Fusarium fujikuroi is a pathogenic fungus that causes rice bakanae disease. Historically, this pathogen has been known as Fusarium moniliforme, along with many other species based on a broad species concept. Gibberellin, which is currently known as a plant hormone, is a virulence factor of F. fujikuroi. Fumonisin is a carcinogenic mycotoxin posing a serious threat to food and feed safety. Although it has been confirmed that F. fujikuroi produces gibberellin and fumonisin, production varies among strains, and individual production has been obscured by the traditional appellation of F. moniliforme, difficulties in species identification, and variation in the assays used to determine the production of these secondary metabolites. In this study, we discovered two phylogenetic subgroups associated with fumonisin and gibberellin production in Japanese F. fujikuroi.
KEYWORDS: fumonisin, Fusarium fujikuroi, gibberellin
ABSTRACT
Fusarium fujikuroi is a pathogenic fungus that infects rice. It produces several important mycotoxins, such as fumonisins. Fumonisin production has been detected in strains of maize, strawberry, and wheat, whereas it has not been detected in strains from rice seedlings infested with bakanae disease in Japan. We investigated the genetic relationships, pathogenicity, and resistance to a fungicide, thiophanate-methyl (TM), in 51 fumonisin-producing strains and 44 nonproducing strains. Phylogenetic analyses based on amplified fragment length polymorphism (AFLP) markers and two specific genes (a combined sequence of translation elongation factor 1α [TEF1α] and RNA polymerase II second-largest subunit [RPB2]) indicated differential clustering between the fumonisin-producing and -nonproducing strains. One of the AFLP markers, EATMCAY107, was specifically present in the fumonisin-producing strains. A specific single nucleotide polymorphism (SNP) between the fumonisin-producing and nonproducing strains was also detected in RPB2, in addition to an SNP previously found in TEF1α. Gibberellin production was higher in the nonproducing than in the producing strains according to an in vitro assay, and the nonproducing strains had the strongest pathogenicity with regard to rice seedlings. TM resistance was closely correlated with the cluster of fumonisin-nonproducing strains. The results indicate that intraspecific evolution in Japanese F. fujikuroi is associated with fumonisin production and pathogenicity. Two subgroups of Japanese F. fujikuroi, designated G group and F group, were distinguished based on phylogenetic differences and the high production of gibberellin and fumonisin, respectively.
IMPORTANCE Fusarium fujikuroi is a pathogenic fungus that causes rice bakanae disease. Historically, this pathogen has been known as Fusarium moniliforme, along with many other species based on a broad species concept. Gibberellin, which is currently known as a plant hormone, is a virulence factor of F. fujikuroi. Fumonisin is a carcinogenic mycotoxin posing a serious threat to food and feed safety. Although it has been confirmed that F. fujikuroi produces gibberellin and fumonisin, production varies among strains, and individual production has been obscured by the traditional appellation of F. moniliforme, difficulties in species identification, and variation in the assays used to determine the production of these secondary metabolites. In this study, we discovered two phylogenetic subgroups associated with fumonisin and gibberellin production in Japanese F. fujikuroi.
INTRODUCTION
The Fusarium fujikuroi species complex, which mostly corresponds to Fusarium section Liseola, includes more than 50 phylogenetic species (1, 2), including 13 independent mating populations (MPs) (3, 4). Fusarium fujikuroi Nirenberg, corresponding to MP-C in the F. fujikuroi complex, causes rice bakanae disease. Diseased rice plants are slender, have pale yellow leaves, and are taller than nondiseased plants (5). In Japan, the disease has been controlled with benzimidazole fungicides, although it is now also controlled using other types of fungicides, such as ipconazole, or by disinfestation by hot-water immersion. One benzimidazole fungicide, thiophanate-methyl (TM), has been used extensively for seed disinfection, but TM-resistant strains have emerged since 1984 in Japan (6). Historically, F. fujikuroi has been known as Fusarium moniliforme, along with many other species in the F. fujikuroi complex. The precise characteristics and diversity of F. fujikuroi have been obscured by the traditional appellation of F. moniliforme, although this broad species concept is no longer used (7). F. fujikuroi has been found in rice, wheat, maize, strawberry (8), and tomato (9) plants, in sugarcane, and in grape vines (10), although F. moniliforme has been reported in a wider variety of plant species.
F. fujikuroi is known to produce a large variety of secondary metabolites, and 45 putative secondary metabolite gene clusters have been found in its genome (11). Among such metabolites, gibberellins and fumonisins have been studied intensively. Gibberellin, which is a well-known plant hormone, was originally identified as a virulence factor in F. fujikuroi rice infestation. Gene disruption studies have indicated the contribution made by gibberellin to initial fungal colonization in the host roots (11). Fumonisin is a carcinogenic mycotoxin that was first discovered in Fusarium verticillioides, a relative of F. fujikuroi (12, 13). Fumonisin contamination of maize poses a serious global threat to food and feed safety.
Although it has been confirmed that F. fujikuroi produces gibberellin and fumonisin, production varies among strains, and individual production has been obscured by confusion over species classification, difficulties in species identification (14), and variation in the assays used to determine the production of these secondary metabolites. Furthermore, many researchers have focused only on F. fujikuroi strains in rice and on either gibberellin or fumonisin production. In a previous investigation of Japanese F. fujikuroi in a wide variety of host plants, fumonisin production was detected in 46 strains obtained from maize, strawberry, wheat, and rice seeds, whereas it was undetectable in 43 strains obtained from bakanae-diseased rice, rice seeds, and unknown sources (8). In the present study, strains unable to produce fumonisins at detectable levels are referred to as fumonisin nonproducers, although they may have been able to produce low levels of fumonisins. Although both fumonisin producers and nonproducers were isolated from rice seeds, all 21 strains from bakanae-diseased rice were fumonisin nonproducers (8). Eighteen out of 31 strains (58%) of F. fujikuroi from rice in the Philippines were fumonisin nonproducers (15).
Combined investigations of fumonisin and gibberellin production in F. fujikuroi from rice have been carried out by several researchers (16–18). In these studies, gibberellin production was detected in all 75 F. fujikuroi strains, as reported by Malonek et al. (19): “all so-far-analyzed rice isolates of the species F. fujikuroi (MP-C) produced significant amounts of biologically active GAs without any exclusion,” demonstrating gibberellin production in 5 F. fujikuroi strains. In contrast, fumonisin production in F. fujikuroi from rice tends to be minimal; none of the 4 strains investigated in one study (20) and 2 of the 7 strains investigated in another study were fumonisin producers (16); 2 of the 10 strains produced 0.1 to 1 ppm fumonisin, but the remaining 8 strains produced less than 0.1 ppm in one study (17); 8 out of 58 strains were fumonisin producers in one study (18); and none of the 21 strains from bakanae-diseased rice were fumonisin nonproducers in another study (8).
Whereas most F. fujikuroi strains isolated from plants other than rice are fumonisin producers, all 4 strains from wheat (21), 4 out of 5 strains from maize (22), all 4 strains from strawberry plants (8), and all 50 strains from grape vines were fumonisin producers (10). However, in case of F. fujikuroi from plants other than rice, only fumonisin production has been a focus and their gibberellin production has not been investigated (8, 10, 21, 22). Therefore, there has not yet been a precise assessment of the biological hazard posed by F. fujikuroi as a mycotoxin producer and rice pathogen. All F. fujikuroi strains from bakanae-diseased rice are fumonisin nonproducers, and the presence of a specific single nucleotide polymorphism (SNP) in the translation elongation factor 1 α (TEF1α) gene suggests possible genetic differences between fumonisin producers and nonproducers in Japanese F. fujikuroi (8). Phylogenetic analyses of the F. fujikuroi complex from rice and maize in South Korea revealed a subclade consisting mostly of rice isolates in the F. fujikuroi clade (23). Recently, two distinctive pathotypes, the bakanae type and the stunting type, corresponding to gibberellin and fumonisin production, respectively, have been identified in F. fujikuroi (24).
In the present study, we carried out a comprehensive investigation of gibberellin production, TM resistance, and genetic variability in Japanese F. fujikuroi strains that had been studied previously with regard to fumonisin production (8). The results indicate that fumonisin and gibberellin production is associated with genetic differentiation in Japanese F. fujikuroi.
RESULTS
SNP analysis.
The authors of a previous study reported an SNP (TEF_T618G) that distinguishes fumonisin producers from nonproducers (8). Additional SNPs were investigated to elucidate the genetic differences between fumonisin producers and nonproducers. First, seven SNPs that distinguish a fumonisin producer (Gfc0825009) from a nonproducer (Gfc0801001) were assessed with an additional four fumonisin producers (MAFF235463, Mo78, Gfc0825007, and Gfc0009105) and four nonproducers (MAFF235949, Gfc9424702, Gfc0625008, and APF06083) using PCR-restriction fragment length polymorphism (PCR-RFLP) or PCR sequencing. The seven SNPs were TEF_C447A, FUM1_G423A, FUM78_C41T (the intergenic region between FUM7 and FUM8), FUM8_G2834A, and FUM18_G51T in a fumonisin biosynthesis gene cluster; and CPR_C1152A and P4504_C842T in the genes involving gibberellin production. In the case of FUM18_G51T, the DNA was not amplified from three fumonisin nonproducers (Gfc9424702, Gfc0625008, and APF06083), and their SNPs could not be determined. The results indicated that FUM78_C41T, FUM8_G2834A, CPR_C1152A, and P4504_C842T were associated with differences in fumonisin production in the tested strains. The specificities of these SNPs with regard to fumonisin production compared with TEF_T618G were further investigated in all 95 strains using the Luminex assay. All the SNPs except P4504_C842T were associated with a difference in fumonisin production (Table 1).
TABLE 1.
Fusarium fujikuroi strains used in the present study and their characteristicsa
| Fumonisin productiona | Strain | Isolation sourceb | Geographic originc | Translation elongation factor 1α sequence (GenBank accession no.) | RNA polymerase II second-largest subunit |
SNP data (TEF_T618G, FUM78_C41T, FUM8_G2834A, CPR_C1152A, P4504_C842T)e |
AFLP haplotype | Thiophanate-methyl resistance |
Gibberellin spot on TLCf |
Reference or sourceg |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence (GenBank accession no.) |
SNP RPB2_C3250Td | GA4/7 | GA3 | |||||||||
| Nonproducer | ||||||||||||
| MAFF235949h | 91 rice | N | JN695731 | LC133055 | D (T) | G, T, A, A, T | 1 | S | − | ± | ||
| Gfc0801001h | 08 bakanae-diseased rice | P-I | JN695733 | LC133056 | D (T) | (G, T, A, A, T) | 2 | R | + | + | ||
| Gfc0801002 | 08 bakanae-diseased rice | P-I | D | G, T, A, A, T | 3 | R | + | + | ||||
| Gfc0801003 | 08 bakanae-diseased rice | P-I | D | G, T, A, A, T | 4 | R | + | + | ||||
| MYG92-10 | 92 bakanae-diseased rice | J | D | G, T, A, A, T | 5 | R | + | + | Tateishi and Chida (48) |
|||
| SMN86-2h | 86 bakanae-diseased rice | u | D | G, T, A, A, T | 6 | R | + | + | Tateishi and Chida (48) |
|||
| MIE92-4 | 92 bakanae-diseased rice | A | D | G, T, A, A, T | 7 | R | + | + | Tateishi and Chida (48) |
|||
| Gfc9424702h | 94 bakanae-diseased rice | A | JN695735 | LC133057 | D (T) | G, T, A, A, T | 8 | R | + | + | H. Suzuki | |
| Gfc9424703 | 94 bakanae-diseased rice | A | D | G, T, A, A, T | 9 | R | + | + | H. Suzuki | |||
| Gfc9424707 | 94 bakanae-diseased rice | A | D | G, T, A, A, T | 10 | R | + | + | H. Suzuki | |||
| Gfc0625001h | 06 bakanae-diseased rice | B-I | D | G, T, A, A, T | 11 | R | + | + | T. Kitazawa | |||
| Gfc0625002 | 06 bakanae-diseased rice | B-II | D | G, T, A, A, T | 12 | R | + | + | T. Kitazawa | |||
| Gfc0625004 | 06 bakanae-diseased rice | B-III | D | G, T, A, A, T | 12 | R | + | + | T. Kitazawa | |||
| Gfc0625005h | 06 bakanae-diseased rice | B-IV | D | G, T, A, A, T | 12 | R | + | ± | T. Kitazawa | |||
| Gfc0625006 | 06 bakanae-diseased rice | B-V | D | G, T, A, A, T | 12 | R | + | + | T. Kitazawa | |||
| Gfc0625007 | 06 bakanae-diseased rice | B-VI | D | G, T, A, A, T | 12 | R | + | + | T. Kitazawa | |||
| Gfc0625008 | 06 bakanae-diseased rice | B-IV | D | G, T, A, A, T | 13 | R | + | + | T. Kitazawa | |||
| Gfc0625010 | 06 bakanae-diseased rice | B-IV | D | G, T, A, A, T | 14 | R | + | + | T. Kitazawa | |||
| Gfc0925005h | 09 bakanae-diseased rice | B-IV | D | G, T, A, A, T | 15 | R | + | ± | ||||
| Gfc0925010 | 09 bakanae-diseased rice | B-VII | D | G, T, A, A, T | 16 | R | + | + | ||||
| Gfc0925011 | 09 bakanae-diseased rice | B-VIII | D | G, T, A, A, T | 17 | R | + | + | ||||
| Gfc0925012 | 09 bakanae-diseased rice | B-IX | D | G, T, A, A, T | 18 | R | + | + | ||||
| APF06083h | Unknown | Unknown | JN695736 | LC133058 | D (T) | G, T, A, A, T | 19 | R | + | ± | S. Fuji | |
| Gfc8707123 | Unknown | Unknown | D | G, T, A, A, T | 20 | R | + | + | H. Hamamura | |||
| Gfc8707182 | Unknown | Unknown | D | G, T, A, A, T | 20 | R | + | + | H. Hamamura | |||
| Gfc8707117 | Unknown | Unknown | D | G, T, A, A, T | 21 | R | + | + | H. Hamamura | |||
| Gfc8707249 | Unknown | Unknown | D | G, T, A, A, T | 22 | S | + | + | H. Hamamura | |||
| Gfc8707642 | Unknown | Unknown | D | G, T, A, A, T | 23 | R | + | + | H. Hamamura | |||
| Gfc0825001h | 05 rice seed | B-VII | JN695734 | LC133059 | D (T) | G, T, A, A, T | 24 | R | + | + | ||
| Gfc0825002 | 05 rice seed | B-VII | D | G, T, A, A, T | 24 | R | + | + | ||||
| Gfc0825003 | 05 rice seed | B-VII | D | G, T, A, A, T | 24 | R | + | + | ||||
| Gfc0825004 | 05 rice seed | B-VII | D | G, T, A, A, T | 24 | R | + | + | ||||
| Gfc0825005 | 05 rice seed | B-VII | D | G, T, A, A, T | 24 | R | + | + | ||||
| Gfc0825006 | 05 rice seed | B-VII | D | G, T, A, A, T | 24 | R | + | + | ||||
| Gfc0901002 | 07 rice seed | P-II | D | G, T, A, A, T | 25 | R | + | + | ||||
| Gfc0901005 | 07 rice seed | P-II | D | G, T, A, A, T | 25 | R | + | + | ||||
| Gfc0901009h | 07 rice seed | P-II | D | G, T, A, A, T | 26 | R | + | + | ||||
| Gfc1004001 | 09 rice seed | J | D | G, T, A, A, T | 27 | R | + | + | ||||
| Gfc1004002 | 09 rice seed | J | D | G, T, A, A, T | 28 | R | + | + | ||||
| Gfc1034001h | 09 rice seed | y | D | G, T, A, A, T | 29 | R | + | + | ||||
| GL24h | Rice | California, USA | LC133052 | LC133060 | D (T) | G, T, A, A, T | 19 | S | + | + | Proctor et al. (49) |
|
| GL25 | Rice | Unknown | D | G, T, A, A, T | 30 | S | + | + | Proctor et al. (49) |
|||
| GL27 | Rice | Unknown | D | G, T, A, A, T | 31 | S | + | + | Proctor et al. (49) |
|||
| GL28 | Rice | California, US | D | G, T, A, A, T | 32 | R | + | + | Proctor et al. (49) |
|||
| Producer | ||||||||||||
| MAFF235463h | 90 wheat | N | JN695737 | LC133061 | (C) | T, C, G, C, C | 33 | S | − | − | ||
| Gfc1004003h | 09 rice seed | J | LC133053 | LC133062 | (C) | T, C, G, C, T | 34 | S | − | − | ||
| Gfc1006001 | 09 rice seed | m | C | T, C, G, C, C | 35 | S | − | − | ||||
| Gfc1006002 | 09 rice seed | m | C | T, C, G, C, C | 36 | S | − | − | ||||
| Gfc1006007 | 09 rice seed | m | C | T, C, G, C, C | 37 | S | − | − | ||||
| Gfc1019001 | 10 rice seed | g-I | C | T, C, G, C, C | 38 | S | − | − | ||||
| Gfc1019003h | 10 rice seed | g-I | C | T, C, G, C, C | 39 | S | − | − | ||||
| Gfc1019004 | 10 rice seed | g-I | C | T, C, G, C, C | 40 | S | − | − | ||||
| Gfc1016022 | 09 rice seed | H | C | T, C, G, C, C | 41 | S | − | − | ||||
| Gfc1016024 | 09 rice seed | H | C | T, C, G, C, C | 42 | S | − | − | ||||
| Gfc1016025 | 09 rice seed | H | C | T, C, G, C, C | 43 | S | − | − | ||||
| Gfc1016026 | 09 rice seed | H | C | T, C, G, C, C | 43 | S | − | − | ||||
| Gfc0821004h | 08 rice seed | K-I | JN695740 | LC133063 | (C) | T, C, G, C, T | 44 | S | − | − | ||
| Gfc0921001 | 09 rice seed | K-II | C | T, C, G, C, C | 45 | S | − | − | ||||
| Gfc0921002 | 09 rice seed | K-II | C | T, C, G, C, C | 46 | S | − | − | ||||
| Gfc0921009 | 09 rice seed | K-II | C | T, C, G, C, C | 47 | S | − | − | ||||
| Gfc0921014 | 09 rice seed | K-II | C | T, C, G, C, C | 48 | S | − | − | ||||
| Gfc0921034 | 09 rice seed | K-III | C | T, C, G, C, C | 49 | S | − | − | ||||
| Gfc0921039h | 09 rice seed | K-III | C | T, C, G, C, C | 50 | S | + | − | ||||
| Gfc0921040 | 09 rice seed | K-III | C | T, C, G, C, C | 51 | S | − | − | ||||
| Gfc0921041 | 09 rice seed | K-III | C | T, C, G, C, C | 52 | S | − | − | ||||
| Gfc1025029 | 09 rice seed | B-VII | C | T, C, G, C, C | 53 | S | − | − | ||||
| Gfc1025037 | 09 rice seed | B-VII | C | T, C, G, C, C | 53 | S | − | − | ||||
| Gfc1025091 | 09 rice seed | B-VII | C | T, C, G, C, T | 54 | S | − | − | ||||
| Gfc1034002 | 09 rice seed | y | C | T, C, G, C, C | 55 | S | − | − | ||||
| Gfc1041003 | 10 rice seed | p-I | C | T, C, G, C, C | 56 | S | − | − | ||||
| Gfc1041010 | 10 rice seed | p-I | C | T, C, G, C, C | 57 | S | − | − | ||||
| Gfc1041011 | 10 rice seed | p-I | C | T, C, G, C, C | 56 | S | − | − | ||||
| Gfc1043032h | 09 rice seed | R-I | C | T, C, G, C, C | 58 | S | − | − | ||||
| Gfc1043035 | 09 rice seed | R-I | C | T, C, G, C, C | 59 | S | − | − | ||||
| Gfc1043037 | 09 rice seed | R-I | C | T, C, G, C, C | 60 | S | − | − | ||||
| Gfc1043045 | 09 rice seed | R-I | C | T, C, G, C, C | 60 | S | − | − | ||||
| Gfc1043046 | 09 rice seed | R-I | C | T, C, G, C, C | 58 | S | − | − | ||||
| Gfc1043047 | 09 rice seed | R-I | C | T, C, G, C, C | 58 | S | − | − | ||||
| Mo78 | 06 forage paddy rice for livestock | C-I | C | T, C, G, C, C | 61 | S | − | − | T. Tsukiboshi | |||
| Mo80 | 06 forage paddy rice for livestock | C-I | C | T, C, G, C, C | 62 | S | − | − | T. Tsukiboshi | |||
| Mo141 | 07 silage of forage paddy rice for livestock | f-I | C | T, C, G, C, T | 63 | S | − | − | T. Tsukiboshi | |||
| Mo136 | 07 silage of forage paddy rice for livestock | f-I | C | T, C, G, C, C | 64 | S | − | − | T. Tsukiboshi | |||
| Mo309 | Maize | C | C | T, C, G, C, C | 65 | S | − | − | ||||
| IBR89-1 | Unknown | N | C | T, C, G, C, C | 66 | S | − | − | Tateishi and Chida (48) |
|||
| Gfc0825007h | 07 maize | B-IV | JN695738 | LC133064 | (C) | T, C, G, C, C | 67 | S | − | − | ||
| Gfc0825009h | 07 maize | B-IV | JN695739 | LC133065 | (C) | (T, C, G, C, C) | 68 | S | − | − | ||
| Gfc0825011h | 07 maize | B-IV | C | T, C, G, C, C | 69 | S | + | − | ||||
| Gfc0009063h | 00 strawberry | C-II | C | T, C, G, C, C | 70 | S | − | − | M. Morishima | |||
| Gfc0009105h | 00 strawberry | C-III | JN695742 | LC133066 | (C) | T, C, G, C, C | 71 | S | − | − | M. Morishima | |
| Gfc0009110 | 00 strawberry | C-IV | C | T, C, G, C, C | 72 | S | − | − | M. Morishima | |||
| Gfc0009117 | 00 strawberry | C-V | C | T, C, G, C, C | 73 | S | − | − | M. Morishima | |||
| 41-79h | Wheat | USA | LC133054 | LC133067 | (C) | T, C, G, C, C | 74 | S | − | − | Busman et al. (21) |
|
| 41-84 | Wheat | USA | C | T, C, G, C, C | 75 | S | − | − | Busman et al. (21) |
|||
| 41-116 | Wheat | USA | C | T, C, G, C, C | 76 | S | − | − | Busman et al. (21) |
|||
| 41-108 | Wheat | USA | C | T, C, G, C, C | 77 | S | − | − | Busman et al. (21) |
|||
| F. fujikuroi (whole genome, secondary metabolite profiles, and pathogenicity to ricei | ||||||||||||
| (Nonproducer)j | IMI58289 | Infected rice | China | (T) | (G, C, G, C, T) | Producer | ||||||
| Nonproducer | m567 | Infected rice | Japan | (T) | (G, C, G, C, T) | Producer | ||||||
| Nonproducer | MRC2276 | Infected rice | Philippines | (T) | (G, C, A, C, T) | Producer | ||||||
| Nonproducer | C1995 | Infected rice | Taiwan | (T) | (G, C, G, A, T) | Producer | ||||||
| Producer | B14 | Infected rice | South Korea | (C) | (T, C, G, C, C) | (Nonproducer)j | ||||||
| (Nonproducer)i | B20 | Infected rice | South Korea | (T) | (G, C, G, A, T) | Producer | ||||||
| Nonproducer | E282 | Infected rice | Italy | (T) | (G, T, A, A, T) | Producer | ||||||
| Nonproducer | FSU48 | Maize | Germany | (T) | (G, T, A, A, T) | Producer | ||||||
| Nonproducer | NCIM 1100 | Infected rice | India | (T) | (G, T, A, A, T) | Producer | ||||||
All strains used in this study were previously investigated in Suga et al. (8).
First two digits indicate harvest year.
Letters indicate differences in geographic origin rather than actual geographic names. Prefecture-city or town (if the city or town is known) was indicated by insignia.
Single nucleotide polymorphisms were determined by dCAPs. Nucleotide determined by sequencing is indicated in parentheses, but the uncut type is indicated as D (i.e., not C).
Single nucleotide polymorphisms were determined by Luminex.
Ten microliters of culture filtrate was spotted on TLC. +, clear spot detected; ±, ambiguous spot; −, no spot detected.
All strains were used in the previous fumonisin analysis (Suga et al. [8]), and the references only of strains not originally isolated in the previous analysis were indicated.
Strains used to fumonisin and gibberellin quantification by UPLC-MS/MS and pathogenicity test.
Conditions for fumonisin and gibberellin detection were different from those of this study, and SNP data were obtained from the whole-genome sequences in the study by Niehaus et al. (24).
Results of triplicates were not stable, and a small amount was occasionally detected (Niehaus et al. [24]).
We also compared the whole-genome sequences, secondary metabolite profiles, and pathogenicity with regard to rice resulting from the SNPs in nine F. fujikuroi strains that had already been investigated (24) (Table 1). They were isolated from geographically diverse regions. One of the nine strains (B14) is apparently a fumonisin producer, and the other strains are fumonisin nonproducers, although a small amount of fumonisin is occasionally detected in strains IMI58289 and B20 (24). Strain B14 has the same SNP combination patterns as Japanese fumonisin producers (Table 1). Four strains had the same SNP combination patterns as the Japanese fumonisin nonproducers, and the other five strains, including Japanese isolate m657, had none of the SNP combination patterns of the Japanese fumonisin producers or nonproducers (Table 1). Among the SNPs, TEF_T618G was associated with a difference in fumonisin production in the Japanese strains and also in the nine strains investigated by Niehaus et al. (24).
Phylogenetic tree.
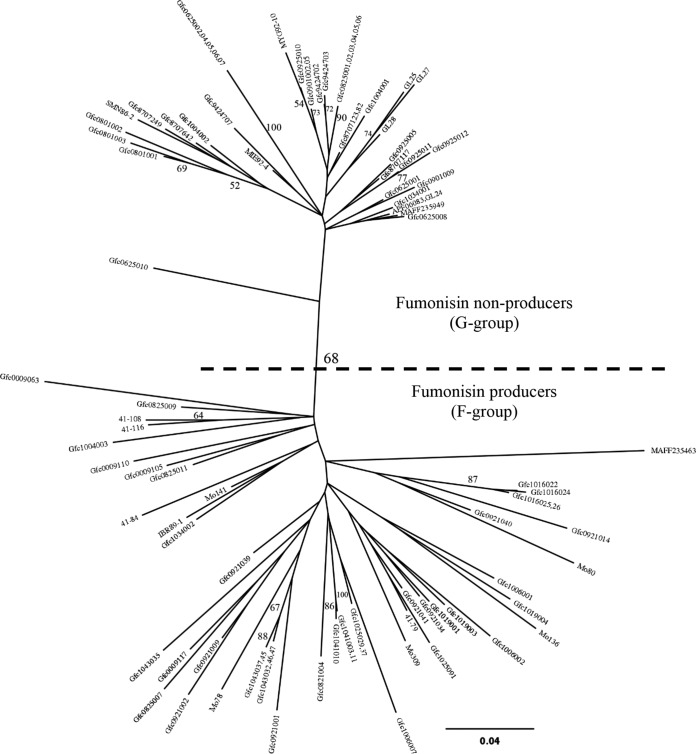
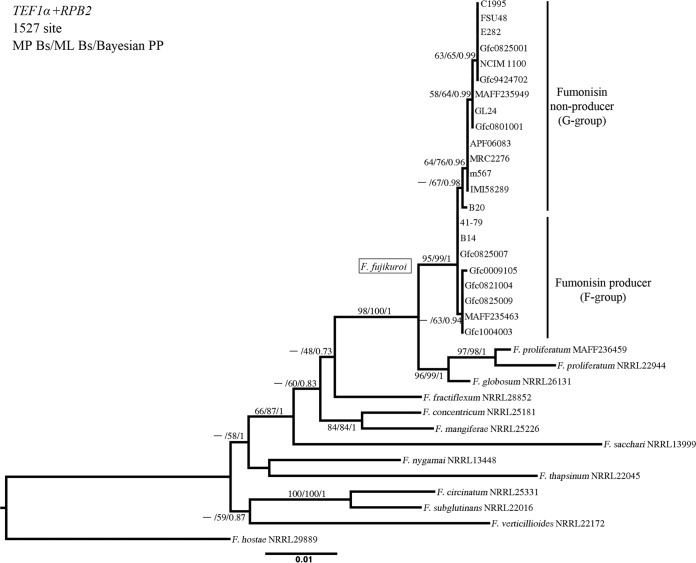
Based on the result of the SNP analyses, we conducted phylogenetic analyses on 66 AFLP markers (see Table S3 in the supplemental material) to demonstrate the genetic relationships between the individual strains. One of the markers (EATMCAY107) was specific to fumonisin producers. Fumonisin producers and nonproducers were separated by a bootstrap value of 68% in the phylogenetic tree (Fig. 1). It was reproduced in the phylogenetic tree based on the combined sequences TEF1α and RPB2 of the representative strains and the nine strains investigated by Niehaus et al. (24) (Fig. 2). The aligned data matrix of 1,527 sequences consisted of 1,248 characters, of which 279 characters were variable and 121 were phylogenetically informative for parsimony analysis. The MP analysis using PAUP* generated 30 equally parsimonious trees with 431 steps (consistency index [CI], 0.76; retention index [RI], 0.76; rescaled consistency index [RC], 0.58; homoplasy index [HI], 0.24). Although we observed slight differences in the small branching orders of the terminal branches and in branch lengths, the tree topologies were generally consistent among all 30 trees (data not shown). Moreover, the topology of the tree generated by ML analysis (Fig. 2) was similar to the topology of the MP and Bayesian phylogenetic trees.
FIG 1.
Phylogenetic tree of Fusarium fujikuroi based on amplified fragment length polymorphism (AFLP) data inferred by unrooted neighbor-joining analysis. Node support given for >50 neighbor-joining bootstrap values is shown above or below each branch. F. fujikuroi fumonisin producers and nonproducers are indicated as the F group and G group mentioned in the Discussion, respectively.
FIG 2.
Phylogenetic tree of Fusarium fujikuroi species complex based on the sequences of the translation elongation factor 1 α (TEF1α) and the second largest subunits of RNA polymerase II (RPB2) genes inferred by maximum likelihood analysis. The sequence of Fusarium hostae strain NRRL29889 was used as an outgroup. Node supports given for >55 maximum-parsimony bootstrap values (MP Bs), maximum likelihood bootstrap values (ML Bs), and >0.80 Bayesian posterior probabilities (Bayesian PP) are shown above or below each branch. F. fujikuroi fumonisin producers and nonproducers are indicated as the F group and G group mentioned in the Discussion, respectively.
We detected an SNP (RPB2_C3250T) between the fumonisin producers and nonproducers, including the nine strains investigated by Niehaus et al. (24), in the RPB2 sequence. We developed a diagnostic PCR-RFLP using a derived cleaved amplified polymorphic sequence (dCAPS) primer for the SNP and applied it to all 95 strains. Fifty-one fumonisin producers displayed the cut type (i.e., position 3250 was occupied by cytosine), whereas 44 nonproducers were of the uncut type (i.e., position 3250 was not occupied by cytosine) (Table 1).
Fumonisin production.
The fumonisin production of all strains in Table 1 as previously investigated by enzyme-linked immunosorbent assay (ELISA) though the presence or absence of fumonisins in 10 strains was confirmed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (8). In order to compare fumonisin production levels among the strains, FB1, FB2, and FB3 of 12 representative strains from each fumonisin producer and nonproducer were quantified by ultraperformance LC-MS/MS (UPLC-MS/MS). The production of FB1 and FB2 of higher than 50 mg/liter and FB3 of higher than 10 mg/liter was observed in 10 out of 12 fumonisin producers (Table 2). Although the production valance of FB1, FB2, and FB3 could not be determined in these fumonisin producers because their concentrations exceeded the high limit of quantification, two remaining fumonisin producers showed highest production in FB1 (Table 2). In the case of the fumonisin nonproducers, neither FB1, FB2, or FB3 was detected with LOQs, at FB1/FB2 of 0.1 mg/liter and FB3 of 0.05 mg/liter.
TABLE 2.
Fumonisin and gibberellin production and pathogenicity of Fusarium fujikuroi strains
| Fumonisin productiona | Strain | Fumonisin production (ppm)b |
Gibberellin production (ppm)b |
Pathogenicityc | |||||
|---|---|---|---|---|---|---|---|---|---|
| FB1 | FB2 | FB3 | GA3 | GA7 | GA4 | GA1 | |||
| Nonproducer | |||||||||
| MAFF235949 | ND | ND | ND | 4.08 (±0.32) | ND | ND | ND | 2.1 (±0.3) | |
| Gfc0801001 | ND | ND | ND | 31.79 (±10.92) | 2.02 (±0.49) | 1.20 (±0.14) | 1.11 (±0.25) | 2.0 (±0.4) | |
| SMN86-2 | ND | ND | ND | 12.14 (±5.06) | ND | ND | ND | 1.2 (±0.1) | |
| Gfc9424702 | ND | ND | ND | 21.93 (±15.00) | 3.96 (±1.69) | 1.53 (±0.26) | ND | 2.0 (±0.3) | |
| Gfc0625001 | ND | ND | ND | 15.93 (±5.00) | 2.02 (±1.66) | ND | 0.61 (±0.07) | 1.7 (±0.2) | |
| Gfc0625005 | ND | ND | ND | 19.89 (±5.34) | 2.24 (±0.29) | ND | ND | 1.1 (±0.1) | |
| Gfc0925005 | ND | ND | ND | 6.34 (±1.70) | 3.39 (±0.82) | 0.77 (±0.04) | ND | 2.1 (±0.4) | |
| APF06083 | ND | ND | ND | 4.15 (±1.02) | 2.29 (±2.36) | 1.16 (±1.11) | ND | 1.5 (±0.1) | |
| Gfc0825001 | ND | ND | ND | 10.46 (±4.91) | 5.01 (±0.95) | 2.64 (±0.21) | 0.64 (±0.15) | 1.8 (±0.3) | |
| Gfc0901009 | ND | ND | ND | 12.46 (±6.91) | 6.47 (±2.58) | 5.96 (±0.71) | 1.32 (±0.64) | 2.1 (±0.1) | |
| Gfc1034001 | ND | ND | ND | 12.51 (±2.83) | 5.82 (±0.57) | 3.80 (±1.23) | 0.76 (±0.26) | 2.1 (±0.6) | |
| GL24 | ND | ND | ND | 17.75 (±1.48) | 3.22 (±0.36) | 1.54 (±0.13) | 0.84 (±0.06) | 1.6 (±0.4) | |
| Producer | |||||||||
| MAFF235463 | 15.02 (±3.49) | 1.59 (±0.19) | 1.82 (±0.43) | ND | ND | ND | ND | 1.1 (±0.0) | |
| Gfc1004003 | >50.00 | >50.00 | >10.00 | 1.34 (±0.49) | ND | ND | ND | 0.9 (±0.1) | |
| Gfc1019001 | >50.00 | >50.00 | >10.00 | 1.20 (±0.45) | ND | ND | ND | 1.2 (±0.2) | |
| Gfc0821004 | >50.00 | >50.00 | >10.00 | 0.93 (±ND) | ND | ND | ND | 0.9 (±0.0) | |
| Gfc0921039 | >50.00 | >50.00 | >10.00 | 0.83 (±0.11) | ND | ND | ND | 0.9 (±0.0) | |
| Gfc1043032 | >50.00 | >50.00 | >10.00 | 2.90 (±ND) | ND | ND | ND | 1.0 (±0.2) | |
| Gfc0825007 | >50.00 | >50.00 | >10.00 | 0.93 (±0.08) | ND | ND | ND | 1.1 (±0.1) | |
| Gfc0825009 | >50.00 | >50.00 | >10.00 | 2.21 (±1.18) | ND | ND | ND | 1.1 (±0.1) | |
| Gfc0825011 | >50.00 | >50.00 | >10.00 | 2.31 (±1.70) | 1.31 (±0.54) | ND | ND | 1.0 (±0.0) | |
| Gfc0009063 | >50.00 | >50.00 | >10.00 | ND | ND | ND | ND | 1.0 (±0.1) | |
| Gfc0009105 | >50.00 | >50.00 | >10.00 | 2.30 (±ND) | ND | ND | ND | 1.0 (±0.1) | |
| 41-79 | >50.00 | 9.99 (±1.85) | 5.28 (±0.52) | ND | ND | ND | ND | 0.9 (±0.0) | |
Fumonisin production determined in Suga et al. (8).
Mean of 3 replicates. Standard deviation is indicated in parentheses. ND, below the limits of quantification (LOQs) for the fumonisin analysis, as follows: FB1and FB2, 0.1 ppm; FB3, 0.05 ppm; and for gibberellin analysis; GA1, GA3, GA4, and GA7, 0.5 ppm.
Ratio of mean height of inoculated plants to the mean height of control (uninoculated) plants. Standard deviation is indicated in parentheses.
Gibberellin production and pathogenicity in rice seedling.
All strains from bakanae-diseased rice are fumonisin nonproducers, according to a previous analysis (8). This suggests that gibberellin production is different between fumonisin producers and nonproducers. We made a preliminary investigation of gibberellin production in the strains by thin-layer chromatography (TLC) (Table 1). TLC detected GA3 in fumonisin nonproducers but not in fumonisin producers (Table 1). We used UPLC-MS/MS to determine the concentration of gibberellin in the 12 representative strains from each fumonisin producer and nonproducer and carried out a pathogenicity assay on the rice seedlings (Table 2). The concentration of GA3 in the fumonisin nonproducers was 4.08 to 31.79 mg/liter (Table 2). We did not detect GA3 in the fumonisin producers using TLC and detected less than 3 mg/liter in 9 out of the 12 strains using UPLC-MS/MS. The pathogenicity values of the 12 fumonisin nonproducers ranged from 1.1 to 2.1, and the pathogenicity values of the 12 fumonisin producers ranged from 0.9 to 1.2 (Fig. 3 and Table 2). The fumonisin producers did not induce typical bakanae symptoms in the rice seedlings, although they were reisolated from the stems after surface sterilization.
FIG 3.
Symptoms of rice seedlings infected with Fusarium fujikuroi strains. The figure indicates part of the pathogenicity assays conducted on the fumonisin-producing and nonproducing strains. F. fujikuroi fumonisin producers and nonproducers are indicated as the F group and G group mentioned in the Discussion, respectively. Strain names are indicated under each picture.
TM resistance.
TM resistance in F. moniliforme isolated from bakanae-diseased rice has been well known in Japan, but preliminary experiments on several F. fujikuroi strains in a study by Suga et al. (8) indicate that fumonisin producers are sensitive to TM. Therefore, we investigated the sensitivity to TM in all 95 strains by measuring growth in medium containing 100 ppm of TM. None of the 51 fumonisin producers grew on the medium, and all were considered to be TM sensitive (TMS) (Table 1), whereas 39 of the 44 fumonisin nonproducers grew on the medium and were considered to be TM resistant (TMR) (Table 1).
DISCUSSION
In the present study, we revealed genetic differences between fumonisin producers and nonproducers in Japanese F. fujikuroi by SNP and phylogenetic analyses. Fumonisin nonproducers produce more gibberellin and have higher pathogenicity with regard to rice seedlings than do fumonisin producers (Fig. 3 and Tables 1 and 2). Therefore, we designated fumonisin producers and nonproducers fumonisin (F) and gibberellin (G) groups, respectively.
The ability to produce certain secondary metabolites may have an impact on the evolution and ecological adaptations of fungi (25). F. fujikuroi competes with various microorganisms during its life cycle, and fumonisin is a potential antibiotic. Keyser et al. (26) demonstrated that the MIC values of fumonisin B1 for four fungal species other than F. fujikuroi complex species were 0.25 to 10 mM, whereas they were higher than 40 mM for four F. fujikuroi complex species and Aspergillus flavus. Gibberellin production contributes to initial fungal colonization in rice roots (11). F-group strains have been detected in various plant species, including rice, but G-group strains have only been isolated in rice so far.
The cause of low-level or zero fumonisin production in G-group strains remains unclear. Recently, Rösler et al. (27) demonstrated that the low level of FUM21 transcription, a local transcription factor in the fumonisin biosynthesis gene cluster, is a cause of low fumonisin production in the IMI58289 strain from Taiwan. Although IMI58289 retains the ability to produce fumonisin, a lack of the genes that are essential for fumonisin biosynthesis could cause a complete loss of fumonisin production. Fumonisin-nonproducing F. verticillioides strains isolated from bananas lack most of the fumonisin biosynthesis gene cluster and have been reclassified as a new sister species, Fusarium musae (28–30). Chiara et al. (31) also discovered the absence of seven genes from the fumonisin biosynthesis gene cluster in a fumonisin nonproducer strain of F. fujikuroi (FGSC 8932). In the present study, PCR amplification for FUM18_G51T was successful in all five F-group strains but failed in three out of the five G-group strains tested. PCR amplification was attempted repeatedly, but no amplification was observed for the three strains, even at low annealing temperatures (data not shown). These results suggest that a part of the fumonisin biosynthesis gene cluster that includes the FUM18 gene may be absent in these strains.
In the present study, we determined the genetic differences between the G and F groups of F. fujikuroi (Fig. 1 and 2). The F group is closer to Fusarium proliferatum, which is a fumonisin-producing species, than the G group, and is more genetically diverse than the G group. However, the bootstrap values between the groups in the phylogenetic trees were not large (Fig. 1 and 2), and reproductive isolation between the groups seems to have been absent, because crossing between an F-group strain (Gfc0825009) and a G-group strain (Gfc0801001) succeeded (data not shown). Recently, Niehaus et al. (24) indicated that two pathotypes of F. fujikuroi associated with secondary metabolites, a bakanae type, producing gibberellin but little or no fumonisin, and a stunting type, producing fumonisin but not gibberellin, are phylogenetically different, as observed in the F and G groups in the present study (Fig. 1 and 2).
TM resistance corresponded closely to the cluster of G-group strains in the phylogenetic tree (Fig. 1 and Table 1). In Japan, TM has been used intensively to control bakanae disease, and benzimidazole-resistant strains have emerged since 1984 in Japan (6). In China, benzimidazole-resistant strains of F. fujikuroi have also emerged, and amino acid mutations E198V and F200Y, which promote resistance, have been found in β2-tubulin (32). The prevalence of benzimidazole-resistant strains was 83% (432 out of 518 strains) in isolates from Japanese rice seeds between 1985 and 1989 (33) and 79% (46 out of 58 strains) in bakanae-diseased rice plants in 1993 (18), although at the time they were identified as F. moniliforme, not F. fujikuroi. These results suggest that TM-resistant strains prevail in the F. fujikuroi population in rice plants, possibly as a result of seed treatment. In the present study, 89% (39 out of 44 strains) of the G-group strains were TM resistant (Table 1). Most of the exceptional TM-sensitive strains in the G group were not Japanese, but were U.S. isolates, whereas none of the 51 F-group strains were TM resistant (Table 1). Therefore, strong selection could be exerted on the F. fujikuroi population in rice plants. This selection pressure could result in the comparatively low levels of genetic variation and branching found in the G group in the phylogenetic trees (Fig. 1), generally known as the bottleneck effect.
Our results show the tendency that fumonisin nonproducers have gibberellin high production and are pathogenic to rice seedlings (Table 2). This is consistent with the following published observations. Fumonisin production was not detected in 86% (50 out of 58 strains) of the F. moniliforme isolates with gibberellin production (1 to 74 ppm) (18). Little or no fumonisin was detected in seven strains of F. fujikuroi isolated from Nepalese rice with gibberellin production (240 to 1,590 ppm) (16). Little or no fumonisin production and high gibberellin production were observed in all 10 strains of F. fujikuroi isolated from Asian rice (from Nepal, Vietnam, China, and Bangladesh) (17) and from 8 strains of geographically diverse origin in the whole-genome sequences investigated (24). However, with regard to pathogenicity, caution and prudence were required for an assessment. Pathogenicity was assessed by plant height in the present study. It has been reported that fumonisin producers do not induce the typical symptoms induced by gibberellin in rice, such as stem elongation, but instead cause root rot and stunting (24, 34). Gene disruption of Fusarium cyclin C1 (FCC1) or FUM1, which play an important role in fumonisin biosynthesis in the fumonisin-producing strain B14, resulted in a significant reduction in stunting-type symptoms (24, 35). These studies indicate that fumonisin production contributes to stunting-type symptoms. Although the F-group strains with fumonisin production (8) did not incite typical stunting of rice in the present study, stunting-type symptoms may have been detected if another pathogenicity assay method had been used.
Among the SNPs investigated in the present study, FUM78_C41T, FUM8_G2834A, and CPR_C1152A were specific only to the F and G groups in the Japanese strains, whereas two SNPs (TEF_T618G and RPB2_2C3250T) were specific to the Japanese strains and to the eight strains from other countries (24). Niehaus et al. (24) demonstrated the specific PCR detection of NRPS31 and PKS51 in bakanae-type strains and stunting-type strains. These are possible markers for differentiation between the two types of F. fujikuroi, although their robustness should be investigated in further studies because it has been reported that TEF_T618G is not associated with fumonisin production in F. fujikuroi isolated from grapes in the southern United States (10).
The significant bias of TM resistance and the low genetic variability observed in the G-group strains suggest that selection pressure by TM could have been exerted on the F. fujikuroi population in Japan. However, the discovery of genetically different bakanae and stunting types in F. fujikuroi by Niehaus et al. (24) suggests that the genetic differentiation of F. fujikuroi is not exclusive to Japan. It is possible that the F group/stunting type and G group/bakanae type of F. fujikuroi may have different host plant preferences, although they overlap in rice. This overlap makes the difference in their host plant preferences ambiguous. Both F- and G-group strains were isolated from rice in the present study, and PCR detection of PKS51 was indicated in the B14 stunting type and the B20 bakanae type isolated from rice in South Korea (24). In some cases, we isolated both the F-group and G-group strains from a single rice plant with bakanae symptoms (data not shown). However, when crops other than rice were used as isolation sources, none of the G-group strains were isolated in the present study, and none of the bakanae-type strains were detected when maize isolates from South Korea were investigated (24). Further studies of the F. fujikuroi population in various crops from geographically diverse regions accompanied by the identification of the two types would reveal the ongoing global intraspecific evolution of F. fujikuroi.
MATERIALS AND METHODS
Fungal strains.
The 95 F. fujikuroi strains investigated in the present study were the same as those previously subjected to fumonisin production analysis (Table 1) (8). All except 5 fumonisin producers and 5 nonproducers were Japanese isolates. Fifty-one strains, including MAFF235463 isolated from maize, strawberries, wheat, and rice, were fumonisin producers, and 44 strains, including MAFF235949 isolated from rice seeds, rice seedlings carrying bakanae disease, and from unknown sources, were fumonisin nonproducers.
Genomic DNA extraction.
Genomic DNA was extracted from 3- to 4-day-old mycelia cultured in potato dextrose broth (PDB), as described previously (36). Each of the obtained DNA pellets was dissolved in 200 μl of water, and the DNA concentration was adjusted to 5 ng/μl based on a comparison with DNA of a known concentration conducted by agarose gel electrophoresis.
AFLP analysis.
We used an AFLP microbial fingerprinting kit (Life Technologies, Carlsbad, CA, USA). Preselective and selective PCRs were performed in an iCycler thermal cycler (Bio-Rad Laboratories Hercules, CA, USA). Hi-Di formamide (9.95 μl; Life Technologies) and standard-size GeneScan 500 dye (ROX; 0.05 μl; Life Technologies) were added to a 1.5-μl sample after selective PCR. The samples were analyzed with a 3100 genetic analyzer, and the obtained data were processed using the GeneMapper software (Life Technologies) to produce the binary matrix for phylogenetic analysis. Detailed information for this analysis is available in the supplemental material.
PCR and sequencing.
PCRs for CPR and P450-4 were performed using AmpliTaq DNA polymerase (Life Technologies) and the following cycling parameters: 94°C for 2 min and 30 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 1 min, using the P138-5/P138-6 and P450-4-GD1/P450-4-GD2 primer pairs, respectively (Table S1) (19). PCRs for FUM1, FUM8 including the FUM7/FUM8 intergenic region, and FUM18 including the FUM18/FUM19 intergenic region were performed using the HS398/HS399, HS576/HS577, and HS506/HS519 primer pairs, respectively (Table S1), under the same conditions described above except that the annealing temperatures were 63°C, 58°C, and 60°C, respectively, instead of 68°C. The PCR for RPB2 was performed using rTaq (TaKaRa Bio, Inc., Ootsu, Japan) and the same cycling parameters described above except that the annealing temperature was 53°C instead of 68°C and the primer pair was 7cf/11ar (37). The PCR for TEF1α was performed according to Suga et al. (36). The PCR products were directly sequenced as previously described (36). We used BigDye Terminator version 3.1 with cycle sequencing kits (Life Technologies) and both or either of the primers used for the PCR, and the sequences were obtained using an ABI 3100 genetic analyzer (Life Technologies). The nucleotide sequence data were processed using ChromasPro (Technelysium Pty., Tewantin, Queensland, Australia) and Genetyx (Tokyo, Japan).
Phylogenetic analysis.
The binary data for a total of 66 AFLP markers from 95 taxa were compiled into a single data matrix. Neighbor-joining (NJ) analysis (38) of the binary data was performed using PAUP* (version 4.0β10) (39).
The sequence data sets combined the data for TEF1α and RPB2 from 35 taxa, including an outgroup (Fusarium hostae NRRL29889). The sequences were aligned with MAFFT (version 7). Phylogenetic trees were obtained using maximum parsimony (MP), maximum likelihood (ML), and Bayesian phylogenetic analyses. The best-fit evolutionary model was determined for each data set by comparing different evolutionary models for MP and ML analyses, and using the Bayesian information criterion (BIC) (40) for the Bayesian analysis. As a result of the calculations, the combined data set of the TEF1α and the RPB2 regions was fitted to a J2ef model with a gamma-shaped distributed rate. MP analysis was carried out using PAUP*. The best tree topology of the MP trees was established using the Kishino-Hasegawa likelihood test (41) on PAUP*. The ML analysis was performed by the likelihood ratchet method (42). The strength of the internal branches from the resulting tree was tested by bootstrap (BS) analysis (43) using 10,000 replications in NJ, MP, and ML analyses. Bayesian phylogenetic analyses with the selected evolutionary model were carried out using MrBayes (version 3.2.5) (44). Detailed information for this analysis is available in the supplemental material.
Single nucleotide polymorphism analysis.
PCR-restriction fragment length polymorphism (PCR-RFLP) experiments for TEF_C447A, FUM1_G423A, FUM78_C41T, FUM8_G2834A, FUM18_G51T, and RPB2_C3250T were performed under conditions similar to those used for HIS PCR-RFLP (8). The derived cleaved amplified polymorphic sequence (dCAPS) primers (HS435, HS482, and HS834) for PCR-RFLP were designed using dCAPS Finder 2.0 (Table S1) (45). PCR sequencing for CPR_C1152A and P4504_C842T was performed as described above.
A multiplex PCR comprising TEF1α/FUM or CPR/P450-4 was performed to prepare template DNA for allele-specific primer extension (ASPE) reactions. The PCR was performed in an iCycler thermal cycler (Bio-Rad Laboratories). The ASPE reactions were performed according to the manufacturer’s instructions using Platinum Genotype Tsp DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA). The biotin-labeled products were sorted by hybridization with polystyrene microspheres coated with the anti-tag sequences. The microspheres were finally resuspended in buffer containing streptavidin-R-phycoerythrin (Invitrogen Life Technologies). The median fluorescence intensity (MFI) of 100 microspheres was measured with a Luminex 100 flow cytometer (Luminex Corporation, Austin, TX, USA). SNPs were determined from more than 100 MFI values after subtracting the background MFI value obtained using water instead of the extension product. SNPs were determined based on ratios of more than twice the MFI values between the paired microspheres corresponding to SNPs. The SNPs of the F. fujikuroi strains used by Niehaus et al. (24) were obtained from the BioProject at the NCBI, as described below. Detailed information for this analysis is available in the supplemental material.
Pathogenicity assay.
A dwarf rice cultivar, Tanginbozu, was used to assess the pathogenicity of the strains. The rice seeds were soaked in water at 60°C for 10 min to disinfest them of possible natural pathogens and then were cooled with running water immediately before use. The strains were cultured in potato dextrose agar (PDA) for 1 week at 25°C, and a conidial suspension in sterile water was obtained. The rice seeds (weighting 1 g before hot-water disinfestation) were soaked in a petri dish containing a 1 × 105/ml conidial suspension and incubated at 30°C for 48 h in the dark. The control seeds were soaked in sterile water instead of the conidial suspension. Ten budding seeds were transferred to a filter paper, and superfluous mycelia that had stuck to the seed surfaces were removed. The seeds were then sown in pots (5.5 cm diameter, 7 cm height) containing wetted nursery soil (Grand-sol no. 11; Sumitomo Chemical, Tokyo, Japan). The pots were placed in a transparent plastic box covered with cellophane. The box was placed in an MIR-154 incubator (Panasonic Healthcare, Tokyo, Japan), and maintained at 30°C with continuous lighting for 4 days. The cellophane was then removed from the box, and the plants were watered. The incubator conditions were changed to 23°C with a 16-h light/8-h dark cycle. The heights (in millimeters) of the three tallest individual plants per pot were measured 3 days after the change of incubator conditions. The disease level was scored as the ratio of the mean height of the inoculated plants to the mean height of the control (noninoculated) plants, and the mean result from three pots (replicates) was considered to represent the pathogenicity of the strain.
Fumonisin analysis.
The strains were grown on sterile rice. Five grams of rice grain was soaked in 4 ml water prior to autoclaving in an Erlenmeyer flask. Each flask was inoculated with a piece of culture grown on PDA and kept at 25°C for 10 days. The experimental cultures were repeated in triplicate. Fumonisins were extracted with 25 ml methanol-water (3:1 [vol/vol]) by reciprocal shaking for 30 min. The extract filtered through paper was purified by a strong anion-exchange column (Sep-Pak Accell Plus QMA short cartridge [360 mg]; Waters, Milford, MA, USA), and the concentrations of the fumonisins (FB1, FB2, and FB3) were quantified by ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) analysis using an Acquity UPLC system coupled to a Xevo quadrupole time of flight (QTof) mass spectrometer (Waters). The fumonisins FB1 (Enzo Life Sciences, Lausen, Switzerland), FB2 (Enzo Life Sciences), and FB3 (Iris Biotech, Marktredwitz, Germany) were used as a standard. Working solutions containing FB1, FB2, and FB3 at concentrations between 0.05 and 5.0 mg/liter (FB1/FB2, 0.1, 0.2, 0.5, 1.0, 2.0, and 5.0 mg/liter, and FB3, 0.05, 0.1, 0.2, 0.4, 0.6, and 1.0 mg/liter, respectively, in 6 bottles) were used to create a calibration curve. The concentration (x) (FB1, FB2, and FB3) and corresponding peak area ratio (y) were plotted for the 6 bottles of the working solution. The limits of quantification (LOQs) for the fumonisin analysis were the lowest concentration (FB1/FB2, 0.1 mg/liter; FB3, 0.05 mg/liter) on the calibration curves for FB1, FB2, and FB3. In cases in which the concentration of the sample exceeded the range given by the calibration curve, the sample was diluted 10 times and reanalyzed. Detailed information for this analysis is available in the supplemental material.
Gibberellin analysis.
The strains were grown in 10% ICI (Imperial Chemical Industries Ltd., UK) medium (46) for 7 days on a reciprocal shaker. The gibberellins in the culture filtrate were analyzed by thin-layer chromatography (TLC) or by UPLC-MS/MS. The gibberellins GA3 (Wako Pure Chemicals Ind., Ltd., Osaka, Japan), GA1, GA4, and GA7 (Olchemim Ltd., Olomouc, Czech Republic) were used as standards. TLC for GA3 and GA4/7 was performed according to Linnemannstöns et al. (47).
The concentrations of the gibberellins (GA1, GA3, GA4, and GA7) were quantified by UPLC-MS/MS analysis with the same equipment as fumonisin analysis. The experimental cultures were repeated in triplicate, and the resulting broths were subjected to UPLC-MS/MS analysis. Working solutions containing GA1, GA3, GA4, and GA7 at concentrations of 0.5 to 5.0 mg/liter (GA3/GA7, 0.5, 1.0, 2.0, and 5.0 mg/liter; GA1/GA4, 0.5, 1.0, 1.5, and 2.0 mg/liter, in four bottles) were used to create a calibration curve. The concentration (x) (GA1, GA3, GA4, and GA7) and corresponding peak area ratio (y) were plotted for the four bottles of the working solution. The LOQ for the gibberellin analysis was the lowest concentration (0.5 mg/liter) on the calibration curves for GA1, GA3, GA4, and GA7. In cases in which the concentration of the sample exceeded the range given by the calibration curve, the sample was diluted 4 and 10 times and reanalyzed. Detailed information for this analysis is available in the supplemental material.
TM resistance.
The strains were grown on PDA for 1 week. Disks (4 mm in diameter) were transferred to PDA amended with TM (water-soluble Topsin M powder containing 70.0% TM; Nihon Nohyaku, Tokyo, Japan) at a final concentration of 100 μg/ml to discriminate between TM-resistant (TMR) and TM-sensitive (TMS) strains. This was done because the MIC of benomyl that is a benzimidazole fungicide differs between resistant (MIC, ≥100 μg/ml) and sensitive (MIC, ≤12.5 μg/ml) strains of Fusarium moniliforme (33).
Data availability.
The sequences obtained in the present study are available with the accession numbers LC133052 to LC133070 at the DDBJ/EMBL/GenBank database. The sequences of the TEF1α and RPB2 genes of the F. fujikuroi strains used by Niehaus et al. (24) were obtained from the BioProject PRJEB14872 (ID: 412609) at the National Center for Biotechnology Information (NCBI), and the other sequences were obtained from the DDBJ/EMBL/GenBank database with the accession numbers shown in Table S2.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Suzuki (Mie Prefecture Agricultural Research Institute) for providing rice cultivar Tanginbozu and M. Funasaka (Gifu University) for technical support.
This work was supported in part by Japan Society for the Promotion of Science (JSPS) KAKENHI grant JP24580474 and research project for improving food safety and animal health.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02414-18.
REFERENCES
- 1.Nirenberg HI, O’Donnell K. 1998. New Fusarium species and combination within the Gibberella fujikuroi species complex. Mycologia 90:434–458. doi: 10.1080/00275514.1998.12026929. [DOI] [Google Scholar]
- 2.O’Donnell K, Cigelnik E, Nirenberg HI. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465–493. doi: 10.1080/00275514.1998.12026933. [DOI] [Google Scholar]
- 3.Leslie JF. 1995. Gibberella fujikuroi: available populations and variable traits. Can J Bot 73:S282–S291. doi: 10.1139/b95-258. [DOI] [Google Scholar]
- 4.Lima CS, Pfenning LH, Costa SS, Abreu LM, Leslie JF. 2012. Fusarium tupiense sp. nov., a member of the Gibberella fujikuroi complex that causes mango malformation in Brazil. Mycologia 104:1408–1419. doi: 10.3852/12-052. [DOI] [PubMed] [Google Scholar]
- 5.Sun SK, Snyder WC. 1981. The bakanae disease of the rice plant, p 104–113. In Nelson PE, Toussoun TA, Cook RJ (ed), Fusarium disease, biology, and taxonomy. Pennsylvania State University Press, University Park, PA. [Google Scholar]
- 6.Yoshino R. 1988. Current status of bakanae disease occurrence and its control. Shokubutsu Boueki 42:321–325. (In Japanese.) [Google Scholar]
- 7.Seifert KA, Aoki T, Baayen RP, Brayford D, Burgess LW, Chulze S, Gams W, Geiser D, de Gruyter J, Leslie JF, Logrieco A, Marasas WFO, Nirenberg HI, O’Donnell K, Rheeder J, Samuels GJ, Summerell BA, Thrane U, Waalwijk C. 2003. The name Fusarium moniliforme should no longer be used. Mycol Res 107:643–644. doi: 10.1017/S095375620323820X. [DOI] [Google Scholar]
- 8.Suga H, Kitajima M, Nagumo R, Tsukiboshi T, Uegaki R, Nakajima T, Kushiro M, Nakagawa H, Shimizu M, Kageyama K, Hyakumachi M. 2014. A single nucleotide polymorphism in the translation elongation factor 1alpha gene correlates with the ability to produce fumonisin in Japanese Fusarium fujikuroi. Fungal Biol 118:402–412. doi: 10.1016/j.funbio.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Imazaki I, Kadota I. 2015. Molecular phylogeny and diversity of Fusarium endophytes isolated from tomato stems. FEMS Microbiol Ecol 91:fiv098. doi: 10.1093/femsec/fiv098. [DOI] [PubMed] [Google Scholar]
- 10.Bolton SL, Brannen PM, Glenn AE. 2016. A novel population of Fusarium fujikuroi isolated from Southeastern U.S. winegrapes reveals the need to re-evaluate the species’ fumonisin production. Toxins 8:254. doi: 10.3390/toxins8090254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiemann P, Sieber CMK, von Bargen KW, Studt L, Niehaus E-M, Espino JJ, Huß K, Michielse CB, Albermann S, Wagner D, Bergner SV, Connolly LR, Fischer A, Reuter G, Kleigrewe K, Bald T, Wingfield BD, Ophir R, Freeman S, Hippler M, Smith KM, Brown DW, Proctor RH, Münsterkötter M, Freitag M, Humpf H-U, Güldener U, Tudzynski B. 2013. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog 9:e1003475. doi: 10.1371/journal.ppat.1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bezuidenhout SC, Gelderblom WCA, Gorst-Allman CP, Horak RM, Marasas WFO, Spiteller G, Vleggaar R. 1988. Structure elucidation of the fumonisins, mycotoxins from Fusarium moniliforme. J Chem Soc Chem Commun 0:743–745. doi: 10.1039/c39880000743. [DOI] [Google Scholar]
- 13.Gelderblom WCA, Marasas WFO, Horak RM, Vlegaar R, Kriek NPJ, Legaar R. 1988. Fumonisins–novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl Environ Microbiol 54:1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desjardins AE. 2006. Selected mycotoxigenic Fusarium species, p 145–194. In Desjardins AE. (ed), Fusarium mycotoxins, chemistry genetics, and biology. American Phytopathological Society Press, St. Paul, MN. [Google Scholar]
- 15.Cruz A, Marin P, Gonzalez-Jaen MT, Aguilar KG, Cumagun CJ. 2013. Phylogenetic analysis, fumonisin production and pathogenicity of Fusarium fujikuroi strains isolated from rice in the Philippines. J Sci Food Agric 93:3032–3039. doi: 10.1002/jsfa.6136. [DOI] [PubMed] [Google Scholar]
- 16.Desjardins AE, Manandhar HK, Plattner RD, Manandhar GG, Poling SM, Maragos CM. 2000. Fusarium species from Nepalese rice and production of mycotoxins and gibberellic acid by selected species. Appl Environ Microbiol 66:1020–1025. doi: 10.1128/AEM.66.3.1020-1025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wulff EG, Sørensen JL, Lübeck M, Nielsen KF, Thrane U, Torp J. 2010. Fusarium spp. associated with rice bakanae: ecology, genetic diversity, pathogenicity and toxigenicity. Environ Microbiol 12:649–657. doi: 10.1111/j.1462-2920.2009.02105.x. [DOI] [PubMed] [Google Scholar]
- 18.Yoshizawa T, Yamasaki M, Nanba N, Yamashita A, Ueda S, Shibata H. 1994. Fumonisin producibility of fungicide-resistant Gibberella fujikuroi, a pathogen of bakanae disease. Proc Jpn Assoc Mycotoxicol 1994:33–37. doi: 10.2520/myco1975.1994.40_33. [DOI] [Google Scholar]
- 19.Malonek S, Bömke C, Bornberg-Bauer E, Rojas MC, Hedden P, Hopkins P, Tudzynski B. 2005. Distribution of gibberellin biosynthetic genes and gibberellin production in the Gibberella fujikuroi species complex. Phytochemistry 66:1296–1311. doi: 10.1016/j.phytochem.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Desjardins AE, Plattner RD, Nelson PE. 1997. Production of fumonisin B1 and moniliformin by Gibberella fujikuroi from rice from various geographic areas. Appl Environ Microbiol 63:1838–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busman M, Desjardins AE, Proctor RH. 2012. Analysis of fumonisin contamination and the presence of Fusarium in wheat with kernel black point disease in the United States. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 29:1092–1100. doi: 10.1080/19440049.2012.671787. [DOI] [PubMed] [Google Scholar]
- 22.Uegaki R, Kobayashi H, Tohno M, Tsukiboshi T. 2012. Identification of mycotoxin-producing Fusarium spp. isolated from corn and the changes in concentration of fumonisin during the cultivation period. Grassl Sci 58:121–126. doi: 10.1111/j.1744-697X.2012.00255.x. [DOI] [Google Scholar]
- 23.Kim J-H, Kang M-R, Kim H-K, Lee S-H, Lee T, Yun S-H. 2012. Population structure of the Gibberella fujikuroi species complex associated with rice and corn in Korea. Plant Pathol J 28:357–363. doi: 10.5423/PPJ.OA.09.2012.0134. [DOI] [Google Scholar]
- 24.Niehaus E-M, Kim H-K, Munsterkotter M, Janevska S, Arndt B, Kalinina SA, Houterman PM, Ahn I-P, Alberti I, Tonti S, Kim D-W, Sieber CMK, Humpf H-U, Yun S-H, Guldener U, Tudzynski B. 2017. Comparative genomics of geographically distant Fusarium fujikuroi isolates revealed two distinct pathotypes correlating with secondary metabolite profiles. PLoS Pathog 13:e1006670. doi: 10.1371/journal.ppat.1006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox EM, Howlett BJ. 2008. Secondary metabolism: regulation and role in fungal biology. Curr Opin Microbiol 11:481–487. doi: 10.1016/j.mib.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Keyser Z, Vismer HF, Klaasen JA, Snijman PW, Marasas WFO. 1999. The antifungal effect of fumonisin B1 on Fusarium and other fungal species. S Afr J Sci 95:455–458. [Google Scholar]
- 27.Rösler SM, Sieber CMK, Humpf H-U, Tudzynski B. 2016. Interplay between pathway-specific and global regulation of the fumonisin gene cluster in the rice pathogen Fusarium fujikuroi. Appl Microbiol Biotechnol 100:5869–5882. doi: 10.1007/s00253-016-7426-7. [DOI] [PubMed] [Google Scholar]
- 28.Glenn AE, Zitomer NC, Zimeri AM, Williams LD, Riley RT, Proctor RH. 2008. Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol Plant Microbe Interact 21:87–97. doi: 10.1094/MPMI-21-1-0087. [DOI] [PubMed] [Google Scholar]
- 29.Moretti A, Mulè G, Susca A, González-Jaén MT, Logrieco A. 2004. Toxin profile, fertility and AFLP analysis of Fusarium verticillioides from banana fruits. Eur J Plant Pathol 110:601–609. doi: 10.1023/B:EJPP.0000032399.83330.d7. [DOI] [Google Scholar]
- 30.Van Hove F, Waalwijk C, Logrieco A, Munaut F, Moretti A. 2011. Gibberella musae (Fusarium musae) sp. nov., a recently discovered species from banana is sister to F. verticillioides. Mycologia 103:570–585. doi: 10.3852/10-038. [DOI] [PubMed] [Google Scholar]
- 31.Chiara M, Fanelli F, Mulè G, Logrieco AF, Pesole G, Leslie JF, Horner DS, Toomajian C. 2015. Genome sequencing of multiple isolates highlights subtelomeric genomic diversity within Fusarium fujikuroi. Genome Biol Evol 7:3062–3069. doi: 10.1093/gbe/evv198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, Gao T, Liang S, Liu K, Zhou M, Chen C. 2014. Molecular mechanism of resistance of Fusarium fujikuroi to benzimidazole fungicides. FEMS Microbiol Lett 357:77–84. doi: 10.1111/1574-6968.12504. [DOI] [PubMed] [Google Scholar]
- 33.Wada T, Kuzuma S, Takenaka M. 1990. Sensitivity of Fusarium moniliforme isolates to pefurazoate. Jpn J Phytopathol 56:449–456. doi: 10.3186/jjphytopath.56.449. [DOI] [Google Scholar]
- 34.Hwang IS, Ahn I-P. 2016. Multi-homologous recombination-based gene manipulation in the rice pathogen Fusarium fujikuroi. Plant Pathol J 32:173–181. doi: 10.5423/PPJ.OA.12.2015.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim W-B, Woloshuk CP. 2001. Regulation of fumonisin B1 biosynthesis and conidiation in Fusarium verticillioides by a cyclin-like (C-type) gene, FCC1. Appl Environ Microbiol 67:1607–1612. doi: 10.1128/AEM.67.4.1607-1612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suga H, Karugia GW, Ward T, Gale LR, Tomimura K, Nakajima T, Miyasaka A, Koizumi S, Kageyama K, Hyakumachi M. 2008. Molecular characterization of the Fusarium graminearum species complex in Japan. Phytopathology 98:159–166. doi: 10.1094/PHYTO-98-2-0159. [DOI] [PubMed] [Google Scholar]
- 37.O’Donnell K, Sutton DA, Rinaldi MG, Sarver BAJ, Balajee SA, Schroers H-J, Summerbell RC, Robert VARG, Crous PW, Zhang N, Aoki T, Jung K, Park J, Lee Y-H, Kang S, Park B, Geiser DM. 2010. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J Clin Microbiol 48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony and other methods, 4.0β10. Sinauer, Sunderland, MA. [Google Scholar]
- 40.Schwarz G. 1978. Estimating the dimension of a model. Ann Stat 6:461–464. doi: 10.1214/aos/1176344136. [DOI] [Google Scholar]
- 41.Kishino H, Hasegawa M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 42.Vos RA. 2003. Accelerated likelihood surface exploration: the likelihood ratchet. Syst Biol 52:368–373. doi: 10.1080/10635150390196993. [DOI] [PubMed] [Google Scholar]
- 43.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 44.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 45.Neff MM, Turk E, Kalishman M. 2002. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet 18:613–615. doi: 10.1016/S0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- 46.Geissman TA, Verbiscar AJ, Phinney BO, Cragg G. 1966. Studies on the biosynthesis of gibberellins from (−)-kaurenoic acid in cultures of Gibberella fujikuroi. Phytochemistry 5:933–947. doi: 10.1016/S0031-9422(00)82790-9. [DOI] [Google Scholar]
- 47.Linnemannstöns P, Voss T, Hedden P, Gaskin P, Tudzynski B. 1999. Deletions in the gibberellin biosynthesis gene cluster of Gibberella fujikuroi by restriction enzyme-mediated integration and conventional transformation-mediated mutagenesis. Appl Environ Microbiol 65:2558–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tateishi H, Chida T. 2000. Sensitivity of Fusarium moniliforme isolates to ipconazole. J Gen Plant Pathol 66:353–359. doi: 10.1007/PL00012977. [DOI] [Google Scholar]
- 49.Proctor RH, Plattner RD, Brown DW, Seo JA, Lee YW. 2004. Discontinuous distribution of fumonisin biosynthetic genes in the Gibberella fujikuroi species complex. Mycol Res 108:815–822. doi: 10.1017/S0953756204000577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences obtained in the present study are available with the accession numbers LC133052 to LC133070 at the DDBJ/EMBL/GenBank database. The sequences of the TEF1α and RPB2 genes of the F. fujikuroi strains used by Niehaus et al. (24) were obtained from the BioProject PRJEB14872 (ID: 412609) at the National Center for Biotechnology Information (NCBI), and the other sequences were obtained from the DDBJ/EMBL/GenBank database with the accession numbers shown in Table S2.