Marine polysaccharides, found in the cell walls of seaweeds and other marine macrophytes, represent a vast sink of photosynthetically fixed carbon. As such, their breakdown by marine microbes contributes significantly to global carbon cycling. Pectin is an abundant polysaccharide found in the cell walls of terrestrial plants, but it has recently been reported that some marine bacteria possess the genetic capacity to degrade it. In this study, we biochemically characterized seven key enzymes from a marine bacterium that, together, fully degrade the backbone of pectin into its constituent monosaccharides. Our findings highlight the importance of pectin as a marine carbon source available to bacteria that possess this pathway. The characterized enzymes also have the potential to be utilized in the production of biofuels from plant waste.
KEYWORDS: glycoside hydrolase, pectin, polysaccharide lyase, zosterin, carbon metabolism, marine microbiology, pectic enzymes
ABSTRACT
Pectin is a complex uronic acid-containing polysaccharide typically found in plant cell walls, though forms of pectin are also found in marine diatoms and seagrasses. Genetic loci that target pectin have recently been identified in two phyla of marine bacteria. These loci appear to encode a pectin saccharification pathway that is distinct from the canonical pathway typically associated with phytopathogenic terrestrial bacteria. However, very few components of the marine pectin metabolism pathway have been experimentally validated. Here, we biochemically reconstructed the pectin saccharification pathway from a marine Pseudoalteromonas sp. in vitro and show that it results in the production of galacturonate and the key metabolic intermediate 5-keto-4-deoxyuronate (DKI). We demonstrate the sequential de-esterification and depolymerization of pectin into oligosaccharides and the synergistic action of glycoside hydrolases (GHs) to fully degrade these oligosaccharides into monosaccharides. Furthermore, we show that this pathway relies on enzymes belonging to GH family 105 to carry out the equivalent chemistry afforded by an exolytic polysaccharide lyase (PL) and KdgF in the canonical pectin pathway. Finally, we synthesize our findings into a model of marine pectin degradation and compare it with the canonical pathway. Our results underline the shifting view of pectin as a solely terrestrial polysaccharide and highlight the importance of marine pectin as a carbon source for suitably adapted marine heterotrophs. This alternate pathway has the potential to be exploited in the growing field of biofuel production from plant waste.
IMPORTANCE Marine polysaccharides, found in the cell walls of seaweeds and other marine macrophytes, represent a vast sink of photosynthetically fixed carbon. As such, their breakdown by marine microbes contributes significantly to global carbon cycling. Pectin is an abundant polysaccharide found in the cell walls of terrestrial plants, but it has recently been reported that some marine bacteria possess the genetic capacity to degrade it. In this study, we biochemically characterized seven key enzymes from a marine bacterium that, together, fully degrade the backbone of pectin into its constituent monosaccharides. Our findings highlight the importance of pectin as a marine carbon source available to bacteria that possess this pathway. The characterized enzymes also have the potential to be utilized in the production of biofuels from plant waste.
INTRODUCTION
Polysaccharides are abundant and chemically diverse macromolecules that are found in all ecological habitats. They represent an immense store of photosynthetically fixed carbon and source of energy to those organisms that possess the appropriate degradative and metabolic machinery to release it. Due to the complexity and chemical diversity of polysaccharides found in different environments, the carbohydrate-active enzymes (CAZymes), transporters, and other metabolic processing enzymes employed by heterotrophic organisms are usually specific for a particular polysaccharide and, therefore, ecological habitat (1). In bacteria, the genes encoding these specific proteins are often grouped together in the genome into coregulated regions known as polysaccharide utilization loci (PULs). These PULs define the metabolic capacity of the organism to utilize polysaccharides and thus greatly contribute to determining its environmental niche (2, 3).
Pectin is a complex and abundant family of polysaccharides found primarily in the cell walls of terrestrial plants (4). As such, its degradation and metabolism by microbes constitute an important component of the natural turnover of biomass, infection by plant pathogens, and digestion of dietary fiber in the mammalian gut (5–7). The simplest and most common pectin is homogalacturonan (HG), which comprises a linear backbone of α(1, 4)-linked d-galacturonate (GalUA) residues. These GalUA residues are commonly methylesterified at C-6 and are occasionally acetylesterified at O-2 or O-3 (8). Pectins can also contain more complex structural motifs, such as rhamnogalacturonan I (RGI) and rhamnogalacturonan II (RGII), which contain variable backbone structures and extensive side chain modifications. Given the abundance and structural nature of pectin in plant cell walls, a wide variety of microbes have acquired enzymatic pathways for the degradation and metabolism of this polysaccharide (5, 7, 9–13).
Because of its importance to phytopathogenesis, the metabolism of HG has been extensively studied over the last 50 years, allowing the construction of what we refer to as the canonical HG metabolism pathway, which is applicable to a wide variety of pectinolytic bacteria (13–18). Depolymerization is typically initiated extracellularly by one or more secreted endo-acting polysaccharide lyases (PLs), which cleave the HG backbone via a β-elimination mechanism, thereby generating oligouronates with 4,5-unsaturated nonreducing ends. Extracellular depolymerization is often assisted by other CAZymes, namely, carbohydrate esterases (CEs) and endo-acting α-galacturonidases belonging to glycoside hydrolase (GH) family 28. Saturated and unsaturated oligouronates then diffuse into the periplasm through porins belonging to the KdgM family. Once in the periplasm, oligouronates are further degraded into di- and trisaccharides by a different set of endo- and exo-acting PLs and an exo-acting GH28. Finally, these small oligogalacturonates are imported into the cytoplasm via the ATP-dependent transporter TogMNAB and degraded into saturated GalUA and Δ-4,5-unsaturated GalUA (ΔGalUA) by an intracellular oligogalacturonate lyase belonging to PL family 22 (13). In the cytoplasm, ΔGalUA undergoes KdgF-catalyzed linearization into 5-keto-4-deoxyuronate (DKI) (16) before it is further processed by an isomerase (KduI) and a reductase (KduD) into the key metabolite 2-keto-3-deoxygluconate (KDG) (19). GalUA is also converted into KDG by the actions of UxaC, UxaB, and UxaA, consecutively (20).
The discovery of GH105 enzymes, which are unsaturated uronyl hydrolases that cleave the 4,5-unsaturated residues from the nonreducing ends of PL-generated oligouronates (21, 22), has hinted at the potential for an alternate model of pectin metabolism. The mechanism of GH105 catalysis is thought to parallel that of GH88 members, which results in release of the linearized monosaccharide directly (23, 24). The direct release of DKI by GH105 enzymes would obviate the need for a PL22 (or potentially an equivalent) and KdgF to convert the unsaturated nonreducing ends of PL-generated oligouronates into DKI. The recent extensive characterization of complex pectin metabolism pathways in human gut microbiome Bacteroides spp. has suggested that GH105 enzymes would play a role in the complete saccharification of dietary pectin (7, 25).
Pectin is typically considered to be a terrestrial polysaccharide utilized by phytopathogens, plant cell wall-degrading saprophytes, and some members of the gut microbiome. However, the ability of some marine bacteria to grow on pectin and the secretion of pectin/pectate lyases by these organisms (26–30), as well as the recent identification of pectin-responsive PULs in a number of marine bacteria (29, 31), are shifting this view. These marine organisms are thought to target pectic substances found in marine diatoms and the cell walls of seagrasses (31–33). Zosterin, or marine pectin, has been isolated and characterized from a number of species of seagrass belonging to the Zosteraceae family (33–36). It consists predominantly of apiogalacturonan (AGU), which is an HG backbone decorated with relatively frequent substitutions at O-3 of single residues, or short oligosaccharides, of d-apiose. RGI stretches, as well as methyl- and acetylesterification, have also been detected in zosterin (35). The marine Bacteroidetes member Gramella flava JLT2011 and the marine Gammaproteobacteria member Pseudoalteromonas haloplanktis ANT/505 both have the ability to degrade terrestrial pectin (27, 29). Proteomic and/or transcriptomic analysis has identified PULs within their genomes that are responsive to pectin and encode seemingly complete pectin degradation and catabolism pathways (29, 31). However, based on the genetic content of these PULs, these organisms appear to employ an enzymatic strategy for pectin degradation that differs from the canonical model (which relies on an arsenal of endo- and exolytic PLs and GH28s to fully degrade the HG backbone into GalUA and ΔGalUA and on KdgF to subsequently convert ΔGalUA into DKI). Rather, these marine microbes have genes encoding putative GH105 enzymes, and therefore it is anticipated, but not yet validated, that these organisms use a combination of PLs and GH105s to degrade HG to the key DKI intermediate.
We have previously isolated a marine species of Pseudoalteromonas, which we refer to as PS47, that is able to utilize multiple marine polysaccharides for growth (37). Here we report the identification and functional characterization of a pectin PUL from PS47 that shares regions of synteny and homology with those found in G. flava and P. haloplanktis ANT/505. Using the Pseudoalteromonas sp. strain PS47 pectin PUL as a model, we experimentally demonstrate the individual activities of seven key CAZymes from this marine pectin PUL. By biochemically reconstituting the complete enzymatic pathway in vitro, we show that together these enzymes degrade the HG backbone of pectin, liberating GalUA and DKI. Notably, the GH105 enzymes are demonstrated to play a critical role in the complete saccharification process, thus providing key biochemical support for the alternative pathway of pectin metabolism. On this basis, we propose a model of marine pectin degradation based on our experimental findings and discuss the presence of GH105s and an associated domain of unknown function among bacteria from distinct phyla.
RESULTS
Pseudoalteromonas sp. PS47 possesses a pectin utilization locus and produces pectinases.
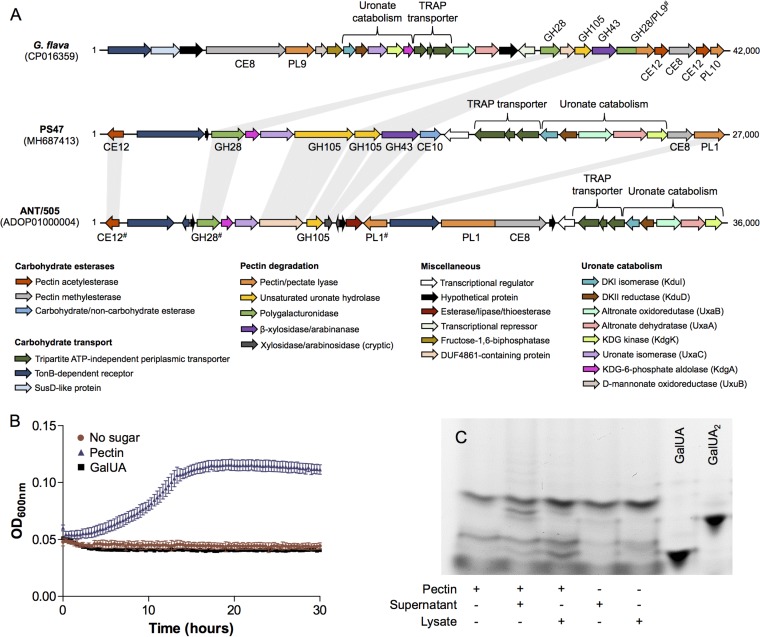
Pseudoalteromonas sp. PS47 was isolated from the intertidal zone of Victoria, British Columbia, Canada, and found to grow on a number of marine polysaccharides. In agreement with this observation, CAZyme-specific annotation of its sequenced genome (with dbCAN2 [38] supplemented with manual curation) resulted in the identification of many genes encoding putative CAZymes targeted toward marine polysaccharides, including agarose, alginate, and carrageenan. On further analysis, we also identified an ∼27,000-bp locus that includes a pair of genes encoding putative GH105 enzymes and likely genes encoding the metabolic machinery necessary to process both GalUA and DKI (Fig. 1A). This locus also encodes three putative CEs, a PL1 (putative pectin/pectate lyase), a GH28, a GH43 (putative β-xylosidase/arabinanase), a TonB-dependent transporter, and a tripartite ATP-independent periplasmic (TRAP) transporter. As expected, the transport and metabolic processing enzymes encoded in this locus are homologous to those encoded in the previously identified P. haloplanktis pectin PUL; however, based on the reported annotations, the CAZyme compositions in these two species initially appeared to be quite different (Table 1). Our reannotation of the P. haloplanktis PUL and sequence comparisons with proteins from the PS47 PUL actually revealed these two PULs to be very similar (Fig. 1A). The main points of difference are the addition of a multimodular PL1/CE8 and the reduction of a putative β-xylosidase/arabinanase into two cryptic genes in the P. haloplanktis PUL. The CAZyme annotation of the G. flava genome is publicly available through the Carbohydrate Active Enzymes database (www.cazy.org), and although the pectin PUL in G. flava PUL is more expansive in its CAZyme composition than the Pseudoalteromonas sp. PULs, the general complements of CAZyme functions are all similar (Fig. 1A and Table 1). Furthermore, all four GHs present in the G. flava PUL are homologous to one or more GHs from the PS47 PUL (Table 1). Given the genetic composition of the PS47 locus and its regions of homology and synteny with the G. flava and P. haloplanktis PULs, we hypothesized that the PS47 PUL also enables the degradation, uptake, and metabolism of marine pectin.
FIG 1.
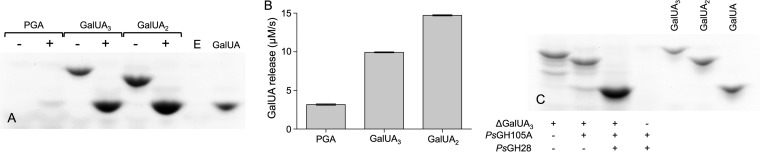
Pseudoalteromonas sp. PS47 contains a pectin utilization locus and produces pectinases. (A) Schematic depiction of the pectin utilization loci found in the genomes of Gramella flava JLT2011, Pseudoalteromonas sp. PS47, and Pseudoalteromonas haloplanktis ANT/505. Open reading frames (ORFs) are shown to scale (within a locus) and colored according to putative function. CAZyme family classifications, according to www.cazy.org and/or the dbCAN2 server, are given underneath or above; some of these classifications have been updated (indicated by #) and therefore differ from those originally reported by Tang et al. (29) and Hehemann et al. (31). Light gray bars between CAZyme-encoding ORFs indicate amino acid sequence identities of >40% (see Table 1 for exact sequence identities). Abbreviations: CE, carbohydrate esterase; GH, glycoside hydrolase; PL, polysaccharide lyase; DKI, 5-keto-4-deoxyuronate; DKII, 2,5-diketo-3-deoxygluconate; KDG, 2-keto-3-deoxygluconate. (B) Growth of PS47 on citrus pectin as the sole carbon source in minimal marine medium, with a no-sugar control shown for comparison. Absence of growth on galacturonate (GalUA) is also shown. Data shown are the mean of two biological replicates; error bars represent the SEM. (C) FACE gel showing the presence of pectinases in both the culture supernatant and cell lysate of PS47 grown in Zobell marine medium containing 0.2% pectin. + and −, presence and absence, respectively, of substrate or cell fraction. GalUA and digalacturonate (GalUA2) were included on the gel as standards.
TABLE 1.
Comparison of proteins present in pectin PULs from Pseudoalteromonas sp. PS47, Pseudoalteromonas haloplanktis ANT/505, and Gramella flava JLT2011
| Protein name in PS47 | dbCAN2 HMMER E value | Protein name in P. haloplanktis and locus taga | % amino acid identity (PS47 vs P. haloplanktis) | Protein name in G. flava and locus tagb | % amino acid identity (PS47 vs G. flava) |
|---|---|---|---|---|---|
| PsCE12 | 3.5e−72 | RhgT (transcriptional regulator; GNTR) | 63.5 | Rga (CE12) | |
| PH505_ap00680 | GRFL_2179 | 5.6 | |||
| GRFL_218) | 12.4 | ||||
| PsGH28 | 1e−87 | GhyB (amylomaltase; GH77) | 75.3 | PelL (GH28) | |
| PH505_ap00640 | GRFL_2174 | 46.0 | |||
| GRFL_2178 | 33.0 | ||||
| PsGH105A | 1.1e−106 | DNA-directed RNA polymerase, α-subunit | 64.5 | RhiN (GH105) | 38.4d |
| PH505_ap00610 | GRFL_2176 | ||||
| PsGH105B | 4.2e−113 | GhyA (GH105) | 63.6 | RhiN (GH105) | 43.6 |
| PH505_ap00600 | GRFL_2176 | ||||
| PsGH43 | 2.1e−71 | XylA/B (xylosidase/arabinase) | XynB (GH43) | 46.4 | |
| PH505_ap00580 | 56.8c | GRFL_2177 | |||
| PH505_ap00590 | 69.7c | ||||
| PsCE10 | 5.9e−44 | Esterase/lipase/thioesterase | 52.8 | NAf | NA |
| PH505_ap00560 | |||||
| PsCE8 | 1.5e−31 | PelA (pectate lyase/methylesterase; PL1; CE8)e | 21.6d | PemA (CE8) | |
| PH505_ap00520 | GRFL_2158 | 5.4 | |||
| GRFL_2180 | 14.1 | ||||
| PsPL1 | 9.5e−66 | PelA (pectate lyase/methylesterase; PL1; CE8)e | NA | NA | |
| PH505_ap00520 | 15.0d | ||||
| PelC (pectate lyase; PL3) | |||||
| PH505_ap00550 | 51.6 |
From reference 31.
From reference 29.
Sequence identity based upon only partial coverage, as XylA (118 residues) and XylB (76 residues) are encoded by cryptic genes.
For fusion proteins or proteins with additional domains, the sequence identity shown applies only to the homologous domain, not the entire protein sequence.
The annotation of this protein as a bifunctional pectate lyase/methylesterase has been experimentally validated.
NA, not applicable.
We initially addressed this hypothesis by investigating whether PS47 produced pectinases and could utilize pectin for growth. In general, growth of PS47 in minimal marine medium was limited, even on its preferred substrate (d-galactose) (data not shown), but it did exhibit some growth on citrus pectin (Fig. 1B). Interestingly, PS47 was unable to grow on GalUA. We also detected the presence of pectinases in the supernatant and cell lysate from a culture of PS47 grown in pectin-supplemented Zobell broth by fluorophore-assisted carbohydrate electrophoresis (FACE) (Fig. 1C). A faint ladder of large labeled oligosaccharides and a strong band corresponding to approximately a disaccharide were visible following exposure of pectin to PS47 culture supernatant. Treatment of pectin with cell lysate produced smaller labeled products, including a band that likely corresponds to GalUA.
PsPL1 is an endolytic pectate lyase active on a range of pectin-related substrates.
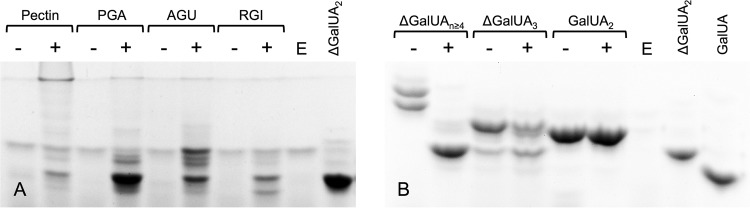
Having confirmed the presence of pectinase activity in cultures of PS47, we set out to biochemically identify and characterize the CAZymes responsible for this activity. Degradation of the HG backbone of pectin is typically initiated by a calcium-dependent endolytic PL; therefore, we cloned the putative PL1 encoded by the PS47 pectin PUL (PsPL1) without its signal peptide, expressed it in Escherichia coli, and tested it for activity against a range of GalUA-containing substrates by FACE (Fig. 2A). PsPL1 produced a faint ladder of oligosaccharides from citrus pectin and RGI, whereas it exhibited more extensive degradation of polygalacturonate (PGA) and AGU. In particular, PsPL1 appeared to produce predominantly unsaturated digalacturonate (ΔGalUA-GalUA [ΔGalUA2]) from PGA. The high rate of production of oligouronates bearing a 4,5-double bond from PGA by PsPL1 was also observed as an increase in absorbance at 230 nm (data not shown). The increased activity of PsPL1 against PGA compared with pectin suggests that it is hindered by the decorations found on pectin and is therefore a pectate, rather than a pectin, lyase. We also used the increase in absorbance at 230 nm against PGA to determine the pH optima of PsPL1 (≥9) (see Fig. S1 in the supplemental material).
FIG 2.
PsPL1 displays endolytic lyase activity against a range of pectin-related substrates. (A) FACE gel of pectin and related polysaccharides following overnight incubation with 1 μM PsPL1 in the presence of Ca2+. + and −, presence and absence, respectively, of PsPL1; E, enzyme-only sample. Abbreviations: PGA, polygalacturonate; AGU, apiogalacturonan; RGI, rhamnogalacturonan I; ΔGalUA2, unsaturated digalacturonate (included as a standard). (B) Activity of PsPL1 against oligosaccharides of galacturonate with various degrees of polymerization. + and −, presence and absence, respectively, of PsPL1; E, enzyme-only sample. ΔGalUA2 and galacturonate (GalUA) are included as standards.
In order to determine the minimum degree of polymerization (DP) of GalUA oligosaccharides required by PsPL1 for activity and to assist in our characterization of downstream enzymes, we used PsPL1 to generate a range of unsaturated oligogalacturonates from PGA. PsPL1 was incubated with PGA in the absence of calcium in an attempt to slow the degradation of larger oligosaccharides by PsPL1 into ΔGalUA2 and obtain a panel of larger glycans. The resulting oligosaccharides were purified and their DPs estimated following comparison with standards labeled and separated by FACE (Fig. 2B). A relatively (∼80%) pure unsaturated oligogalacturonate estimated to be unsaturated trigalacturonate (ΔGalUA-GalUA-GalUA [ΔGalUA3]) was obtained, as well as a mixture of larger unsaturated oligouronates with an estimated DP of ≥4. PsPL1 was tested for activity against these purified glycans, in addition to GalUA2, in the presence of calcium (Fig. 2B). PsPL1 was able to completely degrade the larger oligosaccharides into ΔGalUA2, but activity against ΔGalUA3 was limited and GalUA2 did not act as a substrate for PsPL1. We suggest that these results are most consistent with PsPL1 having endolytic pectate lyase activity with a minimum substrate DP requirement of 4.
Demethylation of pectin by PsCE8 increases PsPL1 activity.
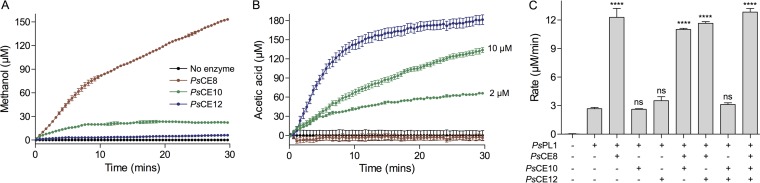
Our FACE results suggest that the activity of PsPL1 is hindered by the esterification of citrus pectin. The PS47 pectin PUL encodes three putative CEs belonging to families 8, 10, and 12 (PsCE8, PsCE10, and PsCE12). We cloned and expressed these three CAZymes without their signal peptides and tested them for pectin methyl- and acetylesterase activities using enzyme-coupled assays. These assays provide a read-out of liberated methanol and acetate, respectively. PsCE8 exhibited methylesterase activity against pectin, while PsCE12 demonstrated acetylesterase activity (Fig. 3A and B). PsCE10 appeared to demonstrate both weak methyl- and acetylesterase activities, but only the acetylesterase activity was increased when the concentration of enzyme was increased; therefore, the apparent methylesterase activity is likely artifactual.
FIG 3.
Terrestrial pectin is de-esterified by PsCE8, PsCE10, and PsCE12. (A and B) The ability of the three putative pectin esterases to liberate methanol (A) or acetic acid (B) from citrus pectin was tested in enzyme-coupled assays. The weak acetylesterase activity of PsCE8 was confirmed at two enzyme concentrations (2 μM and 10 μM); the apparent weak methylesterase activity of PsCE10 seen in panel A was not increased in magnitude at 10 μM enzyme (data not shown), so it appears to be artifactual. The symbol legend applies to both panels. (C) The rate of unsaturated oligouronate production from pectin by PsPL1 was monitored at 230 nm in the presence and absence of PsCE8, PsCE10, and PsCE12 (indicated by + and −). Asterisks indicate statistically significant differences between enzyme combinations and the PsPL1-only sample as determined by a one-way analysis of variance (ANOVA) with Bonferroni posttest (****, P ≤ 0.0001; ns, P > 0.05). In all panels, data shown are the mean of three replicates and error bars represent the SEM.
Given the presence of genes in the PUL encoding active esterases, we reasoned that their role would be to remove pectin modifications and thereby potentially aid depolymerization by PsPL1. To test this, we measured the rate of 4,5-double bond production by PsPL1 in the presence and absence of a single esterase or esterases in combination (Fig. 3C). Demethylation of pectin by PsCE8 increased the rate of PsPL1 activity by ∼4-fold; deacetylation by PsCE10 and/or PsCE12 had no statistically significant effect. Despite the significant increase in rate observed in the presence of PsCE8, the rate of PsPL1 activity against pectin in the presence of PsCE8 is still ∼20-fold lower than that against unesterified PGA (data not shown).
PS47 possesses two Δ-4,5-unsaturated α-galacturonidases with different catalytic efficiencies.
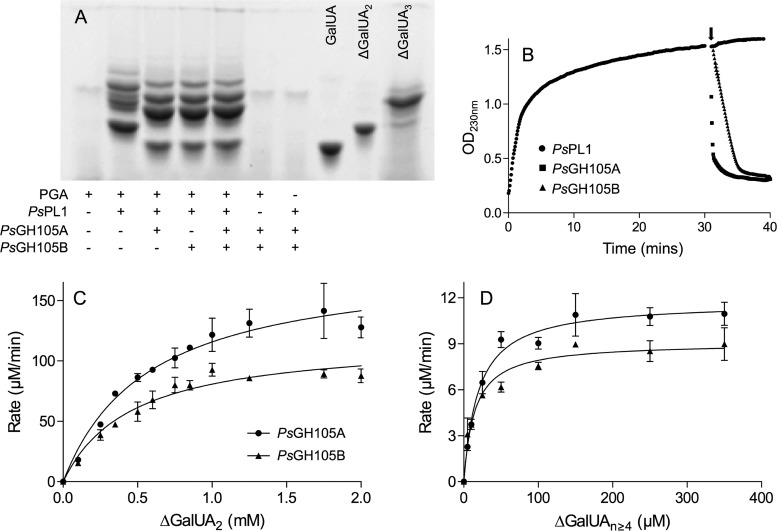
The action of PsPL1 results in the production of small unsaturated oligogalacturonates. In pectin degradation pathways, these oligosaccharides can be further degraded by either exolytic pectate lyases or unsaturated galacturonyl hydrolases belonging to the GH105 family. As the PS47 pectin PUL encodes only a single PL (PsPL1) but two GH105s (PsGH105A and PsGH105B), we cloned and expressed these two putative hydrolases and tested them for activity against the products of PsPL1 (Fig. 4). PGA was treated with PsPL1 in the absence of calcium so as to produce a ladder of products, the reactions were stopped by heating, and then the mixtures were split into aliquots to which the different GH105s (or no enzyme) were added (Fig. 4A). By FACE analysis, we observed a shift down in the banding pattern produced by PsPL1 together with either GH105 compared with PsPL1 alone and also the appearance of a product migrating to the same point as the GalUA standard. While the unsaturated galacturonyl residue released by the GH105 would not be labeled by the fluorophore (due to its lack of a free aldehyde), cleavage of ΔGalUA2 by a GH105 would also result in the release of GalUA. The banding patterns produced by PsGH105A and PsGH105B, as well as both enzymes together, were not distinguishable from one another.
FIG 4.
PsGH105A and PsGH105B act on the unsaturated oligogalacturonates produced by PsPL1 with different efficiencies. (A) FACE gel showing the shift in the banding patterns of oligogalacturonates produced by PsPL1 from polygalacturonate (PGA) following treatment with PsGH105A, PsGH105B, or both. Galacturonate (GalUA), unsaturated digalacturonate (ΔGalUA2), and unsaturated trigalacturonate (ΔGalUA3) were included as standards. + and −, presence and absence, respectively, of substrate or enzyme. (B) Removal of unsaturated uronyl residues from the nonreducing end of PsPL1-generated oligogalacturonates by GH105As as detected by a decrease in OD230. PsPL1 was added to PGA in all reaction mixtures at time zero, and either PsGH105A, PsGH105B, or buffer was added to separate reaction mixtures after 30 min (indicated by the arrow). Data shown are the mean of three replicates, and error bars, where visible, represent the SEM. (C and D) Michaelis-Menten kinetics for PsGH105A and PsGH105B against ΔGalUA2 (C) and a mixture of unsaturated oligogalacturonates with an estimated degree of polymerization of ≥4 (D). Data shown are the mean of three replicates, and error bars represent the SEM. The enzyme concentrations used were 250 nM for both enzymes against ΔGalUA2 and 1 nM PsGH105A and 10 nM PsGH105B against ΔGalUAn≥4.
We also observed the activity of the GH105s on the products of PsPL1 as a decrease in absorbance at 230 nm (Fig. 4B). This linked assay is essentially the same as we have previously used to demonstrate the double-bond depletion activity of KdgF (16). PsPL1 was added to PGA, and the increase in absorbance at 230 nm was monitored for 30 min prior to the addition of PsGH105A or PsGH105B. In agreement with the FACE analysis, the addition of either GH105 resulted in an immediate and rapid decrease in absorbance. This rapid decrease in absorbance is consistent with removal of the terminal ΔGalUA and its direct conversion into DKI, which does not absorb at 230 nm; however, KdgF has previously been hypothesized to play a role in post-GH105 processing of oligouronates (31). We tested the effect of adding Yersinia enterocolitica KdgF (YeKdgF), an enzyme that is known to catalyze the conversion of ΔGalUA to DKI (16), to our post-PsPL1 reaction mixtures simultaneously with either GH105 and observed no alteration in the rate of double-bond depletion (data not shown). Therefore, the results support the direct release of the linear molecule DKI from unsaturated oligogalacturonides by PsGH105A and PsGH105B.
Our FACE analysis did not suggest any difference in substrate specificity between PsGH105A and PsGH105B; however, PsGH105A did exhibit a higher rate of double-bond depletion than PsGH105B against the pool of PGA-generated oligouronates (Fig. 4B). To further investigate this possible difference, we determined the Michaelis-Menten kinetics of both enzymes against ΔGalUA2 and ΔGalUA≥n = 4 at their pH optima (pH 6.5 and 5.7 for PsGH105A and PsGH105B, respectively) (Fig. S1). The kinetics for PsGH105A and PsGH105B were very similar against ΔGalUA2 (Km = 0.58 ± 0.1 mM and 0.46 ± 0.1 mM, kcat = 12.2 ± 0.9 s−1 and 7.9 ± 0.5 s−1, and kcat/Km = 21.0 ± 3.9 s−1 mM−1 and 17.2 ± 3.9 s−1 mM−1 for PsGH105A and PsGH105B, respectively [mean ± standard error of the mean {SEM}]), and both enzymes exhibited much higher affinities for the larger oligosaccharides (21.2 ± 3.7 μM and 15.8 ± 3.4 μM for PsGH105A and PsGH105B, respectively) (Fig. 4C and D); however, the kcat for PsGH105A against ΔGalUA≥n = 4 was >10-fold higher than that determined for PsGH105B (200.8 ± 7.3 s−1 versus 15.5 ± 0.7 s−1). These data indicate that both enzymes have considerably higher affinities for unsaturated oligogalacturonates larger than ΔGalUA2, but PsGH105A is able to process these large glycans faster than PsGH105B. As such, the catalytic efficiency (kcat/Km) of PsGH105A against ΔGalUA≥n = 4 is 9.5 ± 1.7 s−1 μM−1, compared with 1.0 ± 0.2 s−1 μM−1 for PsGH105B.
Sequence analysis of PsGH105A and PsGH105B reveals that PsGH105A is approximately twice the size of PsGH105B due to the presence of an N-terminal domain of unknown function (DUF) belonging to family 4861. The DUF4861 family is uncharacterized, but its members are often found upstream of unsaturated uronyl hydrolases belonging to GH family 88 members. Therefore, it has been speculated that they may be involved in carbohydrate binding (http://pfam.xfam.org/family/PF16153 ). In order to test the potential carbohydrate binding function of the DUF of PsGH105A and whether it contributes to the increased turnover rate of this enzyme compared with PsGH105B, we cloned the DUF and catalytic domain of PsGH105A separately. The boundaries for these constructs were determined based on domain annotations in BLAST (39) and dbCAN2 (38), a PHYRE2 homology model (40) of the catalytic domain, and secondary structure predictions for loop regions between the two domains (41–43). The DUF was successfully expressed and purified, and binding of this protein to PGA and ΔGalUA≥n = 4 was evaluated using differential scanning fluorimetry (see Fig. S2 in the supplemental material). No change in the melting curve of the DUF was observed in the presence of either ligand at any concentration or pH. Unfortunately, the catalytic domain of PsGH105A was expressed insolubly in E. coli; therefore, a comparison of catalytic parameters with and without the DUF could not be performed.
PsGH28 is an exo-acting α-galacturonidase that completes the depolymerization of oligouronates.
The limited activity of PsPL1 against oligogalacturonates with a DP of <4 hints toward a requirement by PS47 for an exolytic α-galacturonidase in order to fully utilize all the GalUA in the HG backbone of pectin. We cloned and expressed the GH28 encoded in the PS47 pectin PUL (PsGH28) and investigated its activity against PGA and small saturated galacturonate oligosaccharides. FACE analysis showed that PsGH28 released a small amount of a product with the same mobility as GalUA from PGA, and no detectable oligosaccharides, but was able to fully convert GalUA2 and GalUA3 into what appeared to be GalUA (Fig. 5A). To confirm the production of GalUA, we used an enzyme-linked assay in which a uronate dehydrogenase processes GalUA into d-galactarate with concomitant reduction of NAD+. This revealed release of GalUA from PGA, GalUA2, and GalUA3 (Fig. 5B). PsGH28 did not display any capability to degrade ΔGalUA2 (see Fig. S3 in the supplemental material), suggesting that PsGH28 has strict exo-α-galacturonidase activity against saturated oligosaccharides.
FIG 5.
PsGH28 is an exo-α-galacturonidase that liberates galacturonate from oligouronates. (A and B) Activity of PsGH28 against polygalacturonate (PGA), trigalacturonate (GalUA3), and digalacturonate (GalUA2) as shown by FACE analysis (A) and turnover of released galacturonate by uronate dehydrogenase in an enzyme-coupled assay (B). + and −, presence and absence of PsGH28, respectively; E, enzyme-only sample. Abbreviation: GalUA, galacturonate. Data shown in panel B are the mean initial rates from three replicates; error bars indicate the SEM. (C) FACE gel showing complete degradation of unsaturated trigalacturonate (ΔGalUA3) into GalUA by PsGH105A and PsGH28. GalUA3, GalUA2, and GalUA were included as standards. + and −, presence and absence of substrate or enzyme, respectively.
The demonstrated exo-α-galacturonidase activity of PsGH28 suggests that it would work synergistically with the GH105s to process the unsaturated oligogalacturonide products of PsPL1, and we examined this by FACE. This analysis revealed that only cotreatment of ΔGalUA3 with PsGH105A and PsGH28 enabled complete depolymerization of the oligosaccharide, likely through the GH105-catalyzed release of GalUA2 which is then a substrate for PsGH28 (Fig. 5C). The same complete conversion of ΔGalUA3 into monosaccharides was also observed with PsGH105B in place of PsGH105A (data not shown).
PS47 enzymes work in concert to liberate GalUA and DKI.
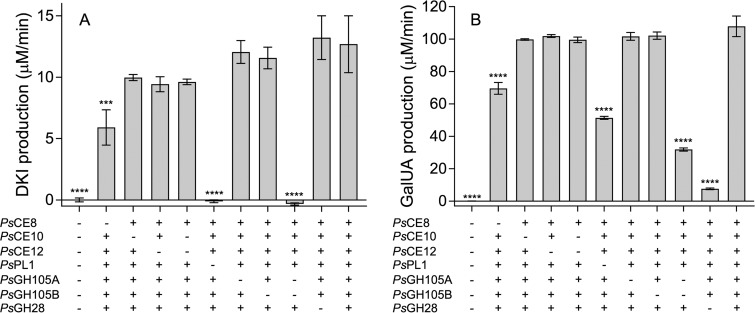
With its complement of CEs, GHs, and a PL, PS47 appears to possess all the enzymatic activities required to release both GalUA and DKI from pectin. We tested this theory by reconstituting the enzyme pathway in vitro and assessing the effect of omitting one or more enzymes on the rates of GalUA and DKI production. The rate of DKI production was measured as described previously (16) using a linked assay in which released DKI is processed by YeKduI and YeKduD into KDG with concomitant oxidation of NADH. The fully reconstituted enzyme pathway released significant amounts of both GalUA and DKI (Fig. 6). We also observed distinct differences in the importance of certain enzymes to the production of each monosaccharide. As expected, omission of both GH105s from the pathway completely abolished production of DKI, and PsPL1 was also essential for DKI production, as it generates the unsaturated oligouronates upon which the GH105s act (Fig. 6A). Consistent with its isolated enzymatic activity, PsGH28 had no significant effect on the DKI production rate but its omission had a profound effect on GalUA production. Exclusion of PsPL1 and the two GH105s also had a significant effect on the rate of GalUA production (albeit not as pronounced as their effect on DKI release), as together these enzymes generate more saturated nonreducing ends for PsGH28 to act on. In agreement with the observed effect of the esterases on the rate of PsPL1 activity (Fig. 3C), only PsCE8 had a significant effect on the production of either monosaccharide. We also observed redundancy among PsGH105A and PsGH105B, as omission of either one in isolation did not have a significant effect. In contrast, because the effect of excluding both acetylesterases was not significant, we were unable to distinguish whether PsCE10 and PsCE12 are redundant in their activities. Finally, we observed that addition of YeKdgF to the reconstituted pathway did not increase the rate of DKI production (data not shown), thereby confirming the lack of a role for this enzyme in pectin degradation by PS47.
FIG 6.
In vitro reconstitution of the pectin degradation pathway from Pseudoalteromonas sp. PS47. Linked assays in which the rates of DKI (A) and GalUA (B) production from pectin by PS47 enzymes were quantified by following their downstream processing into KDG (by YeKduI and YeKduD, with concomitant oxidation of NADH) and galactarate (by uronate dehydrogenase, with concomitant reduction of NAD+), respectively, were performed. Data shown indicate the effect of omitting one or more enzyme. Data are the means of four replicates and have been corrected for background changes in absorbance in the presence of pectin only. Error bars represent the SEM. Asterisks indicate statistically significant differences between enzyme combinations and the “all enzymes” sample as determined by a one-way ANOVA with Dunnett’s posttest (***, P ≤ 0.001; ****, P ≤ 0.0001; absence of asterisks, P > 0.05).
DISCUSSION
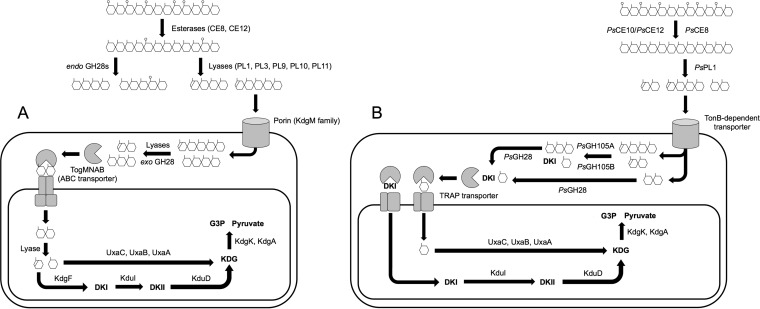
The classical pathway of pectin degradation and metabolism employed by terrestrial bacteria has been studied for more than 50 years, and its roles in biomass turnover and plant-pathogen interactions are well understood (14, 15, 44). Similarly, the presence of a pectin-like polysaccharide in marine seagrass was identified 50 years ago (33, 45). However, unlike in the case of terrestrial pectin metabolism, it was only recently that the capacity of marine bacteria to metabolize pectin was uncovered. Using the PS47 pectin PUL as a model, we have experimentally reconstructed an alternate pectin degradation pathway in vitro that utilizes a GH105 to carry out the equivalent chemistry typically afforded by an exolytic PL22 and KdgF in the canonical pathway used, for example, by bacterial phytopathogens (Fig. 7). The extracellular steps of the phytopathogen and marine pathways are essentially parallel, with de-esterification by CEs followed by backbone cleavage by an endolytic PL. The phytopathogen pathway also employs an endolytic GH28 to assist with backbone cleavage (this may be a function fulfilled by the apparent GH28/PL9 fusion encoded in the G. flava PUL). The resulting pool of small saturated and unsaturated oligogalacturonides is then imported into the periplasm for further degradation. In marine bacteria, this transport is likely carried out by a TonB-dependent receptor; these are transport proteins found in the outer membranes of Gram-negative bacteria and widely associated with carbohydrate utilization in marine microbes (46, 47). Once in the periplasm, marine bacteria employ one or more GH105s to remove the unsaturated galacturonyl residues and release DKI (and GalUA if the unsaturated substrate is a disaccharide) and an exolytic GH28 to cleave the remaining saturated oligosaccharides into GalUA. Based on signal peptide and localization predictions (48, 49), we propose that PsGH105A, PsGH105B, and PsGH28 are periplasmic and, therefore, that all monosaccharide production would occur in the periplasm. This is supported by the fact that PS47 is unable to grow on extracellular GalUA as the sole carbon source, indicating an inability to transport this monosaccharide (Fig. 1B). Import of the released GalUA and/or DKI from the periplasm into the cytoplasm is likely via the TRAP transporter that is encoded in all the known marine pectin PULs. Characterized TRAP transporters bind and transport a range of linear and cyclic acidic sugars, including GalUA (50, 51). We anticipate that the single TRAP transporter encoded in the PS47 pectin PUL may be able to import both GalUA and DKI into the cytoplasm, but this remains to be experimentally tested. In the canonical HG degradation pathway, a single transporter, TogMNAB, imports both saturated and unsaturated oligogalacturonides (52). These imported oligouronates are then further degraded by an exolytic PL (PL22), and KdgF linearizes the released ΔGalUA into DKI. The requirement for the PL22-KdgF pair is a key distinction between the phytopathogen and marine bacterial pectin degradation pathways. Finally, in both pathways, the intracellular processing of GalUA and DKI is catalyzed by an orthologous set of enzymes.
FIG 7.
Comparison of the canonical pathway of pectin degradation and metabolism (A) and the pathway employed by Pseudoalteromonas sp. PS47 (B). Panel A is adapted from reference 17. Abbreviations: ABC, ATP binding cassette; TRAP, tripartite ATP-independent periplasmic; DKI, 5-keto-4-deoxyuronate; DKII, 2,5-diketo-3-deoxygluconate; KDG, 2-keto-3-deoxygluconate; G3P, glyceraldehyde-3-phosphosphate.
The PS47 PUL encodes two GH105 proteins with Δ-4,5-unsaturated α-galacturonidase activity. Homologs of these are also encoded in the P. haloplanktis and G. flava pectin PULs, though the functions of these enzymes have not been demonstrated (Table 1). Nevertheless, a hallmark of the marine pectin pathways studied to date is the presence of GH105-encoding genes, supporting the concept that utilizing a GH105 enzyme in a pectinolytic pathway avoids the requirement for an exolytic PL-KdgF pair.
The two GH105s, PsGH105A and PsGH105B, encoded in the PS47 pectin PUL each have homologs encoded in the P. haloplanktis PUL (Table 1). Based on the data presented here, PsGH105A and PsGH105B appear to be redundant in function. However, PsGH105A possesses a DUF4861 and, with the data available, we were unable to evaluate whether the difference in catalytic efficiency between these two enzymes against longer oligouronates is due to the presence of DUF4861 or to variation within the GH105 domain itself. The G. flava pectin PUL encodes only a single GH105, which is not a fusion with a DUF4861; however, an adjacent gene encodes a putative protein that consists almost entirely of a DUF4861 (Fig. 1A). A GenBank search for DUF4861-containing proteins reveals that this domain is most commonly found as part of a larger protein in Proteobacteria, such as Pseudoalteromonas spp., but exists as a stand-alone protein most often in Bacteroidetes. Therefore, this may represent an example of the fusion of protein functions among marine bacteria during the evolution of marine pectin degradation, as proposed by Hehemann et al. (31). Irrespective of whether they are partnered with another protein, DUF4861s are a common occurrence among pectin degradation PULs (see Fig. S4 in the supplemental material). Therefore, while the function of DUF4861 remains unknown, we can speculate that its presence is beneficial to efficient pectin catabolism in some way.
We have biochemically characterized seven key CAZymes from the PS47 PUL, and given the high amino acid sequence identity between these proteins and their homologs in P. haloplanktis ANT/505 (Table 1), we can infer that the P. haloplanktis PUL also encodes these activities. There remains one putative CAZyme from the PS47 PUL, PsGH43, which we were unable to study due to its insoluble expression in E. coli. Characterized members of the GH43 family display β-xylosidase, α-arabinanase, and/or β-galactanase activity (www.cazy.org). As RGI fragments bearing xylose and arabinose side chains have been detected in zosterin (35, 53), it is reasonable to expect that the activity of PsGH43 would be in line with its family classification. However, the presence of two cryptic genes for GH43s in the P. haloplanktis ANT/505 PUL, in place of the complete PsGH43 gene found in PS47, may imply that a functional GH43 is not essential for marine pectin degradation. Other questions still remaining regarding the degradation of pectin by PS47 include the apparent absence of an apiosidase and inessentiality of the acetylesterases. Given that zosterin possesses a backbone of AGU, we would expect efficient marine pectin degradation to require an apiosidase. An endolytic apiosidase that is active on RGII was recently identified in Bacteroides thetaiotaomicron and represents the founding member of the new GH140 family (7). The PS47 genome does not encode any homologs of the B. thetaiotaomicron apiosidase; however, that does not mean that an apiosidase belonging to an as-yet-unidentified family is not present. In terms of the apparent lack of contribution of PsCE8 and PsCE10 to efficient pectin breakdown, this may be an effect of the pectin source used. The degree of acetylation of citrus pectin is reported to be <10% (54); therefore, the use of a pectin with more acetyl groups may reveal a more important role for these enzymes. Further investigation would also be required to determine whether PsCE10 and PsCE12 are redundant in function or whether they target acetyl groups attached to different oxygen atoms on GalUA.
In summary, we have reconstructed the complete biochemical pathway for pectin saccharification in a marine bacterium and demonstrated that it differs from the canonical pathway by its use of a GH105 and lack of a requirement for KdgF. Given the promise that microbes engineered to contain the canonical pectin degradation pathway have shown for the production of bioethanol from plant waste (55, 56), this alternate enzymatic pathway also has the potential to be exploited for the same purpose and may prove to be more efficient due to the requirement for fewer enzymes. In terms of the ecological niche occupied by PS47, this bacterium is capable of degrading alginate, agar, and carrageenan; therefore, it seems counterintuitive that it would acquire and maintain the capability to degrade pectin, a polysaccharide that has historically been considered terrestrial. Our experimental validation of the pectin degradation pathway employed by PS47 acts to highlight the importance of marine pectin as a carbon source for suitably adapted marine heterotrophs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and materials.
The isolation, genome sequencing, and annotation of Pseudoalteromonas sp. PS47 have been described previously (37). PS47 was routinely grown in Zobell marine broth or agar (HiMedia Laboratories, PA, USA) at 25°C. Polygalacturonic acid, citrus pectin, digalacturonic acid (GalUA2), and trigalacturonic acid (GalUA3) were purchased from Sigma-Aldrich (St. Louis, MO). Zosterin (apiogalacturonan) and rhamnogalacturonan I were obtained from Elicityl (Crolles, France) and Megazyme (Bray, Ireland), respectively. Unsaturated digalacturonate was a kind gift from Wade Abbott, Agriculture and Agri-Food Canada. All other reagents were from Sigma-Aldrich unless otherwise stated.
Growth of PS47 on pectin and detection of pectinase activity.
Cells of PS47 grown overnight in Zobell marine broth were pelleted, washed, and diluted in complex minimal marine medium (57) containing no carbon source and then were used to inoculate 0.5 ml minimal medium containing either 1% (wt/vol) filtered citrus pectin, galactose, or no sugar in a 24-well tissue culture plate. One well for each carbon source was left uninoculated as a control. Plates were sealed with Breathe-Easy sealing membrane and incubated at 25°C in a SpectraMax M5 plate reader (Molecular Devices, San Jose, CA) for 60 h. Plates were shaken and readings of optical density at 600 nm (OD600) taken every 20 min. OD readings were blanked against the absorbance of the uninoculated control well. For the detection of pectinases in cultures of PS47, the bacterium was grown overnight in Zobell marine broth plus 0.5% (wt/vol) citrus pectin at 25°C. A sample of culture was pelleted, the supernatant retained, and the pellet washed once with minimal marine medium containing no sugar before being resuspended in BugBuster plus DNase I. Following lysis at room temperature for 20 min, cellular debris was removed by centrifugation. Overnight digests (30 μl total) were set up containing 1.5% (wt/vol) citrus pectin in binding buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl) with 5 μl of either culture supernatant or cleared lysate. Following digestion, reaction products were dried and labeled for fluorophore-assisted carbohydrate electrophoresis (FACE) as detailed below.
Cloning, protein expression, and purification.
The genes encoding PsPL1, PsCE8, PsCE10, PsCE12, PsGH43, PsGH105A, PsGH105B, and PsGH28 were cloned, without any predicted signal peptides, into pET28a between the NdeI and BamHI sites using the In-Fusion HD cloning kit (Clontech Laboratories Inc., CA, USA) and the primers listed in Table 2. The cloning of YeKdgF, YeKduI, and YeKduD has been reported previously (16). The catalytic domain and DUF4861 of PsGH105A were also cloned separately into pET28a; the DUF construct consisted of residues 30 to 461 and the catalytic construct of residues 462 to 821. All cloning was confirmed by bidirectional sequencing (Sequetech Corp., CA, USA). Constructs were transformed into Escherichia coli BL21(DE3) for expression, and proteins were expressed in autoinduction medium at 16°C for 72 h or in Luria-Bertani broth with IPTG (isopropyl-β-d-thiogalactopyranoside) induction at 16°C for 18 h. Recombinant proteins were released from cells by chemical lysis and purified by immobilized Ni2+ affinity chromatography in binding buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl) with an imidazole gradient. All proteins were assessed for purity by SDS-PAGE analysis, dialyzed against binding buffer overnight at 4°C, and concentrated using an Amicon stirred ultrafiltration unit fitted with either a 5- or 10-kDa-molecular-mass cutoff cellulose membrane, as appropriate, prior to use.
TABLE 2.
Primers used in this study
| Primer name | Primer sequence (5′→3′)a |
|---|---|
| PsPL1 fwd | GCCGCGCGGCAGCCAACTCGACTCAAATTTAGCCTTTAAAAATGC |
| PsPL1 rev | GCTCGAATTCGGATCGATTACTCCGTAATCGAATTTATATAAGC |
| PsCE8 fwd | GCCGCGCGGCAGCCATTGCAACATCCACTCAAAAACACC |
| PsCE8 rev | GCTCGAATTCGGATCTTATAAGCTAAAGCTATCAGTAGC |
| PsCE10 fwd | GCCGCGCGGCAGCCAGAAAGAGTACACGTTGC |
| PsCE10 rev | GCTCGAATTCGGATCTTATAGCTGTTTTTTTAAAAATAACG |
| PsCE12 fwd | GCCGCGCGGCAGCCAGAAACAAGGTACAACACA |
| PsCE12 rev | GCTCGAATTCGGATCTCAACCTTTTAGTTTTAAATGC |
| PsGH105A fwd | GCCGCGCGGCAGCCAAAAAGAAAGCACTGCAGCAT |
| PsGH105A rev | GCTCGAATTCGGATCTTACTTTACTGAGTTATCTATATC |
| PsGH105B fwd | GCCGCGCGGCAGCCACGAGCCATTACCAAATAAAAATTTATG |
| PsGH105B rev | GCTCGAATTCGGATCTTAACGCATCAAAGCAAGGCTAGC |
| PsGH28 fwd | GCCGCGCGGCAGCCAATGTAAATCAGCCCCCCTTG |
| PsGH28 rev | GCTCGAATTCGGATCTTACTGGGTTATTTTAGCTAA |
| PsGH43 fwd | GCCGCGCGGCAGCCAAATGCGACCAATAAATATAATAATACTG |
| PsGH43 rev | GCTCGAATTCGGATCTTAATTTTGCTCTGTTGGTGTGAAACG |
| PsGH105A CAT fwd | GCCGCGCGGCAGCCAAGATGCAAAAAGTGCTTTAAAGTGG |
| PsGH105A DUF rev | CTCGAATTCGGATCTTACAGTGGTGCTTTGAATTGAG |
Vector sequences used for In-Fusion cloning are underlined.
Production and purification of unsaturated galacturonate oligosaccharides.
Polygalacturonate (1 g) was digested with 0.3 μM PsPL1 in 20 mM Tris-HCl (pH 8.0) at 25°C and 70 rpm for 24 h. Any remaining full-length polysaccharide was removed by passing the digest through a 10-kDa-molecular-mass cutoff cellulose membrane using an Amicon stirred ultrafiltration unit. The flowthrough was lyophilized, resuspended in distilled water (dH2O), and passed over a size exclusion column containing Bio-Gel P-2 (Bio-Rad, Hercules, CA) in 20 mM ammonium carbonate. Fractions containing unsaturated oligosaccharides were identified by measuring the absorbance at 230 nm in UV-Star 96-well microplates (Grenier Bio-One, Monroe, NC) in a SpectraMax M5 plate reader (Molecular Devices, San Jose, CA). Oligouronate size and separation were assessed by thin-layer chromatography as previously described (17). Any residual salts in fractions containing unsaturated oligouronates of a defined size were eliminated by repeating the size exclusion step with dH2O before use.
FACE.
Polysaccharide digests for fluorophore-assisted carbohydrate electrophoresis (FACE) analysis contained 0.5% (wt/vol) polysaccharide, 1 μM enzyme, and 1 mM CaCl2 (when PsPL1 was present) in binding buffer in 10-μl reaction mixtures and were incubated at 25°C overnight. Oligosaccharide digests were set up in the same way but contained approximately 10 μg substrate. All reaction products were dried, labeled with 20 mM 2-aminonaphthalene trisulfonic acid (ANTS), and separated in 35% polyacrylamide gels as previously described (58).
Esterase assays.
All real-time enzyme assays were performed at 25°C in a SpectraMax M5 plate reader. PsCE8, PsCE10, and PsCE12 were tested for acetylesterase activity using the Megazyme acetic acid assay kit according to the manufacturer’s instructions. Reaction mixtures contained 0.3% (wt/vol) citrus pectin and 150 mM NaCl in kit buffer, and reactions were initiated by the addition of esterase to 2 or 10 μM. Data were processed according to the manufacturer’s instructions and corrected for background changes in absorbance observed when binding buffer was added in place of esterase. Methylesterase activity was tested using an enzyme-coupled assay as described by Grsic-Rausch and Rausch (59). Reaction mixtures contained 0.2% (wt/vol) citrus pectin, 0.9 mM NAD+, 1 U alcohol oxidase from Pichia pastoris, 0.02 U formaldehyde dehydrogenase from Pseudomonas sp., and 150 mM NaCl in 50 mM Tris (pH 7.5), and reactions were initiated by the addition of esterase to 5 or 10 μM. Data were corrected for background as described above and converted into concentrations of methanol using an extinction coefficient for NADH of 6,220 M−1 cm−1. All statistical analyses were performed in GraphPad Prism 5 or 7.
Lyase assays.
Lyase activity exhibited by PsPL1 was observed in real time as an increase in OD230 in UV-Star 96-well microplates. The approximate pH optimum of PsPL1 was initially determined using McIlvaine buffers (pH 4 to 8), and then the determination was repeated in 200 mM Tris-phosphate (pH 7 to 9). Reaction mixtures contained 0.1% (wt/vol) PGA and 1 mM CaCl2, and reactions were initiated by the addition of PsPL1 to 250 nM. The effect of the esterases on the rate of unsaturated oligouronate production from pectin by PsPL1 was determined in the presence of 0.3% (wt/vol) citrus pectin, 50 mM Tris (pH 8.0), and 150 mM NaCl. Reactions were initiated by simultaneous addition of PsPL1 and esterase, each to a final concentration of 1 μM. Initial rates were converted into μM/min using an extinction coefficient for unsaturated uronates of 5,200 M−1 cm−1 as previously reported (6, 16).
Unsaturated galacturonyl hydrolase assays.
Cleavage of unsaturated galacturonyl residues from the nonreducing end of oligouronates by PsGH105A and PsGH105B was observed in real time as a decrease in OD230. pH profiles were determined for PsGH105A and PsGH105B in McIlvaine buffers (pH 4 to 8) containing 1 mM ΔGalUA2 with 250 nM enzyme. The PsPL1-PsGH105 linked assay mixture contained 0.3% (wt/vol) PGA and 500 mM NaCl in 20 mM Tris (pH 8.0). PsPL1 was added to 1 μM and the reaction was allowed to proceed for 30 min, at which point OD230 readings were paused for approximately 1 min to allow for the addition of either PsGH105A or PsGH105B to 200 nM. Michaelis-Menten kinetics were determined for PsGH105A and PsGH105B at their pH optima (pH 6.5 and 5.7, respectively) in 50 mM MES (morpholineethanesulfonic acid) buffer and 200 mM NaCl with 1 to 250 nM enzyme from initial rates in the presence of various concentrations of either ΔGalUA2 or a mixture of large unsaturated oligosaccharides with an estimated degree of polymerization of ≥4. The effect of YeKdgF on the rate of unsaturated oligouronate depletion by PsGH105A and PsGH105B was tested using 1 mM large unsaturated oligosaccharides as the substrate in 50 mM Tris (pH 7.5) with 150 mM NaCl. Reactions were initiated by simultaneous addition of YeKdgF and/or a single PsGH105, each to a final concentration of 1 μM. No-enzyme control reaction mixtures contained binding buffer or 20 mM Tris (pH 8.0) in place of enzyme. The preparation of YeKdgF was confirmed to be active by testing it against ΔGalUA2 in a linked assay with GalUA2 and YeOgl as described previously (16).
Galacturonate release assay.
Liberation of free GalUA from PGA or saturated oligosaccharides by PsGH28 was observed in real time using the Megazyme d-glucuronic/d-galacturonic acid assay kit according to the manufacturer’s instructions. Reaction mixtures contained 0.5% (wt/vol) PGA, or 1 mM GalUA2 or GalUA3, and 150 mM NaCl in kit buffer, and reactions were initiated by the addition of PsGH28 to 1 μM. Data were processed according to the manufacturer’s instructions and corrected for background changes in absorbance observed when binding buffer was added in place of PsGH28.
In vitro reconstitution of enzyme pathway.
The rates of GalUA and DKI production from pectin by the complete set of PS47 pectinases and the effect of omitting one or more enzymes were determined using enzyme-coupled assays. GalUA release was quantified using the Megazyme d-glucuronic/d-galacturonic acid assay kit, according to the manufacturer’s instructions. In addition to the standard kit components, reaction mixtures contained each pectinase at 1 μM and 150 mM NaCl. Reactions were initiated by the addition of citrus pectin to a final concentration of 0.2% (wt/vol), and OD340 readings were taken every 5 to 20 seconds. DKI release was quantified using the isomerase YeKduI and dehydrogenase YeKduD as previously described (16). Reaction mixtures contained 1 mM NADH, 150 mM NaCl, 50 mM Tris (pH 7.5), and 1 μM all enzymes, including YeKduI and YeKduD. As for GalUA release, reactions were initiated by the addition of citrus pectin to a final concentration of 0.2% (wt/vol), and OD340 readings were taken. For both coupled assays, initial rates were determined, blanked using the no-enzyme control, and converted into μM/min using either the manufacturer’s instructions (for GalUA) or an extinction coefficient of 6,220 M−1 cm−1 (for DKI).
Carbohydrate binding assays.
The binding of the DUF of PsGH105A to PGA and ΔGalUA≥n = 4 was evaluated using differential scanning fluorimetry. Samples contained DUF at 1 mg/ml, 10× SYPRO Orange (Invitrogen, Carlsbad, CA), 500 mM NaCl, 20 mM buffer (MES [pH 5.7 and 6.5], HEPES [pH 7.5], Tris-HCl [pH 8.0], or Tris-phosphate [pH 9.0]), and 0.1 or 0.2% PGA or ΔGalUA≥n = 4. Reaction mixtures were incubated in a Bio-Rad CFX96 Touch real-time PCR machine at 25 to 95°C with a ramp rate of 1°C/min and read using the HEX channel. Controls included samples of DUF to which dH2O was added in place of ligand or binding buffer was added in place of DUF.
Accession number(s).
The complete sequence of the Pseudoalteromonas sp. PS47 pectin PUL has been submitted to GenBank under accession number MH687413.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (FRN 04355).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02114-18.
REFERENCES
- 1.Barbeyron T, Thomas F, Barbe V, Teeling H, Schenowitz C, Dossat C, Goesmann A, Leblanc C, Oliver Glöckner F, Czjzek M, Amann R, Michel G. 2016. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans DsijT. Environ Microbiol 18:4610–4627. doi: 10.1111/1462-2920.13584. [DOI] [PubMed] [Google Scholar]
- 2.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. 2017. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol 199:e860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemsworth GR, Dejean G, Davies GJ, Brumer H. 2016. Learning from microbial strategies for polysaccharide degradation. Biochem Soc Trans 44:94–108. doi: 10.1042/BST20150180. [DOI] [PubMed] [Google Scholar]
- 4.Voragen AGJ, Coenen G-J, Verhoef RP, Schols HA. 2009. Pectin, a versatile polysaccharide present in plant cell walls. Struct Chem 20:263–275. doi: 10.1007/s11224-009-9442-z. [DOI] [Google Scholar]
- 5.Chung D, Pattathil S, Biswal AK, Hahn MG, Mohnen D, Westpheling J. 2014. Deletion of a gene cluster encoding pectin degrading enzymes in Caldicellulosiruptor bescii reveals an important role for pectin in plant biomass recalcitrance. Biotechnol Biofuels 7:147. doi: 10.1186/s13068-014-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shevchik VE, Condemine G, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. 1999. The exopolygalacturonate lyase PelW and the oligogalacturonate lyase Ogl, two cytoplasmic enzymes of pectin catabolism in Erwinia chrysanthemi 3937. J Bacteriol 181:3912–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A, Terrapon N, Buffetto F, Nepogodiev S, Xiao Y, Field RA, Zhu Y, O’Neill MA, Urbanowicz BR, York WS, Davies GJ, Abbott DW, Ralet M-C, Martens EC, Henrissat B, Gilbert HJ. 2017. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544:65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridley BL, O'Neill MA, Mohnen D. 2001. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57:929–967. doi: 10.1016/S0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- 9.Schink B, Ward JC, Zeikus JG. 1981. Microbiology of wetwood: importance of pectin degradation and clostridium species in living trees. Appl Environ Microbiol 42:526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benoit I, Coutinho PM, Schols HA, Gerlach JP, Henrissat B, de Vries RP. 2012. Degradation of different pectins by fungi: correlations and contrasts between the pectinolytic enzyme sets identified in genomes and the growth on pectins of different origin. BMC Genomics 13:321. doi: 10.1186/1471-2164-13-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tayi L, Maku RV, Patel HK, Sonti RV. 2016. Identification of pectin degrading enzymes secreted by Xanthomonas oryzae pv. oryzae and determination of their role in virulence on rice. PLoS One 11:e0166396. doi: 10.1371/journal.pone.0166396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Yeoman CJ, Miller MEB, Fields CJ, Terrapon N, Le Bourvellec C, Renard CMGC, Henrissat B, White BA, Mosoni P. 2016. Unraveling the pectinolytic function of Bacteroides xylanisolvens using a RNA-seq approach and mutagenesis. BMC Genomics 17:147. doi: 10.1186/s12864-016-2472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abbott DW, Boraston AB. 2008. Structural biology of pectin degradation by Enterobacteriaceae. Microbiol Mol Biol Rev 72:301–316. doi: 10.1128/MMBR.00038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preiss J, Ashwell G. 1963. Polygalacturonic acid metabolism in bacteria. I. Enzymatic formation of 4-deoxy-L-threo-5-hexoseulose uronic acid. J Biol Chem 238:1571–1583. [PubMed] [Google Scholar]
- 15.Preiss J, Ashwell G. 1963. Polygalacturonic acid metabolism in bacteria. II. Formation and metabolism of 3-deoxy-D-glycero-2, 5-hexodiulosonic acid. J Biol Chem 238:1577–1583. [PubMed] [Google Scholar]
- 16.Hobbs JK, Lee SM, Robb M, Hof F, Barr C, Abe KT, Hehemann J-H, McLean R, Abbott DW, Boraston AB. 2016. KdgF, the missing link in the microbial metabolism of uronate sugars from pectin and alginate. Proc Natl Acad Sci U S A 113:6188–6193. doi: 10.1073/pnas.1524214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbott DW, Gilbert HJ, Boraston AB. 2010. The active site of oligogalacturonate lyase provides unique insights into cytoplasmic oligogalacturonate β-elimination. J Biol Chem 285:39029–39038. doi: 10.1074/jbc.M110.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Condemine G, Robert-Baudouy J. 1991. Analysis of an Erwinia chrysanthemi gene cluster involved in pectin degradation. Mol Microbiol 5:2191–2202. doi: 10.1111/j.1365-2958.1991.tb02149.x. [DOI] [PubMed] [Google Scholar]
- 19.Condemine G, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. 1986. Isolation of Erwinia chrysanthemi kduD mutants altered in pectin degradation. J Bacteriol 165:937–941. doi: 10.1128/jb.165.3.937-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. 1987. Hexuronate catabolism in Erwinia chrysanthemi. J Bacteriol 169:1223–1231. doi: 10.1128/jb.169.3.1223-1231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh T, Ochiai A, Mikami B, Hashimoto W, Murata K. 2006. A novel glycoside hydrolase family 105: the structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide. J Mol Biol 360:573–585. doi: 10.1016/j.jmb.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 22.Collén PN, Jeudy A, Sassi J-F, Groisillier A, Czjzek M, Coutinho PM, Helbert W. 2014. A novel unsaturated β-glucuronyl hydrolase involved in ulvan degradation unveils the versatility of stereochemistry requirements in family GH105. J Biol Chem 289:6199–6211. doi: 10.1074/jbc.M113.537480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jongkees SAK, Yoo H, Withers SG. 2014. Mechanistic investigations of unsaturated glucuronyl hydrolase from Clostridium perfringens. J Biol Chem 289:11385–11395. doi: 10.1074/jbc.M113.545293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jongkees SAK, Withers SG. 2011. Glycoside cleavage by a new mechanism in unsaturated glucuronyl hydrolases. J Am Chem Soc 133. doi: 10.1021/ja209067v. [DOI] [PubMed] [Google Scholar]
- 25.Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet M-C, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ensor L, Stosz S, Weiner R. 1999. Expression of multiple complex polysaccharide-degrading enzyme systems by marine bacterium strain 2-40. J Ind Microbiol Biotechnol 23:123–126. doi: 10.1038/sj.jim.2900696. [DOI] [PubMed] [Google Scholar]
- 27.Truong LV, Tuyen H, Helmke E, Binh LT, Schweder T. 2001. Cloning of two pectate lyase genes from the marine Antarctic bacterium Pseudoalteromonas haloplanktis strain ANT/505 and characterization of the enzymes. Extremophiles 5:35–44. doi: 10.1007/s007920000170. [DOI] [PubMed] [Google Scholar]
- 28.Ramaiah N, Chandramohan D. 1992. Densities, cellulases, alginate and pectin lyases of luminous and other heterotrophic bacteria associated with marine algae. Aquat Bot 44:71–81. doi: 10.1016/0304-3770(92)90082-T. [DOI] [Google Scholar]
- 29.Tang K, Lin Y, Han Y, Jiao N. 2017. Characterization of potential polysaccharide utilization systems in the marine Bacteroidetes Gramella Flava JLT2011 using a multi-omics approach. Front Microbiol 8:220. doi: 10.3389/fmicb.2017.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imran M, Pant P, Shanbhag YP, Sawant SV, Ghadi SC. 2017. Genome aequence of Microbulbifer mangrovi DD-13T reveals its versatility to degrade multiple polysaccharides. Mar Biotechnol 19:116–124. doi: 10.1007/s10126-017-9737-9. [DOI] [PubMed] [Google Scholar]
- 31.Hehemann J-H, Truong L, Van Unfried F, Welsch N, Kabisch J, Heiden SE, Junker S, Becher D, Thürmer A, Daniel R, Amann R, Schweder T. 2017. Aquatic adaptation of a laterally acquired pectin degradation pathway in marine gammaproteobacteria. Environ Microbiol 19:2320–2333. doi: 10.1111/1462-2920.13726. [DOI] [PubMed] [Google Scholar]
- 32.Desikachary TV, Dweltz NE. 1961. The chemical composition of the diatom frustule. Proc Indian Acad Sci Sect B 53:157–165. [Google Scholar]
- 33.Ovodova RG, Vaskovsky VE, Ovodov YS. 1968. The pectic substances of zosteraceae. Carbohydr Res 6:328–332. doi: 10.1016/S0008-6215(00)81454-8. [DOI] [Google Scholar]
- 34.Gloaguen V, Brudieux V, Closs B, Barbat A, Krausz P, Sainte-Catherine O, Kraemer M, Maes E, Guerardel Y. 2010. Structural characterization and cytotoxic properties of an apiose-rich pectic polysaccharide obtained from the cell wall of the marine phanerogam Zostera marina. J Nat Prod 73:1087–1092. doi: 10.1021/np100092c. [DOI] [PubMed] [Google Scholar]
- 35.Lv Y, Shan X, Zhao X, Cai C, Zhao X, Lang Y, Zhu H, Yu G. 2015. Extraction, isolation, structural characterization and anti-tumor properties of an apigalacturonan-rich polysaccharide from the sea grass Zostera caespitosa Miki. Mar Drugs 13:3710–3731. doi: 10.3390/md13063710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khotimchenko Y, Khozhaenko E, Kovalev V, Khotimchenko M. 2012. Cerium binding activity of pectins isolated from the seagrasses Zostera marina and Phyllospadix iwatensis. Mar Drugs 10:834–848. doi: 10.3390/md10040834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hettle AG, Vickers C, Robb CS, Liu F, Withers SG, Hehemann J-H, Boraston AB. 2018. The molecular basis of polysaccharide sulfatase activity and a nomenclature for catalytic subsites in this class of enzyme. Structure 26:747–758.e4. doi: 10.1016/j.str.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin K, Simossis VA, Taylor WR, Heringa J. 2005. A simple and fast secondary structure prediction method using hidden neural networks. Bioinformatics 21:152–159. doi: 10.1093/bioinformatics/bth487. [DOI] [PubMed] [Google Scholar]
- 43.Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Condemine G, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. 1984. An enzyme in the pectinolytic pathway of Erwinia chrysanthemi: 2-keto-3-deoxygluconate oxidoreductase. Microbiology 130:2839–2844. doi: 10.1099/00221287-130-11-2839. [DOI] [Google Scholar]
- 45.Maeda M, Koshikawa M, Nisizawa K, Takano K. 1966. Cell wall constituents, especially pectic substance of a marine phanerogam Zostera marina. Shokubutsugaku Zasshi 79:422–426. doi: 10.15281/jplantres1887.79.422. [DOI] [Google Scholar]
- 46.Tang K, Jiao N, Liu K, Zhang Y, Li S. 2012. Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PLoS One 7:e41204. doi: 10.1371/journal.pone.0041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanvillain S, Meyer D, Boulanger A, Lautier M, Guynet C, Denancé N, Vasse J, Lauber E, Arlat M. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juncker AS, Willenbrock H, von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen H. 2017. Predicting secretory proteins with SignalP. Methods Mol Biol 1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 50.Mulligan C, Fischer M, Thomas GH. 2011. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol Rev 35:68–86. doi: 10.1111/j.1574-6976.2010.00236.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhao J, Binns AN. 2016. Involvement of Agrobacterium tumefaciens galacturonate tripartite ATP-independent periplasmic (TRAP) transporter GaaPQM in virulence gene expression. Appl Environ Microbiol 82:1136–1146. doi: 10.1128/AEM.02891-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbott DW, Boraston AB. 2007. Specific recognition of saturated and 4,5-unsaturated hexuronate sugars by a periplasmic binding protein involved in pectin catabolism. J Mol Biol 369:759–770. doi: 10.1016/j.jmb.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 53.Shibaeva VI, Ovodova RG, Ovodov YS. 1971. Pectin substances of seaweeds. VII. Acetolysis of zosterin. Chem Nat Compd 7:240–242. doi: 10.1007/BF00568987. [DOI] [Google Scholar]
- 54.Schmidt US, Koch L, Rentschler C, Kurz T, Endreß HU, Schuchmann HP. 2015. Effect of molecular weight reduction, acetylation and esterification on the emulsification properties of citrus pectin. Food Biophys 10:217–227. doi: 10.1007/s11483-014-9380-1. [DOI] [Google Scholar]
- 55.Edwards MC, Doran-Peterson J. 2012. Pectin-rich biomass as feedstock for fuel ethanol production. Appl Microbiol Biotechnol 95:565–575. doi: 10.1007/s00253-012-4173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doran JB, Cripe J, Sutton M, Foster B. 2000. Fermentations of pectin-rich biomass with recombinant bacteria to produce fuel ethanol. Appl Biochem Biotechnol 84-86:141–152. doi: 10.1385/ABAB:84-86:1-9:141. [DOI] [PubMed] [Google Scholar]
- 57.Therkildsen M, Isaksen M, Lomstein B. 1997. Urea production by the marine bacteria Delaya venusta and Pseudomonas stutzeri grown in a minimal medium. Aquat Microb Ecol 13:213–217. doi: 10.3354/ame013213. [DOI] [Google Scholar]
- 58.Robb M, Hobbs JK, Boraston AB. 2017. Separation and visualization of glycans by fluorophore-assisted carbohydrate electrophoresis. Methods Mol Biol 1588:215–221. doi: 10.1007/978-1-4939-6899-2_17. [DOI] [PubMed] [Google Scholar]
- 59.Grsic-Rausch S, Rausch T. 2004. A coupled spectrophotometric enzyme assay for the determination of pectin methylesterase activity and its inhibition by proteinaceous inhibitors. Anal Biochem 333:14–18. doi: 10.1016/j.ab.2004.04.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.