The huge diversity of non-Saccharomyces yeasts in grapes is dominated by the apiculate genus Hanseniaspora. Two native strains of Hanseniaspora vineae applied to winemaking because of their high oenological potential in aroma and fermentation performance were selected to obtain high-quality genomes. Here, we present a phylogenetic analysis and the complete transcriptome and aroma metabolome of H. vineae during three fermentation steps. This species produced significantly richer flavor compound diversity than Saccharomyces, including benzenoids, phenylpropanoids, and acetate-derived compounds. The identification of six proteins, different from S. cerevisiae ATF, with diverse acetyltransferase domains in H. vineae offers a relevant source of native genetic variants for this enzymatic activity. The discovery of benzenoid synthesis capacity in H. vineae provides a new eukaryotic model to dilucidate an alternative pathway to that catalyzed by plants’ phenylalanine lyases.
KEYWORDS: flavor compounds, genome, Illumina, metabolome, transcriptome, wine aroma
ABSTRACT
Hanseniaspora is the main genus of the apiculate yeast group that represents approximately 70% of the grape-associated microflora. Hanseniaspora vineae is emerging as a promising species for quality wine production compared to other non-Saccharomyces species. Wines produced by H. vineae with Saccharomyces cerevisiae consistently exhibit more intense fruity flavors and complexity than wines produced by S. cerevisiae alone. In this work, genome sequencing, assembling, and phylogenetic analysis of two strains of H. vineae showed that it is a member of the Saccharomyces complex and it diverged before the whole-genome duplication (WGD) event from this clade. Specific flavor gene duplications and absences were identified in the H. vineae genome compared to 14 fully sequenced industrial S. cerevisiae genomes. The increased formation of 2-phenylethyl acetate and phenylpropanoids such as 2-phenylethyl and benzyl alcohols might be explained by gene duplications of H. vineae aromatic amino acid aminotransferases (ARO8 and ARO9) and phenylpyruvate decarboxylases (ARO10). Transcriptome and aroma profiles under fermentation conditions confirmed these genes were highly expressed at the beginning of stationary phase coupled to the production of their related compounds. The extremely high level of acetate esters produced by H. vineae compared to that by S. cerevisiae is consistent with the identification of six novel proteins with alcohol acetyltransferase (AATase) domains. The absence of the branched-chain amino acid transaminases (BAT2) and acyl coenzyme A (acyl-CoA)/ethanol O-acyltransferases (EEB1) genes correlates with H. vineae’s reduced production of branched-chain higher alcohols, fatty acids, and ethyl esters, respectively. Our study provides sustenance for understanding and potentially utilizing genes that determine fermentation aromas.
IMPORTANCE The huge diversity of non-Saccharomyces yeasts in grapes is dominated by the apiculate genus Hanseniaspora. Two native strains of Hanseniaspora vineae applied to winemaking because of their high oenological potential in aroma and fermentation performance were selected to obtain high-quality genomes. Here, we present a phylogenetic analysis and the complete transcriptome and aroma metabolome of H. vineae during three fermentation steps. This species produced significantly richer flavor compound diversity than Saccharomyces, including benzenoids, phenylpropanoids, and acetate-derived compounds. The identification of six proteins, different from S. cerevisiae ATF, with diverse acetyltransferase domains in H. vineae offers a relevant source of native genetic variants for this enzymatic activity. The discovery of benzenoid synthesis capacity in H. vineae provides a new eukaryotic model to dilucidate an alternative pathway to that catalyzed by plants’ phenylalanine lyases.
INTRODUCTION
It is well known that yeast transforms grape sugars to ethanol and CO2 as the main wine fermentation products; however, cell secondary metabolism generates the highest impact compounds that dramatically affect the final flavor of wine. Flavor traits matter most in fermented beverages and should be considered the key properties when developing yeast selection within food biotechnology industries (1, 2). In wine, non-Saccharomyces yeast strains that account for more than 99% of the grape native flora are still poorly explored (2), and their impact on flavor richness will require multidisciplinary studies from genetics to metabolomic analyses of yeast cells. The limited numbers of commercial yeast strains applied by today’s winemakers are not contributing to flavor diversity, decreasing the possibilities to obtain more differentiated wine styles. Besides the grape selection and viticulture and vinification technologies used, which have been traditionally emphasized for quality wine production, yeast aspects should be taken into account. In a highly competitive market with more than one million brands of wines, differentiation and increased flavor diversity will be obtained with the application of increased yeast diversity during the fermentation process. Non-Saccharomyces species of yeast have been reported as beneficial for winemaking because they contribute to the sensory complexity of wines (3, 4). The main non-Saccharomyces genus associated with grapes is Hanseniaspora. Among the species comprising this genus, Hanseniaspora vineae is one of the most promising, with high oenological potential (5). Recently, H. vineae demonstrated the ability to provide increased levels of acetate esters and benzenoids and decreased levels of higher alcohols (except benzyl and 2-phenylethyl alcohols) in wines by pure fermentation or by cofermentation with S. cerevisiae (6–10). An aroma sensory analysis of wines, attributed to H. vineae winemaking, indicated a significant increase in fruit intensity, described as banana, pear, apple, citric fruits, and guava (9). These favorable characteristics for the winemaking industry have turned H. vineae into a species increasingly regarded as a means to improve aroma quality (5). Flavor diversity, including subtle characteristic differences in fine wines, has been described for other non-Saccharomyces species such as Pichia, Metschnikowia, and Torulaspora (2, 4, 11). Various genes have been identified as contributors to higher alcohol, acetate ester, and ethyl ester biosynthesis in S. cerevisiae; however, other species remain uncharacterized in this regard (12).
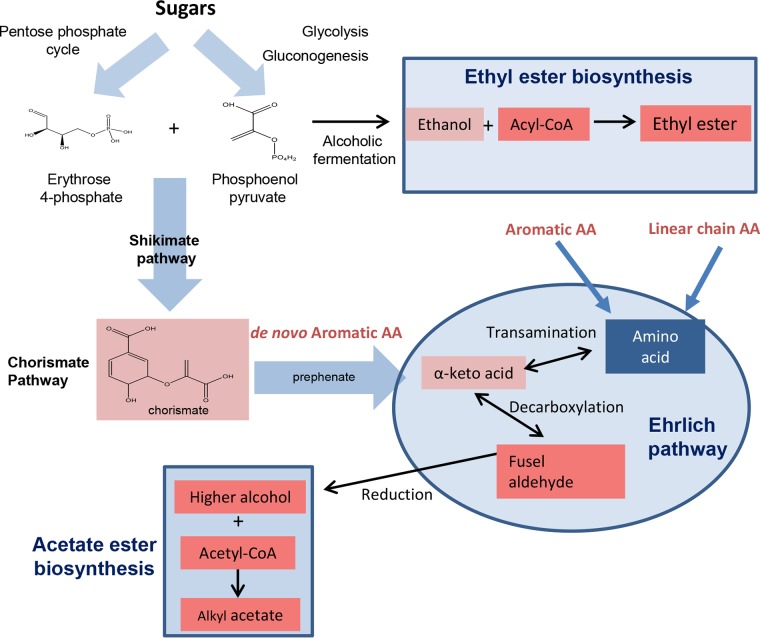
Higher alcohol formation via the Ehrlich pathway is subdivided into three steps: transamination, decarboxylation, and reduction (Fig. 1). In transamination, the key enzymes are the branched-chain amino acid transaminases (encoded by BAT genes) and the aromatic amino acid aminotransferases (encoded by ARO8 and ARO9 genes), which catalyze the transfer of amines between amino acids and their respective α-keto acid. In the second step, the α-keto acids are decarboxylated through pyruvate decarboxylases (encoded mainly by PDC and ARO10 genes) to form the respective aldehydes. Finally, the reduction from aldehydes to alcohols is carried out by alcohol dehydrogenases (encoded by ADH genes) and aryl-alcohol dehydrogenases (encoded by AAD genes). The formation of the fruity- and flowery-like aroma acetate esters is dependent on acetate and alcohols, and they are due to two alcohol acetyltransferases (AATases) encoded by ATF genes in S. cerevisiae. The biosynthesis of ethyl esters is carried out by two acyl coenzyme A (acyl-CoA)/ethanol O-acyltransferases (encoded by the EEB1 and EHT1 genes) and involves ethanol and acyl-CoA units (derived from fatty acid synthesis). Ethyl esters as well as acetate esters contribute fruity-like aromas, although their concentration levels in wine are significantly lower than those of the acetate esters (13–18).
FIG 1.
Metabolic pathways studied in this work involved in wine aroma formation. Ehrlich pathway for higher alcohol production, acetate ester biosynthesis, and ethyl ester biosynthesis from amino acids (AA) and sugars.
In this work, genomic, transcriptomic, and metabolomic analyses of the novel and native yeast for winemaking (H. vineae) was conducted, and the results were compared with those of S. cerevisiae to understand the aroma compound differences produced. We identified several changes in the dosage of key genes involved in higher levels of alcohol, fatty acid, acetate ester, and ethyl ester biosynthesis in H. vineae. We analyzed the expression profiles of these genes through transcriptomics and by assessing the concentrations of several aroma compounds during three different phases of the H. vineae fermentation process. A comparative work that analyzed the genomic, transcriptomic, and metabolomic profiles of a member of the apiculate group of the Saccharomycodaceae yeast family is presented. An understanding of the alternative metabolic pathways of H. vineae compared to those of S. cerevisiae will contribute to an understanding of apiculate yeast biology, which is the main yeast group associated with fruits (16, 17).
RESULTS AND DISCUSSION
The yeasts analyzed in this work are shown in Table 1, and putative genes and codes related to aroma synthesis by S. cerevisiae are described in Data Set S1 in the supplemental material.
TABLE 1.
Yeast strains analyzed in this work
| Species | Strain | Ploidy | Source | BioSample ID from NCBI databasea | Use |
|---|---|---|---|---|---|
| H. vineae | T02/19AF | Haploid | Uruguayan Tannat grape vines | SAMN02644989 | Genomic transcriptomic and phenomic study |
| H. vineae | T02/05AF | Haploid | Uruguayan Tannat grape vines | SAMN04487210 | Genomic study |
| S. cerevisiae | BY4742 | Haploid | Laboratory strain, derived from S288c | SAMN03020230 | FCM analysis |
| S. cerevisiae | BY4743 | Diploid | Laboratory strain, derived from S288c | SAMN01822968 | FCM analysis |
| S. cerevisiae | Montrachet 522 | Diploid | Fortified wines, CA | SAMN03325349 | Flavor compound analysis |
| S. cerevisiae | S288c | Haploid | Laboratory strain, CA | SAMD00065885 | Genomic comparison |
| S. cerevisiae | AWRI1631 | Haploid | Australian derivative of South African commercial wine strain N96 | SAMN02953734 | Genomic comparison |
| S. cerevisiae | AWRI796 | Diploid | South African wine strain | SAMN04286136 | Genomic comparison |
| S. cerevisiae | BC187 | Diploid | Derivative of CA wine barrel isolate | SAMEA687137 | Genomic comparison |
| S. cerevisiae | DBVPG6044 | Diploid | West African isolate | SAMEA687132 | Genomic comparison |
| S. cerevisiae | EC1118 | Diploid | Commercial wine strain | SAMEA2272624 | Genomic comparison |
| S. cerevisiae | L1528 | Diploid | Chilean wine strain | SAMN03020223 | Genomic comparison |
| S. cerevisiae | LalvinQA23 | Diploid | Portuguese Vinho Verde white wine strain | SAMN02981266 | Genomic comparison |
| S. cerevisiae | M22 | Diploid | Italian vineyard isolate | SAMN00189351 | Genomic comparison |
| S. cerevisiae | PW5 | Diploid | Nigerian Raphia palm wine isolate | SAMN00199004 | Genomic comparison |
| S. cerevisiae | RM11-1A | Haploid | Natural isolate collected from a vineyard, CA | SAMN02953602 | Genomic comparison |
| S. cerevisiae | T73 | Near-diploid | Spanish red wine strain | SAMN00198997 | Genomic comparison |
| S. cerevisiae | Vin13 | Diploid | South African white wine strain | SAMN02981268 | Genomic comparison |
| S. cerevisiae | VL3 | Diploid | French white wine strain | SAMN02981289 | Genomic comparison |
| S. cerevisiae | YJM269 | Diploid | Austrian wine from Blauer Portugieser grapes isolate | SAMN02981310 | Genomic comparison |
ID, identifier; NCBI, National Center of Biotechnology Information.
Genome characterization of H. vineae.
The two strains of H. vineae most used at the winemaking level by our group since 2009 were selected for genome sequencing: T02/19AF and T02/05AF. The sequencing of both strains was performed on an Illumina Genome Analyzer IIx platform.
The genome analysis revealed high similarity in both genomes with regard to size and the prediction of genes (Table 2; see also Table S1 and Fig. S1a). Therefore, only the data obtained from T02/19AF genomes are specified below in detail.
TABLE 2.
Genome assembly report of the two strains of H. vineae
| Strain | Genome size assembly (Mb) | Total no. of contigs | No. of ORFsa | No. of predicted proteins homologous to S. cerevisiae |
|---|---|---|---|---|
| H. vineae T02/05AF | 11.37 | 741 | 4,741 | 3,862 |
| H. vineae T02/19AF | 11.33 | 305 | 4,708 | 3,861 |
ORF, open reading frame.
The sequencing run generated a mean of 13,302,566 paired-end reads (2 × 100 cycles). After filtering and removing redundant reads, a final set of 9,203,956 reads was used for the genome assembly. A total of 87 scaffolds with a median length of 76,832 bp were assembled through MaSuRCA software, yielding a genome (haploid) of 11.3 Mb, representing an average coverage of 163-fold, with an N50 of 261 kb and a GC content of 37% (Table 2; Table S1; Fig. S1a). Higher quality data and a more extensive analysis of the genome of H. vineae were obtained than in our previous report (19).
Genome size and ploidy level were also addressed by flow cytometry (FCM) analysis using linear plots of fluorescence intensity of cell populations stained with propidium iodide (PI). This technique discriminated two cellular subpopulations with different DNA contents, namely R1 and R2 (see Fig. S2). All tested samples presented a half-peak coefficient of variation of R1 of less than 10% (data not shown), indicating high-resolution DNA measurements. As the references for genomic DNA estimation, we used both S. cerevisiae haploid (BY4742) and diploid (BY4743) strains, containing genomes of 11.67 and 23.35 Mb, respectively (20). A concurrent FCM analysis of S. cerevisiae haploid and diploid strains revealed three distinct peaks (Fig. S2), corresponding to 1n, 2n, and 4n DNA contents, where the mean PI fluorescence intensity of each peak was directly correlated (r2 > 0.999) to the amount of DNA of its corresponding cell subpopulation (Fig. S2). The genome size of each H. vineae strain was estimated in accordance with the R1 cell subpopulation (Fig. S2). The analysis by FCM revealed a diploid genome size of 16.71 ± 0.79 Mb (Table S1). Regarding gene copy number, we expected that the H. vineae genome would show a certain (but unknown) level of ploidy given its sporulation capacity (21). In any case, diploidy of both strains was confirmed. However, the slight difference in genome size obtained by FCM and our genome assembly-based calculations might be explained by the principles of the technique. H. vineae genome size was estimated using S. cerevisiae as the control strain. Because the cells themselves can act as a lens, changes in cell size or shape can affect the PI fluorescence detected by FCM (22), resulting in differences in genome size estimations obtained by FCM versus sequencing.
A total of 4,708 gene models were predicted using Augustus software, of which 3,855 had at least one Pfam domain of the Pfam platform databases. We identified 3,861 sequences homologous to S. cerevisiae genes and more than 4,141 sequences aligned to the National Center for Biotechnology Information (NCBI) nonredundant protein database (Table 2; Table S1). Due to the presence of a different number of homologous genes than reported for S. cerevisiae strain S288c in H. vineae, an Augustus prediction number (gXXXX.t1) is provided to clarify the putative gene, which was analyzed in each case.
We identified 243 of the 248 core eukaryotic genes (CEGs) and 445 of the 458 CEGs from the Augustus predictions, showing that our genome is ∼98% complete. Interestingly, the protein identity between H. vineae and S. cerevisiae is only 52%, demonstrating a great divergence between these two species. Moreover, a high heterozygosity level was evidenced by single-nucleotide polymorphism (SNP) analysis using different S. cerevisiae strains (23). A total of 56,662 SNPs (1 heterozygous SNP per 200 bp) were found, of which, 30,740 SNPs (54%) were present in coding sequences (Fig. S1b). According to the high genetic similarity found between T02/19AF and T02/05AF, the nucleotide diversity between both H. vineae strains was 1 variant per 179 bp (63,021 SNPs), a similar rate to those found among different S. cerevisiae strains (24, 25).
The genes related to yeast aroma compound synthesis in H. vineae were compared with those reported for S. cerevisiae (see Tables 3 and S2). Absent homologies and repeated genes were found.
TABLE 3.
Comparison of genes involved in biosynthesis routes for key flavor compound production in S. cerevisiae and H. vineae
| Biosynthesis route | Enzymatic activity | Genes identified (% amino acid identity with S. cerevisiae homologous protein)a |
|---|---|---|
| Higher alcohols | Aromatic amino acid transferases | 3×ARO8 (45.51, 59.84, 56.06), 4×ARO9 (42.70, 35.27, 36.08, 36.91) |
| Branched-chain amino acid transferases | BAT1 (78.84), BAT2 | |
| Decarboxylase | 2×ARO10 (34.10, 30.99), 2×PDC1 (80.46, 50.66), PDC5, PDC6, THI3 | |
| Alcohol dehydrogenase | 2×ADH1 (77.71, 78.74), ADH2, 2×ADH3 (74.79, 74.80), ADH4, ADH5, 4×ADH6 (44.74, 44.47, 44.74, 44.06), ADH7, SFA1 (68.16), 4×GRE2 (44.74, 50.73, 47.51, 43.02), YPR1, PAD1, SPE1, 3×OYE2 (55.10, 58.06, 57.25), HOM2 (78.24) | |
| Aryl alcohol dehydrogenase | AAD3, AAD4, AAD6, AAD10, AAD14, AAD15, AAD16 | |
| Regulation | ARO80 (34.80), GAT2, GLN3, GZF3, DAL80 | |
| Acetate esters | Alcohol acetyl transferases | ATF1, ATF2 (26.58), 4×SLI1 (22–24), g4599.t1 |
| Ethyl esters | Ethanol O-acyltransferase and esterase | EEB1, EHT1 (51.35), MGL2 (30.06), AAD, IAH1 (54.67) |
| Volatile organic acids | Aldehyde dehydrogenase | 2×ALD2 (40.55, 44.01), ALD3, ALD4, ALD5 (53.45), ALD6 (55.07) |
| Aromatic amino acid synthesis | Synthesis of chorismate, phenylalanine, tryptophan, and tyrosine | ARO1 (66.79), ARO2 (80.59), ARO3 (77.03), ARO4 (83.51), TRP2 (70.84), TRP3 (69.14), ARO7 (67.97), PHA2 (41.99), TYR1 (62.37) |
| Benzyl alcohol/benzaldehyde synthesis | Mandelate pathway | 2×ARO10 (34.10, 30.99), 2×PDC1 (80.46, 50.66), SCS7 (66.50), ALD6 (55.07), 2xALD2 (40.55, 44.01), DLD1 (53.00), DLD2 (70.00), DLD3 |
Homologous genes in H. vineae are not underlined, and the copy numbers (e.g., 2×) are indicated as prefixes for repeated genes. Predicted amino acid sequences from the genome of H. vineae were compared with protein homologs found in S. cerevisiae. Underlined genes represent absent homologous genes in H. vineae.
H. vineae diverged before the WGD clade of the Saccharomyces complex.
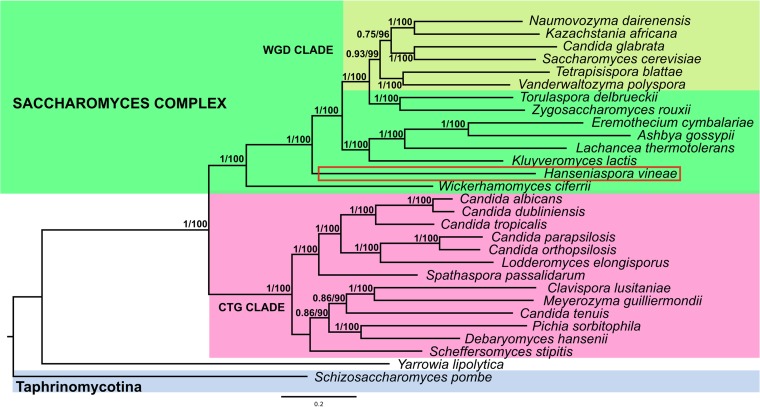
To determine the phylogenetic position of H. vineae, a phylogenetic tree was inferred by concatenating 227 genes from 29 species. The proteins were selected by an orthologous alignment of the predicted proteins from the H. vineae genome compared with those from yeast species obtained from databases. The maximum likelihood phylogeny classified H. vineae as part of the Saccharomyces complex but out of the whole-genome duplication (WGD) clade with very high support (Fig. 2). H. vineae was recovered as the sister taxa to two lineages, one composed of the WGD yeasts Kazachstania africana, Naumovozyma dairenensis, Saccharomyces cerevisiae, Candida glabrata, and Tetrapisispora blattae and the other composed of species diverged before the WGD, including Ashbya (Eremothecium) gossypii, Eremothecium cymbalariae, Lachancea thermotolerans, and Kluyveromyces lactis. Node support for this placement of H. vineae was very high (internode certainty [IC] = 1.0 and bootstrap support [BS] = 100).
FIG 2.
Maximum likelihood phylogeny of Saccharomyces complex species from concatenation of 227 genes. H. vineae is framed in red inside the Saccharomyces complex and outside the whole-genome duplication (WGD) clade. The clade CTG groups yeasts with alternative genetic codes. Numbers close to the node match bootstrap support (BS) for those values above 70 and internode certainty (IC). The scale bar represents units of amino acid substitutions per site. The tree has a midpoint root for easier visualization.
A total of 372 orthologous groups were expanded in S. cerevisiae compared to H. vineae, which involved 427 genes. These genes have a 2:1 relationship between S. cerevisiae and H. vineae, supporting the theory that H. vineae diverged previously to the WGD and arose out of the fungal CTG clade formed by yeasts that present differences in their genetic codes (26). Although this phylogeny presents some differences to that previously reported for the Saccharomyces complex (27, 28), the phylogenetic position of H. vineae presents a high node support and is similar to that obtained by Kurtzman and Robnett (27). The phylogenies inferred by these authors were based on divergence in genes of the ribosomal DNA (rDNA) repeat (18S, 26S, ITS), single copy nuclear genes (translation elongation factor 1α, actin-1, RNA polymerase II), and mitochondrially encoded genes (small-subunit rDNA, cytochrome oxidase II).
Overview of transcriptome dynamics during fermentation.
To perform a comprehensive analysis, we obtained transcriptomic profiles of H. vineae strain T02/19AF along three different days of the fermentation process (see Fig. S3): exponential growth phase (day 1), end of exponential phase (day 4), and end of stationary phase (day 10). Fermentations were carried out in triplicates using 25-ml Erlenmeyer flasks with 125 ml of chemically defined grape (CDG) medium that presents a similar nutrient composition to grape juice. The medium was supplemented with 100 mg N/liter yeast available nitrogen (YAN), 200 g/liter of an equimolar mixture of glucose, and fructose, and the pH was adjusted to 3.5.
These data were analyzed to compare the expression of the key genes related to the flavor compounds present in H. vineae, and, moreover, to those extra copies identified exclusively in H. vineae and not in S. cerevisiae. Transcriptome sequencing of nine libraries was performed in three replicates for the three fermentation points with Illumina. Similar quantities of genes were expressed at the three fermentation points, although their expression levels differed considerably. More than 2,500 (∼56%) genes were differentially expressed according to the false discovery rate (FDR) calculation (FDR < 0.05) between each pair of fermentation time points.
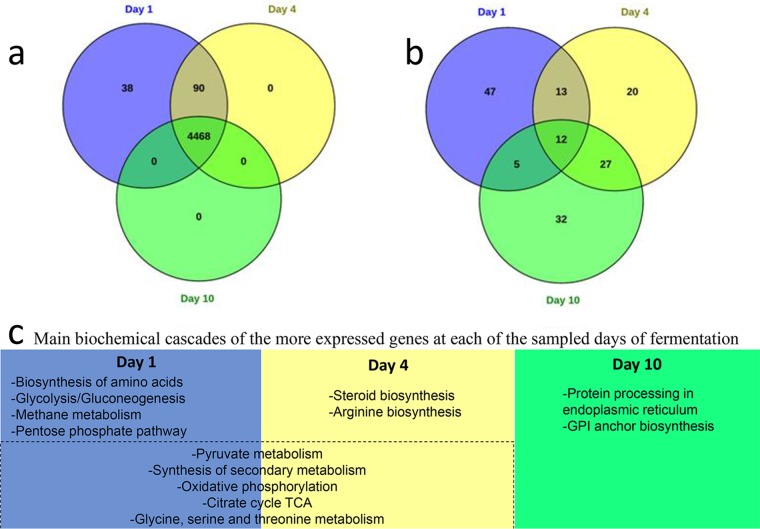
Transcriptome assembly enabled the identification of 15 more genes than those obtained by genome analysis, and almost all paralogous genes identified within the genome were confirmed. The transcriptome analyses for days 1, 4, and 10 presented 4,596, 4,558 and 4,468 expressed genes, respectively, of which 4,468 were in common (Fig. 3a and b). The most significant gene ontology (GO) terms associated with the genes shared between the three fermentation points were tRNA processing for biological processes, GTPase regulator activity for molecular function, and Golgi apparatus for cellular components.
FIG 3.
Overview of transcriptomic analysis. (a) Venn diagram showing the differentially expressed genes shared between each fermentation point. (b) Venn diagram showing the genes shared between each fermentation point for the top 100 most highly expressed genes. (c) Main biochemical cascades of the most expressed genes at each sampled day of fermentation. GPI, glycosylphosphatidylinositol.
High number of differentially expressed genes in H. vineae during fermentation.
For the three fermentation points (1, 4, and 10 days), the differentially expressed genes were analyzed using edgeR software. Important changes in gene expression were detected between any pair of the three fermentation points, while the differences between replicates were minimal (see Fig. S4).
In H. vineae, the large number of differentially expressed genes (DEGs) identified along the fermentation process was remarkable. Of the 4,468 genes shared among the 3 days, more than 2,500 (∼56%) were differentially expressed between each point (FDR < 0.05). However, microarray studies of various S. cerevisiae strains reported a smaller number of DEGs, ranging from 1,000 to 1,500 genes (29). The largest number of DEGs was identified between the first and last point; on the other hand, the fewest numbers were detected between day 4 and day 10. This situation is consistent with the fact that day 4 is the start of stationary phase and at day 10 the stationary phase is ending. As the fermentation process approaches stationary phase, fewer genes are expected to be differentially expressed.
Unique and expanded orthologous groups in H. vineae compared to those in S. cerevisiae.
Using OrthoMCL software, 85 expanded orthologous groups were detected in H. vineae compared to data from the S. cerevisiae S288c sequence.
There was consistently higher expression at days 1 and 4 of genes related to growth biochemical cascades (such as amino acid biosynthesis, the pentose phosphate pathway, oxidative phosphorylation, and tricarboxylic acid [TCA]) and glycolysis (such as pyruvate metabolism and the synthesis of secondary compounds) (Fig. 3c; Table S3). However, at day 10, the protein turnover genes were expressed the most, as at the middle and end of fermentation, amino acids are generally exhausted from the medium. The expression of genes related to protein processing at the end of stationary phase might be related to autophagy processes. Autophagy in yeast is a response to nutrient limitation, and the endoplasmic reticulum and glycosylphosphatidylinositol (GPI) anchor mechanisms are activated under this stress situation for the recovery process of proteins (30, 31). Interestingly, methane metabolism genes were mainly expressed at exponential growth to early stationary phase (days 1 to 4), but this might be specific to Hanseniaspora yeasts, as they are a methylotrophic group that may be active when oxygen is present at the beginning of fermentation (32).
The most complete KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathways were those related to tyrosine and phenylalanine metabolism, both aromatic amino acids that are related to phenylpropanoid synthesis (see Table S4a). The main genes that are exclusive to H. vineae belong to the following KEGG modules: β-lactam resistance and lysine biosynthesis, with five genes, and bacterial proteasome and benzoate degradation, with three genes (Table S4b). Five serine endopeptidases that might be involved in diverse functions were related to the β-lactam resistance module, while for lysine biosynthesis, two aldehyde dehydrogenases (g3618.t1 and g3619.t1), an unknown (4147.t1), one mlo2-like protein (g2280.t1), and ssm4 (g570.t1) proteins were found.
Genomics and yeast flavors.
Several genetic and phenomic characteristics were analyzed to compare H. vineae and S. cerevisiae strains. The comparative genomics analysis included H. vineae and up to 14 wine industry strains of S. cerevisiae whose genomes were analyzed in previous studies (33, 34) (see Table S5). The aroma compound profiles determined by gas chromatography-mass spectrometry (GC-MS) of H. vineae were compared with those of S. cerevisiae strain M522 and are shown in Table 4. On the other hand, in Table 5., the aroma compounds produced by H. vineae at days 4 and 10 are shown, and the differential expression of genes involved in higher alcohol, acetate ester, and ethyl ester metabolism were evaluated (see Fig. S5). These results are discussed in the following sections.
TABLE 4.
Exometabolome of H. vineae flavor compounds produced at days 4 and 10
| Compound | LRIa | Average ± SD content (g/liter)b |
|
|---|---|---|---|
| Day 4 | Day 10 | ||
| Alcohols | |||
| 2-Methyl-2-butanol | 975 | 66 ± 5 | 42 ± 2 |
| 1-Propanol | 996 | 116 ± 5 | 40 ± 5 |
| 2-Methyl-1-propanol | 1,067 | 3,620 ± 268 | 2,990 ± 290 |
| 1-Butanol | 1,128 | 149 ± 51 | 122 ± 10 |
| 3-Methyl-1-butanol | 1,187 | 42,525 ± 1,288 | 36,859 ± 1,693 |
| 2,3-Butanediol | 1,526 | 1,310 ± 74 | 1,450 ± 252 |
| 3-Ethoxy-1-propanol | 1,389 | 13 ± 13 | 177 ± 8 |
| 2-Ethyl-1-hexanol | 1,453 | NDc | 39 ± 2 |
| Methionol | 1,716 | 1,605 ± 60 | 1,925 ± 60 |
| 3-Acethoxy-1-propanol | 1,756 | 1,335 ± 109 | 1,520 ± 50 |
| Benzyl alcohol | 1,822 | 280 ± 9 | 407 ± 33 |
| 2-Phenylethanol | 1,906 | 6,657 ± 317 | 7,587 ± 361 |
| Tyrosol | 3,012 | 33 ± 33 | 2,213 ± 638 |
| Esters | |||
| 3-Methylbutyl acetate | 1,126 | 91 ± 21 | 112 ± 33 |
| Ethyl lactate | 1,341 | ND | 62 ± 3 |
| Ethyl 2-hydroxyhexanoate | 1,650 | ND | 20 ± 10 |
| Benzyl acetate | 1,690 | ND | 10 ± 1 |
| 2-Phenylethyl acetate | 1,813 | 5,862 ± 627 | 10,260 ± 995 |
| Ethyl 4-hydroxy-butoanoate | 1,819 | ND | 1,344 ± 47 |
| Diethyl 2 hydroxy glutarate | 2,202 | ND | 10 ± 2 |
| Fatty acids | |||
| 2-Methylpropanoic acid | 1,588 | 2,366 ± 158 | 3,024 ± 138 |
| Butanoic acid | 1,625 | 57 ± 12 | 97 ± 6 |
| 3-Methylbutanoic acid | 1,650 | 71 ± 11 | 128 ± 5 |
| Hexanoic acid | 1,843 | 50 ± 4 | 110 ± 4 |
| Octanoic acid | 2,070 | 44 ± 12 | 164 ± 17 |
| Decanoic acid | 2,243 | 15 ± 15 | 308 ± 67 |
| Other compounds | |||
| 2,3-Butanedione | 935 | 407 ± 53 | 58 ± 9 |
| 2,3-Pentanedione | 1,046 | 76 ± 25 | 15 ± 3 |
| 3-Hydroxy-2-butanone | 1,270 | 12,691 ± 348 | 9,669 ± 275 |
| 3-Hydroxy-2-pentanone | 1,330 | 1,353 ± 45 | 1,121 ± 184 |
| γ-Butyrolactone | 1,620 | 64 ± 32 | 116 ± 7 |
| N-Formyl tyramine | 2,890 | 727 ± 145 | 8,788 ± 451 |
Linear retention index based on a series of n-hydrocarbons reported according to elution order on Carbowax 20M.
Means and standard deviations from triplicate fermentations at 20°C in chemically defined grape synthetic medium.
ND, not detected.
TABLE 5.
Exometabolome of H. vineae and S. cerevisiae flavor compounds at the end of the fermentation
| Compound | LRIa | Average ± SD content (g/liter)b |
||
|---|---|---|---|---|
|
H. vineae |
S. cerevisiae |
|||
| T02/05AF | T02/19AF | M522 | ||
| Alcohols | ||||
| 2-Methyl-2-butanol | 975 | 168 ± 78 | 159 ± 1 | NDc |
| 1-Propanol | 996 | ND | 2 ± 2 | 42 ± 1 |
| 2-Methyl-1-propanol | 1,067 | 631 ± 490 | 750 ± 22 | 3,488 ± 4 |
| 1-Butanol | 1,128 | 31 ± 14 | 33 ± 5 | 91 ± 2 |
| 3-Methyl-1-butanol | 1,187 | 25,028 ± 3,699 | 28,326 ± 954 | 54,953 ± 41 |
| 2,3-Butanediol | 1,526 | 422 ± 68 | 1,076 ± 65 | ND |
| 3-Ethoxy-1-propanol | 1,389 | 75 ± 17 | 135 ± 6 | 175 ± 1 |
| 2-Ethyl-1-hexanol | 1,453 | 29 ± 4 | 26 ± 6 | 312 ± 1 |
| Methionol | 1,716 | 2,032 ± 230 | 2,601 ± 170 | 4,980 ± 6 |
| Benzyl alcohol | 1,822 | 141 ± 25 | 179 ± 8 | ND |
| 2-Phenylethanol | 1,906 | 8,029 ± 2,067 | 9,879 ± 120 | 18,387 ± 2 |
| Tyrosol | 3,012 | 814 ± 188 | 1,006 ± 11 | 7,683 ± 4 |
| Esters | ||||
| 3-Methylbutyl acetate | 1,126 | 33 ± 19 | 20 ± 4 | 54 ± 1 |
| Ethyl lactate | 1,341 | 66 ± 5 | 81 ± 17 | 116 ± 1 |
| Benzyl acetate | 1,690 | 6 ± 0 | 4 ± 0 | ND |
| 2-Phenylethyl acetate | 1,813 | 10,054 ± 929 | 9,205 ± 1,435 | 1,185 ± 6 |
| Fatty acids | ||||
| 2-Methylpropanoic acid | 1,588 | 301 ± 21 | 668 ± 52 | 168 ± 1 |
| Butanoic acid | 1,625 | 59 ± 6 | 55 ± 6 | 133 ± 1 |
| 3-Methylbutanoic acid | 1,650 | 67 ± 10 | 146 ± 3 | 448 ± 1 |
| Hexanoic acid | 1,843 | 82 ± 19 | 67 ± 4 | 461 ± 1 |
| Octanoic acid | 2,070 | 127 ± 37 | 89 ± 14 | 875 ± 2 |
| Decanoic acid | 2,243 | 170 ± 111 | 81 ± 26 | 96 ± 2 |
| Other compounds | ||||
| 3-Hydroxy-2-butanone | 1,270 | 4,328 ± 1,858 | 5,165 ± 742 | 303 ± 20 |
| γ-Butyrolactone | 1,620 | 90 ± 22 | 153 ± 14 | 338 ± 2 |
LRI, linear retention index based on a series of n-hydrocarbons reported according to their elution order on Carbowax 20M.
Means and standard deviations from triplicate fermentations at 20°C in chemically defined grape (CDG) synthetic medium.
ND, not detected.
Alcohols and 2-phenylethanol.
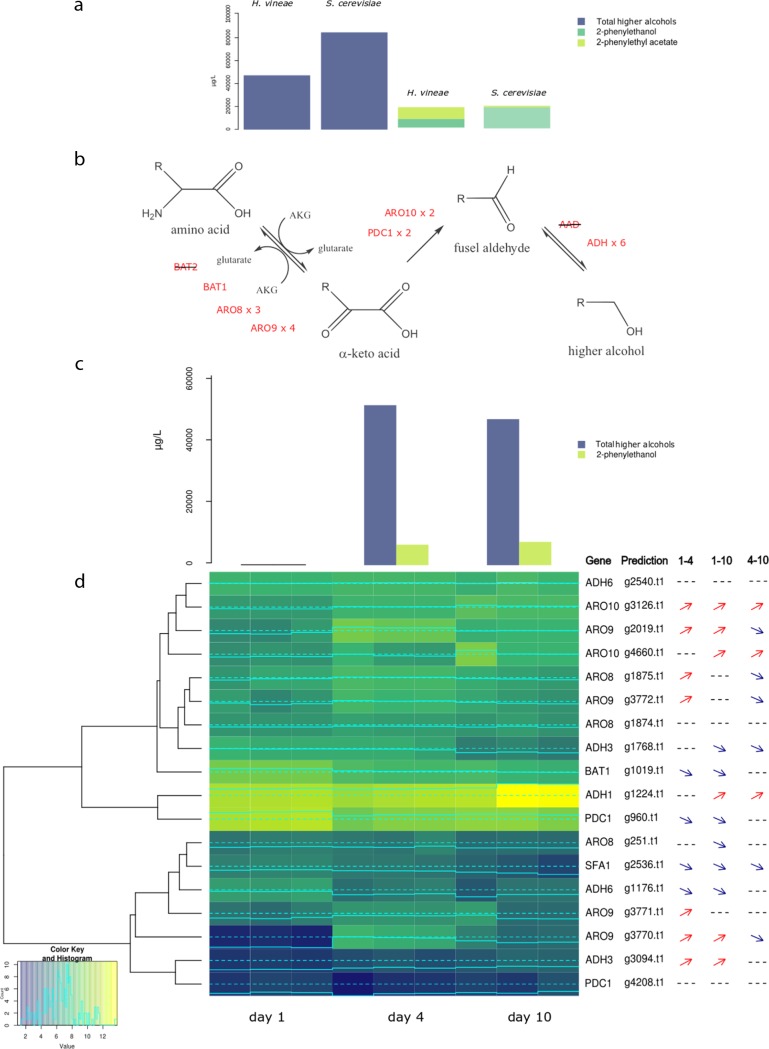
The aroma compound analysis showed that overall, alcohol production was more than twice as high in S. cerevisiae M522 than in H. vineae (Fig. 4a; Table 5). In fact, other studies comparing H. vineae to the wine yeast S. cerevisiae EC1118 have found similar results (6). However, the proportion of 2-phenylethanol in H. vineae with respect to S. cerevisiae M522 is approximately equivalent (Fig. 4a) if 2-phenylethyl acetate is taken into account as a derived compound of 2-phenylethanol.
FIG 4.
Higher alcohols and 2-phenylethanol production and putatively related genes. (a) Comparison of total higher alcohols, 2-phenyelthanol, and 2-phenylethyl acetate produced in H. vineae and S. cerevisiae at day 10 of fermentation. (b) Three steps of metabolic pathway of higher alcohols biosynthesis with putative enzymes involved in H. vineae. (c) Production of total higher alcohols and 2-phenylacetate by H. vineae at 1, 4, and 10 days of fermentation. (d) Expression heatmap of genes putatively involved in higher alcohols and 2-phenylethanol production from H. vineae at 1, 4, and 10 days of fermentation. Lighter colors indicate higher expression values, and data are shown for triplicates. Significant changes in expression of each gene are indicated with arrows to the right of the heatmap as analyzed using the package edgeR (FDR < 0.05). 1–4, differential expression between days 1 and 4; 1–10, differential expression between days 1 and 10; 4–10, differential expression between days 4 and 10.
The three steps of higher alcohol biosynthesis (transamination, decarboxylation, and reduction) (Fig. 4b) were analyzed attending to transcriptomics and phenomic results.
(i) Transamination. In S. cerevisiae, the most important gene involved in transamination leading to the production of higher alcohols is BAT2 (35), which encodes the branched-chain amino acid aminotransferase. BAT2 is absent in the H. vineae genome. This might explain the reduced presence of overall branched-chain higher alcohols in H. vineae fermentations compared to that in S. cerevisiae M522. In this scenario, the BAT1 gene in H. vineae would perform the two reactions of the reversible transamination step. BAT1 showed higher expression levels on day 1 and a decay in expression on days 4 and 10, while overall alcohol levels remained constant (Fig. 4d and S5). Therefore, the production of alcohols occurs early in fermentation, preceded by the expression of this gene.
On the other hand, the amounts of 2-phenylethanol/2-phenylethyl acetate remain constant between days 4 and 10, while the expression of the ARO8 and ARO9 genes reaches a peak by day 4 (Fig. 4c and d; Fig. S5). S. cerevisiae industrial strains present only one copy of these ARO genes (Table S5); however, H. vineae presents three copies of ARO8 and four of ARO9 that are all very similarly expressed during fermentation (Fig. 4d; Fig. S5). ARO8 and ARO9 encode aromatic amino acid transaminases, which act as broad-substrate-specificity amino acid transaminases in the Ehrlich pathway (15) and they are involved in the anabolism and catabolism of the aromatic amino acids phenylalanine and tyrosine. These data are in agreement with the KEGG pathways overrepresented in H. vineae as shown in Table S4a. Therefore, the overexpression of these two expanded genes might explain the larger proportion of 2-phenylethanol in two ways: first, for their incremented specificity for aromatic amino acids present in the medium, and second, for an increased synthesis of phenylalanine that is known as 2-phenylethanol precursor (36).
(ii) Decarboxylation. Five genes are involved in the decarboxylation step in S. cerevisiae (PDC1, PDC5, PDC6, ARO10, and THI3) (15), of which, H. vineae has two copies of ARO10 and two of PDC1. The most highly expressed paralogous copy of PDC1 had an expression pattern similar to that of BAT1 on day 1, prior to alcohol production (Fig. 4c and d; Fig. S5a).
It is possible that ARO10 duplication (Table 3) enables an efficient decarboxylation of aromatic α-keto acids derived from the enhanced transamination step. In fact, this is supported by the expression profiles (Fig. 4d; Fig. S5a) of both ARO10 genes that are very similar to the expression profiles found for ARO8 and ARO9 copies. It should be noted that the cofermentation of H. vineae with S. cerevisiae resulted in an increased intensity of citrusy aromas of which 2-phenylethanol (and therefore the ARO gene duplications) might be responsible (9). All 14 S. cerevisiae industrial strains only showed one copy of ARO10 (Table S5). Further, ARO10 has been shown to be related to the production of benzyl alcohol in a putative metabolic pathway of mandelate (7). Therefore, this decarboxylase activity might be involved in the enhanced (more than two orders of magnitude) synthesis of benzylic alcohol in H. vineae compared to that in S. cerevisiae (Table 5).
(iii) Reduction to higher alcohol. Surprisingly, H. vineae did not contain homologous sequences or any transcriptional evidence of the seven aryl alcohol dehydrogenases (AAD genes) present in the S. cerevisiae S288c sequenced genome (Table 3). This activity catalyzes the chemical reaction between aromatic aldehydes and alcohols. Given the overproduction of benzyl and 2-phenylethyl alcohol (precursor of the 2-phenylethyl acetate) in H. vineae compared to that in S. cerevisiae M522 (Table 5), at least one aryl alcohol dehydrogenase protein would be expected. However, it should be noted that the final step of the Ehrlich pathway (higher alcohol formation) can be catalyzed by any one of the ethanol dehydrogenases (Adh1, Adh2, Adh3, Adh4, and Adh5) or by Sfa1 (a formaldehyde dehydrogenase) in S. cerevisiae (37). In S. cerevisiae, the alcohol dehydrogenases are present in two multigenic families, with four genes each (according to Ensembl): ADH6, ADH7, YAL061W, and YAL060W in one family and ADH1, ADH2, ADH3, and ADH5 in the other. H. vineae presents two copies of ADH1 and ADH3 and four of ADH6, totaling eight genes, as in S. cerevisiae (Table 3). The ADH4 gene does not belong to either of these multigenic families and is absent in H. vineae. In H. vineae, not all paralogous copies of ADH genes showed significant transcriptional activity (many paralogous copies were assembled but they were filtered out before differential expression analysis). Interestingly, two of the four paralogous copies of ADH6 found in H. vineae were not expressed under these conditions (Fig. S5a).
In regard to the expression levels, four of the other five alcohol dehydrogenase genes, as well as SFA1, were significantly more expressed on days 1 and 4, while just one copy of ADH1 and ADH3 were more expressed on days 4 and 10 (Fig. 4d; Fig. S5a). One of the ADH6 copies showed a significant decline in expression levels between days 1 and 4, which is consistent with that previously reported for S. cerevisiae (29). In contrast, one ADH3 gene copy showed a 2-fold increase in expression by day 4 relative to day 1, similar to that for the AAD10 and AAD14 genes during S. cerevisiae wine fermentation (29). As a result, we suggest that the ADH genes that may be replacing the AAD genes might be those that show the same expression profile found in S. cerevisiae. Further biochemical studies will be necessary to confirm this suggestion.
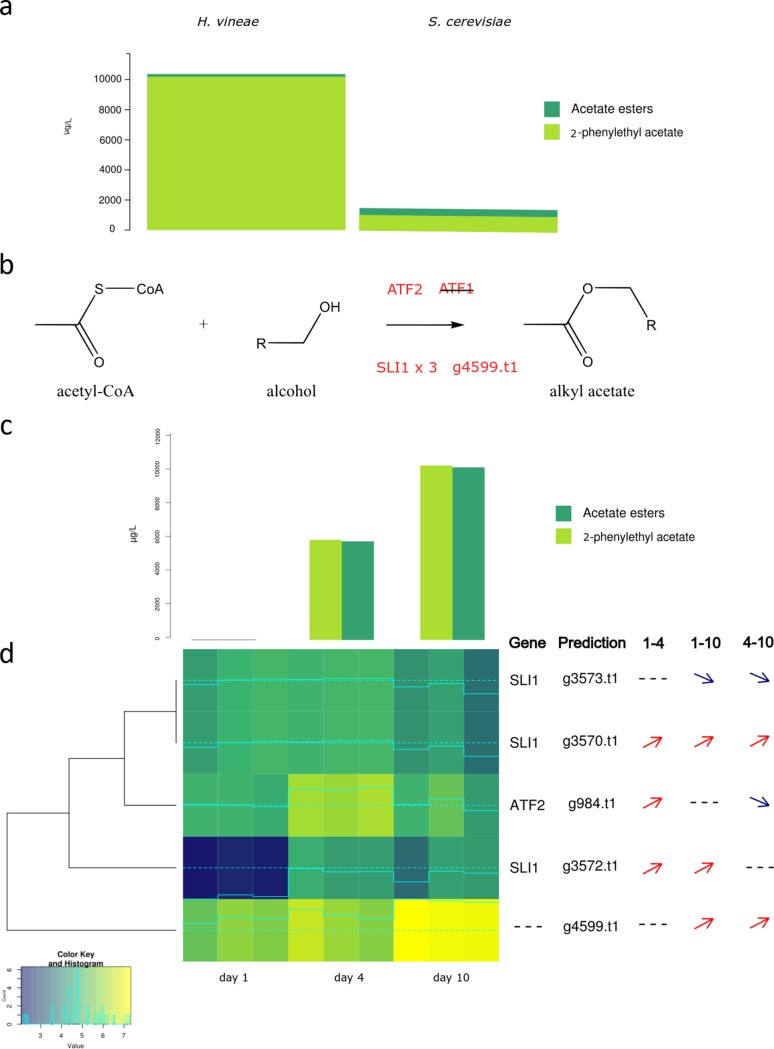
Acetate esters.
H. vineae and S. cerevisiae M522 also showed notable differences in overall acetate production, whereby H. vineae produced concentrations one order of magnitude higher than S. cerevisiae (Fig. 4a). As mentioned, H. vineae also showed a larger turnover from 2-phenylethanol to 2-phenylethyl acetate than S. cerevisiae. For example, 2-phenylethyl acetate only constituted a small fraction in S. cerevisiae of the total 2-phenylethanol produced compared to that in H. vineae (Fig. 4a; Table 5).
With regard to the genes involved in acetate ester formation, the H. vineae genome presented a highly divergent putative ortholog of the S. cerevisiae ATF2 gene, and it did not present any sequences homologous to ATF1. However, there were also five predictions containing the AATase Pfam domain. The four SLI1 N-acetyltransferase homologues are repeated in tandem in the H. vineae genome (one of them is out of the transcriptomic analysis according to threshold evaluation) (Fig. 5d). Three of these genes that were highly expressed in H. vineae (Fig. 5d) have weak similarity (22% to 24% at the amino acid level) with S. cerevisiae SLI1, which is a unique copy gene encoding N-acetyltransferase activity. It is known that SLI1 has wide specificity for aromatic amines, similar to the ATF genes (38). The other H. vineae AATase predicted (g4599.t1) has no homology with any S. cerevisiae gene previously reported; however, it is the most highly expressed gene at the end of stationary phase (Fig. 5d). The ATF2 gene and the most highly expressed SLI1 gene copy were both highly expressed on day 4, which explains the notable 2-fold increment of acetate esters between days 4 and 10 (Fig. 5c; Fig. S5b). Curiously, only S. cerevisiae strain M522 did not present the ATF1 gene, while none of the 14 industrial S. cerevisiae strains showed more than one gene similar to SLI1 (Table S5).
FIG 5.
Acetate ester production and putatively related genes. (a) Comparison of total acetate esters and 2-phenylethyl acetate produced in H. vineae and S. cerevisiae at day 10 of fermentation. (b) Metabolic pathway of acetate esters biosynthesis with putative enzymes involved in H. vineae. (c) Production of total acetate esters and 2-phenylethyl acetate by H. vineae at 1, 4, and 10 days of fermentation. (d) Expression heatmap of genes putatively involved in total acetate esters and 2-phenylethyl acetate production from H. vineae at 1, 4, and 10 days of fermentation. Lighter colors indicate higher expression values, and data are shown for triplicates. Significant changes in expression of each gene are indicated with arrows to the right of the heatmap as analyzed using the package edgeR (FDR < 0.05). 1–4, differential expression between days 1 and 4; 1–10, differential expression between days 1 and 10; 4–10, differential expression between days 4 and 10.
Therefore, the presence of six sequences with AATase domains (one ATF2, four for SLI1, and g4599.t1) might explain why H. vineae produces significantly more acetate esters than S. cerevisiae. The higher turnover of 2-phenylethanol to its corresponding acetate esters in H. vineae compared to that in S. cerevisiae clearly suggests that some of the H. vineae AATases (e.g., SLI1 paralogs) might be specific for this aromatic alcohol. The increased level of acetate esters in H. vineae explains the more intense fruity aroma resulting from the fermentation of H. vineae in Chardonnay (9) and Macabeo (8) wines. In fact, other apiculate yeasts from the Hanseniaspora genus are higher acetate esters producers than S. cerevisiae (39). However, high production of 2-phenylacetate is a particular characteristic of H. vineae compared to other species of this genus (40). Other Hanseniaspora species commonly produce increased levels of ethyl acetate. It is noteworthy that with regard to information about sequenced genomes of other Hanseniaspora species available in databases (41, 42), most do not present SLI1 homologous sequences. The exception is Hanseniaspora osmophila with two putative SLI1 copies. The detection of six AATases in H. vineae provides a relevant higher number of proteins for acetate ester biosynthesis than from the three copies of S. cerevisiae. These variations might contribute to improved functional designs for 2-phenylethanol acetylation and the synthesis of other phenylpropanoid aroma compounds, which are scarce pathways in S. cerevisiae strains.
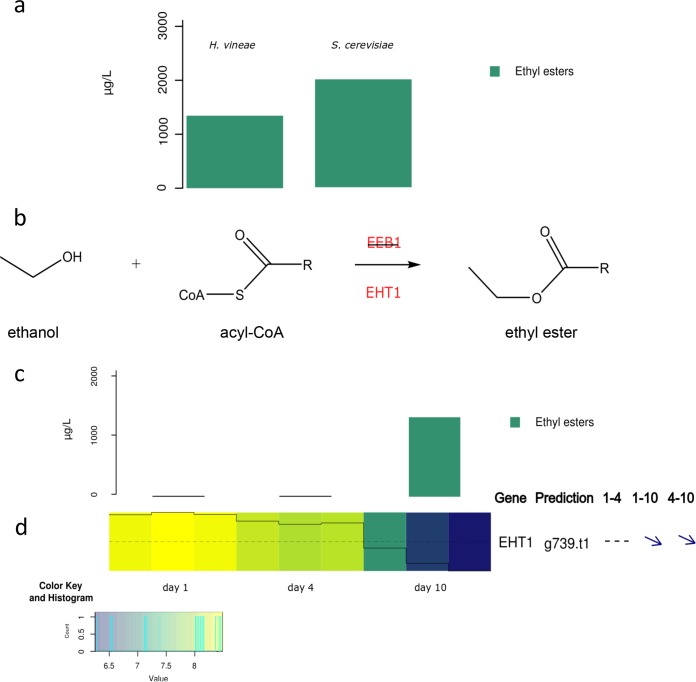
Ethyl esters.
EEB1 and EHT1 code for ethanol O-acyltransferases responsible for medium-chain fatty acid ethyl ester biosynthesis in S. cerevisiae (43). A decrease in the production of ethyl esters was observed in H. vineae compared to that in S. cerevisiae M522 (Fig. 6; Table 5). Furthermore, the absence of one of the main genes involved in ethyl ester production (EEB1) in H. vineae is consistent with this result (Fig. 6b). Only three strains of S. cerevisiae did not present this gene (Table S5). EHT1 is present in the H. vineae genome and it is highly and significantly expressed on days 1 and 4 relative to day 10 (Fig. 6c; Fig. S4). This might be consistent with the fact that esterified fatty acids were quantified on day 10 but were not detected on day 4 (Table 5).
FIG 6.
Ethyl esters production and putatively related genes. (a) Comparison of ethyl esters produced in H. vineae and S. cerevisiae at day 10 of fermentation. (b) Metabolic pathway of acetate ester biosynthesis with putative enzymes involved in H. vineae. (c) Production of ethyl esters by H. vineae at 1, 4, and 10 days of fermentation. (d) Expression heatmap of genes putatively involved in ethyl ester production from H. vineae at 1, 4, and 10 days of fermentation. Lighter colors indicate higher expression values, and data shown are of triplicates. Significant changes in expression of each gene are indicated with arrows to the right of the heatmap as analyzed using the package edgeR (FDR < 0.05). 1–4, differential expression between days 1 and 4; 1–10, differential expression between days 1 and 10; 4–10, differential expression between days 4 and 10.
Even so, an important interstrain difference in the expression of acyltransferases was found during the fermentation process in S. cerevisiae. In general, the expression of EHT1 in S. cerevisiae increased somewhat as fermentation progressed (30), which differs from our findings in H. vineae. Regarding our data, ethyl ester compounds were detectable on day 10 of the fermentation process. Here, it should be noted that our results are consistent with those obtained with this species in wines of Chardonnay (9) and Macabeo (8) fermentations, in which they exhibited decreased levels of ethyl esters compared to those of acetate esters.
Conclusion.
The use of non-Saccharomyces yeasts in winemaking is limited due to the insufficient characterization of many species that naturally participate in these processes. H. vineae has proved to contribute with flavor diversity in winemaking conditions. Here, we present a deep genomic, transcriptomic, and metabolomic analyses and their comparisons with Saccharomyces strain data. On the basis of our results with a synthetic chemically defined grape juice medium, this work represents a relevant contribution to understand the biology and phylogenic relationship of the main yeast genus associated with grapes. The larger production of acetate esters, the increased ratio of 2-phenylethyl acetate to 2-phenylethanol, and the reduced amount of ethyl esters found in H. vineae may be due to the high presence of putative alcohol acetyltransferase proteins and the absence of EEB1. These results are in agreement with previous reports studied in real winemaking conditions. As was shown, H. vineae produced a large amount of phenylpropanoids compared to that by S. cerevisiae and other yeasts, which might be explained by gene duplications and highly expressed ARO genes. This work established that H. vineae may be a potential model eukaryotic species to study benzenoid synthesis pathways, an alternative to the phenylalanine ammonia lyase (PAL) pathway commonly found in plants and Basidiomycetes. These phenolic volatile compounds have several known key functions in plants, such as cell-cell communication, antimicrobial activity, or phytohormone production, that make them highly attractive to the yeast biotechnology industry.
MATERIALS AND METHODS
Yeasts.
Table 1 shows all the yeast strains utilized in this work.
Genomic characterization of H. vineae.
(i) DNA and RNA isolation from H. vineae strains. H. vineae T02/19AF and T02/05AF strains were isolated from the Uruguayan Tannat vineyards. These strains were identified as H. vineae by sequencing the ribosomal D1/D2 region, and the strains were differentiated using the tandem repeats tRNA PCR technique (44). Genomic DNA was obtained from H. vineae cultures grown in yeast extract-peptone (YP) medium (1% yeast extract and 2% peptone, supplemented with 2% glucose) at 30°C, using the Wizard Genomic DNA purification kit (Promega, Madison, WI, USA), according to the manufacturer's instructions. Total RNA was obtained from the H. vineae T02/19AF strain grown under static batch fermentation conditions using the RiboPure RNA purification kit yeast (Life Technologies, Grand Island, USA). The poly(A) mRNA fraction was then isolated using the Oligotex mRNA Minikit (Qiagen, Hilden, Germany) and converted to indexed transcriptome sequencing (RNA-seq) libraries with the ScriptSeq v2 RNA-seq library preparation kit (Epicentre Biotechnologies, Madison, WI, USA).
(ii) Genome length and ploidy estimation by flow cytometry. H. vineae strains were grown in YP medium supplemented with 2% glucose, and 1× 107 cells were pelleted at 3,000 × g for 3 min and washed with ice-cold phosphate-buffered saline (PBS; 138 mM NaCl, 3 mM KCl, 8.1 mM Na2HPO4, and 1.5 mM KH2PO4). To fix cells, 1 ml of 70% cold ethanol was slowly added, and the samples were stored at 4°C overnight. After removing the ethanol by centrifugation, the cell pellet was washed with PBS and resuspended in 700 μl of the same buffer. Each sample was sequentially treated with 250 μl of 1 mg/ml RNase A (Applichem, USA) (1 h at 50°C), 50 μl of proteinase K (20 mg/ml; Sigma-Aldrich, USA) (1 h at 50°C), and incubated overnight at 4°C in the dark with 50 μl of propidium iodide (PI, 1 mg/ml; Life Technologies, USA). The analysis of DNA content by FCM requires staining yeasts with PI, a fluorochrome that binds to DNA.
FCM analyses were performed using a CyAn ADP LX, 7-color flow cytometer (Beckman Coulter, USA). The blue laser (488 nm) was selected to excite the PI fluorophore. The fluorescence area signal was detected with a 575/25-nm (FL2) emission filter and plotted on a linear scale. Data acquisition and analysis were achieved using Summit v4.3 software (DakoCytomation, UK), and 10,000 events per sample were collected. The gating strategy comprised a forward scatter (FSC) versus side scatter (SSC) cell region that excluded cellular debris and irrelevant small particles. This region was applied to a PI histogram so that only gated events were displayed. S. cerevisiae strains BY4742 and BY4743 (Table 1) were used as the controls. The mean fluorescence intensity of stained cells as measured by FCM was taken as indicative of the total DNA content, and a direct correlation between fluorescence intensity measurements and the amount of DNA in each control strain was established. All cultures generated bimodal fluorescence profiles composed of two peaks: one corresponding to a population of a majority of cells in G phase (lower intensity peak) and the other (higher intensity peak) attributed to cells in S phase undergoing DNA synthesis. The genome size of each H. vineae strain was estimated in accordance with the mean fluorescence of the peak subpopulation that showed lower intensity values. Three independent biological experiments were performed, and samples were analyzed in triplicates for each experiment.
(iii) Genome assembly and gene annotation. Genomes were sequenced using an Illumina Genome Analyzer IIx platform in paired-end mode. A shotgun genomic library was generated on the basis of standard methods.
The reads were filtered and trimmed with the QC Toolkit (45). The first 15 bases at the 5ʹ end and the last bases of the 3ʹ end with a Phred quality smaller than 30 were trimmed. The reads with average Phred quality scores smaller than 20 were filtered.
Digital normalization to the paired reads was applied to systematize the coverage, from uneven 200× to 30× across the genome, to gain computation efficiency and to eliminate most of the erroneous kmer (46, 47). The de novo genome assembly was performed using MaSuRCA (48) (insert length, 900). To reduce heterozygosis redundancy and find any potential gene tandem repeats, HaploMerger (49) was applied using default parameters.
Gene prediction was carried out using Augustus (50) trained with S. cerevisiae gene models. Peptide predictions were then annotated through BLASTp (cutoff for E value, 1e−10) against S. cerevisiae proteins obtained from the Saccharomyces genome database (20). The Pfam protein families database (51) was used to predict possible protein domains. To evaluate genome completeness, core eukaryotic genes (CEGs) (52) were sought with BLASTp (cutoff for E value, 1e−10). Gene ontology analysis was carried out using topGO (53).
(iv) SNP identification. Genomic short reads sequences were mapped to the assembled genome of T02/19AF using Bowtie2 in paired-end mode with default conditions (54) and processed using SAMtools (55) and Picard (http://broadinstitute.github.io/picard/). Through the GATK pipeline (56, 57), SNPs were identified using Unified Genotyper applying hard filter (QD < 2.0, FS > 60.0, MQ < 40.0, HaplotypeScore > 13.0, MappingQualityRankSum < −12.5, ReadPosRankSum < −8.0). Base pair coverage was calculated using BEDTools (58). The reads of H. vineae T02/05AF were aligned to those of T02/19AF to estimate the nucleotide divergence between these two strains.
Analysis of 14 S. cerevisiae industrial wine strains.
For several genes with known functions in the biosynthesis of acetate esters, ethyl esters, and higher alcohols, we determined which ones were present, duplicated, or absent in the H. vineae genome compared to S. cerevisiae S288c and an additional 14 S. cerevisiae wine strains. These strains were selectively chosen because they are used in wine fermentation and commercial winemaking studies (Table 1).
Ortholog cluster analysis.
The proteomes of 31 fungal species were downloaded from OrthoDB (59). This web service has the orthologous relationships among a broad group of predefined species. For orthologous identification, we first used pairwise BLASTp against H. vineae and selected the reciprocal best hit. Then, we compared our orthologous group with those present on the OrthoDB database, and if they contained at least one gene not belonging to the corresponding OrthoDB group, they were filtered out. The protein alignment was conducted with MUSCLE v3.8.31 (60). We used PAL2NAL (61) for aligning the nucleotides on the basis of the protein alignment and Gblocks v0.91b (62) to eliminate poorly aligned positions. We finally obtained 227 proteins for 29 species (two species had to be discarded because we could not find the correspondence between their protein and nucleotide sequences) to recover the phylogenetic position of H. vineae.
To establish orthologous clusters between S. cerevisiae S288c and H. vineae T02/19AF, the predicted proteins were analyzed with the OrthoMCL web server (63). Orthologous clusters were classified as expanded in H. vineae if the number of H. vineae genes in one OrthoMCL group was larger than the number of S. cerevisiae genes present in that group. To identify the pathways involved in each group, S. cerevisiae genes were used as input on the DAVID functional annotation pipeline (64). Those orthologous cluster groups exclusive to H. vineae (not containing any S. cerevisiae sequences) were analyzed using the EC enzymes and KEGG modules of the corresponding orthologous group (65) using custom Python scripts.
Phylogenetic analysis.
A supermatrix tree was constructed using a set of 227 genes from 29 species, including H. vineae. First, FASconCAT (62) was used to concatenate the supermatrix of 214,302 bases. The problematic aligned regions were previously removed with Gblocks v0.91b (66). For this supermatrix, the best partition scheme was chosen through PartitionFinder (67). The phylogenetic inference under maximum likelihood was performed with RAxML employing a GTRCAT substitution model for each of the 32 partitions suggested by PartitionFinder and using 200 starting trees. Node support was summarized in RAxML. Bootstrap support (BS) was calculated using extended majority-rule consensus for the bootstrapped trees set. Support is also shown as internode certainty (IC) values, a recently developed metric that considers the frequency of the bipartition defined by the internode in a given set of trees jointly with that of the most prevalent conflicting bipartition in the same tree set (68).
Transcriptome analyses.
Nine transcriptomes, three replicates from three different fermentation stages, at days 1, 4, and 10, were analyzed. Paired-end transcriptome sequencing was performed using Illumina MiSeq. High-quality raw sequencing reads were directly assembled using Trinity (47). They yielded a total of 7.8 Gb of data and 52 million 75-bp paired reads. The transcriptomic reference constructed resulted in 4,725 contigs with an average and median length of 1,982 and 1,683 bp, respectively (Table 6).
TABLE 6.
Transcriptomic assembly reference metrics for H. vineae T02/19AF
| Parameter | Transcriptomic reference |
|---|---|
| Total length (bp) | 9,362,444 |
| Total contig number | 4,725 |
| Contig length (bp) | |
| Maximum | 17,336 |
| Minimum | 226 |
| Mean | 1,982 |
| Median | 1,683 |
| No. of genes annotated | 4,725 |
A transcriptomic reference was constructed using the transcriptome of each sample, and an assembly was constructed by joining all of the reads for the subsequent gene expression analysis. For the construction of the transcriptomic reference, we selected the best reciprocal hit between the contigs among the 10 assembled transcriptomes and the subject sequences (19). The subject sequences were constructed using H. vineae T02/19AF protein predictions and S. cerevisiae proteins from the OMA browser (69). The alignments were carried out using reciprocal BLASTx (E value cutoff, 1e−10).
The reads were aligned against the transcriptomic reference implementing RSEM (default settings) (70). The obtained expected counts for each gene were then used for the differential gene expression analysis carried out with edgeR (71). Genes with cpm of <5 in 2 samples or more per each fermentation point were removed from the differential expression analysis. Genes with an FDR of <0.05 were considered differentially expressed.
Aroma compound analysis in a synthetic medium.
(i) Fermentation conditions. Chemically defined grape (CDG) fermentation medium (simulating the nutrient components of grape juice but devoid of grape precursors) was prepared with the same composition to study the de novo formation of aroma compounds and for the transcriptome analysis with a previously described process (72) with some variations. The modifications were as follows: the total nitrogen content was adjusted to a basic amount of 50 mg of nitrogen (N)/liter with each amino acid and ammonium component added in the same proportions as indicated previously (72). The final CDG medium used for inoculum preparation and fermentations was made by increasing the basic concentration by supplementing with diammonium phosphate (DAP) up to a yeast available nitrogen (YAN) concentration of 100 mg N/liter. This YAN amount was not a limiting concentration for the complete fermentation of sugars by the yeast strains used. The final pH of the medium was adjusted to 3.5 with HCl. Equimolar concentrations of glucose and fructose were added to reach a total of 200 g/liter, and the mixed vitamins and salts were as described previously (72). Tween 80 was excluded from the medium, because it was not found to be necessary for complete fermentation and it had a negative impact on the sensory characteristics of the resultant wines. Ergosterol was added as the only supplemented lipid at a final concentration of 10 mg/liter.
Inocula were prepared in 10 ml of the same CDG medium by incubating for 12 h in a rotary shaker at 150 rpm and 25°C. Fermentations were carried out in 125 ml of medium contained in 250-ml Erlenmeyer flasks closed with cotton plugs to simulate microaerobic conditions (73). The inoculum size was 1 × 105 cells/ml in the final medium for all strains. Static batch fermentations were conducted at 20°C in triplicates, simulating winemaking conditions. Wine samples for GC analysis were taken at days 4 and 10 during fermentation and at the end of the process. The samples were filtered through 0.45-μm pore membranes; SO2 was added as 50 mg/liter of sodium metabisulfite.
(ii) Aroma volatile compounds. The extraction of aroma compounds was performed using adsorption and separate elution from an Isolute ENV1 cartridge (IST Ltd., Mid Glamorgan, UK) packed with 1 g of a highly cross-linked styrene-divinylbenzene (SDVB) polymer. The treatment of samples and GC-MS analysis were performed as described previously (4) in a Shimadzu-QP 2010 ULTRA (Tokyo, Japan) mass spectrometer equipped with a Stabilwax (30 m by 0.25 mm inside diameter [i.d.], 0.25-µm film thickness; Restek) capillary column.
(iii) Identification and quantification. The components of wine aromas were identified by comparing their linear retention indices with pure standards (Aldrich, Milwaukee, WI). A comparison of mass spectral fragmentation patterns with those stored in databases was also performed. GC-flame ionization detection (GC-FID) and GC-MS instrumental procedures using an internal standard (1-heptanol) were applied for quantitative purposes, as described previously (4). All fermentations and chemical analysis were performed in triplicates. Analyses of variance (ANOVAs) were conducted to determine the differences in aroma compound concentrations among the strains with Statistica 7.0 (StatSoft Inc., Tulsa, OK, USA).
Accession number(s).
This whole-genome shotgun project has been deposited in DDBJ/EMBL/GenBank under the accession number JFAV00000000. The version described in this paper is JFAV03000000.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the Comisión Sectorial de Investigación Científica (CSIC) Group Project no. 656 and the CSIC Productive Sector Project no. 602 of UdelaR, Uruguay (grant no. ANII Postgraduate POS_NAC_2012_1_9099), the Agencia Nacional de Investigación e Innovación (ANII) Hanseniaspora vineae FMV 6956 project and a postdoctoral fellowship (PD_NAC_2016_1_133945), and a Clarín-COFUND postdoctoral fellowship from Principado de Asturias and European Union.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01959-18.
REFERENCES
- 1.Fleet GH. 2003. Yeast interactions and wine flavour. Int J Food Microbiol 86:11–22. doi: 10.1016/S0168-1605(03)00245-9. [DOI] [PubMed] [Google Scholar]
- 2.Carrau F, Gaggero C, Aguilar PS. 2015. Yeast diversity and native vigor for flavor phenotypes. Trends Biotechnol 33:148–154. doi: 10.1016/j.tibtech.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Steensels J, Snoek T, Meersman E, Nicolino MP, Voordeckers K, Verstrepen KJ. 2014. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol Rev 38:947–995. doi: 10.1111/1574-6976.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jolly NP, Varela C, Pretorius IS. 2014. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res 14:215–237. doi: 10.1111/1567-1364.12111. [DOI] [PubMed] [Google Scholar]
- 5.Martin V, Valera MJ, Medina K, Boido E, Carrau F. 2018. Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines–a review. Fermentation 4:76. doi: 10.3390/fermentation4030076. [DOI] [Google Scholar]
- 6.Martin V, Boido E, Giorello F, Mas A, Dellacassa E, Carrau F. 2016. Effect of yeast assimilable nitrogen on the synthesis of phenolic aroma compounds by Hanseniaspora vineae strains. Yeast 33:323–328. doi: 10.1002/yea.3159. [DOI] [PubMed] [Google Scholar]
- 7.Martin V, Giorello F, Fariña L, Minteguiaga M, Salzman V, Boido E, Aguilar PS, Gaggero C, Dellacassa E, Mas A, Carrau F. 2016. De novo synthesis of benzenoid compounds by the yeast Hanseniaspora vineae increases the flavor diversity of wines. J Agric Food Chem 64:4574–4583. doi: 10.1021/acs.jafc.5b05442. [DOI] [PubMed] [Google Scholar]
- 8.Lleixà J, Martín V, Portillo M. d C, Carrau F, Beltran G, Mas A. 2016. Comparison of fermentation and wines produced by inoculation of Hanseniaspora vineae and Saccharomyces cerevisiae. Front Microbiol 7:338. doi: 10.3389/fmicb.2016.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medina K, Boido E, Fariña L, Gioia O, Gomez ME, Barquet M, Gaggero C, Dellacassa E, Carrau F. 2013. Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem 141:2513–2521. doi: 10.1016/j.foodchem.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 10.Viana F, Belloch C, Vallés S, Manzanares P. 2011. Monitoring a mixed starter of Hanseniaspora vineae-Saccharomyces cerevisiae in natural must: impact on 2-phenylethyl acetate production. Int J Food Microbiol 151:235–240. doi: 10.1016/j.ijfoodmicro.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Ciani M, Comitini F, Mannazzu I, Domizio P. 2009. Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res 10:123–133. doi: 10.1111/j.1567-1364.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- 12.Curtin CD, Pretorius IS. 2014. Genomic insights into the evolution of industrial yeast species Brettanomyces bruxellensis. FEMS Yeast Res 14:997–1005. doi: 10.1111/1567-1364.12198. [DOI] [PubMed] [Google Scholar]
- 13.Cordente AG, Curtin CD, Varela C, Pretorius IS. 2012. Flavour-active wine yeasts. Appl Microbiol Biotechnol 96:601–618. doi: 10.1007/s00253-012-4370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazelwood LA, Daran J-M, van Maris AJA, Pronk JT, Dickinson JR. 2008. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pires EJ, Teixeira JA, Brányik T, Vicente AA. 2014. Yeast: the soul of beer’s aroma–a review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl Microbiol Biotechnol 98:1937–1949. doi: 10.1007/s00253-013-5470-0. [DOI] [PubMed] [Google Scholar]
- 16.Rosini G, Federici F, Martini A. 1982. Yeast flora of grape berries during ripening. Microb Ecol 8:83–89. doi: 10.1007/BF02011464. [DOI] [PubMed] [Google Scholar]
- 17.Loureiro V, Ferreira MM, Monteiro S, Ferreira RB. 2012. The microbial community of grape berry, p 241–268. In Geros H, Chaves M, Delrot S, Atta-ur-Rehman, Chaudhary MI (ed), The biochemistry of the grape berry, Bentham Science Publishers, Emirate of Sharjah, United Arab Emirates. [Google Scholar]
- 18.Bisson LF, Karpel JE. 2010. Genetics of yeast impacting wine quality. Annu Rev Food Sci Technol 1:139–162. doi: 10.1146/annurev.food.080708.100734. [DOI] [PubMed] [Google Scholar]
- 19.Giorello FM, Berná L, Greif G, Camesasca L, Salzman V, Medina K, Robello C, Gaggero C, Aguilar PS, Carrau F. 2014. Genome sequence of the native apiculate wine yeast Hanseniaspora vineae T02/19AF. Genome Announc 2:e00530-14. doi: 10.1128/genomeA.00530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goffeau a, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. 1996. Life with 6000 Genes. Science 274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 22.Haase SB, Lew DJ. 1997. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol 283:322–332. doi: 10.1016/S0076-6879(97)83026-1. [DOI] [PubMed] [Google Scholar]
- 23.Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. 2011. Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet 7:e1001287. doi: 10.1371/journal.pgen.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borneman AR, Forgan AH, Pretorius IS, Chambers PJ. 2008. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res 8:1185–1195. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 25.Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, Cao Z, Gu Z, Bruno D, Miranda M, Nguyen M, Wilhelmy J, Komp C, Tamse R, Wang X, Jia P, Luedi P, Oefner PJ, David L, Dietrich FS, Li Y, Davis RW, Steinmetz LM. 2007. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci U S A 104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos MAS, Gomes AC, Santos MC, Carreto LC, Moura GR. 2011. The genetic code of the fungal CTG clade. Comptes Rendus - Biol 33:607–611. doi: 10.1016/j.crvi.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Kurtzman CP, Robnett CJ. 2003. Phylogenetic relationships among yeasts of the “Saccharomyces complex” determined from multigene sequence analyses. FEMS Yeast Res 3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 28.Suh S-O, Blackwell M, Kurtzman CP, Lachance M-A. 2006. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 98:1006–1017. doi: 10.1080/15572536.2006.11832629. [DOI] [PubMed] [Google Scholar]
- 29.Rossouw D, Næs T, Bauer FF. 2008. Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics 9:530. doi: 10.1186/1471-2164-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. 2006. Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulick MG, Bertozzi CR. 2008. The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry 47:6991–7000. doi: 10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negruţa O, Csutak O, Stoica I, Rusu E, Vassu T. 2010. Methylotrophic yeasts: diversity and methanol metabolism. Rom Biotechnol Lett 15:5369–5375. [Google Scholar]
- 33.Borneman AR, Forgan AH, Kolouchova R, Fraser JA, Schmidt SA. 2016. Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 (Bethesda) 6:957–971. doi: 10.1534/g3.115.02569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauser NC, Fellenberg K, Gil R, Bastuck S, Hoheisel JD, Pérez-Ortín JE. 2001. Whole genome analysis of a wine yeast strain. Comp Funct Genomics 2:69–79. doi: 10.1002/cfg.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimoto H, Fukushige T, Yonezawa T, Sone H. 2002. Genetic and physiological analysis of branched-chain alcohols and isoamyl acetate production in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 59:501–508. doi: 10.1007/s00253-002-1041-5. [DOI] [PubMed] [Google Scholar]
- 36.Trinh TTT, Woon WY, Yu B, Curran P, Liu SQ. 2010. Effect of l-isoleucine and l-phenylalanine addition on aroma compound formation during longan juice fermentation by a co-culture of Saccharomyces cerevisiae and Williopsis saturnus. S Afr J Enol Vitic 31:116–124. [Google Scholar]
- 37.Dickinson JR, Salgado LEJ, Hewlins MJE. 2003. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem 278:8028–8034. doi: 10.1074/jbc.M211914200. [DOI] [PubMed] [Google Scholar]
- 38.Momoi M, Tanoue D, Sun Y, Takematsu H, Suzuki Y, Suzuki M, Suzuki A, Fujita T, Kozutsumi Y. 2004. SLI1 (YGR212W) is a major gene conferring resistance to the sphingolipid biosynthesis inhibitor ISP-1, and encodes an ISP-1 N-acetyltransferase in yeast. Biochem J 381:321–328. doi: 10.1042/BJ20040108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreira N, Mendes F, Hogg T, Vasconcelos I. 2005. Alcohols, esters and heavy sulphur compounds production by pure and mixed cultures of apiculate wine yeasts. Int J Food Microbiol 103:285–294. doi: 10.1016/j.ijfoodmicro.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 40.Medina K. 2014. Biodiversidad de levaduras no-Saccharomyces: efecto del metabolismo secundario en el color y el aroma de vinos de calidad. PhD dissertation. Universidad de la República, Montevideo, Uruguay. [Google Scholar]
- 41.Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH, Aerts AL, Barry KW, Choi C, Clum A, Coughlan AY, Deshpande S, Douglass AP, Hanson SJ, Klenk H-P, LaButti KM, Lapidus A, Lindquist EA, Lipzen AM, Meier-Kolthoff JP, Ohm RA, Otillar RP, Pangilinan JL, Peng Y, Rokas A, Rosa CA, Scheuner C, Sibirny AA, Slot JC, Stielow JB, Sun H, Kurtzman CP, Blackwell M, Grigoriev IV, Jeffries TW. 2016. Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci 113:9882–9887. doi: 10.1073/pnas.1603941113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sternes PR, Lee D, Kutyna DR, Borneman AR. 2016. Genome Sequences of three species of Hanseniaspora isolated from spontaneous wine fermentations. Genome Announc 4:e01287-16. doi: 10.1128/genomeA.01287-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verstrepen KJ, Van Laere SDM, Vanderhaegen BMP, Derdelinckx G, Dufour JP, Pretorius IS, Winderickx J, Thevelein JM, Delvaux FR. 2003. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol 69:5228–5237. doi: 10.1128/AEM.69.9.5228-5237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barquet M, Martín V, Medina K, Pérez G, Carrau F, Gaggero C. 2012. Tandem repeat-tRNA (TRtRNA) PCR method for the molecular typing of non-Saccharomyces subspecies. Appl Microbiol Biotechnol 93:807–814. doi: 10.1007/s00253-011-3714-4. [DOI] [PubMed] [Google Scholar]
- 45.Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown CT, Howe A, Zang Q, Pyrksz AB, Brom THA. 2012. A reference-free algorithm for computational normalization of shotgun sequencing data. ArXiv 1203:4802. [Google Scholar]
- 47.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimin AV, Marçais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. 2013. The MaSuRCA genome assembler. Bioinformatics 29:2669–2677. doi: 10.1093/bioinformatics/btt476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S, Chen Z, Huang G, Yu T, Yang P, Li J, Fu Y, Yuan S, Chen S, Xu A. 2012. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res 22:1581–1588. doi: 10.1101/gr.133652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 53.Alexa A, Rahnenführer J. topGO: enrichment analysis for gene ontology. R package version 2.28.0. http://www.bioconductor.org.
- 54.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterhouse RM, Zdobnov EM, Kriventseva EV. 2011. Correlating traits of gene retention, sequence divergence, duplicability and essentiality in vertebrates, arthropods, and fungi. Genome Biol Evol 3:75–86. doi: 10.1093/gbe/evq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 63.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane H, Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 65.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. 1999. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kück P, Meusemann K. 2010. FASconCAT: Convenient handling of data matrices. Mol Phylogenet Evol 56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 67.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 68.Salichos L, Rokas A. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497:327–331. doi: 10.1038/nature12130. [DOI] [PubMed] [Google Scholar]
- 69.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. 2011. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res 39:D289–D294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Henschke PA, Jiranek V. 1993. Yeast: metabolism of nitrogen compounds, p 77–164. In Fleet GH. (ed), Wine microbiology and biotechnology. Harwood Academic Publishers, Amsterdam, The Netherlands. [Google Scholar]
- 73.Fariña L, Medina K, Urruty M, Boido E, Dellacassa E, Carrau F. 2012. Redox effect on volatile compound formation in wine during fermentation by Saccharomyces cerevisiae. Food Chem 134:933–939. doi: 10.1016/j.foodchem.2012.02.209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.