Summary
Specifying the primitive streak (PS) guides stem cell differentiation in vitro; however, much remains to be learned about the transcription networks that direct anterior and posterior PS cells (APS and PPS, respectively) to differentiate to distinct mesendodermal subpopulations. Here, we show that APS genes are predominantly induced in YAP1−/− human embryonic stem cells (hESCs) in response to ACTIVIN. This finding establishes the Hippo effector YAP1 as a master regulator of PS specification, functioning to repress ACTIVIN-regulated APS genes in hESCs. Moreover, transient exposure of wild-type hESCs to dasatinib, a potent C-SRC/YAP1 inhibitor, enables differentiation to APS-derived endoderm and cardiac mesoderm in response to ACTIVIN. Importantly, these cells can differentiate efficiently to normal beating cardiomyocytes without the cytoskeletal defect seen in YAP1−/− hESC-derived cardiomyocytes. Overall, we uncovered an induction mechanism to generate APS cells using a cocktail of ACTIVIN and YAP1i molecules that holds practical implications for hESC and induced pluripotent stem cell differentiation into distinct mesendodermal lineages.
Keywords: human embryonic stem cells, iPSC, primitive streak, cardiomyocyte differentiation, YAP, ACTIVIN
Highlights
-
•
YAP represses the anterior primitive streak (APS) fate in hPSCs
-
•
YAP-null-derived APS cells progress into cardiac mesoderm and endoderm
-
•
The PS cells are not fully committed and can be reprogrammed to alternate cell fates
-
•
Co-treatment of Activin + YAP1 inhibitor in hPSCs induces cardiomyocyte differentiation
In this report, Estarás, Jones and colleagues show that YAP suppresses the anterior primitive streak gene signature in hPSCs. As a consequence, YAP-null hPSCs differentiate into endoderm and cardiac mesoderm after a short Activin exposure. Authors also provide a one-step strategy to direct cardiomyocyte differentiation using a combined YAP inhibitor and Activin treatment in hPSCs.
Introduction
During embryonic development, gastrulation is marked by the formation of the primitive streak (PS). Within the PS, cells are directed to specific fates based on their spatiotemporal position. Thus, cells from the anterior part of the PS (APS) form the endoderm and cardiac mesoderm, whereas posterior PS (PPS) derivatives are destined to populate the extraembryonic and somitic mesoderm (Murry and Keller, 2008, Wang and Chen, 2016). As a consequence, the anterior and posterior regions of the PS have unique differentiation potentials and depend on different transcription networks to guide their exit from pluripotency along the anterior-posterior PS axis.
The induction of the PS in mouse embryos or cultured human epiblast cells is directed by ACTIVIN/NODAL and WNT signaling (Brennan et al., 2001, Gadue et al., 2006, Liu et al., 1999, Martyn et al., 2018, Sumi et al., 2008). In epiblast cells, WNT/β-CATENIN and ACTIVIN/SMAD2,3 cooperate to transcribe pan-PS lineage genes, such as MIXL1 and EOMES. Relatively high levels of ACTIVIN direct APS differentiation toward endoderm (SOX17+), both in vitro and in vivo, while inhibiting posterior fates through the repression of PPS genes, such as CDX2 (D'Amour et al., 2005, Gritsman et al., 2000, Mendjan et al., 2014). In turn, posterior gene markers, such as CDX2, MSGN1, and TBX6 are targets of WNT/β-CATENIN signaling. The expression of these latter genes correlates with WNT levels, and consequently stronger WNT signals contribute to the PPS fate (Amin et al., 2016, Wittler et al., 2007, Yamaguchi et al., 1999).
PS patterning also depends on other factors. For example, the DNA demethylase TET activates Lefty to downregulate NODAL levels along the streak in mouse embryos (Dai et al., 2016), whereas the transcription factor ZFP281 represses TET expression to activate genes important for NODAL signaling, such as Nodal and Foxh1 (Fidalgo et al., 2016, Huang et al., 2017). In addition, the tumor suppressor P53 regulates PS differentiation by repressing Wnt3 expression in mouse embryonic stem cells (ESCs) (Wang et al., 2017). Pluripotency factors are also key players of PS specification. For instance, early downregulation of SOX2 is required to exit pluripotency, whereas OCT4 regulates SOX17 expression, and NANOG cooperates with SMAD2,3 to induce mid-anterior and APS fates via repression of CDX2 (Aksoy et al., 2013, Mendjan et al., 2014, Rao et al., 2016). These findings suggest that factors contributing to fine-tune WNT and NODAL signaling are key to pattern the PS.
Extending these findings, we recently discovered that the Hippo effector YAP1 (Fu et al., 2017) strongly regulates the activity of WNT and ACTIVIN signaling in human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) (Estaras et al., 2015, Estaras et al., 2017). Binding of the YAP1:TEAD complex to WNT3 and NODAL enhancers in hESCs represses transcription of these ligands and block expression of WNT-ACTIVIN co-induced pan-PS genes. Importantly, long-term analysis of YAP1−/− hESCs revealed that these cells differentiate selectively along the cardiac lineage in response to ACTIVIN. These findings suggest that YAP1 regulates genes important for differentiation toward the APS fate, which is the precursor of cardiac mesoderm.
To further elaborate the role of YAP1 in PS differentiation, we used both CRISPR-engineered YAP1 knockout hESCs and an inhibitor to temporally disrupt YAP1 activity in hESCs. We show that YAP1−/− hESCs specify into APS progenitors (LHX1+/SOX17+/CDX2−/TBX6−) in response to ACTIVIN. The YAP−/−-derived APS cells further progress into cardiac mesoderm and endoderm. Importantly, we demonstrate that transient inactivation of YAP1 in hESCs using the dasatinib C-SRC/YAP1 inhibitor efficiently gave rise to functional cardiomyocytes using a one-step strategy. These findings demonstrate that YAP1 is a key regulator of APS specification, and provide a coherent strategy to guide stem cell differentiation along the endoderm and cardiac lineages.
Results
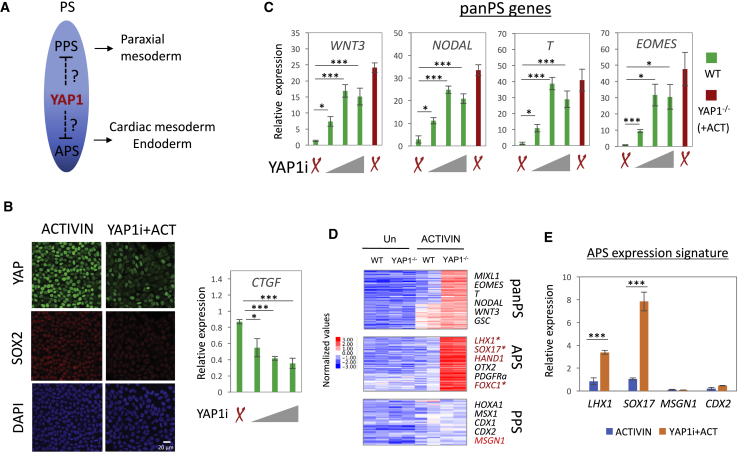
Transient Inhibition of YAP1 Induces hESC Differentiation along the Anterior PS Cell Fate
We have shown that YAP1−/− hESCs progress toward cardiac lineage in response to ACTIVIN (Estaras et al., 2017). The cardiac lineage derives from the mid-anterior part of the PS, and therefore we hypothesize that inhibition of YAP1 activity in hESCs favors differentiation to anterior-like progenitors in response to ACTIVIN. Thus, we aimed to explore whether transient inactivation of YAP1 in hESCs affects the specification of PS progenitors (Figure 1A). To begin, we treated wild-type (WT) hESCs with dasatinib, a small-molecule C-SRC kinase inhibitor shown to affect the actin cytoskeleton and compromise the stability of YAP1 (Zanconato et al., 2016). Figure 1B shows that 24 hr dasatinib (YAP1i) treatment in hESCs effectively decreased YAP1 protein levels along with a dose-dependent reduction of the YAP1 target gene, CTGF. We next tested whether transient exposure to YAP1i is sufficient to induce widely expressed pan-PS genes. As shown in Figure 1C, the combined ACTIVIN + YAP1i treatment in WT cells induced expression of MIXL1, EOMES, and T genes to levels comparable with those seen in YAP1−/− hESCs treated with ACTIVIN. These results were consistent among two different hESCs lines (H1 and H9), also the EC11 iPSC line (Figure S1).
Figure 1.
YAP1 Depletion in hESCs Induces APS Fate in Response to ACTIVIN
(A) A diagram illustrating the potential role of YAP1 in specifying PS fate.
(B) Left: immunostaining of YAP1 and SOX2 in H1 hESCs treated with ACTIVIN or ACTIVIN + dasatinib (YAP1i) for 24 hr. Right: CTGF mRNA abundance (qRT-PCR) in H1 hESCs treated with dasatinib for 24 hr. The final concentration of dasatinib varied from 0, 5, and 10 to 20 nM.
(C) mRNA abundance of tested pan-PS genes in WT and YAP1−/− H1 hESCs at 24 hr following the indicated treatments.
(D) An RNA-seq heatmap for two biological independent replicates of WT or YAP1−/− H1 hESCs, untreated or treated with ACTIVIN for 24 hr.
(E) mRNA abundance of APS (LHX1 and SOX17) and PPS (MSGN1 and CDX2) genes. The final concentration of dasatinib (YAP1i) was 10 nM. n ≥ 3 biological independent experiments, mean ± SD.
Statistical significance of difference by paired two-tailed t test; ∗p < 0.05; ∗∗∗p < 0.001. Non-significant differences are not marked.
hESCs can differentiate into anterior- and posterior-like PS cells, which are partially committed precursors that give rise to distinct mesoderm and endoderm tissues. To further investigate the identity of the YAP1-depleted PS cells, we examined their transcriptome by RNA sequencing (RNA-seq) (Estaras et al., 2017). Surprisingly, we found that ACTIVIN-treated YAP1−/− hESCs express high levels of APS markers, such as SOX17, LHX1, and HAND1, but low levels of PPS markers (MSGN1, CDX2, and HOXA1) compared with ACTIVIN-treated WT hESCs. Moreover, we found that hESCs treated with ACTIVIN + YAP1i also express high levels of APS, but not PPS markers (Figure 1E). We conclude that ACTIVIN treatment in YAP1-null human pluripotent stem cells (hPSCs) induces APS fate. Overall, these data show that YAP1 regulates the A-P specification of PS, and provides a specific approach to generate APS cells from hPSCs in vitro, through a brief exposure to ACTIVIN and dasatinib (YAP1i).
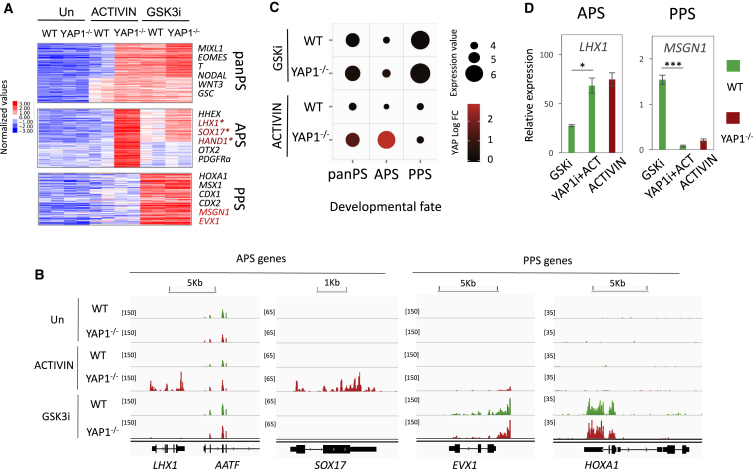
YAP1 Does Not Regulate Differentiation to Posterior PS Cell Fate
In hESCs, a strong WNT/β-CATENIN signal promotes the induction of PPS fate. In agreement with this finding, RNA-seq analysis of WNT signaling in WT hESCs treated with a GSK3 inhibitor (GSK3i) (factor XV), revealed strong expression of pan-PS and PPS genes, including MSGN1, EVX1, and HOXA1, but no significant induction of the APS transcriptome (Figure 2A). Interestingly, loss of YAP1 did not significantly affect the induction of the PPS genes by GSK3i. Neither inhibition nor loss of YAP1 altered the induction of PPS genes in response to ACTIVIN (Figures 2 and S2). In contrast, YAP1 inactivation potentiated the expression of some APS genes under GSK3i treatments, including HHEX and LHX1 (Figures 2A–2C), but the effect was less dramatic relative to that seen in ACTIVIN-treated cells. Taken together, these results indicate that YAP1 selectively represses the APS genes in hESCs by counteracting ACTIVIN signaling.
Figure 2.
Differentiation to PPS Fate Is Independent of YAP1
(A) An RNA-seq heatmap of WT or YAP1−/− H1 hESCs, untreated, or treated with either ACTIVIN or GSK3i for 24 hr, as indicated (two biological independent replicates).
(B) Representative loci of APS and PPS genes from RNA-seq.
(C) A bubble chart to summarize the dependency of pan-PS, APS, and PPS induction on YAP1 activity. Bubble sizes represent the log2 scale average gene expression. Bubble colors represent YAP1 logFC, defined as the average logFC of YAP1−/− over WT expression under each treatment (GSKi or ACTIVIN).
(D) mRNA abundance of LHX1 and MSGN1 in WT and YAP1−/− H1 hESCs treated as indicated, for 24 hr. The final concentration of dasatinib (YAP1i) was 10 nM. n = 3 biological independent experiments, mean ± SD.
∗p < 0.05; ∗∗∗p < 0.001.
ACTIVIN + Dasatinib (YAP1i)-Derived PS Cells Can Differentiate along the Endoderm and Cardiac Cell Lineages
These data show that exposure of hESCs to a combination of ACTIVIN with dasatinib (YAP1i) leads to strong induction of APS genes, whereas WNT/GSK3i induces the PPS genes. These divergent roles of ACTIVIN and WNT in specifying PS cell fates prompted us to assess possible differences in their developmental potential. We first evaluated the ability of the ACTIVIN + YAP1i-induced PS cells to differentiate toward two anterior derivatives, endoderm and the cardiac lineages. Following 24 hr incubation with ACTIVIN and YAP1i, the medium was replaced and the cells were analyzed following a total of 5 days in culture. As shown in Figures 3A and 3B, the ACTIVIN + YAP1i-induced PS cells expressed high levels of the cardiac precursor marker, NKX2.5 (Paige et al., 2015), indicating that these cells acquired a cardiac lineage without any additional treatment (condition C). To test whether the ACTIVIN + YAP1i-derived PS cells can differentiate into endoderm, the cells were exposed to a prolonged ACTIVIN signaling (days 2–5) (Mallanna and Duncan, 2013). Importantly, the ACTIVIN + YAP1i-exposed PS cells express high levels of SOX17, strongly suggesting that these precursors had differentiated to endoderm cells (Figures 3A and 3B, condition D).
Figure 3.
ACTIVIN + Dasatinib (YAP1i)-Derived PS Cells Differentiate into Endoderm and Cardiac Lineages
(A) The diagram illustrates the different procedures that were tested for hESC differentiation.
(B) mRNA abundance of MEOX1, NKX2.5, and SOX17 on day 5 from the initial treatments in H1 hESCs, as indicated in diagram. n = 3 biological independent experiments, mean ± SD. ∗p < 0.05; ∗∗∗p < 0.001.
(C) The diagram illustrates the various signaling cues that direct PS fate specification to mesendodermal lineages.
Finally, we examined the capacity of the ACTIVIN + YAP1i-treated cells to differentiate into posterior derivatives by analyzing the expression of the paraxial gene MEOX1 (Mankoo et al., 2003) at day 5 following the initial treatment. Interestingly, we found that MEOX1 expression remained low in the ACTIVIN + YAP1i-treated cells (Figure 3B, condition C) in contrast to the high expression in the GSK3i-treated cells (Figure 3B, condition B). In accordance with their posterior nature, the GSK3i-induced PS cells failed to differentiate into anterior cardiac lineages or endoderm (Figure 3B, condition A). Overall, we conclude that unlike GSK3i, a treatment combining ACTIVIN and YAP1i is effective to generate PS precursors with a differentiation capacity toward anterior and mid-anterior derivatives.
PS Progenitors Can Be Reprogrammed
We have demonstrated that GSK3i-induced PPS cells are prone to acquire a paraxial fate. Therefore, it seems contrary to employ WNT ligands or GSK3i as the most common practice for inducing cardiac differentiation. Noteworthy, the GSK3i-dependent cardiomyocyte differentiation requires an additional step at 3 days that is designed to block further WNT signaling (referred to as GiWi protocol in Lian et al., 2013, Loh et al., 2016).
It is shown that the addition of a WNT inhibitor is necessary to cease the expression of key cardiac inhibitors such as CDX2 and MSX1, allowing the cells to adopt a cardiac fate (Rao et al., 2016) (Figure S3). We hypothesize that the WNT inhibitor may also serve to suppress the expression of paraxial markers, and therefore redirect the posterior PS cells toward cardiac mesoderm. To test this possibility, we analyzed the expression of paraxial and cardiac genes in the presence or absence of WNT inhibitor, which was added 2 days following the GSK3i treatment (Figure 3A, conditions A and B). As predicted, Figure 3B (condition B) shows that the exposure to a WNT inhibitor strongly favors the induction of the cardiac lineage marker NKX2.5 in GSK3i-treated cells. More importantly, the WNT inhibitor also impairs the induction of the paraxial gene, MEOX1. Consequently, blocking of WNT signaling reprograms the PPS precursors from a paraxial to a cardiac cell fate (Figure 3B, conditions A and B). These findings demonstrate that APS and PPS cells are not fully committed precursors, but rather can be redirected to alternate fates in response to appropriate developmental cues.
ACTIVIN + YAP1i Treatment Provides an Alternative Strategy for Cardiac Induction
Thus far, these data strongly indicate that combined treatment of ACTIVIN and YAP1i during the first 24 hr of differentiation in WT hESCs and hiPSCs is sufficient to generate APS cells that efficiently differentiate into cardiac mesoderm (NKX2.5+/GATA4+/CDX2−) (Figures 3 and S3). Thus, we reasoned that transient ACTIVIN + YAP1i exposure might provide an alternative strategy to generate cardiomyocytes in WT hESCs (Figure 4A). To test this possibility, we investigated the expression of cardiac markers following a full 14 days of differentiation. Indeed, ACTIVIN + YAP1i-treated hESCs express high levels of the CTNT, MLC2-A, and NKX2.5 cardiac markers (Figure 4B). Importantly, fluorescence-activated cell sorting analysis revealed that approximately 70% of the ACTIVIN + YAP1i-treated PS cells were CTNT positive, which is similar to the cardiomyocyte yield from the GiWi protocol (Lian et al., 2013) (Figures 4C and S4B). In addition, active beating cardiomyocytes derived from ACTIVIN + YAP1i treatment were observed in culture from day 8 onward (Video S1). As shown in the Venn diagram of Figure 4D, the transcriptome of the ACTIVIN + YAP1i-derived cardiomyocytes shows extensive overlap with that of cardiomyocytes derived using the GiWi protocol, and the heatmap further establishes that key cardiac marker genes are strongly expressed in cardiac cells derived from both protocols.
Figure 4.
ACTIVIN + Dasatinib (YAP1i) Treatment Is an Alternative Strategy for Cardiomyocyte Differentiation
(A) Steps of the GiWi and one-step protocols for cardiomyocyte differentiation.
(B) Immunoblots of MLC-2A, CTNT, and NKX2.5 expression on day 14 of H1 hESC treatment.
(C) Fluorescence-activated cell sorting analysis of CTNT+ cells on day 14 of treatments. n = 3 biological independent experiments, mean ± SD. ∗∗∗p < 0.001.
(D) Left: a Venn diagram summarized the RNA-seq data showing upregulated genes in cardiomyocytes derived from GiWi or one-step protocols. Right: a heatmap of the top 50 upregulated cardiac genes in H1 hESCs and cardiomyocytes generated from the GiWi or one-step protocols, respectively. Two biological independent experiments of hESCs and GiWi-derived cardiomyocytes (CMs), and three biological independent experiments of YAP1i-derived CMs.
(E) Immunostaining of CTNT in cardiomyocytes derived from WT or YAP1−/− H1 hESCs following the GiWi or one-step protocols.
(F) The schematic depicts the strategy of APS specification for cardiac mesoderm and endoderm differentiation.
We have recently reported that YAP1−/−-derived cardiomyocytes are viable; however, the organization of the actin cytoskeleton is significantly compromised (Estaras et al., 2017), as shown by immunostaining with CTNT (Figure 4E). Importantly, analysis of the ACTIVIN + YAP1i-derived cardiomyocytes revealed an intact actin cytoskeleton that resembles cardiomyocytes obtained using the GiWi strategy. We conclude that the one-step strategy (ACTIVIN + dasatinib [YAP1i]) is an effective approach for the generation of functional cardiomyocytes. In summary, this work shows that ACTIVIN + YAP1i co-treatment in hESCs provides a robust method to generate APS cells that effectively differentiate into endoderm and cardiac lineages (Figure 4F).
Discussion
Transcription factors involved in primitive streak patterning play an essential role in early organogenesis and stem cell differentiation. Here, we show that YAP1 is a key regulator of PS cell specification through its ability to selectively repress the expression of genes for APS fate. We find that YAP1 represses ACTIVIN target genes, including SOX17, without affecting the expression of PPS genes. As a result, the loss of YAP1 in hESCs exposed to ACTIVIN enables efficient differentiation toward the APS cell fate. We previously showed that YAP1 inhibits hESC differentiation to cardiac mesoderm (Estaras et al., 2017). Here, we extend these findings by showing that YAP1 inhibits APS gene expression to restrain the cardiac fate. Moreover, we further discovered that APS cells generated from hESCs treated with ACTIVIN and the YAP1-inhibitor dasatinib can further differentiate to endoderm cells capable of further development to lung, liver, or pancreatic β cells.
How does YAP1 repress APS fate in hESCs? Several lines of evidence suggest that ACTIVIN acts in a dose-dependent manner to induce APS cell fate. In mouse models and ESCs, high ACTIVIN levels induce the APS markers Sox17, Foxa2, and Cer1, and repress the PPS marker Cdx2 (Brennan et al., 2001, Estaras et al., 2017, Mendjan et al., 2014). Our observation that YAP1 represses NODAL in hESCs raises the possibility that elevated SMAD2,3 signaling in YAP1−/− cells may contribute to this phenotype (Estaras et al., 2017). Nevertheless, high ACTIVIN/NODAL signaling is not sufficient to induce an APS fate in WT hESCs, indicating the existence of other key YAP1 targets. Consistent with this idea, we find that YAP1 counteracts the ability of ACTIVIN/SMAD2,3 to repress the WNT3 gene in hESCs. In the absence of YAP1, SMAD2,3 activate endogenous WNT3 and nuclear β-CATENIN to initiate the PS program. Consequently, exposure of hESCs to WNT or GSK3i can overcome the ability of YAP1 to repress pan-PS genes in response to ACTIVIN. However, GSK3i treatment or WNT signaling does not bypass the need for YAP1 to repress the APS genes. Consequently, the APS gene program is induced in response to ACTIVIN only in cells that lack YAP1 activity. What are the key YAP1 targets? RNA-seq analysis of YAP1−/− hESCs exposed to ACTIVIN revealed SOX17, a master regulator of endoderm and cardiac mesoderm specification (Liu et al., 2007, Viotti et al., 2014). In addition to pan-PS genes, we speculate that YAP1 also counteracts SMAD2,3-dependent transcription on APS genes, such as SOX17, to regulate PS specification. Further studies are needed to examine whether YAP1 and SMAD2,3 bind differentially to APS and PPS genes. Taken together, we conclude that YAP1 depletion in hESCs unlocks the ability of ACTIVIN to induce differentiation by activating endogenous WNT3 signaling and allowing SMAD2,3 to induce pan-PS and APS genes, such as SOX17.
hPSC differentiation is an excellent model to identify new regulators and mechanisms of cell differentiation in embryos (Ang et al., 2016). Accordingly, the phenotype of YAP1−/− embryos recapitulates the misregulation of the PS gene expression that we observe in YAP1−/− hESCs. YAP1−/− embryos die soon after the onset of gastrulation, indicating a crucial role of YAP1 in the specification of three germ layers in vivo. Analyses of embryos at the peri-gastrulation stage reveals that the PS in YAP1−/− embryos appears shorter and broader than in WT embryos (Morin-Kensicki et al., 2006). We speculate that the anterior-posterior axis defects in YAP1−/− embryos reflect the inability of YAP1 to repress APS genes. These findings suggest that the PS abnormality in YAP1−/− embryos might include an expanded APS domain (SOX17+/CDX2−) with a curtailed or absent PPS domain (CDX2+), similar to YAP1−/− hESCs that preferentially induce the APS but not PPS program. In addition, YAP1 expression in developing embryos is lower within the region of the PS destined to become endoderm later in development. Thus, low levels of YAP1 likely favor the in vivo specification of APS precursors, as we observed in hESCs. Altogether, these findings suggest that distinct levels of YAP1 along the anterior-posterior axis could serve as a sensor to enable cells to exit pluripotency and adopt specific PS fates.
Lastly, we extend our findings to an alternative strategy for directing hESC and iPSC differentiation using a combined ACTIVIN and dasatinib (YAP1i) treatment. We show that a transient exposure of WT hESCs to ACTIVIN and dasatinib is sufficient to generate beating cardiomyocytes. Of note, we find that including ACTIVIN and dasatinib in the stem cell medium, without additional supplements, is sufficient to initiate hESC specification along the APS lineage. This simple procedure illuminates the critical control of ACTIVIN signaling and YAP1 activity in APS induction. Most importantly, the transient inhibition of YAP1 eliminates the severe cytoskeletal defects observed in YAP1−/− hESC-derived cardiomyocytes, and bypasses the need for cell engineering. Consequently, the use of ACTIVIN and dasatinib to induce hESCs and iPSCs to undergo APS differentiation along the cardiomyocyte lineage may provide a useful step toward the therapeutic goals of regenerative medicine.
Experimental Procedures
ACTIVIN One-Step Cardiomyocyte Differentiation
WT hESC cultures were disaggregated into single cells using Accutase. Cells were seeded at 200,000 cells/well in Matrigel-coated 24-well plates and grown until reaching 80% confluence. At day 0, hESCs were simultaneously treated with dasatinib (10 nM, Sigma) and ACTIVIN (100 ng/mL) in the mTeSR1 medium for 24 hr. Then, the medium was aspirated and replenished with RPMI/B-27 minus insulin and replaced every 2 days. From day 7, differentiated cells were maintained in RPMI/B-27 containing insulin and replenished in every 3 days. Spontaneous beating phenotype should occur from day 7 to 9 onward.
RNA-Seq Analysis
Author Contributions
H.-T.H. and C.E. conceived the study, designed and performed the experiments, and wrote the manuscript. L.H. performed the bioinformatics analysis of the sequencing data. K.A.J. designed the experiments and wrote the manuscript.
Acknowledgments
We acknowledge support from Salk Bioinformatics and Stem Cell Cores. This work is funded by the California Institute for Regenerative Medicine (GC1R-06673-B) and NIH/NCI RO1CA125535 (to K.A.J.). Conchi Estarás was supported by Salkexcellerators Program.
Published: November 15, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, three tables, and one video and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.10.013.
Contributor Information
Conchi Estarás, Email: cestarasibanez@salk.edu.
Katherine A. Jones, Email: jones@salk.edu.
Accession Numbers
Analyzed data are provided in Supplemental Tables. Genome-wide datasets are deposited at the GEO under accession number GEO: GSE121877 and GSE99202.
Supplemental Information
References
- Aksoy I., Jauch R., Chen J., Dyla M., Divakar U., Bogu G.K., Teo R., Leng Ng C.K., Herath W., Lili S. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 2013;32:938–953. doi: 10.1038/emboj.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S., Neijts R., Simmini S., van Rooijen C., Tan S.C., Kester L., van Oudenaarden A., Creyghton M.P., Deschamps J. Cdx and T Brachyury co-activate growth signaling in the embryonic axial progenitor niche. Cell Rep. 2016;17:3165–3177. doi: 10.1016/j.celrep.2016.11.069. [DOI] [PubMed] [Google Scholar]
- Ang Y.S., Rivas R.N., Ribeiro A.J.S., Srivas R., Rivera J., Stone N.R., Pratt K., Mohamed T.M.A., Fu J.D., Spencer C.I. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167:1734–1749.e22. doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J., Lu C.C., Norris D.P., Rodriguez T.A., Beddington R.S., Robertson E.J. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- Dai H.Q., Wang B.A., Yang L., Chen J.J., Zhu G.C., Sun M.L., Ge H., Wang R., Chapman D.L., Tang F. TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016;538:528–532. doi: 10.1038/nature20095. [DOI] [PubMed] [Google Scholar]
- Estaras C., Benner C., Jones K.A. SMADs and YAP compete to control elongation of beta-CATENIN: LEF-1-recruited RNAPII during hESC differentiation. Mol. Cell. 2015;58:780–793. doi: 10.1016/j.molcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estaras C., Hsu H.T., Huang L., Jones K.A. YAP repression of the WNT3 gene controls hESC differentiation along the cardiac mesoderm lineage. Genes Dev. 2017;31:2250–2263. doi: 10.1101/gad.307512.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M., Huang X., Guallar D., Sanchez-Priego C., Valdes V.J., Saunders A., Ding J., Wu W.S., Clavel C., Wang J. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell. 2016;19:355–369. doi: 10.1016/j.stem.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu V., Plouffe S.W., Guan K.L. The Hippo pathway in organ development, homeostasis, and regeneration. Curr. Opin. Cell Biol. 2017;49:99–107. doi: 10.1016/j.ceb.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P., Huber T.L., Paddison P.J., Keller G.M. WNT and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2006;103:16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsman K., Talbot W.S., Schier A.F. Nodal signaling patterns the organizer. Development. 2000;127:921–932. doi: 10.1242/dev.127.5.921. [DOI] [PubMed] [Google Scholar]
- Huang X., Balmer S., Yang F., Fidalgo M., Li D., Guallar D., Hadjantonakis A.K., Wang J. Zfp281 is essential for mouse epiblast maturation through transcriptional and epigenetic control of Nodal signaling. Elife. 2017;6 doi: 10.7554/eLife.33333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating WNT/beta-CATENIN signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Wakamiya M., Shea M.J., Albrecht U., Behringer R.R., Bradley A. Requirement for WNT3 in vertebrate axis formation. Nat. Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Liu Y., Asakura M., Inoue H., Nakamura T., Sano M., Niu Z., Chen M., Schwartz R.J., Schneider M.D. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Chen A., Koh P.W., Deng T.Z., Sinha R., Tsai J.M., Barkal A.A., Shen K.Y., Jain R., Morganti R.M. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell. 2016;166:451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallanna S.K., Duncan S.A. Differentiation of hepatocytes from pluripotent stem cells. Curr. Protoc. Stem Cell Biol. 2013;26:1G.4.1–1G.4.13. doi: 10.1002/9780470151808.sc01g04s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoo B.S., Skuntz S., Harrigan I., Grigorieva E., Candia A., Wright C.V., Arnheiter H., Pachnis V. The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development. 2003;130:4655–4664. doi: 10.1242/dev.00687. [DOI] [PubMed] [Google Scholar]
- Martyn I., Kanno T.Y., Ruzo A., Siggia E.D., Brivanlou A.H. Self-organization of a human organizer by combined WNT and Nodal signalling. Nature. 2018;558:132–135. doi: 10.1038/s41586-018-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., Mascetti V.L., Ortmann D., Ortiz M., Karjosukarso D.W., Ng Y., Moreau T., Pedersen R.A. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15:310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki E.M., Boone B.N., Howell M., Stonebraker J.R., Teed J., Alb J.G., Magnuson T.R., O'Neal W., Milgram S.L. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol. Cell. Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Paige S.L., Plonowska K., Xu A., Wu S.M. Molecular regulation of cardiomyocyte differentiation. Circ. Res. 2015;116:341–353. doi: 10.1161/CIRCRESAHA.116.302752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J., Pfeiffer M.J., Frank S., Adachi K., Piccini I., Quaranta R., Arauzo-Bravo M., Schwarz J., Schade D., Leidel S. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell. 2016;18:341–353. doi: 10.1016/j.stem.2015.11.019. [DOI] [PubMed] [Google Scholar]
- Sumi T., Tsuneyoshi N., Nakatsuji N., Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical WNT/beta-CATENIN, ACTIVIN/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- Viotti M., Nowotschin S., Hadjantonakis A.K. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat. Cell Biol. 2014;16:1146–1156. doi: 10.1038/ncb3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen Y.G. Signaling control of differentiation of embryonic stem cells toward mesendoderm. J. Mol. Biol. 2016;428:1409–1422. doi: 10.1016/j.jmb.2015.06.013. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zou Y., Nowotschin S., Kim S.Y., Li Q.V., Soh C.L., Su J., Zhang C., Shu W., Xi Q. The p53 family coordinates WNT and Nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell. 2017;20:70–86. doi: 10.1016/j.stem.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittler L., Shin E.H., Grote P., Kispert A., Beckers A., Gossler A., Werber M., Herrmann B.G. Expression of Msgn1 in the presomitic mesoderm is controlled by synergism of WNT signalling and Tbx6. EMBO Rep. 2007;8:784–789. doi: 10.1038/sj.embor.7401030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T.P., Takada S., Yoshikawa Y., Wu N., McMahon A.P. T (Brachyury) is a direct target of WNT3a during paraxial mesoderm specification. Genes Dev. 1999;13:3185–3190. doi: 10.1101/gad.13.24.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Battilana G., Cordenonsi M., Piccolo S. YAP/TAZ as therapeutic targets in cancer. Curr. Opin. Pharmacol. 2016;29:26–33. doi: 10.1016/j.coph.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.