Abstract
The splicing of mRNA is dependent on serine-arginine (SR) proteins that are mobilized from membrane-free, nuclear speckles to the nucleoplasm by the Cdc2-like kinases (CLKs). This movement is critical for SR protein-dependent assembly of the macromolecular spliceosome. Although CLK1 facilitates such trafficking through the phosphorylation of serine-proline dipeptides in the prototype SR protein SRSF1, an unrelated enzyme known as SR protein kinase 1 (SRPK1) performs the same function but does not efficiently modify these dipeptides in SRSF1. We now show that the ability of SRPK1 to mobilize SRSF1 from speckles to the nucleoplasm is dependent on active CLK1. Diffusion from speckles is promoted by the formation of an SRPK1-CLK1 complex that facilitates dissociation of SRSF1 from CLK1 and enhances the phosphorylation of several serine-proline dipeptides in this SR protein. Down-regulation of either kinase blocks EGF-stimulated mobilization of nuclear SRSF1. These findings establish a signaling pathway that connects SRPKs to SR protein activation through the associated CLK family of kinases.
Keywords: kinase, phosphorylation, speckles, splicing, SR protein
Summary Statement
SRPKs induce SR protein mobilization in the nucleus through the CLK kinases.
INTRODUCTION
The splicing of precursor mRNA (pre-mRNA) is a fundamental co-transcriptional event that enormously expands the number of proteins generated from a limited gene pool. Splicing occurs through the regulated inclusion of protein-coding exons and exclusion of non-coding introns within a gene. The splicing mechanism is initiated by the phosphorylation of an essential family of splicing factors known as SR proteins, so named owing to a C-terminal domain rich in arginine-serine dipeptide repeats (RS domains). Phospho-SR proteins attach to pre-mRNA through one or two RNA recognition motifs (RRMs) that guide the sequential assembly of the spliceosome, a macromolecular machine composed of five snRNPs (U1–6) and over 100 proteins [1]. Two principal families of protein kinases phosphorylate and regulate SR protein subcellular localization, mRNA association and splicing function. The serine-arginine protein kinases (SRPKs) are mostly cytoplasmic with a smaller nuclear presence in unstimulated cells. SRPKs phosphorylate Arg-Ser repeats in RS domains facilitating transport of SR proteins from the cytoplasm to nuclear storage speckles via coupling to an SR-specific transportin (TR-SR) [2–4]. Speckles are dynamic, membrane-free structures that harbor SR proteins along with other RNA processing factors for transcription and splicing [5]. While concentrated in nuclear speckles, SR proteins can still undergo rapid speckle-nucleoplasmic exchange (t1/2 < 10 sec) based on FRAP studies [6]. The CDC2-like kinases (CLKs) are localized to the nucleus and further modify the RS domains, mobilizing SR proteins from speckles to the nucleoplasm where they support co-transcriptional splicing [7, 8]. The latter modification facilitates the binding of several early spliceosomal components including the U1 snRNP and the U2AF heterodimer that mark the correct 5’ and 3’ splice sites and define exon boundaries [9–11]. These regulated events furnish the critical, initial steps that generate multiple, mature mRNA strands from a single pre-mRNA. In addition to phosphorylation, SR protein dephosphorylation is also important for later steps in spliceosome catalysis and for the export of mature mRNA that is translated in the cytoplasm [12].
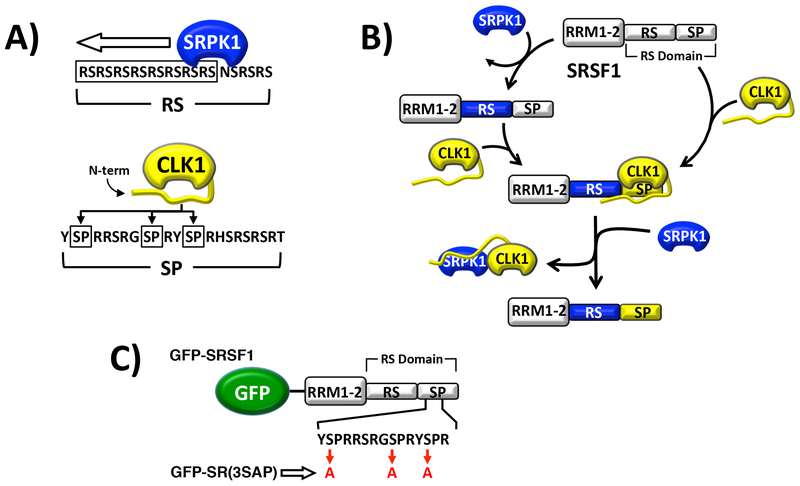
Much of what we know about the SR protein phosphorylation cycle comes from studies on the prototype SR protein SRSF1 (aka ASF/SF1). SRPK1 phosphorylates about 8–10 serines in an Arg-Ser repeat toward the N-terminus of the SRSF1 RS domain (Fig. 1A). This reaction occurs in a C-to-N-terminal, processive manner that is promoted by a docking groove in SRPK1 and structural features in the SR protein [13–15]. Upon modification, the phosphorylated SR protein readily dissociates from SRPK1 [16], an event that allows TR-SR binding and localization in nuclear speckles [4, 17, 18]. The nuclear CLK1 then phosphorylates Ser-Pro dipeptides toward the C-terminus of the RS domain mobilizing SRSF1 from speckles to the nucleoplasm (Fig. 1A). CLK1 also phosphorylates Arg-Ser dipeptides throughout the RS domain of SRSF1 [3], a feature that may balance the actions of nuclear phosphatases that return SRSF1 to the cytoplasm. These studies describe a compartmentalized, sequential pathway where cytoplasmic SRPKs layer phosphates on the Arg-Ser repeats in the RS domain and nuclear CLKs then phosphorylate Ser-Pro dipeptides (Fig. 1B). However, recent studies suggest that SRPK1 and CLK1 can function together in a synergistic manner in the nucleus for splicing activation. We showed recently that EGF stimulation induces translocation of SRPK1 from the cytoplasm to the nucleus where it forms a complex with CLK1 [19]. Unlike SRPK1, CLK1 stays tightly bound to phospho-SRSF1 inhibiting splicing function owing to strong contacts between the CLK1 N-terminus and the SRSF1 RS domain [19, 20]. The formation of the SRPK1-CLK1 complex results in dissociation of phospho-SRSF1 from CLK1 and engagement of the SR protein with U1 snRNP for splicing propagation [19]. This new function explains how signals that induce translocation of SRPK1 to the nucleus can cause large, broad changes in splicing patterns [21].
Figure 1: Phosphorylation of SRSF1 by SRPK1 and CLK1.
A) Kinase phosphorylation regiospecificity in RS domain. B) Sequential phosphorylation of RS domain in SRSF1 by SRPK1 and CLK1. C) GFP-fused forms of SRSF1.
It is widely accepted that phosphorylation mobilizes SR proteins from nuclear speckles to the nucleoplasm where they engage the spliceosome and catalyze pre-mRNA splicing. Nonetheless, how phosphorylation orchestrates this critical trafficking event is not well understood. For example, while it is clear that CLK1 over-expression mobilizes SRSF1 from speckles to the nucleoplasm by phosphorylating several Ser-Pro dipeptides in the C-terminus of the RS domain [22], SRPK1 can perform the same function [23, 24] although this kinase poorly phosphorylates this region in SRSF1 [3, 25]. Such findings suggest that bulk increases in RS domain phosphorylation rather than specific Ser-Pro phosphorylation may induce mobilization of SRSF1 and mRNA splicing. On the other hand, the new observations that SRPK1 can enter the nucleus of stimulated cells and form a complex with CLK1 raises the possibility that SRPK1-induced mobilization of SR proteins may be triggered through such a complex. In this new study we investigated the role of the SRPK1-CLK1 complex in phosphorylating and mobilizing SRSF1 in the nucleus. We found that SRPK1-dependent diffusion of nuclear SRSF1 from nuclear speckles can be blocked by down-regulation of CLK1. We discovered that formation of the SRPK1-CLK1 complex dissociates SRSF1 from CLK1 and greatly enhances the phosphorylation state of Ser-Pro dipeptides in the RS domain of this splicing factor. This event provides a strong chemical stimulus for enhanced nuclear dynamics and increased transition of SRSF1 from speckles to the nucleoplasm. The data can be used to support a new signaling pathway that connects nuclear SRPKs to SR protein activation through the associated CLK family of kinases.
EXPERIMENTAL
Materials:
ATP, Mops, HEPES, Tris, MgCl2, MnCl2, NaCl, EDTA, Brij 35, glycerol, acetic acid, lysozyme, DNAse, RNAse, Phenix imaging film, BSA, Ni-resin and liquid scintillant were obtained from Fisher Scientific. γ-32P-ATP was obtained from NEN Products. SRPIN340 was obtained from Sigma. FuGene reagent was obtained from Promeg and Lipofectamine 2000 was obtained from ThermoFisher. Protease inhibitor cocktail, EGF, and TG003 were obtained from Roche. Anti-SRPK1 monoclonal antibody was purchased from BD Biosciences. Anti-CLK1 polyclonal antibody was purchased from Aviva. Anti-SRSF1 monoclonal antibody was purchased from Life Tech. Anti-GFP monoclonal antibody, anti-HA monoclonal antibody, anti-His monoclonal antibody and Protein G beads were purchased from Cell Signaling. Anti-GST monoclonal antibody was purchased from BioLegend. InstantBlue was purchased from Expedeon.
Expression and Purification of Recombinant Proteins:
All forms of recombinant SRPK1 and SRSF1 were expressed in E.coli and purified from pET19b vectors containing an N-terminal His tag as previously described [26]. CLK1 virus was transfected and expressed in Hi5 insect cells. CLK1 was purified with a nickel resin and a previously described procedure [20]. GFP-SRSF1 constructs were expressed from a pcDNA3.1+N-eGFP vector, kdCLK1-RFP was expressed from a pcDNA3-mRFP vector, CLK1-HA was expressed from a pcDNA3.1+C-HA vector and SRPK1 was expressed from a pUHD vector [27].
Phosphorylation & Dephosphorylation Reactions:
Phosphorylation of all forms of SRSF1 by SRPK1 and/or CLK1 were carried out in the presence of 100 mM Mops (pH 7.4), 50 mM HEPES, 100 mM NaCl, 2 mM DTT, 0.01% Brij 35, 1 mM Mn2+, 10 mM Mg2+ and 5 mg/ml BSA at 37°C, using either 50 μM 32P-ATP for SRPK1 or 100 μM 32P-ATP for CLK1 both with a specific activity of 4000–8000 cpm/pmol, and in the absence or presence of PP1γ. All reactions were carried out in a total volume of 10 μl and quenched with 10 μl of SDS/PAGE loading buffer. Phosphorylated SR proteins were separated from unreacted 32P-ATP and other protein constituents by SDS-PAGE (10% or 12%), cut from the dried gel and quantified on the 32P channel in liquid scintillant.
Immunoprecipitation Experiments:
Whole cell or nuclear lysates (200 μL) of HeLa cells were incubated overnight at 4°C with 25 μL Protein G beads and 2 μg primary antibody. Beads were spun at 2,052 × g and washed 2× with 200 μL lysis buffer followed by the addition of 2× SDS loading buffer and heated to 90°C. The heated slurry was run on either a 10% or 12% SDS-PAGE followed by immunoblot analysis. HeLa cell visualization and quantitation analyses were performed using ImageJ software and cell fractionations were performed using the cell signaling fractionation kit (#9038).
Confocal Imaging Experiments:
For live-cell confocal imaging, HeLa cells were plated on 2.5-cm2 MatTek poly-D-lysine plates and transfected with GFP-, RFP-, or Myc-tagged constructs (2 μg) for 24 hr. EGF stimulation was performed by replacing the growth media with OptiMem supplemented with 200 ng/mL EGF and incubated for 1 hour. TG003 and SRPIN340 treatment were performed by replacing the growth media with OptiMem supplemented with 20 μM TG003 or 50 μM SRPIN340 for 1 hour. For knockdown experiments, Lipofectamine 2000 was used to transfect 50 pmol CLK1 siRNA in a 12 well poly-D-lysine plate. The transfected cells were washed with 1× PBS and analyzed using an Olympus FV1000 as described previously [22]. For fixed-cell imaging, cells were washed with 1× PBS, fixed with 1% paraformaldehyde (PFA) for 20 minutes at room temperature, permeabilized with PBS-0.5% Triton X-100 for 10 minutes at 4°C, and then blocked with 20% goat serum in PBS (blocking buffer). Samples were incubated with primary antibody (overnight in blocking buffer), washed 3 times with 1× PBS, incubated with secondary antibodies for 1 hour at room temperature (goat anti-mouse AlexaFluor 647 - Life), washed 3 times with 1× PBS, and mounted in DAPI-containing mounting medium (Vector Laboratories). Confocal images were acquired using an Olympus FV1000 with 405, 488, 555, and 647 laser lines. Images were linearly analyzed and pseudo-colored using ImageJ analysis software.
RESULTS
CLK1 Activity Is Required For SRPK1-Induced Nuclear Mobilization of SRSF1:
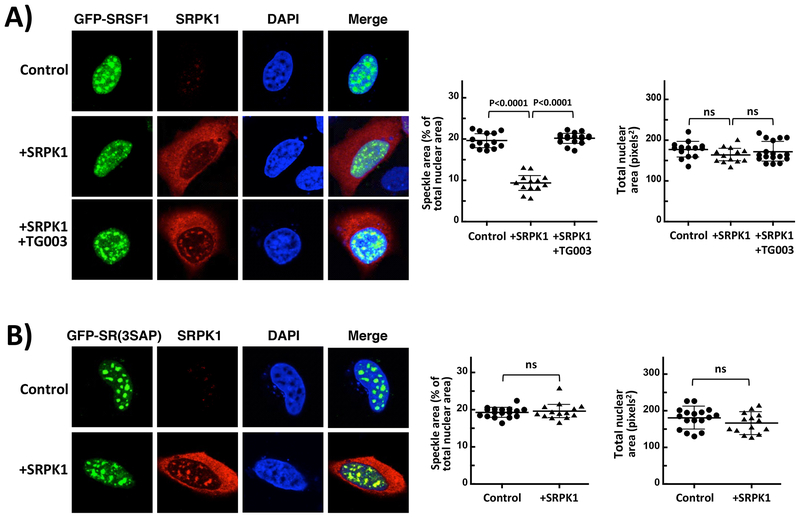
Although CLK1 mobilizes SRSF1 from speckles to the nucleoplasm through Ser-Pro dipeptide phosphorylation [2, 22], SRPK1 expression elicits similar changes although it does not efficiently phosphorylate these dipeptides in the RS domain of SRSF1 [3, 16, 28]. We wished to further investigate this phenomenon by studying the nuclear trafficking of SRSF1 expressed from a pcDNA3.1-N-eGFP vector containing an N-terminal GFP tag (GFP-SRSF1) as a function of SRPK1 expression and CLK1 activity using confocal microscopy (Fig. 1C). Prior studies demonstrated that GFP-SRSF1 possesses the identical, observable features of endogenous SR proteins including similar subnuclear localization in speckles, multiple phosphorylation states in vivo, ability to induce mRNA splicing of reporter genes and ability to migrate to sites of active gene transcription outside the speckle [29]. We first confirmed previous findings [20, 22, 30] that over-expression of CLK1 using a pcDNA3.1+C-HA vector that places an HA tag at the C-terminus (CLK1-HA) alters GFP-SRSF1 localization from nuclear speckles to the nucleoplasm in HeLa cells, a process that can be blocked by the CLK1 inhibitor TG003 (Supplementary Fig. S1). Over-expression of SRPK1 from a pUHD vector [27] also mobilizes SRSF1 in accordance with previous studies [23, 31], but we now demonstrate that this phenomenon is blocked by TG003 (Fig. 2A). To quantify this mobilization phenomenon, we integrated speckle area for a representative population of cells and found that SRPK1 over-expression approximately halves the speckle area without affecting the total nuclear area. We also confirmed that GFP-SRSF1 co-localizes with the speckle marker SC35 in the absence and presence of TG003 (Supplementary Figure S2). To identify cells that over-express SRPK1 in these studies, we used low SRPK1 antibody levels (Fig. 2A) but showed that, at higher levels, endogenous SRPK1 could be readily detected in cells (Supplementary Figure S3). In these imaging studies we fixed cells to monitor both GFP-SRSF1 using direct fluorescence and SRPK1 over-expression using immunofluorescence but also showed in control experiments that such expression also diffuses GFP-SRSF1 from speckles in live cells (Supplementary Figure S1). To rule out the possibility that TG003 could also inhibit SRPK1 in our hands, we monitored the phosphorylation of recombinant, His-tagged SRSF1 with recombinant, His-tagged CLK1 and SRPK1. Using in vitro kinetic assays we showed that TG003 is a potent inhibitor of CLK1 at concentrations used for imaging (20 μM) but does not affect SRPK1 activity toward SRSF1 (Supplementary Fig. S4). We showed that treatment of cells expressing GFP-SRSF1 with TG003 in the absence of either SRPK1 or CLK1 over-expression had no significant effect on speckles (Supplementary Fig. S5). Overall, these findings suggest that CLK1 activity is necessary for SRPK1-induced diffusion of nuclear speckles.
Figure 2: SRPK1 Over-expression Influences SRSF1 Subnuclear Localization.
A) Confocal microscopy of HeLa cells expressing GFP-SRSF1 with and without co-expression of SRPK1. B) Confocal microscopy of GFP-SR(3SAP) in the absence and presence of SRPK1 expression. All speckle areas and total nuclear areas for cells (n=13–16 in (A) and (B)) were calculated from replicate experiments using Image J and displayed as a dot plot with mean values ± S.D. All P-values were calculated using a one-way ANOVA test. P-values above 0.05 are considered nonsignificant (ns). Control reflects cells over-expressing GFP-SRSF1 or GFP-SR(3SAP) but not transfected with the SRPK1 vector.
We next wished to determine whether the observed effects on localization induced by SRPK1 are linked to Ser-Pro dipeptides in SRSF1 as would be expected for a CLK1-dependent phenomenon. To test this, we studied the subcellular localization of GFP-SRSF1 containing alanine mutations in 3 serines flanking prolines in the RS domain (GFP-SR(3SAP)) (Fig. 1C). We found that GFP-SR(3SAP) is localized to nuclear speckles as previously described [22] but, unlike the wild-type SR protein, SRPK1 over-expression does not diffuse these speckles suggesting that Ser-Pro dipeptides are required for mobilization by SRPK1 (Fig. 2B). We also confirmed that GFP-SR(3SAP) like GFP-SRSF1 co-localizes with SC35 in nuclear speckles in the absence and presence of TG003 (Supplementary Figure S2). Although these cells were fixed to monitor the expression of SRPK1 using immunofluorescence, we also replicated these results using live-cell imaging (Supplemental Figure S6). In total, these findings suggest that SRPK1-induced trafficking of SRSF1 in the nucleus requires both CLK1 catalytic activity and Ser-Pro dipeptides.
SRPK1 Enhances SRSF1 Phosphorylation Status:
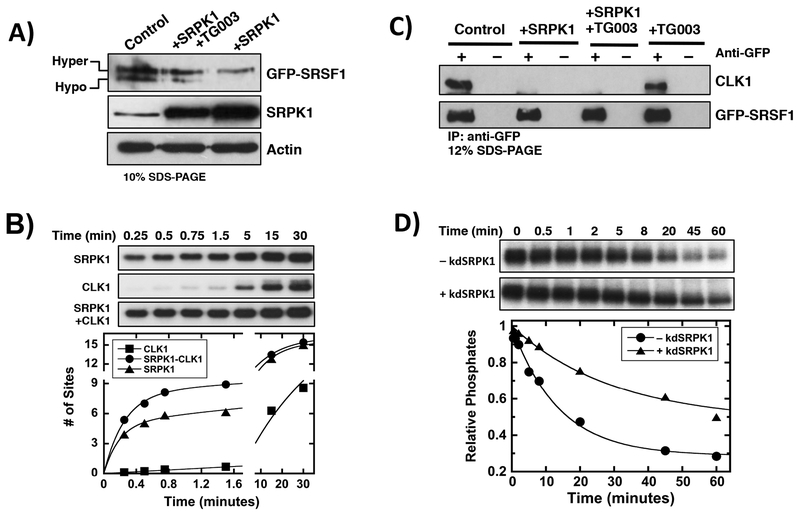
Having shown in a prior report that SRPK1 forms a complex with nuclear CLK1 [19], we addressed whether this complex impacts phosphorylation efficiency, thereby offering a link to changes in nucleoplasmic levels. We ran HeLa cell lysates expressing GFP-SRSF1 and SRPK1 on a 10% SDS-PAGE gel to analyze the phosphorylation state of the SR protein. Consistent with previous data [32, 33], under these conditions, GFP-SRSF1 appears as two discrete bands that correspond to a slow-migrating, hyper-phosphorylated species and a fast-migrating, hypo-phosphorylated species (Fig. 3A). SRPK1 over-expression led to an increase in the ratio of slow/fast migrating bands whereas treatment with TG003 diminished this ratio. We also showed that exogenous SRPK1 is over-expressed in these cells relative to the endogenous kinase (Fig. 3A). We confirmed previous findings [22] that the observed bands correlate with phosphorylated forms of GFP-SRSF1 since phosphatase treatment increases their migration on SDS-PAGE (Supplementary Figure S7). These findings suggest that SRPK1 over-expression increases SRSF1 phosphorylation status, an event that can be reversed by chemical inhibition of CLK1. Using in vitro, single-turnover experiments we next showed that formation of the SRPK1-CLK1 complex facilitates increased phosphorylation of SRSF1 in the first minute of the reaction compared to the individual kinases alone (Fig. 3B & Supplementary Table S1). In a prior study we showed that SRPK1 and CLK1 form a stoichiometric, high affinity complex (Kd = 20 nM) ensuring that these kinetic experiments are performed with little free kinase in the assay [19]. These results demonstrate that the efficiency of SR protein phosphorylation can be enhanced through the physical coupling of SRPK1 and CLK1.
Figure 3: Regulating SRSF1 Phosphorylation Through The SRPK1-CLK1 Complex.
A) Effects of SRPK1 over-expression on GFP-SRSF1 phosphorylation state in HeLa cell extracts in the absence and presence of TG003. GFP-SRSF1, SRPK1 and actin were detected using anti-GFP, SRPK1 and actin antibodies. B) Single-turnover kinetic assay for SRSF1 phosphorylation by SRPK1, CLK1 and the SRPK1-CLK1 complex. Y-axis data are normalized to total SRSF1 concentration to obtain # of sites phosphorylated. SRPK1=CLK1= 1 μM, SRSF1= 100 nM, ATP= 50 μM. Data fits are presented in Supplementary Table S1. C) Co-immunoprecipitation of GFP-SRSF1 and endogenous CLK1 in HeLa cell extracts in the absence and presence of SRPK1 over-expression. GFP-SRSF1 and CLK1 were detected using anti-GFP and CLK1 antibodies. D) PP1-dependent dephosphorylation of CLK1-phosphorylated SRSF1 in absence and presence of kdSRPK1. CLK1=PP1=kdSRPK1= 2 μM, SRSF1= 200 nM, TG003= 50 μM, ATP= 100 μM. Data fits are presented in Supplementary Table S2. Controls reflect cells over-expressing GFP-SRSF1 but not transfected with the SRPK1 vector.
Since CLK1 forms a high-affinity complex with SRSF1 that can be broken by SRPK1 in vitro [19], we wished to investigate whether SRPK1 expression alters the bound state of the SR protein in cells. Using co-immunoprecipitation assays, we showed that endogenous CLK1 interacts with GFP-SRSF1 in HeLa cell extracts in the absence or presence of TG003 and that SRPK1 over-expression disrupts this complex (Fig. 3C). To determine whether SRPK1-induced release from CLK1 stabilizes the RS domain against dephosphorylation, we pre-phosphorylated SRSF1 with CLK1 in vitro, inhibited the kinase with TG003 and then monitored the rate of PP1-dependent dephosphorylation in the absence and presence of a kinase inactive version of SRPK1 (kdSRPK1). This mutant removes a critical, conserved lysine in the active site (K109M) rendering the kinase inactive but still capable of binding SR proteins and CLK1 [19]. Although SRPK1 binds tightly to SRSF1 (Kd = 100 nM) [34], binding is very poor to the phosphorylated product so there is no residual kdSRPK1 associated with phospho-SRSF1 [16]. We found that the addition of kdSRPK1 noticeably reduced the dephosphorylation rate of SRSF1 (Fig. 3D & Supplementary Table S2). These studies reveal that the RS domain of SRSF1 is more readily phosphorylated by the SRPK1-CLK1 complex than the individual kinases alone and more resistant to dephosphorylation upon SRPK1-induced release of CLK1. Both effects correlate with an increase in the overall phosphorylation state of SRSF1.
SRPK1-CLK1 Complex Accelerates Ser-Pro Dipeptide Phosphorylation:
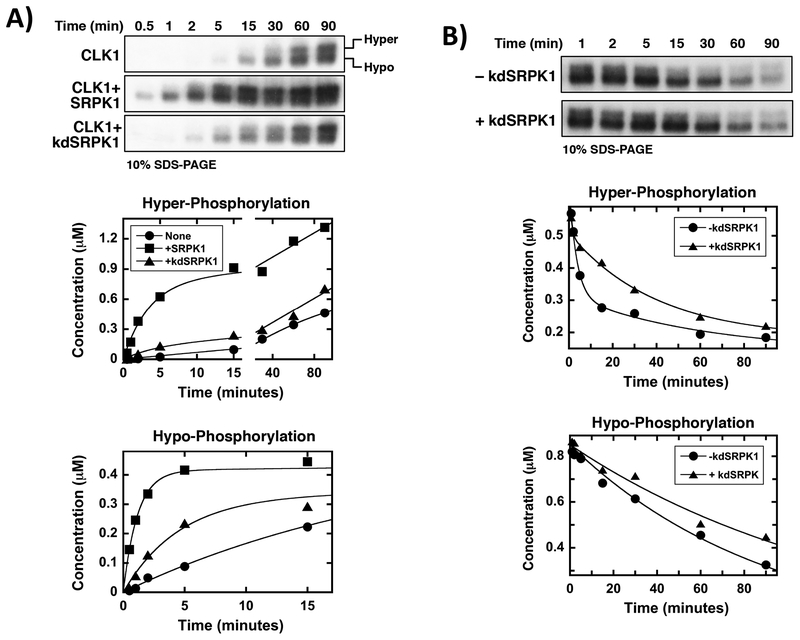
CLK1 is known to phosphorylate Ser-Pro dipeptides in the RS domain of SRSF1 inducing a unique gel shift on low percentage SDS-PAGE [3, 22]. We showed that in vitro phosphorylation by CLK1 reduces the migration of SRSF1 on a 10% SDS-PAGE gel generating a hyper-phosphorylated band (Fig. 4A). In control experiments, removal of three serines flanking prolines in SRSF1 generates a mutant protein, SR(3SAP), that does not undergo this gel shift on 10% SDS-PAGE confirming that CLK1 induces a hyper-phosphorylated state by modifying Ser-Pro dipeptides at the RS domain C-terminus (Supplementary Fig. S8). In prior studies we also showed that GFP-SR(3SAP) expressed in HeLa cells does not display a hyper-phosphorylated species compared to the wild-type SR protein suggesting that Ser-Pro phosphorylation is responsible for this observed gel shift [22]. Given this unique CLK1-dependent modification, we wished to determine whether complex formation with SRPK1 accelerates this event thereby providing a chemical means for improved mobilization of nuclear SRSF1. Using in vitro kinetic assays, we found that addition of SRPK1 (100 nM) to CLK1 (1 μM) substantially increased the appearance rate of the hyper-phosphorylated form of SRSF1 (Fig. 4A & Supplementary Table S3) although SRPK1 alone does not induce a gel shift (Supplementary Fig. S8). We found that after 15 minutes of reaction time, SRPK1 enhanced overall CLK1-dependent Ser-Pro phosphorylation (i.e.- hyper-phosphorylation) by about 10-fold relative to CLK1 alone (Fig. 4A). In these experiments, we used substoichiometric amounts of SRPK1 (relative to CLK1) since this kinase is much faster than CLK1 and would otherwise dominate the autoradiogram and make observation of the doublet difficult. Thus, we suspect that the 10-fold enhancement in hyper-phosphorylation (after 15 minutes) may be a lower limit on the true enhancement. To determine whether the gel shift is induced by SRPK1 phosphorylation priming of the RS domain, we repeated this experiment using a stoichiometric amount of kdSRPK1 and found that the inactive kinase also accelerated the appearance of the hyper-phosphorylated species although not to the same extent as SRPK1 (Fig. 4A). Furthermore, kdSRPK1 appears to be a general enhancer of CLK1 activity since it also accelerated the appearance rate of the hypo-phosphorylated band. These experiments were performed under identical specific activities of 32P-ATP and all gels were exposed for the same time to aid in direct visual comparisons of the autoradiograms. These results demonstrate that the SRPK1-CLK1 complex increases the rate of Ser-Pro dipeptide phosphorylation, a posttranslational modification associated with increased SRSF1 mobility in the nucleus.
Figure 4: SRPK1-CLK1 Complex Stabilizes Ser-Pro Dipeptide Phosphorylation.
A) SRPK1-CLK1 complex formation enhances the rate of Ser-Pro dipeptide phosphorylation. SRPK1=kdSRPK1= 1 μM, SRSF1= 100 nM, ATP= 100 μM. Data fits are presented in Supplementary Table S3. B) SRPK1-dependent dissociation of CLK1 from SRSF1 reduces the rate of Ser-Pro dipeptide dephosphorylation by PP1. CLK1=kdSRPK1=PP1= 2 μM, ATP= 100 μM, TG003= 50 μM. Data fits are presented in Supplementary Table S4.
Ser-Pro Phosphates Are Stabilized By SRPK1-Induced Release of SRSF1:
Since SRPK1-induced release of SRSF1 from CLK1 impedes PP1-dependent dephosphorylation, we wished to ask whether this reduction in dephosphorylation rate is due to the protection of Ser-Pro phosphates. To address this, we initially phosphorylated SRSF1 (100 nM) with CLK1 (2 μM) in vitro, inhibited the kinase with TG003 (50 μM) and then added PP1 (2 μM) in the absence and presence of kdSRPK1 (2 μM). We found that kdSRPK1 slowed the disappearance of the upper, hyper-phosphorylated band but had little effect on the lower band (Fig. 4B & Supplementary Table S4). After 15 minutes of reaction time, kdSRPK1 reduced the PP1-dependent dephosphorylation of the hyper-phosphorylated species by about 2-fold. Thus, within this time frame, complex formation enhances CLK1-dependent phosphorylation of Ser-Pro dipeptides by about 10-fold while reducing dephosphorylation by 2-fold. As a control, we showed that the disappearance of the single band for SR(3SAP) was unaffected by kdSRPK1 (Supplementary Fig. S9). Overall, these findings suggest that formation of the SRPK1-CLK1 complex and subsequent release of CLK1 from SRSF1 stabilizes Ser-Pro phosphates in the RS domain, a phenomenon that correlates with greater diffusion of SRSF1 from nuclear speckles.
EGF Stimulation Mobilizes Nuclear SRSF1 Through Ser-Pro Dipeptides:
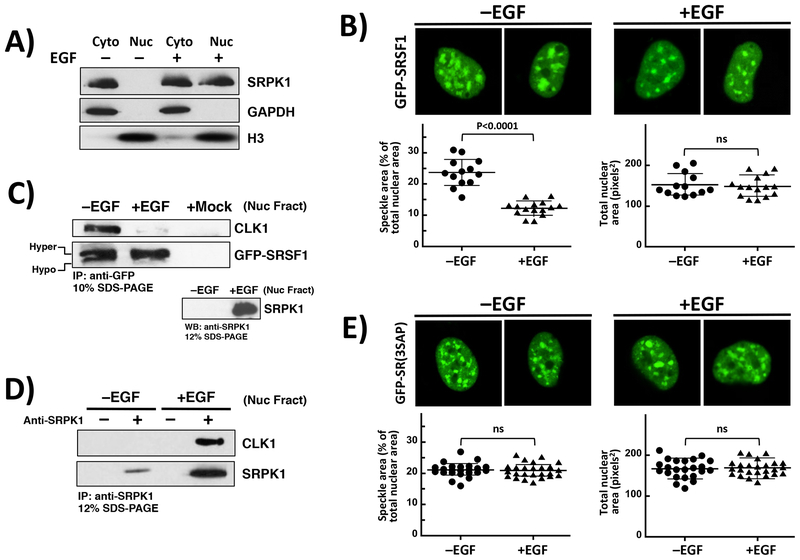
Having shown that mobilization of SRSF1 can be induced by SRPK1 over-expression through active CLK1, we next wished to determine whether such mobilization could occur under physiological signaling conditions. We showed previously that EGF stimulation induces migration of SRPK1 from the cytoplasm to the nucleus [19, 21] and wished to now investigate whether this stimulation increases SRSF1 dynamics in the nucleus through CLK1. We first confirmed in fractionation studies that EGF treatment induced translocation of endogenous SRPK1 from the cytoplasm to the nucleus as expected (Fig. 5A). We then imaged live cells expressing GFP-SRSF1 and showed that EGF stimulation decreased speckle area similar to that for SRPK1 expression (Fig. 5B). In general, EGF stimulation led to an approximate halving of the speckle area with no change in total nuclear area. Using immunofluorescence, we replicated the results from live-cell imaging and also showed that EGF induced SRPK1 enrichment in the nucleus (Supplementary Figure S10A). To control for any potential effects of the fluorescent tag on subnuclear localization, we monitored endogenous SRSF1 by immunofluorescence and showed that EGF stimulation of HeLa cells reduced speckle area as well as induced migration of endogenous SRPK1 into the nucleus as expected (Supplementary Figure S11). To determine whether this mobilization correlates with SRPK1-induced release of SRSF1 from CLK1, we performed co-immunoprecipitation assays and found that EGF treatment disrupts GFP-SRSF1 binding to endogenous CLK1 in nuclear extracts (Fig. 5C). Increases in the relative amounts of slow- versus fast-migrating bands for GFP-SRSF1 were also observed, consistent with EGF-induced hyper-phosphorylation (Fig. 5C). These results are similar to the observed effects for exogenous SRPK1 expression (Fig. 3). We confirmed that EGF stimulation causes an increase in SRPK1 in the nucleus and also ran a mock control (no plasmid) to show that no endogenous CLK1 is co-immunoprecipitated in the absence of GFP-SRSF1 expression (+Mock, Fig 5C). We next showed in co-immunoprecipitation assays of these nuclear extracts that EGF-induced release of SRSF1 from CLK1 is accompanied by increases in nuclear SRPK and formation of the SRPK1-CLK1 complex (Fig. 5D). To verify that the mobilization of SRSF1 is likely due to Ser-Pro dipeptide phosphorylation, as in the case of SRPK1 expression, we stimulated live cells expressing GFP-SR(3SAP) with EGF and did not observe any significant changes in nuclear speckles (Fig. 5E). We then performed immunofluorescence experiments to show the nuclear enrichment of SRPK1 upon EGF stimulation of cells expressing GFP-SR(3SAP) (Supplementary Figure S10B). These studies confirm that the absence of an effect on GFP-SR(3SAP) is not due to a loss of EGF-stimulated nuclear entry of SRPK1. Overall, these studies demonstrate that extracellular signals can mobilize SRSF1 from speckles to the nucleoplasm by disrupting CLK1-SRSF1 interactions and stabilizing a hyper-phosphorylated RS domain.
Figure 5: Effects of EGF Stimulation on Speckles and SRSF1 Phosphorylation.
A) Fractionation of HeLa cells. Endogenous SRPK1 is monitored in nuclear and cytoplasmic fractions as a function of EGF using an anti-SRPK1 antibody. B) Confocal microscopy of live HeLa cells expressing GFP-SRSF1 in the absence and presence of EGF. C) Co-immunoprecipitation of GFP-SRSF1 and endogenous CLK1 in HeLa cell nuclear extracts in the absence and presence of EGF. Mock lane represents cells lacking GFP-SRSF1 plasmid. GFP-SRSF1, CLK1 and SRPK1 were detected using anti-GFP, CLK1 and SRPK1 antibodies. D) Co-immunoprecipitation of endogenous SRPK1 & CLK1 in HeLa cell nuclear extracts in the absence and presence of EGF. E) Confocal microscopy of HeLa cells expressing GFP-SR(3SAP) in the absence and presence of EGF. All speckle areas and total nuclear areas for cells (n=12–14 in (B) and 23–25 in (E)) were calculated from replicate experiments using Image J and displayed as a dot plot with mean values ± S.D. P-values were calculated using a one-way ANOVA test. P-values above 0.05 are considered nonsignificant (ns).
SRSF1 Dynamics Are Regulated By An EGF-Stimulated SRPK1-CLK1 Axis:
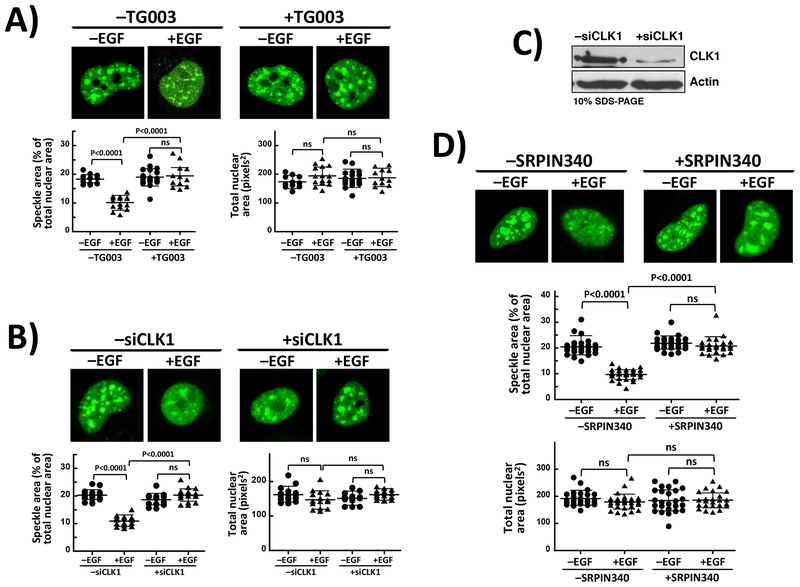
Since EGF increases SRSF1 dynamics in the nucleus, we wished to investigate whether this phenomenon is dependent on both SRPK1 and CLK1 activities. We imaged live cells expressing GFP-SRSF1 and showed that treatment with TG003 reversed the effects of EGF stimulation and induced increases in speckle area (Fig. 6A). The observed elevation in speckle area approaches the level for GFP-SRSF1 in unstimulated cells (compare Fig. 5B). To verify that specific chemical inhibition of CLK1 and not an off-pathway target is responsible for this effect, we treated HeLa cells expressing GFP-SRSF1 with CLK1 siRNA and used confocal microscopy to monitor any effects on nuclear speckles upon EFG stimulation. We found that whereas EGF treatment reduced speckle area as already observed in Figures 5B & 6A, siRNA treatment blocked this phenomenon (Fig. 6B). In control experiments, we showed that siRNA effectively depleted the endogenous levels of CLK1 suggesting that this phenomenon is linked to CLK1 knockdown (Fig. 6C). We also showed that co-expression of an inactive form of CLK1 expressed from a pcDNA3-mRFP vector that places an RFP tag on the C-terminus (kdCLK1-RFP) blocked EGF-induced speckle diffusion in HeLa cells (Supplementary Fig. S12). In these studies, kdCLK1-RFP co-localized with GFP-SRSF1 in nuclear speckles indicating that CLK1 activity is, indeed, required for SRSF1 release to the nucleoplasm. Since EGF promotes SRPK1 nuclear translocation through Akt activation [21] and Akt has been reported to phosphorylate and activate CLK1 [35], we wished to determine whether the observed effects on speckles occurs through SRPK1. To accomplish this, we used the ATP-competitive isonicotinamide compound SRPIN340 which has been shown to specifically inhibit SRPK1 in cells with no activity against CLKs or Akt [36]. We found that treatment of HeLa cells with SRPIN340 (50 μM) blocked EGF-stimulated mobilization of GFP-SRSF1 in a similar manner as TG003 (Fig. 6D). For these experiments, we confirmed that EGF elicited speckle diffusion in the absence of SRPIN340. We showed using in vitro kinetic assays that SRPIN340 specifically inhibits the catalytic activity of SRPK1 (KI = 0.6 μM) without affecting CLK1 and does not impede formation of the SRPK1-CLK1 complex in pull-down assays (Supplementary Fig. S13). These findings show that both SRPK1 and CLK1 activities are required for SRSF1 subnuclear mobility and that such phenomena likely occur through an SRPK1-CLK1 complex and signaling axis.
Figure 6: SRSF1 Mobilization Requires Both SRPK1 and CLK1 Activities.
A) Confocal microscopy of EGF-treated live HeLa cells expressing GFP-SRSF1 in the absence and presence of TG003. B) Effects of CLK1 knockdown using siRNA on EGF-stimulated mobilization of GFP-SRSF1. C) Treatment of HeLa cells with CLK1 siRNA. Endogenous CLK1 is probed using anti-CLK1 antibody in cell lysates. D) Confocal microscopy of live HeLa cells expressing GFP-SRSF1 in the absence and presence of SRPIN340 and EGF. All speckle areas and total nuclear areas for cells (n=11–18 in (A), 14–20 in (B), and 25–33 in (D)) were calculated using Image J and displayed as a dot plot with mean values ± S.D. P-values were calculated using a one-way ANOVA test. The data in (A) and (D) were generated from replicate experiments. P-values above 0.05 are considered nonsignificant (ns).
DISCUSSION
All RS domains in SR proteins contain two types of serines defined by their flanking amino acids. In phosphopeptide mapping and kinetic studies of the prototype SR protein SRSF1 (aka ASF/SF2), SRPK1 was shown to vigorously phosphorylate Arg-Ser dipeptides in long repeats whereas CLK1 modifies these along with isolated Ser-Pro dipeptides in the RS domain [22, 37] (Fig. 1). The latter modifications have been a subject of recent interest since it was shown that CLK1-dependent phosphorylation of these dipeptides triggers release of SRSF1 from nuclear speckles to the nucleoplasm [22], a critical trafficking event that leads to attachment of the SR protein to the nascent spliceosome and broad changes in alternative mRNA splicing. Indeed, CLK1 activity was shown in prior studies to mobilize GFP-fused SRSF1 from the interior of speckles to sites of transcription and pre-mRNA splicing outside the speckle [29]. Nonetheless, how Ser-Pro phosphorylation induces such movement is still not clear. CLK1 creates a migration change in SRSF1 on low percentage SDS-PAGE (gel shift) consistent with conformational changes that might alter interactions within the speckle. Although biological scaffolds have not been categorically identified as yet, several studies suggest RNA or the Son protein may provide a structural frame for assembly of nuclear speckles [38, 39]. SRSF1 is also known to self-associate, a process that is thought to be disrupted by RS domain phosphorylation [40]. This feature could be beneficial for regulating aggregation and speckle dynamics with or without a putative scaffold. Whatever the nature of this physicochemical signal, it has always been problematic that SRPK1 and CLK1 can induce diffusion of SRSF1 from speckles to the nucleoplasm although only one of the kinases can efficiently trigger Ser-Pro phosphorylation and both display different phosphorylation regiospecificities within the RS domain (Fig. 1).
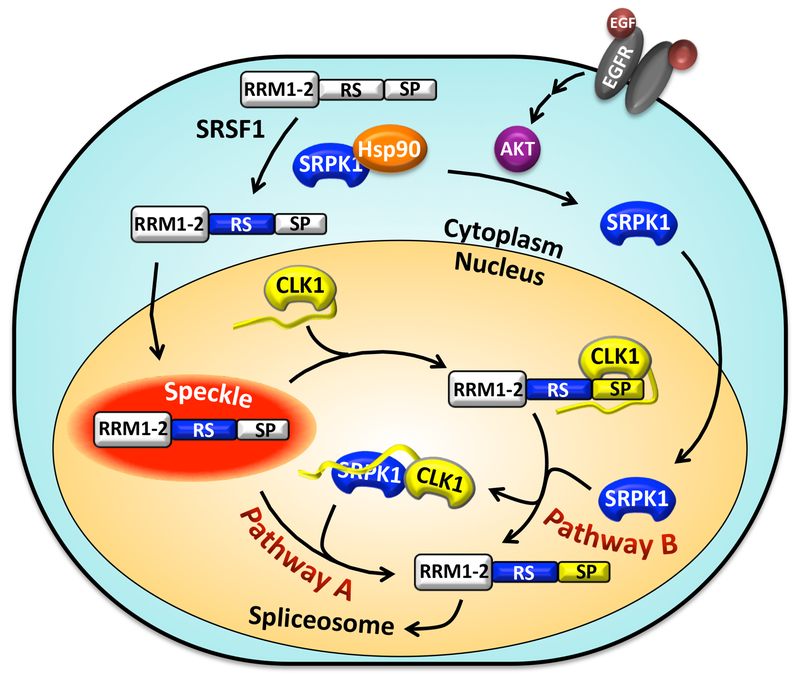
Using detailed kinetic, immunological and imaging techniques we can now assemble a mechanism whereby SRPK1 can induce SRSF1 mobilization from speckles to the nucleoplasm through the CLK1 kinase. In this scheme, CLK1 remains the principle catalyst for Ser-Pro phosphorylation and increased SRSF1 mobility, but formation of the SRPK1-CLK1 complex increases the probability of modification through two discrete pathways (Fig. 7). First, the SRPK1-CLK1 complex synergistically increases the overall phosphorylation rate of Ser-Pro dipeptides in SRSF1, the primary chemical means of SR protein mobilization (pathway A, Fig. 7). Interestingly, we found that active SRPK1 appears to enormously accelerate Ser-Pro phosphorylation by priming the RS domain with some phosphates prior to specific CLK1-dependent modification. Second, nuclear SRPK1 can dissociate SRSF1 from CLK1 protecting the RS domain from dephosphorylation by nuclear phosphatases such as PP1 (pathway B, Fig. 7). This is a surprising result since it is expected that the release of CLK1 should increase both solvent accessibility of the RS domain and phosphatase sensitivity. Such findings suggest that SRPK1-induced release allows SRSF1 to adopt a unique conformation that is more resistant to PP1. NMR studies have shown that RS domains are highly disordered in solution but phosphorylation induces some changes resembling an “arch-like” conformation [41]. Computational studies likewise predict that Arg-Ser repeats adopt a random conformation although phosphorylation may instead induce a “claw-like” structure [42]. We showed in previous studies that separate portions of the RS domain interact directly with RRM1 and RRM2 in SRSF1 but phosphorylation disrupts these interactions [13, 33]. Such changes may promote a unique SR protein conformation that breaks specific protein-protein interactions important for diffusion from speckles. Although SRPK1 is depicted as an activating agent outside the speckle in our model, we also observed SRPK1 in speckles particularly when CLK1 is down-regulated (Fig. 2A). Such findings suggest that SRPK1 may enter the speckle possibly helping to recruit CLK1 to the nucleoplasm through enhanced Ser-Pro phosphorylation. What drives this entry is not well understood but it is possible that interactions with hypo-phosphorylated SR proteins and/or CLK1 may be important for this. Whatever the structural underpinnings, our studies demonstrate that SRPK1 appears to play a vital role in stabilizing RS domain phosphates that promote SRSF1 dynamics in the nucleus.
Figure 7: Model for SRPK1-Stimulated Phosphorylation and Nuclear Mobilization of SRSF1 Through CLK1.
SRPK1 supports movement of SRSF1 from speckles to nucleoplasm using two pathways. In pathway A, the SRPK1-CLK1 complex directly enhances the phosphorylation rate of SRSF1. In pathway B, SRPK1 dissociates CLK1 from phospho-SRSF1 stabilizing Ser-Pro phosphates on the RS domain.
SR proteins are essential factors that initiate splice-site selection through the binding of key early spliceosomal components. RS domain phosphorylation is critical for the attachment of SR proteins to exonic sequences in precursor mRNA and for the recruitment of U1 snRNP to the 5’ splice site in the spliceosome [10, 11]. A recent advance to this model came with the observation that phospho-SRSF1 is less efficient for splicing activation owing to a tightly bound CLK1 that inhibits attachment to the 70K subunit of U1 snRNP [19]. Nuclear SRPK1 severs this inhibitory interaction by forming a complex with CLK1 freeing phospho-SRSF1 for splicing activation. At the molecular level, SRPK1 peals away the N-terminus of CLK1 from the RS domain of SRSF1, thereby breaking strong CLK1 interactions with the splicing factor [19] (Fig. 7). New findings presented herein suggest that SRPK1 provides a means for generating the free phospho-SRSF1 in the nucleoplasm for splicing function. Importantly, this mechanism not only can explain older observations regarding the role of expressed SRPK1 in diffusing SRSF1 from speckles but also can provide a more general means for explaining how physiological signals that activate SRPK1 in the nucleus (e.g., EGF) broadly impact mRNA splicing. We showed previously that chaperone contacts maintaining SRPK1 in the cytoplasm (e.g., Hsp90) are broken by EGF stimulation [21] (Fig. 7). Activated Akt displaces molecular chaperones from SRPK1, thereby removing the kinases’ cytoplasmic tethers. CLK1 then serves as a nuclear anchor until Akt is down-regulated and chaperones in the cytoplasm re-establish the resting equilibrium for SRPK1. Thus, cell signals that modulate the cytoplasmic and nuclear levels of SRPK1 are expected to strongly affect splicing through changes in the SRPK1-CLK1 complex and the phosphorylation states of SR proteins such as SRSF1. In addition to physiological signals that regulate SRPK1 and CLK1 for mRNA splicing in normal cells, these splicing-related kinases are often over-expressed in cancers and have been targeted with small molecule inhibitors for chemotherapeutic intervention [43, 44]. This new model offers potential insights into designing SRPK- and CLK-specific inhibitors for cancer therapy and interpreting such effects in light of not only the isolated kinase but also the kinase-kinase complex and its signaling implications.
Supplementary Material
Acknowledgments
Funding Sources:
This work was supported by NIH grants GM67969 and GM95828.
Abbreviations:
- CLK1
cdc2-like kinase 1
- PP1
protein phosphatase 1
- RRM
RNA recognition motif
- RS domain
domain rich in arginine-serine dipeptide repeats
- SR protein
splicing factor containing arginine-serine dipeptide repeats
- SRPK1
serine-arginine-specific protein kinase 1
- SRSF1
SR protein splicing factor 1 (aka ASF/SF2)
Footnotes
Competing Interests:
The Authors declare that there are no competing interests associated with the manuscript.
REFERENCES
- 1.Jurica MS and Moore MJ (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12, 5–14 [DOI] [PubMed] [Google Scholar]
- 2.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD and Ghosh G (2005) Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell 20, 77–89 [DOI] [PubMed] [Google Scholar]
- 3.Velazquez-Dones A, Hagopian JC, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD and Adams JA (2005) Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J. Biol. Chem 280, 41761–41768 [DOI] [PubMed] [Google Scholar]
- 4.Lai MC, Lin RI and Tarn WY (2001) Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. U. S. A 98, 10154–10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spector DL and Lamond AI (2011) Nuclear speckles. Cold Spring Harb. Perspect. Biol 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phair RD and Misteli T (2000) High mobility of proteins in the mammalian cell nucleus. Nature. 404, 604–609 [DOI] [PubMed] [Google Scholar]
- 7.Sacco-Bubulya P and Spector DL (2002) Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol 156, 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall LL, Smith KP, Byron M and Lawrence JB (2006) Molecular anatomy of a speckle. Anat. Rec. A Discov. Mol. Cell. Evol. Biol 288, 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR and Ghosh G (2011) Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U. S. A 108, 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA and Manley JL (1994) Protein-protein interactions and 5’-splice-site recognition in mammalian mRNA precursors. Nature. 368, 119–124 [DOI] [PubMed] [Google Scholar]
- 11.Wu JY and Maniatis T (1993) Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 75, 1061–1070 [DOI] [PubMed] [Google Scholar]
- 12.Mermoud JE, Cohen P and Lamond AI (1992) Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res 20, 5263–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serrano P, Aubol BE, Keshwani MM, Forli S, Ma CT, Dutta SK, Geralt M, Wuthrich K and Adams JA (2016) Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif. J. Mol. Biol 428, 2430–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA and Ghosh G (2008) A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell 29, 563–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu XD and Adams JA (2008) Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J. Mol. Biol 376, 55–68 [DOI] [PubMed] [Google Scholar]
- 16.Aubol BE and Adams JA (2011) Applying the brakes to multisite SR protein phosphorylation: substrate-induced effects on the splicing kinase SRPK1. Biochemistry. 50, 6888–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kataoka N, Bachorik JL and Dreyfuss G (1999) Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol 145, 1145–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun CY, Velazquez-Dones AL, Lyman SK and Fu XD (2003) Phosphorylation-dependent and -independent nuclear import of RS domain-containing splicing factors and regulators. J. Biol. Chem 278, 18050–18055 [DOI] [PubMed] [Google Scholar]
- 19.Aubol BE, Wu G, Keshwani MM, Movassat M, Fattet L, Hertel KJ, Fu XD and Adams JA (2016) Release of SR Proteins from CLK1 by SRPK1: A Symbiotic Kinase System for Phosphorylation Control of Pre-mRNA Splicing. Mol. Cell 63, 218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keshwani MM, Hailey KL, Aubol BE, Fattet L, McGlone ML, Jennings PA and Adams JA (2015) Nuclear protein kinase CLK1 uses a non-traditional docking mechanism to select physiological substrates. Biochem. J 472, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG and Fu XD (2012) The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 47, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshwani MM, Aubol BE, Fattet L, Ma CT, Qiu J, Jennings PA, Fu XD and Adams JA (2015) Conserved proline-directed phosphorylation regulates SR protein conformation and splicing function. Biochem. J 466, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui JF, Lane WS and Fu XD (1994) A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 369, 678–682. [DOI] [PubMed] [Google Scholar]
- 24.Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC and Fu XD (1998) SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol 140, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T and Fu XD (1996) SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem 271, 24569–24575 [DOI] [PubMed] [Google Scholar]
- 26.Aubol BE and Adams JA (2014) Recruiting a Silent Partner for Activation of the Protein Kinase SRPK1. Biochemistry. 53, 4625–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding JH, Zhong XY, Hagopian JC, Cruz MM, Ghosh G, Feramisco J, Adams JA and Fu XD (2006) Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol. Biol. Cell 17, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC and Duncan PI (1996) The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J 15, 265–275. [PMC free article] [PubMed] [Google Scholar]
- 29.Misteli T, Caceres JF and Spector DL (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature. 387, 523–527 [DOI] [PubMed] [Google Scholar]
- 30.Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura H, Furuichi K, Shibuya H, Krainer AR, Suzuki M and Hagiwara M (2004) Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem 279, 24246–24254 [DOI] [PubMed] [Google Scholar]
- 31.Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR and Hagiwara M (1999) The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs). J. Biol. Chem 274, 11125–11131. [DOI] [PubMed] [Google Scholar]
- 32.Yeakley JM, Tronchere H, Olesen J, Dyck JA, Wang HY and Fu XD (1999) Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol 145, 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aubol BE, Hailey KL, Fattet L, Jennings PA and Adams JA (2017) Redirecting SR Protein Nuclear Trafficking through an Allosteric Platform. J. Mol. Biol 429, 2178–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aubol BE, Plocinik RM, Hagopian JC, Ma CT, McGlone ML, Bandyopadhyay R, Fu XD and Adams JA (2013) Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J. Mol. Biol 425, 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M and Cooper DR (2009) Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology. 150, 2087–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA and Hagiwara M (2006) Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. U. S. A 103, 11329–11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma CT, Hagopian JC, Ghosh G, Fu XD and Adams JA (2009) Regiospecific phosphorylation control of the SR protein ASF/SF2 by SRPK1. J. Mol. Biol 390, 618–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma A, Takata H, Shibahara K, Bubulya A and Bubulya PA (2010) Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 21, 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shevtsov SP and Dundr M (2011) Nucleation of nuclear bodies by RNA. Nat. Cell Biol 13, 167–173 [DOI] [PubMed] [Google Scholar]
- 40.Xiao SH and Manley JL (1998) Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J 17, 6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang S, Gapsys V, Kim HY, Bessonov S, Hsiao HH, Mohlmann S, Klaukien V, Ficner R, Becker S, Urlaub H, Luhrmann R, de Groot B and Zweckstetter M (2013) Phosphorylation drives a dynamic switch in serine/arginine-rich proteins. Structure. 21, 2162–2174 [DOI] [PubMed] [Google Scholar]
- 42.Hamelberg D, Shen T and McCammon JA (2007) A proposed signaling motif for nuclear import in mRNA processing via the formation of arginine claw. Proc. Natl. Acad. Sci. U. S. A 104, 14947–14951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gammons MV, Lucas R, Dean R, Coupland SE, Oltean S and Bates DO (2014) Targeting SRPK1 to control VEGF-mediated tumour angiogenesis in metastatic melanoma. Br. J. Cancer 111, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Araki S, Dairiki R, Nakayama Y, Murai A, Miyashita R, Iwatani M, Nomura T and Nakanishi O (2015) Inhibitors of CLK protein kinases suppress cell growth and induce apoptosis by modulating pre-mRNA splicing. PLoS One. 10, e0116929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.