This work provides insights on the mechanisms involved in raw beef spoilage during refrigerated storage and on the selective pressure exerted by the packaging conditions. We highlighted the presence of different microbial metagenomes during the spoilage of beef packaged aerobically or under vacuum. The packaging condition was able to select specific Pseudomonas fragi strains with distinctive genomic repertoires. This study may help in deciphering the behavior of different biomes directly in situ in food and in understanding the specific contribution of different strains to food spoilage.
KEYWORDS: Pseudomonas, meat spoilage, metagenomics, pangenomics
ABSTRACT
Microbial spoilage of raw meat causes huge economic losses every year. An understanding of the microbial ecology associated with the spoilage and its dynamics during the refrigerated storage of meat can help in preventing and delaying the spoilage-related activities. The raw meat microbiota is usually complex, but only a few members will develop during storage and cause spoilage upon the pressure from several external factors, such as temperature and oxygen availability. We characterized the metagenome of beef packed aerobically or under vacuum during refrigerated storage to explore how different packaging conditions may influence the microbial composition and potential spoilage-associated activities. Different population dynamics and spoilage-associated genomic repertoires occurred in beef stored aerobically or in vacuum packaging. Moreover, the pangenomes of Pseudomonas fragi strains were extracted from metagenomes. We demonstrated the presence of specific, storage-driven strain-level profiles of Pseudomonas fragi, characterized by different gene repertoires and thus potentially able to act differently during meat spoilage. The results provide new knowledge on strain-level microbial ecology associated with meat spoilage and may be of value for future strategies of spoilage prevention and food waste reduction.
IMPORTANCE This work provides insights on the mechanisms involved in raw beef spoilage during refrigerated storage and on the selective pressure exerted by the packaging conditions. We highlighted the presence of different microbial metagenomes during the spoilage of beef packaged aerobically or under vacuum. The packaging condition was able to select specific Pseudomonas fragi strains with distinctive genomic repertoires. This study may help in deciphering the behavior of different biomes directly in situ in food and in understanding the specific contribution of different strains to food spoilage.
INTRODUCTION
Meat spoilage involves bacterial growth to unacceptable levels, which causes changes to the meat structure and appearance, as well as the production of off-flavors. The initial level of microbial load mainly depends on the contamination at the slaughtering, but subsequent microbial transfer may occur during transport, manipulation, and retail. Indeed, the meat processing environment harbors a resident microbiota that may be considered a primary source of microbial spoilers and pathogens (1–4). The main microbial players that are recognized as meat spoilers are several genera of the Enterobacteriaceae, Pseudomonas spp., Brochothrix thermosphacta, and lactic acid bacteria (LAB) (2, 5–7). However, the spoiling microbiota is selected by both intrinsic factors (i.e., meat composition) and extrinsic factors applied during storage, such as temperature, composition of the gaseous atmosphere, and oxygen availability (6, 8). Therefore, only a fraction of the initial microbial population can develop during storage and become responsible for spoilage (so-called specific spoilage organisms [SSO] [9]).
The packaging of meat under vacuum (VP) or a CO2-modified atmosphere (MAP) results in the extension of the shelf life compared to that with aerobic storage, creating hurdles to the development of aerobic bacteria while enabling the growth of facultative and strict anaerobes. This change in packaging conditions determines a shift from aerobic bacteria, such as Pseudomonas spp., to facultative anaerobic species, such as Brochothrix thermosphacta and LAB, or sometimes strict anaerobes (Clostridia), that dominate during vacuum or MAP refrigerated storage (5, 6, 10–12). Psychrotrophic Pseudomonas species, namely Pseudomonas fragi, Pseudomonas lundensis, and Pseudomonas putida, are commonly isolated in meat, with Pseudomonas fragi becoming usually dominant during spoilage (6). Although members of the genus Pseudomonas are commonly considered aerobic, they were often found also in meat stored VP and in a MAP (5, 10, 13–15).
The apparently contrasting results might be explained by the activity of different strains of this species under diverse packaging conditions. Therefore, the occurrence and role of Pseudomonas fragi as meat spoilers deserve further investigation.
The SSO metabolize meat substrates, with subsequent changes in its texture and the production of volatile organic compounds (VOCs) responsible for off-flavors (16). VOCs produced during beef spoilage have been thoroughly studied to find key odor-active molecules to be used as molecular markers of spoilage (16). High variability regarding the storage temperature and time, the type of packaging, and the meat microbiota is reported (16). In addition, different strains of the same species might coexist or succeed during storage and show specific behaviors as meat spoilers. Indeed, different strains may show distinctive patterns of VOC production, differently affecting the flavor profile (16–19).
Here, we used a shotgun metagenomics approach for in situ monitoring of microbial genes and metabolic pathways involved in the spoilage of beef stored aerobically or under vacuum. In addition, using a pangenomic analysis of Pseudomonas fragi genomes retrieved from beef metagenomes, we characterized the strain-level diversity of this important beef spoiler, investigating whether different packaging conditions select Pseudomonas fragi strains with potentially different behaviors.
RESULTS
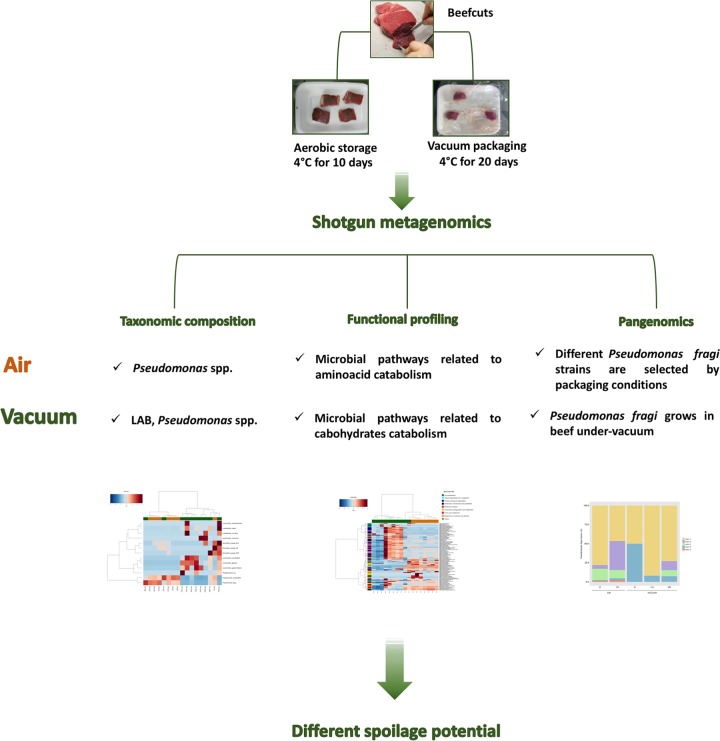
In this study, we analyzed the metagenomes of beef stored either aerobically or under vacuum to highlight the effect of storage conditions on the meat microbiome and its potential spoilage-related activities. The study design is reported in Fig. 1. Unfortunately, we failed to construct the libraries for samples with low microbial counts (all samples at time zero and samples At03.vac, F03.air, and F03.vac). Thus, we only sequenced 17 of the 24 processed samples.
FIG 1.
Study design and summary of the main findings.
Beef microbiome and spoilage-related potential activities are affected by storage conditions.
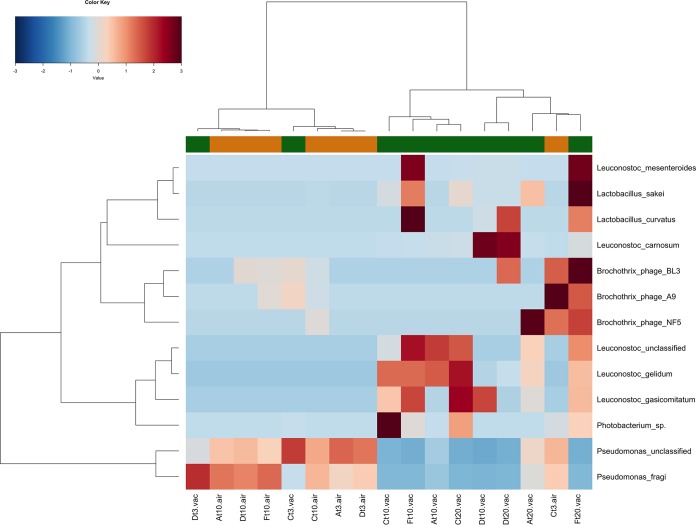
The metagenomes of beef samples stored aerobically and VP showed different microbiota profiles (Fig. 2). Aerobically stored samples were characterized by the dominance of Pseudomonas fragi and unclassified Pseudomonas spp., while most of the VP samples clustered together with a high abundance of LAB and Photobacterium. Nevertheless, few of them grouped closer to those stored aerobically, due to the higher levels of Pseudomonas spp. Indeed, Pseudomonas spp. were found in all the samples, regardless of the type of packaging, although their levels were higher in aerobically stored beef. Finally, some Brochothrix thermosphacta phages were identified, although the chromosome sequence belonging to this microorganism was not found.
FIG 2.
Different taxonomic compositions in the microbiomes of beef stored aerobically and VP. Ward-linkage clustering based on the Spearman’s correlation coefficients of the proportion of the top 13 taxa identified in the beef samples. The color scale represents the scaled abundance of each variable, denoted as Z-score, with red indicating high abundance and blue indicating low abundance. Columns are colored according to the type of packaging: aerobic (orange) or under vacuum (green). In the sample labels, A, C, D, and F indicate meat chops from the four different retails; air, meat stored aerobically; vac, meat stored under vacuum.
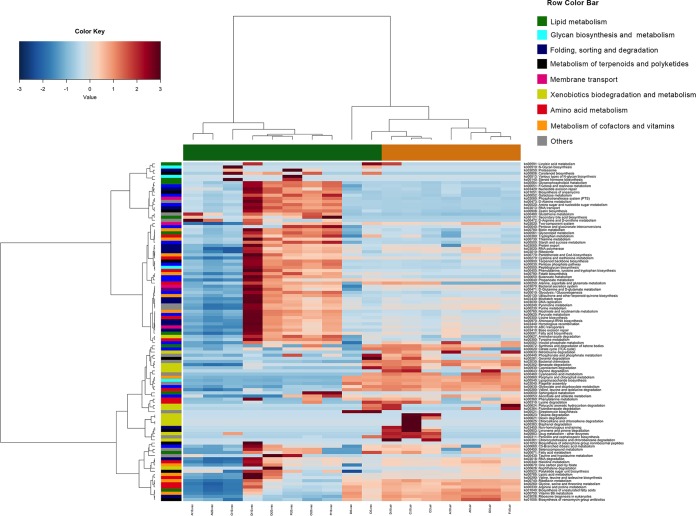
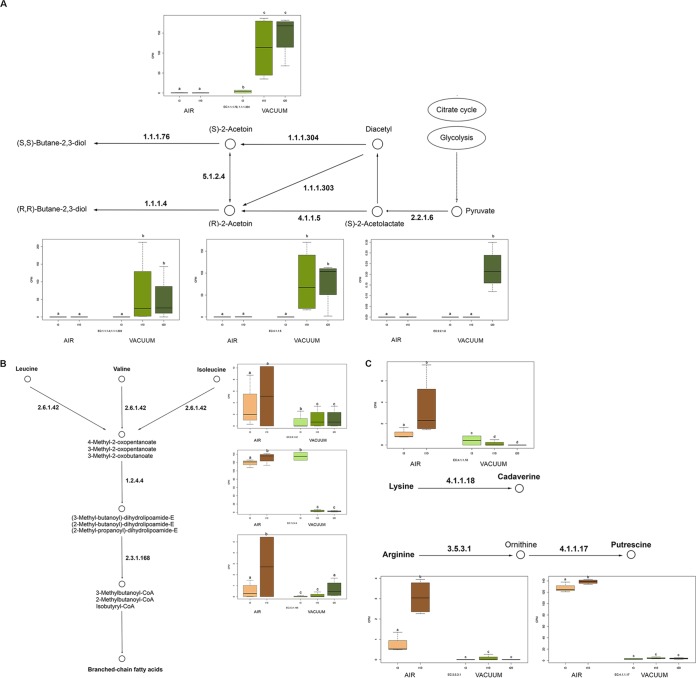
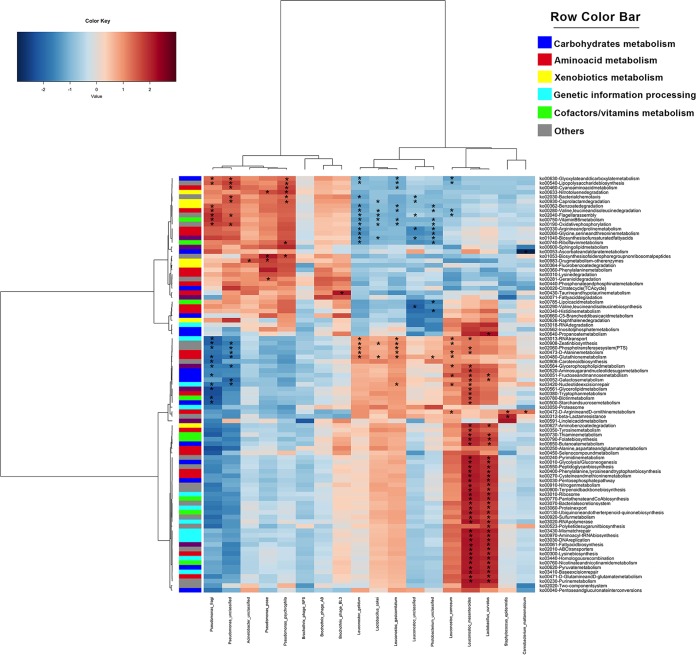
Differences in the taxonomic compositions of the beef microbiomes reflected its potential activities, which were clearly distinct according to the type of packaging. The metagenome of VP samples was dominated by genes involved in the KEGG metabolism of carbohydrates, at higher levels than in those stored aerobically (Fig. 3). On the contrary, the microbiome of samples stored aerobically showed a higher potential for the metabolism of amino acids and xenobiotics (Fig. 3). (R,R)-Butanediol dehydrogenase (EC 1.1.1.4 and 1.1.1.303), (S,S)-butanediol dehydrogenase (EC 1.1.1.76 and 1.1.1.304), acetolactate decarboxylase (EC 4.1.1.5), and acetolactate synthase (EC 2.2.1.6), involved in the production of acetoin and diacetyl from pyruvate, were all enriched in VP beef at 10 and 20 days (Fig. 4A), while the metagenomes of aerobically stored beef showed higher levels of several protease/peptidase genes (see Fig. S1 in the supplemental material), which are related to branched-chain amino acid degradation and biogenic amine production (Fig. 4B and C).
FIG 3.
Different genomic repertoires are associated with beef spoilage under aerobic conditions or under vacuum. Ward-linkage clustering based on the Spearman’s correlation coefficients of the proportion of the KEGG pathways identified in the beef samples. The color scale represents the scaled abundance of each variable, denoted as Z-score, with red indicating high abundance and blue indicating low abundance. Columns are colored according to the type of packaging: aerobic (orange) or under vacuum (green). Rows are colored according to the higher KEGG classification. In the sample labels, A, C, D, and F indicate meat chops from the four different retails; air, meat stored aerobically; vac, meat stored under vacuum.
FIG 4.
Spoilage-related pathways in VP- and aerobically stored beef. Abundance of KEGG genes involved in pyruvate catabolism pathways (A) and branched-chain amino acid (B) and biogenic amine (C) biosynthesis. CPM, copies per million reads. Different lowercase letters indicate significantly different values (P < 0.05).
Accordingly, microbial species with higher abundances in aerobically stored meat positively correlated with KEGG pathways involved in amino acid and xenobiotic metabolism, while LAB showed positive correlations with carbohydrate metabolism pathways (Fig. 5).
FIG 5.
Correlations between microbial taxa and KEGG pathways. Asterisks indicate significant correlations after correction for multiple comparisons (FDR < 0.05). Rows and columns are clustered by Euclidean distance and Ward-linkage hierarchical clustering. The intensity of the colors represents the degree of association as measured by Spearman’s correlations. Rows are colored according to the higher KEGG classification.
Pseudomonas spp. in beef stored aerobically and under vacuum.
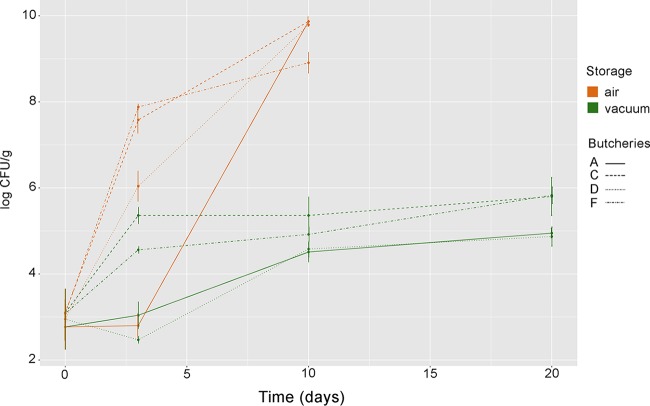
The loads of Pseudomonas spp. on fresh beef were around 103 CFU/g in all the samples, regardless of the type of retail (labeled A, C, D, and F), and increased during storage (Fig. 6). Multiplex PCR of the carA gene was carried out on colonies collected in bulk from countable plates. Pseudomonas fragi was found in all the beef samples. However, Pseudomonas lundensis was identified in the samples from 2 butcheries at time zero and detected until 20 days of storage only in VP samples (data not shown).
FIG 6.
Growth of Pseudomonas spp. under aerobic conditions and under vacuum. Average microbial loads (log CFU/g) of Pseudomonas spp. in beef stored aerobically or under vacuum. Values reported are the averages from three replicates. A, C, D, and F indicate meat chops from the four different retails.
Storage conditions select potentially different Pseudomonas fragi strains.
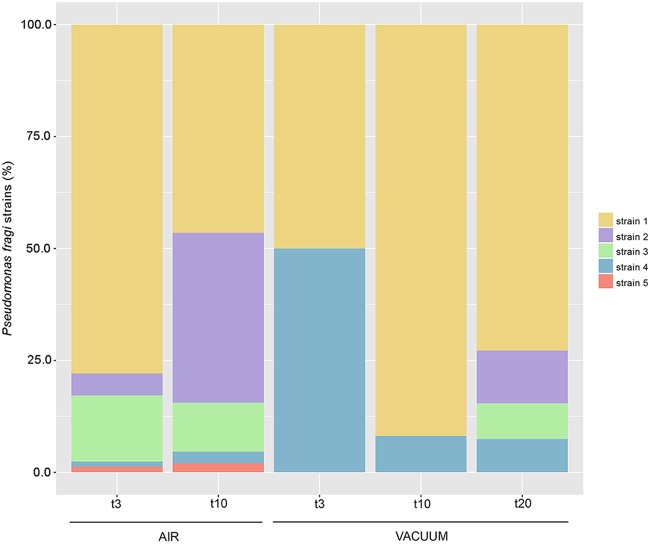
Five potentially different strains of Pseudomonas fragi were identified by StrainEst, with different abundance patterns according to the type of storage (Fig. 7). Pseudomonas fragi strain 1 was the most abundant in all the samples. However, strains 2 and 3 reached more than 50% of the total Pseudomonas fragi abundance in samples stored aerobically for 10 days but started developing in VP samples only at 20 days. Finally, strain 5 was detected only in aerobically stored beef, while higher levels of strain 4 were found in VP samples.
Fig. 7.
Different P. fragi strains are selected by packaging conditions. Stacked-bar chart showing the average abundances of different P. fragi strains (as defined by StrainEst) in aerobically stored and vacuum-packed beef samples.
The pangenome of Pseudomonas fragi was computed by using PanPhlAn. With some exceptions, the pangenomes of VP samples showed higher numbers of genes (see Fig. S2). The genes differentially present according to the storage type are shown in Table 1 (Fisher’s test, P < 0.05). Indeed, the Pseudomonas fragi pangenome of samples stored aerobically showed a higher prevalence of several genes involved in respiratory metabolism coding for subunits of the NADH-quinone oxidoreductase (electron transport chain), as well as genes related to oxidative stress response (oxyR, hydrogen peroxide-inducible genes activator; MsrPQ system). In addition, Pseudomonas fragi strains of aerobically stored beef contained higher counts of several lipases and the gene coding for the biofilm dispersion protein, bldA. On the contrary, the Pseudomonas fragi pangenome of VP beef was enriched in genes related with amino acid and protein degradation and transport, antibiotic resistance (tolC and mdtB), iron transport, and flagellar and lipopolysaccharide biosynthesis (Table 1).
TABLE 1.
P. fragi pangenes with significantly different occurrence in aerobically and vacuum-packed beef samplesa
| Gene annotation | Prevalence (% of samples) |
|
|---|---|---|
| Aerobic | Under vacuum | |
| Amino acid ABC transporter_g001843 | 28.57 | 87.50 |
| Amino acid ABC transporter permease_g000271 | 42.86 | 100.00 |
| Arginine N-succinyl transferase, subunit beta_g006675 | 14.29 | 87.50 |
| Aromatic amino acid transport protein AroP_g006499 | 14.29 | 87.50 |
| Branched-chain amino acid ABC transporter permease_g001018 | 42.86 | 100.00 |
| Channel protein TolC_g001045 | 42.86 | 100.00 |
| Colicin I receptor_g007007 | 42.86 | 100.00 |
| Electron transfer flavoprotein-ubiquinone oxidoreductase_g005516 | 42.86 | 0.00 |
| Electron transfer flavoprotein, subunit alpha_g005518 | 28.57 | 25.00 |
| Electron transfer flavoprotein, subunit beta_g005517 | 57.14 | 0.00 |
| Esterase/lipase/thioesterase_g000542 | 100.0 | 12.50 |
| Esterase/lipase/thioesterase_g000542 | 100.0 | 12.50 |
| Flagellar biosynthesis protein FlhA_g006471 | 14.29 | 100.00 |
| Flagellar biosynthetic protein FliP_g006475 | 57.14 | 100.00 |
| Flagellar biosynthetic protein FliR_g006473 | 14.29 | 87.50 |
| Hemin transport system permease protein HmuU_g004720 | 71.43 | 12.50 |
| Hemin transport system permease protein HmuU_g007012 | 14.29 | 100.00 |
| Hydrogen peroxide-inducible genes activator_g004882 | 42.86 | 0.00 |
| Hydrogen peroxide-inducible genes activator_g006647 | 100.0 | 25.00 |
| Hydroperoxy fatty acid reductase gpx2_g006838 | 71.43 | 0.00 |
| Inner membrane amino-acid ABC transporter permease protein YhdY_g006813 | 42.86 | 100.00 |
| Iron(3+)-hydroxamate-binding protein FhuD_g006324 | 14.29 | 75.00 |
| Iron(3+)-hydroxamate import system permease protein FhuB_g006325 | 14.29 | 75.00 |
| Iron transporter_g001368 | 28.57 | 87.50 |
| l-Threonine dehydratase biosynthetic IlvA_g004942 | 42.86 | 100.00 |
| Leucine, isoleucine, valine, threonine, and alanine-binding protein_g005368 | 28.57 | 100.00 |
| Lipopolysaccharide export system protein LptC_g006847 | 42.86 | 100.00 |
| Long-chain-fatty-acid–CoA ligase_g006700 | 85.71 | 12.50 |
| Long-chain-fatty-acid–CoA ligase_g006701 | 85.71 | 25.00 |
| Methionine amino peptidase_g006769 | 28.57 | 100.00 |
| Multidrug resistance protein MdtB_g005252 | 42.86 | 100.00 |
| Murein dd-endopeptidase MepH_g004974 | 42.86 | 100.00 |
| NADH-quinone oxidoreductase, subunit F_g005645 | 71.43 | 37.50 |
| NADH-quinone oxidoreductase, subunit F_g005646 | 85.71 | 0.00 |
| NADH-quinone oxidoreductase, subunit G_g005643 | 71.43 | 37.50 |
| NADH-quinone oxidoreductase, subunit L_g005641 | 100.00 | 12.50 |
| NADH-quinone oxidoreductase, subunit M_g005639 | 57.14 | 0.00 |
| NADH-quinone oxidoreductase, subunit N_g005638 | 42.86 | 0.00 |
| Nicotinate phosphoribosyltransferase 2_g007029 | 42.86 | 100.00 |
| Oligopeptide-binding protein AppA_g006420 | 28.57 | 87.50 |
| Peptidase_g000364 | 42.86 | 100.00 |
| Proline-specific permease ProY_g007110 | 42.86 | 100.00 |
| Protease HtpX_g001735 | 42.86 | 100.00 |
| Protein-methionine-sulfoxide reductase catalytic subunit MsrP_g006893 | 100.00 | 0.00 |
| Protein-methionine-sulfoxide reductase catalytic subunit MsrP_g006893 | 100.00 | 0.00 |
| Sensor protein QseC_g006731 | 42.86 | 87.50 |
| Stress response protein SCP2_g006950 | 42.86 | 100.00 |
| Succinate-semialdehyde dehydrogenase [NADP(+)]_g007028 | 42.86 | 100.00 |
| Tail-specific protease_g005617 | 57.14 | 100.00 |
| Thiol:disulfide inter-change protein_g001662 | 28.57 | 12.50 |
| TolC family protein_g000069 | 0.00 | 100.00 |
| Urease accessory protein UreF_g001013 | 42.86 | 100.00 |
| Uvr ABC system protein B_g006309 | 57.14 | 100.00 |
| Biofilm dispersion protein BdlA_g006694 | 100.00 | 12.50 |
Significant at a P value of <0.05.
DISCUSSION
We characterized the metagenomes of beef during spoilage under different packaging conditions. The initial microbial community was strongly affected by the type of packaging. As previously reported, facultative anaerobic LAB dominate during refrigerated storage under vacuum (7), while Pseudomonas spp. belong to the dominant microbiota under aerobic conditions (6). However, Pseudomonas spp. were also found in beef stored under vacuum, although at a lower extent (5, 10, 13–15). In our study, the results from metagenomics were supported by microbial counts and a carA gene assay of cultivable communities, showing that Pseudomonas spp. were viable and metabolically active under both packaging conditions.
We did not find Brochothrix thermosphacta, usually reported as dominant in meat-spoiling microbiota (1, 5, 10), while we highlighted the presence of three different Brochothrix thermosphacta phages. The possible activity of lytic bacteriophages for this species might explain the absence of Brochothrix thermosphacta observed here.
We highlighted the presence of several microbial pathways and genes possibly involved in meat spoilage. The metagenome of VP beef, where LAB represented the dominant microbiome, was enriched in genes related to carbohydrate fermentation. Several psychrotrophic LAB species ferment sugars under low oxygen conditions, leading to the production of CO2 and off-flavors, such as acetoin and diacetyl (7, 19, 20), thus conferring buttery and other undesirable smells (16). In our study, Leuconostoc carnosum, Leuconostoc gasicomitatum, and Leuconostoc gelidum were the LAB species most likely to be involved in carbohydrate fermentation. Accordingly, we found higher levels of genes related to the production of acetoin and diacetyl in VP metagenomes. Besides their production by LAB, these compounds have also been reported as associated with other microbial spoilers, such as Brochothrix thermosphacta (18) and Pseudomonas spp. (21). Moreover, acetoin may be further reduced to butane-2,3-diol by the fermentation activity of some Enterobacteriaceae and Leuconostoc gasicomitatum (22, 23).
Branched-chain fatty acids were previously found only in aerobically spoiled meat and derive from leucine, isoleucine, and valine degradation (16). 2-Methylbutanoic acid has pungent, acid, and Roquefort cheese odors, while 3-methylbutanoic acid has sour, stinky, foot, sweaty, and cheese odors (16). Their presence was often associated with Brochothrix thermosphacta (18). This species was not detected in the present study, suggesting that other microbes may potentially harbor the genomic repertoire needed to produce these off-flavors. In addition, the metagenome of aerobically stored beef was enriched in genes involved in the production of the biogenic amines (BAs) cadaverine and putrescine. These BAs are not responsible of off-flavors but, in addition to the safety implications, may be considered a marker of freshness (24), and their presence in spoiled meat was previously linked to Enterobacteriaceae and Pseudomonas spp. (24, 25). The results obtained here highlight that these metabolites and the microbial genes involved in their production may be present, thus pointing out the poor quality and freshness of the meat considered in this study and supporting their usefulness as markers of meat quality and freshness.
Pseudomonas fragi is considered the main spoiler of refrigerated meat stored aerobically (10). Due to its fast growth ability and the production of impactful spoilage-related off-odors, it quickly leads to the unacceptability of the meat (16). Moreover, it was often reported also in VP meat (5, 10, 13–15), suggesting the presence of different strains with likely specific genomic repertoires. We dissected beef metagenomes to highlight the differences in the pangenomes of Pseudomonas fragi strains possibly driven by the packaging conditions. Differences in the pangenomes were found in beef stored aerobically or VP, suggesting the presence of strains with distinctive genomic potential. Indeed, previous works emphasized that a strain-level phenotypical diversity in the main beef spoilers exists and that different strains may have distinctive roles in meat spoilage (17–19, 26). Although Pseudomonas spp. are usually considered aerobic, we demonstrated that potentially different strains of Pseudomonas fragi may be selected by the packaging conditions and may act differently during beef spoilage. Indeed, strains developing in VP beef showed a lower prevalence of genes involved in the electron transport chain and in oxidative stress response. This might explain why they are outcompeted by other strains during aerobic storage, although competition experiments would be necessary to validate this hypothesis. Aerobic storage selected Pseudomonas fragi strains with higher lipolytic potential, while strains present under vacuum showed a higher prevalence of genes involved in proteolysis and amino acid degradation, suggesting a different potential for spoilage-related activities. We previously showed that only few Pseudomonas fragi isolates displayed proteolytic activity, but this difference might be explained by the origin of the strains, all isolated from aerobically stored meat (17). On the contrary, lipolytic activity may lead to the biosynthesis of ethyl and methyl esters, considered the main VOCs produced by Pseudomonas fragi and responsible for fruity and sweet off-flavors (16, 17, 26). Finally, the Pseudomonas fragi pangenome of VP beef was characterized by a higher prevalence of genes coding for proteins involved in xenobiotic efflux (tolC and mdtB) and therefore possibly linked with drug and antibiotic resistance (27, 28).
Although the study is limited by the low number of samples involved, we provided an in-depth description of the genomic repertoire associated with beef spoilage and demonstrated that different packaging conditions may lead to different potential spoilage patterns. In addition, we highlighted the presence of specific storage-driven strain-level profiles of Pseudomonas fragi, one of the most important bacteria involved in meat spoilage. The genetic repertoire of Pseudomonas fragi that has been shown in this study may be responsible for distinctive features with possibly different metabolic activities that influence meat quality during storage. The results shown here are easily extensible to other food-associated microbes, either alterative or pathogens, that may be selected by storage conditions. Pangenomics based on the reconstruction of genomes retrieved from metagenomic reads will help to understand the behavior of different strains directly in situ in the food environment and to decipher their contribution in food fermentation or spoilage (2). Such new knowledge on the microbial ecology of fresh meat may be useful in understanding the mechanisms leading to spoilage and how to counteract it. This may help in the development of future strategies for the prevention of spoilage to reduce food waste and loss.
MATERIALS AND METHODS
Beef storage.
Beef cuts (brisket) were bought from four different local shops (Campania Region, Southern Italy; indicated as A, C, D, and F), portioned in 40-g chops and transported refrigerated to the laboratory. Beef chops to be stored aerobically were placed in polystyrene trays sealed with an oriented polypropylene (OPP) low-density polyethylene (LDPE) film (pO2, 2,500 cm3 · m−2 · h−1). Beef chops from the same cut were vacuum packed using bags (200 mm by 300 mm) of plastic barrier film (low-density polyethylene, Cryovac BB3050, oxygen transmission rate of 0.83 cm3 · m−2 · h−1 at 23°C; Cryovac Sealed Air S.r.l., Milan, Italy). Two chops were placed in each tray or bag and used for DNA extraction and microbiological analysis, respectively. The samples were stored at 4°C, and analyses were carried out at 0, 3, and 10 or 0, 3, 10, and 20 days for beef stored aerobically or under vacuum, respectively.
Pseudomonas sp. enumeration and carA gene multiplex PCR essay.
For each sampling point, viable Pseudomonas spp. were counted on Pseudomonas agar base containing cetrimide-fucidin-cephaloridine (CFC) selective supplement (both from Oxoid, Milan). Twenty-five grams of beef was homogenized in 225 ml of quarter-strength Ringer’s solution (Oxoid) for 2 min in a stomacher (LAB Blender 400; PBI, Milan, Italy). Decimal dilutions were prepared, and 0.1-ml aliquots of the appropriate dilutions were spread in triplicates. Plates were incubated at 20°C for 48 h, and typical Pseudomonas colonies were confirmed by the oxidase test (Sigma-Aldrich Co., St. Louis. MO, USA). After plate counts, all the colonies present on the surfaces of countable plates were collected in bulk. DNA was extracted as previously described (29) and used as the templates in a multiplex PCR assay for the identification of Pseudomonas fragi, Pseudomonas lundensis, and Pseudomonas putida by targeting the carA gene (29).
DNA extraction and sequencing.
For each sampling point, microbial cells on the surfaces of the beef chops (2 beef chops for each shop and sampling point) were collected with sterile cotton-tipped swabs moistened with phosphate-buffered saline (PBS). Swabs from the replicates of the same sample were pooled, placed in 5 ml of PBS, and vortexed thoroughly. The buffer was centrifuged (12,000 × g for 2 min), and DNA extraction was carried out on the pellet by using the Biostic Bacteremia DNA isolation kit (MO BIO Laboratories Inc., Carlsbad, CA). DNA was quantified using the Qubit dsDNA HS assay kit (Invitrogen). The Nextera XT library preparation kit (Illumina) was used for library preparation and sample multiplexing. Sequencing was carried out on an Illumina HiSeq sequencer (Illumina, San Diego, CA), yielding 2 × 150-bp paired-end reads. We failed to prepare the libraries for 7 samples that were excluded from the analysis (see Results).
Metagenomics data analysis.
(i) Read filtering. Host contamination was removed by mapping raw reads against the Bos taurus genome (UMD 3.1.1) using bmtagger (ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/). Raw reads were quality trimmed (Phred score < 25), and reads shorter than 60 bp were discarded with the SolexaQA++ software (30). The numbers of reads per sample resulting after filtering are reported in Table S1 in the supplemental material.
(ii) Taxonomic and functional profiling. Taxonomic profiling was carried out by using MetaPhlAn2 (version 2.6; 34), while HUMAnN2 (31) was used for functional annotation.
(iii) Pseudomonas fragi pangenomics. Strain-level analysis of Pseudomonas fragi was performed through a gene-content-based profiling using PanPhlAn (32) with parameters –min_coverage 1, –left_max 1.70, and –right_min 0.30. To avoid biases due to different sequencing depths, the reads were subsampled at the lowest number of reads per sample. A Pseudomonas fragi pangenome database was created according to the developer’s instructions, using P. fragi genomes available on the NCBI website (see Table S2). Pangenome representative sequences were extracted from the PanPhlAn database and aligned to the NCBI nonredundant (NR) database using BLASTx (E value cutoff of 1e−5, requiring a hit to display >90% of identity over at least 30% of the query length) for gene annotation. The same procedure was used to explore species belonging to the most abundant genera and normally involved in meat spoilage: Pseudomonas putida, Pseudomonas lundensis, Lactobacillus curvatus, Lactobacillus sakei, Leuconostoc carnosum, Leuconostoc citreum, Leuconostoc gelidum, Leuconostoc mesenteroides, and Leuconostoc pseudomesenteroides.
The differential occurrence of pangenes between samples stored aerobically or under vacuum was determined by Fisher’s exact test, carried out in the R environment (fisher.test).
Moreover, StrainEst (33) was used to explore Pseudomonas fragi strain patterns. StrainEst is a reference-based method that uses the single-nucleotide variant (SNV) profiles of the available genomes of the species of interest to determine the number and identity of coexisting strains and their relative abundances in mixed metagenomic samples.
Accession number(s).
Raw sequence reads were deposited in the Sequence Read Archive (SRA) of the NCBI under accession number SRP144630 (https://trace.ddbj.nig.ac.jp/DRASearch/study?acc=SRP144630).
Supplementary Material
ACKNOWLEDGMENTS
D.E. was responsible for the research design and contributed resources for sequencing data generation. F.V. contributed reagents and materials for the study and critically assessed the experimental design. F.D.F. carried out bioinformatics data analyses, wrote the paper, and prepared the figures and tables. F.D.F. and A.L.S. performed the experiments.
The authors declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02212-18.
REFERENCES
- 1.De Filippis F, La Storia A, Villani F, Ercolini D. 2013. Exploring the sources of bacterial spoilers in beefsteaks by culture-independent high-throughput sequencing. PLoS One 8:e70222. doi: 10.1371/journal.pone.0070222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Filippis F, Parente E, Ercolini D. 2018. Recent past, present and future of the food microbiome. Annu Rev Food Sci Technol 9:589–608. doi: 10.1146/annurev-food-030117-012312. [DOI] [PubMed] [Google Scholar]
- 3.Hultman J, Rahkila R, Ali J, Rousu J, Björkroth J. 2015. Meat processing plant microbiome and contamination patterns of cold-tolerant bacteria causing food safety and spoilage risks in the manufacture of vacuum-packaged cooked sausages. Appl Environ Microbiol 81:7088–7097. doi: 10.1128/AEM.02228-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stellato G, La Storia A, De Filippis F, Borriello G, Villani F, Ercolini D. 2016. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl Environ Microbiol 82:4045–4054. doi: 10.1128/AEM.00793-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ercolini D, Ferrocino I, Nasi A, Ndagijimana M, Vernocchi P, La Storia A, Laghi L, Mauriello G, Guerzoni ME, Villani F. 2011. Monitoring of microbial metabolites and bacterial diversity in beef stored under different packaging conditions. Appl Environ Microbiol 77:7372–7381. doi: 10.1128/AEM.05521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doulgeraki AI, Ercolini D, Villani F, Nychas GJ. 2012. Spoilage microbiota associated to the storage of raw meat in different conditions. Int J Food Microbiol 157:130–141. doi: 10.1016/j.ijfoodmicro.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Pothakos V, Devlieghere F, Villani F, Björkroth J, Ercolini D. 2015. Lactic acid bacteria and their controversial role in fresh meat spoilage. Meat Sci 109:66–74. doi: 10.1016/j.meatsci.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Chaillou S, Chaulot-Talmon A, Caekebeke H, Cardinal M, Christieans S, Denis C, Desmonts MH, Dousset X, Feurer C, Hamon E, Joffraud JJ, La Carbona S, Leroi F, Leroy S, Lorre S, Macé S, Pilet MF, Prévost H, Rivollier M, Roux D, Talon R, Zagorec M, Champomier-Vergès MC. 2015. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME J 9:1105–1118. doi: 10.1038/ismej.2014.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gram L, Ravn L, Rasch M, Bruhn JB, Christensen AB, Givskov M. 2002. Food spoilage–interactions between food spoilage bacteria. Int J Food Microbiol 78:79–97. doi: 10.1016/S0168-1605(02)00233-7. [DOI] [PubMed] [Google Scholar]
- 10.Pennacchia C, Ercolini D, Villani F. 2011. Spoilage-related microbiota associated with chilled beef stored in air or vacuum pack. Food Microbiol 28:84–93. doi: 10.1016/j.fm.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Húngaro HM, Caturla MYR, Horita CN, Furtado MM, Sant’Ana AS. 2016. Blown pack spoilage in vacuum-packaged meat: a review on clostridia as causative agents, sources, detection methods, contributing factors and mitigation strategies. Trends Food Sci Technol 52:123–138. doi: 10.1016/j.tifs.2016.04.010. [DOI] [Google Scholar]
- 12.Kaur M, Shang H, Tamplin M, Ross T, Bowman JP. 2017. Culture-dependent and culture-independent assessment of spoilage community growth on VP lamb meat from packaging to past end of shelf-life. Food Microbiol 68:71–80. doi: 10.1016/j.fm.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Kiermeier A, Tamplin M, May D, Holds G, Williams M, Dann A. 2013. Microbial growth. communities and sensory characteristics of vacuum and modified atmosphere packaged lamb shoulders. Food Microbiol 36:305–315. doi: 10.1016/j.fm.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Botta C, Ferrocino I, Cavallero MC, Riva S, Giordano G, Cocolin L. 2018. Potentially active spoilage bacteria community during the storage of vacuum packaged beefsteaks treated with aqueous ozone and electrolyzed water. Int J Food Microbiol 266:337–345. doi: 10.1016/j.ijfoodmicro.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Jääskeläinen E, Hultman J, Parshintsev J, Riekkola M-L, Björkroth J. 2016. Development of spoilage bacterial community and volatile compounds in chilled beef under vacuum or high oxygen atmospheres. Int J Food Microbiol 223:25–32. doi: 10.1016/j.ijfoodmicro.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Casaburi A, Piombino P, Nychas GJ, Villani F, Ercolini D. 2015. Bacterial populations and the volatilome associated to meat spoilage. Food Microbiol 45:83–102. doi: 10.1016/j.fm.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Ercolini D, Casaburi A, Nasi A, Ferrocino I, Di Monaco R, Ferranti P, Mauriello G, Villani F. 2010. Different molecular types of Pseudomonas fragi have the same overall behaviour as meat spoilers. Int J Food Microbiol 142:120–131. doi: 10.1016/j.ijfoodmicro.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Casaburi A, De Filippis F, Villani F, Ercolini D. 2014. Activities of strains of Brochothrix thermosphacta in vitro and in meat. Food Res Int 62:366–374. doi: 10.1016/j.foodres.2014.03.019. [DOI] [Google Scholar]
- 19.Casaburi A, Nasi A, Ferrocino I, Di Monaco R, Mauriello G, Villani F, Ercolini D. 2011. Spoilage-related activity of Carnobacterium maltaromaticum strains in air-stored and vacuum-packed meat. Appl Environ Microbiol 77:7382–7393. doi: 10.1128/AEM.05304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jääskeläinen E, Johansson P, Kostiainen O, Nieminen T, Schmidt G, Somervuo P, Mohsina M, Vanninen P, Auvinen P, Björkroth J. 2013. Significance of heme-based respiration in meat spoilage caused by Leuconostoc gasicomitatum. Appl Environ Microbiol 79:1078–1085. doi: 10.1128/AEM.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrocino I, La Storia A, Torrieri E, Musso SS, Mauriello G, Villani F, Ercolini D. 2013. Antimicrobial packaging to retard the growth of spoilage bacteria and to reduce the release of volatile metabolites in meat stored under vacuum at 1°C. J Food Prot 76:52–58. doi: 10.4315/0362-028X.JFP-12-257. [DOI] [PubMed] [Google Scholar]
- 22.Jääskeläinen E, Vesterinen S, Parshintsev J, Johansson P, Riekkola ML, Björkroth J. 2015. Production of buttery-odor compounds and transcriptome response in Leuconostoc gelidum subsp. gasicomitatum LMG18811T during growth on various carbon sources. Appl Environ Microbiol 81:1902–1908. doi: 10.1128/AEM.03705-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radoš D, Turner DL, Catarino T, Hoffart E, Neves AR, Eikmanns BJ, Blombach B, Santos H. 2016. Stereospecificity of Corynebacterium glutamicum 2,3-butane-diol dehydrogenase and implications for the stereochemical purity of bioproduced 2,3-butanediol. Appl Microbiol Biotechnol 100:10573–10583. doi: 10.1007/s00253-016-7860-6. [DOI] [PubMed] [Google Scholar]
- 24.Galgano F, Favati F, Bonadio M, Lorusso V, Romano P. 2009. Role of biogenic amines as index of freshness in beef meat packed with different biopolymeric materials. Food Res Int 42:1147–1152. doi: 10.1016/j.foodres.2009.05.012. [DOI] [Google Scholar]
- 25.De Filippis F, Pennacchia C, Di Pasqua R, Fiore A, Fogliano V, Villani F, Ercolini D. 2013. Decarboxylase gene expression and cadaverine and putrescine production by Serratia proteamaculans in vitro and in beef. Int J Food Microbiol 165:332–338. doi: 10.1016/j.ijfoodmicro.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Stanborough T, Fegan N, Powell SM, Singh T, Tamplin M, Chandry PS. 2018. Genomic and metabolic characterization of spoilage-associated Pseudomonas species. Int J Food Microbiol 268:61–72. doi: 10.1016/j.ijfoodmicro.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 28.Zgurskaya HI, Krishnamoorthy G, Ntreh A, Lu S. 2011. Mechanism and function of the outer membrane channel TolC in multidrug resistance and physiology of enterobacteria. Front Microbiol 2:189. doi: 10.3389/fmicb.2011.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ercolini D, Russo F, Blaiotta G, Pepe O, Mauriello G, Villani F. 2007. Simultaneous detection of Pseudomonas fragi, P. lundensis, and P. putida from meat by use of a multiplex PCR assay targeting the carA gene. Appl Environ Microbiol 73:2354–2359. doi: 10.1128/AEM.02603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methé B, Schloss PD, Gevers D, Mitreva M, Huttenhower C. 2012. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol 8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F, Truong DT, Tett A, Morrow AL, Segata N. 2016. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods 13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 33.Albanese D, Donati C. 2017. Strain profiling and epidemiology of bacterial species from metagenomic sequencing. Nat Commun 8:2260. doi: 10.1038/s41467-017-02209-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.