Abstract
Meiotic resumption‐coupled degradation of maternal transcripts occurs during oocyte maturation in the absence of mRNA transcription. The CCR4–NOT complex has been identified as the main eukaryotic mRNA deadenylase. In vivo functional and mechanistic information regarding its multiple subunits remains insufficient. Cnot6l, one of four genes encoding CCR4–NOT catalytic subunits, is preferentially expressed in mouse oocytes. Genetic deletion of Cnot6l impaired deadenylation and degradation of a subset of maternal mRNAs during oocyte maturation. Overtranslation of these undegraded mRNAs caused microtubule–chromosome organization defects, which led to activation of spindle assembly checkpoint and meiotic cell cycle arrest at prometaphase. Consequently, Cnot6l −/− female mice were severely subfertile. The function of CNOT6L in maturing oocytes is mediated by RNA‐binding protein ZFP36L2, not maternal‐to‐zygotic transition licensing factor BTG4, which interacts with catalytic subunits CNOT7 and CNOT8 of CCR4–NOT. Thus, recruitment of different adaptors by different catalytic subunits ensures stage‐specific degradation of maternal mRNAs by CCR4–NOT. This study provides the first direct genetic evidence that CCR4–NOT‐dependent and particularly CNOT6L‐dependent decay of selective maternal mRNAs is a prerequisite for meiotic maturation of oocytes.
Keywords: CCR4–NOT, maternal mRNA decay, maternal‐to‐zygotic transition, meiotic maturation, oocyte
Subject Categories: Cell Cycle, Development & Differentiation, RNA Biology
Introduction
Most of mature mRNAs in eukaryotic cells contain a 5ʹ‐cap structure and a 3ʹ‐poly (A) tail. In mRNA turnover, shortening of the poly(A) tail by deadenylation is the initial and often rate‐limiting step (Doidge et al, 2012). In all model systems examined, including yeast, nematode, fruit fly, zebra fish, frog, and human cells, the multisubunit CCR4–NOT complex has been identified as the main deadenylase (Collart & Panasenko, 2012; Miller & Reese, 2012; Winkler & Balacco, 2013). Nonetheless, in vivo functional and mechanistic information regarding CCR4–NOT remains to be described in detail.
In yeast, the CCR4–NOT complex contains two different catalytic subunits: CCR4 and CAF1, which deadenylate mRNA via their 3ʹ‐5ʹ‐poly(A)‐specific exoribonuclease activity (Collart & Panasenko, 2017). CCR4 belongs to the endonuclease–exonuclease–phosphatase family of enzymes, and CAF1 is a member of the RNase D family of nucleases, which contain a conserved acidic catalytic motif, DEDD (Wang et al, 2010). In yeast as well as other invertebrates, a single CCR4 subunit is present. By contrast, two CCR4 paralogs called CNOT6 and CNOT6L are present in vertebrates. Both CNOT6 and CNOT6L can associate with the CCR4–NOT complex, but the two paralogs cannot coexist in the same complex (Winkler & Balacco, 2013; Villanyi & Collart, 2016). The duplication of the genes encoding CCR4 in vertebrates prompted the suggestion that the paralogs may have tissue‐specific and/or target‐specialized roles in mRNA deadenylation. Moreover, the coexistence of deadenylases CCR4 and CAF1 as well as the presence of two paralogs for each of them (CNOT6 and CNOT6L as well as CNOT7 and CNOT8, respectively) in vertebrates raises the question whether their functions are complementary or redundant. So far, these long‐standing questions have not been addressed physiologically.
Although this complex is conserved in all eukaryotes, most knowledge about the function of CCR4–NOT has emerged from research on yeast and on cultured human cell lines, with many contradictory results (Ukleja et al, 2016). Many inconsistencies arise because mRNA transcription and degradation proceed concurrently in most cells. Therefore, inhibition of CCR4–NOT subunits may indirectly affect transcription and make it difficult to evaluate their direct participation in the regulation of mRNA stability. On the other hand, the fully grown mammalian oocyte is emerging as an ideal model for studying the mechanism and physiological importance of CCR4–NOT‐mediated mRNA degradation.
In mammalian species including humans, fully grown oocytes in preovulatory follicles are transcriptionally silent during the period from before meiotic resumption until after fertilization, when most of the transcriptional reactivation takes place at the 2‐ to 8‐cell stage (Li et al, 2013a; Yu et al, 2013, 2016a). Maternal mRNAs transcribed during oocyte growth before genome silencing are stored for use during oocyte maturation and early stages of preimplantation embryonic development (Tadros & Lipshitz, 2009). Massive but target‐specific and temporally selective destruction of transcripts occurs during meiotic maturation of an oocyte and is a prerequisite for maternal‐to‐zygotic transition (MZT) after fertilization (Su et al, 2007; Barckmann & Simonelig, 2013; Ma et al, 2015). Nevertheless, little is known about the biochemical mechanism that mediates the large‐scale maternal mRNA degradation in maturing oocytes. Nor do we understand the physiological role of this meiotic resumption‐coupled, hierarchical degradation of maternal transcripts. Answering these questions should provide insights into the processes occurring during oocyte maturation and help to improve clinical practice of human assisted reproduction.
In vertebrate oocytes, meiotic progression is driven by sequential translational activation of dormant maternal mRNAs stored in the cytoplasm (Ivshina et al, 2014; Sha et al, 2017). This activation is mainly induced by cytoplasmic elongation of their poly(A) tails (Curinha et al, 2014). Recent studies reveal that BTG4 is a meiotic cell cycle‐coupled MZT‐licensing factor in mouse oocytes (Liu et al, 2016; Wu & Dean, 2016; Yu et al, 2016b). BTG4 triggers maternal mRNA decay by recruiting the CCR4–NOT catalytic subunit CNOT7 or CNOT8 to actively translated mRNAs. A mitogen‐activated protein kinase (MAPK) cascade couples meiotic cell cycle progression to translational activation of maternal mRNAs, including those encoding BTG4 and CNOT7, and therefore also initiates mRNA degradation as a negative feedback loop (Sha et al, 2017). Nonetheless, it remains unknown whether CNOT6 and/or CNOT6L, the other catalytic subunit of the CCR4–NOT complex, is also regulated by the MAPK cascade and involved in the negative feedback regulation of maternal mRNA decay.
In this study, we addressed the functional significance of the CNOT6L nuclease by means of a mouse knockout model. The only unusual feature observed in Cnot6l −/− mice is the severe subfertility of females. Genetic deletion of Cnot6l but not Btg4 impairs the deadenylation and degradation of a subset of maternal mRNAs during oocyte maturation. As a result, overtranslation of these undegraded mRNAs caused microtubule–chromosome organization defects, which led to activation of the spindle assembly checkpoint and meiotic cell cycle arrest at prometaphase. Guided by the phenotypes of Cnot6l knockout mice, we sequentially assessed the stability, polyadenylation, translation, and polysome binding of maternal transcripts affected by Cnot6l KO. As logical extension of these findings, we investigated the regulation of CNOT6L during mouse oocyte maturation: We provided evidence that the RNA‐binding protein ZFP36L2 functions as a CNOT6L adaptor in targeting a subset of maternal transcripts; and the translation of Cnot6l itself is coupled to meiotic resumption by MAPK cascade.
Results
Cnot6l is highly expressed in mouse oocytes and is essential for female fertility
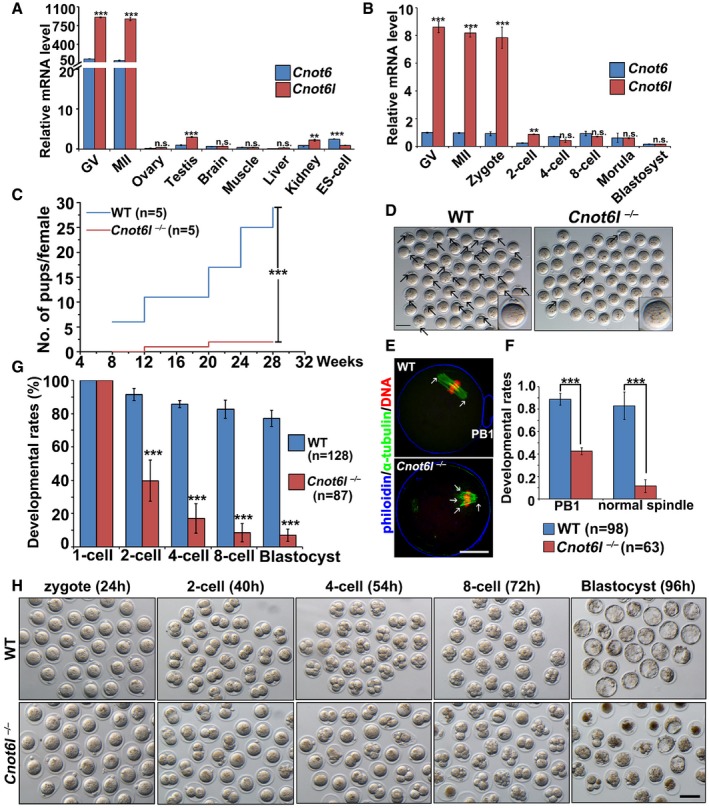
Previous transcriptome analyses have shown Cnot6 and/or CNOT6L mRNA is highly expressed in murine and human oocytes (Yan et al, 2013; Yu et al, 2016b). By quantitative RT–PCR, we confirmed that Cnot6 and Cnot6l mRNA levels were significantly higher in mouse oocytes (germinal vesicle (GV) stage and metaphase II (MII) stage) than in other cell types (Fig 1A). While the Cnot6 mRNA levels remained relatively stable in oocytes and preimplantation embryos, the Cnot6l levels were high in oocytes and the zygote but dramatically decreased after the 2‐cell stage. Moreover, the Cnot6l mRNA amount was much higher than that of Cnot6 mRNA in oocytes and zygotes (Fig 1B). This is in agreement with a previous report that Cnot6l transcripts were approximately 3‐fold more abundant than Cnot6 transcripts in mouse oocytes based on microarray data (Ma et al, 2015). Therefore, Cnot6l instead of its homolog Cnot6 is preferentially expressed during murine oocyte maturation and fertilization.
Figure 1. Cnot6l expression pattern and phenotype of Cnot6l knockout mice.
-
A, BQuantitative RT–PCR results showing relative expression levels of mouse Cnot6 and Cnot6l in oocytes (GV and MII), somatic tissues, ES cells (A), and preimplantation embryos (B). n = 3 biological replicates.
-
CCumulative numbers of pups per female showing fertility of WT and Cnot6l −/− female mice. n = 5 females for each genotype.
-
DRepresentative image results of oocytes collected from oviducts of WT and Cnot6l −/− mice at 16 h after hCG injection. Scale bar, 100 μm. Arrows indicate polar body‐1 (PB1).
-
EConfocal microscopy results of oocytes collected from oviducts of WT and Cnot6l −/− mice at 16 h after hCG injection. Scale bar, 20 μm. Arrows indicate spindle poles.
-
FRates of PB1 emission and normal spindle formation in oocytes ovulated by WT and Cnot6l −/− female mice. The numbers of analyzed oocytes are indicated (n).
-
GQuantification of preimplantation embryos derived from WT and Cnot6l −/− females that develop to the indicated stages after hCG administration and mated with adult WT males. The numbers of analyzed embryos are indicated (n).
-
HRepresentative images of embryos collected from the oviducts or uteri of WT and Cnot6l −/− female at indicated time points after hCG administration. Scale bar, 100 μm.
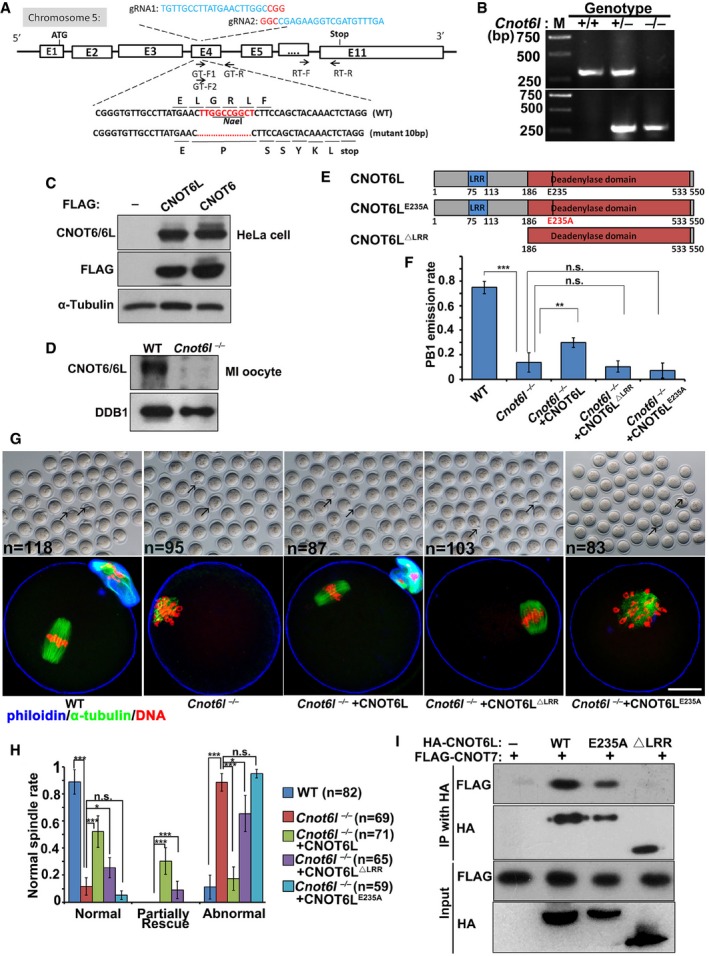
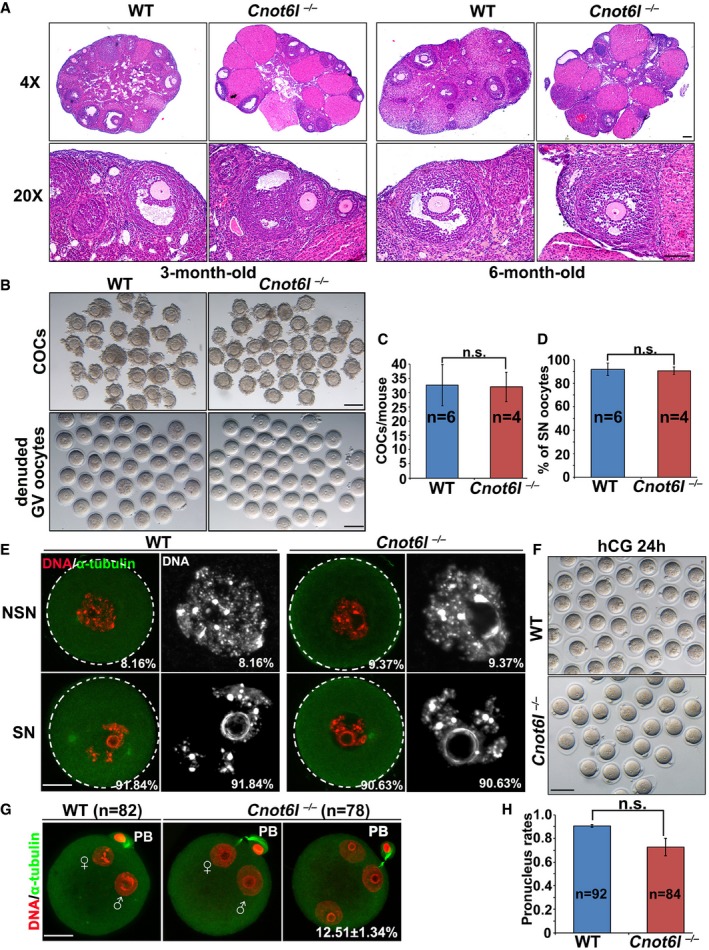
To study the in vivo function of Cnot6l, we generated a Cnot6l knockout mouse strain using the CRISPR‐Cas9 system (Fig EV1A). We obtained a mouse strain in which 10 nucleotides located in exon 4 of the Cnot6l gene are deleted downstream of the translation start site. This deletion caused a reading frame shift, which created a premature stop codon (Fig EV1A and B). There are no CNOT6L‐specific antibodies available. We have purchased a commercially available polyclonal CNOT6 antibody (Abcam, ab86209). This antibody recognizes both mouse CNOT6 and CNOT6L ectopically expressed in HeLa cells due to their high homology (Fig EV1C). Based on the fact that expression of Cnot6l is more abundant than Cnot6 in mouse oocytes at the mRNA level, we used this antibody to detect CNOT6L protein expression in WT and Cnot6l null oocytes. The Western blot result showed that the expected CNOT6/CNOT6L band was clearly detected in WT oocytes, but its intensity was greatly reduced in Cnot6l null oocytes (Fig EV1D), indicating that CNOT6L protein was successfully depleted in these oocytes. The Cnot6l −/− mice were found to be viable and healthy, and the males have normal fertility. In contrast, Cnot6l −/− females were found to be severely subfertile (Fig 1C). In a 32‐week fertility test, the females produced only 0.8 ± 0.24 (n = 5) pups per litter, whereas their wild‐type (WT) littermates produced 6.02 ± 0.17. Moreover, Cnot6l −/− females manifested a progressive loss of fertility because they gave birth to 1–3 pups in the first 1–2 litters and became infertile thereafter (Fig 1C). Nevertheless, sexually mature Cnot6l −/− females have normal ovarian histology. We did the histological analyses for ovaries of 3‐ and 6‐month‐old Cnot6l null mice. Hematoxylin and eosin staining results showed that all these ovaries contain multiple developing follicles and corpus lutea (Fig EV2A). Therefore, the Cnot6l null mice do not have a premature ovarian failure phenotype as the oocyte‐specific Ddb1 and Dcaf1 knockout mice we reported before (Yu et al, 2013, 2015b). We collected cumulus–oocyte complexes (COCs) containing fully grown GV oocytes from antral follicles of PMSG‐primed Cnot6l null mice by needle puncturing under a stereoscope, and determined the configuration of chromatin in these oocytes by DAPI staining. Similar numbers of COCs were harvested from ovaries of WT and Cnot6l null mice (Fig EV2B and C); more than 90% of GV oocytes from COCs of WT and Cnot6l null mice have a surrounded nucleolus (SN) as previously reported (Fig EV2D and E). These results further suggested that the oocyte development of Cnot6l KO mice is normal before meiotic maturation.
Figure EV1. Rescuing effect of ectopically expressed CNOT6L in Cnot6l −/− oocytes.
-
AGene targeting strategy for CRISPR/Cas9‐based mouse Cnot6l knockout. F, forward; R, reverse; Stop, stop codon; gRNA, guide RNA.
-
BPCR results using tail genomic DNA as the template. The WT allele (“+”) and 10‐nucleotide‐deletion allele (“−”) were amplified with the primer pairs GT‐F1, GT‐F2, and GT‐R. Primer sequences were provided in Appendix Table S1.
-
C, DWestern blot of CNOT6 and CNOT6L in HeLa cells transfected with plasmids expressing Flag‐CNOT6 or Flag‐CNOT6L (C) and in oocytes (WT and Cnot6l null) at the MI stage (D).
-
EA diagram showing the major functioning domains of mouse CNOT6L protein.
-
F–HRates of PB1 emission (F), representative images and immunofluorescent staining of α‐tubulin (G), and rates of normal spindle assembly (H) in oocytes cultured for 16 h. Arrows indicate polar body‐1 (PB1). Fully grown GV oocytes were microinjected with mRNAs encoding CNOT6L, CNOT6LΔLRR, or CNOT6LE235A and were released from meiotic arrest at 12 h after microinjection. Rescue: oocytes contain normal shaped bipolar spindle and aligned chromosomes; partial rescue: oocytes contain normal shaped bipolar spindle but chromosomes were not properly aligned; abnormal: both spindle assembly and chromosome alignment failed in oocytes. Scale bar, 20 μm. Error bars, SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant. The numbers of analyzed oocytes are indicated (n) in (H). n = 3 biological replicates in (F).
-
ICo‐IP results showing interactions of CNOT6L, CNOT6LE235A, and CNOT6LΔLRR with CNOT7. HeLa cells were co‐transfected with plasmids expressing HA‐CNOT6L and FLAG‐CNOT7 for 48 h before immunoprecipitation.
Source data are available online for this figure.
Figure EV2. Phenotypic analyses of Cnot6l −/− female mice.
-
AHematoxylin and eosin staining showing ovarian histology of WT and Cnot6l −/− mice. Scale bar, 100 μm.
-
B, CRepresentative images (B) and numbers (C) of cumulus–oocyte complexes (COCs) and denuded oocytes collected from antral follicles of WT and Cnot6l −/− mice. Scale bar, 100 μm. Error bars, SEM. n.s.: non‐significant. The numbers of analyzed mice are indicated (n).
-
DPercentage (%) of oocytes containing a surrounded nucleolus (SN) among oocytes in (B). Error bars, SEM. n.s.: non‐significant. The numbers of analyzed mice are indicated (n).
-
EConfocal microscopy results showing representative oocytes containing a non‐surrounded nucleolus (NSN) or a surrounded nucleolus (SN) among oocytes in (B). Percentages of NSN and SN oocytes among the total oocytes being collected are indicated in the corner. Scale bar, 20 μm.
-
FRepresentative images of zygotes collected from oviducts of WT and Cnot6l −/− female mice at 24 h after hCG injection and were mated with WT male mice. Scale bar, 100 μm.
-
GConfocal microscopy results showing representative zygotes containing 2 or 3 pronuclei. Scale bar, 20 μm.
-
HQuantification of pronucleus formation rates in zygotes shown in (F). Error bars, SEM. n.s.: non‐significant. The numbers of analyzed zygotes are indicated (n).
The Cnot6l knockout oocytes developed to the fully grown GV stage in vivo and could be ovulated by superovulation treatment. Nonetheless, nearly 40% of ovulated Cnot6l −/− oocytes were found to have no polar body 1 (PB1; Fig 1D and F). We collected these oocytes from oviducts and analyzed their potential defects in oocyte meiotic maturation at 16 h after hCG injection. Immunofluorescence and confocal microscopy results showed that most of ovulated Cnot6l −/− oocytes contained distorted multipolar spindles (Fig 1E and F). We superovulated the 3‐ to 4‐week‐old Cnot6l −/− females and then mated them with WT males. Fertilization rates were determined by visualizing the formation of pronuclei under a stereoscope, at 24 h after hCG injection (Fig EV2F). In addition, confocal microscopy of zygotes was performed to observe the pronuclei more clearly (Fig EV2G). It appears that although the Cnot6l null oocytes have severe defects of spindle assembly, most of them can be fertilized and form 2–3 PNs. The successful PN formation rate was slightly lower than that of control zygotes but was statistically non‐significant (Fig EV2H). However, more than 80% of the zygotes derived from Cnot6l −/− females underwent a cell cycle arrest at 2‐cell and 4‐cell stages. Only 10% of these embryos developed to the blastocyst stage (Fig 1G and H).
Oocyte meiotic maturation is impaired in Cnot6l −/− females
To examine the meiotic maturation process more closely, we isolated fully grown GV oocytes from pregnant mare serum gonadotropin (PMSG)‐primed Cnot6l −/− females and cultured these oocytes in vitro. Cnot6l −/− oocytes showed a GVBD rate similar to that of WT oocytes (Fig 2A), but the PB1 emission rate of Cnot6l −/− oocytes was significantly lower (Fig 2A). Although a small proportion of Cnot6l −/− oocytes released PB1 and developed to the MII stage (Fig 2B), they have a higher rate of aneuploidy than normal (Fig 2C and D). Similar to the oocytes collected from in vivo, the in vitro matured Cnot6l −/− oocytes contained distorted and multipolar spindles and failed to assemble bipolar spindles at both the MI and MII stage; chromosomes were not aligned at the equatorial plates at both stages (Fig 2E and F). BTG4 is a recently identified MZT‐licensing factor and function as an adaptor protein of CCR4–NOT deadenylase (Liu et al, 2016; Yu et al, 2016b). Another study reported that RNAi depletion of Btg4 in in vitro developed oocytes impaired meiotic maturation (Pasternak et al, 2016). Therefore, we also analyzed the maturation process in oocyte isolated from the Btg4 −/− mice. Btg4 −/− oocytes have GVBD and PB1 emission rates similar to those of WT oocytes (Fig 2B and E) and assembled normal bipolar spindles (Fig 2F). In contrast, most in vitro matured Cnot6l −/− oocytes contained distorted and multipolar spindles (Fig 2F). This phenotype involved frequent occurrence of spindles with MTOC‐clustering defects. In WT oocytes, pericentrin, the key component of MTOCs, localized as several clusters in the polar area of the MI spindle and merged into a single dot at each pole of the MII spindle (Fig 2G). On the other hand, pericentrin clusters did not localize in the polar area and failed to merge in Cnot6l −/− oocytes (Fig 2G). Thus, CNOT6L but not BTG4 is required for oocyte meiotic maturation.
Figure 2. Effect of Cnot6l and Btg4 knockout on oocyte meiotic maturation.
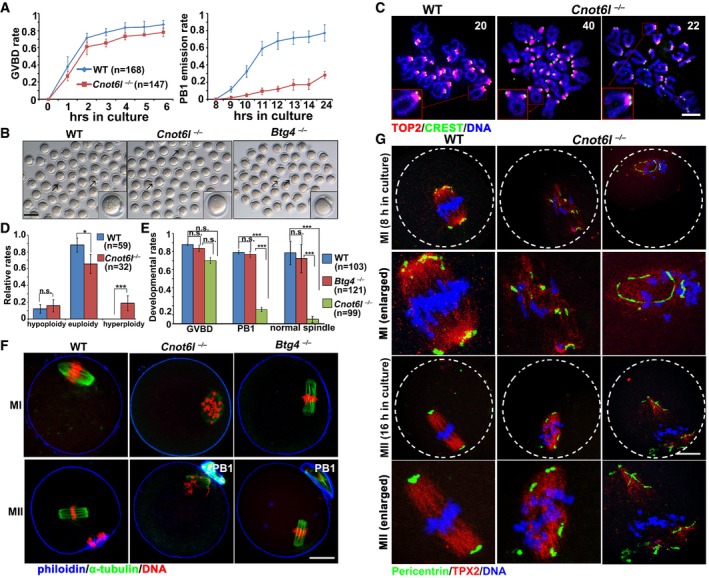
- Rates of germinal vesicle breakdown (GVBD) and PB1 emission in oocytes cultured in vitro. Fully grown GV oocytes were collected from PMSG‐primed (44 h) WT and Cnot6l −/− mice. PB1: polar body‐1. Error bars, SEM. The numbers of analyzed oocytes are indicated (n).
- Representative images of WT, Cnot6l −/−, and Btg4 −/− oocytes showing PB1 emission at 16 h after culture. Arrows indicate PB1. Scale bar, 100 μm.
- Representative images of chromosome spreads made from WT and Cnot6l −/− oocytes after 16 h of in vitro maturation culture. Immunofluorescent staining of topoisomerase II (TOP2) and the centromere antigen CREST were performed to indicate chromosome arms and centromeres, respectively. Numbers of paired sister chromatids are indicated. Scale bar, 5 μm.
- Percentage (%) of aneuploidy among in vitro cultured WT and Cnot6l −/− oocytes that have released PB1s. Error bars, SEM. *P < 0.05; ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant. The numbers of analyzed oocytes are indicated (n).
- Rates of GVBD, PB1 emission, and normal spindle assembly in WT, Cnot6l −/−, and Btg4 −/− oocytes cultured in vitro. Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant. The numbers of analyzed oocytes are indicated (n).
- Confocal microscopy results showing spindle assembly in WT, Cnot6l −/−, and Btg4 −/− oocytes at metaphase I (MI) and metaphase II (MII). Scale bar, 20 μm.
- Pericentrin immunofluorescence showing MTOCs in cultured WT, Cnot6l −/−, and Btg4 −/− oocytes at MI and MII stages. Spindle and DNA were labeled by microtubule nucleation factor (TPX2) and DAPI, respectively. Scale bar, 20 μm.
Cnot6l −/− oocytes were arrested at prometaphase I by spindle assembly checkpoints
Employing live‐cell imaging microscopy, we next compared the dynamic spindle assembly and meiotic cell cycle progression between WT and Cnot6l −/− oocytes. GV‐arrested oocytes were microinjected with mRNA encoding GFP‐tagged α‐tubulin (to label spindles) and were cultured in a medium containing Hoechst 33342 (to label DNA). Injected oocytes were released from milrinone and subjected to live‐cell imaging. Results in Fig 3A mean that WT oocytes finished meiosis I and developed to MII at the end of cultivation. In contrast, bipolar‐spindle formation, equatorial chromosome alignment, and PB1 emission all failed in Cnot6l −/− oocytes.
Figure 3. Meiotic cell cycle of Cnot6l −/− oocytes is arrested at metaphase I.
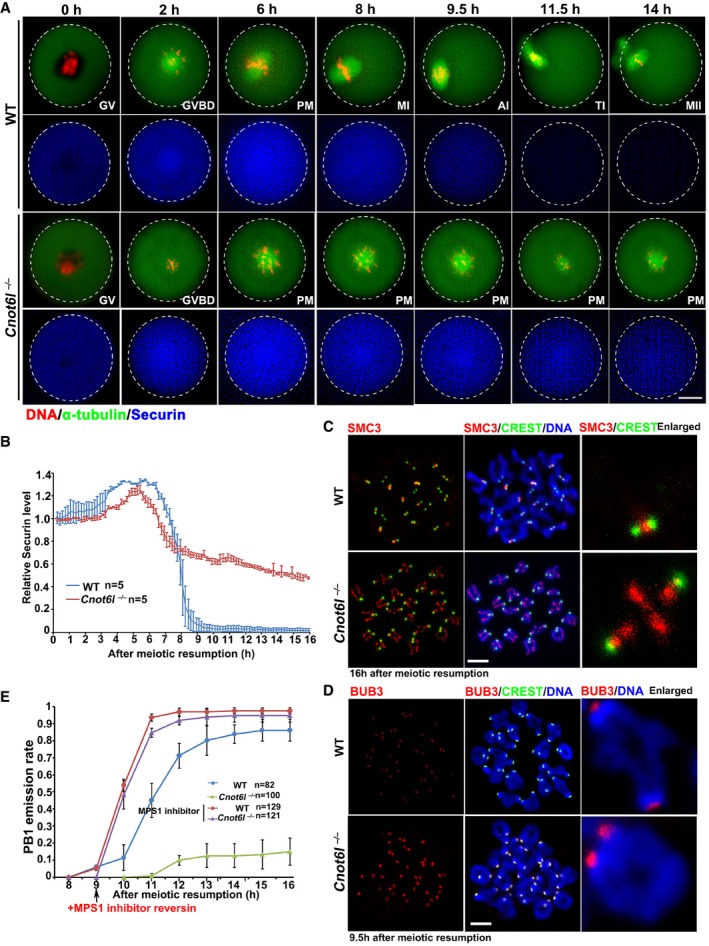
- Live‐cell imaging results showing in vitro meiotic division of WT and Cnot6l −/− oocytes. Abbreviations: PM, prometaphase; MI, metaphase I; AI, anaphase I; TI, anaphase I‐metaphase II transition; MII, metaphase II; PB1: polar body‐1. Hours after released from GV arrest are indicated. Scale bar, 20 μm.
- Data represent the mean and standard deviations of mCherry‐securin fluorescence intensity in WT and Cnot6l −/− oocytes at each time point. Values from individual oocytes are normalized relative to that at 0 h after released from GV arrest. The numbers of analyzed oocytes are indicated (n).
- SMC3 immunofluorescence showing cohesin on chromosomes of WT and Cnot6l −/− oocytes at 16 h after culture. Centromeres and DNA were labeled by CREST and DAPI, respectively. Scale bar, 5 μm.
- BUB3 immunofluorescence on chromosome spreads made from WT and Cnot6l −/− oocytes at 9.5 h after culture. More than 8 oocytes were observed for each genotype with similar results. Scale bar, 5 μm.
- PB1 emission rates in WT and Cnot6l −/− oocytes cultured with or without MPS1 inhibitor reversin (5 μm). Reversin is added at 9 h after culture. Error bars, SEM. The numbers of analyzed oocytes are indicated (n).
During meiotic division I, sister chromatids are attached to one another by cohesin complexes when homologous chromosomes form tetrads. During the MI‐to‐anaphase I (AI) transition, accurate homologous chromosome segregation is achieved by cohesion removal from chromosome arms by separase, which is a cysteine protease that cleaves cohesin. Separase is kept inactive for most of the cell cycle by binding to an inhibitory chaperone called securin. Securin is only removed at the metaphase–anaphase transition by APC‐mediated proteolysis. To assess the kinetics of MI‐to‐AI transition affected by Cnot6l deletion, we microinjected mRNA encoding mCherry‐securin into WT and Cnot6l −/− oocytes and monitored the dynamics of mCherry fluorescence. As shown in Fig 3A and quantified in Fig 3B, securin degraded in WT oocytes approximately 9–10 h after meiotic resumption but remained stable in Cnot6l −/− oocytes up to 16 h. During the MI‐to‐AI transition of WT oocytes, structural maintenance of chromosomes 3 (SMC3), a protein component of the cohesin complex, was removed from chromosome arms and exclusively localized to centromeres at the MII stage (Yu et al, 2015a). Nonetheless, in Cnot6l −/− oocytes, SMC3 was retained on the chromosome axis and more concentrated on centromeres when WT oocytes had extruded PB1 (Fig 3C). All these results indicated that Cnot6l −/− oocytes failed to enter AI.
At prometaphase I, the active spindle assembly checkpoint (SAC) inhibits premature activation of the anaphase‐promoting complex (APC) before chromosome alignment at the equatorial plate (Jones, 2011). The SAC is inactivated upon entry into AI. In Cnot6l −/− oocytes, BUB3, a component of the SAC, stayed on centromeres when BUB3 left centromeres 9.5 h after meiotic resumption in WT oocytes (Fig 3D). When we added MPS1 kinase inhibitor reversin (5 μM), which inactivates the SAC, the PB1 emission rate reached 94% in Cnot6l −/− oocytes, similar to that in WT oocytes (Fig 3E). These results indicated that chromosome alignment defects caused activation of the SAC, which led to MI arrest of Cnot6l −/− oocytes.
Ectopic expression of CNOT6L in Cnot6l −/− oocytes by mRNA microinjection partially reversed their meiosis defects: PB1 emission rates increased and the formation of normal MI spindles improved (Fig EV1F–H). In contrast, a catalytic site‐mutated CNOT6L (CNOT6LE235A) did not have any rescue effects (Fig EV1F–H). Coimmunoprecipitation experiments suggested that the leucine‐rich region (LRR) close to the N‐terminus of CNOT6L mediated its interaction with CNOT7 (Fig EV1I). Nevertheless, an N‐terminus–lacking CNOT6L still partially reversed the defects of MI spindle assembly (Fig EV1F). These results implied that although the catalytic activity was essential for CNOT6L functioning, some functions of CNOT6L in oocyte meiotic maturation were independent of the interaction with other subunits of the CCR4–NOT complex.
CNOT6L deletion impaired maternal mRNA clearance during oocyte maturation
As a RNA deadenylase, CNOT6L may directly regulate mRNA stability. A previous study has shown that microinjection of siRNAs targeting Cnot6l partially inhibited deadenylation of selective transcripts in cultured mouse oocytes (Ma et al, 2015). However, the effect of Cnot6l depletion on oocyte transcriptome as a whole was not reported. Therefore, we next investigated the effects of Cnot6l deletion on mRNA metabolism in oocytes. We subjected WT and Cnot6l −/− oocytes at 0, 8, and 16 h after in vitro culture (corresponding to GV, MI, and MII stages in WT oocytes) as well as the derived zygotes to RNA‐seq analyses with exogenous mRNA encoding mCherry as a spike‐in control. The oocytes that failed underwent GVBD within 2 h after culture were discarded.
Gene expression levels were assessed as fragments per kilobase of transcript per million mapped reads (FPKM), and the relative mRNA copy number was evaluated using the mCherry spike‐in. All samples were analyzed in duplicate, and all replicates showed high correlations (average r s = 0.955; Fig EV3A). As expected, maternal mRNAs were drastically degraded during the meiotic maturation of WT oocytes. 59 and 76% of maternal mRNAs were degraded at the MI and MII stages, respectively, and after fertilization, only 17% of mRNAs remained (Fig 4A). However, mRNA degradation was significantly impaired in Cnot6l −/− oocytes at all stages we examined, with the most dramatic difference observed at the MI stage (Fig 4A). These results indicated that CNOT6L is crucial for maternal mRNA clearance during meiotic maturation of an oocyte.
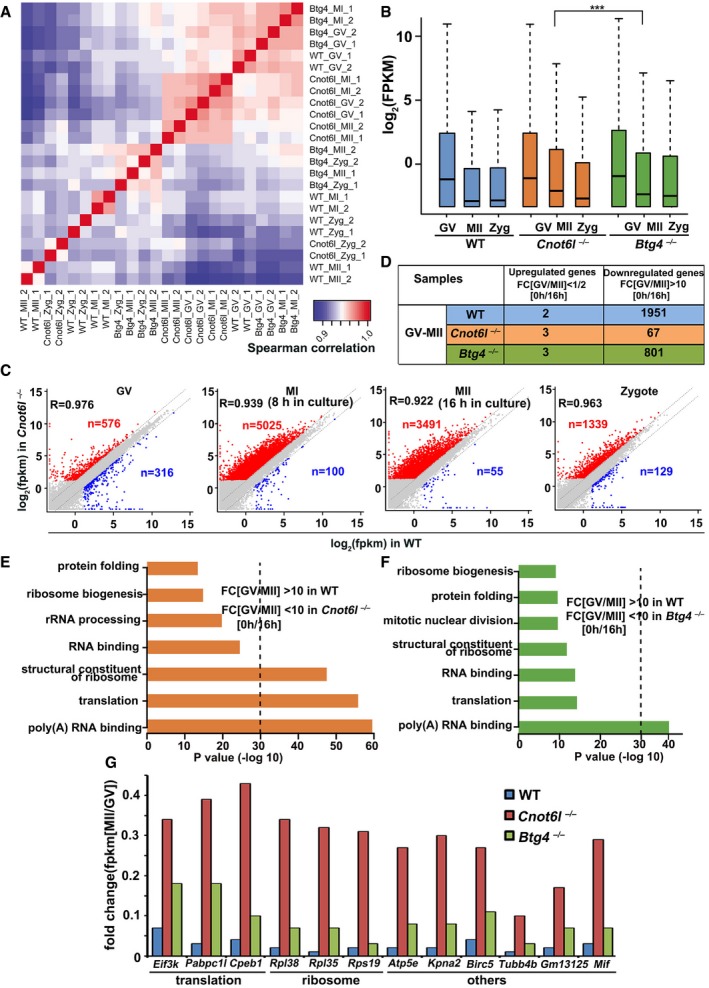
Figure EV3. RNA sequencing analyses of oocytes and zygotes derived from WT, Cnot6l −/−, and Btg4 −/− female mice.
-
AHeatmap of Spearman correlation coefficients of total transcripts among WT, Cnot6l −/−, and Btg4 −/− oocytes and embryos at different stages.
-
BGlobal mRNA levels in WT, Cnot6l −/−, and Btg4 −/− oocytes at different stages. The expression level of each transcript is normalized by mRNA encoding mCherry, which was in vitro transcribed and equally added to each sample before RNA extraction. The box indicates upper and lower quantiles, the thick line in the box indicates the median, and the whiskers represent 2.5th and 97.5th percentiles. ***P < 0.001 by two‐tailed Student's t‐test.
-
CScatter plot comparing transcripts between WT and Cnot6l −/− oocytes (at GV, MI, and MII stages) and zygotes derived from these oocytes. Transcripts decreased or increased more than 2‐fold in Cnot6l −/− oocyte samples were highlighted with blue or red, respectively.
-
DNumber of genes up‐ and downregulated during GV‐to‐MII transitions.
-
E, FGene ontology analysis of transcripts significantly degraded during GV‐to‐MII transition (GV/MII > 10) in WT oocytes but were stabilized in Cnot6l −/− (E) or Btg4 −/− oocytes (F).
-
GRNA‐seq results showing the relative expression levels (MII/GV) of representative transcripts in WT, Cnot6l −/−, and Btg4 −/− oocytes.
Figure 4. RNA sequencing analyses of oocytes and zygotes derived from WT, Cnot6l −/−, and Btg4 −/− female mice.
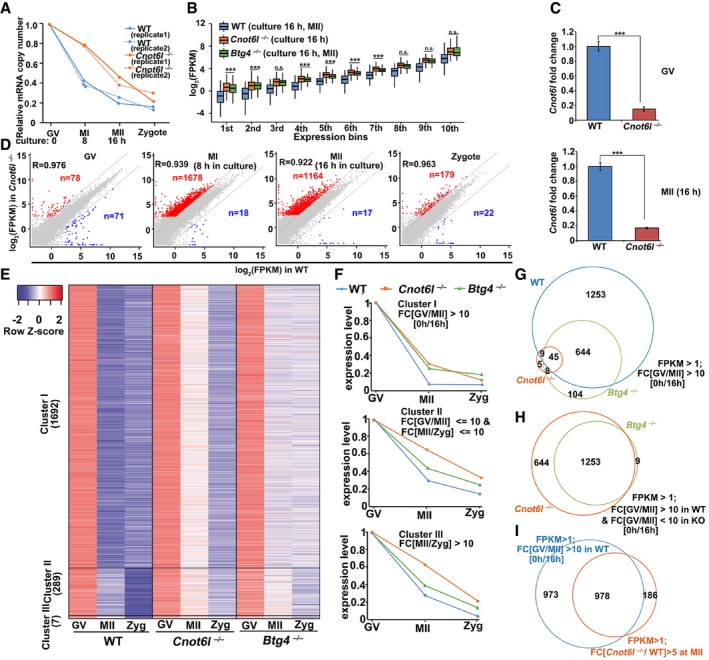
- Relative mRNA copy number dynamics in WT and Cnot6l −/− samples at the indicated stages. Error bars indicate values of the two biological replicates.
- Box plot showing gene expression levels of WT, Cnot6l −/−, and Btg4 −/− oocytes at the MII stage. Genes were divided into 10 bins according to their expression levels in the WT MII oocytes. The box indicates upper and lower quantiles, the thick line in the box indicates the median, and the whiskers represent 2.5th and 97.5th percentiles. ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant.
- qRT–PCR results showing the relative expression levels of Cnot6l in oocytes collected from WT and Cnot6l −/− mice (Cnot6l −/−/WT). Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test. n = 3 biological replicates.
- Scatter plot comparing transcripts between WT and Cnot6l −/− oocytes (at GV, MI, and MII stages) and zygotes derived from these oocytes. Transcripts decreased or increased more than 5‐fold in Cnot6l −/− oocyte samples were highlighted with blue or red, respectively.
- Heatmap showing transcripts that were degraded during meiotic maturation (GV/Zygote > 10) in WT oocytes. The definition of cluster I‐III is described in the text.
- Relative mRNA copy number dynamics of the three gene clusters in WT, Cnot6l −/−, and Btg4 −/− samples.
- Venn diagrams showing the overlap of transcripts that were significantly degraded during GV‐to‐MII transition in WT, Cnot6l −/−, and Btg4 −/− oocytes (GV/MII > 10).
- Venn diagrams showing the overlap of transcripts that were stabilized during GV‐to‐MII transition in Cnot6l −/− and Btg4 −/− oocytes (GV/MII > 10 in WT, but < 10 in KO).
- Venn diagrams showing the overlap of transcripts that were destabilized in GV–MII transition (FPKM > 1; fold change (GV/MII) > 10) of WT oocytes and the genes upregulated in this process after Cnot6l knockout (FPKM > 1; fold change (Cnot6l −/−/WT at MII) > 10).
To explore whether Cnot6l deletion compromised global mRNA clearance or only affected specific transcripts, we divided all expressed genes (FPKM > 1 in at least one sample) into 10 bins according to their expression levels in WT MII oocytes and found that CNOT6L impaired global mRNA clearance regardless of transcript abundance (Fig 4B). Although both CNOT6L and BTG4 deficiency impaired mRNA clearance, the median mRNA level in Cnot6l −/− MII oocytes is slightly higher than that in Btg4 −/− MII oocytes (Figs 4B and EV3B), suggesting that by the MII stage CNOT6L plays a more important role than BTG4 in global mRNA clearance. Quantitative RT–PCR (qRT–PCR) results showed dramatic decreases of Cnot6l mRNA levels in GV and MII oocytes collected from Cnot6l −/− mice, indicating that the gene knockout is effective (Fig 4C). Relatively very small numbers of transcripts were up‐ or downregulated for more than 5‐fold in Cnot6l null oocytes at the GV stage; the numbers of upregulated and downregulated genes are not significantly different (78 versus 71) (Fig 4D). In contrast, remarkably more transcripts were upregulated than downregulated in Cnot6l null oocytes at the MI (1,678 versus 18) and MII (1,164 versus 17) stages, as well as in zygotes derived from Cnot6l null oocytes (179 versus 22) (Fig 4D). This trend still holds true when we reduced the threshold of analyses to transcripts with fold changes > 2 (Fig EV3C). These results suggest that CNOT6L specifically regulates transcripts during meiotic maturation, instead of oocyte development up to GV stage. During GV‐to‐MII transition, 1,951 genes were degraded (GV/MII > 10) in WT oocytes, 801 in Btg4 −/− oocytes, whereas only 67 in Cnot6l −/− oocytes (Fig EV3D). We then divided the genes that were degraded during the meiotic maturation and fertilization of WT oocytes (GV/zygote > 10 in WT) into three clusters (Fig 4E): Cluster I comprised transcripts that were markedly degraded during GV–MII transition in WT oocytes (GV/MII > 10; n = 1,692); cluster II transcripts gradually decayed from meiotic resumption to zygote formation (1 < GV/MII ≤ 10 or 1 < MII/zygote ≤ 10; n = 289); cluster III represented transcripts that were relatively stable during meiotic maturation but markedly degraded after fertilization in WT oocytes (MII/zygote > 10; n = 7). Among all three clusters, transcripts were accumulated in Cnot6l −/− MII oocytes, comparing to WT and Btg4 −/− MII oocytes (Fig 4F).
Gene set enrichment analysis of the maternal transcripts revealed that fewer transcripts were degraded during GV–MII transition (GV/MII > 10) in Cnot6l −/− oocytes, when compared with WT and Btg4 −/− oocytes (Figs 4G and EV3D). Among the significantly degraded transcripts during GV‐to‐MII transition in WT oocytes, 644 (33.95%) specifically stabilized in Cnot6l −/− oocytes but not in Btg4 −/− oocytes at the MII stage (Fig 4H). Furthermore, we showed the overlap of genes destabilized in GV–MII transition (FPKM > 1; fold change (GV/MII) > 10) of WT oocytes and the genes upregulated in this process after Cnot6l knockout (FPKM > 1; fold change (Cnot6l −/−/WT at MII) > 10) (Fig 4I). The result indicated that the transcripts being stabilized in Cnot6l −/− oocytes are those being destabilized in WT oocytes in GV–MII transition. Therefore, CNOT6L plays a more important role than BTG4 in mRNA degradation during GV–MII transition.
CNOT6L mediates the deadenylation of maternal transcripts encoding translation‐related proteins during oocyte maturation
We further performed gene ontology (GO) analyses on the transcripts that are degraded in WT oocytes (GV/MII > 10) but not in Cnot6l −/− and Btg4 −/− oocytes. Interestingly, genes stabilized in Cnot6l −/− and Btg4 −/− oocytes were enriched for translation‐related functions, such as structural constituents of ribosomes, translation, and poly(A) RNA binding (Fig EV3E and F). Among the 96 transcripts that encode the 60S and 40S ribosome subunits, 90 accumulated in Cnot6l −/− MII oocytes (Fig 5A). By qRT–PCR, we confirmed the RNA‐seq results in Fig EV3G and demonstrated that transcripts that were implicated in translation (Eif3k, Pabpc1l, and Cpeb1) and ribosomes (Rpl38, Rpl35, and Rps19) in GO analysis degraded during maturation of WT oocytes but were stabilized by the Cnot6l knockout (Fig 5B). These genes were tested because the RNA sequencing results showed that they were significantly accumulated in Cnot6l but not in Btg4 knockout oocytes. The mRNA levels in different samples were normalized to the housekeeping gene Gapdh. The levels of Gapdh did not fluctuate in oocytes at different stages when compared by spiking with exogenous mRNA encoding GFP (25 pg/μl; Fig EV4A).
Figure 5. Transcripts related to mRNA translation were degraded during oocyte maturation but were stabilized by Cnot6l knockout.
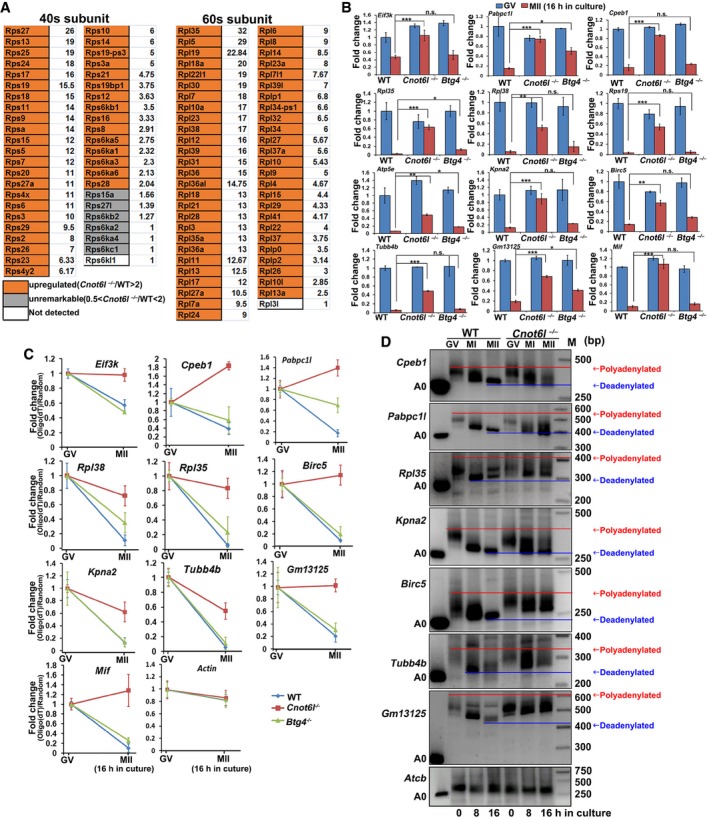
- Level changes of transcripts encoding ribosomal proteins in Cnot6l −/− oocytes at MII stage. The fold changes (Cnot6l −/−/WT) are listed to the right of the boxes.
- Quantitative RT–PCR results showing the relative levels of indicated transcripts in WT, Cnot6l −/−, and Btg4 −/− oocytes at GV and MII stages. n = 3 biological replicates. Error bars, SEM. Statistical analysis was performed using Student's t‐test. *P < 0.05, **P < 0.01, ***P < 0.001. n.s.: non‐significant.
- Changes of qRT–PCR results obtained from oligo‐dT‐ versus random primer‐mediated RT reactions reflecting the poly(A) tail length changes of given maternal transcripts during GV‐to‐MII transitions in WT, Cnot6l −/−, and Btg4 −/− oocytes. n = 3 biological replicates. Error bars, SEM.
- Poly(A) tail assay results showing changes in poly(A)‐tail length of indicated transcripts in WT and Cnot6l −/− oocytes at GV, MI, and MII stages. Experiments were performed three times with reproducible results; a representative result is shown.
Figure EV4. Transcriptome analyses of maturing oocytes derived from WT and Cnot6l −/− mice.
-
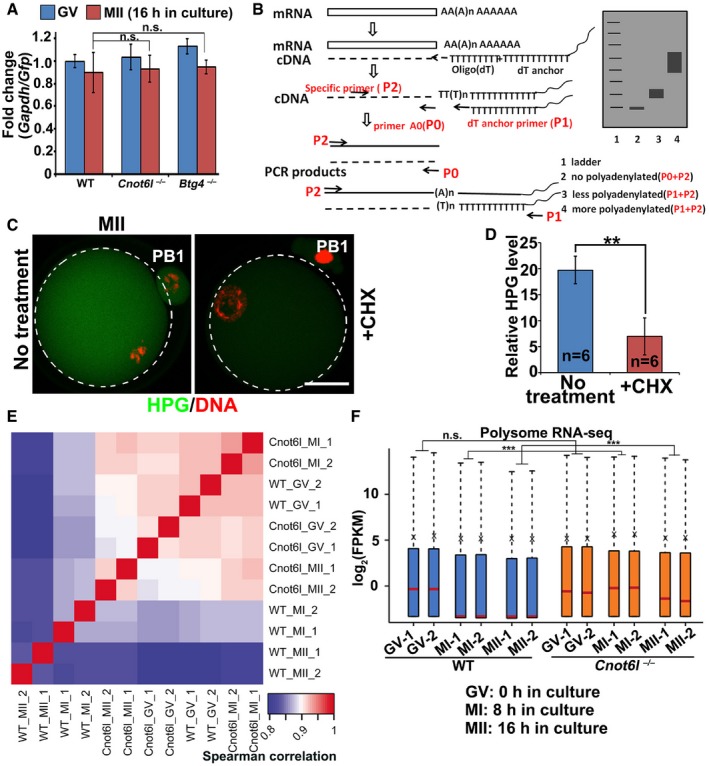
AQuantitative RT–PCR (qRT–PCR) showing the relative levels between Gapdh and Gfp in GV and MII oocytes collected from WT, Cnot6l −/−, and Btg4 −/− mice. mRNAs encoding GFP were in vitro transcribed and equally added to each sample (0.1 ng/μl) before RNA extraction. Error bars, SEM. n.s.: non‐significant. n = 3 technical replicates.
-
BStrategy of the mRNA poly(A) tail assay. P0, primer A0 that located at the end of mRNA 3′TUR without poly (A); P1, dT anchor primer (GCGAGCTCCGCGGCCGCGT12); P2, gene‐specific primer.
-
C, DImmunofluorescence (C) and quantification (D) of HPG showing the overall translation levels of MII oocytes cultured in medium with or without cycloheximide (CHX, 20 μM). Scale bar, 20 μm. **P < 0.01 by two‐tailed Student's t‐test. n = 3 biological replicates.
-
EHeatmap of Spearman correlation coefficients of transcripts associated with polysomes in WT and Cnot6l −/− oocytes at GV, MI, and MII stages.
-
FLevels of polysome‐bound transcripts in WT and Cnot6l −/− oocytes at indicated stages (two biological repeats). The expression level of each transcript was normalized by the mCherry spike‐in, which was in vitro transcribed and equally added to each sample before RNA extraction. The box indicates upper and lower quantiles, the red line in the box indicates the median, and the whiskers represent 2.5th and 97.5th percentiles, respectively. ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant.
In cytoplasmic mRNA turnover, deadenylation (shortening) of the poly(A) tail is the initial and rate‐limiting step. To evaluate the poly(A) tail length, we reverse‐transcribed the transcripts stabilized in Cnot6l −/− MII oocytes using oligo‐dT or random primers and conducted qRT–PCR. Oligo‐dT promotes the reverse transcription (RT) of transcripts containing long poly(A) tails, but random primers do not have this selective effect. Therefore, the ratio changes in qRT–PCR results obtained in oligo‐dT–mediated versus random primer‐mediated RT reactions reflect the poly(A) tail length changes of given maternal transcripts. As presented in Fig 5C, oligo‐dT–mediated RT efficacy of these transcripts decreased during GV–MII transition in WT oocytes. This trend was significantly compromised or even reversed (Cpeb1, Pabpc1l, Birc5, and Mif) in Cnot6l −/− oocytes. On the other hand, the RT efficacy of these transcripts was not affected or only moderately (Pabpc1l, Rpl38) affected by the Btg4 knockout. As a negative control, actin mRNA stayed stable in all experimental and control groups.
Using a poly(A) tail assay (Fig EV4B), we directly measured the poly(A) tail length in several transcripts studied in Fig 5B and C. The poly(A) tails remarkably shortened during meiotic maturation of WT oocytes, but this process was blocked in Cnot6l −/− oocytes (Fig 5D). These results revealed that CNOT6L triggers maternal mRNA clearance by driving the deadenylation of their poly(A) tails, especially for genes related to translation.
CNOT6L deletion caused overtranslation of the accumulated maternal mRNAs
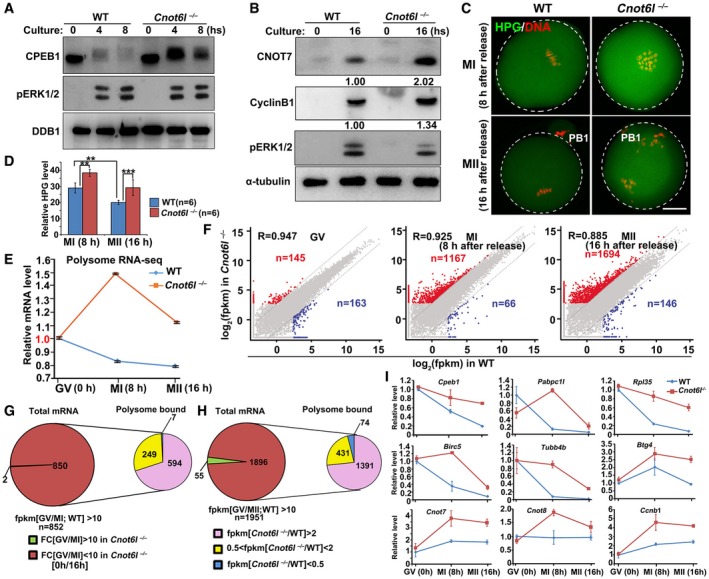
Among the transcripts stabilized in Cnot6l −/− oocytes, Cpeb1 mRNA is of particular interest because the activation and partial degradation of CPEB1 through ERK1‐ and ERK2‐mediated phosphorylation is essential for meiotic maturation (Sha et al, 2017). In Cnot6l −/− oocytes, ERK1 and ERK2 were normally phosphorylated and activated after meiotic resumption (Fig 6A). On the other hand, CPEB1 was phosphorylated (as indicated by a band upshift) in Cnot6l −/− oocytes after meiotic resumption, but its protein level did not decrease as in WT oocytes (Fig 6A). In maturing oocytes, CPEB1 stimulated translation of some cytoplasmic polyadenylation element (CPE)‐containing maternal mRNAs, such as CNOT7 and cyclin B1 transcripts (Pique et al, 2008; Sha et al, 2017). Western blot results showed that CNOT7 and cyclin B1 protein levels moderately increased in Cnot6l −/− MII oocytes (Fig 6B).
Figure 6. Meiotic resumption‐triggered deadenylation of maternal transcripts was blocked in Cnot6l −/− oocytes.
-
A, BWestern blot results showing the levels of CPEB1 in WT and Cnot6l −/− oocytes at the indicated time points after meiotic resumption. Total proteins from 50 (A) or 100 (B) oocytes are loaded in each lane. DDB1 (A) or α‐tubulin (B) is blotted as a loading control. Experiments were performed three times with reproducible results; a representative result is shown.
-
C, DImmunofluorescence (C) and quantification (D) of L‐homopropargylglycine (HPG) showing the overall translation levels of MI and MII oocytes collected from WT and Cnot6l −/− mice. Scale bar, 20 μm. Error bars, standard deviations (n = 6 oocytes for each genotype). **P < 0.01, ***P < 0.001.
-
ERelative mRNA copy numbers of polysome‐bound transcripts in WT and Cnot6l −/− oocytes at indicated stages. Error bars indicate values of the replicates.
-
FScatter plot comparing polysome‐bound transcripts between WT and Cnot6l −/− oocytes at GV, MI, and MII stages, respectively. Transcripts decreased or increased more than 5‐fold in Cnot6l −/− oocyte samples were highlighted with blue or red, respectively.
-
GPie chart in left depicts total transcripts significantly degraded during GV‐MI transitions in WT oocytes. Pie chart in right shows the polysome association of transcripts that were deemed to be degraded during GV‐MI transitions in WT oocytes, but were stabilized after Cnot6l knockout.
-
HPie chart in left depicts total transcripts significantly degraded during GV–MII transitions in WT oocytes. Pie chart in right shows the polysome association of transcripts that were deemed to be degraded during GV–MII transitions in WT oocytes, but were stabilized after Cnot6l knockout.
-
IQuantitative RT–PCR results showing the relative levels of indicated transcripts in association with polysomes in WT and Cnot6l −/− oocytes at GV, MI, and MII stages. Error bars, standard deviations (n = 3 biological repeats).
Source data are available online for this figure.
To test whether the stabilized maternal mRNAs actively participated in translation, we incubated oocytes in a medium containing L‐homopropargylglycine (HPG, a methionine analog that is incorporated into nascent proteins during active protein synthesis). The HPG signal is indicative of the overall level of translation, as its intensity significantly decreased in oocytes treated with translation inhibitor cycloheximide (CHX) (Fig EV4C and D). The HPG signals diminished after MI‐to‐MII transition in WT oocytes. In Cnot6l −/− oocytes however, HPG signals were modestly stronger than those in WT oocytes at both MI and MII stages (Fig 6C and D), suggesting overtranslation of the accumulated maternal mRNAs.
We then sought to provide more direct evidence that the accumulated maternal mRNAs were being actively translated. The position of an mRNA in a sucrose gradient depends on the number of ribosomes associated with this mRNA (Chen et al, 2011); therefore, we isolated the mRNA bound by multiple ribosomes (i.e., the polysome) from whole oocytes by sucrose gradient centrifugation. The polysome‐bound mRNAs at GV, MI, and MII stages were then extracted and subjected to RNA‐seq analyses, using exogenous mRNA encoding mCherry as the spike‐in control. All samples were analyzed in duplicate, and all replicates showed high correlations (average r s = 0.898; Fig EV4E). In WT oocytes, polysome‐bound mRNAs gradually decreased during meiotic maturation as evaluated by the mCherry spike‐in (Figs 6E and EV4F). In contrast, polysome‐bound mRNAs increased in Cnot6l −/− oocytes during GV‐to‐MI transition and remained at a level higher than that in WT oocytes even at the MII stage (Figs 6E and EV4F). We also compared polysome‐bound transcripts between WT and Cnot6l −/− oocytes at GV, MI, and MII stages at the gene level. Although the numbers of genes that gain or lose polysome binding after Cnot6l knockout were comparable at the GV stage, significantly more genes showed increased polysome binding in Cnot6l −/− oocytes at MI and MII stages (Fig 6F).
Among genes that were remarkably degraded in WT oocytes during GV–MI or GV–MII transitions (i.e., GV/MI > 10 or GV/MII > 10 in WT), most were stabilized (i.e., GV/MI < 10 or GV/MII < 10) in Cnot6l −/− oocytes (850 out of 852 genes for the GV–MI transition, and 1,896 out of 1,951 genes for the GV‐to‐MII transition) (Fig 6G and H). Importantly, approximately two‐thirds (594/850 in MI and 1,391/1,896 in MII) of these stabilized transcripts manifested increased polysome binding in Cnot6l −/− oocytes (Fig 6G and H), indicating that they were actively translated.
Using qRT–PCR, we confirmed the results in Fig 6G and H and demonstrated that the accumulated transcripts related to translation (Cpeb1 and Pabpc1l), ribosomes (Rpl35), spindle formation, and chromosome alignment (Birc5 and Tubb4b) in Cnot6l‐null oocytes tend to concentrate on polysomes during meiotic maturation of oocytes. During mitosis and meiosis, survivin protein encoded by Birc5 assembles with the chromosomal passenger complex and regulates chromosomal segregation. Survivin also plays an essential role in proper amphitelic kinetochore‐spindle assembly (Jiang et al, 2014). Tubb4b encodes a tubulin protein that is a structural protein of spindle microtubules (Wawro et al, 2017). Other transcripts that did not degrade but were actively translated in WT oocytes (Btg4, Cnot7, Cnot8, and Ccnb1 mRNA) also showed increased polysome binding during oocyte meiotic maturation after Cnot6l deletion (Fig 6I). Collectively, we conclude that after CNOT6L deletion, overall translation increased including the synthesis of proteins that should be degraded during oocyte meiotic maturation.
ZFP36L2 recruits the CCR4–NOT complex to maternal transcripts that are scheduled to be degraded during meiotic maturation
CNOT6L and other subunits of the CCR4–NOT complex do not contain any known RNA‐binding domain. CNOT6L must rely on an RNA‐binding protein partner to target maternal transcripts that are scheduled for degradation. In Xenopus oocytes, (A+U)‐rich element (ARE)‐binding protein C3H‐4 recruits the CCR4–NOT deadenylase complex to ARE‐containing mRNAs and causes shortening of poly(A) tails (Belloc & Mendez, 2008). ARE with sequence AUUUA is characteristic of mRNAs regulated by deadenylation (Hudson et al, 2004). Our RNA‐seq results showed that AREs were enriched in the 3′‐UTR of transcripts that degraded during the GV–MII transition (Fig 7A). Moreover, at the MII stage, AREs were also enriched in maternal transcripts that were stabilized in Cnot6l −/− oocytes (Fig 7B).
Figure 7. Role of (A+U)‐rich element (ARE)‐binding protein ZFP36L2 in CNOT6L‐mediated maternal mRNA decay.
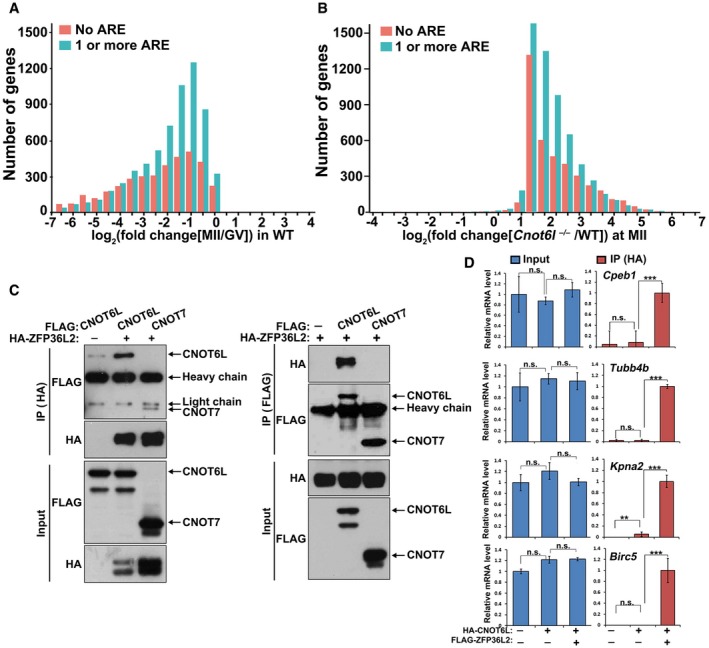
- Presence of putative AREs (AUUUA) in transcripts of WT oocyte. Transcripts containing no ARE or transcripts containing 1 or more AREs were subdivided into frequency groups according to the log2 fold change (MII/GV) in WT oocytes.
- Presence of putative AREs in oocyte transcripts stabilized by Cnot6l −/− knockout. Transcripts containing no ARE or transcripts containing 1 or more AREs were subdivided into frequency groups according to the log2 fold change (Cnot6l −/−/WT) at MII stage.
- Co‐IP results showing interaction of ZFP36L2 with CNOT6L and CNOT7. HeLa cells were co‐transfected with plasmids expressing HA‐ZFP36L2 and FLAG‐CNOT6L/7 for 48 h before immunoprecipitation. Experiments were performed three times with reproducible results; a representative result is shown.
- RNA immunoprecipitation results showing interaction of CNOT6L with indicated transcripts, with or without the presence of ZFP36L2. HeLa cells were co‐transfected with plasmids expressing FLAG‐ZFP36L2 and HA‐CNOT6L for 48 h before immunoprecipitation using an anti‐HA antibody. mRNAs recovered from the immunoprecipitates were subjected to qRT–PCR. Error bars, standard deviations (n = 3 biological repeats). ***P < 0.001 by two‐tailed Student's t‐test.
Source data are available online for this figure.
ZFP36L2 (zinc finger protein 36‐like 2) is a murine homolog of Xenopus C3H‐4. A natural mutation of Zfp36l2 in mice that the N‐terminal 29 amino acid residues were deleted led to oocyte maturation and ovulation defects (Ball et al, 2014). A new study published during the submission of our paper further demonstrated that oocyte‐specific loss of Zfp36l2 causes oocyte maturation and fertilization defects by preventing global transcriptional silencing in GV oocytes (Dumdie et al, 2018). We performed coimmunoprecipitation experiments and revealed that ZFP36L2 preferentially binds to CNOT6L rather than CNOT7 in HeLa cells overexpressing these proteins (Fig 7C). Based on current knowledge, people believe that CNOT6L and CNOT7 should be in the same functional complexes with full deadenylation activity (Doidge et al, 2012). However, it is difficult to evaluate how stable the complex is. An explanation of our result is that: (i) ZFP36L2 directly binds with CNOT6L but not CNOT7; (ii) the interaction between CNOT6L and ZFP36L2 is stronger than that between CNOT6L and CNOT7. As a result, more CNOT6L proteins than CNOT7 proteins were coimmunoprecipitated with ZFP36L2.
Moreover, small‐interfering RNA‐mediated Zfp36l2 depletion in mouse oocytes did not affect GVBD but impaired PB1 emission (Fig EV5A and B) and meiotic spindle assembly (Fig EV5C). These phenotypes were similar to those caused by the Cnot6l knockout. To test whether CNOT6L binds with mRNAs through ZFP36L2, we ectopically expressed HA‐CNOT6L and FLAG‐ZFP36L2 in HeLa cells and performed a RNA immunoprecipitation assay using an anti‐HA antibody. The result indicated that CNOT6L effectively interacted with mRNAs encoding CPEB1, TUBB4B, BIRC5, and KPNA2 only in the presence of ZFP36L2 (Fig 7D). These transcripts are known to be degraded during meiotic maturation and were found to stabilize in Cnot6l −/− oocytes at the MII stage, as uncovered by RNA‐seq and qRT–PCR results. All of them contained 1–5 AREs in their 3′‐UTR. Collectively, these results indicated that ZFP36L2 recruits CNOT6L to target ARE‐containing maternal mRNAs during murine oocyte maturation.
Figure EV5. Role of Zfp36l2 and ERK1/2 in regulating meiotic maturation of mouse oocyte.
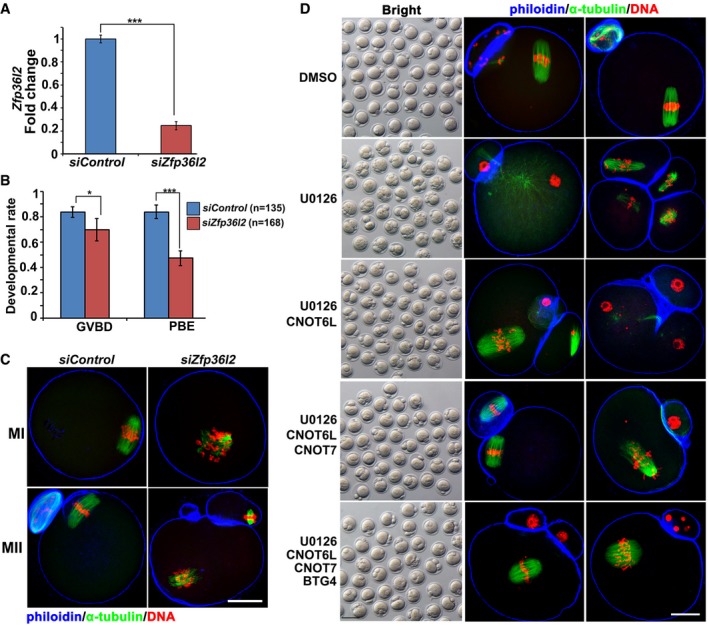
- qRT–PCR results showing the efficiency of Zfp36l2 RNA interference in oocytes at the GV stage. n = 3 technical replicates. Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test.
- GVBD and PB1 emission rates of oocytes microinjected control and Zfp36l2‐targeted siRNAs. Error bars, SEM. *P < 0.05; ***P < 0.001 by two‐tailed Student's t‐test. The numbers of analyzed oocytes are indicated (n).
- Confocal microscopy results showing spindle assembly at MI and MII stages in oocytes microinjected with control siRNAs or siZfp36l2. Scale bar, 20 μm.
- PB2 emission and spindle assembly in oocytes cultured with or without U0126 treatment. Fully grown GV oocytes were microinjected with mRNAs encoding CNOT6L, CNOT7, and/or BTG4 and are released from milrinone at 12 h after microinjection. Then, the oocytes were further culture for 24 h with or without adding U0126 (20 μM) to the medium. Scale bar, 100 μm in brightfield images; Scale bar, 20 μm in immunofluorescence staining images.
CNOT6L partially reversed the meiotic maturation defects in ERK1‐ and ERK2‐inhibited oocytes
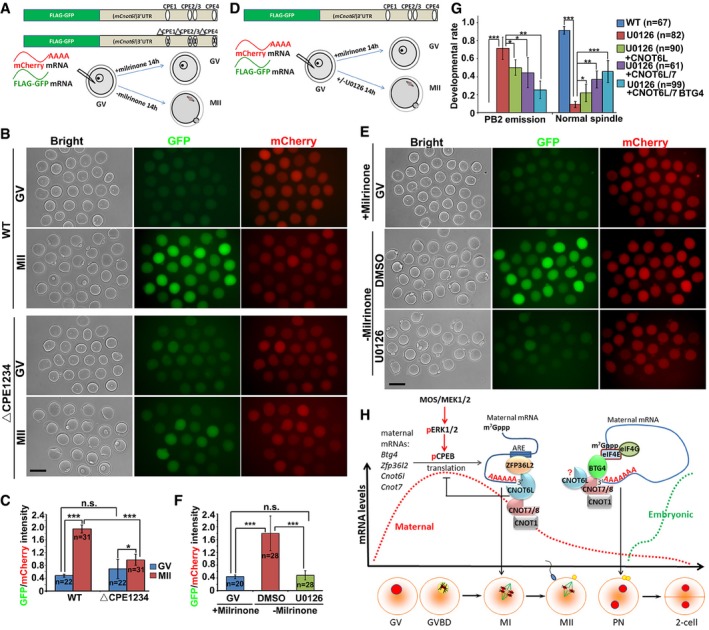
Cnot6l mRNA levels were high in both GV and MII oocytes. By contrast, maternal mRNAs remained stable at the GV stage and started to degrade only after GVBD. We cloned the mouse Cnot6l 3′‐UTR into a GFP reporter plasmid, transcribed it into mRNA in vitro, and microinjected it into GV oocytes (Fig 8A). The in vitro polyadenylated transcript encoding mCherry was coinjected as a positive control. After 14 h, we detected weak GFP signals in GV‐arrested oocytes and strong GFP signals in MII oocytes, whereas the mCherry signals were equal in the GV‐ and MII‐arrested oocytes (Fig 8B and C). This result pointed to meiotic resumption‐coupled translational activation of Cnot6l mRNA mediated by its 3′‐UTR, and confirmed a similar observation being reported (Ma et al, 2015).
Figure 8. CNOT6L is a downstream effector of ERK1/2 in regulating oocyte meiotic cell cycle progression.
-
A, BIllustration (A) and fluorescence microscopy results (B) showing the translation activities of the Cnot6l 3′‐UTR (WT and CPE mutated) in GV‐arrested (maintained by 2 μM milrinone) or MII‐arrested (released from milrinone) oocytes. GFP signal indicated translational activation of Cnot6l 3′‐UTR. An in vitro transcribed and polyadenylated mCherry mRNA was co‐microinjected as a positive control. Scale bar, 100 μm.
-
CRelative intensity of GFP signal in (B) after normalization by mCherry signal in the same oocyte. Error bars, SEM. *P < 0.05; ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant. The numbers of analyzed oocytes are indicated (n).
-
D, EIllustration (D) and fluorescence microscopy results (E) showing the expression of GFP‐fused Cnot6l 3′‐UTR in GV and MII oocytes with or without U0126 treatment (20 μM). Scale bar, 100 μm.
-
FRelative intensity of GFP signal in (E) after normalization by mCherry signal in the same oocyte. Error bars, SEM. ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant. The numbers of analyzed oocytes are indicated (n).
-
GRates of PB2 emission and normal spindle assembly in oocytes cultured with or without U0126. Fully grown GV oocytes were microinjected with mRNAs encoding CNOT6L, CNOT7, and/or BTG4 and are released from milrinone at 12 h after microinjection. Then, the oocytes were further culture for 24 h with or without adding U0126 to the medium. Error bars, SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by two‐tailed Student's t‐test. n.s.: non‐significant. The numbers of analyzed oocytes are indicated (n).
-
HA diagram showing role of CCR4–NOT deadenylase complex in targeting maternal mRNA decay. During the onset of oocyte meiotic resumption, MAPK cascade and CPEB1 triggers translational activation of maternal transcripts including those encoding BTG4, CNOT7, CNOT6L, and ZFP36L2 (Belloc & Mendez, 2008; Ma et al, 2015; Yu et al, 2016b; Sha et al, 2017). CNOT6L and other CCR4–NOT components are important downstream effectors of ERK1 and ERK2 in the regulation of spindle assembly and meiotic cell cycle progression in oocytes. During oocyte maturation, RNA‐binding protein ZFP36L2 associates with CNOT6L and functions as a CCR4–NOT adaptor in triggering the degradation of ARE‐containing transcripts. At a later stage of MZT, an alternative adaptor BTG4 binds to translation initiation factor eIF4E and thereby recruits the CCR4–NOT complex to the actively translated mRNAs. The stepwise recruitment of different adaptors by different catalytic subunits mediates stage‐specific degradation of maternal mRNAs by the CCR4–NOT deadenylase. The question mark means that the direct involvement of CNOT6L in BTG4‐mediated MZT process remains inconclusive because Cnot6l null oocytes had severe meiosis defects before MZT.
CPEB1 targets and mediates cytoplasmic polyadenylation of various CPE‐containing mRNAs (Chen et al, 2011; Ivshina et al, 2014). ERK1‐ and ERK2‐mediated CPEB1 phosphorylation and degradation represent a major mechanism of maternal mRNA translational activation and are crucial for murine oocyte maturation (Sha et al, 2017). The murine Cnot6l 3′‐UTR contains four putative CPEs (Fig 8A). In the study by Ma et al (2015), they used a truncated 3′‐UTR fragment (402 bp, which contained 3 CPEs) in the reporter experiment. We have noticed that there is an additional CRE close to the translation stop codon, which was not included in the Cnot6l 3′‐UTR fragment cloned by Ma et al Therefore, we used the 1,597 bp Cnot6l 3′‐UTR fragment containing 4 CREs in our reporter experiment. Mutation of these CPEs abrogated meiotic maturation‐coupled translational activation of Cnot6l 3′‐UTR (Fig 8B and C). Furthermore, we investigated the potential involvement of ERK1 and ERK2 in regulating Cnot6l mRNA translation during meiotic maturation. U0126, an inhibitor of ERK1 and ERK2 activation, completely blocked the translational activation of microinjected mRNA containing Gfp and Cnot6l 3′‐UTR sequences (Fig 8D–F).
ERK1‐ and ERK2‐inhibited oocytes were found to have poorly assembled spindles, to spontaneously release PB2s, and to be arrested at another type of metaphase called metaphase III (MIII) (Zhang et al, 2015; Sha et al, 2017). Because our results so far indicated that CNOT6L is a novel oocyte meiosis regulator downstream of ERK1 and ERK2, we tested whether overexpression of CNOT6L by mRNA microinjection during oocyte maturation can partially reverse the defects caused by ERK1 and ERK2 inhibition. In vitro matured oocytes cultured in the presence of U0126 had severe spindle assembly defects and parthenogenetically released PB2 at a high frequency (Figs 8G and EV5D). Microinjection of mRNAs encoding CNOT6L into these oocytes moderately reduced PB2 emission rates and improved (normalized) spindle assembly. Nevertheless, overexpression of CNOT7 and BTG4 (which are also proteins downstream of ERK1 and ERK2 and are involved in maternal mRNA decay, in addition to CNOT6L) enhanced the rescue effects (Figs 8G and EV5D). In conclusion, CNOT6L and other CCR4–NOT components are important downstream effectors of ERK1 and ERK2 in the regulation of spindle assembly and meiotic cell cycle progression in oocytes (Fig 8H).
Discussion
This study provides direct genetic evidence that the CCR4–NOT complex is crucial for maternal mRNA decay in mammalian oocytes during meiotic maturation. Our study is the first report of gene knockout of Cnot6l, which encodes one of the two mammalian homologs of the evolutionarily conserved yeast CCR4 RNA deadenylase. A study by Ma et al described that RNAi depletion of Cnot6l in cultured mouse oocytes impaired deadenylation of several maternal transcripts, but did not report the requirement of CNOT6L in female fertility (Ma et al, 2015). Of note, oocyte maturation failure is the only obvious phenotype in Cnot6l −/− mice, indicating that CNOT6 can compensate for the loss of CNOT6L in most cell types and in most physiological events. Cnot6l is more strongly expressed in mature murine oocytes, when compared with its homolog Cnot6. This distinct expression pattern suggests that Cnot6l is indispensable only in oocytes. Results of Cnot6l 3′‐UTR reporter assay showed that the translation of maternal Cnot6l transcripts was activated after meiotic resumption, by an MAPK cascade‐dependent mechanism in maturing oocytes (Ma et al, 2015; and our current study). This is a key mechanism that accelerates the mRNA turnover during oocyte meiotic maturation.
RNA sequencing results indicated that more than 800 maternal mRNAs are affects 2‐fold or more by the GV stage. While this is much fewer than the numbers of transcripts being affected during oocyte maturation, it may cause some potential effect during folliculogenesis. On the other hand, we have presented results showing that follicle growth defects were not observed in Cnot6l knockout mice (Fig EV2). Therefore, although Cnot6l deletion has affected transcriptome of fully grown GV oocytes to some extent, the physiological abnormalities only appear after meiotic resumption, at the end of folliculogenesis.
The in vivo matured Cnot6l null oocytes have a higher PB1 emission rate than those matured in vitro. This might be caused by two reasons: (i) The in vivo microenvironment supports oocyte meiotic maturation better than the in vitro culture system does; (ii) superovulation is an oocyte selection step by itself. Therefore, the oocytes that are less defective have a better chance to be ovulated by the Cnot6l KO mice. We have used 5 Cnot6l null females in the 32‐week fertility test. Four of them were completely infertile. Only 1 female give birth to 2 pups during this period. Although the majority of the Cnot6l null females were completely infertile, redundancy of Cnot6 could be one mechanism that caused only subfertility in some Cnot6l null mice.
Because the degradation of so many maternal mRNAs was blocked or delayed after Cnot6l knockout, and because the functions of many proteins encoded by these maternal mRNAs were unknown, we could not specifically connect the meiotic defects with the decay of defined mRNAs. Nor could we rule out the possibility that some of the eliminated mRNAs might be via downstream/indirect effects instead of direct targets of CNOT6L. For instance, accumulation/stabilization of certain RNA‐binding proteins (PABPC1L, CPEB1, MSY2, etc.) after Cnot6l knockout might indirectly prevent the decay of a subset of maternal mRNAs. In addition, theoretically it is possible that some maternal transcripts were deadenylated but not completely degraded by CNOT6L at certain stages of oocyte maturation. Technically, we have not shown a direct effect of CNOT6L on stability (i.e., by stability assays) of the eliminated transcripts. Nonetheless, in the absence of transcription during oocyte meiotic maturation, this is the most likely that the CNOT6L‐targeted mRNAs were destabilized and completely degraded after deadenylation.
Although both CNOT6L and BTG4 accumulate in maturing oocytes and trigger maternal mRNA decay, it appears that their knockouts yield different phenotypes in oocytes: Although most Cnot6l −/− oocytes are arrested at prometaphase I by an active SAC, Btg4 −/− oocytes complete meiotic maturation and are arrested after fertilization. There is a report of spindle assembly defects in mouse oocytes after RNA interference‐based depletion of Btg4 (Pasternak et al, 2016), but we never observed this phenotype in Btg4 −/− oocytes. It is acknowledged that phenotypes of knockout mouse models reflect gene functions more accurately than those obtained by in vitro RNA interference approaches.
At the molecular level, the Cnot6l knockout stabilized a greater number of maternal transcripts than the Btg4 knockout did, particularly during GV‐to‐MII transition. This phenomenon is consistent with the observation that the Cnot6l knockout led to an earlier phenotype (meiotic maturation failure) than the Btg4 knockout did (MZT failure) in mouse oocytes. There are two explanations for the different in vivo functions of CNOT6L and BTG4.
First, our results suggest that BTG4 is not the only CCR4–NOT adaptor protein in maturing oocytes. The in vivo function of BTG4 relies on its binding to the CCR4–NOT catalytic subunit CNOT7 or CNOT8. During MZT, BTG4 binds to translation initiation factor eIF4E and thereby recruits the CCR4–NOT complex to the actively translated mRNAs (Yu et al, 2016b). At an earlier stage of oocyte maturation, literally prometaphase in this study, RNA‐binding protein ZFP36L2 associates with CNOT6L and functions as an alternative CCR4–NOT adaptor in triggering the degradation of ARE‐containing transcripts. During the preparation of this manuscript, decreased oocyte maturation with ZFP36L2 depletion has been shown and published (Dumdie et al, 2018). This study further indicated that ZFP36L2 is a key regulator of maternal transcriptome during the oocyte growing stage, and is crucial for the genome transcription silence in fully grown GV oocytes. Zfp36l2 KO mice were reported to be infertile due to oocyte maturation defects, but the phenotype was not linked with mRNA degradation (Ramos et al, 2004). As a complementation to these findings, our results indicated that ZFP36L2 is a key adaptor of CNOT6L in mediating mRNA decay at the final stage of oocyte meiosis. The stepwise recruitment of different adaptors by different catalytic subunits mediates stage‐specific degradation of maternal mRNAs by the CCR4–NOT deadenylase, as summarized in Fig 8H.
Second, CNOT6L may have some in vivo functions distinct from those of CNOT7 and CNOT8. In vitro biochemical assays indicated that purified CNOT6L protein was fully functional in terms of RNA deadenylation in the absence of other CCR4–NOT subunits (Wang et al, 2010). Moreover, the partial crystal structure of the yeast CCR4–NOT subcomplex shows that the catalytic centers of CCR4 and CAF1 are pointed in different directions (Collart & Panasenko, 2017). Functional analyses of CNOT7 and CNOT8 (CAF1 homologs) and CNOT6 and ‐6L in human cells also indicate that the expression profiles of CNOT7 and CNOT8 knockdown cells and those of CNOT6 and ‐6L knockdown cells have qualitative differences in differentially regulated mRNA sets (Mittal et al, 2011). The Cnot7 knockout in mice causes infertility only in males (Berthet et al, 2004), in contrast to the Cnot6l knockout. In our study, WT, but not catalytically dead CNOT6L, partially reversed the meiotic maturation defects of Cnot6l −/− oocytes. On the other hand, the LRR‐lacking CNOT6L, which failed to interact with CNOT7 and CNOT8, also exerted a rescue effect in Cnot6l −/− oocytes although weaker than the effect of full‐length CNOT6L. These data imply that CNOT6L is partially functional independently of the other CCR4–NOT subunits.
In maturing mammalian oocytes, massive maternal mRNA degradation is initiated as early as the stage of meiotic resumption (Schier, 2007). The oocyte maturation‐accompanied mRNA decay is considered a prologue of MZT in mammals, but its cellular function and physiological importance have been inconclusive. Our study clearly shows that CNOT6L‐dependent degradation of a subset of maternal transcripts is crucial for spindle assembly and meiosis I completion. Particularly, the abundant transcripts encoding translation‐related proteins (CPEB1, PABPC1L), ribosomal subunits, and cell division regulators (survivin, TUBB4B) are significantly degraded during oocyte maturation. Notably, these transcripts stabilized in Cnot6l‐null but not Btg4‐null oocytes. Again, these molecular details are consistent with the phenotypes of the corresponding knockout mice. Our RNA‐seq analyses of polysome‐binding transcripts revealed that the stabilized transcripts remained polyadenylated and underwent active translation in maturing Cnot6l‐null oocytes, when the translation of these transcripts already decreased in control oocytes. Therefore, it seems that the biological function of immediate mRNA degradation after meiotic resumption is to downregulate the protein translation machinery as well as its regulatory factors in an oocyte, thereby keeping the ooplasm in a state conducive to meiotic cell cycle progression, particularly the acentriolar spindle assembly and homologous chromosome separation, which are unique events of meiosis (Schatten & Sun, 2015). Insufficient maternal mRNA decay and abnormally high translation activity indirectly cause SAC activation and lead to prometaphase arrest of the oocyte cell cycle.
Consistent with our findings, a recent study that was published while this manuscript was in revision reported decreased female fertility of Cnot6l‐null mice. The study showed that in oocytes and zygotes derived from Cnot6l −/− mice, the timely degradation of maternal mRNA is perturbed (Horvat et al, 2018). However, the oocyte maturation process of Cnot6l‐null oocytes was not observed in this study. In contrast, our results suggest that the defects of MZT in the maternal Cnot6l‐deleted embryos are a phenotype secondary to the oocyte meiotic maturation failure.
Materials and Methods
Animals
Cnot6l −/− mice were generated using CRISPR‐CAS9 system as illustrated in Fig EV1A. Btg4 −/− mice were previously reported (Yu et al, 2016b). All mouse strains had a C57B6 background. Wild‐type C57BL6 mice were obtained from the Zhejiang Academy of Medical Science, China. Animal care and experimental procedures were conducted in accordance with the Animal Research Committee guidelines of Zhejiang University.
Production of Cas9 and sgRNAs for Cnot6l knockout
The codon optimized Cas9 expression construct, Cas9‐N‐NLS‐flag‐linker (Addgene No. 44758), was synthesized and inserted into a pST1374 vector as previously described (Shen et al, 2013). The pUC57‐sgRNA expression vector used for in vitro transcription of sgRNAs has been described (Zhou et al, 2014). Oligos for the generation of mCnot6l‐targeting sgRNA expression plasmids (Appendix Table S1) were annealed and cloned into the BsaI sites of pUC57‐sgRNA.
The pST1374‐Cas9‐N‐NLS‐flag‐linker vector was linearized using the AgeI enzyme and in vitro transcribed using the T7 Ultra Kit (Invitrogen, AM1345). SgRNA oligos were annealed into a pUC57‐sgRNA expression vector with a T7 promoter. Then, expression vectors were linearized by DraI and transcribed in vitro using the MEGAshortscript kit (Invitrogen, AM1354). sgRNAs were purified using the MEGAclear Kit (Invitrogen, AM1908) and recovered by alcohol precipitation.
In vitro transcription and preparation of mRNAs for microinjections
To prepare mRNAs for microinjection, expression vectors were linearized and subjected to phenol/chloroform extraction and ethanol precipitation (Sha et al, 2017). The linearized DNAs were in vitro transcribed using the SP6 message mMACHINE Kit (Invitrogen, AM1340). Transcribed mRNAs were added with poly(A) tails (~200–250 bp) using the mMACHINE Kit (Invitrogen, AM1350), were recovered by lithium chloride precipitation, and resuspended in nuclease‐free water.
Microinjection of oocytes
For microinjection, fully grown GV oocytes were incubated in M2 medium with 2 μM milrinone to inhibit spontaneous GVBD. All injections were performed using an Eppendorf transferman NK2 micromanipulator. Denuded oocytes were injected with 5–10 pl samples per oocyte. The concentration of all injected RNAs was adjusted to 500 ng/μl. After injection, oocytes were washed and cultured in M16 medium at 37°C with 5% CO2.
Superovulation and fertilization
As describe previously (Sha et al, 2017), female mice (21–23 days old) were intraperitoneally injected with 5 IU of PMSG (Ningbo Sansheng Pharmaceutical Co., ltd., P.R China). After 44 h, mice were then injected with 5 IU of hCG (Ningbo Sansheng Pharmaceutical Co., ltd., P.R China). After an additional 16 h, oocyte–cumulus complexes were removed from the oviducts and the numbers of oocytes were counted after digestion with 0.3% hyaluronidase (Sigma‐Aldrich). To obtain early embryos, female mice were mated with 10‐ to 12‐week‐old WT males. Successful mating was confirmed by the presence of vaginal plugs. Embryos were harvested from oviducts at the indicated time points after hCG injection.
Oocyte culture
The 21‐day‐old female mice were injected with 5 IU of PMSG and humanely euthanized 44 h later. Oocytes at the GV stage were harvested in M2 medium (M7167; Sigma‐Aldrich) and cultured in mini‐drops of M16 medium (M7292; Sigma‐Aldrich) covered with mineral oil (M5310; Sigma‐Aldrich) at 37°C in a 5% CO2 atmosphere. In some experiments, milrinone (2 μM) was added to the culture media to inhibit spontaneous GVBD. U0126 (20 μM) was added to the culture media to inhibit ERK1/2 activation (Sha et al, 2017).
Immunofluorescence and Confocal microscopy
Oocytes were fixed with 4% paraformaldehyde in PBS. They were then permeabilized with 0.3% Triton X‐100 in PBS. Antibody staining was performed using standard protocols described previously (Li et al, 2013b). Antibodies used in the experiments are described in Appendix Table S1. Imaging was performed on a Zeiss LSM710 confocal microscope. Semi‐quantitative analysis of the fluorescence signals was conducted using the NIH Image program ImageJ, as previously described (Akiyama et al, 2011).
Chromosome spreading and immunofluorescence
ZP‐free oocytes were fixed in a solution containing 1% paraformaldehyde, 0.15% Triton X‐100, and 3 mM DTT (Sigma‐Aldrich) on glass slides for 30 min and air dried. Immunofluorescent staining was performed as in oocytes described above.
Live‐cell imaging
For live imaging, mRNAs encoding for GFP‐tubulin and mCherry‐securin were microinjected into WT and CNOT6L‐deleted oocytes and released from milrinone after 8 h. Oocytes were cultured in M16 medium containing Hoechst‐33342 (to label DNA). The oocytes underwent GVBD within 2 h after being released from milrinone were subjected to live‐cell imaging. Images were acquired on a DV ELITE High Resolution Invented Living Cell Work station. Image acquisition was performed using Zeiss LSM‐780 confocal microscope (Zeiss) equipped with PC‐Apochromat 20×/0.8 NA objective lenses at 6‐min intervals for 16 h.
Western blot analysis
Oocytes were lysed in protein loading buffer and heated at 95°C for 5 min. SDS–PAGE and immunoblots were performed following standard procedures using a Mini‐PROTEAN Tetra Cell System (Bio‐Rad, Hercules, CA). The primary antibodies and dilution factors used are listed in Appendix Table S2.
Cell culture, plasmid transfection, and immunoprecipitation
HeLa cells were obtained from ATCC and were recently authenticated and tested for contamination. Cells were cultured in DMEM (Invitrogen) plus 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin–streptomycin solution (Gibco) at 37°C in a humidified 5% CO2 incubator. Mouse Cnot6l, Cnot7, and Zfp36l2 cDNAs were PCR amplified from a mouse ovarian cDNA pool and ligated into pcDNA‐based eukaryote expression vectors. Transient plasmid transfection was accomplished using Lipofectamine 2000 (Invitrogen). After a 48‐h transfection, cells were harvested in a lysis buffer containing 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10% glycerol, and 0.5% NP‐40. After centrifugation, the supernatant was subjected to immunoprecipitation with different affinity gels (Sigma). After incubation at 4°C for 4 h, beads were washed with lysis buffer. The bead‐bound proteins were eluted using SDS sample buffer for Western blot analysis (Sha et al, 2017).
Immunoprecipitation of mRNA–protein complexes
HeLa cells were obtained from ATCC and were recently authenticated and tested for contamination. Cells were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Hyclone) and 1% penicillin–streptomycin solution (Gibco) at 37°C in a humidified 5% CO2 incubator. Transient plasmid including mouse Cnot6l, Zfp36l2 cDNA transfection was done using Lipofectamine 2000 (Invitrogen). After a 48‐h transfection, cells were lysed in lysis buffer (50 mM Tris–HCl (pH 7.4), 1% Triton X‐100, 150 mM NaCl, 5 mM EDTA, protease inhibitor cocktail, and RNase inhibitor). After centrifugation, the supernatant was subjected to immunoprecipitation with different affinity gels (Sigma). After incubation at 4°C for 4 h, beads were washed with washing buffer (50 mM Tris–HCl (pH 7.4), 0.1% Triton X‐100, 500 mM NaCl, 5 mM EDTA, protease inhibitor cocktail, and RNase inhibitor). Bead‐bound RNA was extracted using RNeasy Mini kit (Qiagen, 74106) according to the manufacturer's instructions. RNAs were reverse‐transcribed with MLV (Invitrogen). The fold change was analyzed through RT–PCR.
Poly(A) tail assay
Total RNA was isolated from 100 oocytes at indicated stages using the RNeasy Mini kit (Qiagen, 74106). P1 (5′‐GCGAGCTCCGCGGCCGCGT12‐3′) was anchored to Oligo(dT) by T4 DNA ligase. Reverse transcription was performed using the SuperScript IV (Invitrogen) with Oligo(dT) anchored P1. The products were used in a PCR product with gene‐specific primers P2 (Appendix Table S1) and dT anchor primer P1 (5′‐GCGAGCTCCGCGGCCGCGT12‐3′). A0 was PCR amplified by using gene‐specific primers P2 (Appendix Table S1) and gene‐specific primers P0 (Appendix Table S1). The PCR conditions were as follows: 30 s at 94°C, 20 s at 58°C, and 40 s at 72°C. PCR products were analyzed on a 2% agarose gel.
RNA‐seq library preparation
Oocytes and zygotes were collected from indicated genotypes (10 oocytes or embryos per sample). Each sample was directly lysed with 4 μl lysis buffer (0.2% Triton X‐100, RNase inhibitor, dNTPs, oligo‐dT primers, and 100 pg mCherry mRNA spike‐in) and immediately used for cDNA synthesis using the Smart‐seq2 method as described previously (Picelli et al, 2014). For RNA‐seq libraries of polysome‐bound mRNA, 2 μl purified RNA was used per sample, with 50 pg mCherry spike‐in added before cDNA synthesis.
Sequencing libraries were constructed from 500 pg of amplified cDNA using TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme, TD503) according to manufacturer's instructions. Barcoded libraries were pooled and sequenced on the Illumina HiSeq X Ten platform with 150 bp paired‐end reads.
Polysome isolation and library construction
Polysomes were isolated from oocytes as reported (Chen et al, 2011). Fully grown GV oocytes are collected from PMSG‐primed (44 h) 23‐day‐old mice of indicated genotypes. GV, MI, and MII oocytes were collected at 0, 8, and 16 after cultured. 500 oocytes of every sample were lysed with PLB (30 mM Tris–HCl at pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1% Triton, 1 mM DTT, 0.25 mM Na3VO4, 20 mM beta‐glycerophosphate, 40 U/ml RNase inhibitor (Takara), 100 μg/ml cycloheximide, plus protease inhibitor cocktail). Oocyte lysates were loaded on a 10‐ml 15–50% sucrose gradient and centrifuged at 200,000 g for 120 min at 4°C. RNAs were precipitated adding with 1/10 volume of NaAc and 3× volume of ethanol at −80°C. Polysome‐bound RNAs were purified with RNeasy Mini kit (Qiagen, 74106). Library construction for polysome‐bound RNAs was similar to the method used for RNA‐seq.
RNA‐seq data analysis
RNA‐ was performed with biological replicates for all samples. Raw reads were trimmed with Trimmomatic (v0.36) to 50 bp and mapped to the mouse genome (mm9) with TopHat (v2.0.11) with default parameters. Uniquely mapped reads were subsequently assembled into transcripts guided by reference annotation (University of California at Santa Cruz [UCSC] gene models) with Cufflinks (v2.2.1). The expression level of each gene was quantified as fragments per kilobase of transcript per million mapped reads (FPKM) and was further normalized with the mCherry spike‐in. Samples prepared in different batches were normalized by the WT GV‐stage oocyte sample in each batch. Only expressed genes (FPKM > 1 in at least one sample) were considered in all analyses unless otherwise specified. Functional annotation was performed with DAVID (https://david.ncifcrf.gov/).
Statistical analyses were implemented with R (http://www.rproject.org). The Spearman correlation coefficient (r s) was calculated using the cor function, and the complete linkage hierarchical algorithm was used to cluster the genes. Quality controls of RNA‐seq and polysome RNA‐seq results were provided as Appendix Tables S3 and S4, respectively.
RNA isolation and real‐time RT–PCR
Total RNA was extracted using RNeasy Mini kit (Qiagen, 74106) according to the manufacturer's instructions. Real‐time RT–PCR analysis was performed using a Power SYBR Green PCR Master Mix (Applied Biosystems, Life technologies) and an Applied Biosystems 7500 Real‐Time PCR System. Relative mRNA levels were calculated by normalizing to the levels of endogenous Gapdh mRNA (internal control) or encoded exogenous Gfp mRNA using Microsoft EXCEL®. The relative transcript levels of samples were compared to the control, and the fold changes are demonstrated. For each experiment, qPCR products were done in triplicate. Primer sequences are listed in Appendix Table S1.
Statistical analysis
Results are given as means ± SEM. Most experiments included at least three independent samples and were repeated at least three times. Results for two experimental groups were compared by two‐tailed unpaired Student's t‐tests. Statistically significant values of P < 0.05, P < 0.01, and P < 0.001 by two‐tailed Student's t‐test are indicated by asterisks (*), (**), and (***), respectively. “n.s.” indicates non‐significant.
Data availability
RNA‐seq data have been deposited in the NCBI Gene Expression Omnibus database under accession code GSE118564.
Author contributions
H‐YF, LS, and X‐HO conceived the project. H‐YF, Q‐QS, and J‐XG designed and analyzed experiments. CY, S‐YJ, Q‐QS, YJ, J‐LY, J‐CJ, Y‐LZ, and X‐XD performed experiments. X‐XD, S‐YZ, and J‐CJ assisted in microinjection of mouse embryos. Q‐QS and H‐YF wrote the paper. Q‐QS, J‐LY, and J‐XG contributed equally to this work.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
This study is funded by the National Key Research and Developmental Program of China (2016YFC1000600, 2017YFC1001500), National Natural Science Foundation of China (31528016, 31371449, 31671558), and The Key Research and Development Program of Zhejiang Province (2017C03022).
The EMBO Journal (2018) 37: e99333
References
- Akiyama T, Suzuki O, Matsuda J, Aoki F (2011) Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet 7: e1002279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CB, Rodriguez KF, Stumpo DJ, Ribeiro‐Neto F, Korach KS, Blackshear PJ, Birnbaumer L, Ramos SB (2014) The RNA‐binding protein, ZFP36L2, influences ovulation and oocyte maturation. PLoS ONE 9: e97324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barckmann B, Simonelig M (2013) Control of maternal mRNA stability in germ cells and early embryos. Biochim Biophys Acta 1829: 714–724 [DOI] [PubMed] [Google Scholar]
- Belloc E, Mendez R (2008) A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature 452: 1017–1021 [DOI] [PubMed] [Google Scholar]
- Berthet C, Morera AM, Asensio MJ, Chauvin MA, Morel AP, Dijoud F, Magaud JP, Durand P, Rouault JP (2004) CCR4‐associated factor CAF1 is an essential factor for spermatogenesis. Mol Cell Biol 24: 5808–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M (2011) Genome‐wide analysis of translation reveals a critical role for deleted in azoospermia‐like (Dazl) at the oocyte‐to‐zygote transition. Genes Dev 25: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO (2012) The Ccr4–not complex. Gene 492: 42–53 [DOI] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO (2017) The Ccr4‐Not complex: architecture and structural insights. Subcell Biochem 83: 349–379 [DOI] [PubMed] [Google Scholar]
- Curinha A, Oliveira Braz S, Pereira‐Castro I, Cruz A, Moreira A (2014) Implications of polyadenylation in health and disease. Nucleus 5: 508–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doidge R, Mittal S, Aslam A, Winkler GS (2012) Deadenylation of cytoplasmic mRNA by the mammalian Ccr4‐Not complex. Biochem Soc Trans 40: 896–901 [DOI] [PubMed] [Google Scholar]
- Dumdie JN, Cho K, Ramaiah M, Skarbrevik D, Mora‐Castilla S, Stumpo DJ, Lykke‐Andersen J, Laurent LC, Blackshear PJ, Wilkinson MF, Cook‐Andersen H (2018) Chromatin modification and global transcriptional silencing in the oocyte mediated by the mRNA decay activator ZFP36L2. Dev Cell 44: 392–402 e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat F, Fulka H, Jankele R, Malik R, Jun M, Solcova K, Sedlacek R, Vlahovicek K, Schultz RM, Svoboda P (2018) Role of Cnot6l in maternal mRNA turnover. Life Sci Alliance 1: e201800084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez‐Yamout MA, Dyson HJ, Wright PE (2004) Recognition of the mRNA AU‐rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264 [DOI] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD (2014) Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol 30: 393–415 [DOI] [PubMed] [Google Scholar]
- Jiang ZZ, Hu MW, Wang ZB, Huang L, Lin F, Qi ST, Ouyang YC, Fan HY, Schatten H, Mak TW, Sun QY (2014) Survivin is essential for fertile egg production and female fertility in mice. Cell Death Dis 5: e1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KT (2011) Anaphase‐promoting complex control in female mouse meiosis. Results Probl Cell Differ 53: 343–363 [DOI] [PubMed] [Google Scholar]
- Li L, Lu X, Dean J (2013a) The maternal to zygotic transition in mammals. Mol Aspects Med 34: 919–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Yu C, Wang ZW, Zhang YL, Liu XM, Zhou D, Sun QY, Fan HY (2013b) DNA topoisomerase II is dispensable for oocyte meiotic resumption but is essential for meiotic chromosome condensation and separation in mice. Biol Reprod 89: 118 [DOI] [PubMed] [Google Scholar]
- Liu Y, Lu X, Shi J, Yu X, Zhang X, Zhu K, Yi Z, Duan E, Li L (2016) BTG4 is a key regulator for maternal mRNA clearance during mouse early embryogenesis. J Mol Cell Biol 8: 366–368 [DOI] [PubMed] [Google Scholar]
- Ma J, Fukuda Y, Schultz RM (2015) Mobilization of dormant Cnot7 mRNA promotes deadenylation of maternal transcripts during mouse oocyte maturation. Biol Reprod 93: 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JE, Reese JC (2012) Ccr4‐Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 47: 315–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S, Aslam A, Doidge R, Medica R, Winkler GS (2011) The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4‐Not complex contribute to the prevention of cell death and senescence. Mol Biol Cell 22: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak M, Pfender S, Santhanam B, Schuh M (2016) The BTG4 and CAF1 complex prevents the spontaneous activation of eggs by deadenylating maternal mRNAs. Open Biol 6: 160184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R (2014) Full‐length RNA‐seq from single cells using Smart‐seq2. Nat Protoc 9: 171–181 [DOI] [PubMed] [Google Scholar]
- Pique M, Lopez JM, Foissac S, Guigo R, Mendez R (2008) A combinatorial code for CPE‐mediated translational control. Cell 132: 434–448 [DOI] [PubMed] [Google Scholar]
- Ramos SB, Stumpo DJ, Kennington EA, Phillips RS, Bock CB, Ribeiro‐Neto F, Blackshear PJ (2004) The CCCH tandem zinc‐finger protein Zfp36l2 is crucial for female fertility and early embryonic development. Development 131: 4883–4893 [DOI] [PubMed] [Google Scholar]
- Schatten H, Sun QY (2015) Centrosome and microtubule functions and dysfunctions in meiosis: implications for age‐related infertility and developmental disorders. Reprod Fertil Dev 27: 934–943 [DOI] [PubMed] [Google Scholar]
- Schier AF (2007) The maternal‐zygotic transition: death and birth of RNAs. Science 316: 406–407 [DOI] [PubMed] [Google Scholar]
- Sha QQ, Dai XX, Dang Y, Tang F, Liu J, Zhang YL, Fan HY (2017) A MAPK cascade couples maternal mRNA translation and degradation to meiotic cell cycle progression in mouse oocytes. Development 144: 452–463 [DOI] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X (2013) Generation of gene‐modified mice via Cas9/RNA‐mediated gene targeting. Cell Res 23: 720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ (2007) Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol 302: 104–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD (2009) The maternal‐to‐zygotic transition: a play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Ukleja M, Valpuesta JM, Dziembowski A, Cuellar J (2016) Beyond the known functions of the CCR4‐NOT complex in gene expression regulatory mechanisms: new structural insights to unravel CCR4‐NOT mRNA processing machinery. BioEssays 38: 1048–1058 [DOI] [PubMed] [Google Scholar]
- Villanyi Z, Collart MA (2016) Building on the Ccr4‐Not architecture. BioEssays 38: 997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Morita M, Yang X, Suzuki T, Yang W, Wang J, Ito K, Wang Q, Zhao C, Bartlam M, Yamamoto T, Rao Z (2010) Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J 29: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawro ME, Sobierajska K, Ciszewski WM, Wagner W, Frontczak M, Wieczorek K, Niewiarowska J (2017) Tubulin beta 3 and 4 are involved in the generation of early fibrotic stages. Cell Signal 38: 26–38 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Balacco DL (2013) Heterogeneity and complexity within the nuclease module of the Ccr4‐Not complex. Front Genet 4: 296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Dean J (2016) BTG4, a maternal mRNA cleaner. J Mol Cell Biol 8: 369–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Li R, Qiao J, Tang F (2013) Single‐cell RNA‐Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Yu C, Zhang YL, Pan WW, Li XM, Wang ZW, Ge ZJ, Zhou JJ, Cang Y, Tong C, Sun QY, Fan HY (2013) CRL4 complex regulates mammalian oocyte survival and reprogramming by activation of TET proteins. Science 342: 1518–1521 [DOI] [PubMed] [Google Scholar]
- Yu C, Ji SY, Sha QQ, Sun QY, Fan HY (2015a) CRL4‐DCAF1 ubiquitin E3 ligase directs protein phosphatase 2A degradation to control oocyte meiotic maturation. Nat Commun 6: 8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Xu YW, Sha QQ, Fan HY (2015b) CRL4DCAF1 is required in activated oocytes for follicle maintenance and ovulation. Mol Hum Reprod 21: 195–205 [DOI] [PubMed] [Google Scholar]
- Yu C, Ji SY, Dang YJ, Sha QQ, Yuan YF, Zhou JJ, Yan LY, Qiao J, Tang F, Fan HY (2016a) Oocyte‐expressed yes‐associated protein is a key activator of the early zygotic genome in mouse. Cell Res 26: 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Ji SY, Sha QQ, Dang Y, Zhou JJ, Zhang YL, Liu Y, Wang ZW, Hu B, Sun QY, Sun SC, Tang F, Fan HY (2016b) BTG4 is a meiotic cell cycle‐coupled maternal‐zygotic‐transition licensing factor in oocytes. Nat Struct Mol Biol 23: 387–394 [DOI] [PubMed] [Google Scholar]
- Zhang YL, Liu XM, Ji SY, Sha QQ, Zhang J, Fan HY (2015) ERK1/2 activities are dispensable for oocyte growth but are required for meiotic maturation and pronuclear formation in mouse. J Genet Genomics 42: 477–485 [DOI] [PubMed] [Google Scholar]
- Zhou J, Shen B, Zhang W, Wang J, Yang J, Chen L, Zhang N, Zhu K, Xu J, Hu B, Leng Q, Huang X (2014) One‐step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int J Biochem Cell Biol 46: 49–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 6
Source Data for Figure 7
Data Availability Statement
RNA‐seq data have been deposited in the NCBI Gene Expression Omnibus database under accession code GSE118564.


