Abstract
Background
The Pacific abalone, Haliotis discus hannai, is the most important cultivated abalone in China. Improving abalone muscle growth and increasing the rate of growth are important genetic improvement programs in this industry. MicroRNAs are important small noncoding RNA molecules that regulate post-transcription gene expression. However, no miRNAs have been reported to regulate muscle growth in H. discus hannai.
Results
we profiled six small RNA libraries for three large abalone individuals (L_HD group) and three small individuals (S_HD group) using RNA sequencing technology. A total of 205 miRNAs, including 200 novel and 5 known miRNAs, were identified. In the L_HD group, 3 miRNAs were up-regulated and 7 were down-regulated compared to the S_HD specimens. Bioinformatics analysis of miRNA target genes revealed that miRNAs participated in the regulation of cellular metabolic processes, the regulation of biological processes, the Wnt signaling pathway, ECM-receptor interaction, and the MAPK signaling pathway, which are associated with regulating growth. Bone morphogenetic protein 7 (BMP7) was verified as a target gene of hdh-miR-1984 by a luciferase reporter assay and we examined the expression pattern in different developmental stages.
Conclusion
This is the first study to demonstrate that miRNAs are related to the muscle growth of H. discus hannai. This information could be used to study the mechanisms of abalone muscle growth. These DE-miRNAs may be useful as molecular markers for functional genomics and breeding research in abalone and closely related species.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5347-9) contains supplementary material, which is available to authorized users.
Keywords: miRNA, Pacific abalone, Haliotis discus hannai, Muscle growth, Hdh-miR-1984
Background
The Pacific abalone, Haliotis discus hannai, is the most important cultivated abalone in China [1, 2]. The foot muscle is the main edible portion of the abalone. Improving abalone muscle growth and increasing the rate of growth are important genetic improvement programs [3]. A better understanding of the molecular mechanisms of muscle growth can provide useful knowledge for programs that aim to improve abalone musculature.
MicroRNAs (miRNAs) are small noncoding RNA molecules (18–22 nt) which regulate post-transcription gene expression by specifically mapping target mRNA 3′ untranslated regions (UTRs) [4, 5]. MicroRNAs have crucial roles in various biological processes, including development [6], sex determination and differentiation [7], apoptosis [8], and immune response [9]. MicroRNAs also play a vital role in regulating muscle growth [10, 11]. For example, miR-133 can regulate skeletal muscle proliferation and differentiation by repressing the serum response factor (SRF) and insulin-like growth factor 1(IGF-1) [12]. Seok et al. reported that miR-155 can repress skeletal muscle differentiation by inhibiting the expression of myocyte enhancer factor 2A (MEF2A) protein [13]. The miR-214 target suppressor of fused (Sufu) regulates the slow muscle phenotype in zebrafish [14]. Pm-miR-133 regulates the expression of RhoA in the pearl oyster Pinctada martensii [15]. Based on these studies, the identification of miRNAs in adductor muscle could provide new insight into the regulatory mechanism of abalone muscle growth.
In this study, we investigated the miRNA profiles of H. discus hannai muscle using an Illumina HiSeq 2500 platform. Differentially expressed miRNAs (DE-miRNAs) related to muscle growth were identified, and the target genes were forecast. The possible roles of the DE-miRNAs and the target genes are discussed. The dynamic expression pattern of hdh-miR-1984 and the predicted target gene bone morphogenetic protein 7 (BMP7) in different developmental stages were examined by quantitative real-time polymerase chain reactions (qRT-PCR). We verified that BMP7 is a target gene of hdh-miR-1984 using the luciferase activities of report vectors method. These data provide new information on the molecular mechanisms of abalone muscle growth.
Materials and methods
Experimental samples
A breeding population of H. discus hannai produced pedigreed offspring. All of the mating was conducted at Fuda Aquaculture in Jinjiang, Fujian Province, China. Adductor muscle tissue from different growth stages (1, 4, 7, 10, 12, and 24 months) of H. discus hannai were acclimated, immediately snap-frozen in liquid nitrogen, and stored at − 80 °C.
Small RNA sequencing
Adductor muscle tissues of three smaller individual H. discus hannai abalones (“S_HD” group) and three larger individuals (“L_HD” group) were used for the sRNA library. The individuals were collected when they were about 2 years old. The total RNA from the abalone samples was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Approximately 3 μg total RNA per sample was used for the small RNA library. We performed the single-end sequencing (50 bp) on an Illumina Hiseq2500 platform at Novogene (Tianjing, China) according to the manufacturer’s protocol.
Small RNA analysis and annotation
After sequencing, clean reads were obtained by removing reads containing the poly-N, poly A/T/G/C, adapter-contaminated tags and low-quality reads from the raw data. Q20, Q30, and GC-content of the raw data were calculated. Then, the downstream analyses were conducted by choosing a certain range of length from clean reads [16]. The small RNA tags were mapped to a reference sequence by Bowtie [17] and then the mapped small RNA tags were used to look for known miRNA. The miRBase20.0 (ftp://mirbase.org/pub/mirbase/20/) was used as reference. Modified software mirdeep2 [18] and srna-tools-cli (http://srna-tools.cm p.uea.ac.uk) were used to obtain the potential miRNA and draw the secondary structures. The software miREvo [19] and mirdeep2 [18] were integrated to predict novel miRNA.
Differentially expressed (DE) miRNAs
Differential expression of the two groups was analyzed using the DESeq R package (1.8.3) [20]. The P-values were adjusted using the Benjamini and Hochberg method [21]. A q-value < 0.05 was set as the threshold for considering differential expression as significant.
Functional analysis
The target gene of miRNA was predicted by psRobot_tar in miRanda [22]. GOSeq software [23] and KOBAS 2.0 software [24] were used to annotate the functions of the predicted target genes. The miRNA-mRNA interaction networks of DE-miRNAs and their corresponding target genes were constructed using Cytoscape (http://www.cytoscape.org/).
qRT-PCR
Reverse transcription of miRNA was carried out using a Mir-X miRNA First-Strand Synthesis Kit (Takara, Dalian, China). Several miRNAs were selected for qRT-PCR using gene-specific primers (Additional file 1) and universal reverse primers and U6 was used as the internal control [15]. The qRT-PCR experiments were conducted on a 7500FAST system (ABI, USA). The PCR amplification was performed in 20 μL reaction mixtures containing the following components: 10 μL FastStart Universal SYBR Green Master (ROX), 1 μL forward and reverse primers (10 μM each), 5 μL of cDNA (100-times diluted), 4 μL distilled water. The cycling parameters used were as follows: 95 °C for 10 min, 40 cycles at 95 °C for 10 s, and 59 °C for 30 s. The fluorescent signal intensities were recorded at the end of each cycle. Relative gene expression levels were quantified using the 2-∆∆CT method [25]. Three independent biological replicates were performed. All of the measurements were made in triplicate.
Luciferase reporter assay
The partial 3′UTR of abalone BMP7 mRNA was amplified by PCR and inserted into the psiCHECK™-2 Vector (Promega, Madison, USA). The primers used to construct plasmids for the luciferase reporter assay are shown in Additional file 1. Hdh-mir-1984 mimic/inhibitor were co-transfected with BMP7 3′UTR psiCHECK™-2 vector into human embryonic kidney 293 T cells by Lipofectamine LTX and PLUS Reagent (Invitrogen, Carlsbad, CA, USA), respectively. At 48 h post-transfection, all of the cells were harvested and normalized firefly luciferase activities (firefly luciferase activities/Renilla luciferase activities) were obtained using the Dual-Glo® Luciferase Assay System (Promega, Madison, USA).
Statistical analysis
All of the qRT-PCR data were expressed as mean ± standard deviation (SD). Statistical significance was evaluated using SPSS 19.0 (IBM, USA).
Results
Analysis and identification of small RNA
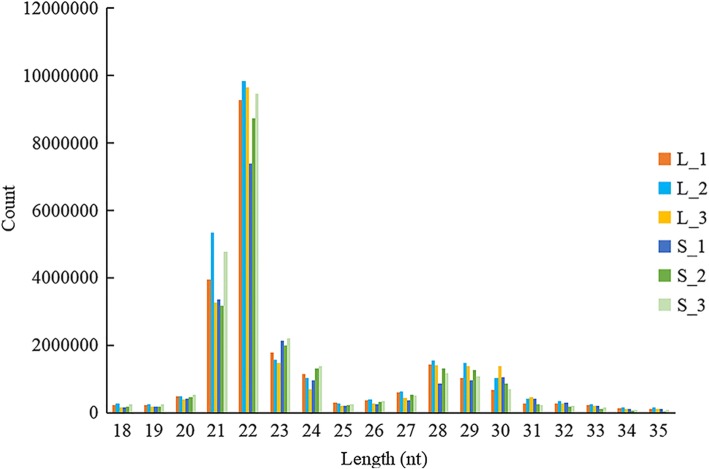
Through high-throughput sequencing, 24.92 million raw reads (from 21.41 M to 28.43 M) were obtained (Additional file 2). A total of 24.25 million clean reads (97.33%) remained after removing the N% > 10 reads, the low-quality reads, the adaptor reads, and the poly A/T/G/C reads. A total of 22.27 million sRNAs were annotated and 91.08% of the total sRNAs were successfully mapped to the H. discus hannai reference genome (Additional file 3). The majority of the reads ranged from 21 to 23 nt in length and the 22 nt small RNA was the most abundant (Fig. 1). These results confirm the reliability of the small RNA sequencing process used in our study.
Fig. 1.
Length comparison of small RNAs from six libraries. Y-axis represents the numbers of small RNA identified in this study. X-axis represents the length of small RNA
To identify the known and novel miRNAs in H. discus hannai, small RNA sequences were mapped to the known mature Haliotis rufescens miRNAs from the miRBase database. After mapping, 5 known miRNAs and 200 novel miRNAs were identified (Additional file 4). To analyze the conservation of H. discus hannai miRNAs, we compared them to all of the species in the miRBase. Only 15 miRNAs were conserved across the different animal species (Additional file 5).
Differential expression of miRNAs among two groups
We identified 10 miRNAs that were significantly differentially expressed (DE-miRNAs) between the L_HD and S_HD specimens (P < 0.05; Table 1). In the L_HD specimens, the novel_353, novel_45, and novel_4 were upregulated compared to the S_HD specimens, while 7 DE-miRNAs were downregulated. Hierarchical clustering analysis (Fig. 2) also suggested that miRNAs were significantly differentially expressed between the two groups.
Table 1.
The information about significantly different expression of miRNAs between the “L_HD” and “S_HD” groups
| sRNA | L_HD_readcount | S_HD_readcount | log2 (Fold Change) | P-value |
|---|---|---|---|---|
| novel_353 | 7.02571687 | 0 | 2.4522 | 5.28E-05 |
| novel_45 | 549.8169218 | 127.5556035 | 1.5764 | 0.0028014 |
| novel_4 | 950,955.0449 | 481,127.1983 | 0.9533 | 4.78E-06 |
| novel_13 | 11,338.62203 | 18,750.9212 | −0.70805 | 0.00016639 |
| novel_18 | 4465.909249 | 8817.185215 | −0.91536 | 0.0026959 |
| novel_9 | 138,675.92 | 318,514.975 | −1.1718 | 1.68E-10 |
| novel_7 | 162,893.4763 | 402,561.4888 | −1.2581 | 3.14E-08 |
| novel_59 | 35.18292072 | 110.4508637 | −1.3563 | 0.0035066 |
| hdh-miR-1984 | 204,523.75 | 636,229.4099 | −1.5182 | 1.59E-06 |
| novel_102 | 1.135605265 | 9.971214003 | −2 | 0.00038411 |
Fig. 2.
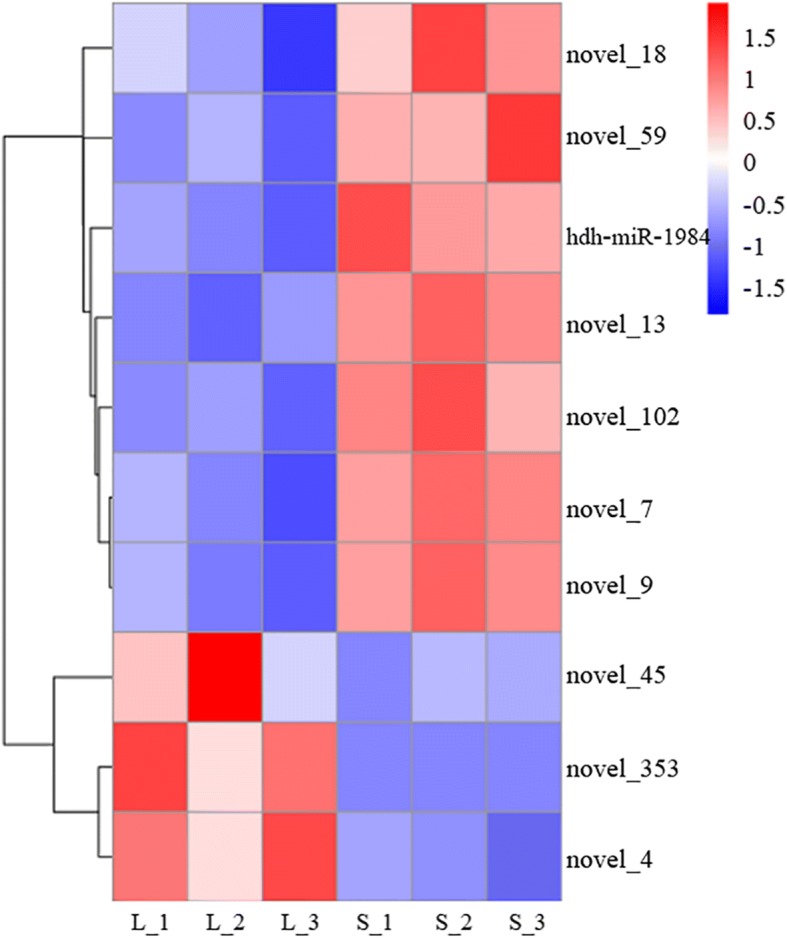
Hierarchical clustering of differentially expressed miRNAs related to the muscle growth. miRNA with a lower expression level is in blue and higher expression in red
Prediction and annotation of differentially expressed miRNAs targets
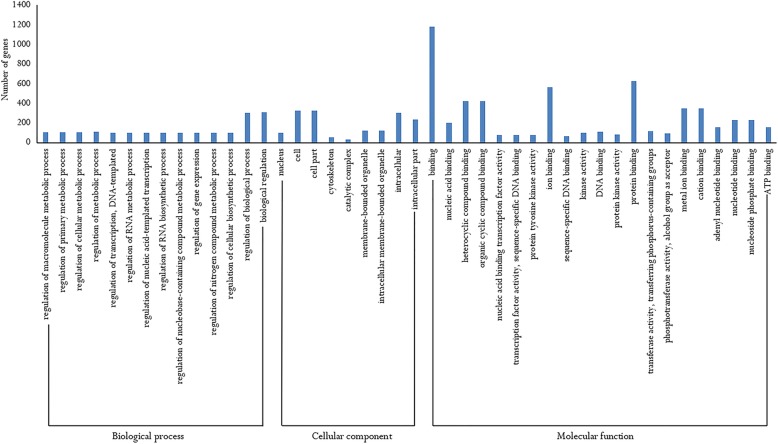
To clarify the molecular functions of the DE-miRNAs in the two groups, we analyzed the target genes of 10 DE-miRNAs. In H. discus hannai, 1091 target genes were predicted for 3 up-regulated miRNAs, and 2039 target genes were predicted for 7 down-regulated miRNAs, respectively. Interestingly, some muscle development-related genes were targeted by DE-miRNAs. For example, BMP7, myosin light chain kinase (MYLK), and myosin heavy chain (MYS), were targeted by the hdh-miR-1984, novel_4, and novel_13, indicating that these miRNAs may regulate muscle growth by targeting these genes. Additional file 6 lists some of the potential miRNA target interactions that may play important roles in the muscle growth of H. discus hannai. Figure 3 shows a complex network consisting of the DE-miRNAs and some of their target genes. The Gene ontology (GO) distribution of the predicted targets is shown in Fig. 4. These analyses show various biological processes between the “L_HD” group and the “S_HD” group, such as regulation of cellular metabolic process and regulation of biological process. Some targets are categorized as cellular components, including nucleus, cell, cell part, cytoskeleton, catalytic complex, membrane-bounded organelle, and intracellular part. The remaining targets are related to important molecular functions, such as binding, ion binding, and protein binding. A KEGG pathway analysis showed 23 significant pathways (P-value < 0.05; Table 2), including the Wnt signaling pathway, MAPK signaling pathway, ECM-receptor interaction, endocrine, and other factor-regulated calcium reabsorption. Several of these terms are primarily involved in growth regulation.
Fig. 3.
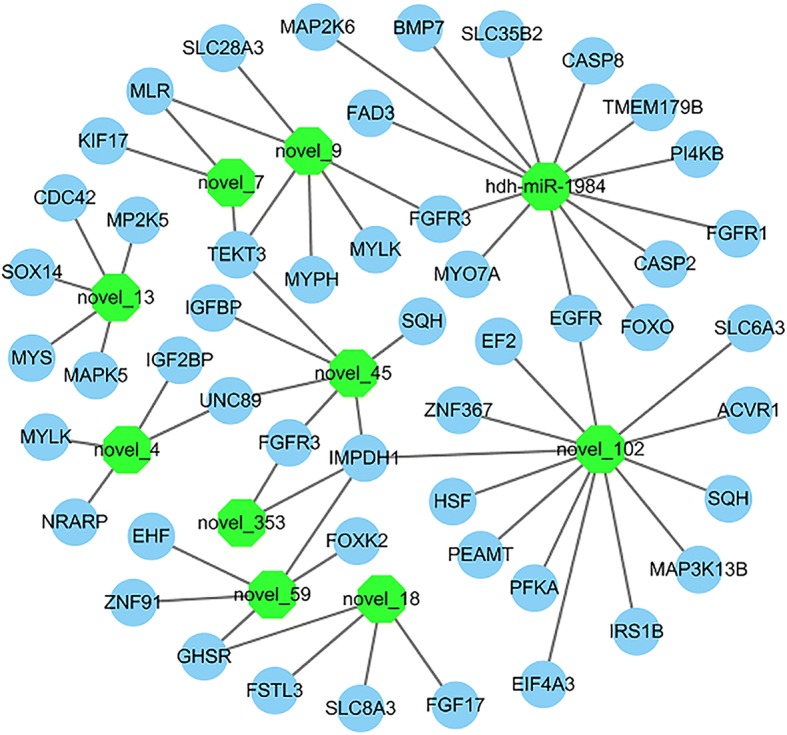
Interaction network of the differentially expressed miRNAs and some of their target genes. The miRNAs are shown in green. Target genes are represented in blue
Fig. 4.
Gene ontology distribution of the target genes for differentially expressed miRNAs
Table 2.
KEGG pathways of targets of differentially expressed miRNAs between the “L_HD” and “S_HD” Haliotis discus hannai
| KEGG pathway | Term | P-value | Gene Counts | Gene number of Pathway |
|---|---|---|---|---|
| map 04310 | Wnt signaling pathway | 1.03E-06 | 37 | 156 |
| map 04010 | MAPK signaling pathway | 4.60E-05 | 54 | 293 |
| map 04120 | Ubiquitin mediated proteolysis | 4.89E-05 | 48 | 253 |
| map 04721 | Synaptic vesicle cycle | 0.000147767 | 22 | 88 |
| map 03010 | Ribosome | 0.000409949 | 6 | 307 |
| map 04512 | ECM-receptor interaction | 0.000409949 | 18 | 491 |
| map 04961 | Endocrine and other factor-regulated calcium reabsorption | 0.000650193 | 15 | 54 |
| map 03015 | mRNA surveillance pathway | 0.001025057 | 29 | 144 |
| map 04962 | Vasopressin-regulated water reabsorption | 0.002088037 | 17 | 70 |
| map 04530 | Tight junction | 0.002088037 | 37 | 207 |
| map 05110 | Vibrio cholerae infection | 0.007001946 | 23 | 116 |
| map 04062 | Chemokine signaling pathway | 0.007001946 | 29 | 159 |
| map 04540 | Gap junction | 0.007001946 | 29 | 159 |
| map 04723 | Retrograde endocannabinoid signaling | 0.007001946 | 21 | 103 |
| map 05203 | Viral carcinogenesis | 0.007001946 | 42 | 259 |
| map 00531 | Glycosaminoglycan degradation | 0.007001946 | 1 | 148 |
| map 04912 | GnRH signaling pathway | 0.007935967 | 30 | 169 |
| map 04913 | Ovarian steroidogenesis | 0.01007854 | 17 | 79 |
| map 00564 | Glycerophospholipid metabolism | 0.034143195 | 21 | 114 |
| map 04977 | Vitamin digestion and absorption | 0.036580765 | 2 | 133 |
| map 04390 | Hippo signaling pathway | 0.036580765 | 28 | 168 |
| map 05120 | Epithelial cell signaling in Helicobacter pylori infection | 0.049867036 | 19 | 103 |
Validation and expression analysis of identified miRNA
The differentially expressed miRNA (novel-353, novel-45, novel-4, hdh-miR-1984, novel-18, and novel-13) were validated using qRT-PCR. The expression patterns of these miRNAs were consistent with the small RNA sequencing (Fig. 5a), suggesting high reliability of the small RNA sequencing analysis. We studied the level of hdh-miR-1984 and its putative target gene BMP7 expression in different age stages of H. discus hannai. The hdh-miR-1984 and BMP7 had different expression levels at different age stages (Fig. 5b and c) and their expression trends were opposite.
Fig. 5.
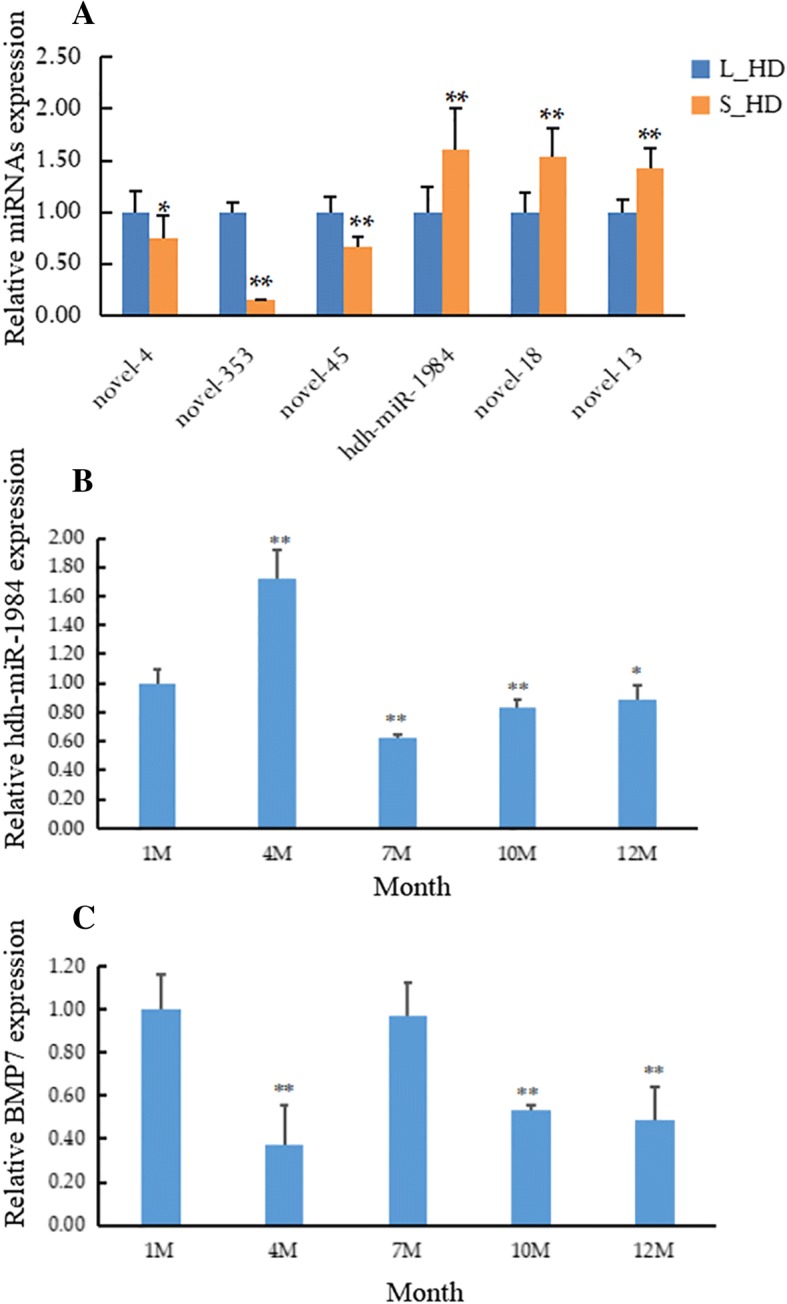
Expression of miRNAs and their target genes quantified with qRT-PCR. (a) Six differentially expressed miRNAs were examined in the muscle of Haliotis discus hannai. The level of (b) hdh-miR-1984 and (c) BMP7 expression was detected in different age stages of Haliotis discus hannai. Values are shown as mean ± SD (n = 3). *, P < 0.05; **, P < 0.01
BMP7 targeted by hdh-miR-1984
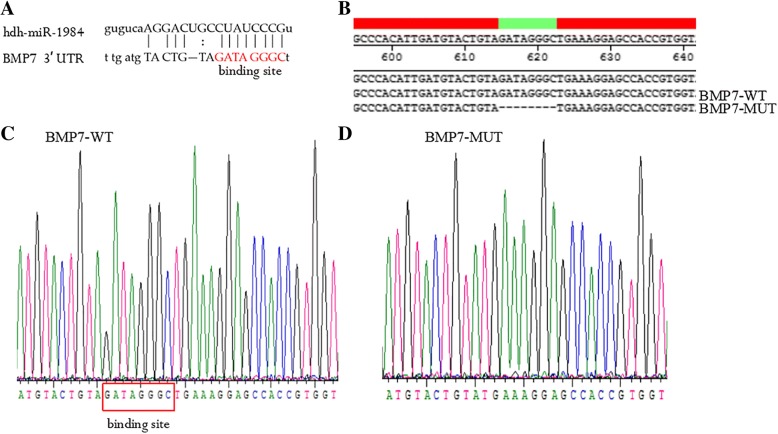
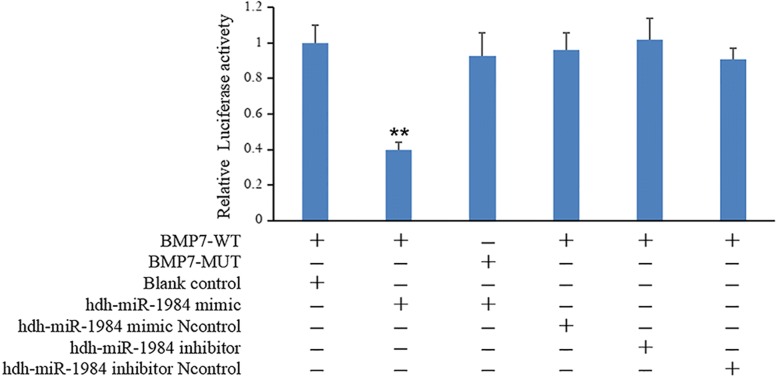
The putative seed sequences for hdh-miR-1984 at the 3’UTR of BMP7 were indicated based on bioinformatics analysis (Fig. 6a). To confirm that BMP7 is a target of hdh-miR-1984, the wild (BMP7-WT) and mutant (BMP7-MUT) forms of the BMP7 3’-UTR recombinant plasmid vectors were constructed (Fig. 6b, c, d). We then transfected hdh-miR-1984 mimic/mimic Ncontrol/inhibitor/inhibitor Ncontrol and BMP7-WT/BMP7-MUT into the 293 T cells. The luciferase activities of BMP7-WT co-transfection with the hdh-miR-1984 mimic were markedly decreased compared to that with Ncontrol or BMP7-MUT with the hdh-miR-1984 mimic (Fig. 7). Luciferase activities were not repressed in the other co-transfection groups. All of these results indicated that BMP7 is a target of hdh-miR-1984.
Fig. 6.
Report of Sequencing results. a The seed sequence for hdh-miR-1984 at the 3’UTR of BMP7. b, c, d BMP7-WT vector contains hdh-miR-1984 binding site, and BMP7-MUT vector with mutated seed region of the predicted hdh-miR-1984 sites
Fig. 7.
Targeting of BMP7 by hdh-miR-1984. Luciferase activity assay of the wild-type (WT) or mutant (MUT) 3′ UTR of BMP7 using a dual luciferase reporter system in 293 T cell lines following co-transfection with hdh-miR-1984 mimic, mimic Ncontrol, inhibitor, inhibitor Ncontrol, and blank control. In addition, luciferase activities significantly decrease in the BMP7-WT plasmid-transfected 293 T cells following co-transfection with hdh-miR-1984 mimic. Effects are blocked in the mutant plasmids transfected 293 T cells and in the BMP7-WT plasmids transfected 293 T cells following co-transfection with hdh-miR-1984 inhibitor. Data are derived from triplicate transfectants of three independent experiments (** P < 0.01)
Discussion
In many eukaryotes, miRNA plays a vital role in biological processes [26]. High-throughput sequencing technologies have enabled large-scale studies on miRNA transcriptome profiles in various mollusks, such as Littorina littorea [27], Crassostrea gigas [28], Pinctada martensii [29], and Haliotis midae [30]. In this study, six small RNA libraries were sequenced to identify the miRNAs in the muscle of H. discus hannai. Length distribution analysis showed that the most abundant reads were 22 nt, which was consistent with other marine animals [16, 31] and demonstrating that the reads from H. discus hannai were reliable and suitable for further analysis. In total, 205 miRNAs were identified in H. discus hannai muscle, among which, 15 miRNAs were conserved and 190 were novel among various animal species. Among all the miRNAs, hdh-miR-1984 and hdh-mir-1986 are the only miRNAs which appear to be mollusk-specific. These results suggest that there were mollusk-specific miRNAs in abalone and those 205 miRNAs expressed in H. discus hannai muscle might be involved in the modulation of muscle growth. However, the abalones displayed significantly different growth rate in the same cage, while the body weight for the L_HD individuals could be 5 times than the S_HD individuals. The reasons for the differences could be internal genetic factors but also external environmental factors. Therefore, samples from different culture environment will be collected and further analyzed to reveal the molecular mechanism of abalone growth in future.
The miRNAs and epigenetic modifications are major components of the myogenic regulatory network [32]. However, the information of myogenic related miRNAs in abalone remains unknown. To study the probable function of miRNA in abalone muscle growth, the expression profile of miRNAs in H. discus hannai muscle was analyzed between the L_HD and S_HD abalones. In the L_HD abalones, 7 miRNAs were down-regulated and 3 miRNAs were up-regulated compared to the S_HD specimens. These results indicate that the 10 DE-miRNAs may be related to growth. Identification of targets can deepen our understanding of the biological roles of miRNA [26]. In this study, the target genes of the 10 muscle growth-related miRNAs were predicted and annotated. The complex network as shown in Fig. 3 suggest that there is a many-to-many interaction relationship between miRNAs and their target genes. The GO terms were associated with many biological functions, such as regulation of gene expression, intracellular membrane-bounded organelle, regulation of cellular biosynthetic process, and binding. The KEGG pathways, including ECM-receptor interaction, Wnt, and the MAPK signaling pathway, were primarily involved in muscle growth regulation. The Wnt signaling pathway plays a important function in the regulation of muscle development [33]. The MAPK signaling pathway is a positive regulator in muscle development [34, 35]. All of the results indicate that these DE-miRNAs might influence the regulation of muscle growth in H. discus hannai by affecting target genes.
Muscle development is a complex system regulated by a cascade of factors containing miRNAs. The miRNAs negatively regulate gene expression by binding to the 3′UTR of the target gene in a sequence-specific manner at the posttranscriptional level [36]. The miR-378 downregulates MyoR, an MyoD inhibitor, by recognizing its 3′UTR [37]. The miR-155 inhibited muscle differentiation by repressing expression of the MEF2A [13]. BMPs, as members of the transforming growth factor β (TGF-β) super family, are usually considered potent inhibitors of muscle differentiation [38–41]. BMP7 stimulates Pax-3 expression in low concentration, but high concentration BMP7 induces muscle loss [42]. The miR-378 could suppress BMP2 by targeting its 3′UTR to regulate myogenesis [43]. In this study, hdh-miR-1984 was downregulated in the L_HD abalones compared to the S_HD individuals. Many muscle growth-related target genes were predicted containing the fibroblast growth factor receptor 3 (FGFR3), Myosin-VIIa, epidermal growth factor receptor (EGFR), and BMP7. The hdh-miR-1984 and BMP7 had different expression levels during different age stages. Interestingly, the expressions of hdh-miR-1984 and BMP7 varied in an inverse manner which suggest BMP7 may be a target gene of the hdh-miR-1984. Furthermore, the dual-luciferase reporter assay results indicate that hdh-miR-1984 directly recognized the BMP7 3′ UTR in abalone. All of these imply that hdh-miR-1984 may be a vital muscle growth-related miRNA that regulates muscle growth by targeting BMP7. However, the function of the other detected miRNAs needs further study.
The miRNA have emerged as important roles in the regulation of gene expression. These DE-miRNAs will further be used as molecular markers to screen for fast-growing strains of abalone. In summary, this research is the first analysis of miRNAs in H. discus hannai using the Illumina HiSeq sequencing platform. A total of 205 miRNAs were identified, among which, 10 DE-miRNAs were closely associated with muscle growth. In addition, we verified the BMP7 is a target gene of hdh-miR-1984 and examined their dynamic expression pattern in different developmental stages. Taken together, our findings provide useful information for understanding the epigenetic regulation of muscle development and also help to reveal the mechanisms of abalone muscle growth. These findings will further be used to improve artificial selection efficiency and contribute to the genetic improvements of the abalone aquaculture.
Additional files
Primers used for PCR and qRT-PCR. (XLSX 10 kb)
Small RNA sequencing Data. (XLSX 11 kb)
Genomic location information of Small RNA. (XLSX 10 kb)
The information of known miRNAs and novel miRNAs. (XLSX 18 kb)
The conserved miRNAs across the different animal species in the miRBase. (XLSX 10 kb)
The potential miRNA-target interactions which may play a vital role in muscle growth of the H. discus hannai. (XLSX 13 kb)
Acknowledgements
We thank all contributors of the present study.
Funding
This work was supported by grants from National Natural Science Foundation of China (No.U1605213 and 31872564); Earmarked Fund for Modern Agro-industry Technology Research System (No. CARS-49); Key S & T Program of Shandong Province (No. 2016GGH4513) and Xiamen Southern Oceanographic Center (No. 18GZY012HJ02).
Availability of data and materials
The raw reads produced in this study were deposited in the NCBI database. Sequence Read Archive under the accession number PRJNA492758. https://www.ncbi.nlm.nih.gov/sra/PRJNA492758.
Abbreviations
- 3′UTRs
3′ untranslated regions
- BMP7
bone morphogenetic protein 7
- DE
differentially expressed
- EGFR
epidermal growth factor receptor.
- FGFR3
fibroblast growth factor receptor 3
- H. discus hannai
Haliotis discus hannai
- IGF-1
insulin-like growth factor 1
- MEF2A
myocyte enhancer factor 2A
- miRNAs
MicroRNAs
- MYLK
myosin light chain kinase
- MYS
myosin heavy chain
- qRT-PCR
quantitative real-time polymerase chain reactions
- SRF
serum response factor
- TGF-β
transforming growth factor β
Authors’ contributions
HJF conceived of this research, performed data analysis, conducted qRT-PCR validation and luciferase reporter assay, and drafted the manuscript. LX, HMQ, and LGM participated in the animal experiments, statistical analysis, and surgical processes. YWW and KCH participated in the design and coordination and helped to draft the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Laboratory Animal Management and Ethics Committee of Xiamen University. The methods were carried out in accordance with the approved guidelines. All experimental procedures involving abalones were performed according to the Regulations for the Administration of Affairs Concerning Experimental Animals (Xiamen University, China; revised in November 2014).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianfang Huang, Email: jianfhuang@foxmail.com.
Xuan Luo, Email: xluo@xmu.edu.cn.
Miaoqin Huang, Email: huangmq@xmu.edu.cn.
Guangmou Liu, Email: xs_xiangmu@163.com.
Weiwei You, Email: wwyou@xmu.edu.cn.
Caihuan Ke, Email: chke@xmu.edu.cn.
References
- 1.Guo YF, Zhao WW, Gao HQ, Wang S, Yu PM, Yu HS, Wang D, Wang Q, Wang JX, Wang ZF, et al. China fishery statistical yearbook. Beijing, China: China agriculture press; 2017. [Google Scholar]
- 2.Luo X, Ke CH, You WW. Estimates of correlations for Shell morphological traits on body weight of interspecific hybrid abalone (Haliotis discus hannai and Haliotis gigantea) J Shellfish Res. 2013;32:115–118. doi: 10.2983/035.032.0117. [DOI] [Google Scholar]
- 3.Elliott NG. Genetic improvement programmes in abalone: what is the future? Aquac Res. 2000;31:51–59. doi: 10.1046/j.1365-2109.2000.00386.x. [DOI] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song YN, Shi LL, Liu ZQ, Qiu GF. Global analysis of the ovarian microRNA transcriptome: implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea: Decapoda) BMC Genomics. 2014;15(1):547. doi: 10.1186/1471-2164-15-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao C, Zhang GS, Yin SW, Li ZC, Wang QT, Chen SQ, Zhou GQ. Integrated analysis of mRNA-seq and miRNA-seq reveals the potential roles of sex-biased miRNA-mRNA pairs in gonad tissue of dark sleeper (Odontobutis potamophila) BMC Genomics. 2017;18:613. doi: 10.1186/s12864-017-3995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu P, Vernooy SY, Guo M, Hay BA. Te drosophila microRNA miR-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/S0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 9.Gong G, Sha Z, Chen S, Li C, Yan H, Chen Y, Wang T. Expression profiling analysis of the microRNA response of Cynoglossus semilaevis to Vibrio anguillarum and other stimuli. Mar Biotechnol. 2015;17(3):338–352. doi: 10.1007/s10126-015-9623-2. [DOI] [PubMed] [Google Scholar]
- 10.Huang CW, Li YH, Hu SY, Chi JR, Lin GH, Lin CC, Gong HY, Chen JY, Chen RH, Chang SJ, Liu FG. Wu JL. Differential expression patterns of growth-related microRNAs in the skeletal muscle of Nile tilapia (Oreochromis niloticus) J Anim Sci. 2012;90:4266–4279. doi: 10.2527/jas.2012-5142. [DOI] [PubMed] [Google Scholar]
- 11.Yan B, Guo JT, Zhao LH, Zhao JL. microRNA expression signature in skeletal muscle of Nile tilapia. Aquaculture. 2012;364-365:240–246. doi: 10.1016/j.aquaculture.2012.08.018. [DOI] [Google Scholar]
- 12.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seok HY, Tatsuguchi M, Callis TE, Hem A, Pu WT, Wang DZ. miR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J Biol Chem. 2011;286(41):35339–35346. doi: 10.1074/jbc.M111.273276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Z, Huang R, Tian R, Jiao Y, Du X. Pm-miR-133 hosting in one potential lncRNA regulates RhoA expression in pearl oyster Pinctada martensii. Gene. 2016;591:484–489. doi: 10.1016/j.gene.2016.06.051. [DOI] [PubMed] [Google Scholar]
- 16.Zhong L, Zhang F, Zhai Y, Cao YH, Zhang S, Chang YQ. Identifcation and comparative analysis of complement C3-associated microRNAs in immune response of Apostichopus japonicus by high-throughput sequencing. Sci Rep. 2015;5:17763. doi: 10.1038/srep17763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen M, Shen Y, Shi S, miREvo TT. An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics. 2010;13:140. doi: 10.1186/1471-2105-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat. 2003;31:2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 22.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young MD, Wakefield MJ, Smyth GK, Oshlack A. goseq: Gene Ontology testing for RNA-seq datasets. 2010. [DOI] [PMC free article] [PubMed]
- 24.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods. 2001;25:402–428. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Ren XY, Cui YT, Gao BQ, Liu P, Li J. Identification and profiling of growth-related microRNAs of the swimming crab Portunus trituberculatus by using Solexa deep sequencing. Mar Genomics. 2016;28:113–120. doi: 10.1016/j.margen.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Biggar KK, Kornfeld SF, Maistrovski Y, Storey KB. MicroRNA regulation in extreme environments: differential expression of MicroRNAs in the intertidal snail Littorina littorea during extended periods of freezing and anoxia. Genomics Proteomics Bioinformatics. 2012;10:302–309. doi: 10.1016/j.gpb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu F, Wang XT, Feng Y, Huang W, Wang W, Li L, Fang XD, Que HY, Zhang GF. Identification of conserved and novel MicroRNAs in the Pacific oyster Crassostrea gigas by deep sequencing. PLoS One. 2014;9(8):e104371. doi: 10.1371/journal.pone.0104371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao Y, Zheng Z, Du XD, Wang QH, Huang RL, Deng YW, Shi SL, Zhao XX. Identification and characterization of MicroRNAs in pearl oyster Pinctada martensii by Solexa deep sequencing. Mar Biotechnol. 2014;16:54–62. doi: 10.1007/s10126-013-9528-x. [DOI] [PubMed] [Google Scholar]
- 30.Picone B, Rhode C, Wilding RR. Identification and characterization of miRNAs transcriptome in the south African abalone, Haliotis midae. Mar Genomics. 2017;31:9–12. doi: 10.1016/j.margen.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Li C, Feng W, Qiu L, Xia C, Su X, Jin C, Zhou T, Zeng Y, Li T. Characterization of skin ulceration syndrome associated microRNAs in sea cucumber Apostichopus japonicus by deep sequencing. Fish Shellfish Immunol. 2012;33:436–441. doi: 10.1016/j.fsi.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Whysall KG, Sweetman D, Munsterberg AE. microRNAs in skeletal muscle differentiation and disease. Clin Sci. 2012;123:611–625. doi: 10.1042/CS20110634. [DOI] [PubMed] [Google Scholar]
- 33.Otto A, Schmidt C, Luke G, Allen S, Valasek P, Muntoni F, LawrenceWatt D, Patel K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J Cell Sci. 2008;121:2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- 34.Keren A, Tamir Y, Bengal E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol Cell Endocrinol. 2006;252:224–230. doi: 10.1016/j.mce.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Zetser A, Gredinger E, Bengal E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J Biol Chem. 1999;274:5193–5200. doi: 10.1074/jbc.274.8.5193. [DOI] [PubMed] [Google Scholar]
- 36.Xi QY, Xiong YY, Wang YM, Cheng X, Qi QE, Shu G, Wang SB, Wang LN, Liu L. Genome-wide discovery of novel and conserved microRNAs in white shrimp (Litopenaeus vannamei) Mol Biol Rep. 2014;42(1):61–69. doi: 10.1007/s11033-014-3740-2. [DOI] [PubMed] [Google Scholar]
- 37.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J Biol Chem. 2011;286:19431–19438. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tzahor E, Kempf H, Mootoosamy RC, Poon AC, Abzhanov A, Tabin CJ, Dietrich S, Lassar AB. Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev. 2003;17:3087–3099. doi: 10.1101/gad.1154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank NY, Kho AT, Schatton T, Murphy GF, Molloy MJ, Zhan Q, Ramoni MF, Frank MH, Kohane IS, Gussoni E. Regulation of myogenic progenitor proliferation in human fetal skeletal muscle by BMP4 and its antagonist gremlin. J Cell Biol. 2006;175:99–110. doi: 10.1083/jcb.200511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biressi S, Tagliafico E, Lamorte G, Monteverde S, Tenedini E, Roncaglia E, Ferrari S, Ferrari S, Cusella-De Angelis MG, Tajbakhsh S, Cossu G. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev Biol. 2007;304:633–651. doi: 10.1016/j.ydbio.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 41.Aoyama K, Yamane A, Suga T, Suzuki E, Fukui T, Mura YN. Bone morphogenetic protein-2 functions as a negative regulator in the differentiation of myoblasts, but not as an inducer for the formations of cartilage and bone in mouse embryonic tongue. BMC Dev Biol. 2011;11:44. doi: 10.1186/1471-213X-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amthor H, Christ B, Doubell FR, Kemp CF, Lang E, Patel K. Follistatin regulates bone morphogenetic Protein-7 (BMP-7) activity to stimulate embryonic muscle growth. Dev Biol. 2002;243:115–127. doi: 10.1006/dbio.2001.0555. [DOI] [PubMed] [Google Scholar]
- 43.Hou X, Tang Z, Liu H, Wang N, Ju H, Li K. Discovery of MicroRNAs associated with Myogenesis by deep sequencing of serial developmental skeletal muscles in pigs. PLoS One. 2012;7(12):e52123. doi: 10.1371/journal.pone.0052123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for PCR and qRT-PCR. (XLSX 10 kb)
Small RNA sequencing Data. (XLSX 11 kb)
Genomic location information of Small RNA. (XLSX 10 kb)
The information of known miRNAs and novel miRNAs. (XLSX 18 kb)
The conserved miRNAs across the different animal species in the miRBase. (XLSX 10 kb)
The potential miRNA-target interactions which may play a vital role in muscle growth of the H. discus hannai. (XLSX 13 kb)
Data Availability Statement
The raw reads produced in this study were deposited in the NCBI database. Sequence Read Archive under the accession number PRJNA492758. https://www.ncbi.nlm.nih.gov/sra/PRJNA492758.