Abstract
The biological processes associated with the onset of schizophrenia remain largely unknown. Current hypotheses favor gene × environment interactions as supported by our recent report about DNA methylation changes during the onset of psychosis. Here, we conducted the first longitudinal transcriptomic analysis of blood samples from 31 at-risk individuals who later converted to psychosis and 63 at-risk individuals who did not. Individuals were followed for a maximum of 1 year. Blood samples were collected at baseline and at the end of follow-up and individuals served as their own controls. Differentially expressed genes between the 2 groups were identified using the RNA sequencing of an initial discovery subgroup (n = 15 individuals). The most promising results were replicated using high-throughput real-time qPCR in the whole cohort (n = 94 individuals). We identified longitudinal changes in 4 brain-expressed genes based on RNAseq analysis. One of these genes (CPT1A) was replicated in the whole cohort. The previously observed hypermethylation in NRP1 and GSTM5 during the onset of psychosis correlated with a decrease in corresponding gene expression. RNA sequencing also identified 2 co-expression networks that were impaired after conversion compared with baseline—the Wnt pathway including AKT1, CPT1A and semaphorins, and the Toll-like receptor pathway, related to innate immunity. This longitudinal study of transcriptomic changes in individuals with at-risk mental state revealed alterations during conversion to psychosis in pathways and genes relevant to schizophrenia. These results may be a first step toward better understanding psychosis onset. They may also help to identify new biomarkers and targets for disease-modifying therapeutic strategies.
Keywords: messenger RNA, gene expression, schizophrenia, at-risk mental state, prodromes, prevention
Introduction
Over the past 20 years, researchers and psychiatrists in the field of psychosis have moved from a conception of a chronic presentation to a more dynamic paradigm.1 Accordingly, schizophrenia is now conceptualized as a progressive illness that typically emerges during late adolescence and transitions between several stages: early vulnerability, at-risk mental state (also called ultra-high risk, abbreviated UHR), first episode of psychosis, and chronic disease.2 Defining criteria to identify UHR individuals has permitted research in the early stages of schizophrenia.3 It has also been possible to conduct longitudinal studies to identify biomarkers and contribute to improve understanding of the peripheral biological changes accompanying conversion from prodromes to full-blown psychosis. Such studies have been instrumental in defining new stage-specific therapeutic strategies that could prevent or delay the onset of this severely disabling disorder. Notably, only one-third of UHR individuals convert to psychosis after 3 years of follow-up, but the reasons why they do are not yet understood.4 Though they could help answer these critical questions, biological investigations have been limited until now.
The molecular mechanisms that trigger illness progression are still largely unknown. Despite undeniable neurodevelopmental and genetic vulnerability, mutations and polymorphisms fail to account for the delayed onset of psychosis, suggesting a role for epigenetic regulation under the influence of environmental and/or maturational processes. We have recently reported longitudinal epigenetic changes in DNA methylation during psychotic conversion,5 suggesting alterations in gene regulation, simultaneous with psychosis onset. DNA methylation is known to regulate gene transcription and quantity of messenger RNA. Traditionally, hypermethylation in gene promoter has been associated with decreased expression, whereas hypomethylation has been associated with increased expression.6 Our hypothesis is that specific changes in gene (or gene network) expression accompany psychotic conversion. Whereas a dozen peripheral whole-transcriptome studies have been published about chronic schizophrenia (for a review see Lai et al.7), only one study—using a candidate gene approach—has analyzed gene expression in UHR individuals.8
Here, we explore the dynamics of longitudinal variations in gene expression during the onset of psychosis. We recruited a longitudinal cohort of individuals with 2 clinical and biological assessments over a 1-year follow-up. Changes in gene expression could be measured and compared across clinical groups with individuals as their own controls. First, we conducted an exploratory analysis using RNA sequencing of blood samples from 15 individuals: 12 converters vs. 3 clinical controls (comparable, help-seeking individuals who did not meet the UHR criteria) following this longitudinal paradigm. RNA sequencing enabled us to thoroughly explore differential gene expression and network preservation during psychotic conversion. Second, best candidate genes from the RNA sequencing exploration and from our previous methylomic study5 underwent a confirmatory study using quantitative-PCR (Q-PCR) in a larger sample—31 converters were compared with 63 non-converters.
Methods
Population
Subjects were selected from the French ICAAR cohort (PHRC AOM-07-118, promoted by Hôpital Sainte-Anne). The cohort was approved by the institutional ethics committee “Comité de protection des personnes, Ile-de-France III, Paris, France” and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki. Informed Consent was obtained from parents when individuals were under the age of 18. The ICAAR collaborative study9 included 16- to-30-year-old help-seeking individuals, who had been consecutively referred to the Adolescent and Young Adult Assessment Centre (Service Hospitalo-Universitaire, Hôpital Sainte-Anne, Paris, France) between 2009 and 2014. Inclusion criteria were alterations in global functioning (Social and Occupational Functioning Assessment Scale score <70) during the past year that were associated with psychiatric symptoms and/or subjective cognitive complaints. All help-seeking individuals were examined with the Comprehensive Assessment of at-risk mental state, CAARMS (Yung et al.,10 in its translated version, Krebs et al.11) by specifically trained psychiatrists followed by a consensus meeting for the best estimate diagnoses. Individuals meeting the CAARMS criteria for at-risk mental state (ultra-high risk, UHR) were included in the study. Exclusion criteria included conspicuous symptoms of psychosis (fulfilling DSM-IV criteria), pervasive developmental disorder, bipolar disorder, or other established diagnoses, such as obsessive-compulsive disorder, severe or nonstabilized somatic and neurological disorders, head injury and an IQ score below 70. Individuals were followed for 1 year at most and follow-up stopped either after a year or after conversion to psychosis. Each individual underwent clinical assessment and blood sampling at 2 time points: at baseline (M0) and at the end of follow-up (MF), that is, after 1 year in the non-converter group and immediately after conversion to psychosis in the converter group. This design enables intraindividual analyses, that is, to detect changes occurring in gene expression between MF and M0, covering conversion to psychosis. Psychotic conversion was characterized using the CAARMS-defined psychosis onset threshold (ie, supra-threshold psychotic symptoms—thought content, perceptual abnormalities and/or disorganized speech—present for more than 1 week as described in supplementary table S1). UHR individuals who reached the threshold during follow-up were considered converters, while UHR individuals who recovered or displayed persistent sub-threshold symptoms were considered non-converters. A description of Sample 1 (RNA sequencing study) and Sample 2 (Q-PCR study) is provided in Table 1. Converters did not differ greatly from non-converters. The only differences were clinical scores at the end of follow-up, as was expected, and follow-up duration because converters were reassessed just after conversion to limit the influence of medication. The rate of conversion was more important than the average rate described in the literature. In the present cohort, one-third of the subjects converted within a year, whereas one-third of the subjects converted within 3 years in a recent meta-analysis.4 This may arise from the built-in specificities of the referral source: an outpatient clinic, which had recently been opened with an adult hospital, with few outreach actions at the time).12 At final assessment, 52% of non-converters had lower symptom intensity, based on the Brief Psychiatric Rating Scale (longitudinal change of BPRS ≤ −10), 40% remained stable (−10 ˂ change in BPRS ˂ 10), and only 8% had slightly higher symptom intensity (change in BPRS ≥ 10; maximum increase by 27 in BPRS in 1 individual. yet not in positive symptoms. None of them/the subjects met the CAARMS-defined criteria for conversion to psychosis).
Table 1.
Clinical and Biological Description of the Samples (Mean ± SD)
| Variables | Sample 1 | Sample 2 | ||||
|---|---|---|---|---|---|---|
| Converters (n = 12) | Non-converters (n = 3) | P-value (Mann- Whitney / Fischer) | Converters (n = 31) |
Non-converters (n = 63) |
P-value | |
| Age | 20.3 ± 3.1 | 24.1 ± 4.1 | .18 | 20.1 ± 3.2 | 21.4 ± 3.7 | .14 |
| Sex ratio (male/female) |
11/1 | 3/0 | .80 | 21/10 | 35/28 | .18 |
| Tobacco (daily use) |
41.7% | 33.3% | .73 | 41.4% | 36.2% | .41 |
| Cannabis (recent use <30 d) |
50% | 33.3% | .55 | 55% | 33.3% | .11 |
| Treatment initiation (antipsychotics) | 50% | 0 | .19 | 61% | 60% | .56 |
| Follow-up (mo) | 11.2 ± 7.6 | 9.8 ± 3.8 | 1 | 10.6 ± 5.8 | 13.9 ± 3.9 | .001 |
| BPRS (initial) | 59.8 ± 13.6 | 44.7 ± 16.2 | .18 | 54.7 ± 12.4 | 50.9 ± 13.4 | .241 |
| BPRS (final) | 58.3 ± 14 | 31.7 ± 4.5 | .007 | 53.2 ± 12.5 | 41.3 ± 11.3 | <10 –3 |
| CGI (initial) | 4.8 ± 0.8 | 3.3 ± 2.1 | .30 | 4.4 ± 0.8 | 4.5 ± 0.9 | .542 |
| CGI (final) | 5.1 ± 0.5 | 2.0 ± 1.7 | .005 | 4.9 ± 0.7 | 3.7 ± 1.4 | <10 –3 |
| Quality of RNA (RIN) | 8.4 ± 0.4 | 8.3 ± 0.6 | .63 | 8.5 ± 0.5 | 8.4 ± 0.5 | .39 |
| Read counts | 95.2 × 106 ± 20.7 × 106 |
94.3 × 106 ± 17.7 × 106 |
.31 | — | — | — |
| Aligned reads % |
89.6 × 106 ± 18.5 × 106 (94.3% ± 1.8%) |
88.7 × 106 ± 16.8 × 106 (94.1% ± 2.3%) |
.31 | — | — | — |
Note: Sample 1 was used for RNAseq; sample 2 included sample 1 and additional subjects and was subjected to Q-PCR analysis. Clinical scales included BPRS (Brief Psychiatric Rating Scale) and CGI (Clinical Global Impressions Scale). Bold value corresponds to P-value <.05. RIN, RNA integrity number.
RNA Sequencing
RNA Extraction, Library, and Sequencing
Total RNA was extracted from blood samples (PAXgene tubes) using a standard protocol with a QIAcube robot and PAXgene Blood RNA kit (QIAGEN). Quality and quantity were checked with a BioAnalyzer (QUANT IT RNA kit—supplementary figure S1). Mean concentration was 59.4 ng/ml and mean RNA Integrity Number (RIN) was 8.4 with no significant difference across groups (table 1).
Total RNA was processed using the mRNA-Seq Sample Prep Kit (Illumina). Poly(A) RNA was isolated from the total RNA with a 2-step magnetic bead protocol. The resulting mRNA was fragmented in a buffer containing divalent cation at 94°C for 5 min. It was also purified by ethanol precipitation. The RNA was re-suspended in water, and a reverse transcription reaction was performed following the manufacturer’s instructions. Thirty libraries (15 individuals, 2 assessments) were prepared using the TruSeq Stranded mRNA kit.
Paired-end 75-bp sequencing runs were performed on the Illumina HiSeq 2000 with more than 80 million reads per sample. During sequencing, each incorporated base was color-coded with a fluorophore. The Illumina software RTA1.12.4.2/HCS1.4.8 converted this fluorophore information to sequence data and FASTA files were provided.
RNAseq Analysis
Quality control was performed using ShortRead package for R 3.3.2 (supplementary figure S2).13 FASTA files were aligned to the reference genome (hg19) using TopHat2 to generate BAM files.14 A matrix of read counts was then created using HTSeq.15 We used 2 pipelines from this matrix: DESeq16 and edgeR17 packages in R 3.3.2 environment according to the recommendations.18 Two pipelines were used to increase confidence in the overlapping results. The contrast matrix was an interaction between time and groups. Only the genes with a sum of counts ≥3 in the 30 samples were kept for further analysis (n = 16096 genes). The level of significance was established at 3.10–6. Due to the small number of genes below this threshold (3 genes with DESeq; 1 gene with edgeR; supplementary table S2), we selected additional genes for further consideration if they had reached top results in the 2 pipelines (nominal P-value < .01). All scripts are available upon request.
Co-expression Network Analysis in Converters
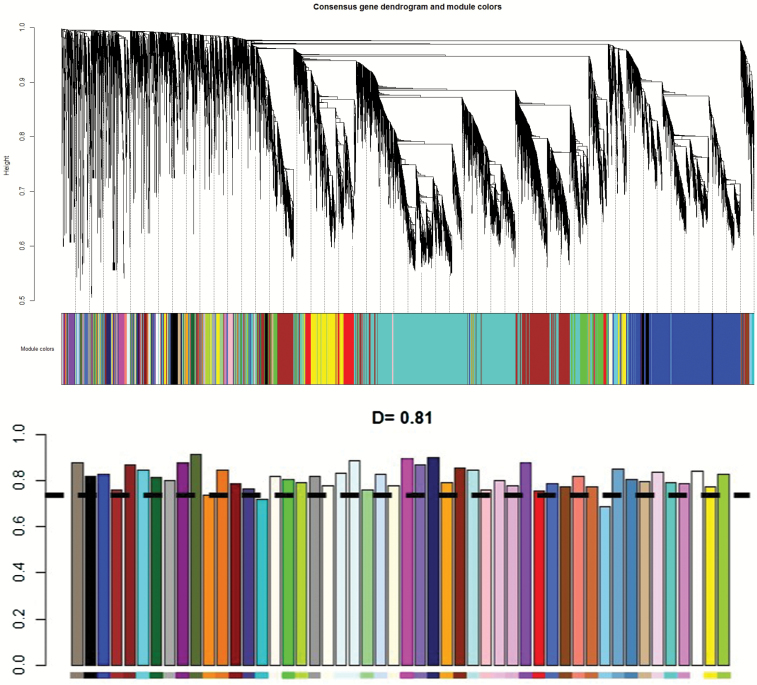
A co-expression network analysis was conducted only in converters (n = 12) with the WGCNA package19 in R 3.3.2 that used correlations between gene expressions to construct the networks. One outlier was identified by clustering and removed using a branch cut at the corresponding height (supplementary figure S3). An automatic, 1-step network construction was used to detect co-expression modules based on baseline data (M0). Then, preservation of the networks was tested after psychotic conversion (MF), and an empirical threshold was set at D = 0.75. The design is supposed to detect the gene networks that are dysregulated during psychotic conversion. A Gene Ontology enrichment analysis of all the genes in the networks was performed to determine the functionality of the modules using the function “GOenrichmentAnalysis” directly implemented in WGCNA.
Cell Count Estimation
Peripheral blood samples are composed of several types of cells. We investigated whether cell population was significantly different between the 2 groups (converters vs. non-converters). We used the CIBERSORT tool20 to assess the abundances of member cell types in the mixed cell population samples, using the gene expression data and the LM22 signature gene file (default file). Relative fractions of cells were computed and are displayed in supplementary figure S4. There was no significant difference in cell count changes between the 2 groups across time (supplementary table S3).
Q-PCR Validation
For confirmation in the whole cohort, we selected genes (1) with a nominal P-value <.01 in the 2 RNA-seq pipelines and (2) that are expressed in any part of the brain at any developmental time point, according to the GTEx database.21 We also selected strong candidate genes with longitudinal methylation changes during conversion to psychosis based on our previous findings.5
We extracted total RNA from PAXgene using the same protocol as we did with RNA sequencing. Quality control was done using LabChip GX (Perkin Elmer, Waltham, United States). Complementary DNA (cDNA) synthesis was performed using Reverse Transcription Master Mix from Fluidigm according to the manufacturer’s protocol with random primers in a final volume of 5 μl containing 100 ng total RNA using a Nexus thermocycler (Eppendorf). cDNA samples were diluted by adding 20 μl of low TE buffer [10 mM Tris; 0.1 mM EDTA; pH = 8.0 (TEKNOVA)]. TaqMan probes were selected for each gene (supplementary table S4a). For specific target pre-amplification, 1.25 μl of each diluted cDNA was used for multiplex pre-amplification with Fluidigm PreAmp Master Mix at 12 cycles. In a total volume of 5 μl, the reaction contained 1 μl of pre-amplification mastermix, 1 μl of PCR water, 1.25 μ of cDNA, 1.25 μ of pooled TaqMan Gene Expression assays (Life Technologies, ThermoFisher) with a final concentration of 180 nM. The cDNA samples were subjected to pre-amplification following the temperature protocol—2 min at 95°C, followed by 12 cycles of 15 s at 95°C and 4 min at 60°C. The pre-amplified cDNA was diluted 5× by adding 20 μl of low TE buffer (TEKNOVA). High-throughput real-time PCR was performed with the qPCR-HD-Genomic Paris Centre platform and was supported by grants from Région Ile-de-France, using the high-throughput BioMark HD System platform and the GE Dynamic Arrays (Fluidigm). Six microliters of sample master mix (SMM) consisted of 1.8 μl of 5× diluted preamplified cDNA, 0.3 μl of 20× GE Sample Loading Reagent (Fluidigm) and 3 μl of TaqMan Gene Expression PCR Master Mix (Life Technologies, ThermoFisher). Each 6 μl assay master mix (AMM) consisted of 3 μl of TaqMan Gene Expression assay 20× (Life Technologies) and 3 μl of 2× Assay Loading Reagent (Fluidigm). Five microliters of SMM and of AMM premixes were added to the dedicated wells. The samples and assays were mixed inside the chip using HX IFC controller (Fluidigm). Thermal conditions for qPCR were as follows: 30 min at 25°C and 60 min at 70°C for thermal mix; 2 min at 50°C and 10 min at 95°C for hot start; 40 cycles of 15 s at 95°C and 1 min at 60°C. Data were processed with an automatic threshold for each assay, with linear derivative baseline correction using BioMark Real-Time PCR Analysis Software 4.0.1 (Fluidigm). The quality threshold was unchanged at the 0.65 default setting. Normalization was conducted using a panel of housekeeping genes with different levels of expression (weak expression: PPIH, HPRT1; moderate expression: PGK1; RPLP0; strong expression: GAPDH, ACTB). The expression of these normalization genes did not differ significantly between the groups whether at M0 or at MF (supplementary table S4b). Livak normalization provided the expression level in each sample with a transformation using the 2ΔΔCT method.22 Scripts are available on GitHub (https://github.com/jpouch/qPCR-Biomark). One gene (GRID2) was expressed in too few individuals and was not analyzed further (supplementary table S4a). Statistical analysis was performed using R 3.3.2 and SPSS Statistics 20 (IBM). Intra-individual mean difference was computed for each gene and intergroup (converters vs. non-converters) comparisons of means were performed using the Wilcoxon test because we had no prior knowledge about data distribution. Secondly, we conducted Shapiro–Wilk tests to determine if the longitudinal changes in gene expression followed a normal distribution. All changes did, except for OSBP2 and GSTM5. Additionally, we used a parametric test (t test) for the other genes (supplementary table S5). Correlation between gene expression data and methylomic data was performed by nonparametric Spearman test due to the small amount of overlapping data.
Results
RNA sequencing analyses with DESeq and edgeR pipelines identified differential gene expressions across time between converters and non-converters. Only concordant results were retained (nominal P-value in each package < .01) resulting in 10 genes (supplementary table S2). Only brain-expressed genes were kept for further analysis, corresponding to 4 candidate genes (table 2). We wanted to test whether some networks were altered during conversion. Thus, a co-expression network analysis was performed using the WGCNA package19 in the converter group only between baseline and after psychotic conversion. This analysis found 50 gene expression networks with a mean preservation coefficient of 0.81 (figure 1; supplementary figure S5). In converters, 2 co-expression networks were less preserved than others (coefficient below 0.75), namely, the Wnt pathway and the Toll-like receptor pathway.
Table 2.
Summary of Findings from RNAseq and Multiplex Q-PCR
| Genes Found With RNAseq (Using edgeR Package) | Genes Found With RNAseq (Using DESeq Package) | Q-PCR Replication of Candidate Genes Identified With RNAseq | ||||
|---|---|---|---|---|---|---|
| Gene | P-value | logFC | P-value | logFC | P-value | logFC |
| CPT1A | .0009 | −0.52 | .0033 | −0.65 | .007 | −2.50 |
| GRID2 | .0074 | −3.93 | .0047 | −3.70 | — | — |
| OSBP2 | .0094 | 0.61 | .0006 | 0.21 | .117 | 0.14 |
| PEG3 | .0038 | −2.12 | .0014 | −4.73 | .112 | −1.00 |
| Candidate genes identified in previous methylomic study | Q-PCR replication of candidate genes identified in methylomic study | |||
|---|---|---|---|---|
| Gene | P-value | Average difference in β-value | P-value | logFC |
| AKT1 | 2.84 E-04 | 1.2% | .432 | −0.47 |
| CHL1 | 6.23 E-05 | 3.1% | .076 | −2.31 |
| GSTM5 | 6.8 E-04 | 14% | .042 | −2.48 |
| IL17RE | 1.37 E-04 | 3.2% | .482 | −1.05 |
| NRP1 | 5.49 E-05 | 2.0% | .026 | −12.6 |
Note: RNAseq, RNA sequencing; Q-PCR, quantitative polymerase chain reaction. “β-Value” referred to the level of methylation in specific CpGs (dinucleotides cytosine-guanine). LogFC refers to the logarithm of the difference between the longitudinal changes in converters and non-converters: logFC = log[(MFconverters − M0converters) − (MFnonconverters − MFnonconverters)]. Negative logFC means longitudinal decrease in gene expression in converters compared with non-converters, whereas positive logFC means longitudinal increase.
Fig. 1.
Graphical representation of network preservation during psychotic conversion using WGCNA (R package) from RNAseq data in converters only. The top figure shows the dendrogram, which corresponds to the graphical representation of constructed gene co-expression. Different gene networks are color-labeled. The bottom figure (histogram) shows the level of preservation of each gene network. The dash line represents the D = 0.75 cutoff (moderate preservation). Two networks are below cutoff (see supplementary figure S5 for more details).
In the following confirmatory study using quantitative-PCR (Q-PCR), 31 converters were compared to 63 non-converters. We selected 4 candidate genes from our RNA-seq analysis and 5 differentially methylated genes from our previously published article about the methylomic changes5 (table 2). Q-PCR identified 3 significantly differentially expressed genes: CPT1A (carnitine palmitoyltransferase 1A), GSTM5 (Glutathion-S-transferase Mu 5), and NRP1 (Neuropilin 1). CPT1A is involved in an essential step in the beta-oxidation of long chain fatty acids in the mitochondria. Its expression significantly increased in non-converters during follow-up (n = 63), whereas it decreased in converters during psychotic conversion (n = 31). NRP1 is part of the Semaphorin Receptor Complex involved in neuronal development and axon guidance. NRP1 interacts with CHL1, whose expression was also modified during psychotic conversion. The change in CHL1 expression only approached significance in the Wilcoxon test (P = .076; table 2) but was significantly different in the t test (supplementary table S3). GSTM5 is a member of the Glutathione-S-transferase family and is involved in the synthesis of glutathione and protection against oxidative stress, which seems to be part of the pathophysiology of schizophrenia.23 Unfortunately, Q-PCR was unable to determine the expression level of GRID2 that is poorly expressed in blood samples.
As the whole cohort also included individuals from the RNAseq analysis, we tested the longitudinal change of CPT1A after discarding overlapping individuals. The result remained significant (Wilcoxon P = .033; fold change = −2.15). This suggested that the effect was not only driven by the 15 overlapping samples. Expression of CPT1A in the RNAseq analysis and Q-PCR was significantly correlated (Spearman’s r = 0.734; P = 4.10–6) in the samples that underwent both techniques confirming the validity of our RNAseq analysis.
We examined whether the longitudinal reduction in GSTM5 and NRP1 expressions was associated with methylation status in the overlapping samples. The correlation was only tested between methylation regions and mRNA level previously identified as dysregulated (see supplementary text). A negative correlation was significant between methylation changes in the previously identified CpG located in NRP1 and longitudinal changes in corresponding gene expression (n = 36; r = −0.338; P = .044). Correlation between methylation in GSTM5 and gene expression in the overlapping samples (n = 45) is significant for 2 CpGs located in its promoter, namely, the CpG located in (hg19) chr1: 110254692 (r = −0.306; P = .041) and the CpG located in chr1: 110254720 (r = −0.337, P = .024). It was not significant for the remaining CpGs in the promoter. The methylation of the previously identified CpG located in CHL1 did not correlate with its expression (P = .187).
Antipsychotic treatment initiation is a potential confounding factor. We analyzed the RNAseq dataset to detect this factor using the same methodology as we did with conversion to psychosis. Two groups were contrasted: individuals who had received antipsychotic treatment (n = 6) and individuals who had been unexposed to antipsychotics (n = 24). Sixteen genes were identified with a nominal P-value <.01 in both edgeR and DESeq packages (supplementary table S6). Only one gene (HSPB7) overlapped with the list of differentially expressed genes during conversion to psychosis, but this gene was not retained in our final list because it is not expressed in the brain. Then, we conducted post hoc analyses of the qPCR data to assess whether medication affected the change in expression of the 3 confirmed genes (CPT1A; GSTM5; NRP1) in the whole cohort. We found no difference in their expression related to psychotropic medication changes across time (supplementary table S7).
Discussion
Our study explores the dynamics of expression changes in peripheral blood samples during the onset of psychosis. RNA sequencing was conducted using 2 pipelines. Only top concordant results from data-driven analyses were retained for further confirmation. Q-PCR was used in a larger sample of individuals to confirm the previous results from RNA sequencing and from previous methylomic results. This first transcriptomic study suggests that psychotic conversion is more specifically associated with differential expression of 3 genes.
First, we found that CPT1A gene under-expression is associated with conversion to psychosis. CPT1A is an enzyme required for long-chain fatty acid oxidation and transport into mitochondria.24 The expression of CPT1A could be related to (ω-3) polyunsaturated fatty acid intake,25 which may prevent psychotic conversion.26,27 The CPT gene family has already been implicated in schizophrenia.28 Moreover, in peripheral samples taken just after psychotic conversion in first-episode psychosis patients, CPT1A was reported hypoexpressed compared with controls29 (P = 7.86E-03; fold change = −1.11). Our results are consistent with these reports. Secondly, we found that NRP1 and GSTM5 expressions decreased in converters over time. These results are consistent and significantly correlate with our previous findings showing hypermethylation of the CpGs located in NRP1 and GSTM5 during conversion to psychosis. NRP1 encodes one of the two human neuropilins, which acts as a cellular receptor to detect guidance cues during neuronal migration30 and axon guidance.31 Neuropilin 1 mediates the response to chemorepulsive class 3 semaphorins32 and its depletion in mice is associated with fewer cortical interneurons.33NRP1 is a close interactor of the neural cell adhesion protein CHL1 gene (cell adhesion molecule L1-like), which codes for the L1CAM2 protein and has been shown to be differentially expressed in the cohort based on parametric testing, though not on nonparametric testing. The L1 family encompasses immunoglobulin-class recognition proteins that promote axon growth and migration in developing neurons.34 In preclinical models, a CHL1-deficit in adult mice impairs working memory,35 social behavior, and synaptic transmission.36 Genetic variants of the CHL1 gene have been found to be associated with schizophrenia.37–39 On the other hand, GSTM5 is involved in glutathione metabolism and in oxidative stress protection. Many studies have repeatedly suggested that oxidative stress might be related to the different stages of schizophrenia.40 GSTM5 is the most commonly expressed member of the GSTM gene family in the brain.41 Its involvement in dopamine metabolism has also been suggested.42 Moreover, lower expression has been reported in the prefrontal cortex of patients with schizophrenia.43
Concordance between methylomic and transcriptomic data was limited to a 22% overlap regarding dysregulated genes. This is not surprising given that the dysregulation of gene expression could be largely modulated by other epigenetic mechanisms, such as posttranslational histone modifications or miRNA regulation, which were not investigated in the study. Moreover, the methylome seemed less dynamic than the transcriptome; some methylomic changes may have occurred several months before conversion, whereas transcriptomic analysis may reflect more rapid changes.
Exploration of gene network preservation identified 2 networks that are dysregulated after psychotic conversion. The first network contained genes from the Wnt pathway including the candidate gene CPT1A and several semaphorins that are close interactors of NRP1 for axon guidance. This biological network has recently been shown to be downregulated in the peripheral blood mononuclear cell transcriptome of subjects with schizophrenia compared with controls.44 The second group corresponded to genes involved in the signaling cascade of Toll-like receptors, which have been involved in innate immunity abnormalities in schizophrenia.45 In addition, mice deficient in Toll-like receptors display “schizophrenia-like” features.46
Because of the tissue specificity of gene expression, modifications seen in peripheral samples only imperfectly reflect changes in brain structures. Genes are expressed differently in the brain, depending on brain region and cell layers. Nevertheless, a direct comparison of gene expressions in different tissues showed a sizeable overlap between blood and many parts of the brain, in particular the prefrontal cortex.47 This correlation is stronger in networks in relation with carbohydrate metabolism (CPT1A), axon guidance (NRP1), and Wnt pathway, reinforcing our corresponding findings. In contrast, the immune response (Toll-like receptors pathway) was not significantly correlated between blood and brain tissues. In previous reports, the estimation of cross-tissue correlation in expression levels was found to range from 0.25 to 0.64 and the blood-brain correlation was greater for genes highly expressed in both tissues.48 Filtering our results for gene expressed in the brain may have strengthened our findings. Alternatively, it has been suggested that network conservation across tissues is more limited.48 Nevertheless, our design remains relevant to identify peripheral biomarkers that could imperfectly mirror the brain processes of the disease.
At this point, it is unknown whether the observed transcriptomic changes play a causal role in the processes leading to psychosis, or if they simply attest to the onset of psychosis. However, the longitudinal changes we observed are unlikely to be due to peripheral cell composition modifications or psychotropic treatment adaptation, as these factors did not differ between groups and are not associated with our results. In our study, HSPB7 was the only gene that was identified as dysregulated both during psychotic conversion and after administration of antipsychotics. We also compared our findings with those identified using RNAseq in schizophrenic patients receiving antipsychotics. None of the dysregulated genes overlapped with our list.49 We cannot exclude that some environmental factors—which may or may not relate to psychosis onset—interfere with gene expression. Nevertheless, intrasubject comparison could minimize this impact. We should also acknowledge the limited sample size though it is one of the largest cohorts in this emerging field. Regarding the co-expression network analysis, the limited number of non-converters prevented us from constructing gene networks. Consequently, we cannot ascertain that the alteration of the 2 networks is specific to psychosis onset. Finally, some non-converters may convert to psychosis after follow-up though most of them (92%) had lower or stable symptom intensity before the second biological and clinical assessment.
In contrast with many studies in the field that conducted transversal comparisons between groups, we assessed the longitudinal transcriptomic changes with individuals as their own controls and replicated data-driven results using a candidate gene approach. This design enables to identify within-subject changes associated with conversion to psychosis. Replication in larger clinical cohorts with repeated blood samplings and brain expression data are needed to refine and confirm our observations and to determine their pathophysiological relevance. Even if analysis of schizophrenia pathogenesis in blood is inherently limited, this study underlines the potential interest of axon guidance, fatty-acid metabolism, redox metabolism, and Wnt pathway in conversion to psychosis. These results open new perspectives to identify peripheral staging biomarkers as well as new phase-specific and disease-modifying therapeutic strategies to prevent psychosis.
Supplementary Material
Acknowledgments
We would like to thank all the patients and parents who participated in the ICAAR study, as well as all the practitioners from the C’JAAD team (Centre d’Evaluation pour Jeunes Adultes et Adolescents), at the Service Hospitalo-Universitaire, Centre Hospitalier Sainte-Anne. Members of the ICAAR Study Group who contributed to this article are as follows: Trial Coordination: MO Krebs.—Coordinating team: Clinical: J Bourgin, M Plaze, M Kazes, G Martinez; Biology: O Kebir, B Chaumette; Brain imaging: M Plaze, O Gay; Cognition: I Amado, E Magaud—Data Management and Statistical Analysis: C Gaillard, B Chaumette—Centers: Coordinating Center, Paris Sainte-Anne Hospital/SHU: MO Krebs, M Plaze, E Magaud, M Kazes, C Mam-Lam-Fook, C Daban, O Gay, J Bourgin, G Martinez; S13: M-N Vacheron, A Viala; Addictology Center: X Laqueille, A Dervaux; S17 F Petitjean, O Canceil; S18 B Garnier, M Fishmann-Mathis; Paris Cochin Hospital Maison des Adolescents: B Gal, JP Benoit, MR Moro; Sainte-Anne Hospital Clinical evaluation and research Center staff (CERC). We would like to thank Pascale Eisenberger from the Centre de Langues in Université Paris Descartes for the English language editing. We also thank the URC Paris Centre Descartes (AP-HP), INSERM, and DRCI for regulatory and technical assistance. This work was supported by a grant by the French Government’s Agence Nationale pour la Recherche (ANR, 08-MNP-007), a grant by the French Ministry of Health’s Programme Hospitalier de Recherche Clinique (PHRC, AOM-07-118), and by ANR-13-SAMA-0010 (SAMENTA 2013—Projet CERBAIS). The Centre Hospitalier Sainte-Anne promoted the study. Additional financial support was obtained from the Institut National de la Santé et de la Recherche Médicale (INSERM), Université Paris Descartes (recurrent funding). The sponsors had no role in the design and conduct of the study, in the collection, management, analysis or interpretation of the data, in the preparation, review or approval of the manuscript, or in the decision to submit the manuscript for publication. All the authors declare they have no conflict of interest.
References
- 1. Nelson B, McGorry PD, Wichers M, Wigman JTW, Hartmann JA. Moving from static to dynamic models of the onset of mental disorder: a review. JAMA Psychiatry. 2017;74:528–534. doi:10.1001/jamapsychiatry.2017.0001 [DOI] [PubMed] [Google Scholar]
- 2. McGorry PD. Early clinical phenotypes, clinical staging, and strategic biomarker research: building blocks for personalized psychiatry. Biol Psychiatry. 2013;74:394–395. [DOI] [PubMed] [Google Scholar]
- 3. Correll CU, Hauser M, Auther AM, Cornblatt BA. Research in people with psychosis risk syndrome: a review of the current evidence and future directions. J Child Psychol Psychiatry. 2010;51:390–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fusar-Poli P, Borgwardt S, Bechdolf A, et al. . The psychosis high-risk state: a comprehensive state-of-the-art review. JAMA Psychiatry. 2013;70:107–120. doi:10.1001/jamapsychiatry.2013.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kebir O, Chaumette B, Rivollier F, et al. . Methylomic changes during conversion to psychosis. Mol Psychiatry. 2017;22:512–518. doi:10.1038/mp.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. [DOI] [PubMed] [Google Scholar]
- 7. Lai CY, Scarr E, Udawela M, Everall I, Chen WJ, Dean B. Biomarkers in schizophrenia: a focus on blood based diagnostics and theranostics. World J Psychiatry. 2016;6:102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Santoro ML, Gadelha A, Ota VK, et al. . Gene expression analysis in blood of ultra-high risk subjects compared to first-episode of psychosis patients and controls. World J Biol Psychiatry Off J World Fed Soc Biol Psychiatry. 2015;18:1–6. doi:10.3109/15622975.2015.1048724. [DOI] [PubMed] [Google Scholar]
- 9. Oppetit A, Bourgin J, Martinez G, et al. . The C’JAAD: a French team for early intervention in psychosis in Paris. Early Interv Psychiatry. September 28, 2016; doi:10.1111/eip.12376. [DOI] [PubMed] [Google Scholar]
- 10. Yung AR, Yuen HP, McGorry PD, Phillips LJ, Kelly D, Dell’Olio M, et al. . Mapping the onset of psychosis: the comprehensive assessment of at-risk mental states. Aust N Z J Psychiatry. 2005;39:964–971. doi:10.1111/j.1440-1614.2005.01714.x. [DOI] [PubMed] [Google Scholar]
- 11. Krebs MO, Magaud E, Willard D, et al. . [Assessment of mental states at risk of psychotic transition: validation of the French version of the CAARMS]. Encephale. 2014;40:447–456. [DOI] [PubMed] [Google Scholar]
- 12. Fusar-Poli P, Rutigliano G, Stahl D, et al. . Deconstructing pretest risk enrichment to optimize prediction of psychosis in individuals at clinical high risk. JAMA Psychiatry. 2016;73:1260–1267. [DOI] [PubMed] [Google Scholar]
- 13. Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anders S, McCarthy DJ, Chen Y, et al. . Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8:1765–1786. [DOI] [PubMed] [Google Scholar]
- 19. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newman AM, Liu CL, Green MR, et al. . Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi:10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 23. Do KQ, Cabungcal JH, Frank A, Steullet P, Cuenod M. Redox dysregulation, neurodevelopment, and schizophrenia. Curr Opin Neurobiol. 2009;19:220–230. [DOI] [PubMed] [Google Scholar]
- 24. Bonnefont J-P, Djouadi F, Prip-Buus C, Gobin S, Munnich A, Bastin J. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol Aspects Med. 2004;25:495–520. doi:10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 25. Radler U, Stangl H, Lechner S, et al. . A combination of (ω-3) polyunsaturated fatty acids, polyphenols and L-carnitine reduces the plasma lipid levels and increases the expression of genes involved in fatty acid oxidation in human peripheral blood mononuclear cells and HepG2 cells. Ann Nutr Metab. 2011;58:133–140. [DOI] [PubMed] [Google Scholar]
- 26. Amminger GP, Schäfer MR, Papageorgiou K, et al. . Long-chain omega-3 fatty acids for indicated prevention of psychotic disorders: a randomized, placebo-controlled trial. Arch Gen Psychiatry. 2010;67:146–154. [DOI] [PubMed] [Google Scholar]
- 27. Amminger GP, Schäfer MR, Schlögelhofer M, Klier CM, McGorry PD. Longer-term outcome in the prevention of psychotic disorders by the Vienna omega-3 study. Nat Commun. 2015;6:7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Virmani A, Pinto L, Bauermann O, et al. . The carnitine palmitoyl transferase (CPT) system and possible relevance for neuropsychiatric and neurological conditions. Mol Neurobiol. 2015;52:826–836. [DOI] [PubMed] [Google Scholar]
- 29. Lee J, Goh LK, Chen G, Verma S, Tan CH, Lee TS. Analysis of blood-based gene expression signature in first-episode psychosis. Psychiatry Res. 2012;200:52–54. [DOI] [PubMed] [Google Scholar]
- 30. Hernández-Miranda LR, Cariboni A, Faux C, et al. . Robo1 regulates semaphorin signaling to guide the migration of cortical interneurons through the ventral forebrain. J Neurosci. 2011;31:6174–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Telley L, Cadilhac C, Cioni J-M, et al. . Dual function of NRP1 in axon guidance and subcellular target recognition in cerebellum. Neuron. 2016;91:1276–1291. doi:10.1016/j.neuron.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 32. He Z, Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell. 1997;90:739–751. [DOI] [PubMed] [Google Scholar]
- 33. Andrews WD, Barber M, Nemitz M, Memi F, Parnavelas JG. Semaphorin3A-neuropilin1 signalling is involved in the generation of cortical interneurons. Brain Struct Funct. 2017;222:2217–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. [DOI] [PubMed] [Google Scholar]
- 35. Kolata S, Wu J, Light K, Schachner M, Matzel LD. Impaired working memory duration but normal learning abilities found in mice that are conditionally deficient in the close homolog of L1. J Neurosci. 2008;28:13505–13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morellini F, Lepsveridze E, Kähler B, Dityatev A, Schachner M. Reduced reactivity to novelty, impaired social behavior, and enhanced basal synaptic excitatory activity in perforant path projections to the dentate gyrus in young adult mice deficient in the neural cell adhesion molecule CHL1. Mol Cell Neurosci. 2007;34:121–136. [DOI] [PubMed] [Google Scholar]
- 37. Chen QY, Chen Q, Feng GY, et al. . Case-control association study of the close homologue of L1 (CHL1) gene and schizophrenia in the Chinese population. Schizophr Res. 2005;73:269–274. [DOI] [PubMed] [Google Scholar]
- 38. Sakurai K, Migita O, Toru M, Arinami T. An association between a missense polymorphism in the close homologue of L1 (CHL1, CALL) gene and schizophrenia. Mol Psychiatry. 2002;7:412–415. [DOI] [PubMed] [Google Scholar]
- 39. Tam GWC, van de Lagemaat LN, Redon R, et al. . Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans. 2010;38:445–451. doi:10.1042/BST0380445. [DOI] [PubMed] [Google Scholar]
- 40. Koga M, Serritella AV, Sawa A, Sedlak TW. Implications for reactive oxygen species in schizophrenia pathogenesis. Schizophr Res. 2016;176:52–71. [DOI] [PubMed] [Google Scholar]
- 41. Knight TR, Choudhuri S, Klaassen CD. Constitutive mRNA expression of various glutathione S-transferase isoforms in different tissues of mice. Toxicol Sci. 2007;100:513–524. [DOI] [PubMed] [Google Scholar]
- 42. Hayes KR, Young BM, Pletcher MT. Expression quantitative trait loci mapping identifies new genetic models of glutathione S-transferase variation. Drug Metab Dispos. 2009;37:1269–1276. [DOI] [PubMed] [Google Scholar]
- 43. Gawryluk JW, Wang JF, Andreazza AC, Shao L, Yatham LN, Young LT. Prefrontal cortex glutathione S-transferase levels in patients with bipolar disorder, major depression and schizophrenia. Int J Neuropsychopharmacol. 2011;14:1069–1074. [DOI] [PubMed] [Google Scholar]
- 44. Wu JQ, Green MJ, Gardiner EJ, et al. . Altered neural signaling and immune pathways in peripheral blood mononuclear cells of schizophrenia patients with cognitive impairment: A transcriptome analysis. Brain Behav Immun. 2016;53:194–206. [DOI] [PubMed] [Google Scholar]
- 45. García Bueno B, Caso JR, Madrigal JL, Leza JC. Innate immune receptor Toll-like receptor 4 signalling in neuropsychiatric diseases. Neurosci Biobehav Rev. 2016;64:134–147. [DOI] [PubMed] [Google Scholar]
- 46. Park SJ, Lee JY, Kim SJ, Choi S-Y, Yune TY, Ryu JH. Toll-like receptor-2 deficiency induces schizophrenia-like behaviors in mice. Sci Rep. 2015;5:8502. doi:10.1038/srep08502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet Part B Neuropsychiatr Genet Off Publ Int Soc Psychiatr Genet. 2006;141B:261–268. doi:10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 48. Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:595–603. [DOI] [PubMed] [Google Scholar]
- 49. Crespo-Facorro B, Prieto C, Sainz J. Schizophrenia gene expression profile reverted to normal levels by antipsychotics. Int J Neuropsychopharmacol. 2015;18. doi:10.1093/ijnp/pyu066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.