Abstract
Genetic factors are known to influence both risk for schizophrenia (SZ) and variation in brain structure. A pressing question is whether the genetic underpinnings of brain phenotype and the disorder overlap. Using multivariate analytic methods and focusing on 1,402 common single-nucleotide polymorphisms (SNPs) mapped from the Psychiatric Genomics Consortium (PGC) 108 regions, in 777 discovery samples, we identified 39 SNPs to be significantly associated with SZ-discriminating gray matter volume (GMV) reduction in inferior parietal and superior temporal regions. The findings were replicated in 609 independent samples. These 39 SNPs in chr6:28308034-28684183 (6p22.1), the most significant SZ-risk region reported by PGC, showed regulatory effects on both DNA methylation and gene expression of postmortem brain tissue and saliva. Furthermore, the regulated methylation site and gene showed significantly different levels of methylation and expression in the prefrontal cortex between cases and controls. In addition, for one regulated methylation site we observed a significant in vivo methylation-GMV association in saliva, suggesting a potential SNP-methylation-GMV pathway. Notably, the risk alleles inferred for GMV reduction from in vivo imaging are all consistent with the risk alleles for SZ inferred from postmortem data. Collectively, we provide evidence for shared genetic risk of SZ and regional GMV reduction in 6p22.1 and demonstrate potential molecular mechanisms that may drive the observed in vivo associations. This study motivates dissecting SZ-risk variants to better understand their associations with focal brain phenotypes and the complex pathophysiology of the illness.
Keywords: PGC, SNP, gray matter volume, angular gyrus, supramarginal gyrus, ICA
Introduction
Schizophrenia (SZ) is a prevalent psychiatric disorder whose pathophysiology remains elusive.1,2 Family and twin studies estimate as much as 80% heritability for SZ, implicating a prominent genetic component in its etiology.1,3 Recent genome-wide association studies (GWAS) provide evidence for a polygenic model where a large number of variants with generally small effect sizes contribute to SZ liability,4,5 and 23% of the variance in this liability might be attributed to common single-nucleotide polymorphisms (SNPs).6 Meanwhile as a brain disease, SZ is associated with alterations in brain structure and function measures, including reduced whole brain and regional gray matter volume (GMV), especially in frontal and temporal cortices, disrupted prefrontal activation in cognitive tasks, as well as disrupted connectivity between brain networks.7,8 These neurobiological traits have also been found to be under genetic influence. Estimated heritability ranges from 0.42 for default-mode functional connectivity,9 to 0.68 for GMV in superior temporal gyrus (STG),10 or higher than 0.80 for volume of the left putamen.11
This raises the question of whether the genetic profiles overlap between SZ and brain phenotypes. A recent study by Franke et al.12 leveraged 2 large-scale GWAS results to explore shared genetic effects on SZ and subcortical brain volumes. Their findings suggest a lack of notable genetic overlap between the occurrence of the disorder and variation in subcortical volumes, at single variant or overall common variants levels. Considering that SZ is a complex polygenic disorder with high heterogeneity, the possibility is expected to be low for all diagnosis-related variants (as identified by GWAS) to converge their effects on a focal brain phenotype. In contrast, given the neurobiological nature of SZ, pleiotropic effects from single variants are more likely to occur.13 The lack of a shared effect at single variant level in Franke et al. might be in part attributable to insufficient statistical power for the brain phenotypes, as noted by the authors.
In light of the observations of Franke et al., we sought to extend this line of research on shared genetic profiles in two directions. First, a further dissection of SZ-risk SNPs might lead to subsets that contribute homogeneously to the variability of focal brain measures. Second, we utilized a multivariate approach, which might be better positioned for capturing moderate shared risks between SZ and brain phenotypes given the sample sizes commonly available in the field. Specifically, we conducted an independent component analysis (ICA)-based analysis14 on SNP and GMV data from 1,386 individuals. For each modality, subsets of variables with covarying patterns were first extracted. Then intermodality associations were assessed based on the multivariate profiles of individual subsets.
Materials and Methods
Participants
A total of 1,386 individuals aggregated from multiple cohorts were employed for this study for discovery and replication analyses. Details regarding data collection and previous publications describing recruitment are listed in table S1. The institutional review board at each site approved the study and all participants provided written informed consents. Each dataset was shared by the individual research group according to their protocol. The discovery sample consisted of 355 SZ patients and 422 controls from cohorts not part of Psychiatric Genomics Consortium (PGC).5 Meanwhile, an aggregated dataset of 294 cases (with 52 schizoaffective disorder [SAD] patients) and 315 controls was borrowed for replication. Diagnosis of SZ or SAD was confirmed using the Structured Clinical Interview for Diagnosis for DSM-IV or DSM-IV-TR. Table 1 provides the cohort-wise demographics.
Table 1.
Demographic Information
| Study | Sample Size | Sites | AFR/AMR/ EUR | Patients | Controls | ||||
|---|---|---|---|---|---|---|---|---|---|
| M/F | Age (mean ± SD) | Age (Min-Max) | M/F | Age (mean ± SD) | Age (Min-Max) | ||||
| Discovery | 777 | 14 | 94/175/508 | 284/71 | 35.18 ± 12.30 | 17–64 | 272/150 | 34.16 ± 12.18 | 16–65 |
| MCIC | 202 | 4 | 17/33/152 | 64/24 | 33.85 ± 10.55 | 18–59 | 69/45 | 32.23 ± 10.83 | 18–58 |
| COBRE | 189 | 1 | 14/81/94 | 77/14 | 37.20 ± 14.24 | 18–64 | 70/28 | 35.88 ± 12.19 | 17–65 |
| FBIRN3 | 172 | 7 | 0/49/123 | 61/12 | 38.70 ± 10.92 | 18–60 | 69/30 | 37.52 ± 11.24 | 19–60 |
| NW | 123 | 1 | 47/0/76 | 49/15 | 32.77 ± 12.68 | 17–61 | 33/26 | 32.78 ± 13.97 | 16–65 |
| OLIN | 91 | 1 | 16/12/63 | 33/6 | 30.59 ± 10.68 | 17–56 | 31/21 | 30.37 ± 12.81 | 16–64 |
| Replication | 609 | 7 | 87/25/497 | 193/101 | 36.81 ± 10.81 | 18–62 | 169/146 | 36.70 ± 10.43 | 18–60 |
| BSNIP | 220 | 5 | 87/25/108 | 88/54 | 35.18 ± 12.30 | 18–62 | 33/45 | 37.91 ± 12.29 | 18–60 |
| TOP | 229 | 1 | 0/0/229 | 45/23 | 33.71 ± 7.75 | 19–54 | 88/73 | 33.95 ± 8.82 | 18–55 |
| HUBIN | 160 | 1 | 0/0/160 | 60/24 | 42.07 ± 7.33 | 24–56 | 48/28 | 41.27 ± 9.78 | 19–56 |
Note: AFR, AMR, and EUR are codes of super populations following 1000 Genomes Project.
Genetic Data
DNA samples drawn from blood or saliva were genotyped with different platforms (see table S1). No significant difference was observed in genotyping call rates between blood and saliva samples. Details regarding genetic preprocessing are provided in Supplemental Information (SI). In brief, a standard preimputation quality control (QC)15 was performed using PLINK.16 In the imputation, SHAPEIT was used for prephasing,17 IMPUTE2 for imputation,18 and the 1,000 Genomes data as the reference panel.19 Only markers with high imputation qualities (INFO score > 0.95) were retained. The standard postimputation QC was done separately for discovery and replication data to avoid losing important SNPs due to platform inconsistency. For discovery, linkage disequilibrium (LD) pruning (r2 > 0.9) was applied and 977,242 SNPs were retained with population structure corrected using principal component analysis.20 For replication, after the same QC without LD pruning, 687,675 out of 977,242 discovery SNPs were available in the replication data, yielding an overlapping rate of 70.37%. By focusing on SNPs residing in the PGC 108 regions and showing relatively strong group differences (P < 1.00 × 10–4) in the PGC report,5 1,402 common SNPs were included for association analyses in discovery, out of which 973 SNPs were available in the replication dataset.
sMRI Data
Whole-brain T1-weighted images were collected with 1.5T and 3T scanners of various models, as summarized in table S1. The discovery images were preprocessed using a standard Statistical Parametric Mapping 12 (SPM12, http://www.fil.ion.ucl.ac.uk/spm) voxel-based morphometry pipeline,21–24 a unified model where image registration, bias correction, and tissue classification are integrated. The resulting modulated images were resliced to 1.5 mm × 1.5 mm × 1.5 mm and smoothed by 6 mm full width at half-maximum Gaussian kernel. We excluded 18 outlier subjects being distant (>3SD) from the average GMV image across all the subjects. A mask (average GMV > 0.2) was applied to include 429,655 voxels. Finally, voxel-wise regression was conducted to eliminate the effects from age, sex, and dummy-coded site covariates.23 While all the scanning parameters (table S1) would yield 93 dummy variables in the discovery data, we chose to correct scanning effects by “site” before association analysis to avoid eliminating too much information due to unknown collinearity. The effects of specific scanning parameters were assessed in the post hoc analysis. See SI for more details. The replication images were preprocessed using the same pipeline.
Multivariate Imaging Genetic Association Analysis
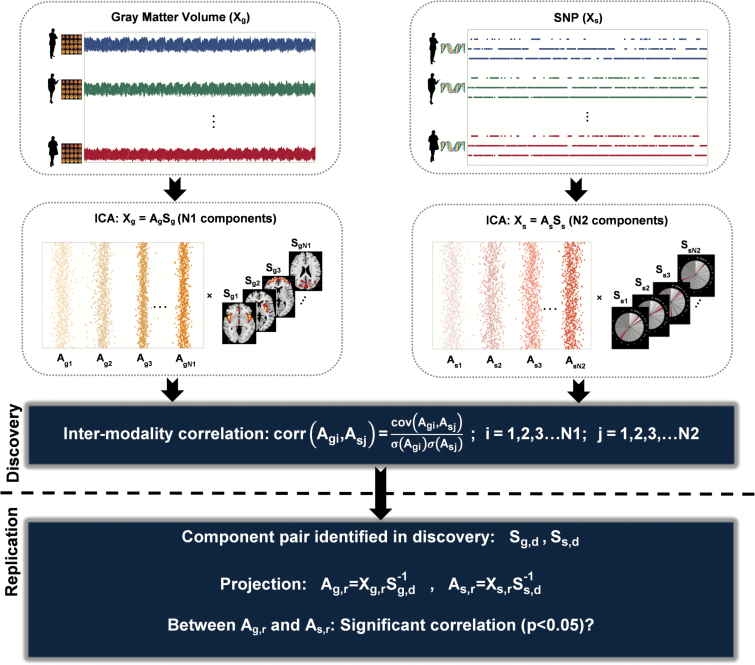
Parallel independent component analysis (pICA)25 (implemented in Fusion ICA Toolbox, http://mialab.mrn.org/software/fit), an analytical method that has been successfully applied to imaging and SNP association analysis,14,26 was used to identify multivariate SNP associations with GMV variation in 777 discovery samples. As shown in figure 1, the SNP and GMV data (Xs and Xg) are separately decomposed into linear combinations of independent components (Ss and Sg) using Infomax ICA.27,28 Then SNP-GMV correlations are evaluated and optimized based on components’ loadings (As and Ag). ICA aggregates variables into components by their contribution to each independent distribution pattern. A component’s loading (a column of A) largely reflects the covariation pattern of the top contributing variables that have high scores in this specific component (a row of S). Loadings (A) are used for assessing intermodality associations, while the conjunct components are used to locate the top contributing variables (ie, voxels or SNPs). ICA has been widely shown to capture consistent and meaningful covarying composite brain regions in structural images.23,29,30 ICA application to SNP data has also been validated,31–33 capturing covariation beyond LD and yielding meaningful biological interpretation.15,34 In ICA, a set of SNPs in LD has a similar chance of being admitted into one component as one single SNP after LD pruning, allowing us to use light LD pruning without overrepresentation in the component level yet avoid missing potential true causal loci. More mathematical details of pICA can be found in the study of Liu et al.25 In this study, the number of components was estimated to be 65 for GMV and 29 for SNP using the minimum description length criterion in discovery.35 The SNP-GMV associations yielded by pICA were reassessed while controlling for age, sex, race, diagnosis, intracranial volume, DNA source, genotyping array and dummy-coded scanning parameters (see SI). Significant associations were Bonferroni corrected for independent component pairs.
Fig. 1.
Graphical presentation of the association analysis framework.
The identified SNP-GMV associations were then evaluated for validity. The primary evaluation with the replication samples used the projection method. As shown in figure 1, for each SNP-GMV pair identified in discovery, the conjunct components (Ss,d and Sg,d) were projected to the replication data Xs,r and Xg,r, yielding the projected loadings As,r = Xs,rSs,d−1 and Ag,r=Xg,rSg,d−1. The discovery SNP-GMV association was considered replicated if a significant association (P < .05) could still be observed between the projected loadings. Note that the projected loadings were computed based on 427,329 overlapping voxels (out of 429,655) and 973 overlapping SNPs (out of 1,402) between discovery and replication. In addition, we also investigated if a pICA analysis on the combined discovery and replication data (1,386 samples, 973 common SNPs, and 427,329 common voxels) would yield a similar pair of SNP and GMV components whose loadings also show a significant association (P < .05).
Analyses on the Identified GMV Component
For the SNP-GMV pairs identified by pICA, the GMV loadings in discovery (extracted by pICA) and replication (projected) were evaluated for group differences while controlling for age and sex. Then we normalized each conjunct component and selected top voxels using the threshold of |z-score| > 2. These voxels were mapped to the Talairach atlas36 for involved brain regions. The GMV loadings were further assessed for associations with cognitive test scores, symptom scores, and equivalent current chlorpromazine dosages in discovery (see SI for calculation) using linear regression adjusted for age, sex, and diagnosis. For cognitive tests, we examined separately the MCIC and COBRE subcohorts for which cognitive data were available however could not be combined (table S3). For the symptom scores, most subcohorts collected PANSS,37 while MCIC and NW collected SAPS/SANS38,39; the latter was converted to PANSS40 and a dummy-coded covariate was further included in the regression to control for the difference. False discover rate correction was used for related cognitive or symptom measures.
Analyses on the Identified SNP Component
For the SNP modality, we first investigated the identified component loadings for group differences using 2-sample t test. Then we normalized each conjunct component and selected top SNPs using the threshold of |z-score| > 2. To explore potential mechanisms of functional impact, we conducted the following analyses to examine these top SNPs for regulatory effects on DNA methylation (DNAm) and gene expression: (1) We located cis-methylation quantitative trait loci (mQTLs) and the targeted methylation sites (distance < 500 Kb) in the top SNPs based on the study of Hannon et al.,41 which investigated mQTLs in fetal and adult postmortem brain samples; (2) The target methylation sites were examined for group differences in DNAm levels of dorsolateral prefrontal cortex (DLPFC) between 184 cases and 230 controls (age ≥ 16) using a dataset contributed by the Lieber Institute (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74193, Lieber’s data)42; (3) In a subcohort of 180 COBRE samples (94 controls and 86 cases) where DNAm in saliva was measured,24 we investigated whether any top SNP presented as mQTL in both brain (Hannon’s data) and saliva (COBRE data) and whether the target methylation site associated with the identified GMV component’s loading to form an SNP-methylation-GMV pathway; (4) We leveraged the Genotype-Tissue Expression (GTEx) Project to locate prefrontal cortex (Brodmann Area BA9) cis-expression quantitative trait loci (eQTLs) and the targeted genes (distance < 500 Kb) in our top SNPs43; (5) We examined the target genes for group differences in expression of prefrontal cortex (BA10) between 28 cases and 23 controls in a dataset contributed by GlaxoSmithKline (GSK’s data, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17612).44 See SI for more details on these tests.
Additional Assessment of the pICA Finding
We further conducted the following tests to assess the validity of the SNP-GMV association: (1) whether the SNP and GMV components were affected by the specific component numbers used in ICA; (2) whether the SNP component was affected by the preselection P-value threshold; (3) whether the populations African (AFR), Mixed-American (AMR), and European (EUR) presented comparable SNP-GMV associations; (4) whether the SNP-GMV association remained to be identified in 508 EUR samples within a range of LD pruning (r2 thresholds: 0.2–1.0); (5) whether a different approach, sparse partial least squares (sPLS)45 might capture a similar multivariate genetic pattern to that identified by ICA.
Univariate and Polygenic Risk Score Analyses
The following series of tests were conducted to compare with pICA: (1) univariate association between SNP and voxel; (2) association between individual SNP and GMV component; (3) association between polygenic risk score (PGRS) for SZ of each of PGC 108 regions4,5 and individual voxel in EUR samples; and (4) association between PGRS and GMV component in EUR samples. See SI for more details.
RESULTS
Multivariate Analysis
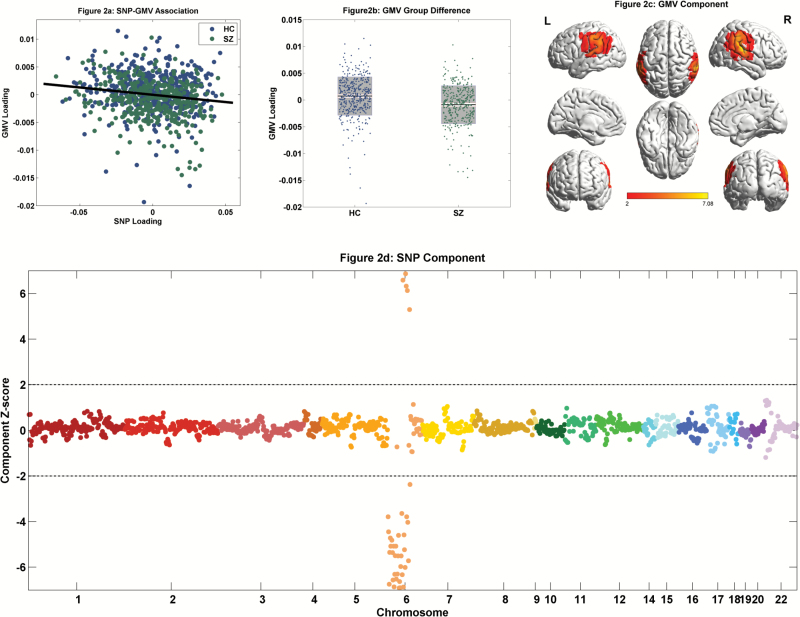
In 777 discovery samples, pICA identified one significantly associated SNP-GMV pair when controlling for confounders of age, sex, race, diagnosis, intracranial volume, DNA source, genotyping array, and dummy-coded scanning parameters (r = −0.16, P = 6.79 × 10–6, figure 2a), passing Bonferroni correction for 1,885 independent SNP-GMV pairs. No significant interaction effect on GMV was noted between diagnosis and SNP. This SNP-GMV association was replicated in 609 independent samples based on the projected loadings (r = −0.08, P = 3.97 × 10–2, controlling for the same confounders). When applying pICA to the combined discovery and replication data, we still observed a highly similar SNP-GMV pair with a significant association (r = −0.11, P = 2.54 × 10–5, see SI for details). The main finding was robust to SNP and GMV component numbers and SNP preselection P-value threshold; showed consistent associations in AFR, AMR, and EUR populations; and largely held in EUR samples with SNPs pruned from 0.2 to 1.0. Particularly when sPLS was used to identify SNP-GMV associations in a nested cross-validation framework, the resulting multivariate genetic pattern highly concurred with the main finding where the sPLS latent variable showed a correlation of 0.95 with the pICA component’s loading. See SI for details.
Fig. 2.
Imaging genetic findings from 777 discovery samples: (a) Scatter plot of the SNP and GMV loadings (r = −0.16, P = 6.79 × 10–6); (b) Group difference of the GMV loading (P = 2.10 × 10–8). The black line reflects mean, the white and gray patches reflect SEM and SD, respectively. (c) Spatial map of the GMV component thresholded at |z-score| > 2; (d) Manhattan plot of the SNP component with the dashed line representing |z-score| = 2.
GMV Component
The identified GMV component loading was significantly lower in cases than controls (P = 2.10 × 10–8, figure 2b). Thresholded at |z-score| > 2, the highlighted regions included inferior parietal lobe (IPL), posterior STG, postcentral and precentral gyri (figure 2c, see table S2 for the Talairach atlas). Collectively, the imaging component presented significant GMV reduction in SZ patients in the aforementioned regions. Furthermore, this SZ-discriminating GMV reduction was replicated in 609 independent samples (P = 4.77 × 10–4). No significant association was observed for current chlorpromazine-equivalent dosages in 203 patients with data available. Meanwhile, the GMV loading significantly negatively associated with PANSS negative score (r = −0.20, P = 2.76 × 10–3) in 239 patients. Regarding cognition, in MCIC subcohort, the GMV loading significantly positively associated with WAIS Block-Design-Total-Score and CalCAP Choice-Reaction-Time Serial-Pattern-Matching (CRT SEQ1) True-Positive (accuracy measure). In COBRE subcohort, significant positive associations were noted for MATRICS domains of Processing-Speed, Attention-Vigilance, and Visual-Learning, as summarized in table S3.
SNP Component
The identified SNP component did not show a significant group difference. Thresholding at |z-score| > 2 yielded 39 top SNPs residing in chr6:28308034-28684183 (6p22.1), as presented in figure 2d and table 2. These SNPs were in LD, with the mean of pairwise correlations being 0.54. Given the negative SNP-GMV association, a positive/negative component z-score indicated the specific allele relating to lower/higher regional GMV. No significant correlation was noted between the top SNPs and those close to complement component 4 genes.46
Table 2.
Top SNPs Derived from the Association Analysis, With rsID, Chromosome, Base Pair Position, Reference Allele in the Local Data, Component z-Score, and Gene Annotation Listed
| ID | Chr | Posi | Allele | z-Score | Gene Annotation |
|---|---|---|---|---|---|
| rs17301128 | 6p22.1 | 28308034 | G | −3.79 | — |
| rs2108926 | 6p22.1 | 28308747 | T | −4.44 | — |
| rs213240 | 6p22.1 | 28315875 | C | −6.75 | — |
| rs6942030 | 6p22.1 | 28315958 | T | −5.35 | — |
| rs9468350 | 6p22.1 | 28319107 | G | −4.72 | ZKSCAN3 |
| rs6903652 | 6p22.1 | 28322120 | G | −5.08 | ZKSCAN3 |
| rs213236 | 6p22.1 | 28324397 | C | −7.11 | ZKSCAN3 |
| rs6921919 | 6p22.1 | 28325201 | G | −4.81 | ZKSCAN3 |
| rs213230 | 6p22.1 | 28330264 | G | −5.35 | ZKSCAN3 |
| rs213228 | 6p22.1 | 28331252 | C | −6.56 | ZKSCAN3 |
| rs9468354 | 6p22.1 | 28337801 | A | −5.07 | — |
| rs10946954 | 6p22.1 | 28340625 | C | −6.86 | — |
| rs9461456 | 6p22.1 | 28343816 | G | −6.30 | — |
| rs7754960 | 6p22.1 | 28346945 | C | −5.50 | ZSCAN12 |
| rs9468365 | 6p22.1 | 28357966 | T | −5.07 | ZSCAN12 |
| rs2859348 | 6p22.1 | 28359170 | G | −6.52 | ZSCAN12 |
| rs4580862 | 6p22.1 | 28367663 | C | −6.30 | — |
| rs13196606 | 6p22.1 | 28370078 | A | −5.51 | — |
| rs71559082 | 6p22.1 | 28372192 | T | −4.60 | — |
| rs2531827 | 6p22.1 | 28373154 | C | −6.91 | — |
| rs1558205 | 6p22.1 | 28382262 | A | −6.62 | — |
| rs2531832 | 6p22.1 | 28389222 | A | −6.90 | — |
| rs2247002 | 6p22.1 | 28397951 | C | −5.96 | — |
| rs9969098 | 6p22.1 | 28398748 | T | −5.51 | — |
| rs7766356 | 6p22.1 | 28400538 | C | −3.64 | ZSCAN23 |
| rs2531804 | 6p22.1 | 28411303 | G | −6.33 | — |
| rs2531805 | 6p22.1 | 28412326 | C | 6.58 | — |
| rs1361387 | 6p22.1 | 28412929 | G | −6.87 | — |
| rs16894116 | 6p22.1 | 28414967 | T | −4.58 | — |
| rs13215804 | 6p22.1 | 28415572 | G | −5.23 | — |
| rs6939966 | 6p22.1 | 28415885 | G | −6.01 | — |
| rs2531806 | 6p22.1 | 28417152 | C | 6.87 | — |
| rs116370852 | 6p22.1 | 28580593 | G | 6.32 | — |
| rs146219985 | 6p22.1 | 28656489 | G | −3.79 | — |
| rs142826538 | 6p22.1 | 28657190 | A | 6.14 | — |
| rs148866241 | 6p22.1 | 28658554 | A | −4.02 | — |
| rs116463813 | 6p22.1 | 28668072 | C | −5.71 | — |
| rs115856117 | 6p22.1 | 28683649 | T | 5.30 | — |
| rs114507210 | 6p22.1 | 28684183 | T | −2.37 | — |
Regulatory Effects of the 39 SNPs
Echoing the five lines of analyses: (1) Out of 39 top SNPs, 31 presented as cis-mQTLs of 6 unique CpG sites in brain (Hannon et al.41), as summarized in table S4; (2) One of these 6 CpG sites, cg23266546 at chr6:28190810, was significantly hypermethylated in cases (P = 1.64 × 10–4, passing Bonferroni correction for 6 CpG sites) in DLPFC in Lieber’s data. Table 3 summarizes the 25 top SNP mQTLs of cg23266546; (3) Three of the 6 CpG sites were profiled in 180 COBRE samples with DNA extracted from saliva. Out of 8 cis-mQTLs of these 3 CpG sites reported for brain by Hannon et al. (highlighted in bold in table S4), rs213240_C significantly positively associated with cg26335602 in saliva (r = 0.25, P = 6.87 × 10–4, passing Bonferroni correction for 8 mQTL-CpG pairs), indicating cross-tissue (brain and saliva) mQTL regulatory effect. Furthermore, cg26335602 DNAm in saliva significantly positively associated with the identified GMV component (r = 0.15, P = 4.62 × 10–2) in COBRE. Although no significant group difference for cg26335602 DNAm in saliva, its relation to GMV reduction in vivo inferred that rs213240_T is the risk allele; (4) 29 out of the 39 top SNPs are cis-eQTLs of 6 unique genes in the prefrontal cortex in GTEx43 (table S5); (5) The rs213240-regulated ZKSCAN3 gene presented a significant downregulation in SZ patients (P = 4.73 × 10–2) in GSK’s data, again implicating rs213240_T as a SZ risk allele, echoing the risk allele for GMV reduction inferred from the imaging data. See SI for additional results.
Table 3.
Methylation Quantitative Trait Loci of cg23266546 Identified in the Top SNPs: Association With GMV and Association With cg23266546 DNAm
| SNP ID | SNP Chr | SNP Posi | Local Data (SZ GMV Reduction) | Jaffe et al. (SZ Hypermethylation) | ||
|---|---|---|---|---|---|---|
| Allele | Effect on GMV (local data) | Allele | Effect on DNAm (Hannon et al.) | |||
| rs2108926 | 6 | 28308747 | T | Higher GMV | T | Lower DNAm |
| rs213240 | 6 | 28315875 | C | Higher GMV | T | Higher DNAm |
| rs6942030 | 6 | 28315958 | T | Higher GMV | T | Lower DNAm |
| rs6903652 | 6 | 28322120 | G | Higher GMV | G | Lower DNAm |
| rs213236 | 6 | 28324397 | C | Higher GMV | C | Lower DNAm |
| rs213228 | 6 | 28331252 | C | Higher GMV | C | Lower DNAm |
| rs9468354 | 6 | 28337801 | A | Higher GMV | A | Lower DNAm |
| rs10946954 | 6 | 28340625 | C | Higher GMV | T | Higher DNAm |
| rs9461456 | 6 | 28343816 | G | Higher GMV | G | Lower DNAm |
| rs7754960 | 6 | 28346945 | C | Higher GMV | C | Lower DNAm |
| rs9468365 | 6 | 28357966 | T | Higher GMV | T | Lower DNAm |
| rs2859348 | 6 | 28359170 | G | Higher GMV | A | Higher DNAm |
| rs4580862 | 6 | 28367663 | C | Higher GMV | C | Lower DNAm |
| rs2531827 | 6 | 28373154 | C | Higher GMV | T | Higher DNAm |
| rs1558205 | 6 | 28382262 | A | Higher GMV | A | Lower DNAm |
| rs2531832 | 6 | 28389222 | A | Higher GMV | G | Higher DNAm |
| rs2247002 | 6 | 28397951 | C | Higher GMV | C | Lower DNAm |
| rs9969098 | 6 | 28398748 | T | Higher GMV | T | Lower DNAm |
| rs2531805 | 6 | 28412326 | C | Lower GMV | C | Higher DNAm |
| rs1361387 | 6 | 28412929 | G | Higher GMV | T | Higher DNAm |
| rs2531806 | 6 | 28417152 | C | Lower GMV | C | Higher DNAm |
| rs116370852 | 6 | 28580593 | G | Lower GMV | G | Higher DNAm |
| rs142826538 | 6 | 28657190 | A | Lower GMV | A | Higher DNAm |
| rs116463813 | 6 | 28668072 | C | Higher GMV | T | Higher DNAm |
| rs115856117 | 6 | 28683649 | T | Lower GMV | T | Higher DNAm |
Univariate and PGRS Analyses
In the univariate analyses, some sporadic SNP-voxel pairs showed significant associations in EUR samples, which, however, could not be replicated at P < 0.05, uncorrected. In the PGRS analyses, although we observed significantly increased risk in cases for different sets of SNPs preselected from PGC, no significant GMV association was noted. See SI for details.
Discussion
In this study, we used multivariate analytic methods to investigate whether genetic variants identified for SZ risk by PGC might relate to variation in GMV. While the univariate and PGRS analyses detected no reliable imaging genetic association, using pICA, we identified a SNP component that correlated with SZ-discriminating GMV reduction in IPL and posterior STG. Both the SNP-GMV association and GMV reduction were independently replicated. The SNP component pinpointed the most significant 6p22.1 region in the PGC report, implicating shared genetic risk between SZ and regional GMV reduction.
The imaging component presented GMV reduction in parietal and temporal regions, roughly corresponding to angular gyrus (AG), supramarginal gyrus (SMG), and part of the somatomotor and associative visual cortices. These regions have been implicated for gray matter reduction, white matter tract abnormalities, aberrant activation, and dysconnectivity in SZ.47–49 These abnormalities have been observed in drug-naïve patients,50,51 thus likely reflect pathological neurobiological deficits rather than medication effects, which is echoed by the absence of association between GMV and current chlorpromazine dosages in our study. Furthermore, a recent work by Lee et al. lends support for anatomical changes in IPL and STG regions correlating with SZ genetic risk.52 In view of brain function, AG and SMG are involved in various high-order cognitive functions, including attention, spatial processes, working memory and episodic memory.53 In line with this, GMV reduction consistently associated with cognitive deficits in this study, including worse performances in MATRICS domain of attention. Moreover, in Bhojraj et al.,54 compared with controls, lower AG and SMG GMV was observed only in SZ patients’ relatives with worse cognitive performances in executive function and attention, not in those relatives with better performances. This observation lends support for IPL’s specific association with cognitive deficits, and a genetic role in GMV variation. Overall, our GMV finding appears to capture characteristic gray matter abnormalities in IPL and posterior STG that may contribute to cognitive deficits in SZ.
The top SNPs pointed to the most significant 6p22.1 major histocompatibility complex (MHC) region in PGC.5 MHC is known for complex LD structure. However, the ICA pattern is not expected to be biased by LD theoretically, which is upheld by the highly consistent findings with a heavy pruning of r2 > 0.2. The pathophysiology remains to be elucidated though. A previous univariate study reported MHC SNP associations with cerebral ventricular volume in SZ.55 Herein we explored potential functional impact through examining regulatory elements. One methylation site cg23266546, regulated by 25 top SNPs (table S4), presented significant SZ hypermethylation in DLPFC in Lieber’s data.42 Notably, risk alleles inferred for in vivo GMV reduction were all consistent with those inferred for SZ from postmortem brain DNAm, providing another line of evidence for shared risk. Using rs2108926 as an example, its negative z-score (table 2) and the negative SNP-GMV in vivo association, stated that rs2108926_T associated with higher GMV, being closer to controls as suggested by the GMV group difference (table 3). In Hannon’s data, rs2108926_T associated with lower DNAm at cg23266546 (table S4), which showed hypermethylation in SZ in Lieber’s data, suggesting rs2108926_T decreases the risk for SZ, coinciding with the in vivo observation.
SNP rs213240 is worthy of particular note. In addition to regulating the aforementioned cg23266546, rs213240 appeared to regulate cg26335602 DNAm in both brain and saliva. Most importantly, the cg26335602 DNAm in saliva positively associated with the identified GMV component in 180 COBRE samples. Thus, cg26335602 DNAm likely bridges between rs213240 and the regional GMV variation, where rs213240_T is the risk allele for lower GMV, complying with the pICA finding in table 2. Besides, GTEx presented rs213240_T associating with lower ZKSCAN3 expression in BA9 (table S5), which, along with the SZ downregulation of ZKSCAN3 in BA10 in GSK’s data, inferred that rs213240_T is the SZ risk allele, echoing both the pICA and methylation results. Though we did not directly examine mQTLs and eQTLs in IPL or STG region, the cross-cohort convergence supports potential regulatory effects of the SNP in 6p22.1.
Our findings motivate further delineation of SZ-risk SNPs for more homogeneous subsets in the sense of impact on brain phenotype. With increasing sample size, GWAS starts to yield converging findings that are generalizable particularly at polygenic level.4,5 However, the associations with diagnosis provide little knowledge on pathophysiology. One initial effort leveraging two large-scale GWAS results by Franke et al. found no notable overlap at either single variant or overall common variants level between SZ and brain volumes of eight subcortical regions. Our results concurred with Franke et al. in that no reliable SNP-GMV association was noted in the univariate or PGRS analyses. However, we identified a set of 39 SNPs using pICA to significantly correlate with GMV variation in IPL and STG. We argue that this is not simply due to including the MHC region in the analysis, as MHC did not stand out in our PGRS analysis; rather a further dissection of diagnosis-associated SNPs is crucial. This finding appeared to be highly robust, proved not to be biased by parameter selection, population stratification, LD structure, or analytic method. Meanwhile, the current finding only explained a small portion of variance in one brain phenotype. Sophisticated data mining techniques are needed to achieve a more complete quantitative model of SZ.
This study should be interpreted in light of several limitations. First, the data were aggregated from studies discrepant in data collection. While we implemented site correction and included scanning parameters as covariates in the post hoc analysis, further evaluation is warranted to confirm the current findings. Second, individuals of different population ancestries were admitted into the study. The observation that the main finding survived in EUR samples and the AFR and AMR samples showed similar associations appears to alleviate this concern. Third, the SNP overlap was moderate (~70%) between the discovery and replication data. However, this should not compromise the validity of the replication, given that top contributing SNPs were in LD. Fourth, no significant group difference was observed in the SNP component, likely due to limited power. More samples are needed to verify the mediation effect. Fifth, the postmortem methylation and gene expression data were not obtained from brain regions highlighted in our work. Focal SNP regulation awaits verification. Sixth, all the current analyses were based on association. While light pruning allows more potential causal loci to be identified in 6p22.1, fine mapping and allele-specific analysis on regulatory effects will be needed to pinpoint the true causal variants.54,55
In conclusion, our study provides support for shared genetic risks between SZ and GMV reduction in IPL and STG and demonstrates potential molecular mechanisms that may drive the observed in vivo associations. The findings highlight the importance of dissecting SZ risk variants to better understand and quantify their impact on neural structure and function, which may in turn help inform an understanding of symptomatology and functional disability evident in SZ.
Funding
This project was funded by the National Institutes of Health (P20GM103472, R01MH094524, R01EB005846, 1R01EB006841, R01MH056584, P50MH071616, and U01MH097435); National Science Foundation (1539067); National Natural Science Foundation (81471367 and 61773380); and The Strategic Priority Research Program of the Chinese Academy of Sciences (XDB02060005).
Author contributions: Drs. Chen, Calhoun, Turner, and Liu designed research; Dr. Chen conducted analyses and wrote the paper. The remaining authors contributed to the recruitment, data collection, or processing for the participating cohorts of the study. All authors critically reviewed content and approved final version for publication.
Conflict of interest: The authors declare no conflict of interest.
Supplementary Material
References
- 1. Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52:139–153. [DOI] [PubMed] [Google Scholar]
- 3. Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. [DOI] [PubMed] [Google Scholar]
- 4. Purcell SM, Wray NR, Stone JL, et al. . Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ripke S, Neale BM, Corvin A, et al. . Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee SH, DeCandia TR, Ripke S, et al. . Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keshavan MS, Tandon R, Boutros NN, Nasrallah HA. Schizophrenia, “just the facts”: what we know in 2008 Part 3: neurobiology. Schizophr Res. 2008;106:89–107. [DOI] [PubMed] [Google Scholar]
- 8. van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20:519–534. [DOI] [PubMed] [Google Scholar]
- 9. Glahn DC, Winkler AM, Kochunov P, et al. . Genetic control over the resting brain. Proc Natl Acad Sci U S A. 2010;107:1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winkler AM, Kochunov P, Blangero J, et al. . Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roalf DR, Vandekar SN, Almasy L, et al. . Heritability of subcortical and limbic brain volume and shape in multiplex-multigenerational families with schizophrenia. Biol Psychiatry. 2015;77:137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franke B, Stein JL, Ripke S, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium; ENIGMA Consortium Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat Neurosci. 2016;19:420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer-Lindenberg A. From maps to mechanisms through neuroimaging of schizophrenia. Nature. 2010;468:194–202. [DOI] [PubMed] [Google Scholar]
- 14. Pearlson GD, Liu J, Calhoun VD. An introductory review of parallel independent component analysis (p-ICA) and a guide to applying p-ICA to genetic data and imaging phenotypes to identify disease-associated biological pathways and systems in common complex disorders. Front Genet. 2015;6:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Calhoun VD, Pearlson GD, et al. . Guided exploration of genomic risk for gray matter abnormalities in schizophrenia using parallel independent component analysis with reference. Neuroimage. 2013;83:384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. [DOI] [PubMed] [Google Scholar]
- 18. Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. [DOI] [PubMed] [Google Scholar]
- 19. Altshuler DM, Durbin RM, Abecasis GR, et al. . An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 21. Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. [DOI] [PubMed] [Google Scholar]
- 22. Segall JM, Turner JA, van Erp TG, et al. . Voxel-based morphometric multisite collaborative study on schizophrenia. Schizophr Bull. 2009;35:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gupta CN, Calhoun VD, Rachakonda S, et al. . Patterns of gray matter abnormalities in schizophrenia based on an international mega-analysis. Schizophr Bull. 2015;41:1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin D, Chen J, Ehrlich S, et al. . Cross-tissue exploration of genetic and epigenetic effects on brain gray matter in schizophrenia. Schizophr Bull. 2017;May 17. doi:10.1093/schbul/sbx068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum Brain Mapp. 2009;30:241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meda SA, Ruaño G, Windemuth A, et al. . Multivariate analysis reveals genetic associations of the resting default mode network in psychotic bipolar disorder and schizophrenia. Proc Natl Acad Sci U S A. 2014;111(19):E2066–E2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. [DOI] [PubMed] [Google Scholar]
- 28. Amari S. Natural gradient works efficiently in learning. Neural Comput 1998;10:251–276. [Google Scholar]
- 29. Calhoun VD, Adalı T. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev Biomed Eng. 2012;5:60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Damoiseaux JS, Rombouts SA, Barkhof F, et al. . Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson PM, Stein JL, Medland SE, et al. ; Alzheimer’s Disease Neuroimaging Initiative, EPIGEN Consortium, IMAGEN Consortium, Saguenay Youth Study (SYS) Group The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bogdan R, Salmeron BJ, Carey CE, et al. . Imaging genetics and genomics in psychiatry: a critical review of progress and potential. Biol Psychiat. 2017;82:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu JY, Calhoun VD. A review of multivariate analyses in imaging genetics. Front Neuroinform. 2014;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J, Calhoun VD, Pearlson GD, et al. . Independent component analysis of SNPs reflects polygenic risk scores for schizophrenia. Schizophr Res. 2017;181:83–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rissanen J. Modeling by shortest data description. Automatica. 1978;14(5):465–471. [Google Scholar]
- 36. Lancaster JL, Woldorff MG, Parsons LM, et al. . Automated talairach atlas labels for functional brain mapping. Hum Brain Mapp. 2000;10:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kay SR, Fiszbein A, Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull. 1987;13:261–276. [DOI] [PubMed] [Google Scholar]
- 38. Andreasen NC. The Scale for the Assessment of Negative Symptoms (SANS). Iowa City, IA: The University of Iowa; 1983. [Google Scholar]
- 39. Andreasen NC. The Scale for the Assessment of Negative Symptoms (SAPS). Iowa City, IA: The University of Iowa; 1984. [Google Scholar]
- 40. van Erp TG, Preda A, Nguyen D, et al. . Converting positive and negative symptom scores between PANSS and SAPS/SANS. Schizophr Res. 2014;152:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hannon E, Spiers H, Viana J, et al. . Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat Neurosci. 2016;19:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jaffe AE, Gao Y, Deep-Soboslay A, et al. . Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat Neurosci. 2016;19:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ardlie KG, DeLuca DS, Segre AV, et al. . The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maycox PR, Kelly F, Taylor A, et al. . Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function (vol 14, pg 1083, 2009). Mol Psychiatr 2010;15(4):442–443. [DOI] [PubMed] [Google Scholar]
- 45. Le Floch E, Guillemot V, Frouin V, et al. . Significant correlation between a set of genetic polymorphisms and a functional brain network revealed by feature selection and sparse partial least squares. Neuroimage. 2012;63:11–24. [DOI] [PubMed] [Google Scholar]
- 46. Sekar A, Bialas AR, de Rivera H, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Torrey EF. Schizophrenia and the inferior parietal lobule. Schizophr Res. 2007;97:215–225. [DOI] [PubMed] [Google Scholar]
- 48. Buchanan RW, Francis A, Arango C, et al. . Morphometric assessment of the heteromodal association cortex in schizophrenia. Am J Psychiatry. 2004;161:322–331. [DOI] [PubMed] [Google Scholar]
- 49. Turken A, Whitfield-Gabrieli S, Bammer R, Baldo JV, Dronkers NF, Gabrieli JD. Cognitive processing speed and the structure of white matter pathways: convergent evidence from normal variation and lesion studies. Neuroimage. 2008;42:1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Braus DF, Weber-Fahr W, Tost H, Ruf M, Henn FA. Sensory information processing in neuroleptic-naive first-episode schizophrenic patients: a functional magnetic resonance imaging study. Arch Gen Psychiatry. 2002;59:696–701. [DOI] [PubMed] [Google Scholar]
- 51. Ren W, Lui S, Deng W, et al. . Anatomical and functional brain abnormalities in drug-naive first-episode schizophrenia. Am J Psychiatry. 2013;170:1308–1316. [DOI] [PubMed] [Google Scholar]
- 52. Lee PH, Baker JT, Holmes AJ, et al. Partitioning heritability analysis reveals a shared genetic basis of brain anatomy and schizophrenia. Mol Psychiatr. 2016;21(12):1680–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cabeza R, Nyberg L. Imaging cognition II: an empirical review of 275 PET and fMRI studies. J Cogn Neurosci. 2000;12:1–47. [DOI] [PubMed] [Google Scholar]
- 54. Bhojraj TS, Francis AN, Montrose DM, Keshavan MS. Grey matter and cognitive deficits in young relatives of schizophrenia patients. Neuroimage. 2011;54 (suppl 1):S287–S292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Agartz I, Brown AA, Rimol LM, et al. . Common sequence variants in the major histocompatibility complex region associate with cerebral ventricular size in schizophrenia. Biol Psychiatry. 2011;70:696–698.21514568 [Google Scholar]
- 56. Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 2010;11:533–538. [DOI] [PubMed] [Google Scholar]
- 57. Farh KK, Marson A, Zhu J, et al. . Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.