Abstract
Candidate gene and genome-wide association study based common risk variant identification is being complemented by whole exome sequencing (WES)/whole genome sequencing based rare variant discovery in elucidation of genetic landscape of schizophrenia (SZ), a common neuropsychiatric disorder. WES findings of de novo mutations in case-parent trios have further implied genetic etiology, but do not explain the high genetic risk in general populations. Conversely, WES in multiplex families may be an insightful strategy for the identification of highly penetrant rare variants in SZ and possibly enhance our understanding of disease biology. In this study, we analyzed a 5-generation Indian family with multiple members affected with SZ by WES. We identified a rare heterozygous missense variant (NM_003255: c.506C>T; p.Pro169Leu; MAF = 0.0001) in Tissue Inhibitor of Metalloproteinase 2 (TIMP2, 17q25.3) segregating with all 6 affected individuals but not with unaffected members. Linkage analysis indicated a maximum logarithm of the odds score of 1.8, θ = 0 at this locus. The variant was predicted to be damaging by various in silico tools and also disrupt the structural integrity by molecular dynamics simulations. WES based screening of an independent SZ cohort (n = 370) identified 4 additional rare missense variants (p.Leu20Met, p.Ala26Ser, p.Lys48Arg and p. Ile217Leu) and a splice variant rs540397728 (NM_003255:c.232-5T>C), also predicted to be damaging, increasing the likelihood of contribution of this gene to SZ risk. Extensive biochemical and knockout mouse studies suggesting involvement of TIMP2 in neurodevelopmental and behavioral deficits, together with genetic evidence for TIMP2 conferring SZ risk from this study may have possible implications for new therapeutics.
Keywords: multiplex family, whole exome sequencing, linkage analysis, TIMP gene family, behavioural deficit
Introduction
Schizophrenia (SZ) is a severe and chronic neuropsychiatric disorder characterized by impairment in thought, behavior, and neurocognitive function. Important role of genetic factors in conferring risk to SZ is well documented over several years now, on the basis of twin, adoption and family studies,1 together with high heritability estimates (~60%–80%).2,3 Commensurate efforts are being made to decipher the genetics of psychiatric disorders, particularly SZ. A recent Genome-Wide Association Study (GWAS) meta-analysis of 39,989 individuals with SZ and 113075 controls identified 108 loci associated with SZ with small to modest effects,4 supporting the commonly accepted polygenic nature of the disease. Meanwhile GWAS based common risk variant identification has been complemented by whole exome sequencing (WES) and whole genome sequencing based rare variant discovery. WES findings of de novo mutations in case-parent trios have further implied a genetic etiology.5,7–17 Though these mutations were not enriched in specific genes, they support the overall role of neuronal migration, synaptic transmission, signaling, transcriptional regulation, transport, glutamatergic postsynaptic proteins comprising activity-regulated cytoskeleton-associated protein (ARC) and N-methyl-d-aspartate receptor (NMDAR) complexes in SZ etiology.7,12,18 However, such de novo mutations do not sufficiently explain the high inherited genetic risk observed in general populations. On the other hand, WES in multiplex families may be a fruitful and reliable strategy for the identification of highly penetrant rare variants in common diseases and this may also explain the heritable risk and provide a better understanding of disease biology. Recently, we and others have identified rare variants in a number of functionally relevant genes by WES of multimember affected families. These include UNC13B, SHANK2, SMARCA1, TAAR1 GRM5, LRP1B, and RELN.19–23 Continuing our efforts in discovery genomics, in this study, we analyzed an Indian family with multiple members affected with SZ across 5 generations. We employed the WES approach to identify highly penetrant rare variant(s) that may co-segregate with disease phenotype in the family and identified a rare missense variant in Tissue Inhibitor of Metalloproteinase 2/TIMP metallopeptidase inhibitor 2 (TIMP2). Further, we identified 5 additional rare variants in this functionally relevant gene from among 370 SZ exomes available in the laboratory, thus providing the first ever genetic evidence for the role of this gene in SZ etiology.
Materials and Methods
Recruitment of Study Subjects
The detailed protocol for recruitment of study cohort has been reported previously.24,25 Briefly, the samples were recruited from Dr RML hospital New Delhi, and were mostly of north Indian origin. Consensus diagnosis for SZ was made based on DSM IV criteria. Hindi versions of the Diagnostic Interview for Genetic Studies (DIGS) and the Family Interview for Genetic Studies (FIGS) were also administered to all the affected and unaffected individuals used in the study to get additional information. All study protocols were approved by the institutional ethical committees of both participating institutions and all the participants provided written informed consent. Venous blood was drawn from all those individuals and DNA isolation was done by the routine phenol chloroform method, which was further used for genetic analysis.
Family Used for WES
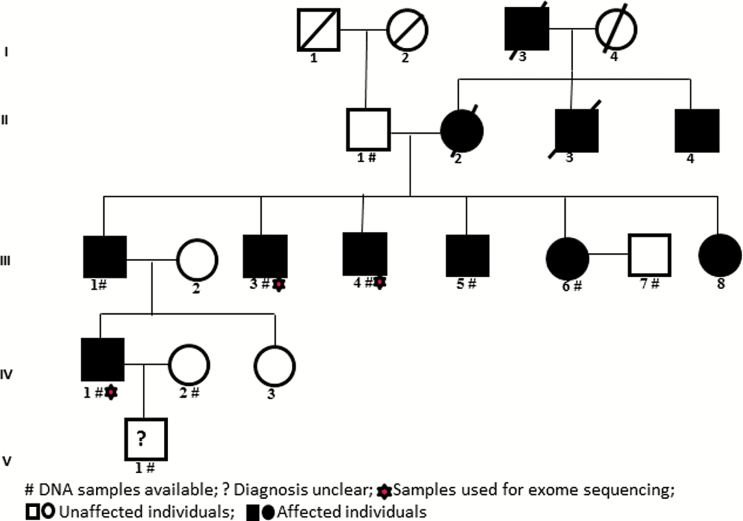
A 5-generation family with 11 members affected with SZ was used in the study (figure 1). Genomic DNA samples from 10 individuals from 4 generations were available, of which DNA from 3 affected members was used for WES. Diagnosis of the single individual from the fifth generation (V.1) could not be confirmed and therefore, that individual was excluded from further analysis.
Fig. 1.
Pedigree of the multiplex SZ family analyzed in the study.
WES and Data Processing
Agilent SureSelect Human All Exon V5 + UTR kit was used for library preparations and sequencing was performed in Illumina HiSeq2000 in paired end sequencing mode, using a commercial facility (Medgenome Labs Pvt. Ltd). Raw data generated were processed using standard tools and software. The protocols used are detailed previously.21
Variant Prioritization
DNA from 3 informative individuals (III.3, III.4 and IV.1) from the family (figure 1) was used for WES. Prioritization of the variants was performed as described previously.26–28 Variants in the combined VCF generated from exome sequence data were annotated using wANNOVAR29 Kggseq30 and nonprotein coding variants removed. As the aim of the study was to identify highly penetrant rare variants, all the variants with minor allele frequency (MAF), MAF > 0.001 in public databases including 1000G, Exome Aggregation Consortium (ExAC r0.3.1) and Genome Aggregation Database (GAD), 6500 exomes, G40,dbSNP along with synonymous variants and those not shared among the 3 affected samples were removed. Variants shared among the 3 affected individuals were screened among the remaining family members by Sanger sequencing/custom target capture sequencing. Subsequently, all variants that were not shared among all the affected individuals in the family, variants from regions with segmental duplications and variants with MAF > 0.001 in non-SZ exome data (n = 150) available in the laboratory were also discounted. Finally, the variants that are present in unaffected individuals in the family were also removed. For the variant annotation and prioritization we used Kggseq software. Based on functional/biological relevance or their prior implication for SZ based on association, linkage, exome sequencing and animal studies, the variants shared among affected individuals were further prioritized. These variants were then confirmed by Sanger sequencing (Primer details in supplementary table 1).
Linkage Analysis.
To evaluate the likely linkage of this gene to SZ, additional analysis using 2 microsatellite markers (D17S949, D17S785) upstream of TIMP2 and 2 downstream markers (D17S784, D17S928) from ABI Linkage Mapping Sets V2.5 was carried out. PCR was performed as per the manufacturer’s protocol and products were run on ABI 3730xl DNA Analyzer at central instrumentation facility at University of Delhi South Campus, and analyzed using peak scanner software. Logarithm of the odds (LOD) scores were calculated.
In silico Analysis of the Prioritized Variant(s)
In silico characterization of the prioritized and Sanger sequencing confirmed variant(s) were performed by 14 different tools SIFT, Polyphen2_HDIV, Polyphen2_HVAR, LRT, MutationAssessor, MutationTaster, FATHOM, PROVEAN, MetaSVM, MetaLR, RadialSVM, LR, Variant Effect Scoring Tool3 (VEST3) and Combined Annotation Dependent Depletion (CADD) score which were all available in dbNSFP2.9.31 In addition we used Predictor of human Deleterious Single Nucleotide Polymorphisms (PhD-SNP)32 and CONsensus DELeteriousness (CONDEL).33 We also examined the evolutionary conservation of the confirmed variants using GERP++_RS phyloP46way_placental, phyloP100way_vertebrate, SiPhy_29way_logOdds, GERP++_NR and for gene level pathogenicity estimation we used haplo-insufficiency estimation and Residual Variation Intolerance Score (RVIS).
Molecular Dynamics Simulation.
The X-ray crystallographic 3D structure of the standalone TIMP2 protein available in the Protein Data Bank (PDB code 1BR9) was used to study the effect of the Pro169Leu variant on the protein structure using Molecular Dynamics simulations (MDs). The Pro169 residue in the wild-type protein structure (present at position 143 in 1BR9 which is missing the first 26 amino acid residues corresponding to a signal peptide) was mutated using the mutagenesis option of the Swiss-PdbViewer (SPDBV) software.34 Both the wild-type and variant structures were subject to a 100-nanosecond simulation run each, using GROMACS 5.1.2 software.35,36 The force-field used for the simulation was CHARMM27 all-atom force field.37,38 Both the structures were subject to the same simulation parameters. The protein was solvated using the TIP3P water model in a dodecahedron solvation box. A net positive charge (+2) on the protein structures at physiological pH was balanced by adding 2 chloride ions in the solvent, bringing the entire system to a neutral charge. Energy minimization was performed for a maximum of 500000 steps using the steepest descent algorithm. After the structures converged to a maximum force < 1000 kJ/mol/nm, a 2-step equilibration using the NVT ensemble (isothermal-isochoric) followed by the NPT ensemble (isothermal-isobaric) was performed at 300K, 1.0 atm and 10,000 ps, 300K, respectively. The covalent bonds were constrained using the LINCS (Linear Constraint Solver) algorithm,39 while Particle Mesh Ewald (PME) was used to treat the electrostatic interactions.40 A coulomb and Vander Waals interactions cut off radii of 10.0 and 14.0 Ǻ were set and MDs were carried out at 300K for 100 ns with the MD trajectories being saved after every 2.0 ps. The MD trajectories were analyzed to obtain the root-mean-square fluctuation (RMSF) as a function of the Cα atoms of protein residues using the Gromacs gmx rmsf utility.36
Results
WES in the Index Family
The mean target depth of the 3 samples used from the family for WES was 54X and on an average, >99% of the target region at 1X and >87% of the target region at 20X were covered. The mean mapping quality of the samples was 47 and detailed statistics are provided in supplementary table 2. Of the 60 variants following prioritization (table 1), only 2 variants one each in TIMP2 and PIWIL3 segregated in the family. Though PIWIL3 is expressed in the brain and could be functionally relevant, due to the predicted tolerant nature of the variant p.Gly31Glu by all the software used, its poor conservation observed across species and an extremely low CADD score of 0.59 (supplementary table 3) it was removed from further analysis.
Table 1.
Prioritization of Variants in the Whole Exome Data from the Index Family
| Filtering Criteria | Number of Remaining Variants |
|---|---|
| Total coding variants | 31573 |
| On removal of variants based on global minor allele frequency (MAF < 0.001) in 1000G, ExAC, Exome Variant Server, CG40, dbSNP, gnomAD browser | 1320 |
| On removal of variants based on population specific frequency (MAF < 0.001) in 1000G, ExAC, Exome Variant Server, gnomAD browser | 850 |
| Total number of protein disrupting variants (Synonymous variants removed) | 605 |
| Total number of homozygous variants shared among 3 affected (Exome sequenced) | 0 |
| Total number of heterozygous variants shared among 3 affected (Exome sequenced) | 128 |
| Total heterozygous variants present in all affected (Screened by target capture sequencing) | 60 |
| On removal of variants from regions with segmental duplication | 45 |
| On removal of >4 variants per gene | 22 |
| On removal of variants present in in-house control sample MAF > 0.001 | 15 |
| Total number of variants segregating with the phenotype | 2 |
| Number of heterozygous segregating variants functionally relevant based on literature | 2 |
On the other hand, the rare heterozygous missense variant in TIMP2 (17q25.3) (NC_000017.10: g.76851906G>A; NM_003255.4: c.506C>T; NP_003246.1:p.Pro169Leu; Exon 5; MAF 0.0001 in ExAC 0.3.1) in TIMP2 present in all 6 affected individuals but absent in the 3 unaffected members (supplementary figure 1) seemed promising. Expression levels of this gene in the different brain regions were checked using 2 different databases namely BrainSpan (http://www.brainspan.org) and Genotype-Tissue Expression (GTEx) portal (https://www.gtexportal.org/). The protein was expressed across the brain regions in all the developmental stages.
Linkage Analysis.
Linkage between D17S785 (2.42 Mb upstream) and D17S784 (0.95 Mb downstream) was estimated with maximum LOD score 1.8 (θ = 0) confirming its linkage.
In silico Analysis
The index variant p.Pro169Leu was highly conserved across species (supplementary figure 2; supplementary table 3), predicted to be damaging by 13 different in silico tools and also predicted to be among the top 1% most deleterious substitutions in the human genome by CADD (supplementary table 3). Gene level analysis revealed TIMP2 to be among the top 31% of intolerant genes and with haploinsufficiency.
Effects of the Index Variant on TIMP2 Structure
The RMSF analysis of MD trajectories showed that although the fluctuations for the WT and the variant structures were nearly concordant for most of the protein’s length, there were notable deviations in the vicinity of the reported variant at the 143rd position (supplementary figure 3). The stretch of residues between positions 130–155, corresponding to the GH loop and its neighboring regions in the C-terminal domain (CTD), showed a marked deviation in the fluctuations of Cα atoms (to the order of ~0.5 nm to 1.0 nm). This indicates that the variant leads to an increased flexibility and disorder in the backbone atoms of CTD of TIMP2, possibly due to disruption of disulphide bridges which stabilize this domain. This structural instability as a consequence of the proline to leucine amino acid change is suggestive of aberrant functionality of the variant protein.
Additional Variants in TIMP2 Identified in an Independent SZ Cohort.
The index variant and additional variants, if any, in TIMP2 exons were screened in an independent SZ cohort comprising of 370 SZ samples, mostly from north India, recruited from Dr. RML hospital, New Delhi. A total of 5 additional coding variants were identified. Three of the novel variants namely p.Leu20Met and p.Ala26Ser in exon 1 and p.Lys48Arg in exon 2 were seen in one individual each. In addition a rare missense variant rs578083142 (NM_003255.4:c.649A>C; p.Ile217Leu) in exon 5 with MAF = 0.0006, in ExAC r0.3.1 and GAD (South Asian population) and a rare splice variant rs540397728 (NM_003255.4:c.232-5T>C) in exon 3 with MAF = 0.0001 in ExAC r0.3.1 and GAD (South Asian population) were observed in 2 other SZ exomes. All these 5 variants were also conserved across species and predicted to be damaging by 1 or more of the in silico tools (supplementary table 4) and were notably absent in 150 non-SZ exome data available in the laboratory.
Discussion
In complex diseases especially in SZ, a likely role of de novo variants in disease development has been evident from previous case-parent studies.41 Though high heritability estimated for this disorder reiterates the role of inherited variants, family based studies are still limited. The multiplex family (figure 1) analyzed by WES in this study is indeed informative and based on the justification provided below, the rare heterozygous missense variant (p.Pro169Leu, exon 5) in TIMP2 seems to be the most promising risk variant in this family. Considering that this variant has segregated with the phenotype in 6 independent meiotic events (of the 10 that could be tested) across 2 generations (figure 1), it may be argued that it is unlikely to be a chance event. This is supported by our suggestive linkage findings [LOD score of 1.8 (θ = 0) with 2 flanking markers at 2.42Mb (D17S785) and 0.95Mb (D17S784)]. Considering the generally accepted polygenic nature of SZ, the role of common variants with MAF > 0.001 in this family cannot be ignored. However, this was not addressed in the current study whose focus was to identify only highly penetrant rare variants in a familial setting. Identification of 5 additional rare variants among 370 exomes derived from an independent SZ cohort (supplementary table 4) increased the likelihood of the possible contribution of this gene to SZ risk. This is further strengthened by the observation of 6 variants with MAF ≥ 0.01 nominally associated (P ≤ 0.05) with SZ in the Psychiatric Genomics Consortium (PGC) data (http://www.med.unc.edu/pgc; supplementary table 5).
TIMP2 is expressed throughout brain tissues and at all stages of brain development and adulthood reiterating its functional relevance. Further, likely functional significance of the rare variant derives support from in silico predictions wherein several tools showed this variant to be damaging (supplementary table 3). These predictions are strengthened by the findings of MD based structural analysis (supplementary figure 3). Structural details of the interactions of TIMP2 protein with some of its metalloproteinase binding partners (pro-MMP2,42 MMP10,43 MMP13,44 and CDMT1-MMP45) are well documented. In general, the N-terminal domain (NTD) of TIMP2 has been implicated in the inhibitory interactions with various MMPs,43–45 with the CTD also possibly involved in some stabilizing interactions.46 This non-synonymous variant can potentially disrupt some disulfide bonds which stabilize the CTD and this disruption in the protein structure can contribute to a loss of function, but this warrants functional validation.
TIMP2 is located at 17q25.3, a region which has been previously implicated in both SZ and bipolar disorder (BD) based on family based linkage studies. In a study of 250 BD pedigrees of mainly Caucasian ancestry, a maximum LOD score of 3.63 was observed at this locus.47 Another study on 22 Canadian pedigrees of Celtic or German descent with SZ identified linkage at the same locus.48 Importance of 17q25.3 locus is also available from another study, which reported a balanced translocation involving chromosome 17 t(9;17)(q33.2;q25.3) in 2 SZ affected members of a small family with diverse psychiatric disorders.49 Interestingly, 4 protein coding genes namely ENDOV, NPTX1, RNF213, and RPTOR are reported to be present in the break point region at 17q25.3. Association studies of SNPs at this locus revealed significant association of NPTX1 with BD (P = 0.004) and ENDOV with SZ (P = 0.007) which withstood multiple testing.50
Substantial literature on TIMP2 is available based on biochemical and animal studies. TIMP2 is a member of TIMP gene family which are natural inhibitors of the matrix metalloproteinases (MMPs). Of note, decreased expression of TIMP2, MMP2, and MMP9 at both mRNA and protein levels in sera samples of subjects with depression and conversely, a direct correlation between elevated expression of these 3 genes and improved cognitive functions have been reported.51
TIMP2 has been demonstrated to be involved in extra cellular matrix remodeling together with MMPs.52 This in turn is notably involved in various neurodevelopmental processes such as neurogenesis, differentiation, axonal growth, formation of axonal tracts, stabilization/maturation of synapses, regulation of synaptic plasticity, remodeling. In addition its involvement in neuro transmission, long-term potentiation (LTP), long-term depression and myelination have also been reported.53–57
TIMP2 inhibits angiogenic factor mediated angiogenesis in vivo58 and possible role of angiogenesis in the pathophysiology of SZ has been suggested previously.59 On the other hand, through the MMP independent activity, TIMP2 is known to induce cell cycle arrest and subsequent neuronal differentiation and neurite outgrowth in PC12 cells.60 Varying roles of TIMP2 have been reported in several pathways namely VEGFR, EGFR, FGFR1, AKT, and Wnt/β-catenin signaling,60–63 all previously implicated in SZ.6,64–70
Besides these studies, TIMP2 has been studied extensively in mouse models including knockout mice and its role in neurodevelopment and behavioral deficits has been reiterated. Role of TIMP2 in neuronal differentiation and neurite outgrowth,60 motor function,71 synaptic plasticity, hippocampal development, cognition and LTP72 have been well documented in these studies on knockout mice. Knockout mice also did not exhibit prepulse inhibition of the startle reflex but showed increased anxiety behavior. These functions have been speculated to be modulated by TIMP2 similar to other ECM proteins like reelin or neural cell adhesion molecule (N-CAM),73 Further, repeated treatment of methamphetamine in rats leads to increase in TIMP2 expression in frontal cortex which has been shown to play an important role in drug induced behavioral sensitization and rewards, confirmed by antisense TIMP2 oligonucleotide treatment. The authors suggested that TIMP2 may be involved in the neural network rearrangement in the mesocorticolimbic dopamine system, particularly through the dopamine D2 receptor.74
Taken together, identification of the rare heterozygous missense variant p.Pro169Leu in TIMP2 in the study family, 5 additional rare protein disturbing variants in an independent SZ cohort and a large number of functional studies reported in literature, support the likely contribution of protein coding variants in TIMP2 to SZ etiology. This first genetic evidence could pave the way for new therapeutic opportunities with implications for personalized medicine.
Funding
Grant #BT/MB/Project-Schizophrenia/2012–2013 and #BT/PR2425/Med13/089/2001 to B.K.T. and S.N.D. from the Department of Biotechnology, Government of India, New Delhi, India; Grant #MH093246, #MH063480, and #TW009114 to V.L.N. from National Institute of Mental Health, the Fogarty International Center, USA; Junior and Senior Research Fellowship (09/045(1166)/2012-EMR-I) to J.J. from Council for Scientific and Industrial Research, New Delhi; Junior and Senior Research Fellowship from the Department of Genetics under University Grants Commission (UGC)-Special Assistance Program Meritorious Award scheme to A.S.; and DSK-PDF (BL/13-14/0404) to P.K. from UGC, New Delhi are gratefully acknowledged.
Supplementary Material
Acknowledgments
We are thankful for the study sample collection by trained and dedicated staff at Dr RML hospital, DNA isolation by Mrs. Anjali Dabral at the University of Delhi South Campus; help with linkage analysis by Mr. Navneesh Yadav and computational facility provided by Central Instrumentation Facility, University of Delhi South Campus. We gratefully acknowledge infrastructure support provided by the UGC, New Delhi, through Special Assistance Programme and Department of Science and Technology, New Delhi, through FIST and DU-DST PURSE programmes to the Department of Genetics, UDSC. B.K.T., S.N.D., and V.L.N. designed the study and obtained research funding; S.N.D. diagnosed and recruited the study samples; J.J. performed all the WES data analysis, interpretation, and confirmation of variant by Sanger sequencing and linkage analysis. T.B. contributed to sample recruitment and phenotype data documentation; P.K. maintained the DNA repository and contributed to data analysis and interpretation; A.S. performed the MD simulations; J.J. and B.K.T. wrote the first draft of manuscript; all authors contributed to and have approved the final manuscript. The authors have declared that there are no conflicts of interest in relation to the subject of this study.
References
- 1. Gejman PV, Sanders AR, Duan J. The role of genetics in the etiology of schizophrenia. Psychiatr Clin North Am. 2010;33:35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cannon TD, Kaprio J, Lönnqvist J, Huttunen M, Koskenvuo M. The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch Gen Psychiatry. 1998;55:67–74. [DOI] [PubMed] [Google Scholar]
- 3. Lichtenstein P, Yip BH, Björk C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambalavanan A, Girard SL, Ahn K, et al. De novo variants in sporadic cases of childhood onset schizophrenia. Eur J Hum Genet. 2016;24:944–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freyberg Z, Ferrando SJ, Javitch JA. Roles of the Akt/GSK-3 and Wnt signaling pathways in schizophrenia and antipsychotic drug action. Am J Psychiatry. 2010;167:388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fromer M, Pocklington AJ, Kavanagh DH, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Girard SL, Gauthier J, Noreau A, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. [DOI] [PubMed] [Google Scholar]
- 9. Guipponi M, Santoni FA, Setola V, et al. Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One. 2014;9:e112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gulsuner S, McClellan JM. De novo mutations in schizophrenia disrupt genes co-expressed in fetal prefrontal cortex. Neuropsychopharmacology. 2014;39:238–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kranz TM, Harroch S, Manor O, et al. De novo mutations from sporadic schizophrenia cases highlight important signaling genes in an independent sample. Schizophr Res. 2015;166:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCarthy SE, Gillis J, Kramer M, et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry. 2014;19:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rees E, Kirov G, Walters JT, et al. ; Taiwanese Trios Exome Sequencing Consortium Analysis of exome sequence in 604 trios for recessive genotypes in schizophrenia. Transl Psychiatry. 2015;5:e607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh T, Kurki MI, Curtis D, et al. ; Swedish Schizophrenia Study; INTERVAL Study; DDD Study; UK10 K Consortium Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takata A, Xu B, Ionita-Laza I, Roos JL, Gogos JA, Karayiorgou M. Loss-of-function variants in schizophrenia risk and SETD1A as a candidate susceptibility gene. Neuron. 2014;82:773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu B, Ionita-Laza I, Roos JL, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu B, Roos JL, Dexheimer P, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gulsuner S, Walsh T, Watts AC, et al. ; Consortium on the Genetics of Schizophrenia (COGS); PAARTNERS Study Group Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Egawa J, Hoya S, Watanabe Y, et al. Rare UNC13B variations and risk of schizophrenia: whole-exome sequencing in a multiplex family and follow-up resequencing and a case-control study. Am J Med Genet Part B Neuropsychiatr Genet. 2016;171:797–805. [DOI] [PubMed] [Google Scholar]
- 20. Homann OR, Misura K, Lamas E, et al. Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Mol Psychiatry. 2016;21:1690–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. John J, Kukshal P, Bhatia T, et al. Possible role of rare variants in Trace amine associated receptor 1 in schizophrenia. Schizophr Res. 2017;189:190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Timms AE, Dorschner MO, Wechsler J, et al. Support for the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry. 2013;70:582–590. [DOI] [PubMed] [Google Scholar]
- 23. Zhou Z, Hu Z, Zhang L, et al. Identification of RELN variation p.Thr3192Ser in a Chinese family with schizophrenia. Sci Rep. 2016;6:24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kukshal P, Bhatia T, Bhagwat AM, et al. Association study of neuregulin-1 gene polymorphisms in a North Indian schizophrenia sample. Schizophr Res. 2013;144:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. John J, Bhatia T, Kukshal P, et al. Association study of MiRSNPs with schizophrenia, tardive dyskinesia and cognition. Schizophr Res. 2016;174:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dashti MJS, Gamieldien J. A practical guide to filtering and prioritizing genetic variants. Biotechniques. 2017;62:18–30. [DOI] [PubMed] [Google Scholar]
- 27. Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li MX, Gui HS, Kwan JS, Bao SY, Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. [DOI] [PubMed] [Google Scholar]
- 33. González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 35. Abraham MJ, Murtola T, Schulz R, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 36. Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. [DOI] [PubMed] [Google Scholar]
- 37. MacKerell AD, Bashford D, Bellott M, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 38. Mackerell AD Jr, Feig M, Brooks CL III. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. [DOI] [PubMed] [Google Scholar]
- 39. Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 40. Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 41. Li J, Cai T, Jiang Y, et al. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry. 2016;21:298. [DOI] [PubMed] [Google Scholar]
- 42. Morgunova E, Tuuttila A, Bergmann U, Tryggvason K. Structural insight into the complex formation of latent matrix metalloproteinase 2 with tissue inhibitor of metalloproteinase 2. Proc Natl Acad Sci U S A. 2002;99:7414–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Batra J, Soares AS, Mehner C, Radisky ES. Matrix metalloproteinase-10/TIMP-2 structure and analyses define conserved core interactions and diverse exosite interactions in MMP/TIMP complexes. PLoS One. 2013;8:e75836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maskos K, Lang R, Tschesche H, Bode W. Flexibility and variability of TIMP binding: X-ray structure of the complex between collagenase-3/MMP-13 and TIMP-2. J Mol Biol. 2007;366:1222–1231. [DOI] [PubMed] [Google Scholar]
- 45. Fernandez-Catalan C, Bode W, Huber R, et al. Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 1998;17:5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olson MW, Gervasi DC, Mobashery S, Fridman R. Kinetic analysis of the binding of human matrix metalloproteinase-2 and -9 to tissue inhibitor of metalloproteinase (TIMP)-1 and TIMP-2. J Biol Chem. 1997;272:29975–29983. [DOI] [PubMed] [Google Scholar]
- 47. Dick DM, Foroud T, Flury L, et al. Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Logue MW, Brzustowicz LM, Bassett AS, Chow EW, Vieland VJ. A posterior probability of linkage-based re-analysis of schizophrenia data yields evidence of linkage to chromosomes 1 and 17. Hum Hered. 2006;62:47–54. [DOI] [PubMed] [Google Scholar]
- 49. Fullston T, Gabb B, Callen D, et al. Inherited balanced translocation t(9;17)(q33.2;q25.3) concomitant with a 16p13.1 duplication in a patient with schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2011;156:204–214. [DOI] [PubMed] [Google Scholar]
- 50. Rajkumar AP, Christensen JH, Mattheisen M, et al. Analysis of t(9;17)(q33.2;q25.3) chromosomal breakpoint regions and genetic association reveals novel candidate genes for bipolar disorder. Bipolar Disord. 2015;17:205–211. [DOI] [PubMed] [Google Scholar]
- 51. Bobińska K, Szemraj J, Gałecki P, Talarowska M. The role of MMP genes in recurrent depressive disorders and cognitive functions. Acta Neuropsychiatr. 2016;28:221–231. [DOI] [PubMed] [Google Scholar]
- 52. Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3:a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Barros CS, Franco SJ, Müller U. Extracellular matrix: functions in the nervous system. Cold Spring Harb Perspect Biol. 2011;3:a005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dityatev A, Schachner M, Sonderegger P. The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat Rev Neurosci. 2010;11:735–746. [DOI] [PubMed] [Google Scholar]
- 55. Lau LW, Cua R, Keough MB, Haylock-Jacobs S, Yong VW. Pathophysiology of the brain extracellular matrix: a new target for remyelination. Nat Rev Neurosci. 2013;14:722–729. [DOI] [PubMed] [Google Scholar]
- 56. Mukhina IV, Korotchenko SA, Dityatev AE. Extracellular matrix molecules, their receptors, and extracellular proteases as synaptic plasticity modulators. Neurochem J. 2012;6:89–99. [Google Scholar]
- 57. Nicholson C, Syková E. Extracellular space structure revealed by diffusion analysis. Trends Neurosci. 1998;21:207–215. [DOI] [PubMed] [Google Scholar]
- 58. Seo DW, Li H, Guedez L, et al. TIMP-2 mediated inhibition of angiogenesis: an MMP-independent mechanism. Cell. 2003;114:171–180. [DOI] [PubMed] [Google Scholar]
- 59. Lopes R, Soares R, Coelho R, Figueiredo-Braga M. Angiogenesis in the pathophysiology of schizophrenia - A comprehensive review and a conceptual hypothesis. Life Sci. 2015;128:79–93. [DOI] [PubMed] [Google Scholar]
- 60. Pérez-Martínez L, Jaworski DM. Tissue inhibitor of metalloproteinase-2 promotes neuronal differentiation by acting as an anti-mitogenic signal. J Neurosci. 2005;25:4917–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim HIe, Lee HS, Kim TH, Lee JS, Lee ST, Lee SJ. Growth-stimulatory activity of TIMP-2 is mediated through c-Src activation followed by activation of FAK, PI3-kinase/AKT, and ERK1/2 independent of MMP inhibition in lung adenocarcinoma cells. Oncotarget. 2015;6:42905–42922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Remillard TC, Bratslavsky G, Jensen-Taubman S, Stetler-Stevenson WG, Bourboulia D. Molecular mechanisms of tissue inhibitor of metalloproteinase 2 in the tumor microenvironment. Mol Cell Ther. 2014;2:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Valacca C, Tassone E, Mignatti P. TIMP-2 interaction with MT1-MMP activates the AKT pathway and protects tumor cells from apoptosis. PLoS One. 2015;10:e0136797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emamian ES. AKT/GSK3 signaling pathway and schizophrenia. Front Mol Neurosci. 2012;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Futamura T, Toyooka K, Iritani S, et al. Abnormal expression of epidermal growth factor and its receptor in the forebrain and serum of schizophrenic patients. Mol Psychiatry. 2002;7:673–682. [DOI] [PubMed] [Google Scholar]
- 66. Iwakura Y, Nawa H. ErbB1-4-dependent EGF/neuregulin signals and their cross talk in the central nervous system: pathological implications in schizophrenia and Parkinson’s disease. Front Cell Neurosci. 2013;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Narla ST, Lee YW, Benson CA, et al. Common developmental genome deprogramming in schizophrenia - Role of Integrative Nuclear FGFR1 Signaling (INFS). Schizophr Res. 2017;185:17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Panaccione I, Napoletano F, Forte AM, et al. Neurodevelopment in schizophrenia: the role of the wnt pathways. Curr Neuropharmacol. 2013;11:535–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xia Y, Wu S. Tissue inhibitor of metalloproteinase 2 inhibits activation of the β-catenin signaling in melanoma cells. Cell Cycle. 2015;14:1666–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zheng W, Wang H, Zeng Z, et al. The possible role of the Akt signaling pathway in schizophrenia. Brain Res. 2012;1470:145–158. [DOI] [PubMed] [Google Scholar]
- 71. Jaworski DM, Soloway P, Caterina J, Falls WA. Tissue inhibitor of metalloproteinase-2(TIMP-2)-deficient mice display motor deficits. J Neurobiol. 2006;66:82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Castellano JM, Mosher KI, Abbey RJ, et al. Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature. 2017;544:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jaworski DM, Boone J, Caterina J, Soloway P, Falls WA. Prepulse inhibition and fear-potentiated startle are altered in tissue inhibitor of metalloproteinase-2 (TIMP-2) knockout mice. Brain Res. 2005;1051:81–89. [DOI] [PubMed] [Google Scholar]
- 74. Mizoguchi H, Yamada K, Mouri A, et al. Role of matrix metalloproteinase and tissue inhibitor of MMP in methamphetamine-induced behavioral sensitization and reward: implications for dopamine receptor down-regulation and dopamine release. J Neurochem. 2007;102:1548–1560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.