Summary
Large numbers of brain regions are active during behaviors. A high-resolution, brain-wide activity map could identify brain regions involved in specific behaviors. We have developed functional ultrasound imaging to record whole-brain activity in behaving mice at a resolution of ∼100 μm. We detected 87 active brain regions during visual stimulation that evoked the optokinetic reflex, a visuomotor behavior that stabilizes the gaze both horizontally and vertically. Using a genetic mouse model of congenital nystagmus incapable of generating the horizontal reflex, we identified a subset of regions whose activity was reflex dependent. By blocking eye motion in control animals, we further separated regions whose activity depended on the reflex’s motor output. Remarkably, all reflex-dependent but eye motion-independent regions were located in the thalamus. Our work identifies functional modules of brain regions involved in sensorimotor integration and provides an experimental approach to monitor whole-brain activity of mice in normal and disease states.
Highlights
-
•
Functional ultrasound enables imaging whole-brain activity during mouse behavior
-
•
Activity in 87 brain regions are modulated during the optokinetic reflex
-
•
Reflex-related regions were identified by perturbing retinal direction selectivity
-
•
A subset of these regions, all in the thalamus, are independent of eye motion
Macé et al. developed an approach for imaging whole-brain activity during mouse behavior using ultrasound. They revealed many brain regions activated during the optokinetic reflex, a visuomotor behavior that stabilizes gaze. They parsed them into functional modules using reflex perturbations.
Introduction
Recent advances in neurotechnology, primarily applied to mice, have revealed insights into the neuronal computations that underlie behavior at the microcircuit level (Callaway and Luo, 2015, Kerr and Denk, 2008, Kim et al., 2017). However, these circuit studies mostly focus on only one or two candidate brain regions at a time and are biased toward regions previously shown or predicted to be involved in a given behavior. An unbiased, brain-wide activity map during mouse behavior could lead to a system-level understanding of how activity drives and is affected by behavior and could guide the selection of brain regions to be studied at the level of microcircuits. However, current whole-brain functional imaging technologies, such as functional magnetic resonance imaging, have limited resolution to reveal activity in small and deep brain nuclei and are difficult to apply to awake and behaving mice. Furthermore, with methods such as FOS and FOS-TRAP (Guenthner et al., 2013), which can be used to identify active regions in the whole brain, it is not possible to follow activity over time, analyze trial-to-trial variation, or detect decreases of activity in brain areas.
We have developed a high-resolution and unbiased approach for studying active regions distributed across the brains of behaving mice. We have combined this approach with genetic, mechanical, and pharmacological perturbations to reveal brain regions involved in visuomotor integration in the context of an innate behavior, the optokinetic reflex, and to group them into functional modules.
The optokinetic reflex is conserved across vertebrates and serves to stabilize images drifting on the retina by moving the eye in the direction of image drift (Distler and Hoffmann, 2012, Masseck and Hoffmann, 2009). The reflex is initiated by motion detectors in the retina, each of which selectively responds to visual motion along one cardinal direction (Hillier et al., 2017, Oyster and Barlow, 1967, Yoshida et al., 2001). Retinal motion detectors drive activity in motor neurons connected to the extraocular muscles via a set of brain regions. Some of these regions have been identified, such as the nuclei of the accessory optic system, the pontine nuclei, the vestibular nuclei, the inferior olive, and the flocculus (Büttner-Ennever, 2006, Distler and Hoffmann, 2012, Portugues et al., 2014, Simpson, 1984). The eye movements during the optokinetic reflex have two alternating phases: tracking of the moving scene followed by fast resetting saccades. Brain regions associated with the reflex can be involved in different ways, such as detection of direction and speed of visual motion, transformation of visual to motor signals, generation of motor commands for tracking, integration of the associated corollary discharge, integration of the sensory feedback signals resulting from eye motion, and finally the initiation and execution of resetting saccades and their associated corollary and feedback signals (Büttner-Ennever, 2006, Distler and Hoffmann, 2012). We refer to all these brain regions as the “reflex pathway” and their activity as the “reflex.” Which brain region is associated with which role is not well understood.
Results
Whole-Brain Functional Ultrasound Imaging
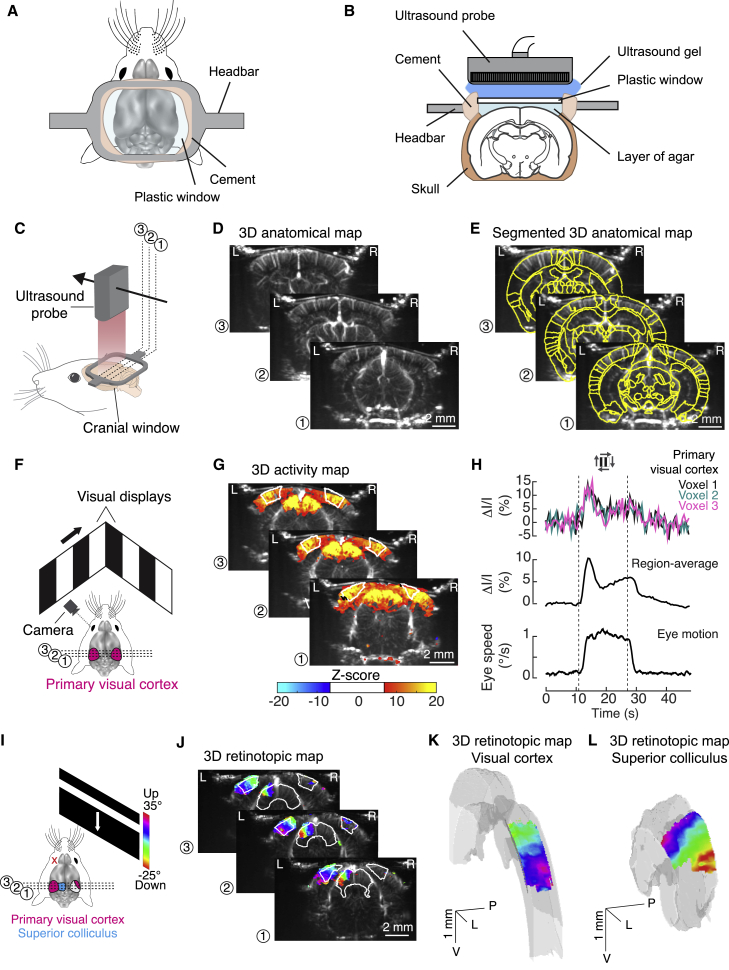
We developed functional ultrasound imaging (Macé et al., 2011), which reports neuronal activity indirectly by monitoring changes in blood volume in the microvasculature (Rubin et al., 1994, Rubin et al., 1995, Macé et al., 2013), to record activity from the whole brain of awake, head-fixed mice. We installed a large cranial window that enabled ultrasound waves to penetrate into the brain for stable chronic imaging (Figures 1A and 1B; Figure S1). We positioned an ultrasound probe above the cranial window and acquired a series of coronal images, each spanning the entire depth of the brain, by stepping the probe along the antero-posterior axis (Figures 1C and 1D). To assign each acquired voxel to a specific brain region, we performed 3D rigid registration of the series of coronal images—obtained in the absence of visual stimulation—to the Allen Mouse Brain Reference Atlas (Figure 1E). Awake mice were then presented with drifting gratings that elicited the optokinetic reflex (Figure 1F). To build a 3D spatial map of brain activity, we compared voxel by voxel the hemodynamic signals (ΔΙ/Ι, referred to as “activity”) obtained during visual motion stimulation and during the presentation of a static gray image (Figure 1G). The hemodynamic signals of all voxels belonging to a given brain region were averaged to obtain a response time course for that brain region (Figure 1H).
Figure 1.
Whole-Brain Functional Ultrasound Imaging in Behaving Mice
(A) Schematic of the chronic cranial window (top view). The head bar is attached to the skull by a ring of dental cement. The craniotomy is stabilized by a layer of agar and closed by a plastic window, transparent to ultrasound.
(B) Schematic of the chronic cranial window (coronal section) and of the ultrasound probe during imaging. Ultrasound gel is applied on top of the window, and the probe is positioned 3 mm above the window.
(C) Schematic of the experimental setup, with an awake head-fixed mouse. Hemodynamic signals are measured through the chronic cranial window using the ultrasonund probe, stepped along the antero-posterior axis (black arrow). Three example image planes are labeled (numbered dashed lines).
(D) Example coronal slices from a 3D anatomical map at three different planes (dashed lines in C). L, left; R, right.
(E) Segmented brain regions (yellow outlines) after 3D registration of the anatomical map to the Allen Mouse Brain Reference Atlas overlaid on the same slices as in (D). L, left; R, right.
(F) Schematic of the experiments for activity mapping in mice during the optokinetic reflex. Drifting gratings (0.05 cpd spatial frequency, 10°/s speed, an example direction is indicated by the black arrow) are presented binocularly on two visual displays. A camera records left eye position. The ultrasound probe is stepped across the brain. Three example imaging planes crossing the primary visual cortex (magenta) are labeled (numbered dashed lines).
(G) Example coronal slices from a 3D activity map at three different planes (dashed lines in F) encompassing the primary visual cortex. Hemodynamic signals during visual motion stimulation relative to the signals during the presentation of a static gray background image—quantified as Z-scores—are color coded (bottom) for all voxels above a significance threshold and superimposed on the anatomical map (gray scale). Left and right primary visual cortices are outlined in white. L, left; R, right.
(H) Top: single-voxel response curves in the primary visual cortex (3 examples). Middle: mean response curve of the primary visual cortex (across voxels from left and right hemispheres). Bottom: mean eye speed curve. Dashed lines indicate start and end of drifting grating stimulus. All signals are averaged across trials and the four motion directions (top, gray arrows). Same animal as in (G).
(I) Schematic of the experiments for retinotopic mapping along the elevation axis. A horizontal bar (20° wide, corrected for planar distortion) is swept across a display monitor in front of the right eye (4°/s). Left eye is covered (red cross). Three example imaging planes crossing the primary visual cortex (magenta) and superior colliculus (cyan) are labeled (numbered dashed lines). See also Figure S3 for the azimuth retinotopic map.
(J) Example coronal slices from a 3D elevation retinotopic map at three different planes crossing the visual cortex and superior colliculus (dashed lines and color scale in I) superimposed on the anatomical map (gray scale). Superior colliculi and primary visual cortex are outlined in white. L, left; R, right.
(K) 3D elevation retinotopic map in the visual cortex. The position of the bar in the visual field eliciting the peak response is color coded (color scale in I) and superimposed on the anatomical structure. Gray surface shows the 3D outline of the cortex. V, ventral; L, left; P, posterior.
(L) 3D elevation retinotopic map in the superior colliculus. The position of the bar in the visual field eliciting the peak response is color coded (color scale in I) and superimposed on the anatomical structure. Gray surface shows the 3D outline of superior colliculi. V, ventral; L, left; P, posterior.
We first determined the spatial resolution of functional ultrasound imaging by measuring the point-spread function (Figure S2). Spatial resolution was 100 ± 25 μm and 113 ± 25 μm in the imaging plane and 293 ± 25μm off plane. We then tested the functional resolution of ultrasound imaging by performing retinotopic mapping with a bar drifting either horizontally or vertically (Kalatsky and Stryker, 2003, Marshel et al., 2011). Activity in both the visual cortex and the superior colliculus showed retinotopic organization (elevation, Figures 1I–1L; azimuth, Figure S3). In the visual cortex, we observed reversals of the retinotopic map corresponding to different visual cortical areas (Figures S3C and S3D; Marshel et al., 2011). Within the primary visual cortex, the functional resolution along the azimuth was 1.9° in visual space and 71 μm in cortical space (Figure S3D). In the superior colliculus, the functional resolution along the azimuth was 2.2° in visual space and 109 μm in collicular space (Figure S3F). Thus, a spatial resolution of ∼100 μm is obtainable with functional ultrasound imaging in mice.
Brain Regions Activated by Visual Motion Eliciting the Optokinetic Reflex
We began the analysis of visuomotor integration by performing whole-brain functional ultrasound imaging in wild-type mice during stimulation with gratings drifting in the four cardinal directions, each of which evoked the optokinetic reflex in the corresponding direction (Figure 2A; Video S1, n = 12 mice). We chose a long stimulus time (16 s drifting grating, 32 s gray background, Figure S4A) to initiate the reflex and measure smooth pursuit and saccadic eye movements. We used a large number of trial repetitions (6 repetitions per stimulus and per slice) and a moderate imaging rate of 1.7 Hz to maximize the sensitivity of the functional ultrasound imaging (Figure S4B). With this stimulation protocol, we acquired data for one brain slice in 20 min and data for the whole-brain in 6.5 hr. The optokinetic reflex, as measured by mean eye speed, was constant over the course of acquiring the whole-brain scan (Figure S6C).
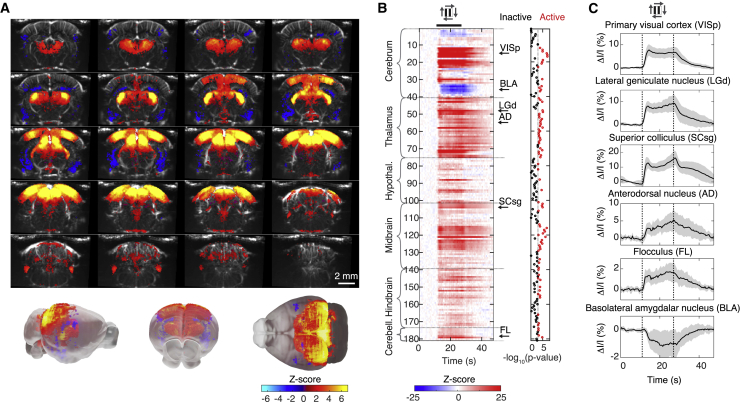
Figure 2.
Brain Regions Activated During Visual Motion Eliciting the Optokinetic Reflex
(A) Top: coronal slices from the mean 3D activity map in wild-type mice (n = 12 mice). Stimulus: drifting gratings. Z-scores (per voxel) are color coded (color scale, bottom) and superposed on the anatomical map of one animal. All voxels significantly active across mice are displayed (p < 0.05, t test). Bottom: 3D views of the same data, presented within a “ghost” surface reference brain in gray (note that the “ghost” is larger, in both rostral and caudal extremities, than the field [6 mm] imaged with functional ultrasound). See also Video S1.
(B) Left: standardized responses of the 181 imaged brain regions, ordered by major brain structures (color scale, bottom). Black thick line, presentation of drifting gratings. Responses are averaged across trials, the four directions of motion (top, gray arrows), and mice (n = 12). Black arrows point to the six regions shown in (C). Right: p values displayed on a log scale. Red circles indicate the 87 active regions (p < 0.001, generalized linear model followed by t test). See also Table S1 for the names of the regions in the same order.
(C) Response curves of the six regions indicated in (B) (black arrows). Curves are averaged across trials and the four directions of motion (top, gray arrows). Thick line, mean over the 12 mice; widths of gray bands, 2 × SD.
Mean 3D activity map for wild-type mice (n = 12 mice) during visual motion eliciting the optokinetic reflex. Active voxels are color coded based on Z-scores (color scale shown in Figure 2A). Transparent gray surface: outer surface of the reference brain. Note that the “ghost” is larger, in both rostral and caudal extremities, than the field (6 mm) imaged with functional ultrasound.
Among the 181 brain regions consistently identified in all animals, 87 regions displayed changes of activity evoked by the stimulus (Figure 2B; Table S1, p < 0.001, t test, corrected for false discovery rate, Figures S4C and S4D). While most regions exhibited an increase in activity during visual stimulation (83/87), a small group showed a decrease in activity (4/87, Figures 2A and 2B). Regions with increased activity were distributed across the whole brain—in the cortex, thalamus, hypothalamus, midbrain, hindbrain, and cerebellum—while regions with decreased activity were located in the amygdalar region (Figures 2B and 2C; Table S1).
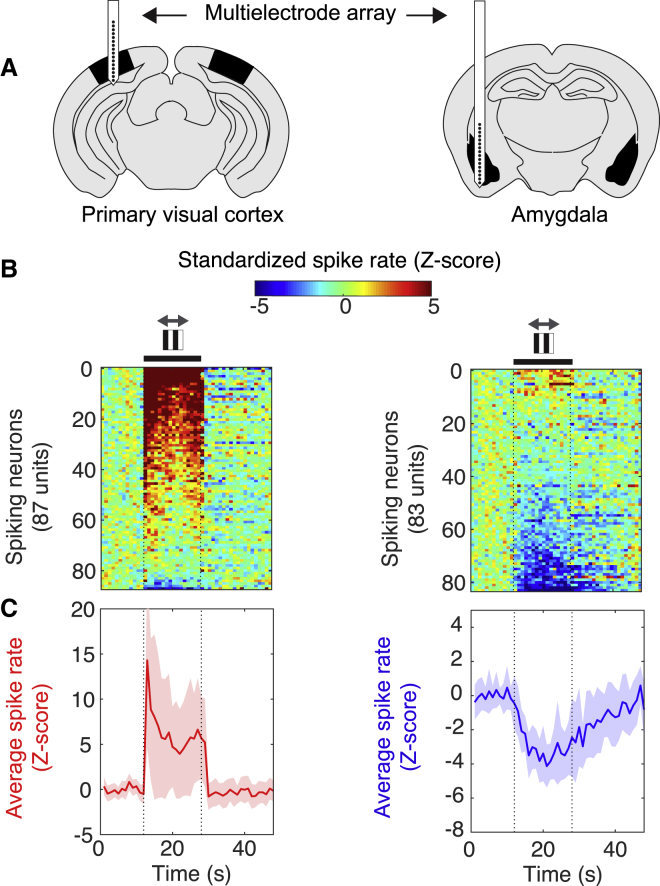
To determine whether the polarity of change in hemodynamic activity correlates with the polarity of change in neuronal activity, we implanted multielectrode probes and recorded spikes from the primary visual cortex and the amygdala, two brain regions that displayed changes in hemodynamic activity with opposite polarity (Figure 3A; Figure S5). Among the neurons that responded to visual stimulation (|Z-score| > 2), 98% showed an increase in spike rate in the primary visual cortex (Figures 3B and 3C). In the amygdala, 100% of responding neurons showed a decrease in spike rate (Figures 3B and 3C). Thus, in these two brain regions, the polarity of change recorded with functional ultrasound imaging matches the polarity of the mean change in spike rate.
Figure 3.
Functional Ultrasound Signals Reflect Modulation of Local Spiking
(A) Schematic of spike recordings with a multielectrode probe positioned in the primary visual cortex (left) or in the amygdala (right) during visual motion eliciting the optokinetic reflex. See also Figure S5.
(B) Standardized spike rate for all neurons recorded in each region (n = 3 mice). Black thick line, presentation of drifting gratings. Gray arrows, axis of visual motion.
(C) Spike rate averaged across all neurons with significant responses in the primary visual cortex (red) and amygdala (blue). Thick line, mean; widths of colored bands, 2 × SD.
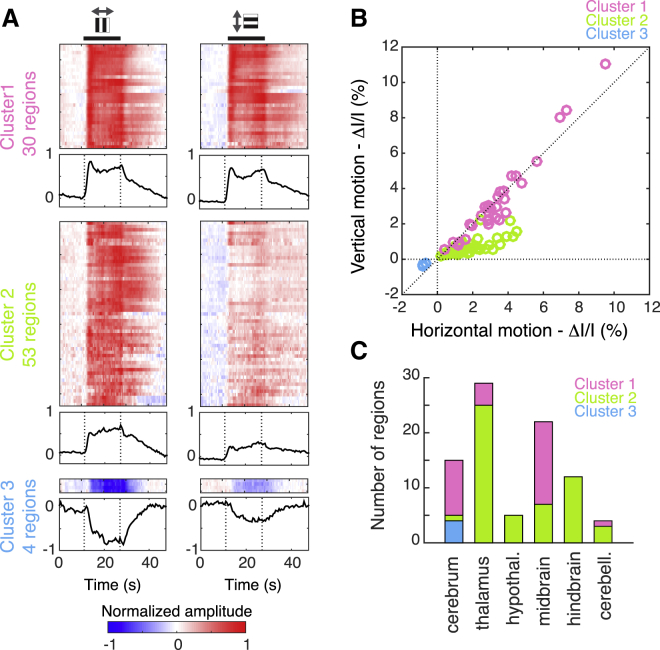
We examined whether the response patterns of brain regions to image drift along the four cardinal directions could be classified into distinct sets. Using k-means clustering, three response clusters emerged (Figure 4). The 30 regions of the first cluster were activated to a similar extent by both horizontal and vertical motion (p = 0.28, Wilcoxon rank-sum test), the 53 regions of the second cluster were activated preferentially during horizontal motion (vertical versus horizontal: −41%, p = 8 × 10−11, Wilcoxon rank-sum test), and the four regions of the third cluster were inhibited preferentially during horizontal motion (vertical versus horizontal: −48%, p = 0.029, Wilcoxon rank-sum test) (Figures 4A and 4B; Table S1). Brain regions in the first cluster were mostly visual areas, including visual cortex, lateral geniculate nucleus, superior colliculus, and pretectal nuclei (Table S1). Regions of the second cluster were distributed in the thalamus, hypothalamus, hindbrain, and cerebellum. The third cluster was exclusively in the amygdalar region (Figure 4C).
Figure 4.
Clusters of Brain Regions Respond Differentially to Horizontal and Vertical Visual Motion
(A) Normalized responses to horizontal visual motion (left panels, average of left and right motion) and vertical visual motion (right panels, average of up and down motion) of the 87 active regions (color scale, bottom) clustered into three sets. Axis of motion, gray arrows on the top. Black thick line, presentation of drifting gratings. The mean response curve of the cluster is shown below each cluster.
(B) Scatterplot of response amplitudes (n = 12 mice) to horizontal and vertical motion for each active region. Each circle represents a brain region, colored according to its cluster (color codes, in legend). Dotted lines, axis of origin lines and unity line.
(C) Distribution of the three clusters within major brain structures.
Brain Regions of the Optokinetic Reflex Pathway
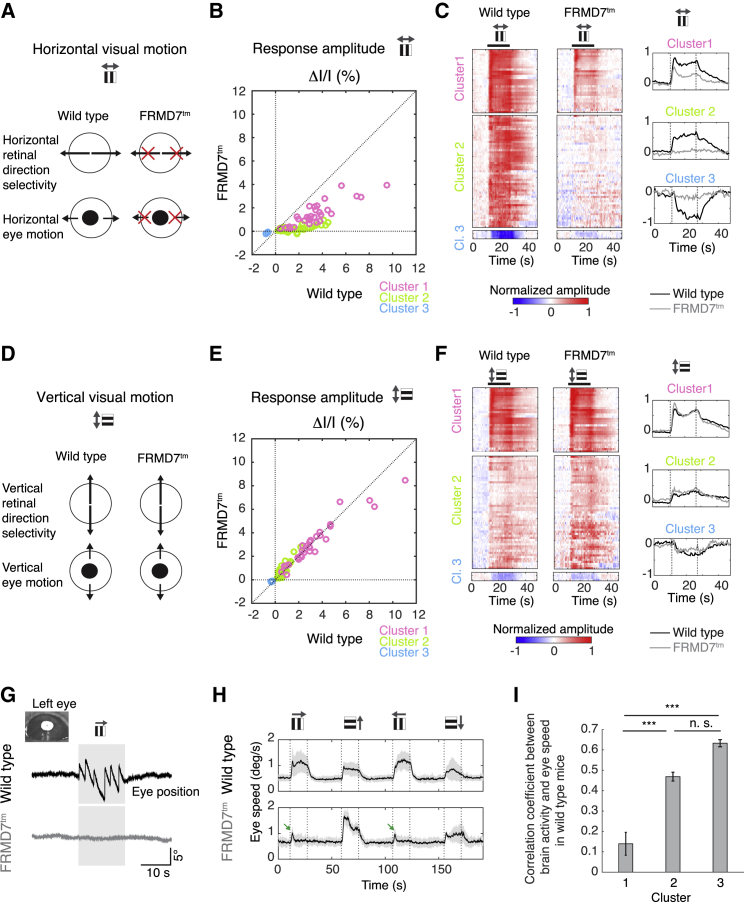
Brain regions responding to the presentation of drifting gratings could be regions that are driven primarily by visual inputs or, alternatively, they could be part of the reflex pathway. To associate brain regions with these two roles, we specifically blocked the generation of the horizontal optokinetic reflex at the level of the retina. The retina contains ∼30 types of output neurons, the ganglion cells (Sanes and Masland, 2015). Many ganglion cell types respond to visual motion, but only a subset, the direction-selective ganglion cells, drives the optokinetic reflex; the direction selectivity of these ganglion cells is necessary for the reflex (Hillier et al., 2017, Yoshida et al., 2001). Each direction-selective ganglion cell shows a preference for one of the four cardinal directions (Oyster and Barlow, 1967, Sabbah et al., 2017). A mouse model of congenital nystagmus, FRMD7tm, lacks direction selectivity in the retina specifically in horizontal directions (Figure 5A), while vertical direction selectivity is normal (Figure 5D) (Yonehara et al., 2016). As a consequence, FRMD7tm mice lack the horizontal optokinetic reflex (i.e., the eyes do not move during the presentation of horizontally drifting gratings, Figure 5A; Figures S6A and S6B), but the vertical reflex is unperturbed (Figure 5D; Figures S6A and S6B; Yonehara et al., 2016). No other oculomotor phenotype has been found in FRMD7tm mice. Although FRMD7tm mice lack horizontal direction selectivity, other types of ganglion cells exhibit normal responses. The lack of direction selectivity can arise from equal responses to visual motion in all directions or from a lack of response to visual motion in any direction (Hillier et al., 2017, Yonehara et al., 2016); the overall effect on the retinal ganglion cell population is slightly elevated spiking activity during visual motion.
Figure 5.
Brain Regions Associated with the Optokinetic Reflex
(A) Schematic illustrating retinal direction selectivity and eye motion in wild-type and FRMD7tm mice during horizontal visual motion. Black arrows, directions of visual and eye motions; red crosses, directions that lack retinal direction selectivity or tracking eye motion.
(B) Scatter plot of response amplitudes to horizontal visual motion in FRMD7tm mice (n = 4 mice) and wild-type mice (n = 12 mice) for each active region. Responses are averaged across trials, two directions of motion (rightward and leftward), and mice. Each circle represents a brain region, colored according to its cluster (color code, in legend). Dotted lines, axis of origin lines and unity line; gray arrows (top), axis of visual motion.
(C) Normalized responses of the 87 active brain regions to horizontal visual motion for wild-type mice (left panels) and FRMD7tm mice (middle panels; color scale, bottom) grouped by their cluster (as in Figure 4A). Responses in wild-type and FRMD7tm mice were normalized by the same values. Black thick line, presentation of drifting gratings. The mean response curve of each cluster is shown in the right panels (black, wild-type; gray, FRMD7tm mice). Dotted lines, start and end of drifting gratings; gray arrows (top), axis of visual motion.
(D) Schematic illustrating retinal direction selectivity and eye motion in wild-type and FRMD7tm mice during vertical visual motion. Black arrows, directions of visual and eye motions.
(E) Scatter plot of response amplitudes to vertical visual motion in FRMD7tm mice (n = 4 mice) and wild-type mice (n = 12 mice) for each active region. Responses are averaged across trials, two directions of motion (upward and downward), and mice. Each circle represents a brain region, colored according to its cluster (color code, in legend). Dotted lines, axis of origin lines and unity line; gray arrows (top), axis of visual motion.
(F) Normalized responses of the 87 active brain regions to vertical visual motion for wild-type mice (left panels) and FRMD7tm mice (middle panels; color scale, bottom) grouped by their cluster (as in Figure 4A). Responses in wild-type and FRMD7tm mice were normalized by the same values. Black thick line, presentation of drifting gratings. The mean response curve of each cluster is shown in the right panels (black, wild-type; gray, FRMD7tm mice). Dotted lines, start and end of drifting gratings; gray arrows (top), axis of visual motion.
(G) Example of eye movements during the optokinetic reflex evoked by visual motion in a horizontal direction (top, gray arrow). Horizontal eye position (y axis) in a single trial shown in a wild-type (top, black) and a FRMD7tm (bottom, gray) mouse. Gray rectangles, presentation of drifting gratings. Inset (top left): example image of the left eye from the eye-tracking camera. White region in middle, pupil.
(H) Mean tracking eye speed over time in wild-type (top, n = 12 mice) and FRMD7tm (bottom, n = 4 mice) mice during the optokinetic reflex. Thick line, mean; gray band, 2 × SD; dotted lines, start and end of drifting gratings. Direction of visual motion, gray arrows on top. Small responses indicated by green arrows are pupil dilation artefacts.
(I) Bar plot (mean ± SEM) of correlation coefficients between paired responses of the three clusters and eye speed of wild-type mice (n = 12 mice).
We acquired whole-brain responses to drifting gratings in FRMD7tm mice (n = 4 mice). During horizontal motion, we observed a brain-wide decrease in activity compared to wild-type mice (Figure 5B; Video S2, p = 7 × 10−13, Wilcoxon rank-sum test). In contrast, the activity during vertical motion was similar between FRMD7tm and wild-type mice (Figure 5E, p = 0.32, Wilcoxon rank-sum test). We further examined changes in activity in FRMD7tm mice cluster by cluster (Figures 5C and 5F). In the first cluster, the response to horizontal motion decreased but was still present (Figure 5C, −56%, p = 2.0 × 10−5, Wilcoxon rank-sum test), whereas in the second and third clusters, the response to horizontal motion was almost completely abolished (Figure 5C, cluster 2, −96%, p = 2.0 × 10−16, cluster 3, −91%, p = 0.029, Wilcoxon rank-sum test). Therefore, regions in the second and third clusters are only active if the retinal input driving the optokinetic reflex is present. Thus, these regions are either part of the reflex pathway or receive direction-selective retinal signals independent of the reflex.
Mean 3D activity map for FRMD7tm mice (n = 4 mice) during horizontal motion eliciting the optokinetic reflex. Voxels are color coded based on Z-scores (color scale shown in Figure 2A, display threshold: −2 < Z-score > 2). Transparent gray surface: outer surface of the reference brain. Note that the “ghost” is larger, in both rostral and caudal extremities, than the field (6 mm) imaged with functional ultrasound.4
If the second and third clusters represent regions of the optokinetic reflex pathway, the motor behavior of wild-type mice should correlate more strongly with the activity of these regions than with the regions of the first cluster. Indeed, the mean eye speed was higher during the horizontal reflex than the vertical reflex (Figures 5G–5I, vertical versus horizontal: −40%, p = 0.0024, Wilcoxon signed-rank test, n = 12 mice), which mirrors the higher activity evoked by horizontal compared to vertical visual motion in the second and third clusters (Figures 4A and 4B). Since the difference between horizontal and vertical eye speed varied from mouse to mouse, we used this variability to correlate eye speed with the response of brain regions in wild-type mice (n = 12 mice). The correlation was significantly higher for the second and third clusters (Figure 5I, Rcluster 2 = 0.47 ± 0.02, Rcluster 3 = 0.63 ± 0.02, mean ± SEM) than for the first cluster (Figure 5I, Rcluster 1 = 0.14 ± 0.06, mean ± SEM, 1/2: p = 1 × 10−8, 1/3: p = 2 × 10−4, 2/3: p = 0.32, one-way ANOVA, Tukey-Kramer correction for multiple comparisons). Together, the observed lack of response to horizontal motion in FRMD7tm mice and the strong correlation with eye movement suggest that regions of the second and third clusters are predominantly part of the reflex pathway and do not simply receive direction-selective signals independent of the reflex. In contrast, the smaller decrease in activity observed in FRMD7tm compared to wild-type mice and the lower correlation with eye movement suggest that regions from the first cluster are primarily visual regions, responsive to visual motion.
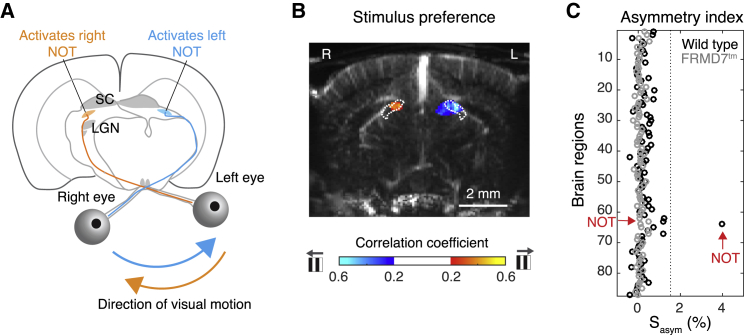
The nucleus of the optic tract (NOT) is a small midbrain nucleus belonging to the accessory optic system that is known to specifically process retinal direction-selective signals and to be indispensable for the generation of the horizontal optokinetic reflex (Gamlin, 2006, Yakushin et al., 2000). In mice, the NOT receives input from temporal-nasal-tuned retinal direction-selective ganglion cells of the contralateral eye (Dhande et al., 2013), and therefore, horizontal image drift in one direction is predicted to cause greater activity in one hemisphere (Figure 6A). Moreover, this hemispheric response asymmetry of NOT should not be present in FRMD7tm mice. To reveal which brain regions exhibit hemisphere asymmetry, we compared responses of brain regions between hemispheres during rightward or leftward visual motion using an asymmetry index. Strikingly, of all the active regions, the NOT was the only one showing strong asymmetry in wild-type mice (Figure 6C, NOT: Sasym = 3.99%, p = 6 × 10−13, t test), with the preferred direction being temporal-nasal (Figure 6B). Asymmetry was completely abolished in FRMD7tm mice (Figure 6C, NOT: Sasym = 0.094%, p = 0.99, t test). Thus, the precision of whole-brain functional ultrasound imaging is sufficient to localize a small and deep midbrain nucleus, the NOT (mean cross section area, 0.29 mm2, mean depth, 2.7 mm). Furthermore, the changes in activity observed in the NOT of FRMD7tm mice were consistent with the retinal and behavioral phenotype of FRMD7tm mice and the known anatomy and physiology of the circuit.
Figure 6.
Direction Selective Responses in the Nucleus of the Optic Tract
(A) Schematic illustrating the logic behind hemisphere-asymmetric responses of the nucleus of the optic tract (NOT). Leftward visual motion (blue arrow) stimulates temporal-to-nasal-tuned retinal direction-selective ganglion cells in the right eye, which then stimulate the NOT in the left hemisphere. The same logic holds for rightward visual motion (orange arrow).
(B) Map of correlation coefficients of single-voxel responses with leftward and rightward stimulus for an example wild-type mouse (color scale at the bottom; gray arrow, direction of visual motion) superposed on the anatomical map. White dotted lines, outline of NOT. L, left; R, right.
(C) Asymmetry index of the 87 active regions in wild-type mice (black, wild-type; gray, FRMD7tm mice). Dotted line, mean + 3 × SD of values in wild-type mice. Red arrows, NOT.
Brain Regions Associated with Eye Motion
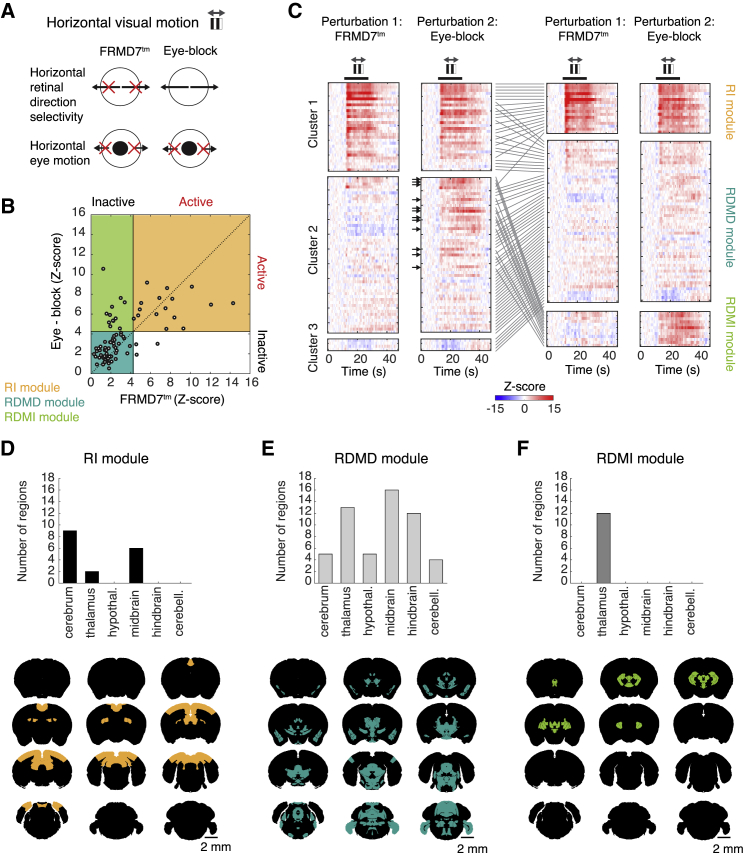
Regions in the optokinetic reflex pathway can be activated at different stages of visuomotor integration. Activity may result from the processing of retinal signals, progressively transformed into motor commands, or could be a consequence of eye motion. In FRMD7tm mice, in which the optokinetic reflex is blocked at the retinal level, the retinal signals driving the reflex and the signals arising from eye motion are both perturbed. To differentiate between these two components, we mechanically blocked eye movement in wild-type mice by depositing a drop of tissue adhesive below each eye. In this perturbation, direction-selective retinal signals are intact but eye motion and associated sensory feedback, together with activities linked to resetting saccades, are blocked. We compared the activity of the 87 visual-motion-activated brain regions during horizontal drifting gratings in FRMD7tm mice (n = 4 mice) and in wild-type mice whose eye movements were blocked (Figure 7A, “eye-block,” n = 4 mice, eye tracking confirmed the abolition of eye movement in eye-block experiments; Figures S6A and S6B). The first and third clusters of FRMD7tm mice and eye-block mice were not significantly different (first cluster: p = 0.6, chi-square test, third cluster: p = 1, chi-square test) (Figure 7C). However, the second cluster was significantly different (p = 1.4 × 10−5, chi-square test): 12 regions of the second cluster responded in eye-block, but not in FRMD7tm, mice (Figure 7C).
Figure 7.
Functional Modules Activated during Visual Motion Eliciting the Optokinetic Reflex
(A) Schematic illustrating retinal direction selectivity and eye motion in FRMD7tm and eye-block mice during horizontal visual motion. Black arrows, directions of visual and eye motions. Red crosses, directions that lack retinal direction selectivity or eye motion.
(B) Scatterplot of the responses of brain regions (quantified by Z-scores) during stimulation with horizontal visual motion in FRMD7tm (n = 4) and eye-block (n = 4) mice. Each circle represents a brain region. Black lines, threshold (p = 0.001, Bonferroni corrected) for defining a brain region as active or inactive. Colored panels indicate the three modules defined by the two threshold lines: RI, RDMD, and RDMI. One region from the RI module is out of range (x = 27.8, y = 10.8).
(C) Standardized responses during horizontal visual motion in both FRMD7tm and eye-block perturbations of the optokinetic reflex (color scale, bottom). Left pair of panels, regions are organized according to clusters (as in Figure 4A). Right pair of panels, regions are organized according to functional modules, defined in (B). Lines between the left and right pairs of panels link the position of each brain region across the two classifications. Black arrows indicate the regions from the RDMI module. Black thick lines, presentation of drifting gratings. Gray arrows (top), axis of visual motion.
(D–F) Top: distribution of brain regions of the RI (D), RDMD (E), and RDMI (F) modules in major brain structures. Bottom: coronal slices showing the spatial distribution of each module region in the reference brain (from bregma + 0.1 mm to bregma + 5.6 mm, 500 μm steps).
Distinct Functional Modules of Brain Areas Activated during the Optokinetic Reflex
Thus, two perturbations of the optokinetic reflex—genetic and mechanical—demarcated three separate functional modules of brain regions activated during visual triggering of the reflex (Figure 7B; Video S3). There is one module (“reflex-independent”: RI, 17/87 active regions) that is active during both perturbations (although less than in unperturbed wild-type mice, Figure S7D), a second module that does not respond when either perturbation is present (“reflex-dependent, motor-dependent”: RDMD, 55/87 active regions), and a third module that is unresponsive during the FRMD7tm perturbation but responsive during the eye-block perturbation (“reflex-dependent, motor-independent”: RDMI, 12/87 active regions) (Figure 7B). The functional modules, obtained by perturbations of the reflex, were linked to the previously characterized response clusters (Figure 7C). Note that clusters are defined based on responses in wild-type mice while modules are defined based on perturbations of behavior. The RI module consists mainly of brain regions in the first cluster (of the 17 regions in the RI module, 16 belong to cluster 1); the RDMD module consists of a large subset of the second and third clusters and a smaller subset of the first cluster (of the 55 regions in the RDMD module, 44 belong to clusters 2 and 3, and 11 to cluster 1); the RDMI module consists of a subset of the second cluster (all 12 regions in the RDMI module belong to cluster 2). Remarkably, while the RI and RDMD modules were distributed across the brain (Figures 7D and 7E), all 12 brain regions of the RDMI module were located in the thalamus (Figure 7F), with six regions in the central thalamus, three in the anterior thalamus, two in the lateral and ventral thalamus, and the remaining region in the reticular nucleus of the thalamus (Table S1).
RI module (left), RDMD module (middle), and RDMI module (right). Voxels of the atlas belonging to the RI, RDMD, and RDMI module regions are colored in purple, red, and cyan, respectively. Transparent gray surface: outer surface of the reference brain. Note that the “ghost” is larger, in both rostral and caudal extremities, than the field (6 mm) imaged with functional ultrasound.5
The optokinetic reflex can also be blocked pharmacologically using anesthesia. Under general anesthesia, visual responses can be recorded in visual areas (Hillier et al., 2017), but motor action, including eye movement, is suppressed (Nair et al., 2011). We obtained whole-brain activity maps during stimulation with horizontal drifting gratings in wild-type mice anesthetized with a mixture of fentanyl, midazolam, and medetomidine (FMM, n = 4 mice, Figure S7). Notably, the input-output conditions of the optokinetic reflex were similar in anesthetized and eye-block mice (Figure S7A): horizontal retinal direction selectivity was present, as confirmed by the preservation of hemisphere asymmetry in the NOT (Figure S7C), but eye motion was absent (Figures S6A and S6B). Despite this similarity, activity under anesthesia was more related to that measured in FRMD7tm than in eye-block mice (Ranesthesia/FRMD7tm = 0.84, Ranesthesia/eye-block = 0.44): regions of the RDMD module were all inactive (anesthesia: 0/12 active, FRMD7tm: 0/12 active, eye-block: 12/12 active, Figure S7B). These results suggest that anesthesia blocks the reflex at an early stage of the visuomotor transformation.
Discussion
We have developed a method for recording and registering whole-brain activity in behaving mice. The spatial resolution (∼100 μm) makes it possible to detect activity in small, deep brain nuclei such as the NOT. Using this method, we produced an unbiased activity map and determined the set of brain regions activated during an innate visuomotor behavior—the optokinetic reflex. Multiple regions throughout the brain responded during the visual stimulus that elicited the behavior. These regions could be divided into three functional modules based on the impact of different perturbations to the reflex.
Functional Modules Activated by Visual Motion
The RI module encompasses visual regions, both retino-recipient regions as well as higher-order regions of major visual pathways. Regions of the RI module responded to the visual stimulus in all perturbations, but responses were higher in wild-type mice (Figure S7D). This suggests that some regions of the reflex pathway modulate visual responses in the regions of the RI module or that the altered image motion on the retina evokes less activity in the absence of eye motion. The RDMD module consists of regions that respond only when the optokinetic reflex is functional. These regions are numerous and distributed across the brain, mainly subcortically. It was expected from previous studies that some of these regions, such as the flocculus in the cerebellum, the pontine and vestibular nuclei in the hindbrain and the NOT in the midbrain, would be involved in the reflex (Büttner-Ennever, 2006, Distler and Hoffmann, 2012, Masseck and Hoffmann, 2009). However, changes in activity recorded for some regions were unexpected, such as decreased activity in the amygdalar nuclei. Finally, the RDMI module consists of regions associated with the optokinetic reflex but independent of eye motion. All of these regions are located in the thalamus, mainly in the central and anterior portions (9/12 regions). The central thalamus, also referred to as the oculomotor thalamus, is known to display eye-motion-related signals in primates (Tanaka and Kunimatsu, 2012, Wurtz et al., 2011). Our results suggest that these regions primarily process visual motion or premotor signals, as opposed to eye position or feedback signals. Interestingly, the anterior thalamus is known to include head-direction cells involved in spatial navigation (Taube, 2007), which suggests a potential link between retinal motion computation and head-direction computation.
Neurovascular Coupling
The link between the hemodynamic response and underlying neuronal activity has been studied in the context of fMRI. It is accepted that positive hemodynamic responses are associated with increases in neuronal activation (Logothetis and Wandell, 2004). Consistent with this view, we detected an increase in local spiking in the primary visual cortex, where we observed a positive functional ultrasound signal. In contrast, the origin of negative hemodynamic responses has not been settled (Kim and Ogawa, 2012) and a variety of mechanisms have been proposed: (1) decrease in neuronal activity, leading to a decrease in blood flow and/or blood volume (Shmuel et al., 2006, Lee et al., 2016); (2) vasoconstriction in the absence of a decrease in neuronal activity (Shih et al., 2009); (3) a “blood-steal” effect close to active regions (Harel et al., 2002); and (4) for BOLD signals, an increase in metabolism with insufficient increase in blood flow and blood volume (Schridde et al., 2008). We detected a decrease in spiking activity in a region where we observed a negative functional ultrasound imaging signal: this supports the first explanation (1).
Applicability of Whole-Brain Functional Ultrasound Imaging
Since the behavior we studied does not habituate (Figure S6C), we used a long stimulation time, many stimulus repetitions, and moderate functional ultrasound imaging rate, which led to a long imaging time (6.5 hr per animal), in order to maximize the sensitivity of the functional ultrasound imaging. However, when imaging time and habituation is a concern, whole-brain functional ultrasound imaging can be done significantly faster by changing the length of the stimulus, the number of repetitions, or the imaging rate. Indeed, we obtained a whole-brain activity map to a short static grating stimulus (1 s) in 14 min by using a higher imaging rate (10 Hz) (Figure S8). Another way to make imaging faster is to acquire all brain slices simultaneously. This could be achieved using 2D ultrasound probes, instead of the 1D probes used here. 2D functional ultrasound probes are currently being developed.
Another potential limitation of functional ultrasound imaging is the need for a large, chronic cranial window. However, large cranial windows are commonly used in wide-field optical imaging (Andermann et al., 2011, Holtmaat et al., 2009, Wekselblatt et al., 2016, Heo et al., 2016), and we have demonstrated the stability of functional ultrasound signal under large cranial windows for at least one month post-surgery (Figure S1). Moreover, a previous study has shown that functional ultrasound imaging can also be performed via a thinned skull, without the need for a cranial window (Urban et al., 2014).
In summary, the region-annotated brain-wide activity map in mice presented here may serve as a starting point for the selection of brain regions for single-cell-resolution recordings and circuit manipulations in attempts to understand the logic of sensorimotor transformations at the level of microcircuits. Moreover, the simplicity, low cost, and ease of use of whole-brain functional ultrasound imaging, together with the established registration and segmentation process, provides a system for obtaining an unbiased view of brain activity in other behaviors, in wild-type mice, and in animal models of neurologic or psychiatric diseases.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fentanyl | Curamed | N/A |
| Medetomidine | Virbac | Medetor |
| Midazolam | Sintetica | N/A |
| Flumazenil | Sintetica | N/A |
| Naloxone | Orpha | N/A |
| Atipamazole | Virbac | Revertor |
| Buprenorphine | Virbac | Buprevet |
| Surgical tissue adhesive | 3M | Vetbond |
| DiI lipophilic tracer | Invitrogen | D3911 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Frmd7tm1a(KOMP)Wtsi | Knockout Mouse Repository | CSD48756 |
| Mouse: C57BL/6J | Charles River Laboratories | N/A |
| Software and Algorithms | ||
| MATLAB | MathWorks | https://mathworks.com/products/matlab.html |
| Blender | Open source | https://www.blender.org |
| Custom-written MATLAB code | This paper | Request from Lead Contact |
| Psychopy2 | Open source | http://www.psychopy.org |
| Spyking Circus | Open source | https://github.com/spyking-circus/spyking-circus |
| Other | ||
| Ultrasound scanner Vantage 128 | Verasonics | http://verasonics.com/vantage-systems/ |
| High-frequency ultrasound probe L22v14 | Verasonics | http://verasonics.com/high-frequency-linear-arrays/ |
| Real-time fUSi data processing and display | AUTC | http://fusi-functional-ultrasound-imaging.com |
| Silicon multielectrode probes, model A1x16-10mm-100-177-A16 | NeuroNexus | http://neuronexus.com/products/neural-probes/ |
| Amplifier, 16 channels, model ME16-FAI-μPA-System | Multi Channel Systems | https://www.multichannelsystems.com/products/portable-me-systems |
| ETL-200 eye tracking system | Iscan | N/A |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Botond Roska (botond.roska@iob.ch).
Experimental Model and Subject Details
Bl6 mice (‘wild-type’, strain: C57BL/6J) were purchased from Charles River laboratories. FRMD7tm mice refers to hemizygous male FRMD7tm1a(KOMP)Wtsi mice obtained from the Knockout Mouse Project (KOMP) Repository and backcrossed with Bl6 mice (Yonehara et al., 2016). Animals were between three and six months old. Only males were used. Animals were housed at 20-24°C, >40% humidity, in a natural light cycle. Water and food pellets (Kliba Nafag) were provided ad libitum. All animal procedures were performed in accordance with standard ethical guidelines (European Communities Guidelines on the Care and Use of Laboratory Animals, 86/609/EEC) and were approved by the Veterinary Department of the Canton of Basel-Stadt, Switzerland.
Method Details
Cranial Window Implantation
Mice were anesthetized with fentanyl-medetomidine-midazolam (FMM, fentanyl 0.05 mg/kg, medetomidine 0.5 mg/kg, midazolam 5.0 mg/kg, injected subcutaneously (s.c.)). Eye gel (Coliquifilm, Allergan) was applied to prevent corneal dehydration during surgery. A custom titanium head bar was attached to the front of the skull with tissue adhesive (Vetbond, veterinary grade, 3M) and dental cement (Paladur, Heraeus). A large craniotomy was made with a drill extending from 0 mm to −8 mm from the bregma along the antero-posterior axis, and from −4 to +4 mm from the midline along the lateral axis. Dura was kept intact. After removal of the skull, a flap of plastic (Polymethylpentene, 250 μm thick, Goodfellow) was cut to fit the cranial window, positioned on top of a layer of 1% agarose, and sealed with tissue adhesive (Figures 1A and 1B). Anesthesia was reversed with atipamezole-flumazenil (s.c., atipamezole 1.25 mg/kg, flumazenil 0.25 mg/kg). Buprenorphine (s.c., 0.1 mg/kg) was administered for post-surgical analgesia. Animals were left to recover for 2 days before the first imaging session. The surgery was adapted from published protocols of large chronic cranial window implantations used for optical imaging with glass (Andermann et al., 2011, Holtmaat et al., 2009, Wekselblatt et al., 2016) or plastic (Heo et al., 2016). To test the stability of the cranial window, we imaged one animal repeatedly for one month post-surgery. Activity maps, response time course in selected regions and behavior (eye speed during reflex) are presented in Figure S1.
Protocol of Functional Ultrasound Imaging
An awake mouse with a cranial window was head-fixed in front of two stimulation monitors and the body was restrained in a foam jacket. To track eye movements, the left eye was recorded with a camera. Acoustic gel (1 mL, Uni’gel, Asept) was applied on the cranial window for ultrasound coupling before placing the ultrasound probe. The acoustic gel was centrifuged to avoid air bubbles, and kept at room temperature. The ultrasound probe (L22-14v, Verasonics) was positioned into the gel ∼3 mm above the cranial window (Figure 1B). Gel was re-applied about every hour without removing the ultrasound probe. The probe holder was mounted on a linear microprecision motor (Zaber) that moved the probe holder along the anterio-posterior axis. The probe was connected to an ultrasound scanner (Vantage 128, Verasonics) controlled by a PC. At the beginning of each imaging session, a high-resolution 3D anatomical scan of the brain microvasculature was acquired (61 coronal slices from bregma + 0 mm to bregma – 6.0 mm, 100 μm steps). This scan was used for registration. Next, a functional scan was acquired (20 coronal slices from bregma − 0.2 mm to bregma − 5.9 mm, 300 μm steps): at each coronal plane, visual stimuli were presented while functional ultrasound images were taken (1.7 Hz) (Figure S4A). Visual stimulation software (PsychoPy 2) triggered ultrasound recording and eye tracking. Visual stimuli were full-field drifting gratings (spatial frequency, 20°; velocity, 10°/s), presented on two monitors (27’’ by ProLite XB2783HSU, Iiyama) placed on each side of the mouse head in portrait orientation. The monitors were placed 18 cm away from each eye at an angle of 45° with regard to the antero-posterior axis of the mouse. A visual stimulation block was composed of alternating gray backgrounds (16 s each, before and after visual motion), and gratings (16 s) moving in one of four different directions (right, up, left or down). The stimulation block was repeated 6 (or 12) times at each coronal plane (Figure S4A).
Eye-Block Experiments
First, anatomical and functional scans were obtained in wild-type mice. After a few days, motion was blocked in both eyes of the recorded mice. For this, mice were anesthetized with isoflurane (4% for induction, 1% for maintenance) and placed on a heating pad. A 30-gauge syringe was positioned inside the eye socket under the eyeball. A drop (∼0.01 mL) of tissue adhesive (Vetbond, veterinary grade, 3M) was deposited. Eye gel (Coliquifilm, Allergan) was applied to prevent corneal dehydration. After recovering from anesthesia, mice were able to blink. Awake, eye-block mice were then head-fixed in the ultrasound setup and underwent the same imaging procedure as before eye-block. Eye-block was confirmed by eye tracking. At the end of the imaging session, mice were euthanized.
Anesthesia Experiments
Mice were anesthetized with injection of FMM s.c. (fentanyl 0.05 mg/kg, medetomidine 0.5 mg/kg, midazolam 5.0 mg/kg). Additional doses (0.25 × initial dose) were supplemented s.c. every 1.5 hr to maintain anesthesia throughout the imaging session. Rectal temperature was monitored and maintained at 37° with a heating pad. Eye gel (Colliquifilm, Allergan) was applied to prevent corneal dehydration. At the end of the imaging session, anesthesia was reversed by an injection of naloxone-atipamezole-flumazenil (naloxone 0.6 mg/kg, atipamezole 1.25 mg/kg, flumazenil 0.25 mg/kg) s.c.
Eye Tracking
Eye tracking was performed using an ETL-200 eye tracking system (Iscan). To visualize the pupil, the left eye was illuminated with IR light and recorded with an IR camera at 120 Hz. Eye position (x and y) and diameter of the pupil were saved as a function of time. After removing outliers generated by blinks or other tracking artifacts, saccades were detected as events with a speed > 20°/s. Eye position during the slow, tracking phase of the optokinetic reflex was obtained by subtracting saccades. Eye-speed during the tracking phase was calculated by taking the first derivative of the eye position trace (Vx and Vy), low pass filtering Vx and Vy (moving average with a period of 1 s), and determining the length of the velocity vector (Vx, Vy). Stimulus-triggered saccade rate and eye speed traces were obtained by averaging over all trials.
Generation of a Functional Ultrasound Image
The principles of functional ultrasound imaging have been described before in the context of rats (Macé et al., 2011, Macé et al., 2013, Urban et al., 2015). We adapted and improved the procedure for generating functional ultrasound images in mice. The sequence for acquiring a functional ultrasound image is illustrated in Figure S4B. A linear array of 128 ultrasound emitters/receivers (ultrasound probe) emitted plane waves (15 MHz, 2 cycles), each followed by the acquisition of echoes by the receivers. Time-gain-compensation has been applied on the received echoes to compensate for attenuation of ultrasound signals with depth (exponential amplification of 1 dB/mm). From this procedure, a single emit-receive image (‘B-mode image’) was obtained in 133 μs. A B-mode image has low contrast and resolution: this was improved by combining B-mode images obtained by emitting waves in five different angles (−6°, −3°, 0°, 3°, 6°) and by averaging three images per angle, resulting in a higher quality image (‘compound B-mode image’). A compound B-mode image was computed from the 15 individual B-mode images in ∼2 ms (500 Hz rate). For Power Doppler imaging, 200 compound B-mode images were acquired consecutively. During this time, the red blood cells move in the microvasculature within a voxel, causing Doppler effects. The Doppler signal was extracted by singular-value-decomposition-based spatiotemporal filtering from the series of 200 compound images (elimination of the first 20 singular values), followed by additional high-pass temporal filtering (cut-off frequency: 20 Hz). The spatiotemporal filtering removes echoes from surrounding tissues as well as motion artifacts, and is critical for imaging awake animals (Urban et al., 2015). The spatiotemporal filtering was performed on a GPU in real-time, as opposed to the CPU used previously, which resulted in a 15% increase in temporal resolution compared to Urban et al. (2015). The value of a voxel in the Power Doppler image was then calculated as the mean intensity of the Doppler signal in a given voxel at a given time: with I, Power Doppler intensity; x,y, coordinates of the voxel in the imaging plane; b, amplitude of the compound B-mode image after filtering; t, time. This value is proportional to the number of ultrasound scatterers (red blood cells) moving in the voxel in that time, which is further proportional to the volume of blood in the voxel in that time (Rubin et al., 1994, Rubin et al., 1995, Macé et al., 2013). Note that Power Doppler, the method which is used in functional ultrasound imaging, is different from Color Doppler, the method more often used in clinical practice. Power Doppler signals are proportional to blood volume, whereas Color Doppler signals are proportional to blood velocity. Power Doppler is independent of blood velocity and more robust to noise to measure hemodynamic changes in the microvasculature (Rubin et al., 1994, Macé et al., 2013). The total time for acquiring a Power Doppler image was 600 ms (1.7 Hz): 400 ms for acquiring the 200 images, and an additional 200 ms for completing the filtering steps and storing the image (Figure S4B). We refer to a Power Doppler image acquired with this procedure as ‘a functional ultrasound image’. Its size was 143 × 128 voxels, each voxel has an x-y physical dimension of 52.5 μm × 100 μm in the coronal plane.
Comparison with Other Methods
Hemodynamic response can be also measured using other methods, such as blood oxygen level dependent fMRI (BOLD-fMRI), cerebral blood volume fMRI (CBV-fMRI) or intrinsic optical imaging (IOS). Here we briefly discuss how functional ultrasound imaging compares to these methods. First, CBV-fMRI and IOS are more comparable to functional ultrasound imaging than BOLD-fMRI, because they measure the same hemodynamic parameter – blood volume, whereas the BOLD signal depends on the interplay between the metabolic rate of oxygen, the blood flow, and the blood volume. The advantage of blood-volume-based methods over BOLD-fMRI is that they are spared of confounds that can arise due to the complex nature of BOLD signals when metabolic and vascular parameters become uncoupled. Second, to compare the specificity of these techniques for detecting underlying neuronal activity, it is important to understand the origin of the blood signal that is monitored by each technique. Experiments using two-photon imaging have shown that hemodynamic changes within arterioles and capillaries are more correlated with local neuronal activity, and provide a more accurate readout of neuronal activity, than hemodynamic changes within veins (O’Herron et al., 2016, Drew et al., 2011). BOLD-fMRI is biased toward monitoring changes in venous blood in superficial cortical layers (Turner, 2002). IOS is also skewed toward the superficial blood vessels due to limited light penetration. CBV-fMRI is more sensitive to arterial and capillary blood in deep cortical layers than BOLD-fMRI, and studies have confirmed that CBV-fMRI methods have a higher specificity to localize neuronal activity (Huber et al., 2017). Similarly, functional ultrasound imaging has a high specificity because it also captures arteriolar and capillary flow (Urban et al., 2014, Macé et al., 2013). Note that there is a major difference in cost of functional ultrasound imaging and fMRI. The price of an ultrafast ultrasound scanner with a probe is ∼125,000 USD (Verasonics, Seattle). In comparison, the cost of a small-animal fMRI system is over 1,000,000 USD and there are substantial maintenance costs.
Generation of an Anatomical Ultrasound Image
To increase resolution and contrast, the acquisition of an anatomical ultrasound image was different from the acquisition of a functional ultrasound image. We acquired compound images by combining B-mode images obtained by emitting waves in 25 different angles (−12° to +12°, 1° steps, no averaging). A compound B-mode image was therefore computed from 25 individual B-mode images. Moreover, we acquired 300 compound B-mode images (instead of 200) to compute a Power Doppler image (the same filtering steps as for functional ultrasound images). We refer to a Power Doppler image acquired with this procedure as an ‘anatomical ultrasound image’. Its size was 143 × 128 voxels, each voxel has an x-y physical dimension of 52.5 μm × 100 μm in the coronal plane. We refer to the stack of 61 anatomical ultrasound images across the brain (inter-slice distance: 100 μm) as a ‘3D anatomical map’.
Protocol and Sequence for Fast, Whole-Brain Functional Ultrasound Imaging
Experimental setup and handling of the awake head-fixed mouse was the same as for other experiments. The visual stimulus was a static vertical grating (spatial frequency, 20°) reversing contrast at 8.5 Hz, presented on the two stimulation monitors (same configuration as for other experiments). A visual stimulation block was composed of alternating gray backgrounds (2 s / 4 s before and after the stimulus, respectively), and grating (1 s) (Figure S8A). The stimulation block was repeated 6 times at each coronal plane (42 s acquisition time per plane, total acquisition time 14 min) (Figure S8A). The acquisition of the 20 coronal planes was randomized. Spatial sampling was 143 × 128 × 20 voxels of size 52.5 μm × 100 μm × 300 μm as for other experiments. Functional ultrasound images were acquired at 10 Hz using a fast sequence (Figure S8B). The fast sequence was achieved by using 50 compound images (instead of 200) for computing a power Doppler image. The parameters for acquiring compound images were the same as for other experiments (5 angles: −6°, −3°, 0°, 3°, 6°, 3 averages per angle, 500 Hz rate). Improving the GPU algorithm for spatiotemporal filtering led to true real-time acquisition (no dead time) (Figure S8B).
Physical Resolution of Power Doppler Imaging
The spatial resolution of a functional ultrasound image (or Power Doppler image) is defined by the size of the point-spread function produced by an infinitely small flow of particles. The point-spread function was measured in two independent ways, illustrated in Figure S2.
Direct Method
The point-spread function of Power Doppler was measured by imaging the flow of a suspension of particles in water in a tube of 80 μm diameter (Figures S2A–S2C). The particles were brewer’s yeast (Saccharomyces cerevisiae), which have similar size and ultrasound scattering characteristics as red blood cells. The spatial resolution was quantified as the full width at half maximum. The spatial resolutions were , , and . Since the size of the tube (80 μm) is comparable to the resolution, a deconvolution was performed. After deconvolution, the adjusted resolutions were , , and .
Indirect Method
The point-spread function of a compound B-mode image was measured by imaging a 20 μm diameter metal wire (Figures S2D–S2F). The Power Doppler point-spread function can be calculated from the compound B-mode point-spread function as
To demonstrate the validity of this equation, the one-dimensional case was considered. If two “point flows” are placed at a position 0 and , respectively, a timeseries of compound B-mode images would give a signal defined as
where are the reflectivity of the particles traveling in the “point tube.” They can be considered a random distribution with zero mean , unit standard deviation , and uncorrelated .
To obtain the Power Doppler image, the mean intensity is computed as
This equation shows that to separate the two “point flows,” the distance must be the size of . Therefore, the point-spread function of Power Doppler is
The resolutions found with the indirect method (full-width at half maximum) were , , and . The resolutions obtained with the direct and indirect methods have a maximal difference of 18 μm, which is below the experimental sampling size (25 μm).
The physical resolutions measured above are relevant for a functional ultrasound image. The physical resolution of an anatomical image is higher because more compound angles and more compound B-mode images were used, which made the point-spread function smaller. The dependence of the point-spread function on the maximal angle and number of images used for Power Doppler imaging has been described before (Macé et al., 2013).
Retinotopic Mapping and Functional Resolution
Mice were imaged as described for anesthesia experiments. Visual stimulation was presented on one monitor (27’’ by ProLite XB2783HSU, Iiyama) placed on the right side of the mouse head in landscape orientation. The monitor was placed 18 cm away from each eye at an angle of 45° with regard to the antero-posterior axis of the mouse. The left eye was covered to ensure monocular stimulation. The stimulus consisted of a 20° wide bar that was periodically swept across the monitor at a velocity of 4°/s. The bar was filled with a checkerboard pattern (25° spatial frequency) reversing at 6 Hz. Spherical stimulus correction was applied to compensate for the flatness of the monitor (Marshel et al., 2011). For azimuth maps, the bar was swept in nasal (leftward) and temporal (rightward) directions successively (6 min / direction / coronal slice). For the elevation maps, the bar was swept in downward and upward directions successively (6 min / direction / coronal slice). Retinotopic maps were computed as described for intrinsic imaging (Kalatsky and Stryker, 2003). Briefly, the position of the bar eliciting the strongest signal in each voxel was computed based on the phase of the hemodynamic signal at the bar sweep frequency. Recordings using bars moving in the opposite directions were combined to obtain the absolute phase without hemodynamic delay. A 3D retinotopic map was reconstructed from a stack of coronal slices. Registration and segmentation procedures were performed on the 3D anatomical map and then were used to extract retinotopic maps of different brain structures (such as in Figure S3B). We used retinotopic mapping to quantify the effective spatial resolution of functional ultrasound imaging in visual brain regions. For this, we repeated azimuth retinotopic mapping five times for one coronal slice on the same animal. After registration, we selected a path along the surface of the visual cortex or the superior colliculus and extracted the five retinotopic curves along these paths (position in the visual field (°) versus distance in the brain (mm), Figures S3D–S3F). The functional resolution was defined as the standard deviation of the five curves around the mean curve in visual degrees and cortical/collicular position.
Multielectrode Recordings
A linear silicon multielectrode probe (Neuronexus, model A1x16-10mm-100-177-A16) connected to an amplifier (ME16-FAI-μPA-System, Mutichannel System) was used. The experimental setup was the same as for ultrasound imaging, except the ultrasound probe holder was replaced with an electrode holder. 2-4 days before recordings a head bar was attached to the head and holes were drilled (0.5 mm drill) in the skull above the basolateral amygdalar nucleus (2.0 mm posterior and 2.8 mm lateral from bregma) and the primary visual cortex (3.8 mm posterior and 2.5 mm lateral from bregma). The holes were protected with a biocompatible silicone sealant (Kwik-Cast, WPI) while the mouse recovered from the surgery. Before the recording, the electrode was placed in a fluorescent dye (DiI) to allow for post hoc confirmation of the recording site (Figure S5A). The mouse was head-fixed in the experimental setup and was anesthetized using isoflurane (4% initiation, 1% maintenance) during electrode positioning. The recording started ∼15 min after the isoflurane was turned off, at a time when the head-fixed mouse was awake and was able to perform the optokinetic reflex. 48-60 trials of visual stimulation eliciting the optokinetic reflex were performed per electrode position (same stimulus as with functional ultrasound: 16 s drifting gratings interleaved by 32 s gray background). Recordings were performed at 25 kHz and were analyzed using a template matching spike-sorting algorithm (Spiking Circus, Yger et al., 2018) to extract single units. Templates were excluded if there were too many violations of the refractory period (> 5%) (indicating that the template matched spikes of multiple cells) or merged if the cross-correlation between templates presented a trough at 0 (indicating that the two templates came from the same cell). Peri-stimulus histograms of the spike rate were computed over all the trials for each cell using 1 s bins (Figure S5D). Peri-stimulus spike rate traces were standardized with respect to the baseline (defined as the 10 s before stimulus onset, as for the ultrasound data). The mean Z-score for each unit was defined as the mean value of the standardized spike trace during stimulation. Units were considered responsive if the absolute value of the mean Z-score was >2.
Quantification and Statistical Analysis
Registration and Segmentation
All processing steps were done in MATLAB and are available upon request. In each mouse, we first obtained a 3D anatomical map. This 3D anatomical map was registered to the reference mouse brain atlas from the Allen Brain Institute (http://atlas.brain-map.org/) using 3D rigid transformations. 3D volumetric data were obtained from http://help.brain-map.org/display/mousebrain/API. Registration of the first anatomical scan of a mouse was done semi-manually based on anatomical landmarks identified on both the 3D ultrasound anatomical map and the reference atlas (hippocampi, corpus callosum, middle cerebral sinus, 4th ventricle). Based on these landmarks, we calculated the rotation and translation in the coronal plane, then the rotation and translation in the horizontal plane, and then the rotation in the sagittal plane. If necessary, the translation in the coronal plane was readjusted at the end. The 3D transformation matrix (from 3D anatomical map to reference atlas) as well as the inverse matrix (from reference atlas to 3D anatomical map) was then calculated. At the beginning of each imaging session of the same mouse we obtained a new 3D anatomical map, which was registered automatically with the first one using a 3D rigid, intensity-based algorithm. Functional ultrasound images were registered automatically based on the 3D anatomical map obtained in the same imaging session. Segmentation of voxels to brain regions was based on the segmentation provided in the Allen Mouse Brain Reference Atlas. 181 brain regions were consistently imaged in all animals included in this study (Table S1).
Response Time Course
First, functional ultrasound images were interpolated, voxel by voxel, in time because the 1.7 Hz acquisition rate varied slightly due to variability in GPU processing time. After interpolation, a constant frame rate of 2 Hz was obtained. Second, frames exhibiting large motion artifacts were automatically detected and removed based on average intensity in a region of interest outside of the brain. Specifically, if the signal in the region of interest outside of the brain crossed a threshold (determined from the distribution of the signal at all the time points recorded for this slice), the time point corresponding to this image was labeled as ‘motion artifact’. The threshold was set to median + 2 × sigma, where sigma was 1.48 × median absolute deviation. Third, signals in each voxel were high-pass filtered to remove slow hemodynamic fluctuations (cut-off frequency 0.0052 Hz). Fourth, the relative hemodynamic time course ΔI/I was computed for each voxel, where I was the mean baseline signal (defined as the 10 s before stimulus onset) and ΔI was the change compared to baseline at each time point. Finally, a response time course was obtained for each voxel by averaging ΔI/I over 6 (or 12) trials. Data from time points labeled as motion artifacts were excluded from the average (with compensation by the effective number of trials). The response time course of a brain region (such as in Figure 1H) was computed by averaging the response time course of each voxel belonging to the brain region (after registration and segmentation). The response amplitude of a brain region was the mean of the response time course in a time window (30 s) after stimulus onset.
3D Activity Map
In each voxel (143 × 128 × 20 voxels) we compared signals along the response time course in a time window before stimulus onset and a time window after stimulus onset (before trial averaging) using two-tailed Wilcoxon rank sum test. Data from time points labeled as motion artifacts were excluded from the test. We obtained the Z-statistics of the test (Z) and the corresponding P-value of the test (P) for each voxel, and consequently a Z-score and P-value map for each coronal slice. A 2D median filtering of 4 × 4 pixels was applied on each P-value map to yield a filtered P-value map. On the filtered P-value map a voxel was considered to be significant if p < 1.4 × 10−7 (which corresponds to p < 0.05 after Bonferoni correction for multiple comparisons). 3D activity maps (Figure 1G) show Z-scores for all voxels across all coronal slices which were significant. Mean 3D activity maps for a group of n animals (Figure 2A) display mean Z-scores in each voxel. Group significance is assessed by a one-sample two-tailed t test of the n Z-scores in each voxel (p < 0.05, after 2D median filtering of 4 × 4 pixels).
Active Brain Regions
To determine if a brain region was activated by a stimulus, response time courses of brain regions were computed for each animal (Figure S4C). Activation was then quantified by a T-score using a general linear model, as commonly used in fMRI (Friston et al., 1995). Next, a one-sample two-tailed t test was performed on the n T-scores obtained for the n animals. The region was considered active if the resulting P-value, adjusted for a false discovery rate (Benjamini and Hochberg FDR procedure), was <0.001 (Figure S4D) (Benjamini and Hochberg, 1995). For display (such as in Figure 2B), response time courses were averaged over the four different visual motion directions and across animals, and then standardized with regard to the values before stimulus onset.
Cluster Analysis
Cluster analysis was based on the time courses of the responses to the four stimuli of the 87 motion-stimulus-activated brain regions. First, each time course was normalized to its maximum value over the four directions of visual motion. Second, the response amplitude (i.e., the mean amplitude in a time window after stimulus onset) was determined for each stimulus direction, resulting in 87 vectors, each with four values. The identity and number of clusters were determined using k-means clustering and the location of the maximum in the silhouette function.
The response time course of a cluster was the mean of the normalized response time courses of the brain regions within the cluster. The response time course of a cluster was computed for horizontal (two directions) or vertical (two directions) motion stimuli separately (such as in Figure 4A). Response time courses of brain regions in FRMD7tm mice were normalized by the maximum value of the wild-type response and parsed into the same clusters (Figures 5C and 5F).
The response amplitude of a cluster was the change in the mean of the response time course of the cluster in a time window before stimulus onset compared to in a time window after stimulus onset. The Wilcoxon rank sum test was used to compare response amplitudes between clusters across two different groups of mice.
Asymmetry Index
The response asymmetry was defined as follows:
where SH(right) and SH(left) are the response time courses of a brain region in the right and left brain hemispheres, respectively. The first term, [SH(right) (t) − SH(left) (t)] rightward visual motion, is determined during stimulation with visual motion to the right, while the second term, [SH(left) (t) − SH(right) (t)] leftward visual motion, is determined during stimulation with visual motion to the left. The asymmetry index (Figure 6C) is the mean of Sasym (t) in a time window after stimulus onset. To determine significance, we calculated the mean and standard deviation of the asymmetry index over the 87 regions active in wild-type mice. The significance threshold was set at mean ± 3 s.d.
Correlation of Brain Activity with Eye Movements
For each mouse, response time courses of active brain regions were correlated with the eye speed time courses. The resulting 87 × n Pearson correlation coefficients were averaged over the n mice. Mean and s.e.m were calculated for each of the three clusters and are shown as absolute values in Figure 5I. A one-way ANOVA, with Tukey-Kramer correction for multiple comparisons, was performed to compare the three clusters.
Module Analysis
Matched data were used to compare FRMD7tm, eye-block, and anesthesia conditions: two stimuli (rightward and leftward visual motion), six trials/slice, 20 slices/mouse, four mice/group. Response time courses of the 87 brain regions were averaged over stimuli, trials, and animals to obtain a mean response time course of each brain region per condition. The resulting mean response time courses were standardized with respect to the values before stimulus onset (Figure 7C), and a Z-score (Figure 7B) was extracted per region and per condition by applying a moving average (16 s) and taking the maximum of each curve. The region was considered active if Z > 4.23, which corresponds to a P-value of 0.001 after Bonferroni correction for multiple comparisons (Figure 7B).
Analysis of Fast Functional Ultrasound Imaging Data
3D activity maps (Figure S8C) were computed as for other experiments using two-tailed Wilcoxon rank sum test. For the 3D activity map from one single scan, the threshold on the filtered P-value map was p < 0.05, uncorrected (absolute Z-score value > 2). For the 3D activity map combining multiple scans, the mean Z-score of the n Z-score maps was displayed and group significance was assessed by a one-sample two-tailed t test of the n Z-scores in each voxel (p < 0.05, after 2D median filtering of 4 × 4 pixels). The 16 fast scans were acquired in four independent imaging sessions, four scans per session (14 min recording time per scan, 1 hr total recording time per mouse for the four successive scans, three mice imaged, one mouse was imaged twice). To show the activity as a function of the number of scans without bias from one imaging session, the data were selected as follow: four scans = first scan of each imaging session, eight scans = first two scans of each imaging session, 16 scans = all scans. Response time courses in the three example brain regions were computed as for other experiments (Figures S8D and S8E). For multiple scan data, mean ± s.d. is displayed.
Data and Software Availability
Data and custom-written software are available upon request.
Acknowledgments
We thank A. Drinnenberg, G. Kosche, D. Hillier, P. King, and S. Oakeley for commenting on the manuscript. We thank P. Argast for assistance with the head and probe holder prototyping. We acknowledge the following grants: Human Frontier Science Program Postdoctoral Fellowship (LT000769/2015) to E.M.; Swiss National Science Foundation Ambizione Grant (PZOOP3_168213) and Canada Research Chair Grant to S.T.; Swiss National Science Foundation grants (3100330B_163457), the National Center of Competence in Research Molecular Systems Engineering grant, European Research Council (669157, RETMUS), and DARPA (HR0011-17-C-0038, Cortical Sight) grants to B.R.
Author Contributions
Experiments were designed by E.M. and B.R. Experimental setup was developed by E.M. Ultrasound software was developed by G.M. and A.U. Experiments were performed by E.M., A.B., and S.T. Data analysis was performed by E.M. Software for 3D rendering was written by C.C. The paper was written by E.M., B.R., and S.T.
Declaration of Interests
E.M and G.M. are inventors on the patent WO2012131418A1. A.U. is a founder and shareholder of AUTC.
Published: December 5, 2018
Footnotes
Supplemental Information includes eight figures, one table, and three videos and can be found with this article online at https://doi.org/10.1016/j.neuron.2018.11.031.
Supplemental Information
The ordering of brain regions is the same as for the rows in Figure 2B. Atlas ID and name columns indicate the region ID and name of imaged regions in the Allen Brain Institute reference atlas. Cluster and module identity of all active regions is indicated.
References
- Andermann M.L., Kerlin A.M., Roumis D.K., Glickfeld L.L., Reid R.C. Functional specialization of mouse higher visual cortical areas. Neuron. 2011;72:1025–1039. doi: 10.1016/j.neuron.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Büttner-Ennever J.A. Neuroanatomy of the oculomotor system. Preface. Prog. Brain Res. 2006;151 doi: 10.1016/S0079-6123(05)51019-7. vii–viii. [DOI] [PubMed] [Google Scholar]
- Callaway E.M., Luo L. Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J. Neurosci. 2015;35:8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande O.S., Estevez M.E., Quattrochi L.E., El-Danaf R.N., Nguyen P.L., Berson D.M., Huberman A.D. Genetic dissection of retinal inputs to brainstem nuclei controlling image stabilization. J. Neurosci. 2013;33:17797–17813. doi: 10.1523/JNEUROSCI.2778-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler C., Hoffmann K.P. The optokinetic reflex. In: Liversedge S.P., Gilchrist I., Everling S., editors. The Oxford Handbook of Eye Movements. Oxford University Press; 2012. pp. 65–83. [Google Scholar]
- Drew P.J., Shih A.Y., Kleinfeld D. Fluctuating and sensory-induced vasodynamics in rodent cortex extend arteriole capacity. Proc. Natl. Acad. Sci. USA. 2011;108:8473–8478. doi: 10.1073/pnas.1100428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston K.J., Holmes A.P., Poline J.B., Grasby P.J., Williams S.C., Frackowiak R.S., Turner R. Analysis of fMRI time-series revisited. Neuroimage. 1995;2:45–53. doi: 10.1006/nimg.1995.1007. [DOI] [PubMed] [Google Scholar]
- Gamlin P.D.R. The pretectum: connections and oculomotor-related roles. Prog. Brain Res. 2006;151:379–405. doi: 10.1016/S0079-6123(05)51012-4. [DOI] [PubMed] [Google Scholar]
- Guenthner C.J., Miyamichi K., Yang H.H., Heller H.C., Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel N., Lee S.-P., Nagaoka T., Kim D.-S., Kim S.-G. Origin of negative blood oxygenation level-dependent fMRI signals. J. Cereb. Blood Flow Metab. 2002;22:908–917. doi: 10.1097/00004647-200208000-00002. [DOI] [PubMed] [Google Scholar]
- Heo C., Park H., Kim Y.-T., Baeg E., Kim Y.H., Kim S.-G., Suh M. A soft, transparent, freely accessible cranial window for chronic imaging and electrophysiology. Sci. Rep. 2016;6:27818. doi: 10.1038/srep27818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier D., Fiscella M., Drinnenberg A., Trenholm S., Rompani S.B., Raics Z., Katona G., Juettner J., Hierlemann A., Rozsa B., Roska B. Causal evidence for retina-dependent and -independent visual motion computations in mouse cortex. Nat. Neurosci. 2017;20:960–968. doi: 10.1038/nn.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A., Bonhoeffer T., Chow D.K., Chuckowree J., De Paola V., Hofer S.B., Hübener M., Keck T., Knott G., Lee W.-C.A. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat. Protoc. 2009;4:1128–1144. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber L., Handwerker D.A., Jangraw D.C., Chen G., Hall A., Stüber C., Gonzalez-Castillo J., Ivanov D., Marrett S., Guidi M. High-resolution CBV-fMRI allows mapping of laminar activity and connectivity of cortical input and output in human M1. Neuron. 2017;96:1253–1263.e7. doi: 10.1016/j.neuron.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalatsky V.A., Stryker M.P. New paradigm for optical imaging: temporally encoded maps of intrinsic signal. Neuron. 2003;38:529–545. doi: 10.1016/s0896-6273(03)00286-1. [DOI] [PubMed] [Google Scholar]
- Kerr J.N.D., Denk W. Imaging in vivo: watching the brain in action. Nat. Rev. Neurosci. 2008;9:195–205. doi: 10.1038/nrn2338. [DOI] [PubMed] [Google Scholar]
- Kim S.-G., Ogawa S. Biophysical and physiological origins of blood oxygenation level-dependent fMRI signals. J. Cereb. Blood Flow Metab. 2012;32:1188–1206. doi: 10.1038/jcbfm.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.K., Adhikari A., Deisseroth K. Integration of optogenetics with complementary methodologies in systems neuroscience. Nat. Rev. Neurosci. 2017;18:222–235. doi: 10.1038/nrn.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Weitz A.J., Bernal-Casas D., Duffy B.A., Choy M., Kravitz A.V., Kreitzer A.C., Lee J.H. Activation of direct and indirect pathway medium spiny neurons drives distinct brain-wide responses. Neuron. 2016;91:412–424. doi: 10.1016/j.neuron.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis N.K., Wandell B.A. Interpreting the BOLD signal. Annu. Rev. Physiol. 2004;66:735–769. doi: 10.1146/annurev.physiol.66.082602.092845. [DOI] [PubMed] [Google Scholar]
- Macé E., Montaldo G., Cohen I., Baulac M., Fink M., Tanter M. Functional ultrasound imaging of the brain. Nat. Methods. 2011;8:662–664. doi: 10.1038/nmeth.1641. [DOI] [PubMed] [Google Scholar]
- Macé E., Montaldo G., Osmanski B.-F., Cohen I., Fink M., Tanter M. Functional ultrasound imaging of the brain: theory and basic principles. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2013;60:492–506. doi: 10.1109/TUFFC.2013.2592. [DOI] [PubMed] [Google Scholar]
- Marshel J.H., Garrett M.E., Nauhaus I., Callaway E.M. Functional specialization of seven mouse visual cortical areas. Neuron. 2011;72:1040–1054. doi: 10.1016/j.neuron.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masseck O.A., Hoffmann K.-P. Comparative neurobiology of the optokinetic reflex. Ann. N Y Acad. Sci. 2009;1164:430–439. doi: 10.1111/j.1749-6632.2009.03854.x. [DOI] [PubMed] [Google Scholar]
- Nair G., Kim M., Nagaoka T., Olson D.E., Thulé P.M., Pardue M.T., Duong T.Q. Effects of common anesthetics on eye movement and electroretinogram. Doc. Ophthalmol. 2011;122:163–176. doi: 10.1007/s10633-011-9271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Herron P., Chhatbar P.Y., Levy M., Shen Z., Schramm A.E., Lu Z., Kara P. Neural correlates of single-vessel haemodynamic responses in vivo. Nature. 2016;534:378–382. doi: 10.1038/nature17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyster C.W., Barlow H.B. Direction-selective units in rabbit retina: distribution of preferred directions. Science. 1967;155:841–842. doi: 10.1126/science.155.3764.841. [DOI] [PubMed] [Google Scholar]
- Portugues R., Feierstein C.E., Engert F., Orger M.B. Whole-brain activity maps reveal stereotyped, distributed networks for visuomotor behavior. Neuron. 2014;81:1328–1343. doi: 10.1016/j.neuron.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J.M., Bude R.O., Carson P.L., Bree R.L., Adler R.S. Power Doppler US: a potentially useful alternative to mean frequency-based color Doppler US. Radiology. 1994;190:853–856. doi: 10.1148/radiology.190.3.8115639. [DOI] [PubMed] [Google Scholar]
- Rubin J.M., Adler R.S., Fowlkes J.B., Spratt S., Pallister J.E., Chen J.F., Carson P.L. Fractional moving blood volume: estimation with power Doppler US. Radiology. 1995;197:183–190. doi: 10.1148/radiology.197.1.7568820. [DOI] [PubMed] [Google Scholar]
- Sabbah S., Gemmer J.A., Bhatia-Lin A., Manoff G., Castro G., Siegel J.K., Jeffery N., Berson D.M. A retinal code for motion along the gravitational and body axes. Nature. 2017;546:492–497. doi: 10.1038/nature22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes J.R., Masland R.H. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu. Rev. Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Schridde U., Khubchandani M., Motelow J.E., Sanganahalli B.G., Hyder F., Blumenfeld H. Negative BOLD with large increases in neuronal activity. Cereb. Cortex. 2008;18:1814–1827. doi: 10.1093/cercor/bhm208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y.-Y.I., Chen C.-C.V., Shyu B.-C., Lin Z.-J., Chiang Y.-C., Jaw F.-S., Chen Y.-Y., Chang C. A new scenario for negative functional magnetic resonance imaging signals: endogenous neurotransmission. J. Neurosci. 2009;29:3036–3044. doi: 10.1523/JNEUROSCI.3447-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel A., Augath M., Oeltermann A., Logothetis N.K. Negative functional MRI response correlates with decreases in neuronal activity in monkey visual area V1. Nat. Neurosci. 2006;9:569–577. doi: 10.1038/nn1675. [DOI] [PubMed] [Google Scholar]
- Simpson J.I. The accessory optic system. Annu. Rev. Neurosci. 1984;7:13–41. doi: 10.1146/annurev.ne.07.030184.000305. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kunimatsu J. Oxford University Press; 2012. Thalamic Roles in Eye Movements. [Google Scholar]
- Taube J.S. The head direction signal: origins and sensory-motor integration. Annu. Rev. Neurosci. 2007;30:181–207. doi: 10.1146/annurev.neuro.29.051605.112854. [DOI] [PubMed] [Google Scholar]
- Turner R. How much cortex can a vein drain? Downstream dilution of activation-related cerebral blood oxygenation changes. Neuroimage. 2002;16:1062–1067. doi: 10.1006/nimg.2002.1082. [DOI] [PubMed] [Google Scholar]
- Urban A., Macé E., Brunner C., Heidmann M., Rossier J., Montaldo G. Chronic assessment of cerebral hemodynamics during rat forepaw electrical stimulation using functional ultrasound imaging. Neuroimage. 2014;101:138–149. doi: 10.1016/j.neuroimage.2014.06.063. [DOI] [PubMed] [Google Scholar]
- Urban A., Dussaux C., Martel G., Brunner C., Macé E., Montaldo G. Real-time imaging of brain activity in freely moving rats using functional ultrasound. Nat. Methods. 2015;12:873–878. doi: 10.1038/nmeth.3482. [DOI] [PubMed] [Google Scholar]
- Wekselblatt J.B., Flister E.D., Piscopo D.M., Niell C.M. Large-scale imaging of cortical dynamics during sensory perception and behavior. J. Neurophysiol. 2016;115:2852–2866. doi: 10.1152/jn.01056.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz R.H., McAlonan K., Cavanaugh J., Berman R.A. Thalamic pathways for active vision. Trends Cogn. Sci. 2011;15:177–184. doi: 10.1016/j.tics.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushin S.B., Gizzi M., Reisine H., Raphan T., Büttner-Ennever J., Cohen B. Functions of the nucleus of the optic tract (NOT). II. Control of ocular pursuit. Exp. Brain Res. 2000;131:433–447. doi: 10.1007/s002219900302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yger P., Spampinato G.L., Esposito E., Lefebvre B., Deny S., Gardella C., Stimberg M., Jetter F., Zeck G., Picaud S. A spike sorting toolbox for up to thousands of electrodes validated with ground truth recordings in vitro and in vivo. eLife. 2018;7:7. doi: 10.7554/eLife.34518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K., Fiscella M., Drinnenberg A., Esposti F., Trenholm S., Krol J., Franke F., Scherf B.G., Kusnyerik A., Müller J. Congenital nystagmus gene FRMD7 is necessary for establishing a neuronal circuit asymmetry for direction selectivity. Neuron. 2016;89:177–193. doi: 10.1016/j.neuron.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Watanabe D., Ishikane H., Tachibana M., Pastan I., Nakanishi S. A key role of starburst amacrine cells in originating retinal directional selectivity and optokinetic eye movement. Neuron. 2001;30:771–780. doi: 10.1016/s0896-6273(01)00316-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mean 3D activity map for wild-type mice (n = 12 mice) during visual motion eliciting the optokinetic reflex. Active voxels are color coded based on Z-scores (color scale shown in Figure 2A). Transparent gray surface: outer surface of the reference brain. Note that the “ghost” is larger, in both rostral and caudal extremities, than the field (6 mm) imaged with functional ultrasound.
Mean 3D activity map for FRMD7tm mice (n = 4 mice) during horizontal motion eliciting the optokinetic reflex. Voxels are color coded based on Z-scores (color scale shown in Figure 2A, display threshold: −2 < Z-score > 2). Transparent gray surface: outer surface of the reference brain. Note that the “ghost” is larger, in both rostral and caudal extremities, than the field (6 mm) imaged with functional ultrasound.4
RI module (left), RDMD module (middle), and RDMI module (right). Voxels of the atlas belonging to the RI, RDMD, and RDMI module regions are colored in purple, red, and cyan, respectively. Transparent gray surface: outer surface of the reference brain. Note that the “ghost” is larger, in both rostral and caudal extremities, than the field (6 mm) imaged with functional ultrasound.5
The ordering of brain regions is the same as for the rows in Figure 2B. Atlas ID and name columns indicate the region ID and name of imaged regions in the Allen Brain Institute reference atlas. Cluster and module identity of all active regions is indicated.