Short abstract
Spinal cord stimulation has become an important modality in pain treatment especially for neuropathic pain conditions refractory to pharmacotherapy. However, the molecular control of inhibitory and excitatory mechanisms observed after spinal cord stimulation are poorly understood. Here, we used RNA-seq to identify differences in the expression of genes and gene networks in spinal cord tissue from nerve-injured rats with and without repetitive conventional spinal cord stimulation treatment. Five weeks after chronic constrictive injury to the left sciatic nerve, male and female rats were randomized to receive repetitive spinal cord stimulation or no treatment. Rats receiving spinal cord stimulation underwent epidural placement of a miniature stimulating electrode and received seven sessions of spinal cord stimulation (50 Hz, 80% motor threshold, 0.2 ms, constant current bipolar stimulation, 120 min/session) over four consecutive days. Within 2 h after the last spinal cord stimulation treatment, the L4-L6 spinal segments ipsilateral to the side of nerve injury were harvested and used to generate libraries for RNA-seq. Our RNA-seq data suggest further increases of many existing upregulated immune responses in chronic constrictive injury rats after repetitive spinal cord stimulation, including transcription of cell surface receptors and activation of non-neuronal cells. We also demonstrate that repetitive spinal cord stimulation represses transcription of several key synaptic signaling genes that encode scaffold proteins in the post-synaptic density. Our transcriptional studies suggest a potential relationship between specific genes and the therapeutic effects observed in patients undergoing conventional spinal cord stimulation after nerve injury. Furthermore, our results may help identify new therapeutic targets for improving the efficacy of conventional spinal cord stimulation and other chronic pain treatments.
Keywords: RNA-seq, gene expression, spinal cord stimulation, nerve injury, pain, spinal cord
Introduction
Increased efforts to avoid the severe side effects known to opioid analgesics are shifting treatment for chronic pain conditions towards non-opioid and interventional therapies. A mounting body of evidence supports the use of spinal cord stimulation (SCS) for its treatment effectiveness and safety.1–5 Conventional SCS was developed based on the seminal “gate control” theory of pain6 and remains a widely used neurostimulation pain therapy. Conventional SCS involves placement of epidural leads, often at a few levels above (i.e., rostral to) the affected spinal segments that receive noxious inputs (e.g. “pain segments”), and delivery of pulsed electricity to stimulate the dorsal column. Conventional SCS activates low-threshold afferents (i.e., Aβ-fibers) which produces the mild paresthesia (i.e., tingling sensation). Thus, pain inhibition from conventional SCS partially acts through antidromic action potentials in dorsal column fibers to activate inhibitory mechanisms in distal “pain segments” via collateral branches.7,8
Pain inhibitory effects by conventional SCS are intricately linked with spinal mechanisms,9–11 as evident by inhibition of neuronal sensitization and nociceptive transmission at spinal level, and changes in release of neurotransmitters and neuromodulators in the spinal cord.11–14 However, the molecular mechanisms which underlie the therapeutic effects of SCS remain unknown. While limited in scope, previous findings suggest that SCS induces broad and prolonged changes in gene expression.15–17 To identify new gene networks and molecular pathways altered after repetitive SCS, we conducted the first RNA-seq study of the lumbar spinal cord after repetitive SCS at the T13-L1 level in rats during the maintenance phase of neuropathic pain. To mimic clinical SCS, we applied bi-polar stimulation through a miniature quadripolar electrode which has been validated in previous studies.12,14,18,19 Our findings are consistent with previous reports of an increased immune response associated with SCS. Notably, we also identified downregulation of several genes encoding scaffold proteins located on the postsynaptic membrane in nerve-injured rats after SCS for the first time, which may impact neurotransmission and synaptic efficacy associated with central sensitization. Such transcriptional studies will help explain physiological changes that occur in the spinal cord following repeated SCS after nerve injury and may identify novel therapeutic targets which improve the efficacy of SCS.
Methods
Animals
Adult male and female Sprague-Dawley rats (n = 12; 12–16 weeks old; Harlan Bioproducts for Science, Indianapolis, IN) were allowed to acclimate for a minimum of 48 h prior to any experimental procedure. The rats were housed separately after implanting the SCS electrode and given access ad libitum to food and water. All procedures involving animals were reviewed and approved by the Johns Hopkins Animal Care and are performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Behavior testing
Mechanical hypersensitivity was measured using von Frey monofilaments as previously described.12,20 Animals were placed in individual plexiglass cages with a wire mesh floor and allowed to acclimate for 1 h. Response to tactile stimulation to the midplantar surface of the hind paw ipsilateral to the nerve lesion was determined with the up-down method using a series of von Frey monofilaments (0.38, 0.57, 1.23, 1.83, 3.66, 5.93, 9.13, and 13.1 g) as described previously.20 Each monofilament was applied for 4 to 6 s to the test area between the footpads on the plantar surface of each hind paw. Monofilaments with increasing force were applied until a positive response was observed (e.g., abrupt paw withdrawal, shaking, and licking). When a positive response was observed, the monofilament with the next lower force was applied. If a negative response was observed, the next higher force was used. The test continued (1) for five filament applications after a positive test was observed or (2) until the upper or lower end of the von Frey monofilament set was reached. The paw withdrawal threshold (PWT) was determined according to the formula provided by Dixon.21 If a rat did not achieve at least a 50% reduction in baseline (BL) PWT after 48 h or on day 14 following nerve injury, then this animal was considered non-allodynic and excluded from the study.
CCI of sciatic nerve
CCI surgery to the left sciatic nerve was performed on all rats as previously described.22 Under 2% to 3% isoflurane, a small incision was made at the level of the mid-thigh. The sciatic nerve was exposed by blunt dissection through the biceps femoris. Previous studies showed that CCI of sciatic nerve with silk ligatures induced similar infiltration of inflammatory cells and changes in function of the nerve-blood barrier,23 and more stable neuropathic pain behaviors,24 as compared to that induced by chromic gut ligature. Accordingly, the nerve trunk proximal to the distal branching point was loosely ligated with four 4-0 silk sutures placed approximately 0.5 mm apart until the epineurium was slightly compressed and minor twitching of the relevant muscles was observed. The muscle layer was closed with 4-0 silk suture, and the wound closed with metal clips.
Electrode placement and SCS treatment
Animals randomized to receive SCS underwent epidural placement of a sterile, quadripolar SCS electrode (Medtronic Inc.) to the dorsal spinal cord (Figure 1(a)). This electrode mimics clinical SCS and was validated in previous studies in rats.12,14,18,19 Under isoflurane anesthesia, a laminectomy was performed at the T13 vertebrae level through which the electrode was inserted epidurally in the rostral direction. The position of the electrode was adjusted so that the contacts were at the T13-L1 spinal cord level which corresponds to the lower thoracic-upper lumbar region. Sutures to the muscle were used to secure the electrode in place, and the proximal end was tunneled subcutaneously and exited the animal at the top of its head for later connection to an external neurostimulator (Model 2100, A-M Systems, Sequim, WA).
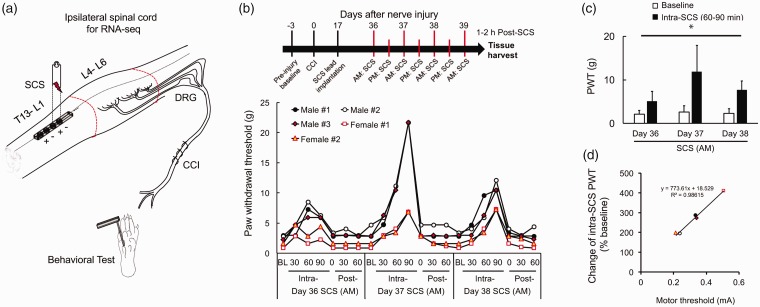
Figure 1.
Experimental setup and pain inhibition by SCS. (a) Schematic diagram illustrating the experimental setup. The miniature SCS lead (Medtronic, Minneapolis, MN) was implanted epidurally over the dorsal spinal cord (midline) at the T13-L1 spinal level. Lumbar spinal cord (L4-L6, marked with red lines) tissues ipsilateral to the side of nerve injury were harvest after the last SCS treatment. (b) Upper: Schematic diagram illustrating the experimental timeline. CCI rats (n = 5) received the same SCS (red bar, 50 Hz, 80% motor threshold, 0.2 ms, constant current, 120 min/session) from days 36 to 38 post-CCI (two sessions/day) and on day 39 post-CCI (one session). Motor thresholds were measured to 4 Hz stimulation (0.2 ms). Lower: On days 36 to 38 post-CCI, PWTs were measured before (baseline, BL), at 30, 60, and 90 min during SCS (intra-SCS), and at 0, 30, and 60 min after completing SCS in the a.m. session. (c) Average PWTs at 60 and 90 min intra-SCS were significantly increased from pre-SCS baseline on each day. Data are expressed as mean + SD. One-way repeated measures ANOVA. *p<0.05 versus pre-SCS baseline. (d) To evaluate the peak inhibitory effect of daily SCS on mechanical hypersensitivity in each animal, we averaged PWTs at 60 and 90 min intra-SCS. Then the “Change of intra-SCS PWT” was calculated as follows: Change of intra-SCS PWT = [(mean intra-SCS PWT) – (baseline PWT)]/(baseline PWT) × 100. Scatterplots showed positive linear correlation between change of intra-SCS PWT and motor threshold.
CCI: chronic constriction injury; PWT: paw withdrawal threshold; SCS: spinal cord stimulation; SD: standard deviation.
In “twin-pairs” SCS, the first and third contacts of the lead from rostral were set as an anode (+), and the second and fourth were set as a cathode (–). Conventional SCS (50 Hz, 0.2 ms, constant current, and 120 min/session) was applied at an intensity that activated low-threshold A-fibers (80% motor threshold (MoT)), as described in previous studies.12,14,18,19 Before SCS, the MoT for each animal was determined by slowly increasing the current amplitude from zero, until muscle contraction in the mid-lower trunk or hind limbs was observed in response to 4 Hz stimulation at 0.2 ms pulse widths. The rats were then acclimated to the testing environment before the pre-SCS BL PWT was measured.
Experimental design
Our primary goal is to examine the changes of gene expression in the spinal cord after repetitive SCS treatments during the maintenance phase of neuropathic pain. All animals developed mechanical hypersensitivity after CCI and were randomized to receive SCS (CCI + SCS group, n = 8) or no treatment (CCI only group, n = 4). Rats randomized to the CCI+SCS group were implanted with a SCS electrode and received SCS (50 Hz, 80% MoT, 0.2 ms, constant current, 120 min/session, twice per day) for three consecutive days on days 36 to 38 post-CCI (Figure 1(b)). PWTs were measured before BL at 30, 60, and 90 min during SCS (intra-SCS) and at 0, 30, and 60 min after completing SCS in each a.m. session. An additional SCS treatment was given on day 39 post-CCI. Within 1 to 2 h following the last SCS treatment, all animals were euthanized by overdose of isoflurane and decapitation. The ipsilateral lumbar spinal cord (L4-L6 spinal segments) ipsilateral to the nerve lesion was harvested and immediately submerged in DNA/RNA shield solution (Zymo, Irvine, CA) for subsequent RNA extraction. We did not separate the dorsal and ventral half of spinal cord, in order to avoid variations due to different dissections of tissue between different animals.
RNA isolation
Total RNA was extracted from the ipsilateral spinal cord with the Quick-RNA MiniPrep Plus kit (Zymo, Irvine, CA) according to manufacturer instructions with on-column DNase I digestion. RNA quantity was measured by the Qubit RNA BR Assay Kit (ThermoScientific, Waltham, MA), and RNA integrity was assessed by the Bioanalyzer RNA Nano Eukaryote kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
RNA-seq library construction and sequencing
Five hundred nanograms of total RNA per sample were used to construct sequencing libraries (n = 1 rat/sample). Strand-specific RNA libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs Inc., Ipswich, MA) after poly(A) selection by the NEBNext poly(A) mRNA Isolation Module (New England Biolabs Inc., Ipswich, MA) according to manufacturer’s instructions. Samples were barcoded using the recommended NEBNext Multiplex Oligos (New England Biolabs Inc., Ipswich, MA). Size range and quality of libraries were verified on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RNA-seq libraries were quantified by quantitative polymerase chain reaction using the KAPA library quantification kit (KAPA Biosystems, Wilmington, MA). Each library was normalized to 2 nM and pooled in equimolar concentrations. Paired-end × 150 sequencing was performed on an Illumina HiSeq4000 (Illumina, San Diego, CA). Libraries were pooled and sequenced using two lanes of one HiSeq4000 flow cell to an average depth of 33.6 million reads per sample.
Data analysis
Sequencing reads were aligned to annotated RefSeq genes of the rat reference genome (rn6) using HISAT2,25 filtered to remove ribosomal RNA, and visualized using the Integrative Genomics Viewer.26 A gene count matrix that contained raw transcript counts for each annotated gene was generated using the featureCounts function of Subread.27 This count matrix was then filtered for low count genes so that only those genes with >0 reads in each sample were retained. To identify genes that were differentially regulated following SCS, transcript counts were normalized and log2 transformed using the default normalization procedures in DESeq2.28 This analysis identified differentially expressed genes between the CCI only and CCI+SCS groups within males or females. The interaction of sex on differential gene expression after injury was evaluated by the interaction term included in the design matrix within DESeq2. All downstream analyses on RNA-seq data were performed on data obtained from DESeq2. Unless otherwise stated an adjusted p-value (i.e., false discovery rate (FDR)) < 0.05 was used to define differentially expressed transcripts between CCI only and CCI+SCS groups. Genes with differential expression between groups were then included in gene ontology (GO) analysis to infer their functional roles and relationships. GO analysis for enriched GO biological processes in each set of differentially enriched genes identified by DESeq2 was performed using ToppGeneSuite (https://toppgene.cchmc.org).29 The International Union of Basic and Clinical Pharmacology database (http://www.guidetopharmacology.org) was used to assign categories to gene products.30
Results
SCS attenuated mechanical hypersensitivity in CCI rats
Rats that developed mechanical hypersensitivity on the ipsilateral hind paw following CCI were randomized to receive SCS (CCI+SCS, n = 8) or not receive SCS treatment (CCI, n = 4). Following implantation of the SCS electrode on day 17 after CCI, one male and two female rats showed impaired motor function that required exclusion from the study. The remaining five rats (i.e., two female rats, three male rats) that received SCS showed no adverse events, and data from these rats were included in all analyses. Each SCS treatment was associated with increases in mechanical PWT in the ipsilateral hind paw from pre-SCS BL (Figure 1(b)). The peak inhibitory effect of SCS often occurred at 60 and 90 min after start of the SCS and returned to the pre-SCS BL within 30 min of cessation of SCS. The averaged PWTs at 60 and 90 min intra-SCS, which reflect the peak effect of SCS, were significantly increased from pre-SCS BL on each day, F(3, 16) = 7.47, p = 0.024; Figure 1(c). The change of intra-SCS PWT in individual animals and MoT show a strong correlation, r(3) = 0.994, p < 0.001, two-tailed test; Figure 1(d).
Differentially regulated genes in the spinal cord after SCS in male and female CCI rats
To determine the effects of SCS on gene expression in the spinal cord that is ipsilateral to the side of nerve injury, we compared RNA-seq data obtained 39 days following CCI to that of rats who received SCS after CCI. Principal component analysis shows segregation of the transcriptomes from CCI rats that received SCS and those that did not receive SCS (Figure 2(a)). The first two principal components accounted for a total of 74%. Compared to CCI only rats, the ipsilateral spinal cord from CCI+SCS rats differentially expressed 1113 (7.9%) genes (FDR<0.05; Figure 2(b)). Of these 1113 differentially expressed genes, 785 (70.5%) were upregulated after SCS and 328 (29.5%) were downregulated (Figure 2(b)). The genes most significantly up- and downregulated with SCS treatment are listed in Table 1 and Table 2, respectively. Of the 1113 differentially expressed genes, 343 genes could be classified into gene classes (i.e., transporters, enzymes, G protein-coupled receptors, ion channels, catalytic receptors, and transcription factors) as defined by International Union of Basic and Clinical Pharmacology (Figure 2(c) and Supplemental Figure 1). Mean normalized counts and relative fold change of specific genes that comprise each of these gene classes is shown in Supplemental Figure 1.
Figure 2.
Differential gene expression between CCI rats with and without SCS. (a) Principal component analysis of libraries sequenced for RNA-seq. (b) Volcano plot showing RNA-seq data of ipsilateral L4-L6 spinal cord from CCI rats with and without SCS treatment. DEGs are designated in red and are defined as differentially expressed genes with a FDR < 0.05. Triangles represent genes with extremely high log10FDR or log2fold change values. (c) Bar plot showing the numbers of genes differentially expressed genes up- and downregulated by gene class as defined by the IUPHAR (top). Relative expression levels for each rat are shown for each gene class represented in the bar plot (bottom). Up- and downregulated genes are colored in yellow and orange, respectively. Horizontal bars indicate group assignment and sex for each rat.
CCI: chronic constriction injury; DEG: differentially expressed gene; FDR: false discovery rate; GPCR= G protein-coupled receptor; IC: ion channel; SCS: spinal cord stimulation.
Table 1.
Top 25 genes upregulated in CCI rats after SCS by FDR.
| Ensembl ID | Gene symbol | Full gene name | Log2 fold change | Standard error | FDR |
|---|---|---|---|---|---|
| ENSRNOG00000046834 | C3 | Complement component 3 | 2.15 | 0.16 | 1.77E-35 |
| ENSRNOG00000046254 | Adgre1 | Adhesion G protein-coupled receptor E1 | 1.81 | 0.18 | 3.16E-19 |
| ENSRNOG00000016294 | Cd4 | CD4 antigen | 1.51 | 0.16 | 2.05E-18 |
| ENSRNOG00000004649 | Il1b | Interleukin 1-beta | 2.46 | 0.26 | 1.29E-17 |
| ENSRNOG00000024899 | Cxcl13 | Chemokine, CXC Motif, Ligand 13 | 4.07 | 0.43 | 1.68E-17 |
| ENSRNOG00000020699 | Cd37 | Leukocyte surface antigen CD37 | 1.07 | 0.12 | 5.84E-15 |
| ENSRNOG00000013886 | Fyb | Fyn-binding protein | 1.54 | 0.18 | 1.06E-14 |
| ENSRNOG00000008816 | Gpnmb | Glycoprotein NMB | 1.42 | 0.17 | 1.24E-13 |
| ENSRNOG00000050430 | Vav1 | VAV1 oncogene | 1.54 | 0.19 | 2.73E-13 |
| ENSRNOG00000042838 | Junb | Oncogene JUN-B | 1.27 | 0.16 | 3.02E-13 |
| ENSRNOG00000008409 | Myo1f | Myosin IF | 1.38 | 0.17 | 6.89E-13 |
| ENSRNOG00000018414 | Csf1r | Colony-stimulating factor 1 receptor | 1.18 | 0.15 | 7.67E-13 |
| ENSRNOG00000043098 | Mt2A | Metallothionein 2A | 1.89 | 0.24 | 7.67E-13 |
| ENSRNOG00000015773 | Il21r | Interleukin 21 receptor | 2.01 | 0.25 | 1.11E-12 |
| ENSRNOG00000038047 | Mt1 | Matrix metalloproteinase 14 | 2.28 | 0.29 | 1.11E-12 |
| ENSRNOG00000028566 | Pld4 | Phospholipase D family, member 4 | 1.16 | 0.15 | 1.45E-12 |
| ENSRNOG00000008465 | Tmem176b | LR8 protein | 1.05 | 0.13 | 2.12E-12 |
| ENSRNOG00000042139 | Clec4a1 | C-type lectin domain family 4, member a1 | 1.71 | 0.22 | 1.63E-11 |
| ENSRNOG00000054964 | Aoah | Acyloxyacyl hydrolase | 2.60 | 0.34 | 2.36E-11 |
| ENSRNOG00000054860 | Clec12a | C-type lectin domain family 12, Member A | 1.79 | 0.24 | 2.36E-11 |
| ENSRNOG00000013564 | Dok3 | Docking protein 3 | 1.93 | 0.26 | 2.81E-11 |
| ENSRNOG00000021161 | Fermt3 | Fermentin family (Drosophila) Homolog 3 | 1.18 | 0.16 | 3.07E-11 |
| ENSRNOG00000007350 | Rac2 | Ras-related C3 Botulinum toxin Substrate 2 | 1.35 | 0.18 | 3.07E-11 |
FDR: false discovery rate.
Table 2.
Top 25 genes downregulated in CCI rats after SCS by FDR.
| Ensembl ID | Gene symbol | Full gene name | Log2 fold change | Standard error | FDR |
|---|---|---|---|---|---|
| ENSRNOG00000007112 | Pcsk1n | Proprotein convertase subtilisin/kexin type 1 inhibitor | −3.05 | 0.36 | 2.93E-14 |
| ENSRNOG00000017932 | St3gal2 | ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | −0.38 | 0.07 | 1.62E-05 |
| ENSRNOG00000007573 | Hoxb9 | Homeobox B9s | −0.69 | 0.14 | 5.53E-05 |
| ENSRNOG00000016897 | Rlbp1 | Retinaldehyde-binding protein 1 | −0.57 | 0.12 | 1.63E-04 |
| ENSRNOG00000000501 | Zfp523 | Zinc finger protein 76 | −0.40 | 0.09 | 1.89E-04 |
| ENSRNOG00000043390 | Samd12 | Sterile alpha motif domain contain 12 | −0.48 | 0.10 | 1.96E-04 |
| ENSRNOG00000006649 | Thrb | Thyroid hormone receptor beta | −0.43 | 0.10 | 3.21E-04 |
| ENSRNOG00000002339 | Mark1 | Microtubule affinity regulating kinase 1 | −0.53 | 0.12 | 3.60E-04 |
| ENSRNOG00000004155 | Samd14 | Sterile alpha motif domain containing 14 | −0.66 | 0.15 | 3.89E-04 |
| ENSRNOG00000058476 | Mast2 | Microtubule associated serine/threonine kinase 2 | −0.53 | 0.12 | 4.24E-04 |
| ENSRNOG00000037793 | Cdk5r2 | Cyclin-dependent kinase 5 activator 2 | −0.48 | 0.11 | 5.32E-04 |
| ENSRNOG00000019958 | Tmem151b | Transmembrane protein 151B | −0.91 | 0.21 | 5.32E-04 |
| ENSRNOG00000009772 | Kirrel3 | Kin of irregular chiasm-like protein 3 | −0.49 | 0.11 | 5.99E-04 |
| ENSRNOG00000048980 | Gng2 | G protein subunit gamma 2 | −0.25 | 0.06 | 6.09E-04 |
| ENSRNOG00000018526 | Dlg4 | Discs large MAGUK scaffold protein 4 | −0.50 | 0.12 | 7.77E-04 |
| ENSRNOG00000023538 | Aldh5a1 | Aldehyde dehydrogenase 5 family member A1 | −0.49 | 0.11 | 7.92E-04 |
| ENSRNOG00000013408 | Npas2 | Neuronal PAS domain protein 2 | −0.65 | 0.15 | 8.05E-04 |
| ENSRNOG00000016653 | Ngef | Neuronal guanine nucleotide exchange factor | −0.46 | 0.11 | 8.31E-04 |
| ENSRNOG00000019404 | Hhatl | Hedgehog acyltransferase-like | −0.53 | 0.12 | 8.52E-04 |
| ENSRNOG00000010938 | Slc7a10 | Solute carrier family 7 member 10 | −0.51 | 0.12 | 9.41E-04 |
| ENSRNOG00000008082 | Rgs6 | Regulator of G protein signaling 6 | −0.55 | 0.13 | 1.07E-03 |
| ENSRNOG00000008145 | Traf3 | TNF receptor-associated factor 3 | −0.69 | 0.16 | 1.07E-03 |
FDR: false discovery rate.
GO analysis of the upregulated genes showed significant enrichment among a variety immune-related biological process (Figure 3(a) and (b)). GO analysis of the downregulated transcripts show significant enrichment among genes involved in synaptic transmission, synaptic organization, and neuron outgrowth (Figure 4(a) and (b)). Molecular functional enrichment analysis identified downregulated differentially expressed genes are involved in protein serine/threonine kinase activity and scaffold protein binding (FDR < 0.005).
Figure 3.
GO biological processes enriched from differentially expressed genes that are upregulated after SCS. (a) The top 25 GO biological processes associated with genes upregulated in CCI+SCS versus CCI only (FDR < 0.05) as ranked by p-value. (b) Heatmap of selected up-regulated genes associated with multiple overrepresented GO biological processes in (a). Data shown are relative expression (i.e., log2FC), mean normalized transcript abundance (i.e., log10(count+1)), and statistical significance level (i.e., log10FDR).
Figure 4.
GO biological processes enriched from differentially expressed genes that are downregulated after SCS. (a) The top 25 GO biological processes associated with genes downregulated in CCI+SCS versus CCI only (FDR < 0.05) as ranked by p value. (b) Heatmap of selected downregulated genes associated with the first five overrepresented GO biological processes in (a). Data shown are relative expression (i.e., log2FC), mean normalized transcript abundance (i.e., log10(count+1)), and statistical significance level (i.e., log10FDR).
Sex differences associated with differentially regulated genes after SCS of CCI rats
Next, we explored sex-specific differential gene expression in the spinal cord associated with repetitive SCS. While both males and females showed a significant increase in PWTs during SCS, the PWTs of the female rats were notably lower than the PWTs of the male rats (Figure 1(b)). To identify sex-specific changes in gene expression associated with SCS treatment, we compared differentially expressed genes between males and females. Following SCS, male CCI+SCS rats differentially expressed 149 genes (Supplemental Figure 2(a)). Of these 149 differentially expressed genes, 28 (18.8%) were downregulated after SCS and 121 (81.2%) were upregulated. GO analysis of the upregulated genes show enrichment in immune and inflammatory pathways (Supplemental Figure 2(b)). In order to perform GO analysis using downregulated genes, we lowered the statistical significance and used the 380 genes which were downregulated after SCS at an unadjusted p <0.05. GO analysis using this subset of genes showed enrichment in genes involved in synaptic signaling (Supplemental Figure 2(b)).
Female CCI + SCS rats differentially expressed 858 genes following SCS at an FDR < 0.05 (Supplemental Figure 2(c)). Of these 858 differentially expressed genes, 192 (22.5%) were downregulated after SCS and 666 (77.5%) were upregulated. Similar to males, GO analysis revealed that the upregulated genes were enriched in immune-related processes and downregulated genes were enriched in synaptic signaling-related processes (Supplemental Figure 2(d)). Hierarchical clustering identified segregation of samples by group and then by sex (Supplemental Figure 2(e)). Two genes (i.e., Eif2s3 and Cpne4) showed significantly increased expression in females versus males at an FDR < 0.05. Expressions of 44 genes were significantly increased in males compared with females (Supplemental Figure 2(f) and Supplemental Table 1).
Discussion
In this study, we identified the effects of multiple sessions of conventional SCS on gene expression in the lumbar spinal cord ipsilateral to the nerve lesion. We administered SCS to rats during the maintenance phase of neuropathic pain using a custom-made quadripolar electrode, which enabled us to use similar parameters as those used clinically to treat chronic pain.12,18,19 We chose to use rats that received CCI only as our comparison group in an effort to capture all changes that occur in the spinal cord as a result of surgical implantation of the stimulation electrode and subsequent SCS. Consistent with previous findings,12,18,19 conventional SCS at the T13-L1 spinal reduced the mechanical hypersensitivity that developed in the ipsilateral hindpaw of CCI rats. The peak inhibitory effect of SCS often occurred 60 to 90 min after starting the SCS. The pain inhibitory effects on each treatment day varied between individual animals and were similar to those observed in other neuropathic pain models.12,18,19 Pain inhibition by SCS was positively correlated with the MoT. However, the correlation coefficient measures only the degree of linear association between two variables and not causal relationships. Although we included both males and females in our study, we chose to report our analyses after pooling data obtained from both sexes. Only a small number of genes were differentially expressed between sexes, and male and female rats showed similar GO biological processes associated with SCS (Supplemental Figure 2). Future investigation should include a larger sample size to determine if meaningful differences exist in pain inhibition and gene expression between males and females in response to SCS.31
Upregulation of immune-related genes
Following nerve injury, a robust immune response is generated as a result of injury and increased neuronal excitability.32 Repetitive SCS at T13-L1 was associated with further increases in the expression of immune-related genes in the lumbar spinal cord of CCI rats (Figure 3). These findings are consistent with the only other transcriptome-wide study which reported upregulation of immune-related genes also after SCS.15 Similarly, SCS was associated with altered expression of proteins involved in a variety of immune-related processes (e.g., wound healing and complement) in cerebrospinal fluid of patients with neuropathic pain.33 Immune response and gliosis in the spinal cord after nerve injury are thought to contribute to the maintenance of pathological pain and hyperexcitability of dorsal horn neurons.34,35 Nevertheless, immune responses can also serve to protect the injured area from further insult, contain pathogens, eliminate damaged cells, and initiate repair mechanisms.36,37 The physiological implications of increased expression of immune-related genes in the spinal cord after SCS of nerve-injured rats warrant further investigation.
Central sensitization underlying chronic pain is associated with persistent N-methyl-D-aspartate receptor (NMDAR) sensitization to maintain neuronal hyperexcitability as well as the upregulation of toll-like receptors (TLRs).38,39 To our surprise, in rats with existing CCI to the sciatic nerve, SCS treatment was associated with upregulated TLRs and markers for activated glia. TLR4 is expressed on the cell surface of neurons and immunocompetent cells and can induce a sterile inflammatory response through transcriptional activation of genes that encode key inflammatory mediators (i.e., CCL2/MCP1) as a result of tissue injury/stress.40 We also found significant upregulation of genes encoding markers for astrocytes (i.e., Gfap and Ccl2) and activated microglia (i.e., Cd68 and Itgam) in the spinal cord following SCS treatments. Activated microglia synthesize and release pro-inflammatory mediators to increase neuronal hyperexcitability following nerve injury.35 Previous studies have reported conflicting evidence regarding the activation of glia in the spinal cord after SCS. Sato et al.16 reported decreased glia activation in the spinal cord following 6 h of SCS for four consecutive days as defined by Itgam and Cd68 protein expression. Recently, increased Tlr2 and Cd68 gene expression provided evidence of SCS-induced microglia activation.15 Our findings are consistent with the latter study. We found upregulation of these genes as well as Gfap which suggests that SCS is associated with increased activation of immune cells in the spinal cord. Whether upregulation of TLRs, glial activation, and immune-related genes may compromise pain inhibition by SCS warrants further investigation.
Downregulation of γ-aminobutyric acid transporters
Despite increased immune responses and glia activation in the spinal cord which may facilitate spinal nociceptive transmission, our animal behavior study found reduction of pain hypersensitivity during each SCS treatment. Thus, the net inhibition of mechanical hypersensitivity by SCS may result from mechanisms other than immune suppression or glial inhibition. The neurochemical mechanisms underlying pain inhibition by conventional SCS include the release of γ-aminobutyric acid (GABA), serotonin, endocannabinoids, acetylcholine, and adenosine into spinal cord.41–44 Uptake of GABA from the presynaptic terminals is required to terminate inhibitory neurotransmission by GABA.45 GAT3 is the GABA transporter expressed on glia that is responsible for the uptake of GABA from the presynaptic terminal and is encoded by Slc6a11. Intriguingly, we found that SCS was associated with decreased expression of Slc6a11. Thus, a decrease of Slc6a11 expression by SCS may be a previously uncharacterized mechanism that promotes pain inhibition through increased availability of GABA within the synaptic cleft.
Downregulation of scaffold genes in the postsynaptic membrane
Changes in synaptic strength between peripheral afferents and second-order neurons underlie central sensitization after nerve injury. This synaptic plasticity is primarily due to activation of NMDAR and localization of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) to the postsynaptic membrane,46 which mediate excitatory synaptic transmission of action potentials from peripheral sensory neurons.47,48 Importantly, we found that several genes involved in neurotransmission and synaptic strength were downregulated in CCI rats following SCS treatment. In particular, among those downregulated were genes encoding scaffold proteins located on the postsynaptic membrane.
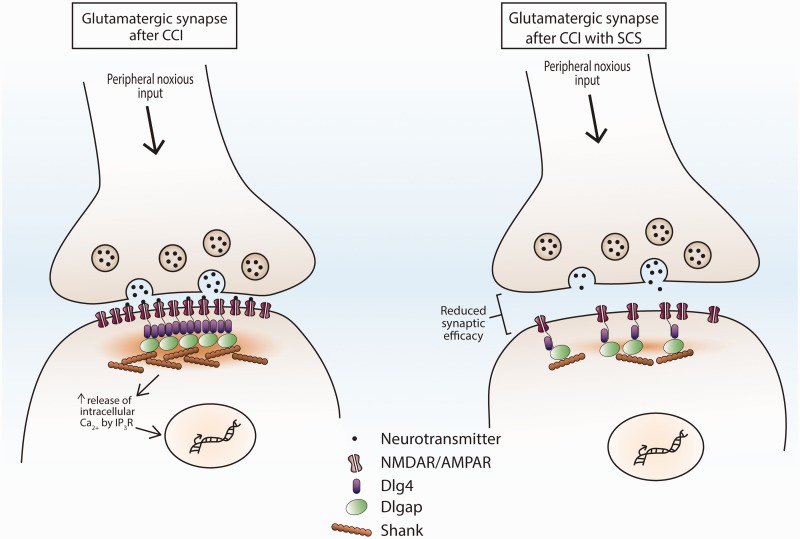
The postsynaptic membrane of glutamatergic synapses contains a dense network of proteins known as the postsynaptic density (PSD) that stabilizes glutamatergic receptors localization,49 prevents lateral diffusion of the receptors in the postsynaptic membrane,49 and physically links the cytoplasmic domains of receptors to intracellular signaling cascades.50 Therefore, scaffold proteins within the PSD directly affect synaptic plasticity. Scaffold proteins are generally organized into three layers with each containing a specific family of proteins (e.g., Dlg4, Dlgap1-4, and Shank1-3; Figure 5). First, Dlg4 encodes the Dlg4 protein which binds to the intracellular tails of NMDARs,51 promotes aggregation of NMDARs and AMPARs in the PSD,52 and stabilizes AMPAR interactions with its auxiliary proteins.53 Intrathecal knockdown of Dlg4 expression reduced mechanical and thermal hyperalgesia in rats following L5 spinal nerve ligation.54,55 In addition, Dlg4-null mice showed decreased glutamate AMPA receptor-mediated synaptic transmission while NMDA receptors were unaffected.56 Second, Dlgap1-4 encodes four Dlgap proteins which contain domains (i.e., 14 amino acid repeat domains, DLC, GH1) that interact directly with Dlg4 and Shank proteins.50,57 Altered expression and function of Dlgap proteins is associated with several neurological disorders (e.g., schizophrenia, obsessive compulsive disorder, and autism).50 Altered Dlgap1-4 gene expression after SCS has not been reported. The third layer contains the Shank family of proteins which are encoded by Shank1-3. Shank proteins are large scaffold proteins that contain many protein binding domains which enables them to connect to other Shank proteins, glutamate receptors, signaling proteins, and cytoskeletal proteins.58 Increased Shank1 protein expression was found after CCI in the ipsilateral dorsal horn.59 On the other hand, inhibition or siRNA knockdown of Shank1 in rats after CCI increased mechanical thresholds to pre-injury levels.60 Our findings are consistent with these studies and suggest that repeated SCS treatment is associated with decreased expression of scaffold proteins that are essential for the stability of NMDA and AMPA receptor aggregation and signaling on the postsynaptic membrane (i.e., Dlg4, Dlgap1, Dlgap3, Shank1, Shank3, Grip2; Figure 5). NMDA and AMPA signaling underlies the increased synaptic efficacy indicative of central sensitization. Therefore, destabilization of the PSD may represent a novel mechanism for SCS to result in inhibition of spinal synaptic transmission and neuropathic pain.
Figure 5.
Illustration of a glutamatergic synapse between the central terminal of primary sensory neuron and a post-synaptic dorsal horn neuron with and without SCS. Left: Nerve injury increases excitatory synaptic transmission. The organization of the PSD by scaffold proteins facilitates this synaptic plasticity which involves AMPAR localization to the post-synaptic membrane, stabilization of membrane receptors, and physical linkage of the cytoplasmic domains of the receptor to intracellular signaling cascades by Dlg4, Dlgap, and Shank proteins. Activation of these intracellular signaling cascades increases intracellular calcium levels and promotes gene transcription. Right: RNA-seq data show downregulation of the scaffold proteins that comprise the PSD (e.g., Dlg4, Dlgap1, Dlgap3, Shank1, Shank3, Grip2), which suggest that repeated SCS treatment is associated with destabilization of the PSD in the spinal cord. Decreased expression of these scaffold genes may reduce NMDAR and AMPAR aggregation at the postsynaptic membrane and hence attenuate excitatory synaptic transmission.
In summary, we showed that gene expression changes in the spinal cord of nerve-injured rats after multiple SCS sessions, and we identify genes and gene networks differentially impacted by conventional SCS under neuropathic pain conditions. Importantly, several key genes that encode scaffold proteins in the PSD are downregulated following SCS which may destabilize the PSD and decrease efficacy of synaptic signaling. The mechanisms leading to changes in gene expression in distal spinal segments after SCS are unknown. During SCS, antidromic action potentials that travel in the dorsal column fibers can reach caudal spinal segments via collateral branches and induce neurochemical changes. SCS may also activate nearby spinal tracts that affect neurons and glial cells in distal spinal segments. Our current findings provide critical insights into transcriptional pathways induced in the spinal cord by repetitive SCS after nerve injury. Future attempts to increase the therapeutic effects of SCS may involve the combination of conventional SCS with other treatments aimed at specific transcriptional and epigenetic targets.
Supplemental Material
Supplemental Material for RNA-seq of spinal cord from nerve-injured rats after spinal cord stimulation by Kimberly E Stephens, Zhiyong Chen, Eellan Sivanesan, Srinivasa N Raja, Bengt Linderoth, Sean D Taverna and Yun Guan in Molecular Pain
Acknowledgments
Electrodes for the spinal cord stimulation were generously provided by Medtronic, Inc. (Minneapolis, MN, USA). We thank Rakel Tryggvadóttir and Colin Callahan for their technical assistance.
Author Contributions
YG designed the experiments; KES, ZC, and ES performed the experiments; KES, SDT, and YG were involved with data analysis; KES, ES, SNR, HL, SDT, BL, and YG were involved in discussion and interpretation of results; KES, SDT, and YG wrote the manuscript. All authors read and approved the final manuscript.
Declaration of Conflicting Interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Dr. Linderoth is a consultant for Medtronic Inc., Minneapolis, Minnesota; St. Jude Medical, Austin, Texas; Boston Scientific, Marlborough, Massachusetts; and Elekta AB, Sweden.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was conducted at the Johns Hopkins University and was supported by grants from National Institutes of Health (Bethesda, Maryland, USA) F32NR015728 (KES), R01NS070814 (YG), R21NS099879 (YG), GM118760 (SDT), and a seed grant from the Johns Hopkins Blaustein Pain Research Fund (SDT). This work was facilitated by the Pain Research Core and funded by the Blaustein Pain Fund and the Neurosurgery Pain Research Institute at the Johns Hopkins University.
Supplemental material
Supplemental material is available for this article online.
References
- 1.Kumar K, Taylor RS, Jacques L, Eldabe S, Meglio M, Molet J, Thomson S, O'Callaghan J, Eisenberg E, Milbouw G, Buchser E, Fortini G, Richardson J, North RB. Spinal cord stimulation versus conventional medical management for neuropathic pain: a multicentre randomised controlled trial in patients with failed back surgery syndrome. Pain 2007; 132: 179–188. [DOI] [PubMed] [Google Scholar]
- 2.Falowski S, Sharan A. A review on spinal cord stimulation. J Neurosurg Sci 2012; 56: 287–298. [PubMed] [Google Scholar]
- 3.Deer TR, Mekhail N, Provenzano D, Pope J, Krames E, Leong M, Levy RM, Abejon D, Buchser E, Burton A, Buvanendran A, Candido K, Caraway D, Cousins M, DeJongste M, Diwan S, Eldabe S, Gatzinsky K, Foreman RD, Hayek S, Kim P, Kinfe T, Kloth D, Kumar K, Rizvi S, Lad SP, Liem L, Linderoth B, Mackey S, McDowell G, McRoberts P, Poree L, Prager J, Raso L, Rauck R, Russo M, Simpson B, Slavin K, Staats P, Stanton-Hicks M, Verrills P, Wellington J, Williams K, North R. and Neuromodulation Appropriateness Consensus C. The appropriate use of neurostimulation of the spinal cord and peripheral nervous system for the treatment of chronic pain and ischemic diseases: the Neuromodulation Appropriateness Consensus Committee. Neuromodulation 2014; 17: 515–550. [DOI] [PubMed] [Google Scholar]
- 4.Krames E, Poree L, Deer T, Levy R. Implementing the SAFE principles for the development of pain medicine therapeutic algorithms that include neuromodulation techniques. Neuromodulation 2009; 12: 104–113. [DOI] [PubMed] [Google Scholar]
- 5.Krames ES, Monis S, Poree L, Deer T, Levy R. Using the SAFE principles when evaluating electrical stimulation therapies for the pain of failed back surgery syndrome. Neuromodulation 2011; 14: 299–311. [DOI] [PubMed] [Google Scholar]
- 6.Melzack R, Wall PD. Pain mechanisms: a new theory. Science 1965; 150: 971–979. [DOI] [PubMed] [Google Scholar]
- 7.Linderoth B, Foreman RD. Conventional and novel spinal stimulation algorithms: hypothetical mechanisms of action and comments on outcomes. Neuromodulation 2017; 20: 525–533. [DOI] [PubMed] [Google Scholar]
- 8.Sdrulla AD, Guan Y, Raja SN. Spinal cord stimulation: clinical efficacy and potential mechanisms. Pain Pract 2018; 18: 1048–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foreman RD, Linderoth B. Neural mechanisms of spinal cord stimulation. Int Rev Neurobiol 2012; 107: 87–119. [DOI] [PubMed] [Google Scholar]
- 10.Linderoth B, Meyerson BA. Spinal cord stimulation: exploration of the physiological basis of a widely used therapy. Anesthesiology 2010; 113: 1265–1267. [DOI] [PubMed] [Google Scholar]
- 11.Guan Y, Wacnik PW, Yang F, Carteret AF, Chung CY, Meyer RA, Raja SN. Spinal cord stimulation-induced analgesia: electrical stimulation of dorsal column and dorsal roots attenuates dorsal horn neuronal excitability in neuropathic rats. Anesthesiology 2010; 113: 1392–1405. [DOI] [PubMed] [Google Scholar]
- 12.Shechter R, Yang F, Xu Q, Cheong YK, He SQ, Sdrulla A, Carteret AF, Wacnik PW, Dong X, Meyer RA, Raja SN, Guan Y. Conventional and kilohertz-frequency spinal cord stimulation produces intensity- and frequency-dependent inhibition of mechanical hypersensitivity in a rat model of neuropathic pain. Anesthesiology 2013; 119: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yakhnitsa V, Linderoth B, Meyerson BA. Spinal cord stimulation attenuates dorsal horn neuronal hyperexcitability in a rat model of mononeuropathy. Pain 1999; 79: 223–233. [DOI] [PubMed] [Google Scholar]
- 14.Yang F, Xu Q, Cheong YK, Shechter R, Sdrulla A, He SQ, Tiwari V, Dong X, Wacnik PW, Meyer R, Raja SN, Guan Y. Comparison of intensity-dependent inhibition of spinal wide-dynamic range neurons by dorsal column and peripheral nerve stimulation in a rat model of neuropathic pain. Eur J Pain 2014; 18: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallejo R, Tilley DM, Cedeno DL, Kelley CA, DeMaegd M, Benyamin R. Genomics of the effect of spinal cord stimulation on an animal model of neuropathic pain. Neuromodulation 2016; 19: 576–586. [DOI] [PubMed] [Google Scholar]
- 16.Sato KL, Johanek LM, Sanada LS, Sluka KA. Spinal cord stimulation reduces mechanical hyperalgesia and glial cell activation in animals with neuropathic pain. Anesth Analg 2014; 118: 464–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tilley DM, Cedeno DL, Kelley CA, Benyamin R, Vallejo R. Spinal cord stimulation modulates gene expression in the spinal cord of an animal model of peripheral nerve injury. Reg Anesth Pain Med 2016; 41: 750–756. [DOI] [PubMed] [Google Scholar]
- 18.Yang F, Xu Q, Shu B, Tiwari V, He SQ, Vera-Portocarrero LP, Dong X, Linderoth B, Raja SN, Wang Y, Guan Y. Activation of cannabinoid CB1 receptor contributes to suppression of spinal nociceptive transmission and inhibition of mechanical hypersensitivity by Aβ-fiber stimulation. Pain 2016; 157: 2582–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda Y, Wacnik PW, Sluka KA. Low frequencies, but not high frequencies of bi-polar spinal cord stimulation reduce cutaneous and muscle hyperalgesia induced by nerve injury. Pain 2008; 138: 143–152. [DOI] [PubMed] [Google Scholar]
- 20.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods 1994; 53: 55–63. [DOI] [PubMed] [Google Scholar]
- 21.Dixon WJ. Efficient analysis of experimental observations. Annu Rev Pharmacol Toxicol 1980; 20: 441–462. [DOI] [PubMed] [Google Scholar]
- 22.Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain 1988; 33: 87–107. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita T, Sakuma Y, Kato Y, Kotani J. Effect of different suture materials on the chronic constriction injury model. J Osaka Dental Universtiy 2004; 38: 89–94. [Google Scholar]
- 24.van der Wal S, Cornelissen L, Behet M, Vaneker M, Steegers M, Vissers K. Behavior of neuropathic pain in mice following chronic constriction injury comparing silk and catgut ligatures. Springerplus 2015; 4: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009; 37: W305–W311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, Catterall WA, Davenport AP, Delagrange P, Dollery CT, Foord SM, Gutman GA, Laudet V, Neubig RR, Ohlstein EH, Olsen RW, Peters J, Pin JP, Ruffolo RR, Searls DB, Wright MW, Spedding M. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res 2009; 37: D680–D685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens KE, Zhou W, Ji Z, He SQ, Ji H, Guan Y, Taverna SD. Sex differences in gene regulation in the dorsal root ganglion after nerve injury. bioRxiv Epub ahead of print 20 June C2017. DOI: 10.1101/152652 [DOI] [PMC free article] [PubMed]
- 32.Gattlen C, Clarke CB, Piller N, Kirschmann G, Pertin M, Decosterd I, Gosselin RD, Suter MR. Spinal cord T-cell infiltration in the rat spared nerve injury model: a time course study. Int J Mol Sci 2016; 17: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lind AL, Emami Khoonsari P, Sjodin M, Katila L, Wetterhall M, Gordh T, Kultima K. Spinal cord stimulation alters protein levels in the cerebrospinal fluid of neuropathic pain patients: a proteomic mass spectrometric analysis. Neuromodulation 2016; 19: 549–562. [DOI] [PubMed] [Google Scholar]
- 34.Ben Achour S, Pascual O. Glia: the many ways to modulate synaptic plasticity. Neurochem Int 2010; 57: 440–445. [DOI] [PubMed] [Google Scholar]
- 35.Tsuda M. Microglia in the spinal cord and neuropathic pain. J Diabetes Investig 2016; 7: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin X, Yamashita T. Microglia in central nervous system repair after injury. J Biochem 2016; 159: 491–496. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein EZ, Church JS, Hesp ZC, Popovich PG, McTigue DM. A silver lining of neuroinflammation: beneficial effects on myelination. Exp Neurol 2016; 283: 550–559. [DOI] [PubMed] [Google Scholar]
- 38.Ma Q, Woolf C. The NMDA receptor, pain and central sensitization In: Sirinathsinghji DJS, Hill RG. (eds) NMDA antagonists as potential analgesic drugs. Basel: Birkhäuser, 2002, pp. 83–103. [Google Scholar]
- 39.Schrepf A, Bradley CS, O'Donnell M, Luo Y, Harte SE, Kreder K, Lutgendorf S. and Multidisciplinary Approach to the Study of Chronic Pelvic Pain Research N. Toll-like receptor 4 and comorbid pain in interstitial cystitis/bladder pain syndrome: a multidisciplinary approach to the study of chronic pelvic pain research network study. Brain Behav Immun 2015; 49: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lacagnina MJ, Watkins LR, Grace PM. Toll-like receptors and their role in persistent pain. Pharmacol Ther 2018; 184: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan Y. Spinal cord stimulation: neurophysiological and neurochemical mechanisms of action. Curr Pain Headache Rep 2012; 16: 217–225. [DOI] [PubMed] [Google Scholar]
- 42.Schechtmann G, Song Z, Ultenius C, Meyerson BA, Linderoth B. Cholinergic mechanisms involved in the pain relieving effect of spinal cord stimulation in a model of neuropathy. Pain 2008; 139: 136–145. [DOI] [PubMed] [Google Scholar]
- 43.Song Z, Meyerson BA, Linderoth B. Spinal 5-HT receptors that contribute to the pain-relieving effects of spinal cord stimulation in a rat model of neuropathy. Pain 2011; 152: 1666–1673. [DOI] [PubMed] [Google Scholar]
- 44.Cui JG, Linderoth B, Meyerson BA. Effects of spinal cord stimulation on touch-evoked allodynia involve GABAergic mechanisms. An experimental study in the mononeuropathic rat. Pain 1996; 66: 287–295. [DOI] [PubMed] [Google Scholar]
- 45.Roth FC, Draguhn A. GABA metabolism and transport: effects on synaptic efficacy. Neural Plast 2012; 2012: 805830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrenko AB, Yamakura T, Baba H, Shimoji K. The role of N-methyl-D-aspartate (NMDA) receptors in pain: a review. Anesth Analg 2003; 97: 1108–1116. [DOI] [PubMed] [Google Scholar]
- 47.Racca C, Stephenson FA, Streit P, Roberts JD, Somogyi P. NMDA receptor content of synapses in stratum radiatum of the hippocampal CA1 area. J Neurosci 2000; 20: 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron 2003; 40: 361–379. [DOI] [PubMed] [Google Scholar]
- 49.Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 2007; 53: 719–734. [DOI] [PubMed] [Google Scholar]
- 50.Rasmussen AH, Rasmussen HB, Silahtaroglu A. The DLGAP family: neuronal expression, function and role in brain disorders. Mol Brain 2017; 10: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 1995; 269: 1737–1740. [DOI] [PubMed] [Google Scholar]
- 52.Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 2000; 408: 936–943. [DOI] [PubMed] [Google Scholar]
- 53.Nicoll RA, Tomita S, Bredt DS. Auxiliary subunits assist AMPA-type glutamate receptors. Science 2006; 311: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 54.Tao F, Tao YX, Gonzalez JA, Fang M, Mao P, Johns RA. Knockdown of PSD-95/SAP90 delays the development of neuropathic pain in rats. Neuroreport 2001; 12: 3251–3255. [DOI] [PubMed] [Google Scholar]
- 55.Tao F, Tao YX, Mao P, Johns RA. Role of postsynaptic density protein-95 in the maintenance of peripheral nerve injury-induced neuropathic pain in rats. Neuroscience 2003; 117: 731–739. [DOI] [PubMed] [Google Scholar]
- 56.Beique JC, Lin DT, Kang MG, Aizawa H, Takamiya K, Huganir RL. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci USA 2006; 103: 19535–19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naisbitt S, Kim E, Weinberg RJ, Rao A, Yang FC, Craig AM, Sheng M. Characterization of guanylate kinase-associated protein, a postsynaptic density protein at excitatory synapses that interacts directly with postsynaptic density-95/synapse-associated protein 90. J Neurosci 1997; 17: 5687–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci 2000; 113: 1851–1856. [DOI] [PubMed] [Google Scholar]
- 59.Miletic G, Miyabe T, Gebhardt KJ, Miletic V. Increased levels of Homer1b/c and Shank1a in the post-synaptic density of spinal dorsal horn neurons are associated with neuropathic pain in rats. Neurosci Lett 2005; 386: 189–193. [DOI] [PubMed] [Google Scholar]
- 60.Miletic G, Dumitrascu CI, Honstad CE, Micic D, Miletic V. Loose ligation of the rat sciatic nerve elicits early accumulation of Shank1 protein in the post-synaptic density of spinal dorsal horn neurons. Pain 2010; 149: 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for RNA-seq of spinal cord from nerve-injured rats after spinal cord stimulation by Kimberly E Stephens, Zhiyong Chen, Eellan Sivanesan, Srinivasa N Raja, Bengt Linderoth, Sean D Taverna and Yun Guan in Molecular Pain