Abstract
Recent remarkable advances in genome sequencing have enabled detailed maps of identified and interpreted genomic variation, dubbed “mutanomes.” The availability of thousands of exome/genome sequencing data has prompted the emergence of new challenges in the identification of novel druggable targets and therapeutic strategies. Typically, mutanomes are viewed as one- or two-dimensional. The three-dimensional protein structural view of personal mutanomes sheds light on the functional consequences of clinically actionable mutations revealed in tumor diagnosis and followed up in personalized treatments, in a mutanome-informed manner. In this review, we describe the protein structural landscape of personal mutanomes and provide expert opinions on rational strategies for more streamlined oncological drug discovery and molecularly targeted therapies for each individual and each tumor. We provide the structural mechanism of orthosteric versus allosteric drugs at the atom-level via targeting specific somatic alterations for combating drug resistance and the “undruggable” challenges in solid and hematologic neoplasias. We discuss computational biophysics strategies for innovative mutanome-informed cancer immunotherapies and combination immunotherapies. Finally, we highlight a personal mutanome infrastructure for the emerging development of personalized cancer medicine using a breast cancer case study.
I. Introduction: Personal Cancer Mutanome Defined
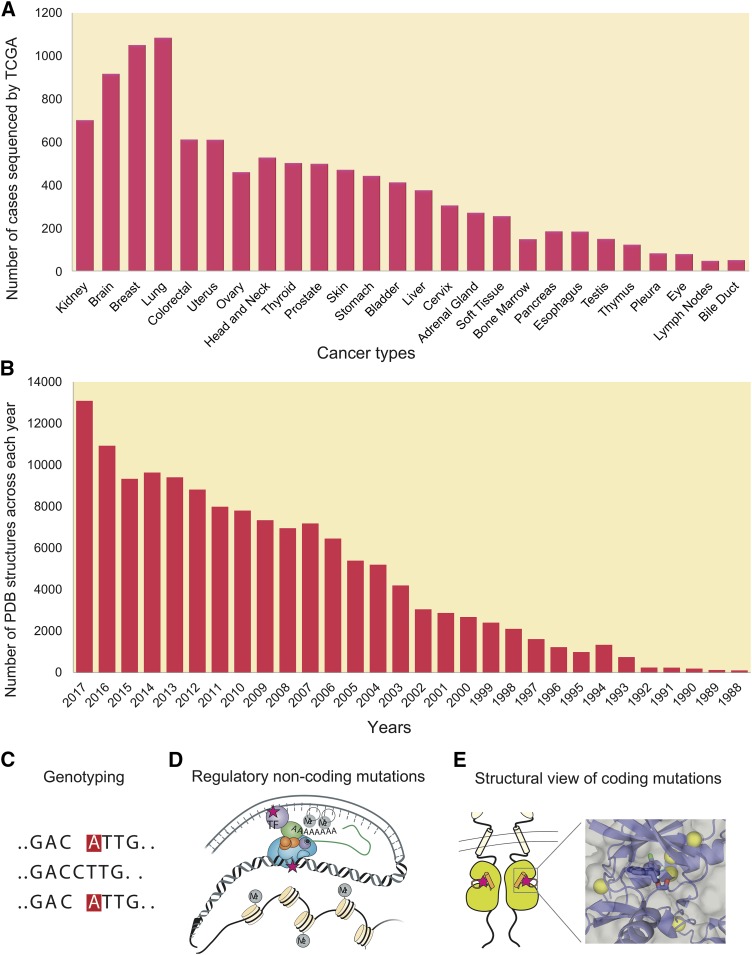
In 2004, the complete sequence of the human genome (NCBI Build 35) containing approximately 2.85 billion nucleotides was reported at an estimated cost of $2.7 billion (International Human Genome Sequencing Consortium, 2004). Owing to remarkable advances of next-generation sequencing technologies, one human genome can now be sequenced at an estimated cost of less than $1000 (Hayden, 2014; Telenti et al., 2016). Multi-center cancer genome projects (Fig. 1A), such as The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC), have significantly improved our understanding of the landscape of somatic alterations by identifying frequently mutated protein-coding genes (Garraway and Lander, 2013; Vogelstein et al., 2013; Martincorena and Campbell, 2015). However, sequencing data from thousands of exome and genomes have now led to newly emerging challenges in the field of oncology, such as how to translate this massive genomic data into recommendations for patient care, how to match the right drugs at the right dose to the right patients rapidly, and how to increase the coverage of molecularly targeted agents that can specifically target actionable mutations. A recent Molecular Analysis for Therapy Choice (NCI-MATCH) trial funded by the US National Cancer Institute suggests that only 2.5% of the patients could be matched with a targeted therapy based on a cohort of 795 enrolled patients with relapsed solid tumors and lymphoma as of May 2016 (http://ecog-acrin.org/nci-match-eay131/interim-analysis). These are compounded by the increase in the number of reported loss-of-function mutations in tumor suppressor genes (e.g., TP53 and PTEN) and gain-of-function mutations in “undruggable” (Table 1) master regulators or oncogenes (e.g., myelocytomatosis and KRAS proto-oncogene, GTPase [KRAS]), underscoring an urgent need to identify new druggable pathways and therapeutic anticancer agents.
Fig. 1.
Diagram illustrating development of structural genomics and cancer genomes. (A) Number of tumor genomes sequenced by The Cancer Genome Atlas across 26 major cancer types from Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov). (B) Number of PDB structures of human proteins from 1988 to 2017 from PDB database (https://www.rcsb.org). (C) Genotyping by next-generation sequence technology. (D) Regulatory non-coding mutations in cancer. (E) Protein structural view of coding mutations in cancer.
TABLE 1.
Definitions of some key words
| Key Word | Definitions |
|---|---|
| Allosteric site | A regulatory site on the protein’s surface is distinct from the substrate, ligand, or partner binding sites (Nussinov and Tsai, 2015). |
| Clinically actionable mutation | A mutation alters clinical responses (e.g., survival or drug responses) in patients harboring this mutation. |
| Driver mutation | A mutation directly or indirectly promotes a selective growth or survival to the cell in which it occurs. |
| Edgetic alleles | Genetic alterations (mutations) alter specific macromolecular interactions (“edges”) rather than affecting folding and stability of proteins (Sahni et al., 2015). Edgetic mutations include “node” removal by truncating mutations (Zhong et al., 2009) and in-frame edgetic mutations that disrupt interactions between proteins, DNA, or RNA. |
| Network-attacking mutations | Mutations alter signaling networks via different types of network perturbations: signaling network dynamics, network structure, and dysregulation of phosphorylation sites (Creixell et al., 2015). |
| Orthosteric site | The primary, unmodulated binding site (on a receptor) of a ligand, such as adenosine triphosphate (ATP) binding site of a kinase. |
| Personal mutanome | A personal mutanome is a portfolio of DNA sequencing data (e.g., whole exome or whole genome), protein structural genomics, and interpretation of mutational landscape of an individual patient. |
| Personalized medication | Interventions or/and products are tailored to the individual patients based on their predicted response or risk (e.g., particularly genomic or molecular profiles as clinically actionable biomarkers) of disease. |
| Spliceosome mutations | Hotspot somatic mutations affect genes encoding RNA splicing factors (Dvinge et al., 2016). |
| Undruggable | A protein could not be targeted pharmacologically. More appropriate terms might be “difficult to drug” or “yet to be drugged.” |
Structural genomics technologies provide an increasing number of high-quality protein three-dimensional structures. The Structural Genomics Consortium successfully demonstrated the integration of the three-dimensional structures of proteins and genomic data for innovative therapeutic strategies for complex diseases, including cancer (http://www.thesgc.org/). The great numbers of deciphered human genomes and innovative computational tools have enabled progress in rapid identification of genetic variation in individual cancer samples.
A personal mutanome (Table 1) offers unique opportunities for understanding the functional consequences of somatic alterations in a single patient and for developing personalized cancer treatment. In this article, we consider a personal mutanome, an emerging personalized medicine infrastructure, for accelerating the development of precision oncology. We will discuss four potential points of clinical utility from deriving personal mutanomes:
Precision Prevention and Diagnostics: From Sequence to Consequence of Personal Mutanome in Cancer. We illustrate the protein structural landscape of druggable cancer proteomes with well-annotated hotspot mutations in cancer, which highlights the successes of the application of genomics in personalized cancer medicine.
Personalized Oncology Drug Discovery. We discuss the protein structural mechanism of orthosteric versus allosteric drugs targeting specific somatic alterations (e.g., KRAS-G12C, EGFR-C797C, and HER2-T798I) for overcoming drug resistance and undruggable challenges in various solid and hematologic malignancies, which inform the usefulness of genomic technologies in personalized oncology drug discovery and development.
Personalized Immunotherapy. We describe computational biophysics approaches that enable development of new neoantigen-focused cancer immunotherapies or combination immunotherapies.
Precision Cancer Medicine and Patient Care. We highlight a personal mutanome infrastructure for patient stratification in accelerating personalized treatment using breast cancer as a case study.
A. Coding Versus Non-coding Personal Mutanomes in Cancer
Although the protein-coding regions of the human genome accounts for ∼2% of the total sequence, variants in protein-coding regions have received the most attention. For instance, whole-exome sequencing is well regarded as a reliable and cost-effective approach to explore somatic mutation profiles in protein-coding genes. Several national and international genome projects have identified numerous coding driver mutations on several highly mutated genes, such as TP53, RAS subfamily, and the ERBB family, which promote tumor initiation and progression. With the remarkable advances of whole-genome sequencing technologies, many studies have demonstrated the importance of non-coding variants in cancer (Weinhold et al., 2014; Califano and Alvarez, 2017; Zhang et al., 2018). Mechanistically, recurrent non-coding mutations commonly can create new binding motifs for transcription factors and lead to dysregulation of gene regulatory networks (Fig. 1D). For example, several earlier studies found frequent promoter mutations on telomerase reverse transcriptase and succinate dehydrogenase complex subunit D in melanoma and pancreatic ductal adenocarcinoma and these mutations are associated with reduced gene expression and poor survival rates (Weinhold et al., 2014; Feigin et al., 2017). Here, we focus on structural mechanisms (Fig. 1E) of protein-coding somatic mutations that are involved in tumor initiation, disease progression, and drug responses. The functional machinery of non-coding variants in cancer can be found in several recent reviews (Albert and Kruglyak, 2015; Khurana et al., 2016).
B. Personal Mutanomes in Modern Oncology Drug Discovery
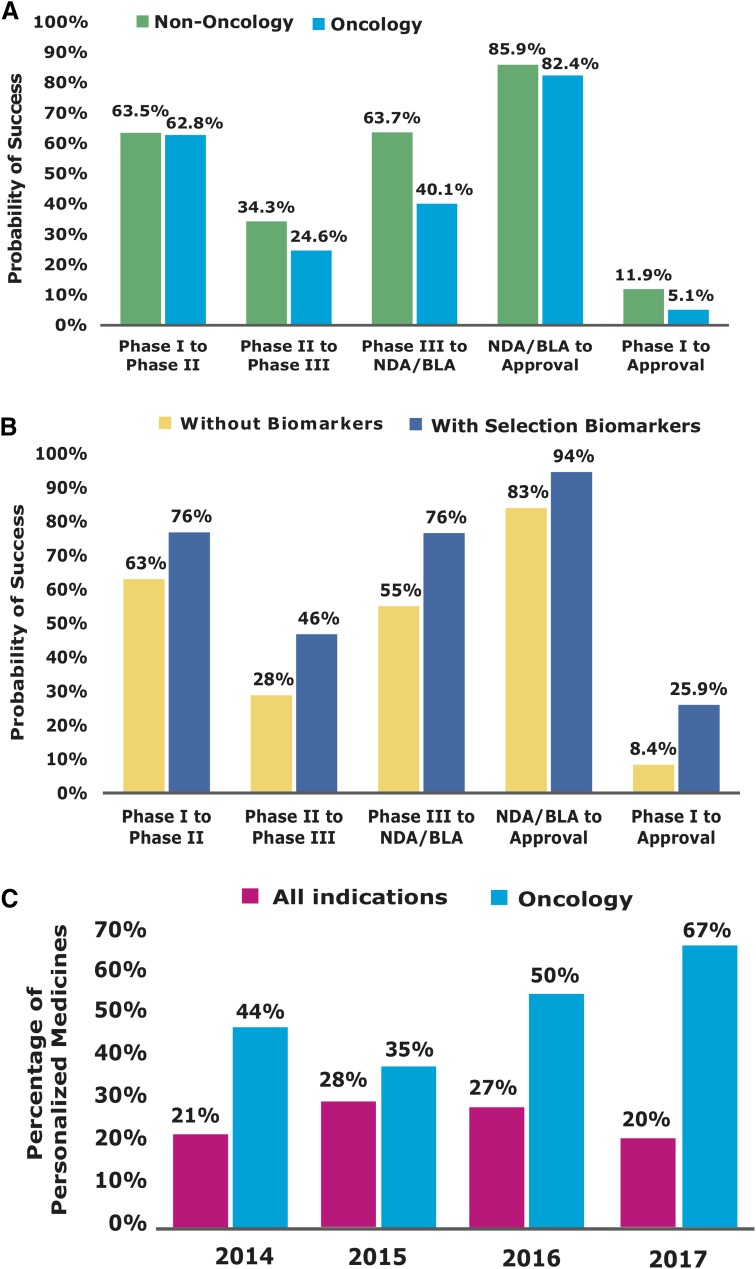
Over the last several decades, the application of genomics to inform drug discovery has generated some successes (Dugger et al., 2018). Getting a drug to the market involves four successfully clinical trial phases (Phases I–IV). The first (Phase I) focuses on drug safety in a small group of patients. The second (Phase II) assesses drug safety and efficacy in a larger group. The third (Phase III), prior to US Food and Drug Administration (FDA) approval, measures efficacy in patient versus placebo populations. Phase IV trials are commonly performed after approval to re-examine safety and efficacy in a larger population as well as potential side effects via postmarketing surveillance. Traditionally, clinical trials survey only few measures from thousands of participants by evaluating the average responses to therapy, which results in high failure rate and soaring cost (Schork, 2015). Figure 2 shows the lowest success rate of traditional drug discovery in oncology compared with other diseases from Phase I to approval. Biomarkers that can be determined in the context of disease diagnosis and clinical intervention have been integrated into the drug discovery protocol and the design of the clinical trials. Recent data show that biomarker-derived personalized clinical trials achieve higher success rate (25.9%) in oncology compared with other therapeutic areas (8.4% success rate shown in Fig. 2B). Table 2 summarizes the US FDA-approved personalized medications for highly targeted therapies in cancer from 2012 to 2017. These approved medications maximize clinical efficacy while minimizing side effects. Half of the FDA-approved oncology drugs in 2016 (Fig. 2C) are personalized medications (Table 1), indicating the great promise of personalized oncology drug discovery.
Fig. 2.
Survey of personalized oncology drug discovery. (A) Oncology vs. non-oncology phase transition success rate. (B) Biomarker-based phase transition success rates. (C) Success rate of personalized medicines approved by FDA from 2014 to 2017. BLA, Biologic License Application; NDA, New Drug Application. Data collected from BIO Industry analysis (https://www.bio.org/bio-industry-analysis-published-reports) and FDA website (https://blogs.fda.gov/fdavoice/index.php/2015/03/fda-continues-to-lead-in-precision-medicine).
TABLE 2.
Lists of US FDA-approved personalized oncological medications during 2012–2017
Data comes from www.fda.gov/drugs.
| Drug Name | Company | Active Ingredient | Approval Date | Indication | Biomarkers |
|---|---|---|---|---|---|
| Verzenio | Eli Lilly | Abemaciclib | 09/28/2017 | Breast cancer | HR+ and HER2- |
| Idhifa | Celgene | Enasidenib | 08/01/2017 | Acute myeloid leukemia (AML) | IDH2 mutation |
| Nerlynx | Puma Biotechnology | Neratinib | 07/17/2017 | Breast cancer | HER2-amplified |
| Rydapt | Novartis | Midostaurin | 04/28/2017 | AML | FLT3+ |
| Alunbrig | Takeda | Brigatinib | 04/28/2017 | Non-small cell lung cancer (NSCLC) | ALK+ |
| Zejula | Tesaro | Niraparib | 03/27/2017 | Recurrent epithelial ovarian, fallopian tube, or primary peritoneal cancer | BRCA1, BRCA2 |
| Kisqali | Novartis | Ribociclib | 03/14/2017 | Metastatic breast cancer | HR+, HER2- |
| Imfinzi | AstraZeneca | Durvalumab | 03/01/2017 | Metastatic urothelial carcinoma | PD-L1 |
| Rubraca | Clovis Oncology | Rucaparib | 12/19/2016 | Ovarian cancer | BRCA1, BRCA2 |
| Lartruvo | Eli Lilly | Olaratumab | 10/19/2016 | Soft tissue sarcoma | PDGFRA+ |
| Tecentriq | Genentech | Atezolizumab | 5/18/2016 | Bladder cancer | PD-L1 |
| Venclexta | AbbVie and Genentech | Venetoclax | 4/11/2016 | Chronic lymphocytic leukemia (CML) | 17p-deleption |
| Alecensa | Roche and Genentech | Alectinib | 12/11/2015 | Metastatic NSCLC | ALK |
| Cotellic | Genentech | Cobimetinib | 11/10/2015 | Metastatic melanoma | BRAF-V600E/K |
| Unituxin | United Therapeutics | Dinutuximab | 3/10/2015 | Neuroblastoma | MYCN-amplification |
| Tagrisso | AstraZeneca | Osimertinib | 11/13/2015 | NSCLC | EGFR-T790M |
| Ibrance | Pfizer | Palbociclib | 2/3/2015 | Metastatic breast cancer | ER+/HR+, HER2- |
| Opdivo | Bristol-Myers Squibb | Nivolumab | 12/22/2014 | Metastatic melanoma | BRAF-V600 |
| Lynparza | AstraZeneca | Olaparib | 12/19/2014 | Metastatic ovarian cancer | BRCA1, BRCA2 |
| Keytruda | Merck | Pembrolizumab | 9/4/2014 | Metastatic melanoma | PD-L1 positive |
| Zykadia | Novartis | Ceritinib | 4/29/2014 | Metastatic NSCLC | ALK+ |
| Imbruvica | Janssen | Ibrutinib | 11/13/2013 | Mantle cell lymphoma (MCL) | 17p-deleption |
| Gazyva | Genentech | Obinutuzumab | 11/1/2013 | Chronic lymphocytic leukemia (CLL) | MS4A1 (CD20+) |
| Gilotrif | Boehringer Ingelheim | Afatinib | 7/12/2013 | Metastatic NSCLC | EGFR |
| Tafinlar | GlaxoSmithKline | Dabrafenib | 5/29/2013 | Metastatic melanoma | BRAF-V600E/K |
| Mekinist | GlaxoSmithKline | Trametinib | 5/29/2013 | Metastatic melanoma | BRAF-V600E/K |
| Kadcyla | Genentech | Ado-trastuzumab emtansine | 2/22/2013 | Metastatic breast cancer | HER2+ |
| Iclusig | Ariad Pharmaceuticals | Ponatinib | 12/14/2012 | CML | BCR-ABL1, T315I |
| Cometriq | Exelixis | Cabozantinib | 11/29/2012 | Medullary thyroid cancer | RET |
| Synribo | Teva Pharmaceutical | Omacetaxine mepesuccinate | 10/26/2012 | CML | BCR-ABL1 |
| Bosulif | Pfizer | Bosutinib | 9/4/2012 | CML | BCR-ABL1 |
| Perjeta | Genentech | Pertuzumab | 6/8/2012 | Metastatic breast cancer | HER2+ |
ALK, anaplastic lymphoma kinase; BRCA, breast cancer; FLT3, fms like tyrosine kinase 3; HR+, hormone receptor-positive; MS4A1, membrane spanning 4-domains A1; MYCN, N-myc proto-oncogene protein; PDGFRA, platelet-derived growth factor receptor A; RET, ret proto-oncogene.
C. Personal Mutanomes in Molecularly Targeted Therapies
In 2003, Francis S. Collins, Director of the National Institutes of Health, penned the concept of personalized medicine: “The sequencing of the human genome, along with other recent and expected achievements in genomics, provides an unparalleled opportunity to advance our understanding of the role of genetic factors in human health and disease, to allow more precise definition of the non-genetic factors involved, and to apply this insight rapidly to the prevention, diagnosis and treatment of disease (Collins et al., 2003).” In January of 2015, US President Barack Obama announced the Precision Medicine Initiative (Collins and Varmus, 2015). The basic idea of precision oncology is to tailor drugs based on the genomic or molecular profile of the tested tumor, such as epidermal growth factor receptor (EGFR)-T790M in non-small cell lung cancer (NSCLC) (Pao and Hutchinson, 2012), BRAF-V600E in melanoma (Sun et al., 2014), HER2 amplification in breast cancer (Arteaga and Engelman, 2014), BCR-ABL fusion oncoprotein in chronic myeloid leukemia (CML), and Philadelphia chromosome-positive acute lymphoblastic leukemia (Shet et al., 2002) (Table 2). Generally speaking, there are two types of small molecule agents marketed for targeted cancer therapies at protein structural perspectives: orthosteric and allosteric. A large number of highly specific small molecule agents have been approved for molecularly targeted therapies to orthosteric ATP-binding sites. For example, Gleevec (Imatinib) is the first FDA-approved drug for CML patients with tumors displaying BCR-ABL fusion, with more than 80% of the patients treated by Imatinib surviving for over 10 years (Kalmanti et al., 2015). Despite promising clinical evidence against mutated tumors in the early months, acquired resistance arises rapidly owing to “gatekeeper” mutations in the ATP-binding sites, such as ABL1-T315I (Pemovska et al., 2015), EGFR-T790M (Ke and Wu, 2016), and HER2-T798I/T798M (Rexer et al., 2013; Hanker et al., 2017). Compared with orthosteric drugs with off-target toxicity resulting from low specificity, allosteric drugs are promising candidates for targeted therapies with a higher specificity (Nussinov and Tsai, 2015; Moslehi, 2016). Recent studies of allosteric drugs pointed to promising preclinical evidence of activity against tumor cells with newly acquired somatic mutations, such as KRAS-G12C (Ostrem et al., 2013), EGFR-C797C (Jia et al., 2016), HER2-T798I (Hanker et al., 2017), and BCR-ABL1 fusion (Wylie et al., 2017). Here we describe structural mechanisms of allosteric drugs at the atomic level for combating the drug resistance and off-target side effects of traditional orthosteric agents in the individuals.
D. Personal Mutanomes in Cancer Immunotherapy
In January of 2016, Obama announced the Cancer Moonshot 2020 initiative, a nationwide effort to accelerate the testing of immunotherapy drug combination to fight cancer by exploiting the promise of precision oncology. Immunotherapies (e.g., immune checkpoint inhibitors), which turn the body’s immune system against cancerous cells, boost clinical outcomes in several cancers, such as advanced melanoma (Ledford, 2016). Ipilimumab is the first FDA-approved anti-cytotoxic T-lymphocyte-associated antigen 4 antibody and nivolumab is the first FDA-approved anti-programmed death-1 (PD-1) antibody (Mahoney et al., 2015). Mechanistically, cytotoxic T-lymphocyte-associated antigen 4 or PD-1 inhibitors do not activate the immune systems to destroy tumor cells; rather, they turn off inhibitory pathways that block effective antitumor T-cell responses (e.g., cytotoxic T lymphocytes) (Mahoney et al., 2015). A recent clinical study showed that ipilimumab and nivolumab dramatically improved the response rates in patients with advanced melanoma from 19% with ipilimumab alone to 58% with a combination (Larkin et al., 2015). However, the overall response rate of immunotherapy alone is highly variable across different cancer types (Sharma and Allison, 2015). Furthermore, the potential immune-related adverse events (e.g., autoimmune lethal cardiotoxicity), especially with combination immunotherapies, have discouraged some investors and cancer immunologists in the cancer immunotherapy community (Ledford, 2016; Cheng and Loscalzo, 2017).
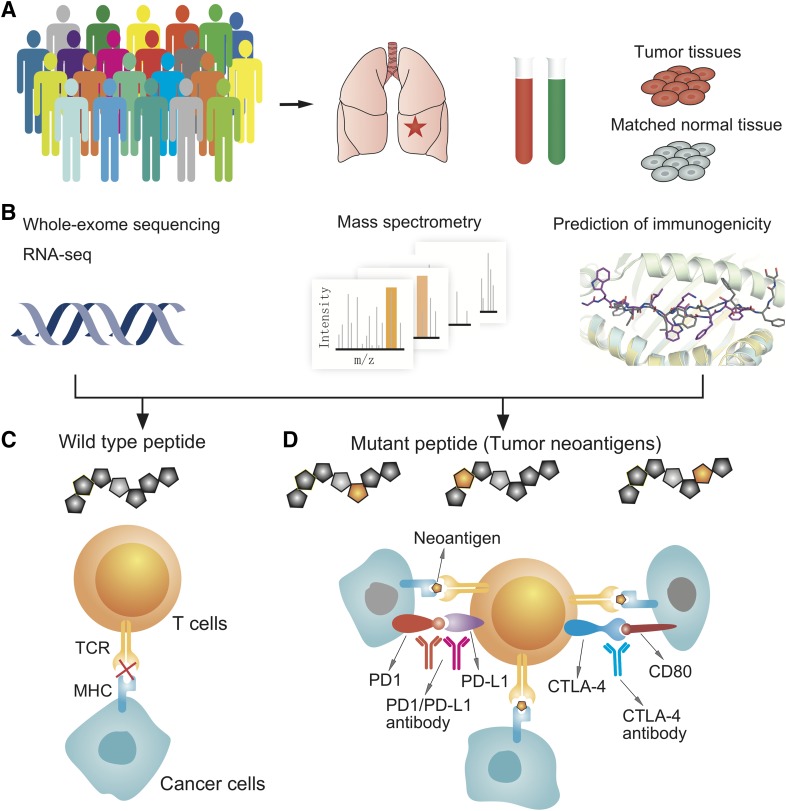
More recently, cancer vaccines have focused on targeting neoantigens, which are new peptides derived from tumor-specific mutations, not found in normal tissues. Compared with traditional tumor-associated self-antigens, neoantigen-focused immunotherapies induce lower toxicity than autoimmune reactions in normal cells. Two recent studies suggested the potential clinical benefits of personalized neoantigen vaccine in melanoma patients by exploiting the personal mutanome (Ott et al., 2017; Sahin et al., 2017). The central question in neoantigen identification is which mutant proteins are processed into 8- to 11-residue peptides by the proteasome and bind to the major histocompatibility complex class I (MHC-I) for recognition by CD8+ T-cells (Fig. 3). Computational biophysics strategies have been reported for identification of new neoantigens, focusing on which peptides bind to the MHC-I molecules (Hackl et al., 2016). Current neoepitope prediction approaches generate a vast number of predicted candidates, of which only a short list is ever found to be immunogenic in patients. Additional research is required to address these challenges, such as that by the Tumor Neoantigen Selection Alliance (Nature Publishing Group, 2017). Recent studies have provided proof-of-principle for computational biophysics approaches, such as neoepitope-focused vaccine design in infectious diseases (Correia et al., 2014). Below, we will briefly discuss recent computational biophysics approaches to develop neoantigen-focused cancer immunotherapies and combination immunotherapies. Detailed description of computational tools for cancer immunotherapies can be found in a recent review (Hackl et al., 2016).
Fig. 3.
A diagram illustrating computational approaches for development of personalized immunotherapies. (A) Collection of patient samples (both tumor samples and matched samples). (B) An integrated approach for identification of actionable biomarkers using innovative genomics approaches, proteomics, and computational biophysics. (C and D) Guiding the application of personalized immunotherapies or combination immunotherapies that highly specifically target neoantigens derived from tumor somatic mutations. CTLA-4, cytotoxic T-lymphocyte-associated protein 4; MHC, major histocompatibility complex; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; TCR, T-cell receptor.
II. Computational Resources and Tools for Personal Mutanomes
Insights into how genes, proteins, and their networks give rise to normal, precancerous, and cancer cellular phenotypes requires large, experimentally derived data sets. Computational investigations are essential, and increasingly, there is a need for collection, storage, and display of the data in robust databases. To understand better the biologic and functional consequences of personal mutanomes, there is a pressing need for computational identification and interpretation of how specific mutations affect tumorigenesis and drug responses. Testable hypotheses derived from these computational models and refinement of models based on new experimental data are essential for development of personalized cancer medicine.
A. Bioinformatics Resources
In 2005, TCGA was launched as the major national cancer genome project for accelerating a comprehensive understanding of the genetic basis and molecular mechanism of cancers, with multidimensional data of >11,000 patients (whole exomes/genomes) across 33 cancer types released by the end of 2016 (Fig. 1A) (Tomczak et al., 2015). In 2008, ICGC launched the biggest international cancer genome project with 88 committed projects to generate a comprehensive catalog of genomic abnormalities of over 25,000 cancer genomes across 50 cancer types/subtypes (Milius et al., 2014). The Catalogue Of Somatic Mutations In Cancer (COSMIC) is the most comprehensive database of somatic mutations in cancer. As of September 2017 (v82), COSMIC contains 4,835,986 coding mutations, 18,809 fusions, and 1,180,789 copy number variants by curating cancer genomics data from 1,326,347 (Forbes et al., 2015; Nakagawa et al., 2015). The cBioPortal was developed by the Memorial Sloan-Kettering Cancer Center to systematically visualize and analyze the cancer genome landscape. As of December 2017, cBioPortal curated genomic data for over 30,000 samples from 164 cancer studies (Gao et al., 2013). The PDB is the global archive for experimentally determined, atomic-level three-dimensional structures of biologic macromolecules (Rose et al., 2017). As of September 2017, the PDB contains 133,759 proteins, DNA, or RNA structures, including 37,284 from Homo sapiens (Fig. 1B). A detailed description of the bioinformatics resources discussed here is summarized in Table 3.
TABLE 3.
Lists of computational tools and bioinformatics resources for analysis of personal mutanomes in cancer
| Names | Description | Website | Refs | |
|---|---|---|---|---|
| Cancer genomics resources | ||||
| TCGA and GDC | The Cancer Genome Atlas and Genomic Data Commons Data Portal | https://portal.gdc.cancer.gov | Chin et al. (2011) | |
| ICGC | The International Cancer Genome Consortium | https://icgc.org | Milius et al. (2014) | |
| CBioPortal | Providing visualization, analysis and download of large-scale cancer genomics data sets. | http://www.cbioportal.org | Gao et al. (2013) | |
| COSMIC | Comprehensive resources for curating somatic mutations | http://cancer.sanger.ac.uk/cosmic | Forbes et al. (2015) | |
| Bioinformatics resources for protein structures | ||||
| PDB | The PDB is the global archive for experimentally determined, atomic-level three-dimensional structures of proteins, DNA, and RNA. | https://www.rcsb.org/ | Rose et al. (2017) | |
| Interactome3D | Manually curated protein-protein interactions with known three-dimensional structure information. | http://interactome3d.irbbarcelona.org | Mosca et al. (2013) | |
| dSysMap | dSysMap is a useful tool to study the network perturbations by genetic variants | http://dsysmap.irbbarcelona.org/ | Mosca et al. (2015) | |
| Interactome INSIDER | An integrative structural and genomic resource for functional exploration of human disease mutations at multiple-scale, proteome-wide human interactome | http://interactomeinsider.yulab.org | Meyer et al. (2018) | |
| Mutational clustering tools on protein structures | ||||
| AlloDriver | Detect druggable mutations via dysregulation of protein allosteric functions. | NA | Shen et al. (2017) | |
| Cancer3D | A useful tool to explore potential cancer drivers or pharmacogenomic biomarkers using protein structure information. | http://cancer3d.org/ | Porta-Pardo et al. (2015) | |
| CLUMPS | Assess the significance of mutational clustering in a given 3D structure. | NA | Kamburov et al. (2015) | |
| HotSpot3D | Detect mutation–mutation and mutation–drug clusters using three-dimensional protein structures. | NA | Niu et al. (2016) | |
| KNMPx | Detect a kinome-wide pharmacogenomic biomarkers via rewiring phosphorylation-related signaling networks and drug sensitivity/resistance | NA | Zhao et al. (2017) | |
| ReKINect | Detect network-attacking mutations in phosphorylation-based signaling networks. | http://rekinect.science/ | Creixell et al. (2015) | |
| SGDriver | A structural genomics-based method that detects mutation clustering on protein–ligand binding site residues via a Bayes inference statistical framework. | NA | Zhao et al. (2016) | |
B. Computational Tools for Personal Mutanomes
1. Mutation Clustering on Protein Structures
CLUMPS (clustering of mutations in protein structures) is a statistical method that detects an overall enrichment of somatic mutant residues that are spatially close to each other in the protein three-dimensional (3D) structure (Kamburov et al., 2015). Applying CLUMPS to 4742 tumors having TCGA whole-exome sequencing data and human 3D protein structures in the PDB identified well-known onco- and tumor-suppressor proteins harboring significant 3D clustering of missense somatic mutations. Several similar computational tools were further developed, such as HotSpot3D, SGDriver, and AlloDriver (Table 3). HotSpot3D extends the 3D clustering hypothesis to detect mutation-mutation and mutation-drug clusters in protein structures and correlates these clusters with known or potentially interacting functional variants, domains, and proteins. HotSpot3D offers a useful tool to identify druggable mutations enriched in ligand-protein binding pockets. SGDriver analysis of 746,631 missense mutations observed in approximately 5000 tumor-normal pairs across 16 cancer types from TCGA, reveals that 13% of the patients might benefit from current targeted therapies (Zhao et al., 2016). This proportion would increase to 31% using drug repositioning strategies (Cheng et al., 2012, 2016, 2018; Jiang et al., 2018), offering a testable strategy to prioritize druggable mutations for personalized treatments in cancer. Finally, AlloDriver (Shen et al., 2017) offers a powerful tool for detection of druggable missense somatic mutations clustered on allosteric sites (Table 1).
2. Mutation Clustering on the Human Interactome Network
Genes and proteins do not function independently. They participate in complex, interconnected networks and pathways in human cells. Perturbations to those networks may promote cells to new disease status, for example, tumorigenesis (Cheng et al., 2014). Understanding how somatic mutations perturb these networks is still missing. A new conceptual paradigm that incorporates both innovative experimental and computational strategies, such as edgetic mutations (Sahni et al., 2015) or “network-attacking” mutations (Cheng et al., 2014; Creixell et al., 2015) (Table 1), may allow better assessment of the intrinsic complexities of cancer cells at their network perturbation level. A pan-cancer network analysis reveals a positive correlation of protein connectivity in the human protein-protein interactome with the number of nonsynonymous somatic mutations, whereas a weaker or insignificant correlation exists with the number of synonymous somatic mutations, suggesting conceptual network-attacking perturbation by somatic mutations (Cheng et al., 2014). The phosphorylation-dependent signaling network plays a crucial role in tumorigenesis and drug responses in cancer. Phosphorylation sites have been clustered to examine the kinome-wide landscape of pharmacogenomic interactions. KNMPx (kinome-wide network module for cancer pharmacogenomics) offers a powerful tool for identifying oncogenic alterations by rewiring phosphorylation-related signaling networks that involve patient survival rate and drug sensitivity/resistance (Zhao et al., 2017). ReKINect detects network-attacking mutations in phosphorylation-based signaling networks (Creixell et al., 2015). By analyzing the exomes and quantifying (phospho-)proteomes of five ovarian cancer cell lines and of the global cancer genome repository, ReKINect identifies several network-attacking mutations that encode specificity switches analogous to de novo appearances of kinases within the kinome (Creixell et al., 2015).
Comprehensive investigation of the functional consequences of somatic alterations on the structurally resolved human protein-protein interactome has offered unexpected opportunities for better understanding the molecular mechanism and the genetic basis of tumorigenesis and tumor progression (Wang et al., 2012). Interactome3D is a comprehensive resource for structural annotation and modeling of the protein-protein interactions in human cells, offering a useful tool for exploring the functional consequences of somatic mutations on the 3D human interactome (Mosca et al., 2013). The dSysMap developed by the same group is a useful tool to study network perturbations by disease mutations, including somatic mutations in cancer (Mosca et al., 2015). However, there are only ∼4000 resolved structural, human protein-protein interactions, limiting global investigation of cellular network perturbations on the proteome scale. Interactome INSIDER is a comprehensive resource for the human protein-protein interactome with well-annotated interaction interface information, including both structurally resolved and large-scale computationally predicted protein-protein interactions using an ensemble machine-learning algorithm (Meyer et al., 2018). Integration of high-throughput experimental assays and computational strategies represent promising strategies for studies of the functional consequences of personal mutanomes from the human protein-protein interactome network prospective. However, understanding biologic networks, pathways, and broadly molecular systems will require dynamic information at multi-dimensional levels, including cells, tissues, organs, and organisms, which are missing in current experimental and computational approaches. Development of new experimental and computational techniques that allow in vivo, real-time, cell/tissue-specific measurements of protein or gene expression, localization, modification, and kinetics are urgently needed.
3. Mutation Clustering on Spliceosome
Recent advances in DNA-seq and RNA-seq data analyses have suggested that hotspot somatic mutations affect genes encoding RNA splicing factors, dubbed “spliceosomal mutations” (Table 1) (Dvinge et al., 2016). Several commonly mutated spliceosomal proteins have been involved in solid and hematologic tumors, such as U2 small nuclear RNA auxiliary factor 1 (U2AF1) (Brooks et al., 2014) and splicing factor 3B, subunit 1 (SF3B1) (Alsafadi et al., 2016; Wang et al., 2016). For example, a pan-cancer analysis has identified splicing changes associated with U2AF1-S34F/Y mutations in lung adenocarcinoma and acute myeloid leukemia (AML). Wang and colleagues examined RNA-seq data from 37 patients with chronic lymphocytic leukemia and found that SF3B1 mutations increased the telomerase activity, Notch signaling, and DNA damage, by promoting alternative 3′ splice sites involved in leukemogenesis (Wang et al., 2016). RNA-seq analysis revealed that SF3B1-R625/K666 mutations affected alternative splicing by promoting alternative branchpoint usage in uveal melanomas (Alsafadi et al., 2016). Although cancer-associated spliceosomal mutations have been recognized, identification and prioritization among hundreds of downstream mis-spliced isoforms, as well as validation and interpretation of the functional role of specific isoforms in cancer are challenging (Dvinge et al., 2016; Yang et al., 2016). For example, KRas4A and KRas4B, two splice variants of KRAS, play essential roles in cancer and other diseases (Nussinov et al., 2016; Cheng and Nussinov, 2018). However, comparison of protein sequences among HRas, NRas, KRas4A, and KRas4B show that the catalytic domains (residues 1–166) are almost identical, indicating the difficult task of determination of isoform-specific functions (Nussinov et al., 2016, 2018). A recent integrative analysis of reverse-transcription polymerase chain reaction assay and splice variant-specific antibodies has shown that among four Ras proteins, KRas4A is unique in possessing a dual membrane-targeting motif that could independently deliver KRas4A to the plasma membrane (Tsai et al., 2015). Analysis of all-atom molecular dynamics simulations (total 5.8 µs) shows a mechanism of unique orientation at the membrane for KRas4A compared with KRas4B (Li and Buck, 2017). Specifically, the electrostatic interaction between the protein’s charged residues and anionic lipids determines the unique KRas4A orientations, consistent with previous experimental observation (Tsai et al., 2015).
Although to date, approaches targeting KRAS have failed, specifically targeting K-Ras4A might play a significant role in KRAS-driven tumors (Tsai et al., 2015). However, experimental measurements of the expression of different protein isoforms in cancer are challenging. The excision repair cross-complementation group 1 (ERCC1) protein with four isoforms by alternative splicing has been demonstrated a potential prognostic biomarker of cancer patient survival and treatment efficacy in various studies, including genomic, transcriptional, and protein levels (Friboulet et al., 2013). However, a recent immunohistochemical analysis using currently available ERCC1 antibodies cannot detect unique functional ERCC1 isoform, limiting therapeutic decision making in clinics (Friboulet et al., 2013). Antibodies can directly inhibit RAS function in cells, and initial results with monoclonal antibody inhibition are promising (Spencer-Smith and O’Bryan, 2017). For example, monoclonal antibody Y13-259 against oncogenic HRAS was first microinjected into NIH-3T3 cells to inhibit HRAS-driven proliferation and oncogenic transformation by binding to the switch 2 region (residues 60–76), blocking RAF interaction. Problems posed by the low solubility of the scFv fragment that resulted in aggregation and the failure of the antibodies to readily cross the cell membrane were subsequently resolved, suggesting that monoclonal antibodies are a viable potential option to block RAS signaling. Monoclonal antibody against a synthetic peptide (anti-p21Ser) related to KRAS (G12S) bound to the nucleotide-free RAS state and inhibited GTP binding, which suggests that the RAS protein picomolar affinity for the nucleotide may not preclude pharmacological targeting. However, whereas normally the GTP-bound form is the active RAS state, nucleotide-free HRAS activates PI3KC2β (Wong et al., 2012), indicating targeting nucleotide-free HRAS may fail to negatively regulate PI3KC2β activation by HRAS (Spencer-Smith and O’Bryan, 2017).
Immunohistochemistry studies with the SP174 antibody were developed to identify Q61R-mutant NRAS as an alternative to genetic testing in melanoma. Two hundred ninety-two malignant melanomas were evaluated, with 29 (10%) presenting positive immunoreactivity. Sequencing revealed a c.182A>G substitution (resulting in Q61R mutation) in NRAS in 22 tumors as well as in HRAS and KRAS. Other NRAS mutations or wild type immunoreacted with the SP174 antibody, suggesting selective specificity to this mutation but cross-reactivity with other RAS isoforms (Turchini et al., 2017; Felisiak-Goląbek et al., 2018). Thus, although the antibody was 100% (14/14) sensitive and specific (83/83) for NRAS Q61R mutation (Massi et al., 2015), its non-selective isoform recognition emphasizes the challenges facing antibody development.
III. Personal Mutanomes for Accelerating Modern Oncology Drug Development
A. Druggable Proteome Informed by Somatic Mutations
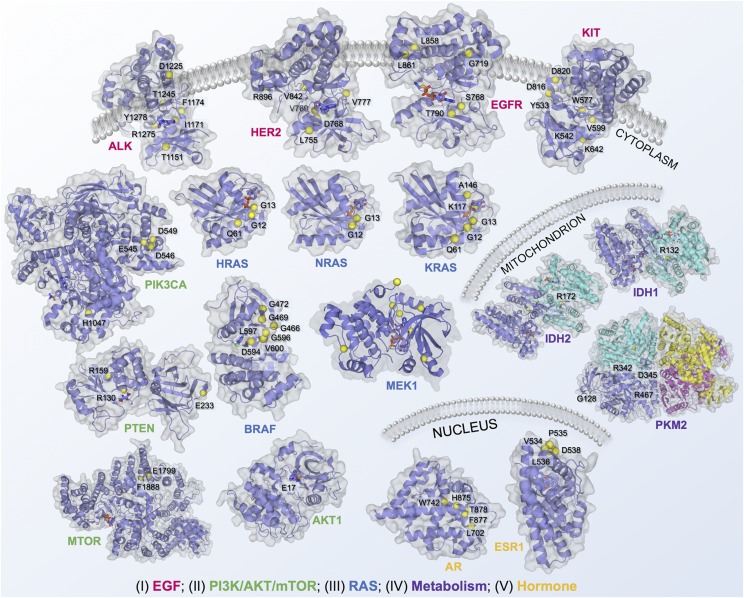
Recent advances in protein structural genomics approaches and computational strategies have provided the detailed structural consequences of somatic alterations through systematic analysis of the positional distribution of protein-altered somatic mutations (Table 3) (Vuong et al., 2014; Kamburov et al., 2015; Araya et al., 2016; Niu et al., 2016; Zhao et al., 2016; Shen et al., 2017). Making the connection between the reams of data coming out of protein structures and sequencing laboratories and the individual cancer patient has sparked excitement for personalized cancer medicine. Here we illustrate the protein structural landscape of the druggable proteome in cancer for 13 selected highly mutated proteins with well-annotated hotspot somatic mutations across various cancer types (Fig. 4).
Fig. 4.
Structural landscape of druggable proteome in oncology drug discovery. In total, 18 selected cancer genes with well-annotated driver mutations and available protein three-dimensional structures across five classic cancer pathways were illustrated: 1) PI3K/AKT/PTEN pathway, 2) EGF/EGFR signaling, 3) RAS pathway, 4) cell metabolism pathways, and 5) hormone (estrogen/androgen) pathways. Cancer driver mutations were collected from My Cancer Genome (https://www.mycancergenome.org/). Protein structures were collected from PDB database (http://www.rcsb.org/pdb/home/home.do).
1. RAS Pathway
More than 30% of all human cancers are driven by mutations of the RAS family of genes with three members: HRAS, KRAS, and NRAS (Nussinov et al., 2016). Mutations in KRAS are estimated to drive over 95% of all pancreatic cancers and 45% of colorectal cancers. The vast majorities of KRAS mutations are recurrent “hot-spot” driver mutations at positions 12, 13, and 61. For example, G12D is the most common KRAS mutation and is estimated to occur in over 50,000 new cases of cancer in the United States every year. Worldwide efforts specifically target mutant RAS genes (Ostrem et al., 2013). Such efforts have met with limited success to date. In-depth review of the structure, dynamics, mutational activation and inactivation, and signaling mechanisms of RAS can be found in several recent reviews (Lu et al., 2016b; Ostrem and Shokat, 2016). RAS small molecule drug discovery has been hampered by the lack of sufficiently deep hydrophobic pockets (Lu et al., 2016a). This has shifted efforts to inhibiting RAS by targeting its pathways (Nussinov et al., 2013), focusing on its plasma membrane anchorage and localization (Cox et al., 2015), its activation by SOS (Winter et al., 2015; Lu et al., 2016c), for example by inducing a modification in the Switch II pocket, such that e.g., ligand 2C07 binding would push Switch I away from the nucleotide, disrupting the essential polar contacts. This inhibits SOS binding and alters nucleotide preference and nucleotide exchange (Gentile et al., 2017). Ras interactions and activation of cofactors were also targeted, although the high affinity of the interaction with RAF has been challenging. Still, recent work has revealed a cyclic peptide that blocked the binding of KRAS with the Ras binding domains (RBDs) of RAF, T-lymphoma invasion and metastasis-inducing protein 1, and Ral guanine nucleotide dissociation stimulator in vitro, albeit with poor membrane permeability (Wu et al., 2013). An upgraded version, Cyclorasin 9A5, potently blocked the RAS-RAF interaction with an IC50 in the low micromolar range. Cyclorasin 9A5 diminished MAPK (ERK), but also AKT serine/threonine kinase 1 (AKT/AKT1) activation and tumor cells lines expressing a mutant EGFR, pointing to its likely toxicity (Upadhyaya et al., 2015). Other strategies include inhibitors to PDEδ-KRAS interaction, which shuttles KRAS to the endomembrane to maintain a pool of KRAS localized at the plasma membrane, and the Arl-2, which helps KRAS release from PDEδ. For example, Deltarasin binds with nanomolar affinity to the farnesyl binding pocket of PDEδ, blocking its association with KRAS (Martin-Gago et al., 2017). Blocking RAS dimerization has been another venue for inhibiting mutant RAS (Spencer-Smith et al., 2017), as well as combinations with drugs targeting frequently observed resistance mutations, such as those in YAP1, the transcriptional coactivator that rescues KRAS-dependent cells upon KRAS inhibition and is required for KRAS-induced cell transformation (Shao et al., 2014). Functional studies established the role of Yap1 and the transcriptional factor Tead2 in driving Kras (G12D)-independent tumor maintenance (Kapoor et al., 2014). Altogether these raise hope that the enigmatic RAS will eventually be subdued (Spencer-Smith and O’Bryan, 2017).
2. Epidermal Growth Factor/Epidermal Growth Factor Receptor Signaling Pathway
The ERBB family consists of four related transmembrane tyrosine kinase receptors: EGFR (ERBB1), HER2/neu (ERBB2), HER3 (ERBB3), and HER4 (ERBB4). EGFR was first discovered by Stanley Cohen, who shared the Nobel Prize in Medicine in 1986 with Rita Levi-Montalcini for their discovery of growth factors. A recent study of >200 lung adenocarcinoma tumors by TCGA has suggested that 14% of the tumors have somatic mutations on EGFR, including substitution mutations, such as T790M and L858R (Cancer Genome Atlas Research Network, 2014). EGFR-related tyrosine kinase inhibitors that target the ATP orthosteric site of the kinases, such as gefitinib and erlotinib, are approved for NSCLC harboring activating mutations on EGFR, affect tumor response rate, survival, and quality of life (Ke and Wu, 2016). The EGFR-T790M is the most common somatic alteration and is detected in approximately 50% of progressing lung tumors (Gazdar, 2009). Osimertinib was approved for the treatment of patients with metastatic NSCLC harboring EGFR-T790M in 2015 (Table 2) (Ke and Wu, 2016). Furthermore, osimertinib and afatinib have been approved for metastatic NSCLC whose tumors have EGFR with second mutations of exon 19 deletions or exon 21 substitution mutations (L858R) (Ke and Wu, 2016). Despite promising clinical evidence of activity against mutated tumors in the early several months, acquired resistance arises rapidly, owing to several new secondary mutations, such as the double EGFR-L858R/T790M as well as the triple EGFR-L858R/T790M/C797S (a mutant resistant to all currently available EGFR-tyrosine-kinase inhibitors) within the ATP orthosteric site of the receptor (Thress et al., 2015).
HER2 was reported to be amplified in multiple cancer types, such as breast, esophageal, and gastric. A recent sequencing study of >5000 recurrent and metastatic breast tumors suggested that 12.5% of the patients (698) harbored HER2 alterations, including 10.6% of tumors (596 patients) with HER2 amplification and 2.4% of tumors (138) with substitution mutations (L755S, D769Y, R768Q, V777L, and V842I in the kinase domain) or indels. Trastuzumab, a humanized immunoglobulin G1 antibody approved by the FDA in 1998, has provided substantial clinical benefit to breast cancer patients with HER2 amplification. Yet, ultimately patients have acquired resistance to trastuzumab and lapatinib owing to various secondary somatic mutations on HER2. Neratinib, a dual irreversible inhibitor of HER2 and EGFR, has shown promising clinical benefits by overcoming resistance of somatic mutations on HER2 (Bose et al., 2013). Hanker et al. (2017) recently identified a new acquired HER2-T798I gatekeeper mutation that induces resistance to neratinib in a metastatic patient with a known HER2-driven mutation L869R. Protein structural modeling suggests that HER2-T798I reduces the binding affinity between HER2 and neratinib (Hanker et al., 2017).
3. Phosphatidylinositol 3-Kinase /AKT/Phosphatase and Tensin Homolog Pathway
The phosphatidylinositol 3-kinase (PI3K) pathway is essential for cancer cell growth, proliferation, survival, and metabolism. PIK3CA activating alterations, such as E542K and E545K (exon9) in the helical domain and H1047R (exon 20) in the kinase domain, are common in human breast cancer (Cancer Genome Atlas, 2012). Two recent studies independently suggested that PIK3CA-H1047R induced multiple-potency and multi-lineage mammary tumors and further drove breast tumor heterogeneity and progression (Koren et al., 2015; Van Keymeulen et al., 2015). A previous sequencing study of 507 primary breast tumors by TCGA suggested that PIK3CA-H1047R was only detected in 17% of HER2 positive tumors (Cancer Genome Atlas, 2012). A recent single cell sequencing study using a specific-to-allele PCR-FISH approach revealed that PIK3CA-activating mutations were present in approximately 20%–40% of HER2-positive tumors, further supporting intratumoral heterogeneity driven by activating mutations on PIK3CA (Janiszewska et al., 2015). Interestingly, activating mutations on PIK3CA are observed in over 30% of ER-positive breast cancer. A preclinical study has suggested that PIK3CA inhibitors (e.g., BYL719) trigger the activation of estrogen receptor (ER) transcriptional activity in ER-positive breast cancer, suggesting that combined PI3K and ER blockage is a rational approach for targeted therapy in breast cancers harboring activating mutations on PIK3CA (Bosch et al., 2015).
PTEN, encoding phosphatase and tensin homolog, is a tumor suppressor gene by negatively regulating the Akt/PKB signaling pathway (Chalhoub and Baker, 2009; Caserta et al., 2015). From a structural perspective, PTEN contains three domains: phosphatase, C2, and PDZ. Several functional mutations on PTEN coding region (Fig. 4) have been involved in multiple cancer types: 1) p.R130*, p.R130G, and p.R130Q mutations on the phosphatase domain and N323fs*2 and N323fs*21 on the C2 domain in ovarian cancer (Chalhoub and Baker, 2009) and 2) p.R159S on the phosphatase domain and the R233* mutation on the C2 domain in breast (Lee et al., 1999) and colorectal cancers (Markman et al., 2010).
4. Hormone Pathways
Hormones (e.g., estrogen and progesterone) are substances that function as chemical messengers in the human body. Overexpression or dysregulation of hormone pathways, such as estrogen or androgen receptors, has played critical roles in tumor growth of various cancers, such as breast and prostate. For example, approximately 80% of breast cancers are ER positive (Davies et al., 2011). ER antagonists (e.g., tamoxifen and toremifene) have demonstrated great success in treatment of ER-positive breast cancer. However, recent genomic studies have suggested that several substitution mutations (e.g., Y537S/C/N or D538G on ER ligand binding domains) promote tumorigenesis and tumor progression without hormone stimulation (Robinson et al., 2013; Toy et al., 2013). Further studies showed that those activating mutations on the ER reduce the efficacy of ER antagonists, such as tamoxifen, thus contribute to acquired endocrine resistance (Li et al., 2013; Robinson et al., 2013; Toy et al., 2013).
5. Cell Metabolism Pathways
Regulation of cancer cell metabolism has attracted much attention over the past several decades (Cairns et al., 2011; Pavlova and Thompson, 2016). The Warburg effect was first reported by Otto Warburg, who received Nobel Prize in Physiology or Medicine in 1931. Warburg (1956) hypothesized that cancer growth is caused by tumor cells generating energy mainly by anaerobic breakdown of glucose. However, metabolic dysregulation in tumors extends beyond the Warburg effect, such as pyruvate kinase and isocitrate dehydrogenases (Cairns et al., 2011). Isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) mutations (e.g., R132H) are activating mutations that lead to production of high levels of 2-hydroxyglutarate from alpha-ketoglutarate in the mitochondria (Mondesir et al., 2016). On August 1, 2017, Enasidenib (AG-221), a first-in-class, oral, selective, small-molecule inhibitor on IDH2-R132H mutant protein was approved by the FDA for the treatment of adult patients with relapsed or refractory acute myeloid leukemia with IDH2-R132H mutations based on the 199-patient phase I/II study (NCT01915498). AG-120 (Ivosidenib), a selective IDH1-R132H inhibitor, was also under Phase I/II for treatment of a number of solid tumors types via targeting cancer cell metabolism (Mullard, 2016). However, recent studies have suggested that some cancer types display elevation in 2-hydroxyglutarate even in the absence of IDH mutations, indicating the unknown clinical outcomes of IDH1/2 mutation-specific inhibitors in those cancer types, such as triple-negative breast cancer (Terunuma et al., 2014) and clear cell renal cell carcinoma (Shim et al., 2014).
B. Mutation-Specific Allosteric Drug Discovery
1. Epithelial Growth Factor Receptor-C797S
Cysteine is the most reactive amino acid, forming covalent bonds with numerous chemical warheads. Several covalent inhibitors have been approved for molecularly targeted therapies in several cancer types, such as Bruton’s tyrosine kinase inhibitor ibrutinib for chronic lymphocytic leukemia (Byrd et al., 2013) and EGFR inhibitor afatinib for non-small cell lung cancer (Yu and Pao, 2013; Yang et al., 2015). However, orthosteric drugs with low specificity often result in off-target toxicity (Moslehi, 2016). Allosteric agents are promising candidates for targeted therapies with a higher specificity (Nussinov and Tsai, 2015). Recently, a novel allosteric inhibitor, EAI045, was revealed to target specifically the conventional tyrosine kinase inhibitor-resistant EGFR mutants by binding an allosteric site created by the displacement of the regulatory C-helix in an inactive conformation of EGFR. EAI045 inhibits the L858R/T790M mutant EGFR with a low nanomolar potency; however, a single agent is not effective in blocking EGFR-driven cell proliferation because of differential potency on the two subunits of the dimeric receptor (Jia et al., 2016). Interestingly, combination of EAI045 and cetuximab that blocks EGFR dimerization synergistically suppresses tumor growth in mouse models of lung cancer driven by the double EGFR-L858R/T790M mutants as well as by triple EGFR-L858R/T790M/C797S mutants, which are resistant to all current EGFR-target therapies (Jia et al., 2016).
2. KRAS-G12C
Inhibitors that act on both wild-type and mutant cancer proteins are more likely to cause substantial toxicity. One merit method of achieving mutant specificity is targeting the mutant residue itself via covalent allosteric inhibitors. KRAS-G12C (c.34G>T) is one of the three most common KRAS driver mutations in various cancer types, with a non-native cysteine residue compared with the wild-type protein. A recent study identified a panel of compounds that irreversibly inhibited KRAS-G12C by forming a covalent attachment to the mutant cysteine (Ostrem et al., 2013). Despite those compounds binding to the KRAS-G12C in the inactive state, they can still be clinically beneficial via a shift of the active mutant population toward the inactive state, with subsequent capture by the covalent inhibitor.
3. Breakpoint Cluster Region Protein-Abelson Murine Leukemia Viral Oncogene Homolog Fusion
The BCR-ABL1 fusion protein promotes tumorigenesis and tumor progression in CML. Several targeted agents (e.g., nilotinib) have been developed for highly specific targeting of ABL1. However, ABL1 kinase inhibitors often elicit acquired resistance in some patients. Using fragment-based nuclear magnetic resonance screens, Wylie et al. (2017) identified allosteric ABL1 inhibitors that specifically bind to the myristoyl pocket to induce the auto-inhibited kinase conformation by supporting a bend in the C-terminal helix. Further lead optimization identified a more potent ABL1 selective allosteric inhibitor ABL001. ABL001 selectively suppresses the growth of BCR-ABL1-driven tumor cells via inhibiting phosphorylation of BCR-ABL1 and signal transducer and activator of transcription 5 during in vitro studies. Structural studies have determined that nilotinib and ABL001 bind to BCR-ABL1 simultaneously by a potential dual targeting effect. In vivo and phase-I studies have suggested that nilotinib and ABL001 combination promote tumor eradication and delay acquired resistance in patients harboring BCR-ABL1-driven tumors through potential dual inhibition of allosteric and catalytic activities of BCR-ABL1.
C. Neoantigen-Driven Personalized Immunotherapies
In contrast to traditional surgery, radiation, chemotherapies, and molecularly targeted therapies, immunotherapy has shown the broadly successful strategies for metastatic tumors in clinic. Pembrolizumab, a humanized antibody, boosts the immune systems against cancer cells by inhibiting the PD-1 receptor. In May 2017, it was approved by the FDA for any unresectable or metastatic solid tumor with mismatch-repair deficiency based on a published phase 2 study (Le et al., 2015). A recent study reported that administering tumor-infiltrating lymphocytes isolated from a 50-year-old woman with metastatic colorectal cancer patient, namely adoptive T-cell transfer immunotherapy, mediate effective antitumor immune responses against tumors that express the KRAS-G12D mutation (Tran et al., 2016). This study firstly implies that the identification of HLA-C*08:02-restricted T-cell receptors that specifically target KRAS-G12D neoantigen (e.g., GADGVGKSAL) would provide an opportunity to develop highly targeted immunotherapies against a common actionable mutation identified in multiple cancers, such as colorectal and pancreatic cancers. A recent study examined the relationship between the objective response rate for anti-PD-1/anti-PD-L1 therapy and the corresponding median tumor mutational burden across 27 cancer types/subtypes (Yarchoan et al., 2017). A significant correlation between the tumor mutational burden and the objective response rate was observed, highlighting a stronger relationship of the tumor mutational burden and the response rate of anti-PD-1 therapies. However, the quality of the neoantigen, rather than the quantity, is reported as actionable biomarkers for guiding the application of personalized immunotherapies, through an integrated approach, including genetic, immunohistochemical and transcriptional immunoprofiling, computational biophysics, and functional assays (Balachandran et al., 2017). Figure 3 illustrates an integrative computational and experimental infrastructure for the emerging development of personalized immunotherapies through incorporating innovative genomics, proteomics technologies, and computational biophysics approaches.
Ott et al. (2017) generated personalized cancer vaccines by identifying tumor-specific mutations and accordingly determining neoantigens from the tumor-normal matched DNA sequencing profiles of six patients with melanoma. They demonstrated proof-of-principle of a vaccine that targets up to 20 computationally predicted tumor neoantigens for eliciting strong T-cell responses against cancer in phase I trial. Sahin et al. (2017) showed a similar framework of identifying the tumor neoantigens from somatic mutations on 13 melanoma patients via computational identification of the binding between small mutant peptides and MHC proteins. Both studies suggested that the predicted neoantigens presented in the vaccine and recognized by CD4 T-cells boosted significantly antitumor immune responses. Although the number of patients who were treated in the two studies were small, they revealed potential durable clinical benefits. Additional controlled, randomized clinical trials will be needed to test the clinical benefits of neoantigens-based personalized immunotherapies in a large cohort study.
A recent study reported a novel computational protein design approach, namely Fold FROM Loops, for the development of epitope-specific neutralizing antibodies for the human respiratory syncytial virus (Correia et al., 2014). Fold FROM Loops generates small, thermally and conformationally stable protein scaffolds that accurately mimic the viral epitope structure. This study provides proof of principle for epitope-driven vaccine design and enables the evaluation and further development of these strategies for cancer vaccine design by targeting specific neoantigens. In addition to somatic mutation-derived neoantigens, nonmutated endogenous proteins, called T-cell epitope associated with impaired peptide process neoantigens, can also act as immunogenic epitopes (van der Burg et al., 2016). A new study provided more unique structural mechanisms of the first identified T-cell epitope associated with impaired peptide process antigens derived from Trh4 proteins using crystallographic investigation, highlighting a significant advance in the search for novel immunotherapeutic targets (Hafstrand et al., 2016).
While these are exciting early results, computational neoantigen prediction is a field still in its infancy. A more disciplined approach to the development of the algorithms might be next for this field. Researchers in other areas of bioinformatics (e.g., DNA variant effect prediction, protein structure prediction, and biomedical text processing) have generated reference datasets on which periodic competitions are held such that more accurate algorithms can be continually refined. The establishment of a reference data set and competition might now be due for neoantigen prediction.
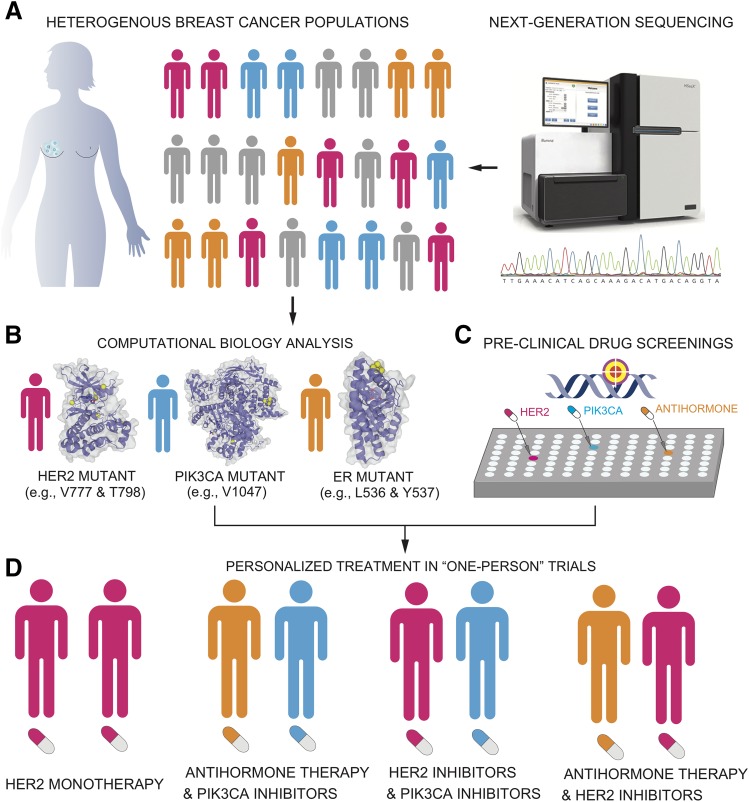
IV. A Personal Mutanome Infrastructure for Precision Oncology
“Precision medicine requires a different type of clinical trial that focuses on individual, not average, responses to therapy,” says Nicholas J. Schork, Director of Human Biology at the J. Craig Venter Institute (Schork, 2015). Studies that focus on “one-person trials” have shown a proof-of-concept for the emerging development of personalized treatment in cancer. A recent study reported that an individual with metastatic bladder cancer with tuberous sclerosis complex 1 somatic mutations showed a super response to Everolimus (Iyer et al., 2012). Thus, exceptional responders can help researchers to predict the responses of many other patients with tumors harboring particular genomic or molecular profiles (Marx, 2015). Currently, cancer therapy is moving from a drug-centered to a patient-centered approach with different levels of personalization. This requires paradigm shifts along the entire drug development process and multiple data integration. A personal mutanome infrastructure that utilizes the wealth of structural genomics would offer a prototype for how to open this territory by developing best-practice blueprints for the implementation of personalized cancer treatments. Fig. 5 illustrates a personal mutanome infrastructure for development of personalized cancer treatment using breast cancer as a case study. The entire infrastructure contains four core components for 1) performing tumor genetic and genomic testing, 2) identifying actionable biomarkers that may guide the personalized therapies using bioinformatics and computational biology tools (e.g., protein structure hotspot clustering as shown in Table 3), 3) in vitro functional assays based on the short list of predicted biomarkers from step 2, and 4) guiding the application of monotherapies or combination therapies based on the identified biomarkers from steps 1–3. Among the promising actionable biomarkers for personalized therapies in breast cancer patients are substitution mutations on HER2 (L755S, D769Y, R768Q, V777L, V842I, and T798I), PIK3CA (E542K, E545K, and H1047R), and ESR1 (Y537S/C/N and D538G), as shown in Fig. 4. However, monotherapies often failed during the emerging drug resistance. Combination therapies that target different pathways have shown some promises. For example, PI3K pathway inhibitors have emerged as promising therapeutic agents for ER-positive breast cancer. A recent study suggested that the epigenetic regulator KMT2D (histone-lysine N-methyltransferase 2D) enhances ERα transcriptional activity in BYL719-treat PIK3CA mutant breast cancer, providing a rationale for targeting the epigenome and PI3K signaling (Toska et al., 2017). Moreover, combination of chemotherapeutic agents or molecularly targeted agents with immunotherapies has also shown promise in clinical trials of breast cancer (Kohrt et al., 2012) (NCT01570036) or advanced triple negative breast cancer (NCT02768701).
Fig. 5.
A personal cancer mutanome infrastructure for the development of personalized treatment using breast cancer as a case study. The entire infrastructure contains four core components: performing tumor genetic and genomic testing (A), identifying actionable biomarkers that may guide the personalized therapies using bioinformatics and computational biology tools (e.g., protein structure hotspot clustering shown in Table 3) (B), pre-clinical validation (in vitro or in vivo functional assays) (C), and guiding the application of monotherapies or combination therapies based on steps 1–3 (D).
V. Conclusions and Perspectives
Recent advances of innovative biomedical technologies and computational tools have made it possible to identify and interpret actionable mutations in individuals to inform drug discovery and personalized treatments. The field of oncology has been a promising pioneer of precision medicine by exploiting the wealth of large-scale sequencing and structural genomic data. Traditionally, chemotherapeutic agents with a broad anticancer spectrum in various cancer types commonly kill both cancerous and healthy cells. Molecular targeted agents that selectively suppress pathways necessary for cancer cell growth work through two primary mechanisms: orthosteric and allosteric site inhibition. Effective orthosteric agents that target the clinically actionable variants in primary binding sites (e.g., the ATP site in kinases) have shown promising clinical benefits against cancer cells in the early months of therapies. However, emerging drug resistance owing to acquired mutations and off-target toxicity have been challenging. Allosteric agents have been demonstrated as promising candidates for the next generation of targeted therapies with a higher specificity in preclinical and clinical studies (Ostrem et al., 2013; Jia et al., 2016; Wylie et al., 2017). New directions are emerging for personalized oncology drug discovery and development in the genomics era. Some highly mutated oncogenes (e.g., RAS and myelocytomatosis), which to date could not be targeted pharmacologically, pose a key challenge in personalized cancer medicine (Dang et al., 2017). Several National Institutes of Health-sponsored national efforts, such as Illuminating the Druggable Genome (http://targetcentral.ws/index) and The RAS Initiative (https://www.cancer.gov/research/key-initiatives/ras), inspire development of new technology platforms to study the “undruggable” genome, which is expected to witness substantial progress. Newer partnerships, for example, the Accelerating Therapeutics for Opportunities in Medicine (ATOM) Project involving the University of California, San Francisco (UCSF), the National Cancer Institute (Fredrick National Laboratory for Cancer Research), the US Department of Energy (Lawrence Livermore National Laboratory), and GlaxoSmithKline, suggest that future innovations are going to require teams of scientific minds powered by significant computer infrastructure and data on potential drug candidates (https://www.ucsf.edu/news/2017/10/408841/public-private-consortium-aims-cut-preclinical-cancer-drug-discovery-six-years).
Cancer immunotherapy that targets T-cells to enhance immune response has generated both excitement and skepticism. Although immunotherapies have generated durable clinical responses and long-term remissions in some cancer types (e.g., melanoma), autoimmune and other immune-related adverse effects associated with cancer immunotherapy tempered the enthusiasm of cancer immunologists for the broad use of these powerful agents (June et al., 2017). There is a pressing need for innovative strategies to improve the clinical benefits for the broad cancer population toward minimizing autoimmune adverse effects. Targeting mutation-specific neoantigens is one of the attractive strategies for patient stratification for improving clinical benefits and mitigating the risk of autoimmunity. Intelligently combining immune checkpoint inhibitors with cytotoxic chemotherapy (e.g., cyclophosphamide) or targeted therapy (e.g., BRAF inhibitors) may provide better control of dosage and cardiotoxicity. It may also help improve long-term cardiovascular outcomes in patients by exploiting immunologic “side” effects of conventional chemotherapeutic agents and targeted antitumor agents (Galluzzi et al., 2015). Future manipulation of the microbiome could further enhance the therapeutic efficacy and diminish immunotoxicity of cancer immunotherapy (Sivan et al., 2015; Honda and Littman, 2016).
Abbreviations
- ABL1
abelson murine leukemia viral oncogene homolog 1
- AKT1
AKT serine/threonine kinase 1
- AML
acute myeloid leukemia
- BCR
breakpoint cluster region protein
- BRAF
proto-oncogene B-Raf
- CML
chronic myeloid leukemia
- 3D
three-dimensional
- EGFR
epidermal growth factor receptor
- ER(α)
estrogen receptor (alpha)
- ERCC1
excision repair cross-complementation group 1
- FDA
Food and Drug Administration
- HER2
human epidermal growth factor receptor 2
- IDH1/IDH2
isocitrate dehydrogenases 1/2
- KRAS
KRAS proto-oncogene, GTPase
- MHC-I
major histocompatibility complex class I
- NSCLC
non-small cell lung cancer
- PD-1/PD-L1
programmed cell death protein 1/programmed death-ligand 1
- PDB
Protein Data Bank
- PDEδ
phosphodiesterase-δ
- PI3K
phosphatidylinositol 3-kinase
- PIK3CA
phosphoinositide-3-kinase, catalytic, alpha polypeptide
- PTEN
phosphatase and tensin homolog
- SF3B1
splicing factor 3B, subunit 1
- TCGA
The Cancer Genome Atlas
- U2AF1
U2 small nuclear RNA auxiliary factor 1
- YAP1
yes-associated protein 1
Authorship Contributions
Participated in research design: Cheng, Eng, Nussinov.
Conducted experiments: Cheng.
Performed data analysis: Cheng, Liang, Butte.
Wrote or contributed to the writing of the manuscript: Cheng, Eng, Nussinov.
Footnotes
This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K99HL138272 and R00HL138272 to F.C. This work has been also supported in part with Federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of NIH, Frederick National Laboratory, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. C.E. is the Sondra J. and Stephen R. Hardis Endowed Chair of Cancer Genomic Medicine at the Cleveland Clinic and an ACS Clinical Research Professor.
References
- Albert FW, Kruglyak L. (2015) The role of regulatory variation in complex traits and disease. Nat Rev Genet 16:197–212. [DOI] [PubMed] [Google Scholar]
- Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S, et al. (2016) Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun 7:10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya CL, Cenik C, Reuter JA, Kiss G, Pande VS, Snyder MP, Greenleaf WJ. (2016) Identification of significantly mutated regions across cancer types highlights a rich landscape of functional molecular alterations. Nat Genet 48:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga CL, Engelman JA. (2014) ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 25:282–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, et al. Australian Pancreatic Cancer Genome Initiative; Garvan Institute of Medical Research; Prince of Wales Hospital; Royal North Shore Hospital; University of Glasgow; St Vincent’s Hospital; QIMR Berghofer Medical Research Institute; University of Melbourne, Centre for Cancer Research; University of Queensland, Institute for Molecular Bioscience; Bankstown Hospital; Liverpool Hospital; Royal Prince Alfred Hospital, Chris O’Brien Lifehouse; Westmead Hospital; Fremantle Hospital; St John of God Healthcare; Royal Adelaide Hospital; Flinders Medical Centre; Envoi Pathology; Princess Alexandria Hospital; Austin Hospital; Johns Hopkins Medical Institutes; ARC-Net Centre for Applied Research on Cancer (2017) Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551:512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A, Li Z, Bergamaschi A, Ellis H, Toska E, Prat A, Tao JJ, Spratt DE, Viola-Villegas NT, Castel P, et al. (2015) PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med 7:283ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, Monsey J, Goel N, Aronson AB, Li S, et al. (2013) Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov 3:224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. (2014) A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One 9:e87361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, et al. (2013) Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med 369:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. (2011) Regulation of cancer cell metabolism. Nat Rev Cancer 11:85–95. [DOI] [PubMed] [Google Scholar]
- Califano A, Alvarez MJ. (2017) The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer 17:116–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014) Comprehensive molecular profiling of lung adenocarcinoma [published correction appears in Nature (2014) 514:262]. Nature 511:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta E, Egriboz O, Wang H, Martin C, Koivisto C, Pecót T, Kladney RD, Shen C, Shim KS, Pham T, et al. (2015) Noncatalytic PTEN missense mutation predisposes to organ-selective cancer development in vivo. Genes Dev 29:1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. (2009) PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 4:127–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Desai RJ, Handy DE, Wang R, Schneeweiss S, Barabási AL, Loscalzo J. (2018) Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun 9:2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Jia P, Wang Q, Lin CC, Li WH, Zhao Z. (2014) Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome. Mol Biol Evol 31:2156–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, Zhou W, Huang J, Tang Y. (2012) Prediction of drug-target interactions and drug repositioning via network-based inference. PLOS Comput Biol 8:e1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Loscalzo J. (2017) Autoimmune cardiotoxicity of cancer immunotherapy. Trends Immunol 38:77–78. [DOI] [PubMed] [Google Scholar]
- Cheng F, Nussinov R. (2018) KRAS activating signaling triggers arteriovenous malformations. Trends Biochem Sci 43:481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Zhao J, Fooksa M, Zhao Z. (2016) A network-based drug repositioning infrastructure for precision cancer medicine through targeting significantly mutated genes in the human cancer genomes. J Am Med Inform Assoc 23:681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Andersen JN, Futreal PA. (2011) Cancer genomics: from discovery science to personalized medicine. Nat Med 17:297–303. [DOI] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS, US National Human Genome Research Institute (2003) A vision for the future of genomics research. Nature 422:835–847. [DOI] [PubMed] [Google Scholar]
- Collins FS, Varmus H. (2015) A new initiative on precision medicine. N Engl J Med 372:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, et al. (2014) Proof of principle for epitope-focused vaccine design. Nature 507:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ, Philips MR. (2015) Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin Cancer Res 21:1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creixell P, Schoof EM, Simpson CD, Longden J, Miller CJ, Lou HJ, Perryman L, Cox TR, Zivanovic N, Palmeri A, et al. (2015) Kinome-wide decoding of network-attacking mutations rewiring cancer signaling. Cell 163:202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Reddy EP, Shokat KM, Soucek L. (2017) Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer 17:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, Wang YC, et al. Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378:771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugger SA, Platt A, Goldstein DB. (2018) Drug development in the era of precision medicine. Nat Rev Drug Discov 17:183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. (2016) RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 16:413–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigin ME, Garvin T, Bailey P, Waddell N, Chang DK, Kelley DR, Shuai S, Gallinger S, McPherson JD, Grimmond SM, et al. (2017) Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma. Nat Genet 49:825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felisiak-Goląbek A, Inaguma S, Kowalik A, Wasąg B, Wang ZF, Zięba S, Pięciak L, Ryś J, Kopczynski J, Sarlomo-Rikala M, et al. (2018) SP174 antibody lacks specificity for NRAS Q61R and cross-reacts with HRAS and KRAS Q61R mutant proteins in malignant melanoma. Appl Immunohistochem Mol Morphol 26:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friboulet L, Olaussen KA, Pignon JP, Shepherd FA, Tsao MS, Graziano S, Kratzke R, Douillard JY, Seymour L, Pirker R, et al. (2013) ERCC1 isoform expression and DNA repair in non-small-cell lung cancer. N Engl J Med 368:1101–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. (2015) Immunological effects of conventional chemotherapy and targeted anticancer agents. Cancer Cell 28:690–714. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. (2013) Lessons from the cancer genome. Cell 153:17–37. [DOI] [PubMed] [Google Scholar]
- Gazdar AF. (2009) Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 28 (Suppl 1):S24–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile DR, Rathinaswamy MK, Jenkins ML, Moss SM, Siempelkamp BD, Renslo AR, Burke JE, Shokat KM. (2017) Ras binder induces a modified switch-II pocket in GTP and GDP states. Cell Chem Biol 24:1455–1466.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl H, Charoentong P, Finotello F, Trajanoski Z. (2016) Computational genomics tools for dissecting tumour-immune cell interactions. Nat Rev Genet 17:441–458. [DOI] [PubMed] [Google Scholar]
- Hafstrand I, Doorduijn EM, Duru AD, Buratto J, Oliveira CC, Sandalova T, van Hall T, Achour A. (2016) The MHC Class I cancer-associated neoepitope Trh4 linked with impaired peptide processing induces a unique noncanonical TCR conformer. J Immunol 196:2327–2334. [DOI] [PubMed] [Google Scholar]
- Hanker AB, Brewer MR, Sheehan JH, Koch JP, Sliwoski GR, Nagy R, Lanman R, Berger MF, Hyman DM, Solit DB, et al. (2017) An acquired HER2T798I gatekeeper mutation induces resistance to neratinib in a patient with HER2 mutant-driven breast cancer. Cancer Discov 7:575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EC. (2014) Technology: the $1,000 genome. Nature 507:294–295. [DOI] [PubMed] [Google Scholar]
- Honda K, Littman DR. (2016) The microbiota in adaptive immune homeostasis and disease. Nature 535:75–84. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431:931–945. [DOI] [PubMed] [Google Scholar]
- Iyer G, Hanrahan AJ, Milowsky MI, Al-Ahmadie H, Scott SN, Janakiraman M, Pirun M, Sander C, Socci ND, Ostrovnaya I, et al. (2012) Genome sequencing identifies a basis for everolimus sensitivity. Science 338:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewska M, Liu L, Almendro V, Kuang Y, Paweletz C, Sakr RA, Weigelt B, Hanker AB, Chandarlapaty S, King TA, et al. (2015) In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet 47:1212–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Yun CH, Park E, Ercan D, Manuia M, Juarez J, Xu C, Rhee K, Chen T, Zhang H, et al. (2016) Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature 534:129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Lu W, Shen X, Wang Q, Lv J, Liu M, Cheng F, Zhao Z, Pang X. (2018) Repurposing sertraline sensitizes non-small cell lung cancer cells to erlotinib by inducing autophagy. JCI Insight 3:e98921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- June CH, Warshauer JT, Bluestone JA. (2017) Is autoimmunity the Achilles’ heel of cancer immunotherapy? Nat Med 23:540–547. [DOI] [PubMed] [Google Scholar]
- Kalmanti L, Saussele S, Lauseker M, Müller MC, Dietz CT, Heinrich L, Hanfstein B, Proetel U, Fabarius A, Krause SW, et al. (2015) Safety and efficacy of imatinib in CML over a period of 10 years: data from the randomized CML-study IV. Leukemia 29:1123–1132. [DOI] [PubMed] [Google Scholar]
- Kamburov A, Lawrence MS, Polak P, Leshchiner I, Lage K, Golub TR, Lander ES, Getz G. (2015) Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci USA 112:E5486–E5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, Zhong Y, Wu CJ, Sadanandam A, Hu B, et al. (2014) Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell 158:185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke EE, Wu YL. (2016) EGFR as a pharmacological target in EGFR-mutant non-small-cell lung cancer: where do we stand now? Trends Pharmacol Sci 37:887–903. [DOI] [PubMed] [Google Scholar]
- Khurana E, Fu Y, Chakravarty D, Demichelis F, Rubin MA, Gerstein M. (2016) Role of non-coding sequence variants in cancer. Nat Rev Genet 17:93–108. [DOI] [PubMed] [Google Scholar]
- Kohrt HE, Houot R, Weiskopf K, Goldstein MJ, Scheeren F, Czerwinski D, Colevas AD, Weng WK, Clarke MF, Carlson RW, et al. (2012) Stimulation of natural killer cells with a CD137-specific antibody enhances trastuzumab efficacy in xenotransplant models of breast cancer. J Clin Invest 122:1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Koren S, Reavie L, Couto JP, De Silva D, Stadler MB, Roloff T, Britschgi A, Eichlisberger T, Kohler H, Aina O, et al. (2015) PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 525:114–118. [DOI] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, et al. (2015) Combined Nivolumab and Ipilimumab or monotherapy in untreated melanoma. N Engl J Med 373:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. (2016) Cocktails for cancer with a measure of immunotherapy. Nature 532:162–164. [DOI] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. (1999) Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99:323–334. [DOI] [PubMed] [Google Scholar]
- Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, et al. (2013) Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep 4:1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZL, Buck M. (2017) Computational modeling reveals that signaling lipids modulate the orientation of K-Ras4A at the membrane reflecting protein topology. Structure 25:679–689.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Jang H, Gu S, Zhang J, Nussinov R. (2016a) Drugging Ras GTPase: a comprehensive mechanistic and signaling structural view. Chem Soc Rev 45:4929–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Jang H, Muratcioglu S, Gursoy A, Keskin O, Nussinov R, Zhang J. (2016b) Ras conformational ensembles, allostery, and signaling. Chem Rev 116:6607–6665. [DOI] [PubMed] [Google Scholar]
- Lu S, Jang H, Zhang J, Nussinov R. (2016c) Inhibitors of Ras-SOS interactions. ChemMedChem 11:814–821. [DOI] [PubMed] [Google Scholar]
- Mahoney KM, Rennert PD, Freeman GJ. (2015) Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 14:561–584. [DOI] [PubMed] [Google Scholar]
- Markman B, Atzori F, Pérez-García J, Tabernero J, Baselga J. (2010) Status of PI3K inhibition and biomarker development in cancer therapeutics. Ann Oncol 21:683–691. [DOI] [PubMed] [Google Scholar]
- Martincorena I, Campbell PJ. (2015) Somatic mutation in cancer and normal cells. Science 349:1483–1489. [DOI] [PubMed] [Google Scholar]
- Martín-Gago P, Fansa EK, Klein CH, Murarka S, Janning P, Schürmann M, Metz M, Ismail S, Schultz-Fademrecht C, Baumann M, et al. (2017) A PDE6δ-KRas inhibitor chemotype with up to seven H-bonds and picomolar affinity that prevents efficient inhibitor release by Arl2. Angew Chem Int Ed Engl 56:2423–2428. [DOI] [PubMed] [Google Scholar]
- Marx V. (2015) Cancer: a most exceptional response. Nature 520:389–393. [DOI] [PubMed] [Google Scholar]
- Massi D, Simi L, Sensi E, Baroni G, Xue G, Scatena C, Caldarella A, Pinzani P, Fontanini G, Carobbio A, et al. (2015) Immunohistochemistry is highly sensitive and specific for the detection of NRASQ61R mutation in melanoma. Mod Pathol 28:487–497. [DOI] [PubMed] [Google Scholar]
- Meyer MJ, Beltrán JF, Liang S, Fragoza R, Rumack A, Liang J, Wei X, Yu H. (2018) Interactome INSIDER: a structural interactome browser for genomic studies. Nat Methods 15:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milius D, Dove ES, Chalmers D, Dyke SO, Kato K, Nicolás P, Ouellette BF, Ozenberger B, Rodriguez LL, Zeps N, et al. (2014) The International Cancer Genome Consortium’s evolving data-protection policies. Nat Biotechnol 32:519–523. [DOI] [PubMed] [Google Scholar]
- Mondesir J, Willekens C, Touat M, de Botton S. (2016) IDH1 and IDH2 mutations as novel therapeutic targets: current perspectives. J Blood Med 7:171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R, Céol A, Aloy P. (2013) Interactome3D: adding structural details to protein networks. Nat Methods 10:47–53. [DOI] [PubMed] [Google Scholar]
- Mosca R, Tenorio-Laranga J, Olivella R, Alcalde V, Céol A, Soler-López M, Aloy P. (2015) dSysMap: exploring the edgetic role of disease mutations. Nat Methods 12:167–168. [DOI] [PubMed] [Google Scholar]
- Moslehi JJ. (2016) Cardiovascular toxic effects of targeted cancer therapies. N Engl J Med 375:1457–1467. [DOI] [PubMed] [Google Scholar]
- Mullard A. (2016) Cancer metabolism pipeline breaks new ground. Nat Rev Drug Discov 15:735–737. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Wardell CP, Furuta M, Taniguchi H, Fujimoto A. (2015) Cancer whole-genome sequencing: present and future. Oncogene 34:5943–5950. [DOI] [PubMed] [Google Scholar]
- Nature Publishing Group (2017) The problem with neoantigen prediction. Nat Biotechnol 35:97. [DOI] [PubMed] [Google Scholar]
- Niu B, Scott AD, Sengupta S, Bailey MH, Batra P, Ning J, Wyczalkowski MA, Liang WW, Zhang Q, McLellan MD, et al. (2016) Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat Genet 48:827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ. (2015) The design of covalent allosteric drugs. Annu Rev Pharmacol Toxicol 55:249–267. [DOI] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ, Chakrabarti M, Jang H. (2016) A new view of Ras isoforms in cancers. Cancer Res 76:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ, Jang H. (2018) Oncogenic Ras isoforms signaling specificity at the membrane. Cancer Res 78:593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R, Tsai CJ, Mattos C. (2013) ‘Pathway drug cocktail’: targeting Ras signaling based on structural pathways. Trends Mol Med 19:695–704. [DOI] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503:548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Shokat KM. (2016) Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov 15:771–785. [DOI] [PubMed] [Google Scholar]
- Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al. (2017) An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Hutchinson KE. (2012) Chipping away at the lung cancer genome. Nat Med 18:349–351. [DOI] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23:27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemovska T, Johnson E, Kontro M, Repasky GA, Chen J, Wells P, Cronin CN, McTigue M, Kallioniemi O, Porkka K, et al. (2015) Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature 519:102–105. [DOI] [PubMed] [Google Scholar]
- Porta-Pardo E, Hrabe T, Godzik A. (2015) Cancer3D: understanding cancer mutations through protein structures. Nucleic Acids Res 43:D968–D973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer BN, Ghosh R, Narasanna A, Estrada MV, Chakrabarty A, Song Y, Engelman JA, Arteaga CL. (2013) Human breast cancer cells harboring a gatekeeper T798M mutation in HER2 overexpress EGFR ligands and are sensitive to dual inhibition of EGFR and HER2. Clin Cancer Res 19:5390–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, et al. (2013) Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 45:1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PW, Prlić A, Altunkaya A, Bi C, Bradley AR, Christie CH, Costanzo LD, Duarte JM, Dutta S, Feng Z, et al. (2017) The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res 45 (D1):D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Löwer M, Bukur V, Tadmor AD, Luxemburger U, Schrörs B, et al. (2017) Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547:222–226. [DOI] [PubMed] [Google Scholar]
- Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, et al. (2015) Widespread macromolecular interaction perturbations in human genetic disorders. Cell 161:647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schork NJ. (2015) Personalized medicine: time for one-person trials. Nature 520:609–611. [DOI] [PubMed] [Google Scholar]
- Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang X, Schinzel AC, Sood S, Rosenbluh J, Kim JW, et al. (2014) KRAS and YAP1 converge to regulate EMT and tumor survival. Cell 158:171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Allison JP. (2015) The future of immune checkpoint therapy. Science 348:56–61. [DOI] [PubMed] [Google Scholar]
- Shen Q, Cheng F, Song H, Lu W, Zhao J, An X, Liu M, Chen G, Zhao Z, Zhang J. (2017) Proteome-scale investigation of protein allosteric regulation perturbed by somatic mutations in 7,000 cancer genomes. Am J Hum Genet 100:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shet AS, Jahagirdar BN, Verfaillie CM. (2002) Chronic myelogenous leukemia: mechanisms underlying disease progression. Leukemia 16:1402–1411. [DOI] [PubMed] [Google Scholar]
- Shim EH, Livi CB, Rakheja D, Tan J, Benson D, Parekh V, Kho EY, Ghosh AP, Kirkman R, Velu S, et al. (2014) L-2-Hydroxyglutarate: an epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov 4:1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. (2015) Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, et al. (2017) Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol 13:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R, O’Bryan JP. (2017) Direct inhibition of RAS: quest for the holy grail? Semin Cancer Biol DOI: 10.1016/j.semcancer.2017.12.005 [published ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C, Haanen J, Blank C, Wesseling J, Willems SM, et al. (2014) Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508:118–122. [DOI] [PubMed] [Google Scholar]
- Telenti A, Pierce LC, Biggs WH, di Iulio J, Wong EH, Fabani MM, Kirkness EF, Moustafa A, Shah N, Xie C, et al. (2016) Deep sequencing of 10,000 human genomes. Proc Natl Acad Sci USA 113:11901–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terunuma A, Putluri N, Mishra P, Mathé EA, Dorsey TH, Yi M, Wallace TA, Issaq HJ, Zhou M, Killian JK, et al. (2014) MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 124:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, Lai Z, Markovets A, Vivancos A, Kuang Y, et al. (2015) Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 21:560–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak K, Czerwińska P, Wiznerowicz M. (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 19 (1A):A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, Dickler MN, Scaltriti M, Leslie CS, Armstrong SA, et al. (2017) PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 355:1324–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, et al. (2013) ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 45:1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. (2016) T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 375:2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, Fiordalisi JJ, Gierut JJ, Cox AD, Haigis KM, Philips MR. (2015) K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci USA 112:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchini J, Andrici J, Sioson L, Clarkson A, Watson N, Toon CW, Shepherd P, Ng D, Dixon-McIver A, Oei P, et al. (2017) NRASQ61R mutation-specific immunohistochemistry is highly specific for either NRASQ61R or KRASQ61R mutation in colorectal carcinoma. Appl Immunohistochem Mol Morphol 25:475–480. [DOI] [PubMed] [Google Scholar]
- Upadhyaya P, Qian Z, Selner NG, Clippinger SR, Wu Z, Briesewitz R, Pei D. (2015) Inhibition of Ras signaling by blocking Ras-effector interactions with cyclic peptides. Angew Chem Int Ed Engl 54:7602–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg SH, Arens R, Ossendorp F, van Hall T, Melief CJ. (2016) Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat Rev Cancer 16:219–233. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Lee MY, Ousset M, Brohée S, Rorive S, Giraddi RR, Wuidart A, Bouvencourt G, Dubois C, Salmon I, et al. (2015) Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525:119–123. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. (2013) Cancer genome landscapes. Science 339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong H, Cheng F, Lin CC, Zhao Z. (2014) Functional consequences of somatic mutations in cancer using protein pocket-based prioritization approach. Genome Med 6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brooks AN, Fan J, Wan Y, Gambe R, Li S, Hergert S, Yin S, Freeman SS, Levin JZ, et al. (2016) Transcriptomic characterization of SF3B1 mutation reveals its pleiotropic effects in chronic lymphocytic leukemia. Cancer Cell 30:750–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H. (2012) Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotechnol 30:159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956) On the origin of cancer cells. Science 123:309–314. [DOI] [PubMed] [Google Scholar]
- Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. (2014) Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 46:1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter JJ, Anderson M, Blades K, Brassington C, Breeze AL, Chresta C, Embrey K, Fairley G, Faulder P, Finlay MR, et al. (2015) Small molecule binding sites on the Ras:SOS complex can be exploited for inhibition of Ras activation. J Med Chem 58:2265–2274. [DOI] [PubMed] [Google Scholar]
- Wong KA, Russo A, Wang X, Chen YJ, Lavie A, O’Bryan JP. (2012) A new dimension to Ras function: a novel role for nucleotide-free Ras in Class II phosphatidylinositol 3-kinase beta (PI3KC2β) regulation. PLoS One 7:e45360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Upadhyaya P, Villalona-Calero MA, Briesewitz R, Pei D. (2013) Inhibition of Ras-effector interaction by cyclic peptides. MedChemComm 4:378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie AA, Schoepfer J, Jahnke W, Cowan-Jacob SW, Loo A, Furet P, Marzinzik AL, Pelle X, Donovan J, Zhu W, et al. (2017) The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 543:733–737. [DOI] [PubMed] [Google Scholar]
- Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, Zhou C, Hu CP, O’Byrne K, Feng J, et al. (2015) Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 16:141–151. [DOI] [PubMed] [Google Scholar]
- Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, et al. (2016) Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164:805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchoan M, Hopkins A, Jaffee EM. (2017) Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 377:2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HA, Pao W. (2013) Targeted therapies: afatinib--new therapy option for EGFR-mutant lung cancer. Nat Rev Clin Oncol 10:551–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Bojorquez-Gomez A, Velez DO, Xu G, Sanchez KS, Shen JP, Chen K, Licon K, Melton C, Olson KM, et al. (2018) A global transcriptional network connecting noncoding mutations to changes in tumor gene expression. Nat Genet 50:613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Cheng F, Wang Y, Arteaga CL, Zhao Z. (2016) Systematic prioritization of druggable mutations in ∼5000 genomes across 16 cancer types using a structural genomics-based approach. Mol Cell Proteomics 15:642–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Cheng F, Zhao Z. (2017) Tissue-specific signaling networks rewired by major somatic mutations in human cancer revealed by proteome-wide discovery. Cancer Res 77:2810–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, Klitgord N, Tam S, Yu H, Venkatesan K, Mou D, et al. (2009) Edgetic perturbation models of human inherited disorders. Mol Syst Biol 5:321. [DOI] [PMC free article] [PubMed] [Google Scholar]