Abstract
Several case series of extreme early-onset obesity due to mutations in the human leptin receptor (LEPR) gene have been reported. In this review we summarize published functional and phenotypic data on mutations in the human LEPR gene causing severe early-onset obesity. Additionally, we included data on six new cases from our obesity center. Literature research was performed using PubMed and OMIM. Functional relevance of mutations was estimated based on reported functional analysis, mutation size, and location, as well as phenotypic characteristics of affected patients. We identified 57 cases with 38 distinct LEPR mutations. We found severe early-onset obesity, hyperphagia, and hypogonadotropic hypogonadism as cardinal features of a complete loss of LEPR function. Other features, for example, metabolic disorders and recurring infections, were variable in manifestation. Obesity degree or other manifestations did not aggregate by genotype. Few patients underwent bariatric surgery with variable success. Most mutations occurred in the fibronectin III and cytokine receptor homology II domains, whereas none was found in cytoplasmic domain. In silico data were available for 25 mutations and in vitro data were available for four mutations, revealing residual activity in one case. By assessing provided information on the clinical phenotype, functional analysis, and character of the 38 mutations, we assume residual LEPR activity for five additional mutations. Functional in vitro analysis is necessary to confirm this assumption.
Keywords: early-onset obesity, leptin melanocortin signaling pathway, leptin receptor deficiency, monogenic obesity
Leptin (LEP) and its receptor (LEPR) are key players in the regulation of body weight and energy homeostasis [1]. LEP is produced mainly in adipocytes, and blood levels are strongly correlated with the amount of body fat [2]. LEP regulates food intake and energy homeostasis by binding to its receptor on neurons in the hypothalamus [3]. Activation of LEPR on proopiomelanocortin (POMC)/cocaine- and amphetamine-regulated transcript neurons results in production of α-melanocyte–stimulating hormone that activates the melanocortin-4 receptor (MC4R) and thereby induces satiety signals. LEPR activation on neuropeptide-Y/agouti-related protein (AgRP) neurons leads to reduced production of orexigenic peptides [4, 5]. In 1998, Clément et al. [6] were the first to describe three subjects with severe early-onset obesity due to a biallelic mutation in the LEPR gene (Clem_1.1 to 1.3). Since then, data on 48 other individuals with biallelic LEPR mutations have been published. The aim of this work is to summarize current knowledge on function and structure of the LEPR and to provide an overview of clinical and laboratory findings in patients with LEPR deficiency. To this end, we reviewed the literature and report six new cases followed at our obesity center.
1. Methods and Patients
We performed a literature search using PubMed and OMIM to find published cases with biallelic LEPR mutations (applied search terms in PubMed included LEPR, human LEPR, leptin-receptor, human leptin-receptor, monogenetic obesity, LEPR and obesity; 1 July 2017). We extracted clinical, biochemical, and functional data. Additionally, we included data obtained from n = 6 subjects with LEPR mutations who presented to our obesity center with severe early-onset obesity.
Ulm_1 is a boy from consanguineous parents from Turkey. He has a homozygous deletion of exons 4 to 20. The 7-year-old boy displayed a BMI of 33.2 kg/m2 (z score +5.4) upon presentation.
Ulm_2 is the son of consanguineous parents of Turkish origin. He has two homozygous mutations in p.P316W and p.W646C. The boy presented with a BMI of 39.3 kg/m2 (z score +6.1) at age 7 years.
Ulm_3 is a German boy born to parents without known consanguinity. He has the homozygous mutation p.H684P. At the age of 9.5 years, he had a BMI of 41.2 kg/m2 (z score +4.5).
Ulm_4 is a German girl born to nonconsanguineous parents. She is compound heterozygous for p.S743P and c.2598-3_2607 deletion. At the age of 15.2 years, she had a BMI of 50.3 kg/m2 (z score +4.0).
Ulm_5 is a 6-year-old girl born to consanguineous Turkish parents. She has the homozygous mutation p.N154Kfs*3. At the age of 5 years, her BMI was 46.1 kg/m2 (z score +7.3)
Ulm_6 is compound heterozygous for p.W625* and p.H684P. At age 14 years, the boy of German origin had a BMI of 53.7 kg/m2 (z score +4.1).
Ulm_1 to 6 or their legal guardian gave written consent to publish their data.
BMI z score calculations were made for subjects were no z score was published but values for age, weight, and height were available. Calculations are based on the World Health Organization standard and are provided for Ulm1-6, Clem_1.1 to 1.3, Maz_1-2, Vau_1, Bey_1, and Gil_1 [7, 8].
Figures were created using GraphPad Prism 6.07 (GraphPad Software, La Jolla, CA) and Inkscape (http://inkscape.org).
We converted all units to the SI system when possible. Using Ensembl.org we standardized and completed genomic information when necessary information was available. Mutation nomenclature is based on the recommendations made by the Human Genome Variation Society when all necessary information was available.
Estimations about functional impact of the respective mutations are based on the literature on LEPR function and by critical investigations of the phenotype, family background, and provided information about functional analysis. We defined five items for functionally relevant criteria: (1) highly suspicious BMI (BMI at 2 years of age >25, 5 years of age >30, and 10 years of age >40); (2) hypogonadotropic hypogonadism; (3) consanguineous parents; (4) highly suspicious variant (large deletion, frameshift, or mutation close to a functionally relevant region; mutation described in other subjects with similar phenotype); and (5) conclusive functional analysis (e.g., in vitro analysis or Sanger sequencing and PCR to detect deletions. In silico analysis were not considered as conclusive analysis. Conclusions on functional relevance are based on the number of fulfilled criteria: “high” indicates high evidence for complete loss of LEPR function (three to five criteria fulfilled); “probably” indicates that the mutation is probably damaging (two to three criteria); “low” indicates low evidence for functional relevance, with in vitro analyses necessary to exclude residual function of LEPR (no to two criteria).
Supplemental Table 4 provides more information about the hypothesized molecular product resulting from the respective mutation: (1) truncated protein, not able to bind LEP; (2) sLEPR-like protein, able to bind LEP in the bloodstream; (3) membrane-anchored LEPR with no ability to bind LEP; and (4) membrane anchored LEPR with no ability for signal transduction.
We based our statements for genetic and protein data on the canonical transcript of the LEPR ENST00000349533.10/Transcript Variant NM_002303.5 using the database UniProt for information about the protein (http://www.uniprot.org/uniprot/P48357) and NCBI for the reference sequence (https://www.ncbi.nlm.nih.gov/nuccore/NM_002303.5).
2. Structure and Function of the LEPR
A. LEPR Isoforms
To date, four membrane-bound and one soluble isoform of the human LEPR have been described [9]. Notably, inconsistent nomenclature is used in the literature between and also within species (Table 1). LEPRb is encoded by the 20-exon canonical transcript. Its translation starts at exon 3 of the transcript. LEPRb is the only isoform with clearly defined functionality and is therefore the focus of most investigations [3]. Further studies are required to better understand the function of the other isoforms.
Table 1.
LEPR Isoforms in Humans and Mice
| Isoforms | Human | Mouse |
|---|---|---|
| Isoform A Alternative splicing | HuB or OB-R219.3: | LEPRa: |
| p.897–1165 missing | p.895–1162 missing | |
| p.892–896: PETFE → RTDIL | p.890–894: PETFE → RTDTL | |
| Isoform B Canonical sequence | HuB or OB-Rb/LEPRb: — | LEPRb: — |
| Isoform C Alternative splicing | HuB or OB-R219.1/LEPRa: | LEPRc: |
| p.907–1165 missing | p.893–1162 missing | |
| p.892–958: PETFEHLFIK ... EKGSVCISDQ | p.890–892: PET → VTV | |
| → MLEGSMFVKS ... KSPSVRNTQE | ||
| Isoform D Alternative splicing | HuB or OB-R219.2: | LEPRd: |
| p.959–1165 missing | p.901–1162 missing | |
| p.892–906: PETFEHLFIKHTASV → KMPGTKELLGGGWLT | p.890–900: PETFEHLFTKH → DISFHEVFIFR | |
| Isoform E Alternative splicing | sLEPR: | LEPRe/sLEPR: |
| p.842–1165 missing | p.806–1162 missing | |
| p.797-805: DNFIPIEKY → GMCTVLFMD |
Source: UniProt (mouse, http://www.uniprot.org/uniprot/P48356; human, http://www.uniprot.org/uniprot/P48357). Alternative nomenclature and protein sequence are given. LEPRb is the canonical sequence of LEPR. It is the most prevalent isoform, highly preserved among species, and has the longest amino acid sequence. LEPRb is the only isoform with clearly defined function and implications for body weight regulations.
Abbreviation: p., amino acid position in the protein.
Structurally, all isoforms share the same extracellular domain with an 820–amino acid (aa) length (Fig. 1). The membrane-bound isoforms have a 21-aa transmembrane domain and a box 1 motif around positions 871 to 879, followed by variable C-terminal lengths due to alternative splicing from position 891 (Table 1). The soluble LEPR is a product of proteolytic ectodomain shedding [10]. It is the only described isoform without intracellular residues and a transmembrane domain and circulates in the bloodstream [11].
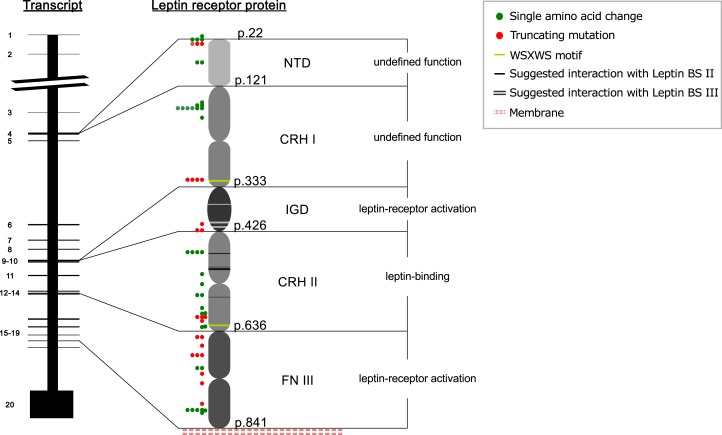
Figure 1.
Simplified depiction of the LEPR gene and the extracellular domain of the mature LEPR protein and visualization of mutations in the human LEPR protein. Transmembrane and cytoplasmatic domains are not depicted, as no human mutations have been described in these domains. Every exon is numbered and assigned to the domain it encodes in the LEPR protein (the structure of the LEPR protein is based on the information of Peelman et al. [12]). The functional relevance of LEPR domains is given. LEP interacting sites are marked by white or black lines in the protein sketch and are positioned in CRHI/immunoglobulin-like domain (p.L372, A409, Y411, H419, H420) and CRHII (p.L471, Y472, F500, IFLL503-506, F563). Positions of WSXWS motifs are depicted as green lines: 319 to 323 and 622 to 626. Colored dots indicate and type affected protein position of human LEPR mutations; mutations result in single amino acid changes (green dots), or a truncated protein (red dots). BS, binding site; IGD, immunoglobulin-like domain; p., amino acid position in the protein.
B. Receptor Structure and Function
The LEPR is a class I cytokine [Janus kinase (JAK)/signal transducer and activator of transcription (STAT)] receptor and shares many similarities with, for example, granulocyte colony-stimulating factor–, erythropoietin-, and oncostatin M–specific receptors [9, 12].
The extracellular domain is divided into six subdomains (Fig. 1). The N-terminal domain is not required for LEP binding or signaling and is currently of undefined function. In vitro studies demonstrate that deletion of the subsequent cytokine receptor homology (CRH)I domain diminishes but does not abolish LEP signaling. In part, this appears to be caused by reduced cell surface expression [12–15]. LEP uses three binding sites denoted I to III to engage its receptor. Binding site I is of undefined function and its mutations affect receptor activation only marginally. Binding site II is required for receptor binding by interacting with the LEPR CRHII domain. Binding site III, in contrast, is responsible for receptor activation by interacting with the LEPR immunoglobulin-like domain (Fig. 1) [12, 16, 17]. LEPR activation furthermore requires the two fibronectin type III (FNIII) domains in membrane proximity [15, 16, 18]. The intracellular domain consists of a box 1 motif without kinase activity for binding of JAK2 and a variable number of tyrosine residues for mutual phosphorylation and activation of signaling cascades. LEPRb has three tyrosine residues (Y986, Y1079, Y1141) and is the only isoform that can induce STAT signaling [12, 19–21].
C. LEPR Signaling Cascade
In vitro studies show ligand-independent homodimerization of LEPR [22]. In the process of LEPR activation, LEP’s binding site II interacts with the CRHII domain of the LEPR [12, 16, 23]. This causes a conformational change that favors formation of 2:2 or even higher complexes [13]. Lacking intrinsic kinase activity, LEPR signaling depends on JAK2 association to the receptor’s box 1 motif [24]. Conformational changes of the LEP–LEPR complex result in the activation of JAK2 by mutual phosphorylation of JAK2 and tyrosine residues of the intracellular receptor domain. Each of the phosphorylated tyrosine residues binds specific proteins that are crucial for further downstream signaling. For example, tyrosine residue Y986 accumulates SOCS3 and initiates a feedback loop mainly by inhibiting phosphorylation of the other tyrosine residues during prolonged signaling [25–27]. Y1079 plays a dominant role in activating STAT5, and Y1141 activates STAT3. STATs are subsequently phosphorylated by JAK2, dimerize, and translocate to the nucleus where they induce SOCS3 and POMC expression while repressing AgRP [15, 28, 29].
3. Identification of Mutations and Inheritance Patterns
We report six new patients with biallelic LEPR mutations and summarize 51 published cases. Reported mutations include 13 mutations that led to single amino acid changes, as well as 25 deletions, duplications, insertions, or nonsense mutations that were predicted to result in truncated LEPR proteins. Most mutations occurred in the FNIII and CRHII domains, whereas none was found in the intracellular domain. Detailed information is given in Table 2 and Table 3 and Fig. 1. Obesity caused by LEPR mutations was inherited in an autosomal-recessive pattern. There was no sex difference in distribution (27 females/30 males) (Supplemental Table 1). Ten subjects had a compound heterozygous mutations (Ulm_2 and Ulm_6) [30–34]. One patient had uniparental disomy, resulting in homoallelism in the LEPR gene inherited from the heterozygous father [35]. Consistent with the inheritance pattern, most patients were born to consanguineous parents, and obesity due to LEPR mutations aggregates in cultures with consanguineous marriages [36, 37].
Table 2.
Overview of Mutations in the Human LEPR
| First Author and Year of Publication | Overview: LEPR Mutations |
||||||
|---|---|---|---|---|---|---|---|
| Number of Cases (n) | Case ID | Patient Nationality | Mutation in the Coding DNA (c.) | Mutation in the Mature Protein (p.) | Affected Domain | Provided Functional Analysis | |
| Clement et al. 1998 [6] | 3 | Clem_1.1, Clem_1.2, Clem_1.3 | Algerian | c.2597 + 1G>A | n.a. | FNIII | PCR and sequencing |
| Farooqi et al. 2007 [30] | 2 | Far_2.2, Far_2.1 | Turkish | n.a. | 11-bp del in codon 70 | NTD | In silico |
| Farooqi et al. 2007 [30] | 3 | Far_4.3, Far_4.2, Far_4.1 | Southern European | n.a. | p.W31* | NTD | n.a. |
| Farooqi et al. 2007 [30] | 1 | Far_3 | Iranian | n.a. | 66-bp del in codon 514 | CRHII | In silico |
| Farooqi et al. 2007 [30] | 1 | Far_5 | Turkish | c.1226C>A | p.A409E | IGD | In vitro |
| Farooqi et al. 2007 [30] | 1 | Far_6 | Norwegian | n.a. | p.W664R | FNIII | In vitro |
| Farooqi et al. 2007; [30] Ulm | 2 | Far_7; Ulm_3 | White (United Kingdom); German | c.2051A>C | p.H684P | FNIII | In vitro |
| Le Beyec et al. [35] 2013 | 1 | Bey_1 | French | c.1871dupA | p.N624Kfs*21 | CRHII + FNIII | In silico |
| Kakar et al. 2013 [40] | 5 | Kak_1.1.1, Kak_1.1.2, Kak_1.2, Kak_1.3. Kak_1.4 | Pakistani | c.1603 + 5G>C | p.R468Sfs*33 | CRHII | In silico |
| Gill et al. 2013 [42] | 2 | Gil_2.1, Gil_2.2 | Sudanese | c.479delA | p.H160Lfs*10 | CRHI | In silico |
| Gill et al. 2013 [42] | 1 | Gil_1 | Guinean | c.556delT | p.C186Afs*28 | CRHI | In silico |
| Saeed et al. 2014 and 2015 [34, 38] | 4 | Sae_2, Sae2_3, Sae2_4, Sae2_5 | Pakistani | c.2396-1G>T | n.a. | FNIII | In silico |
| Saeed et al. 2014 and 2015 [34, 38] | 2 | Sae_1, Sae2_6 | Pakistani | c.1675G>A | p.W558* | CRHII | In silico Ilumina, Sanger |
| Huvenne et al. 2015 [32] | 1 | Huv_2 | French | c.1810T>G | p.C604G | CRHII | In silico |
| Huvenne et al. 2015 [32] | 1 | Huv_3 | Portuguese | c.2357T>C | p.L786P | FNIII | In silico |
| Huvenne et al. 2015 [32] | 1 | Huv_4 | Turkish | c.2491G>A | p.H800_N831del | FNIII | In silico |
| Huvenne et al. 2015 [32] | 5 | Huv_5, Huv_6, Huv_7, Huv_8, Huv_9 | French (Reunion Island) | Del exon 6–8 | p.P166Cfs*7 | CRHI | In silico, PCR |
| Ulm | 1 | Ulm_1 | Turkish | Del exon 4–20 | n.a. | CRHI-NTD | n.a. |
| Ulm | 1 | Ulm_4 | German | Comp. het. c.2227 T>C and c.2598-3_2607delTAGAATGAAAAAG | Comp. het. p.S743P and p.Q865_K870 | FNIII + CRHII | In silico |
| Ulm | 1 | Ulm_5 | Turkish | p.N154Kfs*3 | c.461dupA | CRHI | In silico |
| Ulm | 1 | Ulm_6 | German | Comp. het. c.1874G>A and c.2051A>C | Comp. het. p.W625* and p.H684P | FNIII + CRHII | In vitro (p.H684P; see Farooqi et al. 2007 [30]) |
| Farooqi et al. 2007 [30] | 3 | Far_1.3, Far_1.2, Far_1.1 | Bangladeshi | n.a. | 4-bp del in codon 22 | NTD | In silico |
| Maezen et al. 2011 [37] | 2 | Maz_1, Maz_2 | Egyptian | c.946C>A | p.P316T | CRHI | In silico |
| Andiran et al. 2011 [31]; Ulm | 2 | And_1, Ulm_2 | Turkish | c.946C>A and c. n.a. | p.P316T and p.W646C (both homozygous) | CRHI + FNIII | In silico |
| Huvenne et al. 2015 [32] | 1 | Huv_10 | French (Reunion Island) | Comp. het. c.1604–1G>A and del exon 6–8 | Comp. het. p. n.a and p.P166Cfs*7 | CRHII + CRHI | In silico |
| Hannema et al. 2016 [33] | 1 | Han_2 | Dutch | c.1604–8A>G | K536Sfs*34 and p.V535Dfs*3a | CRHII | In silico |
| Vauthier et al. 2012 [39] | 1 | Vau_1 | French | Del of DNAJC6 and parts of LEPR | n.a. | NTD + CRII | PCR, MPLC |
| Huvenne et al. 2015 [32] | 2 | Huv_11.1, Huv_11.2 | French | Comp. het. c.1264T>C and c.2131dup | Comp. het. p.Y422H and p.T711N fs*18 | IGD + FNIII | In silico |
| Saeed et al. 2015 [34] | 2 | Sae2_2.1, Sae2_2.2 | Pakistani | c.1810T>A | p.C604S | CRHII | In silico |
| Saeed et al. 2015 [34] | 1 | Sae2_1 | Pakistani | Mutation not in transcript | n.a. | — | In silico |
| Hannema et al. 2016 [33] | 1 | Han_1 | Dutch | Comp. het. c.1753–1dupG and c.2168C>T | Comp. het. p.M585Dfs*2 and p.S723F | CRHII | In silico Ilumina, Sanger |
| Farooqi et al. 2007 [30] | 1 | Far_8 | White (United Kingdom) | Comp. het. c. n.a. and c.1835G>A | Comp. het. 1 bp del in codon 15 and p.R612H | NTD + CRHII | In vitro (p.R612H) |
Included are number of cases, case ID and nationality, location of the mutation in the LEPR protein, affected domain, and provided information about functional analysis. Estimation of the functional relevance of the respective mutation was made based on predefined criteria. Criteria for functional relevance were (1) highly suspicious BMI, (2) hypogonadotropic hypogonadism, (3) consanguineous parents, (4) highly suspicious variant, and (5) conclusive functional analysis. Conclusions on functional relevance are based on the number of fulfilled criteria: “High” indicates high evidence for complete loss of LEPR function (three to five criteria fulfilled); “Probably” indicates that the mutation is probably damaging (two to three criteria); “Low” indicates low evidence for functional relevance, with in vitro analyses necessary to exclude residual function of LEPR (two or fewer criteria).
Abbreviations: c., cDNA position in the gene; comp. het., compound heterozygous; del, deletion; fs, frameshift; HH, hypogonadotropic hypogonadism; MPLC, medium pressure liquid chromatography, n.a., no information available; p., amino acid position in the protein; Rf, residual function; *, premature stop codon.
Published as corresponding to p.K597Sfs*34 and p.V596Dfs*3 in the original paper. Based on the experimentally validated changes in the RNA, we assume the correct mutations to be p.K536Sfs*34 and p.V535Dfs*3.
Table 3.
Estimation of Functional Relevance of Mutations in the Human LEPR
| Overview: LEPR Mutations | Estimation of Functional Relevance |
|||||
|---|---|---|---|---|---|---|
| Criteria for Functional Relevance |
Evidence for Functional Relevance | |||||
| Case ID | Suspicious BMI | HH | Consanguineous Parents | Suspicious Variant | Conclusive Functional Analysis | |
| Clem_1.1, Clem_1.2, Clem_1.3 | X | X | X | X | X | High |
| Far_2.2, Far_2.1 | X | X | X | X | High | |
| Far_4.3, Far_4.2, Far_4.1 | X | X | X | X | High | |
| Far_3 | X | X | X | High | ||
| Far_5 | X | X | X | X | High | |
| Far_6 | X | X | X | X | High | |
| Far_7; Ulm_3 | X | X | High | |||
| Bey_1 | X | X | X | X | High | |
| Kak_1.1.1, Kak_1.1.2, Kak_1.2, Kak_1.3. Kak_1.4 | X (not for Kak_1.1.1) | X | X | High | ||
| Gil_2.1, Gil_2.2 | X | X | X | X | High | |
| Gil_1 | X | X | X | High | ||
| Sae_2, Sae2_3, Sae2_4, Sae2_5 | X | X | X | High | ||
| Sae_1, Sae2_6 | X | X | X | X | High | |
| Huv_2 | X | X | X | High | ||
| Huv_3 | X | X | X | High | ||
| Huv_4 | X | X | X | High | ||
| Huv_5, Huv_6, Huv_7, Huv_8, Huv_9 | X | X | X | High | ||
| Ulm_1 | X | X | X | High | ||
| Ulm_4 | X | X | X | High | ||
| Ulm_5 | X | X | X | High | ||
| Ulm_6 | X | X | X | High | ||
| Far_1.3, Far_1.2, Far_1.1 | X (only Far_1.1) | X | X | Probably | ||
| Maz_1, Maz_2 | X | X | X | Probably | ||
| And_1, Ulm_2 | X | X | X | Probably | ||
| Huv_10 | X | X | Probably | |||
| Han_2 | X | X | Probably | |||
| Vau_1 | Low | |||||
| Huv_11.1, Huv_11.2 | X | Low (for p.Y422H) | ||||
| Sae2_2.1, Sae2_2.2 | X | Low | ||||
| Sae2_1 | X | X | Low | |||
| Han_1 | Low (for p.S723F) | |||||
| Far_8 | Rf | |||||
4. Phenotype of Patients With Biallelic LEPR Mutations
A. Weight Gain and Hyperphagia
The most striking symptom of a loss of LEPR function was extreme and early-onset obesity. Birth weight was reported to be normal in all cases with regular pregnancies (data not shown), followed by extreme weight gain in the first months of life [6, 30, 38]. Mean body mass index (BMI, kg/m2) for subjects <16 years of age was 40.3 (n = 31; 13 females, 18 males; mean age, 8 years) (Supplemental Table 1). Median body fat percentage and z score were slightly higher in females compared with males (Fig. 2A and 2B; Supplemental Table 1), but comparison is limited due to differences in age and methodology. In almost all cases, hyperphagia was described except in Gil_2.1, Gil_2.2, and Han_2. Consistently, food impulsivity seems to be a common feature. Aggressive behavior was reported for Clem_1.1 to 1.3 and Ulm_3, 5, and 6 when food was withheld [6]. Lifestyle changes with increased physical activity and caloric restrictions mostly failed to reduce body weight in the long term [6, 35]. Clem_1.1 to 1.3 and Ulm_3 and 6 underwent a period of extreme calorie restriction without successful weight loss. Clem_1.1 to 1.3 had decelerated growth velocity as a result of caloric restriction [6].
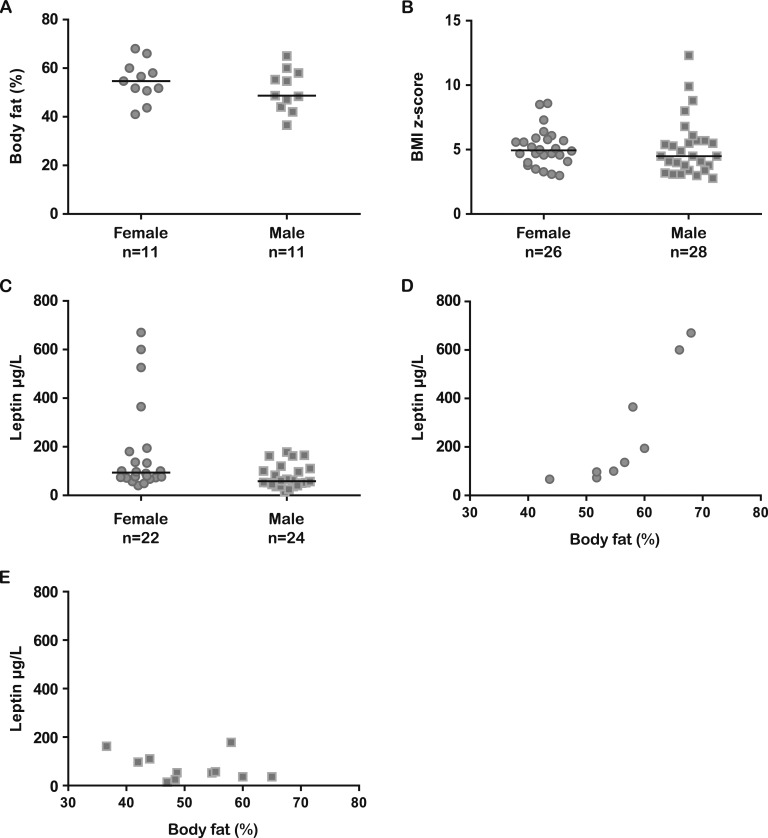
Figure 2.
Body fat percentages, BMI z scores, and serum LEP concentrations in patients with biallelic LEPR mutations. (A) Body fat percentage by sex (mean age females, 18.2 y; mean age males, 10.1 y; median females, 54.7%; median males 48.7%). (B) BMI z score by sex (mean age females, 15.8 y; mean age males, 7.8 y; median females, 4.95; median males, 4.5). (C) Serum LEP concentrations by sex (mean age females, 17.2 y; mean age males, 8.0 y; median females, 93.5 µg/L; median males, 58.4 µg/L). (D) Correlation of body fat percentage and serum LEP concentrations in females (mean age, 19.4 y; n = 9). (E) Correlation of body fat percentage and serum LEP concentrations in males (mean age, 10.1 y; n = 11).
B. Longitudinal Growth
Obese subjects often show accelerated growth with a decreased GH response and IGF-1 concentration. There is some discussion about the effects of impaired LEP signaling on longitudinal growth. Mouse models representing the counterpart of LEP (ob/ob; obese mouse model) and LEPR (db/db; diabetes mouse model) deficiency in humans show decreased linear growth and low GH concentrations [30, 35, 39–41] suggesting a role of LEP signaling for longitudinal growth. Reports on growth in humans with impaired LEPR function are heterogeneous. Farooqi et al. [30] and Kakar et al. [40] reported normal growth in childhood and diminished final height in adulthood due to a diminished pubertal growth spurt. In contrast, Gil_2.1, Sae_1, Ulm_2, Ulm_6, and Clem1.1 to 1.3 showed diminished growth throughout childhood [6, 38, 42] and Maz_1, And_1, Gil_1, Sae_2, and Ulm_3 presented with accelerated growth (Supplemental Table 1) [31, 37, 38, 42].
Likewise, serum concentrations of GH and IGF-1 were heterogeneous. Clément et al. [6], Le Beyec et al. [35], and Huvenne et al. [32] (Huv_2, 6, 8, and 9) reported low GH values. Clément et al. [6] and Andiran et al. [31] found low IGF-1 concentrations. Conversely, Farooqi et al. [30] found normal IGF-1 and GH concentrations. Vau_1, Bey_1, Han_1 and 2, and Ulm_1 to 6 had normal IGF-1 values (Supplemental Table 1). Data for GH serum concentration were scarce and are therefore not listed in a table. Taken together, these data do not allow for a conclusion as to whether LEP signaling plays a role in longitudinal growth.
C. Pubertal Development and Fertility
Comparable to the findings in ob/ob and db/db mice and individuals with LEP deficiency, subjects with impaired LEPR function showed disrupted pubertal development [41, 43, 44]. In healthy individuals, circulating LEP concentrations increase before the onset of puberty [45], and some studies indicate that LEP affects GnRH neuron activity and GnRH secretion by crosstalk with kisspeptin [46]. Accordingly, pulsatile secretion of GnRH (the trigger of increased LH and FSH secretion and gonadal activation) is disturbed in individuals with LEP signaling defects [47, 48].
Von Schnurbein et al. [43] showed that regular subcutaneous injections of a LEP substitute (metreleptin; a recombinant analog of LEP) induced and normalized pulsatile gonadotropin secretion in a LEP-deficient adolescent girl. LEP substitution cannot be considered as a treatment option for patients with LEPR dysfunction. Interestingly, for LEPR-deficient patients >20 years of age, normalization of sex hormones and delayed menses have been reported (see Far_4.1 to 4.3 in Supplemental Tables 1 and 2). This normalization seems to be favored by strict weight control (see Clem_1.1, Bey_1, Huv_5, Ulm_4, and Ulm_6; for details, see “Treatment Outcomes With Bariatric Surgery” below). For instance, Clem_1.1 showed no pubertal development when she was 13.5 years old [6]. At the age of 18 years, Clem_1.1 underwent bariatric surgery (for details, see “Treatment Outcomes With Bariatric Surgery” below) and lost 50 kg in 6 months. After surgery, sex hormones normalized and she was able to conceive. The mother gained 50 kg during pregnancy without any features of gestational diabetes mellitus. The fetus developed regularly [49].
Huv_5, Huv_11.1, and Huv_11.2 showed no signs of hypogonadotropic hypogonadism [32]. It would be interesting to assess whether there was a residual STAT5 phosphorylation by the LEPR in these cases. In mouse models, an exchange of tyrosine residue 1138 to serine induced impaired STAT3 signaling with residual STAT5 function [50]. This resulted in an obese phenotype with normal pubertal development and fertility. Similar mutations with residual STAT5 activity may also occur in humans.
D. Metabolic Features
LEP has been shown to have a direct effect on insulin sensitivity and was suggested as a novel therapeutic agent in metabolic disorders, including insulin resistance and type 2 diabetes [51]. In the db/db mouse model, the incidences of type 2 diabetes and hepatic steatosis are increased [52, 53]. Summarizing the published data in humans, four individuals (Far_4.2, Far_4.1, Gil_1, and Huv_11.1) suffered from type 2 diabetes. Moreover, 17 patients with elevated insulin concentrations were reported. Of these, five were <10 years of age (Supplemental Table 3). This suggests that LEPR-deficient subjects may be at risk for early-onset insulin resistance and diabetes [54].
In 2013, von Schnurbein et al. [55] reported hepatic steatosis in a LEP-deficient patient, a feature that is also common in lipodystrophic subjects who have very low LEP levels. Findings indicative of hepatic steatosis seen upon ultrasound examination were reported for Gil_1 and Ulm_1, 2, 4, and 6 [42]. Fasting triglyceride concentrations were reported in few patients and were increased in Han_1 and Ulm_1. Maz_1 and 2 had triglyceride levels within the reference range.
E. Thyroid Function
Several in vitro and in vivo studies propose an involvement of LEP in thyroid function. A decrease in LEP concentration diminishes TRH expression in the hypothalamic paraventricular nucleus, resulting in decreased TSH and thyroid hormone production. LEP may affect TRH expression directly by activating LEPRb on TRH neurons [56], or indirectly by enhancing α-melanocyte–stimulating hormone and suppressing AgRP production and thus disinhibiting TRH secretion [57].
In concordance with this, Clément et al. [6] and Montague et al. [58] propose a role for LEP in controlling thyroid function in humans. Indeed, there are few reports on impaired function of the thyroid axis in patients with LEPR deficiency (Clem_1.1 to 1.3 and Huv_9, hypothalamic hypothyroidism; Bey_1, low T4 normal basal TSH; Ulm_2, elevated basal TSH; Ulm_4, reduced free thyroxin) (Supplemental Table 3).
F. Immune Function
The effect of LEP or LEPR deficiency in ob/ob or db/db mouse models is well studied. The lack of LEP leads to an abnormal cytokine secretion and thymic hypotrophy that can be remedied by metreleptin administration in ob/ob mice [59]. Patients with LEP deficiency due to a mutation in the LEP gene may also show increased susceptibility to infections and show a decreased ratio of CD4/CD8 T-lymphocytes in these patients [60, 61]. Likewise, evidence for immune dysfunction has been described for subjects with LEPR dysfunction. For example, Far_1-8, Maz_1 and 2, Ulm_6, and Sae_1 and 2 suffered from pulmonary infections at an early age and had frequent episodes of diarrhea. Farooqi et al. [30] reported reduced T-cell number with low proliferation rates and compensatory increased in B-cell counts [30]. Vauthier et al. [39] described a reduced CD4/CD8 T-cell ratio (low CD4 and high CD8), but more natural killer cells in Vau_1.
G. LEP Serum Concentrations
We found a wide range between the reported LEP serum concentrations in subjects with LEPR deficiency (Supplemental Table 1). This might in part be attributable to assay variability. Additionally, truncating LEPR mutations leading to a soluble LEPR (sLEPR)–like product (as is the case for Clem_1.1-1.3) result in highly elevated serum LEP concentrations (measured as bound or total LEP) [62]. Most authors have related the measured LEP concentrations to a BMI- or fat percentage–matched reference population. Thereby, normal serum LEP concentrations were reported by Farooqi et al. [30] in 2007, Saeed et al. [38] in 2014, and Hannema et al. [33] in 2016. Elevated values compared with BMI-matched individuals were reported by Le Beyec et al. [35] in 2013, Gill et al. [42] in 2014, Huvenne et al. [32] in 2015, and Saeed et al. [34] in 2015. Using the published values, LEP concentrations and body fat percentage seem to correlate stronger in females than in males. However, this comparison is limited by the large age difference between the groups (Fig. 2D and 2E). Standardized analytical methods are needed for qualitative statements about LEP concentration in LEPR-deficient subjects.
H. Treatment Outcomes With Bariatric Surgery
Reports about the success of bariatric surgery in subjects with LEPR defects gave conflicting results [32, 35, 49]. We are aware of six subjects with LEPR defects who have undergone bariatric surgery (Table 4). Clem_1.1, Huv_2, and Ulm_4 were not able to maintain reduced body weight. Clem_1.1 regained weight after the above-mentioned unexpected pregnancy (see “Pubertal Development and Fertility” above and Supplemental Table 1) [6, 32, 49]. Ulm_4 had her menarche with six regular cycles during the phase of weight reduction. After weight regain, menses cheesed again.
Table 4.
LEPR-Deficient Patients Who Underwent Bariatric Surgery
| Patient ID | Age at Surgery (y) | Sex | Type of Intervention | Achieved Weight Loss (kg) | Complications/Weight Regain |
|---|---|---|---|---|---|
| Clem_1.1 | 16 | F | Abdominoplasty | n.a. | n.a. |
| 24 | Gastric bypass | 50 | Weight regain after pregnancy (+50 kg) | ||
| Bey_1 | 16 | M | Gastric banding | 46 | Gastric band slippage, weight regain |
| 18 | Gastroplasty | 40 | — | ||
| Huv_2 | n.a. | F | Gastric bypass | 45 | Weight regain (+34 kg) |
| Huv_3 | n.a. | M | Gastroplasty | −44% of body weight | No follow-up data |
| Ulm_4 | 18 | F | Sleeve gastrectomy | 30 | Weight regain (+19 kg) |
| Ulm_6 | 14 | M | Gastric banding | 47 | Gastric band slippage, weight regain (+10 kg) |
We report age at surgery, type of surgery, weight loss in kilograms, reported complications, and the weight regain surgery where information is available.
Abbreviations: F, female; M, male; n.a., no information available.
Bey_1 and Ulm_6 successfully managed to reduce and maintain body weight after surgery, whereby sex hormones normalized [35]. There is no clear evidence whether this was due to the surgery or his advanced age. However, for Ulm_6, malformed sperms, reduced sperm number, and sperm velocity were reported. There are no long-term reports about Huv_3 (Table 4).
In summary, bariatric surgery seems less effective in the female subjects, especially in the long term. The normalization of sex hormones after surgery is in line with outcomes in obese patients without known underlying genetic cause and suggests a mechanism independent of LEP signaling [63, 64].
I. Treatment Outcomes With MC4R Agonist Setmelanotide
Causal treatment of patients with LEPR mutations is not yet available. Setmelanotide, an MC4R agonist, is an interesting candidate. Setmelanotide has been shown to be extremely effective in reducing appetite and body weight in patients with POMC deficiency [65]. It is physiologically plausible that patients with LEPR defects will show a similar response. Preliminary results of a phase 3 clinical trial show substantial weight loss (−19.6%) in one patient with LEPR deficiency after 22 weeks of setmelanotide treatment without reported adverse events [66].
J. Psychomotor Development and Social Behavior
Gil_1, Vau_1, Ulm_3, and Ulm_5 have been reported to be developmentally delayed. However, in the case of Vau_1, this could also be due to a concomitant auxillin-1 deficiency caused by the reported 80-kb deletion, which also affects the DNAJC6 gene [39, 67]. Ulm_3 was diagnosed with hearing impairment and delays in psychomotor, statomotoric, and speech development. These delays occurred in the setting of a complicated pregnancy with gestational diabetes. He was born at 30 weeks with a body weight of 1.6 kg and bilateral hearing impairment. Ulm_5 was diagnosed with cognitive delays. Clément et al. [6] described impulsive behavior, emotional lability, and social disability in Clem_1.1-1.3. In concordance with these findings, we observed defiant behavior and various degrees of psychological abnormalities, including addictive behavior, in the patients Ulm_1 to 6.
5. Functional Analysis of Mutations in Human LEPR
Summarizing clinical data, severe early-onset obesity, hyperphagia, and hypogonadotropic hypogonadism are cardinal features of a complete loss of LEPR function (Supplemental Table 1). In contrast, symptoms such as recurring infections, altered growth, developmental delay, and metabolic disorders showed variable manifestations. Differences in the clinical appearance may be related to residual LEPR activation and may depend on the character and location of the mutation. We therefore investigated whether there is any relationship between genotype and phenotype. For instance, variants affecting solely the N-terminal domain and CRHI domains are thought to be less crucial for receptor function [12]. Furthermore, compound heterozygous variants could have partially preserved function when different domains are affected. Functional in silico analyses were available for 35 mutations (Table 2 and Table 3). Functional in vitro data were available for four mutations. Farooqi et al. [30] confirmed the functional impairment of p.A409E, p.W664R, and p.H684P and reported residual STAT3 phosphorylation in p.R612H. According to our own investigations of the literature, evaluation of the clinical appearance of the described subjects, as well as provided functional analysis, we further expect residual LEPR function in Vau_1, Huv_11.1, Huv_11.2, Sae2_2.1, Sae2_2.2, Sae2_1, and Han_1 (Table 2 and Table 3). These suggestions need to be verified by functional in vitro analysis. Based on the available data, we did not identify an obvious genotype–phenotype relationship, which might be partly due to incomplete information in the literature.
In another approach we categorized mutations in the human LEPR based on the hypothesized impact of respective mutation on the protein product, should translation still occur: (1) truncated protein not able to bind LEP; (2) sLEPR-like molecule; (3) membrane-bound LEPR with no ability to bind LEP; and (4) membrane-bound LEPR with no ability to elicit cellular signaling (Supplemental Table 4). We suggest that mutations belonging to category 2 will result in a sLEPR-like protein resulting in elevated total and bound LEP concentration in the bloodstream, as reported by Lahlou et al. [62]. Furthermore, mutations belonging to categories 3 and 4 might be interesting candidates for further investigations of genotype–phenotype correlations and functional analysis.
6. Conclusion
We present a structured and comprehensive analysis of a large patient cohort with biallelic LEPR mutations comprising all published cases to date. In total, 57 subjects including six yet unpublished patients from our outpatient clinic with 38 distinct LEPR mutations were identified. Most mutations occurred in the FNIII and CRHII domains.
Functional aspects of the mutations were available from in silico or in vitro analyses for 29 mutations, showing residual STAT3 phosphorylation in one mutation. Considering clinical phenotype and character of the respective mutations, we suspect residual function in five additional mutations.
LEPR deficiency causes severe early-onset obesity, hyperphagia, and hypogonadotropic hypogonadism. Association with immune dysfunction, growth restriction, psychomotor delays, and metabolic disorders is variable without any identified genotype–phenotype relationship. Biallelic LEPR mutations should be considered a rare and serious disease associated with severe social and psychological burdens for patients and their families. We propose that findings of LEPR mutations in patients should be harmonized with associated phenotypic characteristics in a structured and comprehensive way by establishing an international registry for this rare disease.
Supplementary Material
Acknowledgments
We acknowledge Prof. Carsten Bergmann (Bioscientia, Center for Human Genetics, Ingelheim am Rhein), Dr. Ulrich Paetow from the University Medical Center Frankfurt am Main, and Dr. Elke Froehlich-Reiterer from the Medical University of Graz for referring patients and for providing genetic and clinical data.
Financial Support: A.N. and J.-B.F. are supported by the International Graduate School in Molecular Medicine Ulm.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- aa
amino acid
- AgRP
agouti-related protein
- BMI
body mass index
- CRH
cytokine receptor homology
- db
diabetes mouse model
- FNIII
fibronectin type III
- JAK
Janus kinase
- LEP
leptin
- LEPR
LEP receptor
- MC4R
melanocortin-4 receptor
- NTD
N-terminal domain
- ob
obese mouse model
- POMC
proopiomelanocortin
- sLEPR
soluble LEPR
- STAT
signal transducer and activator of transcription
References and Notes
- 1. Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372(6505):425–432. [DOI] [PubMed] [Google Scholar]
- 2. Considine RV, Sinha MK, Heiman ML, Kriauciunas A, Stephens TW, Nyce MR, Ohannesian JP, Marco CC, McKee LJ, Bauer TL, et al. Serum immunoreactive-leptin concentrations in normal-weight and obese humans. N Engl J Med. 1996;334(5):292–295. [DOI] [PubMed] [Google Scholar]
- 3. Allison MB, Myers MG Jr. 20 Years of leptin: connecting leptin signaling to biological function. J Endocrinol. 2014;223(1):T25–T35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwartz MW, Woods SC, Porte D Jr, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404(6778):661–671. [DOI] [PubMed] [Google Scholar]
- 5. Dubern B, Clement K. Leptin and leptin receptor-related monogenic obesity. Biochimie. 2012;94(10):2111–2115. [DOI] [PubMed] [Google Scholar]
- 6. Clément K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, Gourmelen M, Dina C, Chambaz J, Lacorte JM, Basdevant A, Bougnères P, Lebouc Y, Froguel P, Guy-Grand B. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392(6674):398–401. [DOI] [PubMed] [Google Scholar]
- 7. WHO Multicentre Growth Reference Study Group WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr Suppl. 2006;450:76–85. [DOI] [PubMed] [Google Scholar]
- 8. de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85(9):660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, Muir C, Sanker S, Moriarty A, Moore KJ, Smutko JS, Mays GG, Wool EA, Monroe CA, Tepper RI. Identification and expression cloning of a leptin receptor, OB-R. Cell. 1995;83(7):1263–1271. [DOI] [PubMed] [Google Scholar]
- 10. Wauman J, De Ceuninck L, Vanderroost N, Lievens S, Tavernier J. RNF41 (Nrdp1) controls type 1 cytokine receptor degradation and ectodomain shedding. J Cell Sci. 2011;124(Pt 6):921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chua SC Jr, Koutras IK, Han L, Liu SM, Kay J, Young SJ, Chung WK, Leibel RL. Fine structure of the murine leptin receptor gene: splice site suppression is required to form two alternatively spliced transcripts. Genomics. 1997;45(2):264–270. [DOI] [PubMed] [Google Scholar]
- 12. Peelman F, Zabeau L, Moharana K, Savvides SN, Tavernier J. 20 Years of leptin: insights into signaling assemblies of the leptin receptor. J Endocrinol. 2014;223(1):T9–T23. [DOI] [PubMed] [Google Scholar]
- 13. Moharana K, Zabeau L, Peelman F, Ringler P, Stahlberg H, Tavernier J, Savvides SN. Structural and mechanistic paradigm of leptin receptor activation revealed by complexes with wild-type and antagonist leptins. Structure. 2014;22(6):866–877. [DOI] [PubMed]
- 14. Carpenter B, Hemsworth GR, Wu Z, Maamra M, Strasburger CJ, Ross RJ, Artymiuk PJ. Structure of the human obesity receptor leptin-binding domain reveals the mechanism of leptin antagonism by a monoclonal antibody. Structure. 2012;20(3):487–497. [DOI] [PubMed]
- 15. Zabeau L, Defeau D, Van der Heyden J, Iserentant H, Vandekerckhove J, Tavernier J. Functional analysis of leptin receptor activation using a Janus kinase/signal transducer and activator of transcription complementation assay. Mol Endocrinol. 2004;18(1):150–161. [DOI] [PubMed] [Google Scholar]
- 16. Peelman F, Iserentant H, De Smet AS, Vandekerckhove J, Zabeau L, Tavernier J. Mapping of binding site III in the leptin receptor and modeling of a hexameric leptin.leptin receptor complex. J Biol Chem. 2006;281(22):15496–15504. [DOI] [PubMed] [Google Scholar]
- 17. Peelman F, Van Beneden K, Zabeau L, Iserentant H, Ulrichts P, Defeau D, Verhee A, Catteeuw D, Elewaut D, Tavernier J. Mapping of the leptin binding sites and design of a leptin antagonist. J Biol Chem. 2004;279(39):41038–41046. [DOI] [PubMed] [Google Scholar]
- 18. Fong TM, Huang RR, Tota MR, Mao C, Smith T, Varnerin J, Karpitskiy VV, Krause JE, Van der Ploeg LH. Localization of leptin binding domain in the leptin receptor. Mol Pharmacol. 1998;53(2):234–240. [DOI] [PubMed] [Google Scholar]
- 19. Lee GH, Proenca R, Montez JM, Carroll KM, Darvishzadeh JG, Lee JI, Friedman JM. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379(6566):632–635. [DOI] [PubMed] [Google Scholar]
- 20. Chen H, Charlat O, Tartaglia LA, Woolf EA, Weng X, Ellis SJ, Lakey ND, Culpepper J, Moore KJ, Breitbart RE, Duyk GM, Tepper RI, Morgenstern JP. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84(3):491–495. [DOI] [PubMed] [Google Scholar]
- 21. Cioffi JA, Shafer AW, Zupancic TJ, Smith-Gbur J, Mikhail A, Platika D, Snodgrass HR. Novel B219/OB receptor isoforms: possible role of leptin in hematopoiesis and reproduction. Nat Med. 1996;2(5):585–589. [DOI] [PubMed] [Google Scholar]
- 22. Bacart J, Leloire A, Levoye A, Froguel P, Jockers R, Couturier C. Evidence for leptin receptor isoforms heteromerization at the cell surface. FEBS Lett. 2010;584(11):2213–2217. [DOI] [PubMed] [Google Scholar]
- 23. Niv-Spector L, Gonen-Berger D, Gourdou I, Biener E, Gussakovsky EE, Benomar Y, Ramanujan KV, Taouis M, Herman B, Callebaut I, Djiane J, Gertler A. Identification of the hydrophobic strand in the A-B loop of leptin as major binding site III: implications for large-scale preparation of potent recombinant human and ovine leptin antagonists. Biochem J. 2005;391(Pt 2):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tartaglia LA. The leptin receptor. J Biol Chem. 1997;272(10):6093–6096. [DOI] [PubMed] [Google Scholar]
- 25. Bjørbæk C, Lavery HJ, Bates SH, Olson RK, Davis SM, Flier JS, Myers MG Jr. SOCS3 mediates feedback inhibition of the leptin receptor via Tyr985. J Biol Chem. 2000;275(51):40649–40657. [DOI] [PubMed] [Google Scholar]
- 26. Dunn SL, Björnholm M, Bates SH, Chen Z, Seifert M, Myers MG Jr. Feedback inhibition of leptin receptor/Jak2 signaling via Tyr1138 of the leptin receptor and suppressor of cytokine signaling 3. Mol Endocrinol. 2005;19(4):925–938. [DOI] [PubMed] [Google Scholar]
- 27. Banks AS, Davis SM, Bates SH, Myers MG Jr. Activation of downstream signals by the long form of the leptin receptor. J Biol Chem. 2000;275(19):14563–14572. [DOI] [PubMed] [Google Scholar]
- 28. Carpenter LR, Farruggella TJ, Symes A, Karow ML, Yancopoulos GD, Stahl N. Enhancing leptin response by preventing SH2-containing phosphatase 2 interaction with Ob receptor. Proc Natl Acad Sci USA. 1998;95(11):6061–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bjørbæk C, El-Haschimi K, Frantz JD, Flier JS. The role of SOCS-3 in leptin signaling and leptin resistance. J Biol Chem. 1999;274(42):30059–30065. [DOI] [PubMed] [Google Scholar]
- 30. Farooqi IS, Wangensteen T, Collins S, Kimber W, Matarese G, Keogh JM, Lank E, Bottomley B, Lopez-Fernandez J, Ferraz-Amaro I, Dattani MT, Ercan O, Myhre AG, Retterstol L, Stanhope R, Edge JA, McKenzie S, Lessan N, Ghodsi M, De Rosa V, Perna F, Fontana S, Barroso I, Undlien DE, O’Rahilly S. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N Engl J Med. 2007;356(3):237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andiran N, Celik N, Andiran F. Homozygosity for two missense mutations in the leptin receptor gene (P316:W646C) in a Turkmenian girl with severe early-onset obesity. J Pediatr Endocrinol Metab. 2011;24(11–12):1043–1045. [PubMed] [Google Scholar]
- 32. Huvenne H, Le Beyec J, Pépin D, Alili R, Kherchiche PP, Jeannic E, Frelut ML, Lacorte JM, Nicolino M, Viard A, Laville M, Ledoux S, Tounian P, Poitou C, Dubern B, Clément K. Seven novel deleterious LEPR mutations found in early-onset obesity: a Δexon6–8 shared by subjects from Reunion Island, France, suggests a founder effect. J Clin Endocrinol Metab. 2015;100(5):E757–E766. [DOI] [PubMed] [Google Scholar]
- 33. Hannema SE, Wit JM, Houdijk ME, van Haeringen A, Bik EC, Verkerk AJ, Uitterlinden AG, Kant SG, Oostdijk W, Bakker E, Delemarre-van de Waal HA, Losekoot M. Novel leptin receptor mutations identified in two girls with severe obesity are associated with increased bone mineral density. Horm Res Paediatr. 2016;85(6):412–420. [DOI] [PubMed] [Google Scholar]
- 34. Saeed S, Bonnefond A, Manzoor J, Shabbir F, Ayesha H, Philippe J, Durand E, Crouch H, Sand O, Ali M, Butt T, Rathore AW, Falchi M, Arslan M, Froguel P. Genetic variants in LEP, LEPR, and MC4R explain 30% of severe obesity in children from a consanguineous population. Obesity (Silver Spring). 2015;23(8):1687–1695. [DOI] [PubMed] [Google Scholar]
- 35. Le Beyec J, Cugnet-Anceau C, Pépin D, Alili R, Cotillard A, Lacorte JM, Basdevant A, Laville M, Clément K. Homozygous leptin receptor mutation due to uniparental disomy of chromosome 1: response to bariatric surgery. J Clin Endocrinol Metab. 2013;98(2):E397–E402. [DOI] [PubMed] [Google Scholar]
- 36. Vonk J, Shackelford TK, eds. The Oxford Handbook of Comparative Evolutionary Psychology. New York, NY: Oxford University Press; 2012. [Google Scholar]
- 37. Mazen I, El-Gammal M, Abdel-Hamid M, Farooqi IS, Amr K. Homozygosity for a novel missense mutation in the leptin receptor gene (P316T) in two Egyptian cousins with severe early onset obesity. Mol Genet Metab. 2011;102(4):461–464. [DOI] [PubMed] [Google Scholar]
- 38. Saeed S, Bonnefond A, Manzoor J, Philippe J, Durand E, Arshad M, Sand O, Butt TA, Falchi M, Arslan M, Froguel P. Novel LEPR mutations in obese Pakistani children identified by PCR-based enrichment and next generation sequencing. Obesity (Silver Spring). 2014;22(4):1112–1117. [DOI] [PubMed] [Google Scholar]
- 39. Vauthier V, Jaillard S, Journel H, Dubourg C, Jockers R, Dam J. Homozygous deletion of an 80 kb region comprising part of DNAJC6 and LEPR genes on chromosome 1P31.3 is associated with early onset obesity, mental retardation and epilepsy. Mol Genet Metab. 2012;106(3):345–350. [DOI] [PubMed] [Google Scholar]
- 40. Kakar N, Ahmad J, Kubisch C, Borck G. Exon skipping and severe childhood-onset obesity caused by a leptin receptor mutation. Am J Med Genet A. 2013;161A(10):2672–2674. [DOI] [PubMed] [Google Scholar]
- 41. Lutz TA, Woods SC. Overview of animal models of obesity. Curr Protoc Pharmacol. 2012;Chapter 5:Unit 5.61. [DOI] [PMC free article] [PubMed]
- 42. Gill R, Cheung YH, Shen Y, Lanzano P, Mirza NM, Ten S, Maclaren NK, Motaghedi R, Han JC, Yanovski JA, Leibel RL, Chung WK. Whole-exome sequencing identifies novel LEPR mutations in individuals with severe early onset obesity. Obesity (Silver Spring). 2014;22(2):576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. von Schnurbein J, Moss A, Nagel SA, Muehleder H, Debatin KM, Farooqi IS, Wabitsch M. Leptin substitution results in the induction of menstrual cycles in an adolescent with leptin deficiency and hypogonadotropic hypogonadism. Horm Res Paediatr. 2012;77(2):127–133. [DOI] [PubMed] [Google Scholar]
- 44. Ozata M, Ozdemir IC, Licinio J. Human leptin deficiency caused by a missense mutation: multiple endocrine defects, decreased sympathetic tone, and immune system dysfunction indicate new targets for leptin action, greater central than peripheral resistance to the effects of leptin, and spontaneous correction of leptin-mediated defects. J Clin Endocrinol Metab. 1999;84(10):3686–3695. [DOI] [PubMed] [Google Scholar]
- 45. Mantzoros CS, Flier JS, Rogol AD. A longitudinal assessment of hormonal and physical alterations during normal puberty in boys. V. Rising leptin levels may signal the onset of puberty. J Clin Endocrinol Metab. 1997;82(4):1066–1070. [DOI] [PubMed] [Google Scholar]
- 46. Elias CF. Leptin action in pubertal development: recent advances and unanswered questions. Trends Endocrinol Metab. 2012;23(1):9–15. [DOI] [PMC free article] [PubMed]
- 47. Parent AS, Lebrethon MC, Gérard A, Vandersmissen E, Bourguignon JP. Leptin effects on pulsatile gonadotropin releasing hormone secretion from the adult rat hypothalamus and interaction with cocaine and amphetamine regulated transcript peptide and neuropeptide Y. Regul Pept. 2000;92(1–3):17–24. [DOI] [PubMed] [Google Scholar]
- 48. Backholer K, Smith JT, Rao A, Pereira A, Iqbal J, Ogawa S, Li Q, Clarke IJ. Kisspeptin cells in the ewe brain respond to leptin and communicate with neuropeptide Y and proopiomelanocortin cells. Endocrinology. 2010;151(5):2233–2243. [DOI] [PubMed] [Google Scholar]
- 49. Nizard J, Dommergues M, Clément K. Pregnancy in a woman with a leptin-receptor mutation. N Engl J Med. 2012;366(11):1064–1065. [DOI] [PubMed] [Google Scholar]
- 50. Bates SH, Stearns WH, Dundon TA, Schubert M, Tso AW, Wang Y, Banks AS, Lavery HJ, Haq AK, Maratos-Flier E, Neel BG, Schwartz MW, Myers MG Jr. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature. 2003;421(6925):856–859. [DOI] [PubMed] [Google Scholar]
- 51. DePaoli AM. 20 Years of leptin: leptin in common obesity and associated disorders of metabolism. J Endocrinol. 2014;223(1):T71–T81. [DOI] [PubMed] [Google Scholar]
- 52. Coleman DL. Effects of parabiosis of obese with diabetes and normal mice. Diabetologia. 1973;9(4):294–298. [DOI] [PubMed] [Google Scholar]
- 53. Anstee QM, Goldin RD. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol. 2006;87(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Farooqi IS, O’Rahilly S. 20 Years of leptin: human disorders of leptin action. J Endocrinol. 2014;223(1):T63–T70. [DOI] [PubMed] [Google Scholar]
- 55. von Schnurbein J, Heni M, Moss A, Nagel SA, Machann J, Muehleder H, Debatin KM, Farooqi S, Wabitsch M. Rapid improvement of hepatic steatosis after initiation of leptin substitution in a leptin-deficient girl. Horm Res Paediatr. 2013;79(5):310–317. [DOI] [PubMed] [Google Scholar]
- 56. Nillni EA, Vaslet C, Harris M, Hollenberg A, Bjørbak C, Flier JS. Leptin regulates prothyrotropin-releasing hormone biosynthesis. Evidence for direct and indirect pathways. J Biol Chem. 2000;275(46):36124–36133. [DOI] [PubMed] [Google Scholar]
- 57. Fekete C, Légrádi G, Mihály E, Huang QH, Tatro JB, Rand WM, Emerson CH, Lechan RM. α-Melanocyte-stimulating hormone is contained in nerve terminals innervating thyrotropin-releasing hormone-synthesizing neurons in the hypothalamic paraventricular nucleus and prevents fasting-induced suppression of prothyrotropin-releasing hormone gene expression. J Neurosci. 2000;20(4):1550–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, Sewter CP, Digby JE, Mohammed SN, Hurst JA, Cheetham CH, Earley AR, Barnett AH, Prins JB, O’Rahilly S. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387(6636):903–908. [DOI] [PubMed] [Google Scholar]
- 59. La Cava A, Matarese G. The weight of leptin in immunity. Nat Rev Immunol. 2004;4(5):371–379. [DOI] [PubMed] [Google Scholar]
- 60. Paz-Filho G, Wong ML, Licinio J. Ten years of leptin replacement therapy. Obes Rev. 2011;12(5):e315–e323. [DOI] [PubMed] [Google Scholar]
- 61. Farooqi IS, Matarese G, Lord GM, Keogh JM, Lawrence E, Agwu C, Sanna V, Jebb SA, Perna F, Fontana S, Lechler RI, DePaoli AM, O’Rahilly S. Beneficial effects of leptin on obesity, T cell hyporesponsiveness, and neuroendocrine/metabolic dysfunction of human congenital leptin deficiency. J Clin Invest. 2002;110(8):1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lahlou N, Clement K, Carel JC, Vaisse C, Lotton C, Le Bihan Y, Basdevant A, Lebouc Y, Froguel P, Roger M, Guy-Grand B. Soluble leptin receptor in serum of subjects with complete resistance to leptin: relation to fat mass. Diabetes. 2000;49(8):1347–1352. [DOI] [PubMed] [Google Scholar]
- 63. Pellitero S, Olaizola I, Alastrue A, Martínez E, Granada ML, Balibrea JM, Moreno P, Serra A, Navarro-Díaz M, Romero R, Puig-Domingo M. Hypogonadotropic hypogonadism in morbidly obese males is reversed after bariatric surgery. Obes Surg. 2012;22(12):1835–1842. [DOI] [PubMed] [Google Scholar]
- 64. Mollar Puchades MA, Gómez RC, del Olmo García MI, Marco JL, Portolés RS, Galiana PA, Piñón Sellés F. Hypogonadotropic hypogonadism in a patient with morbid obesity. Obes Surg. 2007;17(8):1127–1131. [DOI] [PubMed] [Google Scholar]
- 65. Kühnen P, Clément K, Wiegand S, Blankenstein O, Gottesdiener K, Martini LL, Mai K, Blume-Peytavi U, Grüters A, Krude H. Proopiomelanocortin deficiency treated with a melanocortin-4 receptor agonist. N Engl J Med. 2016;375(3):240–246. [DOI] [PubMed]
- 66. Setmelanotide for the treatment of LEPR deficiency obesity. Available at: https://clinicaltrials.gov/ct2/show/NCT03287960. Accessed 1 March 2018.
- 67. Rhythm presents positive initial data for setmelanotide in LepR deficiency obesity. Available at: www.rhythmtx.com/news-resources/press-releases/rhythm-presents-positive-initial-data-setmelanotide-lepr-deficiency-obesity/. Accessed 1st March 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.