi1 reactivates Heliothis zea nudivirus 1 in latently infected cells. J Virol. 2010;84:1057–1065. doi: 10.1128/JVI.01548-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chao YC, et al. A 2.9-kilobase noncoding nuclear RNA functions in the establishment of persistent Hz-1 viral infection. J Virol. 1998;72:2233–2245. doi: 10.1128/jvi.72.3.2233-2245.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu YL, et al. A non-coding RNA of insect HzNV-1 virus establishes latent viral infection through microRNA. Sci Rep. 2011;1:60. doi: 10.1038/srep00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom DC, Giordani NV, Kwiatkowski DL. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta. 2010;1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Asgari S. MicroRNA functions in insects. Insect Biochem Molec. 2013;43:388–397. doi: 10.1016/j.ibmb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Varghese J, Lim SF, Cohen SM. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 2010;24:2748–2753. doi: 10.1101/gad.1995910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varghese J, Cohen S. M. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Gene Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg JK, et al. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav. 2012;11:660–670. doi: 10.1111/j.1601-183X.2012.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asgari S. Role of MicroRNAs in Insect Host-Microorganism Interactions. Front Physiol. 2011;2:48. doi: 10.3389/fphys.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain M, Taft RJ, Asgari S. An insect virus-encoded microRNA regulates viral replication. J Virol. 2008;82:9164–9170. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain M, et al. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 2012;40:2210–2223. doi: 10.1093/nar/gkr848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/S0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 19.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 20.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61:24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 21.Kim KY, et al. Kaposi’s sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A. J Virol. 2013;87:6782–6793. doi: 10.1128/JVI.00011-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toth Z, et al. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 2010;6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 25.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau, B. et al. The Expression of Human Cytomegalovirus MicroRNA MiR-UL148D during Latent Infection in Primary Myeloid Cells Inhibits Activin A-triggered Secretion of IL-6. Sci Rep-Uk6, ARTN 3120510.1038/srep31205 (2016). [DOI] [PMC free article] [PubMed]
- 27.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 29.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 30.Wallrath LL, Elgin SCR. Position Effect Variegation in Drosophila Is Associated with an Altered Chromatin Structure. Gene Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 31.Schotta G, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. Embo J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aagaard L, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. Embo J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Li S, Yin J, Zhong J. Identification and characterization of the Spodoptera Su(var) 3-9 histone H3K9 trimethyltransferase and its effect in AcMNPV infection. PLoS One. 2013;8:e69442. doi: 10.1371/journal.pone.0069442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee K, Fischer R, Vilcinskas A. Histone acetylation mediates epigenetic regulation of transcriptional reprogramming in insects during metamorphosis, wounding and infection. Front Zool. 2012;9:25. doi: 10.1186/1742-9994-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicoll MP, et al. The HSV-1 Latency-Associated Transcript Functions to Repress Latent Phase Lytic Gene Expression and Suppress Virus Reactivation from Latently Infected Neurons. PLoS Pathog. 2016;12:e1005539. doi: 10.1371/journal.ppat.1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu J, Thorley-Lawson DA. EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells. Proc Natl Acad Sci USA. 2014;111:11157–11162. doi: 10.1073/pnas.1406136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perng GC, Jones C. Towards an understanding of the herpes simplex virus type 1 latency-reactivation cycle. Interdiscip Perspect Infect Dis. 2010;2010:262415. doi: 10.1155/2010/262415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/S0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 41.Col E, et al. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 2005;24:2634–2645. doi: 10.1038/sj.emboj.7600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schotta G, Ebert A, Reuter G. SU(VAR)3-9is a conserved key function in heterochromatic gene silencing. Genetica. 2003;117:149–158. doi: 10.1023/A:1022923508198. [DOI] [PubMed] [Google Scholar]
- 43.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 44.Coleman HM, et al. Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression. J Gen Virol. 2008;89:68–77. doi: 10.1099/vir.0.83272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi CK, Ho DN, Hui KF, Kao RY, Chiang AK. Identification of Novel Small Organic Compounds with Diverse Structures for the Induction of Epstein-Barr Virus (EBV) Lytic Cycle in EBV-Positive Epithelial Malignancies. PLoS One. 2015;10:e0145994. doi: 10.1371/journal.pone.0145994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishna BA, et al. Transient activation of human cytomegalovirus lytic gene expression during latency allows cytotoxic T cell killing of latently infected cells. Sci Rep. 2016;6:24674. doi: 10.1038/srep24674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jha, H. C. et al. Kaposi’s sarcoma-associated herpesvirus genome programming during the early stages of primary infection of peripheral blood mononuclear cells. MBio5, 10.1128/mBio.02261-14 (2014). [DOI] [PMC free article] [PubMed]
- 48.Wu YL, et al. Heliothis zea nudivirus 1 gene hhi1 induces apoptosis which is blocked by the Hz-iap2 gene and a noncoding gene, pag1. J Virol. 2011;85:6856–6866. doi: 10.1128/JVI.01843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin CL, et al. Persistent Hz-1 virus infection in insect cells: evidence for insertion of viral DNA into host chromosomes and viral infection in a latent status. J Virol. 1999;73:128–139. doi: 10.1128/jvi.73.1.128-139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Jiang Y, Geiser V, Zhou J, Jones C. Bovine herpesvirus 1 immediate-early protein (bICP0) interacts with the histone acetyltransferasep300, which stimulates productive infection and gC promoter activity. J Gen Virol. 2006;87:1843–1851. doi: 10.1099/vir.0.81766-0. [DOI] [PubMed] [Google Scholar]
- 51.Riviere L, et al. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63:1093–1102. doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 52.Hsu CH, et al. HCMV IE2-mediated inhibition of HAT activity downregulates p53 function. EMBO J. 2004;23:2269–2280. doi: 10.1038/sj.emboj.7600239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu CP, Wang JY, Huang TY, Lo HR, Chao YC. Identification of baculoviral factors required for the activation of enhancer-like polyhedrin upstream (pu) sequence. Virus Res. 2008;138:7–16. doi: 10.1016/j.virusres.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 54.Darling AJ, Boose JA, Spaltro J. Virus assay methods: accuracy and validation. Biologicals. 1998;26:105–110. doi: 10.1006/biol.1998.0134. [DOI] [PubMed] [Google Scholar]
- 55.Lo HR, Chao YC. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol Prog. 2004;20:354–360. doi: 10.1021/bp034132i. [DOI] [PubMed] [Google Scholar]
- 56.Kramer MF. Stem-loop RT-qPCR for miRNAs. Curr Protoc Mol Biol. 2011;Chapter 15:Unit15 10. doi: 10.1002/0471142727.mb1510s95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]