Abstract
The human brain can be represented as a graph in which neural units such as cells or small volumes of tissue are heterogeneously connected to one another through structural or functional links. Brain graphs are parsimonious representations of neural systems that have begun to offer fundamental insights into healthy human cognition, as well as its alteration in disease. A critical open question in network neuroscience lies in how neural units cluster into densely interconnected groups that can provide the coordinated activity that is characteristic of perception, action, and adaptive behaviors. Tools that have proven particularly useful for addressing this question are community detection approaches, which can identify communities or modules: groups of neural units that are densely interconnected with other units in their own group but sparsely interconnected with units in other groups. In this paper, we describe a common community detection algorithm known as modularity maximization, and we detail its applications to brain graphs constructed from neuroimaging data. We pay particular attention to important algorithmic considerations, especially in recent extensions of these techniques to graphs that evolve in time. After recounting a few fundamental insights that these techniques have provided into brain function, we highlight potential avenues of methodological advancements for future studies seeking to better characterize the patterns of coordinated activity in the brain that accompany human behavior. This tutorial provides a naive reader with an introduction to theoretical considerations pertinent to the generation of brain graphs, an understanding of modularity maximization for community detection, a resource of statistical measures that can be used to characterize community structure, and an appreciation of the usefulness of these approaches in uncovering behaviorally-relevant network dynamics in neuroimaging data.
Keywords: community detection, modularity, multi-scale, multi-layer, brain networks
Modularity has been hailed as fundamental property of the human brain dating back to Greek philosophy, and it has continued to inform theories from phrenology [86] and cognitive science [79, 36, 172], to brain mapping and functional localization [168]. The relevance of modularity for brain structure and function stems from its fundamental advantages for evolution and development. Research across the biological sciences suggests that modular organization allows for rapid adaptation [115, 100] and provides robustness to either sudden or gradual perturbations in genes or environment [123, 115, 116]. Unlike homogeneously connected networks, modular networks can effectively buffer the impact of perturbations by keeping their effects relatively local [152] while simultaneously enabling efficient information processing [75, 11], supporting functional specialization [88] and efficient learning [74]. These benefits of modularity are particularly relevant for the human brain, which evolved under evolutionary pressures for adaptability [132], energy efficiency, and cost minimization [60, 49, 173, 55, 34], and which also develops under biological pressures to balance segregation and integration of function [49].
Exactly how modularity is instantiated in the brain is a question that has fascinated neuroscientists for more than a century. The answer to this question is complicated by the fact that the brain is a complex system composed of neural units that communicate with one another in dynamic spatiotemporal patterns [5]. How these patterns of communication are organized, reflecting thought, cognition, and behavior remains a mystery [44]. A particularly appropriate mathematical language to describe these patterns – and to determine the role that modularity might play within them – is network science [155]. In its simplest form, network science summarizes a system by isolating its component parts (nodes) and their pairwise interactions (edges) in a graph [40]. Over the last decade, the application of network science to neuroscience (also known as network neuroscience [16]) has offered intuitions for the fundamental principles of organization and function in the brain [48].
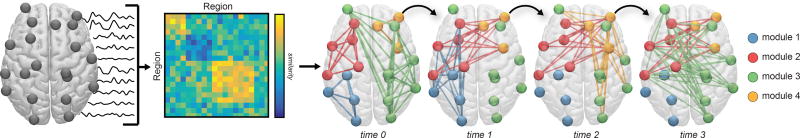
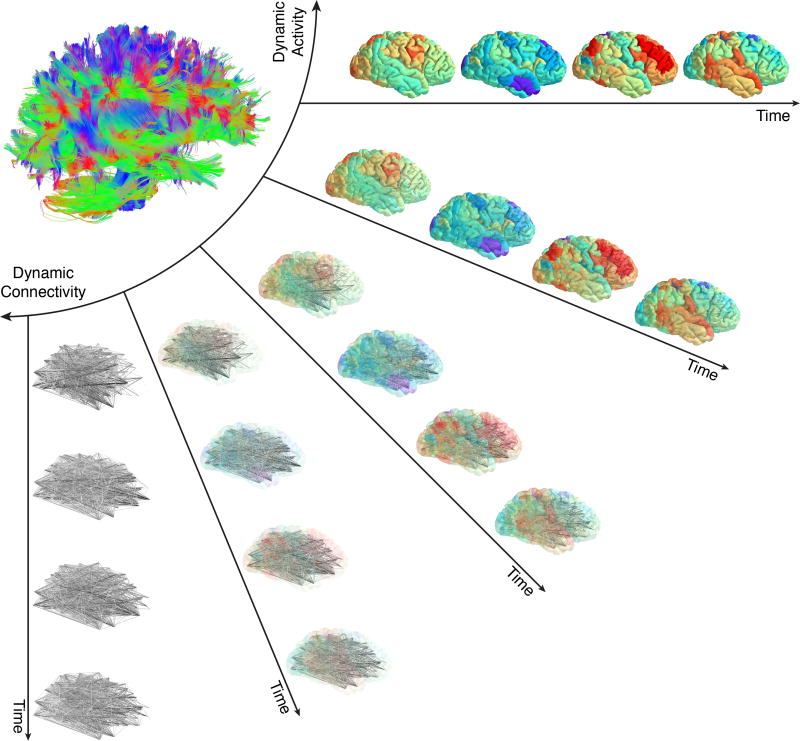
A quintessential concept in network neuroscience is the notion of network modularity, wherein neural units are structurally or functionally connected to one another in clusters or modules [139]. Intuitively, modularity is an architectural design feature that allows system processes to implement local integration of information, while maintaining systems-level adaptability. Graphs that display modular structure [82] can give rise to more complex dynamics than graphs that display random structure [196]. Modular networks of coupled oscillators also promote synchronizability [7] as well as the formation of chimera states, characterized by the coexistence of synchronized and desynchronized elements [216]. Modularity often exists across multiple hierarchical levels [138], enabling rapid responses to fluctuating external input [122, 25, 142], and supporting complex dynamics alongside functional efficiency [187, 113]. The functional properties that modularity confers to a system provide strong motivation for studying modular organization in brain graphs across both health and disease (Fig. 1).
Fig. 1. Brain graphs and communities within them.
One can construct a brain graph in several ways, and subsequently study its modular architecture using community detection techniques developed for graphs. Here, we illustrate an example pipeline in which we use non-invasive neuroimaging in humans to obtain regional timeseries of continuous neural activity (Left). Next, we define a weighted undirected graph and represent that graph in an adjacency matrix, each element of which provides an estimate of the statistical similarity between the time series of region i and the time series of region j (Middle). Finally, we apply community detection techniques to the brain graph to identify modules. Here, a module is composed of nodes (regions) that are more densely interconnected with one another than expected in some appropriate random network null model. If we have temporally extended data, we can also consider defining a temporal graph, and using dynamic community detection techniques to study the temporal evolution of modules and their relation to cognition (Right). In this review, we discuss considerations, methods, statistics, and interpretations relevant to this process.
In this tutorial, we survey the literature pertinent to an understanding of modular structure in brain graphs, and its relevance for human cognition. We begin by discussing common methods for building brain graphs from diverse data sources representing distinct types of neurophysiological signals. We then turn to a description of community detection approaches commonly applied to such graphs, and we place a particular emphasis on the method of modularity maximization. Next, we offer a resource of statistical measures that can be used to characterize community structure in brain graphs, including measures of their topology and embedding into physical space. We complement the discussion of methods and statistics for single graphs with a description of extensions of community detection approaches for time-evolving graphs. Next, we provide a resource of statistical measures that can be used to characterize dynamic community structure in brain graphs. We then review applications of these techniques to questions in neuroscience to give the reader an appreciation of the usefulness of these approaches in uncovering behaviorally-relevant network dynamics in neuroimaging data. Finally, we discuss methodological innovations that are needed to advance our understanding of how patterns of coordinated activity in the human brain account for behavior.
Building brain graphs
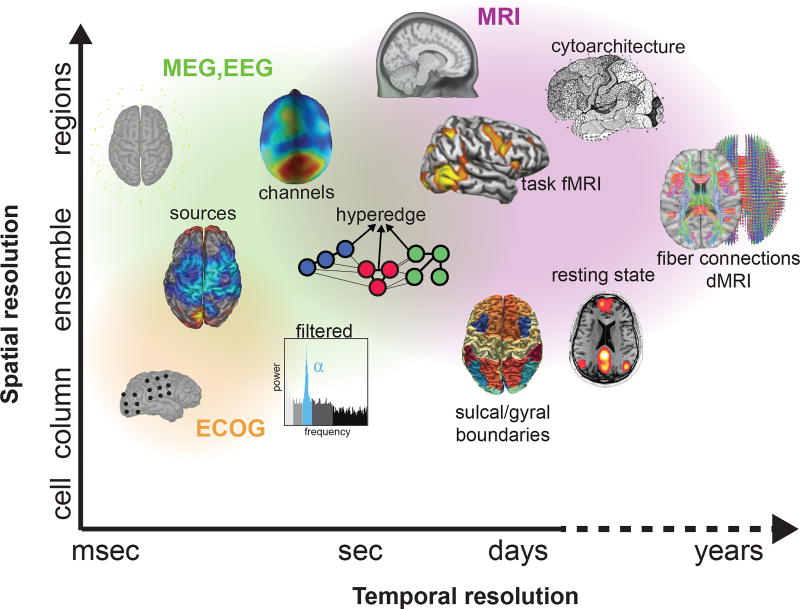
In its general form, a brain graph is composed of (i) a set of nodes characterizing anatomical, functional, or computational units, and (ii) a set of edges representing pairwise relations between two nodes (Fig. 2). Brain graphs can be built from many types of neuroimaging data, including magnetic resonance imaging (MRI), electrocorticography (ECoG), electroencephalography (EEG), and magnetoencephalography (MEG). Across all modalities, one seeks a definition of nodes that distills the brain into its most fundamental pieces relevant to the hypothesis being tested, and a definition of edges that constitutes the fundamental relation relevant to the function under study [51]. The choice of nodes and edges guides the appropriate interpretations that can be drawn from the graph [171, 51, 215].
Fig. 2. Defining brain graphs: common datatypes.
Various neuroimaging techniques can be used to measure brain network dynamics. Due to their prominence in the literature, we focus on direct measurements from implanted electrodes on the cortical surface (ECoG; orange), sensors on or above the scalp (EEG or MEG; green), and indirect measurements from BOLD and diffusion (MRI; purple). Each technique is associated with a specific spatial and temporal scale that can offer different insights into brain structure and function.
a) Defining nodes: In MRI studies, nodes can be defined using anatomical landmarks [1], cytoarchitecture [45, 212], sulcal and gyral landmarks [105, 68, 69, 207], or boundaries of functional activation [167, 222], either in controlled [219] or uncontrolled [223, 127] tasks. Indeed, functional atlases [61, 151] can incorporate activation information across many tasks [71] or parse which regions tend to be activated independently [171]. Nodes at a large-scale can also be defined based on connectivity at a small scale. For example, in connectivity-based parcellation [214, 26], one begins with voxel-level estimates of structural connectivity from diffusion MRI [210], and then applies a clustering technique to extract modules of densely interconnected voxels; each module is then treated as a node in the brain graph [73].
In ECoG, MEG, and EEG studies, graphs are constructed to study synchronized activity of neuronal ensembles [163, 89, 46]. Frequently, nodes are chosen to reflect electrodes in EEG or ECoG (e.g., [119, 50, 118]) and sensors in MEG (e.g., [22, 15, 197]). However, electrode and sensor time series reflect a combination of signals from cortical and subcortical sources, and are susceptible to artifacts from muscles contractions, head movements, and environmental noise [211]. To address these limitations, one can apply source reconstruction [for review, 99, 177], and then use those sources as nodes [191, 147]. One can also use multiple functional neuroimaging measurements such as simultaneous fMRI and EEG recordings [47] to define nodes [148]. Importantly, each method of node determination has implications for network estimates of brain dynamics, as described more thoroughly in several recent reviews [106, 198, 192].
Defining edges
Edges can reflect structural connections across spatial scales [30], such as bundles of axonal fibers between regions (e.g., [218, 146, 112]) or synapses between neurons. In humans, structural connections are measured using diffusion MRI [for review, see 10, 210], and fiber trajectories are estimated using tractography methods [143, 27]. The resultant brain graph can be used to identify specific connections that enable efficient and rapid communication between regions [18], or collections of connections with diverse higher order structures [94, 190, 189].
Edges can also reflect coordinated activity between regions thought to underlie cognition [83, 84, 92], where coordination is quantified by correlation, coherence, phase lag index, or a measure of synchronization between time series [23, 107], or by effective connectivity methods that estimate casual relations [85]. Such functional time series can be derived from MRI [111, 108, 109, 96] or from EEG in cognitively relevant frequency bands [52, 43]. An important caveat for estimates of functional connectivity between two regions is that they can be driven by a third source [37, 98].
When studying relations among edges in time-varying graphs – where a graph is constructed from each of many time windows–, one can use a hypergraph to formalize the idea that groups of edges, rather than single edges alone, represent a fundamental unit of interest [20, 65, 102]. This approach is partially motivated by evidence suggesting that edges can develop differentially in a coordinated fashion over the lifespan [66], leading to architectural features that cannot simply be defined by graphs composed of dyads [20]. Such developmental coordination of functional connections might be driven by intrinsic computations [20], and subsequently have mutually trophic effects on underlying structural connectivity [17]. Co-varying functional connections in early life could support the emergence of cognitive systems observed in adulthood [102]. Hypergraphs can formalize these relationships, and thereby offer a unique perspective on brain graph architecture.
Evaluating community structure in brain graphs
After choosing nodes and edges, we let G = (V, A) be a complex network of N nodes, where V = {1, ⋯, N} is the node set, and A ∈ ℝN×N is the adjacency matrix whose elements Aij give the weight of the edge between node i and node j. In a binary graph, elements are either values of 0 or 1, which indicate whether an edge exists, while in a weighted graph, elements have non-binary values that reflect the strength of their pairwise connection. If the edge weight between a node pair is symmetric, the graph is called undirected, and Aij = Aji for all (i, j); the graph is called directed otherwise. In an undirected binary graph, the degree of a node, ki, is given by the number of its non-zero edges: ki = Σj∈N Aij. To evaluate community structure in the brain graph, we note that a community structure is a partition 𝒞 = {C1, ⋯, CK}, where Ci ⊂ V consists of the nodes in the ith community and K is the number of communities in G. Here we only consider non-overlapping community structure, which means that Ci∩Cj = ∅ if i ≠ j. We note that in the case of a temporal graph with L layers, and where the adjacency matrix of layer l has elements Aijl, one can similarly define a community structure as a set of L partitions.
The overarching goal of community detection is to provide an understanding of how nodes are joined together into tightly knit groups, or modules, and whether such groups are organized into natural hierarchies in the graph. Community structure exists in a variety of real world systems including several social, biological, and political systems [82], and community detection methods can be used to uncover that structure algorithmically. Recent applications of these methods to real-world systems have uncovered segregated committees in the US House of Representatives [169], segregated protein classes in protein-protein interaction networks [56], and segregated functional groups of areas in brain graphs [19]. Uncovering community structure can provide important intuition about the system’s function, and the large-scale functional units that drive the system’s most salient processes [93].
Mathematics of modularity maximization
Many methods exist for community detection [81]. Some draw on notions in physics such as the Potts model [174], while others draw on notions in mathematics such as random walks [225] and spectral properties of the adjacency matrix [153, 158, 154, 224]. Still others more closely track other concepts and techniques in computer science and engineering [e.g., 57]. In this section, we will primarily discuss a single method – modularity maximization – due to its frequent use in the network neuroscience community. However, readers interested in understanding various other algorithms and approaches may enjoy several other recent reviews [82, 170]. Importantly, the summary metrics that we define and discuss for both static and time-varying graphs are applicable to any study of community structure, independent of the specific method used to identify that structure.
Modularity maximization refers to the maximization of a modularity quality function, whose output is a hard partition of a graph’s nodes into communities. The most common modularity quality function studied in network neuroscience to date is
| (1) |
where Aij is the ijth element of the adjacency matrix, i is a node assigned to community Ci and node j is assigned to community Cj. The Kronecker delta δ(Ci, Cj) is 1 if i, j are in the same community and zero otherwise, γ is called a structural resolution parameter [14]. The element Pij is the expected weight of the edge connecting nodes i, j under a specific random network null model. A common null model is the Newman-Girvan null model which is given by kikj/2m where ki is the degree of node i and m is the number of non-zero elements in the upper triangle of the undirected adjacency matrix [157]. This null model encodes the intuition that two nodes of high degree are more likely to share an edge than two nodes of low degree. For a discussion of alternative null models, see [14].
The structural resolution parameter, γ, is often set to unity for simplicity. However, due to a well-known resolution limit [174], this choice will tend to produce a fixed number of communities, even if a stronger community structure could be identified at smaller or larger topological scales. To deal with this limitation, it is common to vary γ over a wide range of values. The benefit of such a parameter sweep is that it can also uncover hierarchical organization in the graph: robust community structure across several topological scales [169]. Some graphs contain a single scale (or several discrete scales) at which community structure is present. For these graphs, it has been suggested that a useful method by which to identify that scale(s) is to search for γ values at which all partitions estimated (from multiple runs of the modularity maximization algorithm) are statistically similar [14].
Maximization of the modularity quality function defined above is NP-hard. Because an exact solution is unknown, various heuristics have been devised to maximize (or nearly maximize) Q without resorting to an exhaustive search of all possible partitions, which for most real-world graphs proves to be computationally intractable [170]. Heuristics vary in terms of their relative speed, fidelity, and appropriateness for large versus small graphs. One common heuristic is a Louvain locally-greedy algorithm [38], which contains two phases: one where modularity is optimized by allowing only local changes of communities, and one where the identified communities are aggregated to build a new network of communities. These two phases are repeated iteratively until modularity no longer increases. A second common heuristic is based on simulated annealing, which appears particularly natural when one realizes that finding the modularity of a network is analogous to finding the ground-state energy of a spin system [175, 104]. In this approach, nodes are combined into communities, and such communities are maintained with some probability dependent on whether they increase or decrease the modularity quality index; the probability of accepting a decrease in modularity slowly decreases as the solution space is explored. A third and complementary approach is based on extremal optimization, and optimizes the global variable (modularity quality index) by improving extremal local variables (contribution of a single node to the modularity quality index) [72]. For example, a simple heuristic randomly partitions the network into two communities, and the node with the lowest fitness (extremal) is moved from its partition to the opposite partition. This procedure is repeated until an optimal division of the network into two components is completed; then each component is iteratively bipartitioned in the same way. Generally speaking, greedy algorithms tend to be relatively swift [59], while simulated annealing [103], extremal optimization [72], and others [160] can be slower yet provide quite stable partitions. With most heuristics, one should perform the optimization many times in order to create an ensemble of partitions, and both understand and report the variability in those solutions.
The modularity landscape is rough, containing many near degeneracies [97]. This means that there are many structurally diverse alternative partitions of nodes into communities with modularity values very close to the optimum. Near degeneracy is particularly prevalent in large binary graphs, and less prevalent in small weighted graphs. Degeneracy becomes especially problematic when the partitions identified by multiple optimizations of the modularity quality function are dissimilar. In these cases, we might wish to identify a single representative partition from the set of partitions observed. One common approach to identify a consensus community structure is similarity maximization [70], where the partition of interest is that which has the greatest similarity to all other observed partitions. A second common approach is an association-recluster method [128, 14, 30], which uses a clustering algorithm to find a consensus partition by exploiting the fact that across an ensemble of partitions, a single node may be affiliated with the same other nodes. Partition degeneracy can also be addressed by expressing the best partition as an average across multiple near-optimal partitions, and by treating the community allegiance of nodes as fuzzy variables [28] or via probabilistic clustering [110].
Summarizing community structure in brain graphs
Topological summary statistics
Several summary statistics that can be derived from community detection methods are reported in neuroimaging studies. Many of these can be defined based on the network’s topology, independent of any embedding of that network into a physical space (Fig. 3). Here we offer a summary of these statistics, and point the reader to a few (certainly not all) recent references that have used them in the neuroimaging literature to address questions of import to neuroscientists.
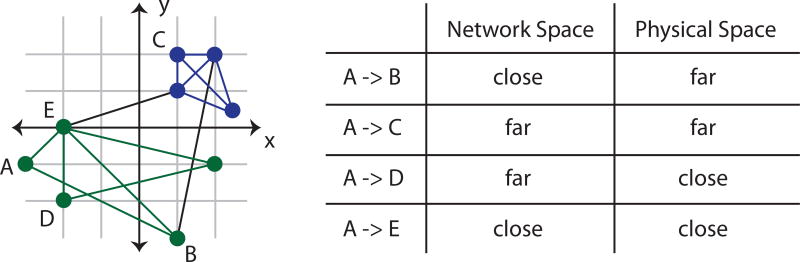
Fig. 3. Distinction between network space and physical space.
Spatial network statistics are important for understanding how a network is embedded into physical space. Here we illustrate the distinction between network space and physical space, motivating the importance of using stastistics in both spaces to understand a system’s organization. (Left) An example graph that has been embedded into a physical (2-dimensional Euclidean) space. (Right) A list of connections between nodes and their respective network and physical distances, demonstrating that long physical distances need not be long network distances, and visa versa.
b) Number of communities:: The number of communities provides an indication of the scale of community structure in a network. Note that NCk = |Ck| is the number of nodes in module Ck. A large number of communities suggests a small scale of structure in the network, while a small number of communities suggests a large scale of structure in the network.
c) Size of communities:: The average size of communities, and the distribution of community sizes are also useful diagnostics of community structure. The number of nodes N divided by the number of communities K gives the mean size of communities in the graph.
d) Modularity quality index:: For community structure identified with modularity-based approaches, the modularity quality index Q serves as a useful measure of the quality of the partition of nodes into communities (see Eq. (1)). To some degree, higher values indicate more optimal partitions than lower values, after accounting for caveats of the roughness of the modularity landscape [97], the size of the graph, and the edge weight distribution, among potentially other confounds. For a recent review of the use of this metric in neuroimaging studies, see [195].
e) Within- and between-module connectivity:: It is also of interest to calculate the strength of edges inside of modules, and the strength of edges between modules. We refer to these notions as within- and between-module connectivity, respectively and define
| (2) |
to be the strength between module Ck1 and module Ck2. When the two modules are identical (k1 = k2), this measure amounts to the average strength of that module, and we interpret it as the recruitment of the module. When the two modules are different (k1 ≠ k2), we might also wish to compute the relative interaction strength
| (3) |
to account for statistical differences in module size. Within- and between-module connectivity has been shown to vary appreciably with learning [21], differ across motor versus working memory function [62], mark neuromodulatory effects [185], track neurodevelopment [101, 24], and change in psychiatric disease [180].
f) Intra-module strength z-score:: One might also wish to quantify how well connected a node is to other nodes in its community, a notion that is formalized in the intra-module strength z-score [103]:
| (4) |
where SCi denotes the strength (i.e., total edge weight) of node i’s edges to other nodes in its own community Ci, the quantity is the mean of SCi over all of the nodes in Ci, and is the standard deviation of SCi in Ci. This statistic was recently applied to brain graphs to study the learning of categories [193].
g) Participation coefficient:: One might also wish to measure how the connections emanating from a node are spread among nodes in the different communities, a notion that is formalized in the participation coefficient [103]:
| (5) |
where SiCk is the strength of edges of node i to nodes in community Ck. This statistic has been used to better understand how learning is impacted by patterns of intermodular connectivity [193], how brain function is altered in antipsychotic-naive first-episode schizophrenia patients [131], and how structural modular segregation mediates the relationship between age and executive function in youth [24].
Spatial summary statistics
It is often interesting to quantify how a network is embedded into physical space (Fig. 3), and specifically the spatial properties of communities. Currently, relatively few measures exist and future work should focus on this important area. Below we present five measures previously proposed to quantify the spatial aspects of community structure. Many of these statistics have not yet been used in the neuroimaging literature, and there is therefore an open opportunity to use them to better understand the spatial embedding of modular structure in the brain.
h) Community average pairwise spatial distance:: The community average pairwise spatial distance, lCk is the average Euclidean distance between all pairs of nodes within a community [76]:
| (6) |
where ri is the position vector of node i. The average pairwise spatial distance of the entire network is given by the same equation calculated over all nodes within the network.
i) Community spatial diameter:: The community spatial diameter, dCk is defined as the maximum Euclidean distance between all pairs of nodes within a community [76]:
| (7) |
The spatial diameter of the entire network is given by the same equation, but calculated over all nodes within the network.
j) Community spatial extent:: The spatial extent of a community is an inverse estimate of the density of a community and quantifies the area or volume of the community, normalized by the number of nodes within the community [76]. Specifically, we can define
| (8) |
where Vh is the volume (3 dimensions) or area (2 dimensions) of the region bounded by the points of the convex hull of nodes within the community. The convex hull is the minimal convex set containing all of the points within the community and is informally described as the polygon created by connecting all points that define the perimeter of the community. It should be noted that in this definition of the spatial extent, the normalization assumes the average size of a region is approximately constant. If this is not the case, the equation could be modified to take into account the boundaries or sizes of individual regions to better estimate the inverse measure of density. This statistic has been used to assess the spatial embedding of synchronized functional clusters estimated from two-photon calcium imaging data acquired in a chronically epileptic dentate gyrus of a mouse model of temporal lobe epilepsy [76].
k) Community radius:: We can define the community radius ρCk as the length of the vector of standard deviations of all nodes in the community [134]:
| (9) |
The average community radius of the entire network is a dimensionless quantity that expresses the average relationship between individual community radii and the network as a whole
| (10) |
where NCk serves to weight every community by the number of nodes it contains, and R is a normalization constant equal to the radius of the entire network: . In the context of human brain graphs, this statistic has previously been used to assess the changes in the spatial extent of modules as they are identified across different levels of the topological hierarchy [134].
l) Community laterality:: Laterality is a property that can be applied to any network in which each node can be assigned to one of two categories, J1 and J2, and describes the extent to which a community localizes to one category or the other.
For an individual community Ck, the laterality ΛCk is defined as [70]:
| (11) |
where NJ1 and NJ2 are the number of nodes located in each category, respectively. The value of ΛCk ranges between zero (i.e., the number of nodes in the community are evenly distributed between the two categories) and unity (i.e., all nodes in the community are located in a single category).
The laterality of a given partition, 𝒞, of a network is defined as:
| (12) |
where 〈ΣkNCkΛCk〉 denotes the expectation value of the laterality under the null model specified by randomly reassigning nodes to the two categories while keeping the total number of nodes in each category fixed. One of the most important functional specializations of the brain is laterality, and prior work has used this statistic to demonstrate subtle interhemispheric discrepancies in functional brain graphs during linguistic processing due to task demands [70].
Strength and significance of communities
When reporting values for either topological or spatial diagnostics, it is important to consider potential sources of error or variation that would inform the confidence in the measured values. For example, there may be error in the estimated weights of individual edges in the network, either from errors in the images themselves, or errors in the statistical estimates of structural or functional connectivity from those images. There may also be variance associated with multiple estimates of a network, either from different subjects, or from the same subject at different instances in time or in different brain states. In each case, it is useful to discuss the potential errors or sources of variance contributing to the estimated diagnostics of community structure, and to quantify them where possible.
In addition to accurately describing the potential sources of error in one’s data, it can also be useful to explicitly measure the significance of a given community structure. In this section, we describe two notions that can be used to quantify the strength and significance of communities. (Note that in this section, we use a few variable names that have been defined differently in earlier sections, largely to remain consistent with the traditional use of these variable names in their relevant subfields.)
m) Normalized persistence probability:: The persistence probability is a measure of the strength of a community in a graph with salient community structure [165]. Given an adjacency matrix A, we construct an N-state Markov chain with transition matrix P by performing a row-normalization on A. Specifically, the transition probability from i to j is given by
| (13) |
Under some mild conditions, there exists a unique equilibrium distribution π ∈ ℝN that satisfies π = πP. Roughly speaking, this implies that if an individual takes a random walk on V with transition probabilities given by P, then — after some sufficiently long period of time — the probability that the individual is on the ith node is πi regardless of where the individual started.
Now, given P and any distribution π on V, we can construct a K-state Markov chain with transition probability
| (14) |
where H is an N×K binary matrix coding the partition 𝒞; that is, hni indicates whether the nth node is in the kth community. We call the K-state Markov chain a lumped Markov chain. We can check that Π = πH is an equilibrium distribution of the lumped Markov chain, which satisfies Π = ΠQ, and therefore the lumped Markov chain can be treated as an approximation of the transition of communities in the original Markov chain. We note that then, the expected escape time of Ck is τk = (1 − qkk)−1, which implies that if now the individual is in Ck, then on average it will take τk jumps for the individual to jump to another community. The persistence probability of the kth comunity is therefore defined as qkk; the larger this value, the longer the expected escape time, and the more significant the community.
In practical applications, the persistence probability is influenced by the size of the community. Larger communities always have larger persistence probabilities. Importantly, this fact can bias empirical results for graphs whose community size distribution is relatively broad. To address this limitation, we can normalize the persistence probability as follows
| (15) |
The kth community is significant if q̃k ≫ 1. Intuitively, this normalization assumes that the graph is fully connected and that the weights of edges are all equal; then, the persistence probability of the k-th cluster is . Whenever a community has a persistence probability that is larger than some threshold α, we will refer to it as an α-community. If all communities are α-communities, we call the entire partition an α-partition.
n) Statistical comparison to a permutation-based null model:: Given a community structure 𝒞, we can in fact compute the contribution of each community to the modularity quality index as follows:
| (16) |
where as before γ is the structural resolution parameter, A is the adjacency matrix, and P is a null model adjacency matrix. Intuitively, Q(Ck|𝒞) measures how strong the kth community is, and it is interesting to ask whether it is stronger than expected under some appropriate null model.
To address this question, we can algorithmically generate a community structure 𝒞r, which has exactly the same number of communities and the same number of nodes in each corresponding community as in 𝒞, by simply permuting the order of nodes in V. We use this permutation-based approach to construct an ensemble of partitions, and for each partition we can calculate Q(Ck|𝒞r). Now, we define
| (17) |
where and . The quantity S(Ck|𝒞) is a normalized measure that provides information about how strong the community is in comparison to what is expected under a permutation-based null model. This method was recently used to identify hyperedges in a set of functional brain graphs ordered over the developmental time period spanning 8 years of age to 22 years of age [102].
Modularity maximization for temporal graphs
The methods described above can be applied to a single graph, or separately to all graphs in a graph ensemble. However, in the study of neural function and its relation to cognition, or its change with age and disease, we often have an ordered set of graphs, where the order is based on time (Fig. 4). In this case, it is useful to consider methods for modularity maximization in temporal graphs — a set of graphs ordered according to time from earliest time to latest time [188]. A recent generalization of modularity maximization for graphs with L layers is given by the multilayer modularity quality function [144]:
| (18) |
where the adjacency matrix of layer l has elements Aijl, and the null model matrix of layer l has elements Pijl, γl is the structural resolution parameter of layer l, ωjlr is the temporal resolution parameter and gives the strength of the inter-layer link between node j in layer l and node j in layer r, δ is the Kronecker delta, and μ is the total edge weight. Small values of the temporal resolution parameter result in greater independence of partitions across neighboring layers, and large values of the temporal resolution parameter result in greater dependence of partitions across neighboring layers. Note that ω can vary from 0 to infinity.
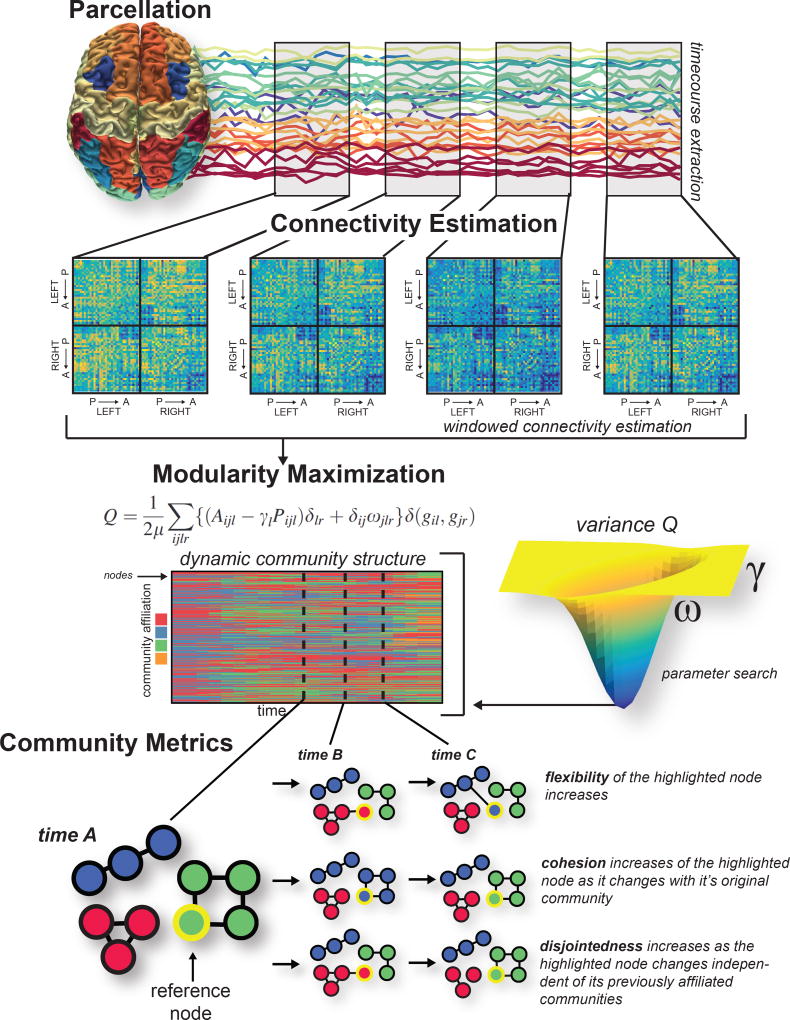
Fig. 4. Building temporal brain graphs and characterizing their dynamic community structure.
Here we illustrate methodological steps to build a temporal brain graph and to estimate its dynamic community structure. First, we define nodes, shown here as a whole-brain parcellation. Next, in each time window, we define edges, shown here as statistical similarities in regional time series. We build a multilayer graph from the ordered set of graphs across all time windows, and we link graphs in neighboring layers by identity links (edges between node i in layer t and itself in layer t − 1 and t + 1). After constructing the multilayer brain graph, we can apply a community detection technique such as the maximization of a multilayer modularity quality function. This process produces a time-dependent partition of nodes into communities or modules. Because multilayer modularity maximization contains tunable parameters, we might also wish to search the 2-dimensional parameter space to find a parameter pair that results in a stable partition (for example, here reflected by a low variance of Qmultilayer across multiple iterations of the maximization algorithm). Finally, the dynamic community structure can be quantitatively characterized with graph statistics (e.g., flexibility, cohesion, and disjointedness).
Determining appropriate choices for the values of the structural (γ) and temporal (ω) resolution parameters is an important enterprise. In some cases, one might have information about the system under study that would dictate the number of communities expected, or their relative size, or their relative variation over time. However, if such information is not available for the system under study, then one must turn to data-driven methods to obtain values for γ and ω that most accurately reflect the spatial and temporal scales of community structure within the data. Several heuristics have been suggested in the literature, including (i) comparison to statistical null models (which we will describe in a later section) [14], (ii) identifying a point in the γ-ω plane where the set of partitions obtained from multiple maximizations of the multilayer modularity quality function are statistically similar [53], or (iii) identifying the point in the γ-ω plane where the dynamic community structure displays certain features [202].
Topological summary statistics for dynamic community structure
Several summary statistics exist which are frequently reported to characterize dynamic community structure in empirical studies. A few particularly simple statistics include (i) the mean and temporal variance of the number of communities, (ii) the mean and temporal variance of the size of communities, and (iii) the multilayer modularity quality index Qmultilayer. In addition to these simple statistics — which have their correlaries in the single-layer case — we can also define several statistics that explicitly capitalize on the temporal nature of the data.
o) Flexibility:: The flexibility of a single node i, ξi, is defined as the number of times a node changes in community allegiance across network layers, normalized by the number of possible changes [19]. Mathematically,
| (19) |
where gi is the number of times that the particle changes its community. The flexibility of the entire multilayer graph is then given by the mean flexibility of all nodes
| (20) |
Intra-individual differences in this metric have been linked to mood [35] and attention [186], while inter-individual differences in this metric have been linked to learning [19], cognitive flexibility and working memory performance [41], and reinforcement learning [91]. The metric has also been shown to be an intermediate phenotype for schizophrenia risk [42], and is altered by an NMDA-receptor antagonist [42].
p) Node disjointedness:: Node disjointedness describes how often a node changes communities independently. Specifically, we are interested in when a node moves from community s to community k, and no other nodes move from community s to community k. If node i makes such changes out of L − 1 possible changes, we define the node disjointedness as follows [203]:
| (21) |
q) Cohesion strength:: The node cohesion can be defined as the number of times a node changes communities mutually with another node. Specifically, node cohesion is a pairwise measure that is expressed as a cohesion matrix, M, where edge weight Mij denotes the number of times a pair of nodes moves to the same community together, divided by L − 1 possible changes. The cohesion strength of node i is then defined as follows [203]:
| (22) |
This metric has been shown to be more sensitive to individual differences in motor skill learning than either node disjointedness or node flexibility [203].
r) Promiscuity:: The promiscuity ψi of node i is defined as the fraction of all communities in which the node participates at least once, across all network layers [162], and importantly can determine whether a node’s flexibility may be high simply because it is switching between two communities or is truly flexible across all communities. The network promiscuity Ψ can be defined as the average promiscuity over all nodes
| (23) |
s) Stationarity:: To define stationarity, we first write the autocorrelation J(Cl, Cl+m) between a given community at layer l, Cl, and the same community at layer l + m, Cl+m, as
| (24) |
where |Cl ∩ Cl+m| is the number of nodes present in community C in layer l and in layer l + m, and |Cl ∪ Cl+m| is the number of nodes present in community Ck at layer l or layer l + m [161]. Then if li is the layer in which community C first appears, and lf is the layer in which it disappears, the stationarity of community Ck is
| (25) |
The stationarity of the entire multilayer network is then given by
| (26) |
While initially defined in the context of social networks [161], the stationarity has also proven useful as a marker of temporal variation in functional brain graphs in healthy adult humans in the process of motor skill acquisition [19].
Statistical Validation and Prediction
After estimating community structure from a single brain graph, or from a multilayer brain graph, one is next faced with the questions of (i) whether and how that community structure is statistically significant, (ii) how to compare community structure in one graph ensemble to community structure in a second graph ensemble, and (iii) how to infer underlying mechanisms driving the observed community structure. Answering these questions requires tools from statistics that are directly informed by network architecture, and tools from generative modeling that can provide insights into possible mechanisms.
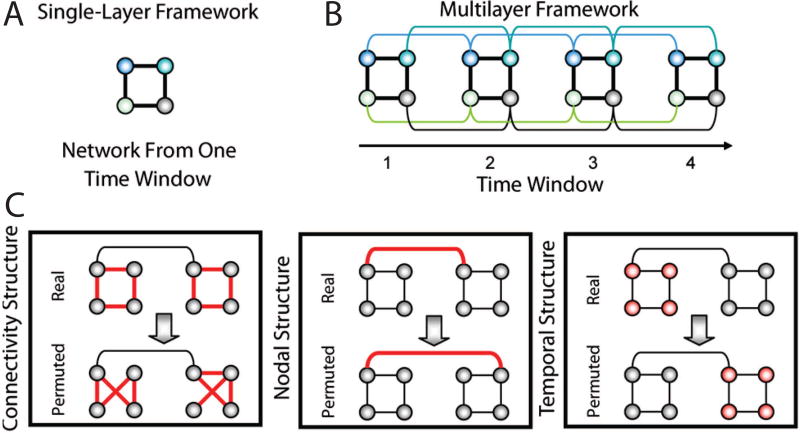
The statistical significance of a community structure can only be determined in relation to a defined null model. One of the most common approaches to defining null models for brain graphs is via permutation: for example, the placement or weight of the edges in the true graph can be permuted uniformly at random (Fig. 5). In prior work, this null model has been referred to both as a connectional null model [19] or a random edges null model [188]. If the graph is a temporal multilayer brain graph, one could also consider permuting the inter-layer links uniformly at random (sometimes referred to as a nodal null model). One could also consider permuting the order of the layers uniformly at random (sometimes referred to as a temporal null model) [19]. For a discussion of related null models specifically for dynamic graphs, see [188, 120].
Fig. 5. Permutation-based null models for statistical testing of community structure.
(A) Schematic of a toy network with four nodes and four edes in a single time window. (B) Multilayer network framework in which the networks from four time windows are linked by connecting nodes in a time window to themselves in the adjacent time windows (colored curves). (C) Statistical framework composed of a connectional null model (Left), a nodal null model (Middle), and a temporal null model (Right) in which intranetwork links, internetwork links, and time windows, respectively, in the real network are randomized in the permuted network. (We show all of the randomized links in red.) Figure reproduced with permission from [19].
When graphs are built from functional data, one can also consider null models that are constructed from surrogate time series [14, 120]. Perhaps the simplest surrogate data technique begins by permuting the elements of each time series uniformly at random and then continues by recomputing the measure of functional connectivity between pairs of time series [205]. This approach is sometimes referred to as a random shuffle null model. While a fundamental benchmark, this null model is quite lenient, and it is commonly complemented by more stringent tests [14]. For example, the Fourier Transform surrogate preserves the linear correlation of the series by permuting the phase of the time series in Fourier space before taking the inverse transform to return the series to temporal space. A related technique – the Amplitude Adjusted Fourier Transform – works similarly except that it also preserves the amplitude distribution of the original time series [204]. For helpful additional discussion of surrogate data time series, see [181, 182].
After confirming that the community structure observed in the empirical graph is unlike that observed in either graph-based or time-series-based null models, one might next wish to compare two sets of empirical graphs. Specifically, one might wish to state that the community structure in one graph ensemble (e.g., healthy brains) is significantly different from the community structure in another graph ensemble (e.g., brains from individuals with disorders of mental health). One simple approach would be to use traditional parametric statistics to determine group differences in a summary measure such as the many defined in the earlier sections of this review. Broadly speaking, this approach assumes the data are normally distributed with a mean that is parameterized by a linear model. Common techniques include t-tests, F-tests, and analyses of variance. However, the assumption that network statistics are normally distributed is often violated in real-world data. In such cases, it is more appropriate to use non-parametric permutation testing, which accounts for the true variation in the empirically observed data. This method is computationally costly since it requires an estimation of the null distribution via unlabeled simulation of the available data. For a recent review of this method as it applies to neuroimaging data, see [159].
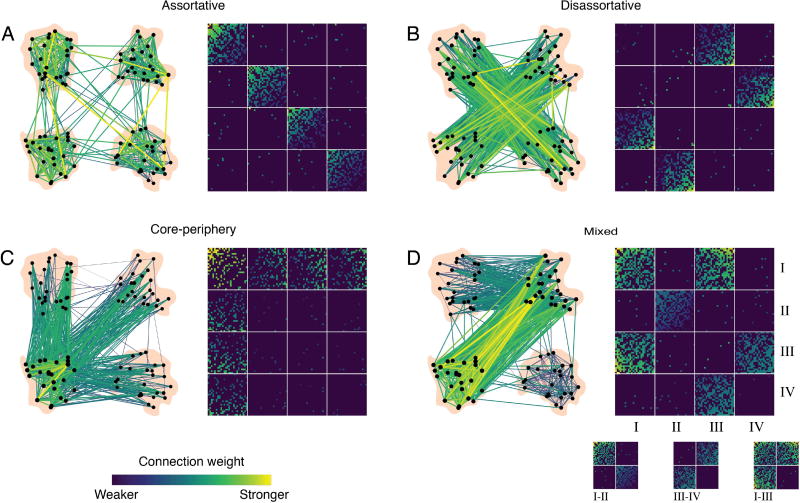
Finally, moving beyond statistical validation, one might also wish to understand the mechanisms by which community structure arises in one’s data of interest, and predict how alterations in those mechanisms could lead to altered community structure. These sorts of topics are particularly important in understanding normative development and aging, and in understanding changes in graph architecture with disease or injury. To begin to build an intuition for possible mechanisms of community structure, it is natural to turn to generative network modeling techniques [31], in which wiring rules are posited and the resultant graph is compared to the empirically observed graphs; if the observed graph displays similar architecture to the modeled graph, then the wiring rule is said to constitute a potential mechanism. Such generative models can be either static or growing models [126], and can be defined either in a deterministic or probabilistic manner [194]. A particularly useful model for mesoscale structure — including but not limited to community structure — is the stochastic blockmodel, which has recently been used in the context of both structural [32] and functional [164] brain graphs (Fig. 6). Importantly, stochastic blockmodels have also recently been extended to multilayer graphs [200], suggesting their potential utility in understanding mechanisms of brain dynamics as well.
Fig. 6. Stochastic blockmodels can detect other types of meso-scale structure unseen by modularity maximization.
Networks can exhibit different types of meso-scale structure. (A) Assortative communities are sub-networks whose internal density of connections exceeds their external density. (B) Disassortative (multi-partite) communities are sub-networks where connections are made preferentially between communities so that communities’ external density exceed their internal density. (C) Core-periphery organization consists of a central core that is connected to the rest of the network and then peripheral nodes that connect to the core but not to one another. (D) These meso-scale structures can be present simultaneously in the same network. For example, communities I–II interact assortatively, III–IV interact disassortatively, while I–III interact as a core and periphery. Reproduced with permission from [32].
Collectively, these statistical approaches provide a rich set of tools to examine the robustness and reliability of brain graphs constructed from neuroimaging techniques sensitive to neural structure and activity across different spatial scales. Furthermore, recent advancements in generative network modeling provide new avenues to examine the mechanisms supporting network modularity that will complement work using community detection to characterize the static and dynamic evolution of these networks.
From modularity in neural systems to behavior
Mounting evidence supports the notion that modularity in brain graphs is important for healthy task-based and resting-state dynamics. Functional network communities correspond to groups of regions that are activated by the performance of specific cognitive and behavioral tasks requiring for example perception, action, and emotion [63]. Interestingly, evidence suggests that the human brain transitions among functional states that maximize either segregation or integration of communities, and the integrated states are associated with faster and more accurate performance on a cognitive task [185, 184]. Several studies have identified relationships between individual differences in modularity and memory performance [209, 58, 3, 185, 78, 199]. Changes in global modularity predict effective memory retrieval [213], account for reaction time on correct responses [209], and relate to individual variability on other measures of behavioral performance [199]. Converging evidence from electroencephalography studies in which community detection approaches are applied [206, 166] further suggest that increased integration is required for successful working memory function [39, 124, 226].
The relationship between network modularity and performance is also expressed in the resting brain. Global network modularity at rest has been shown to predict inter- and intra-individual differences in memory capacity [201]. When network modularity in resting state dynamics decreases following sleep deprivation, it accounts for behavioral performance impairments [29]. Aging brains typically become less modular at the global scale [137, 90, 54], including specific modularity decreases in the executive control network and attention subsystems associated with typical cognitive decline [33]. Similarly, increased modularity is associated with improved learning and neuroplasticity. Patients with brain injury [8] as well as older adults [87] with more modular brain networks at baseline have been shown to exhibit greater improvements following cognitive training.
Importantly, community detection approaches have also revealed the importance of time-evolving changes in modular networks that underlie human behavior. When participants successfully learn a simple motor skill across several days, the community organization and its dynamics change as the skill becomes more automatic [19, 21]. Motor skill learning is also accompanied by a growing autonomy of the sensorimotor system, and by a disengagement of frontal-cingulate circuitry which predicts individual differences in learning rate [21]. Even at much shorter time-scales and over the course of a single session, dynamic community structure can capture changes in task demands and changes in cognitive state [6, 95, 41, 35, 62].
Finally, the importance of modular network organization for healthy brain function is underscored by its alteration in clinical samples [80]. Connectopathy has been documented in patients with several mental health disorders including but not limited to schizophrenia, depression, anxiety, dementia, and autism [140, 136, 220]. Schizophrenia has been characterized by diminished small-world organization [141, 133, 176], altered modular organization [221, 130, 140, 121, 17, 4], and dysmodularity: an overall increase in both structural and functional connectivity that greatly reduces the anatomical specialization of network activity [64]. Other disorders of mental health, such as depression, have also been documented to exhibit altered network modularity [135, 217, 179], and emerging evidence suggests that changes in inter-module connectivity could underlie common reward deficits across both mood and psychotic disorders [183].
Methodological considerations and future directions
There are several methodological considerations that are important to mention in the context of applying modularity maximization techniques to neuroimaging data condensed into brain graphs. Perhaps one of the most fundamental consideration relates to the notion that one might be able to identify an “optimal” structural or temporal resolution parameter with which to uncover the graph’s most salient community structure. Such a notion presupposes that the graph displays strongest community structure at only a single topological or temporal scale. Yet, in many real-world systems — including brain graphs — modules exist across a range of topological scales from small to large, each contributing in a different manner to the system’s overall function. Moreover, such nested modules might display dynamics over different temporal scales, enabling segregation and integration of computational processes from transient control to long-distance synchrony. Thus, while choosing an optimal resolution parameter may not only be difficult, it may also be unfounded, depending on the architecture of the single-layer graph or multilayer graph under study.
Several approaches have been proposed to address the multi-scale organization of brain graphs (Fig. 8; for a recent review, see [30]). One intuitive solution is to sweep across the topological and temporal scales of the system by incrementally changing the resolution parameters [77]. The advantage of this approach is that it allows us to track the stability of partitions across topological scales and identify robust modules. Nevertheless the communities in this approach are identified independently at each scale and thus a secondary algorithm is necessary for the reconstruction of a continuous topological community structure. An explicit multi-scale community detection algorithm can be used to address this limitation, by allowing simultaneous identification of the community organization across several scales [144]. A recent application of this approach to neuroimaging data has uncovered notable topological heterogeneity in the community structure of both structural and functional brain graphs, and in the extent of coupling across these modalities [9] (Fig. 9).
Fig. 8. The multiscale brain.
Brain networks are organized across multiple spatiotemporal scales and also can be analyzed at topological (networks) scales ranging from individual nodes to the network as a whole. Figure reproduced with permission from [30].
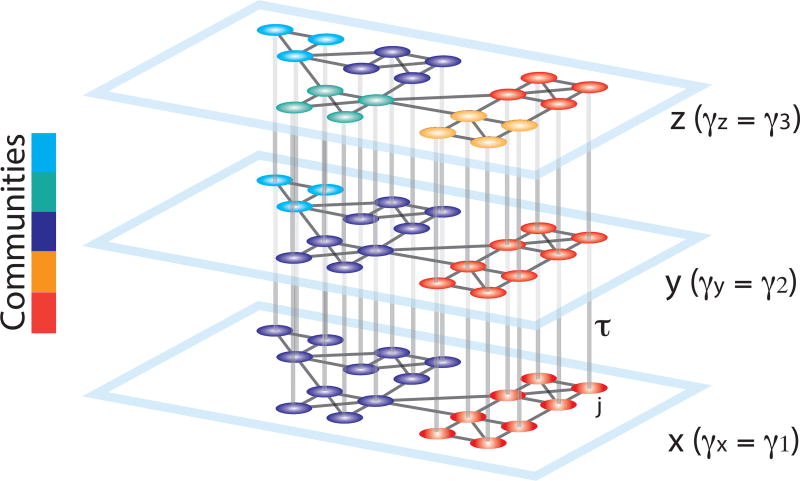
Fig. 9. Schematic representing the construction of a multilayer network for use in multi-scale modularity maximization.
Duplicates of a graph are connected in a multilayer fashion to construct a 3D graph. The smallest resolution parameter γ is assigned to the first layer (x), and it is linearly increased for the neighboring layers (y, z). The topological scale coupling parameter, τ, tunes the strength of dependence of the communities across layers. Since the community assignments are dependent on the adjacent layers, nodes that display high clustering over neighboring topological scales are identified as a single community spanning several scales. In this schematic, the large communities identified at initial layers progressively break into smaller sub-communities, revealing the hierarchical community organization of the graph. Reproduced with permission from [9].
In addition to understanding community structure across different scales in a single data modality, it is becoming increasingly important to identify and characterize community structure across different data modalities. The multilayer network formalism, which we described in this review in the particular context of temporal graphs, can also be used to link graphs from different imaging modalities together [208, 145]. Intuitively, community structure — and the topological or temporal scales at which it is most salient — can differ significantly across imaging modalities. In functional brain graphs estimated over long time scales, the community structure is of neural origin, and thus communities at coarser scales imply higher temporal independence and functional segregation between the communities. By contrast, in structural brain graphs, the community structure can be more reflective of the brain’s spatial organization, constituted by small focal clusters, mesoscale distributed circuits, and gross-scale hemispheres. Since the topological organization of a brain graph can differ across scales in different imaging modalities, it is useful to apply methods that can explicitly compare and contrast community structure across a range of topological and temporal resolutions [9].
Advantages and Disadvantages of the Graph Approach
A key advantage of community detection techniques is their relative simplicity. Nevertheless, this same simplicity can challenge mechanistic understanding of the organizational principles that shape emerging real-time dynamics of the system. This challenge is particularly apparent in the interpretation of the value of the modularity quality index: researchers often interpret higher (lower) modularity values as an increase (decrease) in overall segregation (integration) of brain networks. Yet, it is critical to realize that one can change the structure of a network in a host of ways that all lead to comparable changes in the value of the modularity quality index, but also lead to strikingly different large-scale functional dynamics. Moreover, modularity values themselves are dependent on the resolution parameter at which they are calculated, and direct comparison between modularly values in two graphs using the same resolution parameter hinges on the assumption that both graphs display “optimal” community structure at the same topological scale. Modularity values are also difficult to compare in two graphs that exhibit community structure at different topological scales, as the resolution parameters used for the calculation of the modularity are different. Thus, in general, the interpretability of the modularity value is quite limited.
More generally, it is important to bear in mind that community detection techniques such as modularity maximization assess one specific type of organization in a graph. It may therefore be useful to combine this approach with other techniques to examine complementary types of organization present in the same graph or present in the time series irrespective of the graph [for several recent reviews, see 117, 114, 120]. Within the network science discipline, community detection can be used to examine a specific type of meso-scale organization [for others, see 32], while other graph measures can be used to examine organization at other scales [150]. Examples of these other measures include centralities [13], clustering coefficient [178], path-length [12], and global and local efficiency [129] to name a few. Future work could also use generalizations of these network measures to multilayer data (see [125] for a recent review, and see [188] for a toolbox for use in applying those notions to neuroimaging data). Furthermore, these tools may also provide novel avenues for studying the coupling between the time-varying and multi-scale community structure in functional brain graphs and the underlying hierarchical scaffold in structural brain graphs [9].
Finally, it is important to note that the most common approach used to construct a brain graph treats brain regions as nodes and inter-regional connections as edges. Although this simple graph model has proven useful in advancing our understanding of the organization of brain networks in health and disease, it suffers from an implicit assumption of node homogeneity. That is, each node is distinguished not by any feature of its own, but by its relation to other nodes. Future work could aim to explore and advance community detection methods for annotated graphs [156] in the context of brain networks to account for the heterogeneous function and anatomy of different brain regions [149] (Fig. 10). Moreover, exploring alternative ways to construct brain networks such as hypergraphs [20, 102], and alternative methods to identify community structure such as link-communities [2, 67], could offer important and complimentary information regarding the organizational principles of brain network architecture.
Fig. 10. Mesoscale network methods can address activity, connectivity, or the two together.
In the human brain, the structural connectome supports a diverse repertoire of functional brain dynamics, ranging from the patterns of activity across individual brain regions to the dynamic patterns of connectivity between brain regions. Current methods to study the brain as a networked system usually address connectivity alone (either static or dynamic) or activity alone. Methods developed to address the relations between connectivity and activity are few in number, and further efforts connecting them will be an important area for future growth in the field. In particular, the development of methods in which activity and connectivity can be weighted differently – such as is possible in annotated graphs – could provide much-needed insight into their complimentary roles in neural processing. Figure reproduced with permission from [120].
Conclusion
Here, we have reviewed recent efforts to model brain structure and function using graphs. We focused on describing methods to identify, characterize, and interpret community structure in such graphs, with the goal of better understanding cognitive processes and resulting behavior in health and disease. We began by describing how brain graphs are commonly built, and then we discussed two community detection algorithms based on modularity maximization: one constructed for use on single graphs, and one constructed for use on multilayer graphs. We also offered a collation of summary statistics that can be used to characterize topological features of community structure, spatial features of community structure, and features of dynamic community structure. We closed with a discussion of methodological considerations and future directions, as well as a few comments on the advantages and disadvantages of the graph approach. Our hope is that this review will serve as a useful introduction to the study of community structure in brain graphs, and will spur the development of new tools to more accurately parse and interpret modularity in human brain structure and function.
Fig. 7. From modularity to behavior.
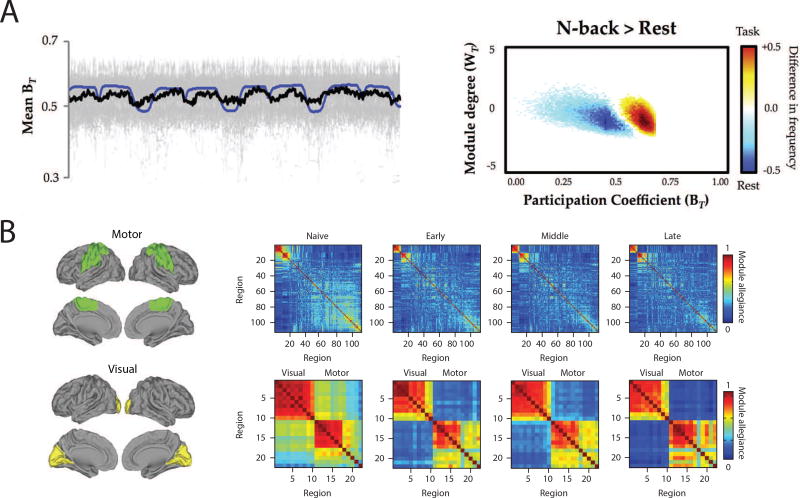
Dynamic changes in the modular organization of functional brain networks capture the short- and long-term network reconfigurations triggered by the requirements of an on-going task or following weeks of training. (A) Dynamic task-related fluctuations of community structure during task performance. (Left) Time series plot demonstrated the close relationship between mean BT (participation coefficient) across 100 subjects (thick black line; individual subject data plotted in gray and task-block repressors plotted in blue). (Right) distinct changes in community structure during N-back task (a common working memory task) compared to the resting state. Note that during N-back performance, the frequency of time points where the network is more integrated significantly increases (red/yellow) compared to the rest blocks (marked by a significant increase in the network segregation) [185]. (B) Learning-induced autonomy of sensorimotor systems captured by a reduction in the probability that motor and visual regions are allied to one another in a single community. The module allegiance matrices are calculated over different phases of learning (naive, early, middle, and late). The bottom row magnifies the visual and motor modules’ allegiance matrices (highlighted with green and yellow brain overlay on the left). Note that the strength of the allegiance between the visual and motor modules decreases as the motor sequences become more automatic, which signifies the increased autonomy of these systems over the course of learning [21]. Figure reproduced with permission from [185] and [21]
Acknowledgments
This work was supported by mission funding to the Army Research Laboratory, as well as research executed under contract number W911NF-10-2-0022. D.S.B. and A.A. would also like to acknowledge support from the John D. and Catherine T. MacArthur Foundation, the Alfred P. Sloan Foundation, the Army Research Office through contract number W911NF-14-1-0679, the National Institute of Health (2-R01-DC-009209-11, 1R01HD086888-01, R01-MH107235, R01-MH107703, R01MH109520, 1R01NS099348, R21-M MH-106799, the Office of Naval Research, and the National Science Foundation (BCS-1441502, CAREER PHY-1554488, BCS-1631550, and CNS-1626008). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies.
References
- 1.Achard Sophie, Salvador Raymond, Whitcher Brandon, Suckling John, Bullmore ED. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. The Journal of neuroscience. 2006;26(1):63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn Yong-Yeol, Bagrow James P, Lehmann Sune. Link communities reveal multiscale complexity in networks. arXiv preprint arXiv:0903.3178. 2009 doi: 10.1038/nature09182. [DOI] [PubMed] [Google Scholar]
- 3.Alavash Mohsen, Thiel Christiane M, Gießing Carsten. Dynamic coupling of complex brain networks and dual-task behavior. Neuroimage. 2016;129:233–246. doi: 10.1016/j.neuroimage.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 4.Alexander-Bloch Aaron, Lambiotte Renaud, Roberts Ben, Giedd Jay, Gogtay Nitin, Bullmore Ed. The discovery of population differences in network community structure: new methods and applications to brain functional networks in schizophrenia. Neuroimage. 2012;59(4):3889–3900. doi: 10.1016/j.neuroimage.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alivisatos A Paul, Chun Miyoung, Church George M, Greenspan Ralph J, Roukes Michael L, Yuste Rafael. The brain activity map project and the challenge of functional connectomics. Neuron. 2012;74(6):970–974. doi: 10.1016/j.neuron.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andric Michael, Hasson Uri. Global features of functional brain networks change with contextual disorder. Neuroimage. 2015;117:103–113. doi: 10.1016/j.neuroimage.2015.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arenas Alex, Diaz-Guilera Albert, Pérez-Vicente Conrad J. Synchronization reveals topological scales in complex networks. Physical review letters. 2006;96(11):114102. doi: 10.1103/PhysRevLett.96.114102. [DOI] [PubMed] [Google Scholar]
- 8.Arnemann Katelyn L, Chen Anthony J-W, Novakovic-Agopian Tatjana, Gratton Caterina, Nomura Emi M, D’Esposito Mark. Functional brain network modularity predicts response to cognitive training after brain injury. Neurology. 2015;84(15):1568–1574. doi: 10.1212/WNL.0000000000001476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashourvan Arian, Telesford Qawi K, Verstynen Timothy, Vettel Jean M, Bassett Danielle S. Multiscale detection of hierarchical community architecture in structural and functional brain networks. arXiv preprint arXiv:1704.05826. 2017 doi: 10.1371/journal.pone.0215520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assaf Yaniv, Pasternak Ofer. Diffusion tensor imaging (dti)-based white matter mapping in brain research: a review. Journal of molecular neuroscience. 2008;34(1):51–61. doi: 10.1007/s12031-007-0029-0. [DOI] [PubMed] [Google Scholar]
- 11.Baldassano Steven N, Bassett Danielle S. Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease. Scientific reports. 2016;6:26087. doi: 10.1038/srep26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barabasi A-L, Albert R. Statistical mechanics of complex networks. Rev. Mod. Phys. 2002;74:47. [Google Scholar]
- 13.Barthelemy M. Betweenness centrality in large complex networks. The European Physical Journal B-Condensed Matter and Complex Systems. 2004;38(2):163–168. [Google Scholar]
- 14.Bassett DS, Porter MA, Wymbs NF, Grafton ST, Carlson JM, Mucha PJ. Robust detection of dynamic community structure in networks. Chaos. 2013;23(1):013142. doi: 10.1063/1.4790830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassett Danielle S, Bullmore Edward T. Small-world brain networks revisited. The Neuroscientist. 2016:1073858416667720. doi: 10.1177/1073858416667720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassett Danielle S, Sporns Olaf. Network neuroscience. Nature neuroscience. 2017;20(3):353. doi: 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bassett Danielle S, Bullmore Edward, Verchinski Beth A, Mattay Venkata S, Weinberger Daniel R, Meyer-Lindenberg Andreas. Hierarchical organization of human cortical networks in health and schizophrenia. Journal of Neuroscience. 2008;28(37):9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassett Danielle S, Brown Jesse A, Deshpande Vibhas, Carlson Jean M, Grafton Scott T. Conserved and variable architecture of human white matter connectivity. Neuroimage. 2011;54(2):1262–1279. doi: 10.1016/j.neuroimage.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Bassett Danielle S, Wymbs Nicholas F, Porter Mason A, Mucha Peter J, Carlson Jean M, Grafton Scott T. Dynamic reconfiguration of human brain networks during learning. Proceedings of the National Academy of Sciences. 2011;108(18):7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bassett Danielle S, Wymbs Nicholas F, Porter Mason A, Mucha Peter J, Grafton Scott T. Cross-linked structure of network evolution. Chaos: An Interdisciplinary Journal of Nonlinear Science. 2014;24(1):013112. doi: 10.1063/1.4858457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassett Danielle S, Yang Muzhi, Wymbs Nicholas F, Grafton Scott T. Learning-induced autonomy of sensorimotor systems. Nature neuroscience. 2015;18(5):744–751. doi: 10.1038/nn.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassett Danielle Smith, Bullmore ED. Small-world brain networks. The neuroscientist. 2006;12(6):512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- 23.Bastos André M, Schoffelen Jan-Mathijs. A tutorial review of functional connectivity analysis methods and their interpretational pitfalls. Frontiers in systems neuroscience. 2015;9 doi: 10.3389/fnsys.2015.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baum GL, Ciric R, Roalf DR, Betzel RF, Moore TM, Shinohara RT, Kahn AE, Vandekar SN, Rupert PE, Quarmley M, Cook PA, Elliott MA, Ruparel K, Gur RE, Gur RC, Bassett DS, Satterthwaite TD. Modular segregation of structural brain networks supports the development of executive function in youth. Curr Biol. 2017;27(11):1561–1572.e8. doi: 10.1016/j.cub.2017.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beggs John M. The criticality hypothesis: how local cortical networks might optimize information processing. Philosophical Transactions of the Royal Society of London A: Mathematical, Physical and Engineering Sciences. 2008;366(1864):329–343. doi: 10.1098/rsta.2007.2092. [DOI] [PubMed] [Google Scholar]
- 26.TEJ Behrens and H Johansen-Berg. Relating connectional architecture to grey matter function using diffusion imaging. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2005;360(1457):903–911. doi: 10.1098/rstb.2005.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behrens Timothy EJ, Johansen Berg H, Jbabdi Saad, Rushworth Matthew FS, Woolrich Mark W. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34(1):144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellec P, Rosa-Neto P, Lyttelton OC, Benali H, Evans AC. Multi-level bootstrap analysis of stable clusters in resting-state fMRI. Neuroimage. 2010;51(3):1126–1139. doi: 10.1016/j.neuroimage.2010.02.082. [DOI] [PubMed] [Google Scholar]
- 29.Simon Eti Ben, Maron-Katz Adi, Lahav Nir, Shamir Ron, Hendler Talma. Tired and misconnected: A breakdown of brain modularity following sleep deprivation. Human Brain Mapping. 2017;38(6):3300–3314. doi: 10.1002/hbm.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betzel RF, Bassett DS. Multi-scale brain networks. Neuroimage. 2016;S1053-8119(16):30615–2. doi: 10.1016/j.neuroimage.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betzel RF, Bassett DS. Generative models for network neuroscience: Prospects and promise. arXiv. 2017;1708:07958. doi: 10.1098/rsif.2017.0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betzel RF, Medaglia JD, Bassett DS. Diversity of meso-scale architecture in human and non-human connectomes. arXiv. 2017;1702:02807. doi: 10.1038/s41467-017-02681-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betzel Richard F, Byrge Lisa, He Ye, Goñi Joaquín, Zuo Xi-Nian, Sporns Olaf. Changes in structural and functional connectivity among resting-state networks across the human lifespan. Neuroimage. 2014;102:345–357. doi: 10.1016/j.neuroimage.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 34.Betzel Richard F, Medaglia John D, Papadopoulos Lia, Baum Graham L, Gur Ruben, Gur Raquel, Roalf David, Satterthwaite Theodore D, Bassett Danielle S. The modular organization of human anatomical brain networks: Accounting for the cost of wiring. Network Neuroscience. 2017 doi: 10.1162/NETN_a_00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betzel Richard F, Satterthwaite Theodore D, Gold Joshua I, Bassett Danielle S. Positive affect, surprise, and fatigue are correlates of network flexibility. Scientific Reports. 2017;7 doi: 10.1038/s41598-017-00425-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilder Robert M, Sabb Fred W, Parker D Stott, Kalar Donald, Chu Wesley W, Fox Jared, Freimer Nelson B, Poldrack Russell A. Cognitive ontologies for neuropsychiatric phenomics research. Cognitive neuropsychiatry. 2009;14(4–5):419–450. doi: 10.1080/13546800902787180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blinowska Katarzyna J, Kamiński Maciej, Brzezicka Aneta, Kamiński Jan. Application of directed transfer function and network formalism for the assessment of functional connectivity in working memory task. Phil. Trans. R. Soc. A. 2013;371(1997):20110614. doi: 10.1098/rsta.2011.0614. [DOI] [PubMed] [Google Scholar]
- 38.Blondel Vincent D, Guillaume Jean-Loup, Lambiotte Renaud, Lefebvre Etienne. Fast unfolding of communities in large networks. Journal of statistical mechanics: theory and experiment. 2008;2008(10):P10008. [Google Scholar]
- 39.Bola Michał, Borchardt Viola. Cognitive processing involves dynamic reorganization of the whole-brain network’s functional community structure. Journal of Neuroscience. 2016;36(13):3633–3635. doi: 10.1523/JNEUROSCI.0106-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bollobas B. Modern Graph Theory. Springer; 2002. [Google Scholar]
- 41.Braun Urs, Schäfer Axel, Walter Henrik, Erk Susanne, Romanczuk-Seiferth Nina, Haddad Leila, Schweiger Janina I, Grimm Oliver, Heinz Andreas, Tost Heike, et al. Dynamic reconfiguration of frontal brain networks during executive cognition in humans. Proceedings of the National Academy of Sciences. 2015;112(37):11678–11683. doi: 10.1073/pnas.1422487112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braun Urs, Schäfer Axel, Bassett Danielle S, Rausch Franziska, Schweiger Janina I, Bilek Edda, Erk Susanne, Romanczuk-Seiferth Nina, Grimm Oliver, Geiger Lena S, et al. Dynamic brain network reconfiguration as a potential schizophrenia genetic risk mechanism modulated by nmda receptor function. Proceedings of the National Academy of Sciences. 2016;113(44):12568–12573. doi: 10.1073/pnas.1608819113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bressler Steven L, Scott Kelso JA. Cortical coordination dynamics and cognition. Trends in cognitive sciences. 2001;5(1):26–36. doi: 10.1016/s1364-6613(00)01564-3. [DOI] [PubMed] [Google Scholar]
- 44.Bressler Steven L, Menon Vinod. Large-scale brain networks in cognition: emerging methods and principles. Trends in cognitive sciences. 2010;14(6):277–290. doi: 10.1016/j.tics.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Brodmann K. Vergleichende lokalisationslehre der grosshirnrinde.[principles of comparative localization in the cerebral cortex presented on the basis of cytoarchitecture. Barth, Leipzig Google Scholar. 1909 [Google Scholar]
- 46.Brooks Justin R, Garcia Javier O, Kerick Scott E, Vettel Jean M. Differential functionality of right and left parietal activity in controlling a motor vehicle. Frontiers in systems neuroscience. 2016;10 doi: 10.3389/fnsys.2016.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown Kevin S, Grafton Scott T, Carlson Jean M. Bicar: A new algorithm for multiresolution spatiotemporal data fusion. PloS one. 2012;7(11):e50268. doi: 10.1371/journal.pone.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bullmore Ed, Sporns Olaf. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature reviews. Neuroscience. 2009;10(3):186. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 49.Bullmore Ed, Sporns Olaf. The economy of brain network organization. Nature Reviews Neuroscience. 2012;13(5):336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- 50.Burns SP, Santaniello S, Yaffe RB, Jouny CC, Crone NE, Bergey GK, Anderson WS, Sarma SV. Network dynamics of the brain and influence of the epileptic seizure onset zone. Proc Natl Acad Sci U S A. 2014;111(49):E5321–30. doi: 10.1073/pnas.1401752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butts Carter T. Revisiting the foundations of network analysis. science. 2009;325(5939):414–416. doi: 10.1126/science.1171022. [DOI] [PubMed] [Google Scholar]
- 52.Buzsaki Gyorgy. Rhythms of the Brain. Oxford University Press; 2006. [Google Scholar]
- 53.Chai LR, Mattar MG, Blank IA, Fedorenko E, Bassett DS. Functional network dynamics of the language system. Cereb Cortex. 2016;26(11):4148–4159. doi: 10.1093/cercor/bhw238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan Micaela Y, Park Denise C, Savalia Neil K, Petersen Steven E, Wig Gagan S. Decreased segregation of brain systems across the healthy adult lifespan. Proceedings of the National Academy of Sciences. 2014;111(46):E4997–E5006. doi: 10.1073/pnas.1415122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Beth L, Hall David H, Chklovskii Dmitri B. Wiring optimization can relate neuronal structure and function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(12):4723–4728. doi: 10.1073/pnas.0506806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J, Yuan B. Detecting functional modules in the yeast protein-protein interaction network. Bioinformatics. 2006;22(18):2283–2290. doi: 10.1093/bioinformatics/btl370. [DOI] [PubMed] [Google Scholar]
- 57.Chen Pin-Yu, Hero Alfred O. Deep community detection. IEEE Transactions on Signal Processing. 2015;63(21):5706–5719. [Google Scholar]
- 58.Chen Tianwen, Cai Weidong, Ryali Srikanth, Supekar Kaustubh, Menon Vinod. Distinct global brain dynamics and spatiotemporal organization of the salience network. PLoS biology. 2016;14(6):e1002469. doi: 10.1371/journal.pbio.1002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Phys. Rev. E. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 60.Clune Jeff, Mouret Jean-Baptiste, Lipson Hod. Proc. R. Soc. B. Vol. 280. The Royal Society; 2013. The evolutionary origins of modularity; p. 20122863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohen Alexander L, Fair Damien A, Dosenbach Nico UF, Miezin Francis M, Dierker Donna, Van Essen David C, Schlaggar Bradley L, Petersen Steven E. Defining functional areas in individual human brains using resting functional connectivity mri. Neuroimage. 2008;41(1):45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cohen Jessica R, D’Esposito Mark. The segregation and integration of distinct brain networks and their relationship to cognition. Journal of Neuroscience. 2016;36(48):12083–12094. doi: 10.1523/JNEUROSCI.2965-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crossley Nicolas A, Mechelli Andrea, Vértes Petra E, Winton-Brown Toby T, Patel Ameera X, Ginestet Cedric E, McGuire Philip, Bullmore Edward T. Cognitive relevance of the community structure of the human brain functional coactivation network. Proceedings of the National Academy of Sciences. 2013;110(28):11583–11588. doi: 10.1073/pnas.1220826110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.David Anthony S. Dysmodularity: A neurocognitive model for schizophrenia. Schizophrenia bulletin. 1993;20(2):249–255. doi: 10.1093/schbul/20.2.249. [DOI] [PubMed] [Google Scholar]
- 65.Davison Elizabeth N, Schlesinger Kimberly J, Bassett Danielle S, Lynall Mary-Ellen, Miller Michael B, Grafton Scott T, Carlson Jean M. Brain network adaptability across task states. PLoS computational biology. 2015;11(1):e1004029. doi: 10.1371/journal.pcbi.1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davison Elizabeth N, Turner Benjamin O, Schlesinger Kimberly J, Miller Michael B, Grafton Scott T, Bassett Danielle S, Carlson Jean M. Individual differences in dynamic functional brain connectivity across the human lifespan. PLoS computational biology. 2016;12(11):e1005178. doi: 10.1371/journal.pcbi.1005178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Reus Marcel A, Saenger Victor M, Kahn René S, van den Heuvel Martijn P. An edge-centric perspective on the human connectome: link communities in the brain. Phil. Trans. R. Soc. B. 2014;369(1653):20130527. doi: 10.1098/rstb.2013.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Desikan Rahul S, Ségonne Florent, Fischl Bruce, Quinn Brian T, Dickerson Bradford C, Blacker Deborah, Buckner Randy L, Dale Anders M, Maguire R Paul, Hyman Bradley T, et al. An automated labeling system for subdividing the human cerebral cortex on mri scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 69.Destrieux Christophe, Fischl Bruce, Dale Anders, Halgren Eric. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53(1):1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doron Karl W, Bassett Danielle S, Gazzaniga Michael S. Dynamic network structure of interhemispheric coordination. Proceedings of the National Academy of Sciences of the United States of America. 2012 Nov;109(46):18661–18668. doi: 10.1073/pnas.1216402109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dosenbach Nico UF, Visscher Kristina M, Palmer Erica D, Miezin Francis M, Wenger Kristin K, Kang Hyunseon C, Burgund E Darcy, Grimes Ansley L, Schlaggar Bradley L, Petersen Steven E. A core system for the implementation of task sets. Neuron. 2006;50(5):799–812. doi: 10.1016/j.neuron.2006.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duch J, Arenas A. Community detection in complex networks using extremal optimization. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;72(2 Pt 2):027104. doi: 10.1103/PhysRevE.72.027104. [DOI] [PubMed] [Google Scholar]
- 73.Eickhoff Simon B, Thirion Bertrand, Varoquaux Gaël, Bzdok Danilo. Connectivity-based parcellation: Critique and implications. Human brain mapping. 2015;36(12):4771–4792. doi: 10.1002/hbm.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellefsen Kai Olav, Mouret Jean-Baptiste, Clune Jeff. Neural modularity helps organisms evolve to learn new skills without forgetting old skills. PLoS computational biology. 2015;11(4):e1004128. doi: 10.1371/journal.pcbi.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Espinosa-Soto Carlos, Wagner Andreas. Specialization can drive the evolution of modularity. PLoS computational biology. 2010;6(3):e1000719. doi: 10.1371/journal.pcbi.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muldoon Sarah Feldt, Soltesz Ivan, Cossart Rosa. Spatially clustered neuronal assemblies comprise the microstructure of synchrony in chronically epileptic networks. Proceedings of the National Academy of Sciences of the United States of America. 2013 Feb;110(9):3567–3572. doi: 10.1073/pnas.1216958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fenn Daniel J, Porter Mason A, McDonald Mark, Williams Stacy, Johnson Neil F, Jones Nick S. Dynamic communities in multichannel data: An application to the foreign exchange market during the 2007–2008 credit crisis. Chaos: An Interdisciplinary Journal of Nonlinear Science. 2009;19(3):033119. doi: 10.1063/1.3184538. [DOI] [PubMed] [Google Scholar]
- 78.Finc Karolina, Bonna Kamil, Lewandowska Monika, Wolak Tomasz, Nikadon Jan, Dreszer Joanna, Duch Włodzisław, Kühn Simone. Transition of the functional brain network related to increasing cognitive demands. Human Brain Mapping. 2017 doi: 10.1002/hbm.23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fodor Jerry A. The modularity of mind: An essay on faculty psychology. MIT press; 1983. [Google Scholar]
- 80.Fornito Alex, Bullmore Edward T. Connectomics: a new paradigm for understanding brain disease. European Neuropsychopharmacology. 2015;25(5):733–748. doi: 10.1016/j.euroneuro.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 81.Fortunato S, Hric D. Community detection in networks: A user guide. Physics Reports. 2016;659:1–44. [Google Scholar]
- 82.Fortunato Santo. Community detection in graphs. Physics reports. 2010;486(3):75–174. [Google Scholar]
- 83.Fries Pascal. A mechanism for cognitive dynamics: neuronal communication through neuronal coherence. Trends in cognitive sciences. 2005;9(10):474–480. doi: 10.1016/j.tics.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 84.Fries Pascal. Rhythms for cognition: communication through coherence. Neuron. 2015;88(1):220–235. doi: 10.1016/j.neuron.2015.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Friston Karl J. Functional and effective connectivity: a review. Brain connectivity. 2011;1(1):13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- 86.Gall Franz Josef. On the functions of the brain and of each of its parts: With observations on the possibility of determining the instincts, propensities, and talents, or the moral and intellectual dispositions of men and animals, by the configuration of the brain and head. Vol. 1. Marsh, Capen & Lyon; 1835. [Google Scholar]
- 87.Gallen Courtney L, Baniqued Pauline L, Chapman Sandra B, Aslan Sina, Keebler Molly, Didehbani Nyaz, D’Esposito Mark. Modular brain network organization predicts response to cognitive training in older adults. PloS one. 2016;11(12):e0169015. doi: 10.1371/journal.pone.0169015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gallos Lazaros K, Makse Hernán A, Sigman Mariano. A small world of weak ties provides optimal global integration of self-similar modules in functional brain networks. Proceedings of the National Academy of Sciences. 2012;109(8):2825–2830. doi: 10.1073/pnas.1106612109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Garcia Javier O, Srinivasan Ramesh, Serences John T. Near-real-time feature-selective modulations in human cortex. Current Biology. 2013;23(6):515–522. doi: 10.1016/j.cub.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geerligs Linda, Renken Remco J, Saliasi Emi, Maurits Natasha M, Lorist Monicque M. A brain-wide study of age-related changes in functional connectivity. Cerebral Cortex. 2014;25(7):1987–1999. doi: 10.1093/cercor/bhu012. [DOI] [PubMed] [Google Scholar]
- 91.Gerraty RT, Davidow JY, Foerde K, Galvan A, Bassett DS, Shohamy D. Dynamic flexibility in striatal-cortical circuits supports reinforcement learning. bioRxiv. 2017:094383. doi: 10.1523/JNEUROSCI.2084-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gilbert Charles D, Sigman Mariano. Brain states: top-down influences in sensory processing. Neuron. 2007;54(5):677–696. doi: 10.1016/j.neuron.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 93.Girvan M, Newman ME. Community structure in social and biological networks. Proc Natl Acad Sci U S A. 2002;99(12):7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Giusti Chad, Ghrist Robert, Bassett Danielle S. Two’s company, three (or more) is a simplex. Journal of computational neuroscience. 2016;41(1):1–14. doi: 10.1007/s10827-016-0608-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Godwin Douglass, Barry Robert L, Marois René. Breakdown of the brain’s functional network modularity with awareness. Proceedings of the National Academy of Sciences. 2015;112(12):3799–3804. doi: 10.1073/pnas.1414466112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goñi Joaquín, van den Heuvel Martijn P, Avena-Koenigsberger Andrea, de Mendizabal Nieves Velez, Betzel Richard F, Griffa Alessandra, Hagmann Patric, Corominas-Murtra Bernat, Thiran Jean-Philippe, Sporns Olaf. Resting-brain functional connectivity predicted by analytic measures of network communication. Proceedings of the National Academy of Sciences. 2014;111(2):833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Good BH, de Montjoye YA, Clauset A. Performance of modularity maximization in practical contexts. Phys Rev E. 2010;81(4 Pt 2):046106. doi: 10.1103/PhysRevE.81.046106. [DOI] [PubMed] [Google Scholar]
- 98.Gramann Klaus, Gwin Joseph T, Ferris Daniel P, Oie Kelvin, Jung Tzyy-Ping, Lin Chin-Teng, Liao Lun-De, Makeig Scott. Cognition in action: imaging brain/body dynamics in mobile humans. Reviews in the Neurosciences. 2011;22(6):593–608. doi: 10.1515/RNS.2011.047. [DOI] [PubMed] [Google Scholar]
- 99.Grech Roberta, Cassar Tracey, Muscat Joseph, Camilleri Kenneth P, Fabri Simon G, Zervakis Michalis, Xanthopoulos Petros, Sakkalis Vangelis, Vanrumste Bart. Review on solving the inverse problem in eeg source analysis. Journal of neuroengineering and rehabilitation. 2008;5(1):25. doi: 10.1186/1743-0003-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gross Thilo, Blasius Bernd. Adaptive coevolutionary networks: a review. Journal of the Royal Society Interface. 2008;5(20):259–271. doi: 10.1098/rsif.2007.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gu S, Satterthwaite TD, Medaglia JD, Yang M, Gur RE, Gur RC, Bassett DS. Emergence of system roles in normative neurodevelopment. Proc Natl Acad Sci U S A. 2015;112(44):13681–13686. doi: 10.1073/pnas.1502829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gu Shi, Yang Muzhi, Medaglia John D, Gur Ruben C, Gur Raquel E, Satterthwaite Theodore D, Bassett Danielle S. Functional hypergraph uncovers novel covariant structures over neurodevelopment. Human Brain Mapping. 2017 doi: 10.1002/hbm.23631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guimera R, Amaral LA. Cartography of complex networks: modules and universal roles. J Stat Mech. 2005;2005:P02001. doi: 10.1088/1742-5468/2005/02/P02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guimera R, Sales-Pardo M, Amaral LAN. Modularity from fluctuations in random graphs and complex networks. Phys. Rev. E. 2004;70:025101. doi: 10.1103/PhysRevE.70.025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hagmann Patric, Cammoun Leila, Gigandet Xavier, Meuli Reto, Honey Christopher J, Wedeen Van J, Sporns Olaf. Mapping the structural core of human cerebral cortex. PLoS biology. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hayasaka Satoru, Laurienti Paul J. Comparison of characteristics between region-and voxel-based network analyses in resting-state fmri data. Neuroimage. 2010;50(2):499–508. doi: 10.1016/j.neuroimage.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.He Bin, Yang Lin, Wilke Christopher, Yuan Han. Electrophysiological imaging of brain activity and connectivity—challenges and opportunities. IEEE transactions on biomedical engineering. 2011;58(7):1918–1931. doi: 10.1109/TBME.2011.2139210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hermundstad Ann M, Bassett Danielle S, Brown Kevin S, Aminoff Elissa M, Clewett David, Freeman Scott, Frithsen Amy, Johnson Arianne, Tipper Christine M, Miller Michael B, et al. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proceedings of the National Academy of Sciences. 2013;110(15):6169–6174. doi: 10.1073/pnas.1219562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hermundstad Ann M, Brown Kevin S, Bassett Danielle S, Aminoff Elissa M, Frithsen Amy, Johnson Arianne, Tipper Christine M, Miller Michael B, Grafton Scott T, Carlson Jean M. Structurally-constrained relationships between cognitive states in the human brain. PLoS computational biology. 2014;10(5):e1003591. doi: 10.1371/journal.pcbi.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hinne M, Ekman M, Janssen RJ, Heskes T, van Gerven MAJ. Probabilistic clustering of the human connectome identifies communities and hubs. PLoS ONE. 2015;10(1):e0117179. doi: 10.1371/journal.pone.0117179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Honey CJ, Sporns O, Cammoun Leila, Gigandet Xavier, Thiran Jean-Philippe, Meuli Reto, Hagmann Patric. Predicting human resting-state functional connectivity from structural connectivity. Proceedings of the National Academy of Sciences. 2009;106(6):2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kahn Ari E, Mattar Marcelo G, Vettel Jean M, Wymbs Nicholas F, Grafton Scott T, Bassett Danielle S. Structural pathways supporting swift acquisition of new visuomotor skills. Cerebral cortex. 2017;27(1):173–184. doi: 10.1093/cercor/bhw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kaiser Marcus, Hilgetag Claus C. Optimal hierarchical modular topologies for producing limited sustained activation of neural networks. Frontiers in neuroinformatics. 2010;4 doi: 10.3389/fninf.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Karahanoğlu Fikret Işık, De Ville Dimitri Van. Dynamics of large-scale fmri networks: Deconstruct brain activity to build better models of brain function. Current Opinion in Biomedical Engineering. 2017 [Google Scholar]
- 115.Kashtan Nadav, Alon Uri. Spontaneous evolution of modularity and network motifs. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13773–13778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kashtan Nadav, Noor Elad, Alon Uri. Varying environments can speed up evolution. Proceedings of the National Academy of Sciences. 2007;104(34):13711–13716. doi: 10.1073/pnas.0611630104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Keilholz Shella, Caballero-Gaudes Cesar, Bandettini Peter, Deco Gustavo, Calhoun Vince. Time-resolved resting-state functional magnetic resonance imaging analysis: Current status, challenges, and new directions. Brain Connectivity. 2017;7(8):465–481. doi: 10.1089/brain.2017.0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Khambhati AN, Davis KA, Lucas TH, Litt B, Bassett DS. Virtual cortical resection reveals push-pull network control preceding seizure evolution. Neuron. 2016;91(5):1170–1182. doi: 10.1016/j.neuron.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Khambhati AN, Bassett DS, Oommen BS, Chen SH, Lucas TH, Davis KA, Litt B. Recurring functional interactions predict network architecture of interictal and ictal states in neocortical epilepsy. eNeuro. 2017;4(1) doi: 10.1523/ENEURO.0091-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Khambhati AN, Sizemore AE, Betzel RF, Bassett DS. Modeling and interpreting mesoscale network dynamics. Neuroimage. 2017;S1053-8119(17):30500–1. doi: 10.1016/j.neuroimage.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim Dae-Jin, Kent Jerillyn S, Bolbecker Amanda R, Sporns Olaf, Cheng Hu, Newman Sharlene D, Puce Aina, O’Donnell Brian F, Hetrick William P. Disrupted modular architecture of cerebellum in schizophrenia: a graph theoretic analysis. Schizophrenia bulletin. 2014;40(6):1216–1226. doi: 10.1093/schbul/sbu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kinouchi Osame, Copelli Mauro. Optimal dynamical range of excitable networks at criticality. arXiv preprint q-bio/0601037. 2006 [Google Scholar]
- 123.Kirschner Marc, Gerhart John. Evolvability. Proceedings of the National Academy of Sciences. 1998;95(15):8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kitzbichler Manfred G, Henson Richard NA, Smith Marie L, Nathan Pradeep J, Bullmore Edward T. Cognitive effort drives workspace configuration of human brain functional networks. Journal of Neuroscience. 2011;31(22):8259–8270. doi: 10.1523/JNEUROSCI.0440-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kivelä Mikko, Arenas Alex, Barthelemy Marc, Gleeson James P, Moreno Yamir, Porter Mason A. Multilayer networks. Journal of complex networks. 2014;2(3):203–271. [Google Scholar]
- 126.Klimm F, Bassett DS, Carlson JM, Mucha PJ. Resolving structural variability in network models and the brain. PLoS Comput Biol. 2014;10(3):e1003491. doi: 10.1371/journal.pcbi.1003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Korhonen Onerva, Saarimäki Heini, Glerean Enrico, Sams Mikko, Saramäki Jari. Consistency of regions of interest as nodes of fmri functional brain networks. Network Neuroscience. 2017 doi: 10.1162/NETN_a_00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lancichinetti Andrea, Fortunato Santo. Consensus clustering in complex networks. Scientific reports. 2012;2:336. doi: 10.1038/srep00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Latora V, Marchiori M. Efficient behavior of small-world networks. Phys Rev Lett. 2001;87(19):198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- 130.Lerman-Sinkoff Dov B, Barch Deanna M. Network community structure alterations in adult schizophrenia: identification and localization of alterations. NeuroImage: Clinical. 2016;10:96–106. doi: 10.1016/j.nicl.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li F, Lui S, Yao L, Ji GJ, Liao W, Sweeney JA, Gong Q. Altered white matter connectivity within and between networks in antipsychotic-naive first-episode schizophrenia. Schizophr Bull. 2017 May 17; doi: 10.1093/schbul/sbx048. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lipson Hod, Pollack Jordan B, Suh Nam P. On the origin of modular variation. Evolution. 2002;56(8):1549–1556. doi: 10.1111/j.0014-3820.2002.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 133.Liu Yong, Liang Meng, Zhou Yuan, He Yong, Hao Yihui, Song Ming, Yu Chunshui, Liu Haihong, Liu Zhening, Jiang Tianzi. Disrupted small-world networks in schizophrenia. Brain. 2008;131(4):945–961. doi: 10.1093/brain/awn018. [DOI] [PubMed] [Google Scholar]
- 134.Lohse Christian, Bassett Danielle S, Lim Kelvin O, Carlson Jean M. Resolving anatomical and functional structure in human brain organization: identifying mesoscale organization in weighted network representations. PLoS Computational Biology. 2014 Oct;10(10):e1003712. doi: 10.1371/journal.pcbi.1003712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lord Anton, Horn Dorothea, Breakspear Michael, Walter Martin. Changes in community structure of resting state functional connectivity in unipolar depression. PloS one. 2012;7(8):e41282. doi: 10.1371/journal.pone.0041282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Menon Vinod. Large-scale brain networks and psychopathology: a unifying triple network model. Trends in cognitive sciences. 2011;15(10):483–506. doi: 10.1016/j.tics.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 137.Meunier David, Achard Sophie, Morcom Alexa, Bullmore Ed. Age-related changes in modular organization of human brain functional networks. Neuroimage. 2009;44(3):715–723. doi: 10.1016/j.neuroimage.2008.09.062. [DOI] [PubMed] [Google Scholar]
- 138.Meunier David, Lambiotte Renaud, Fornito Alex, Ersche Karen D, Bullmore Edward T. Hierarchical modularity in human brain functional networks. Frontiers in neuroinformatics. 2009;3 doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meunier David, Lambiotte Renaud, Bullmore Edward T. Modular and hierarchically modular organization of brain networks. Frontiers in neuroscience. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Micheloyannis Sifis. Graph-based network analysis in schizophrenia. World journal of psychiatry. 2012;2(1):1. doi: 10.5498/wjp.v2.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Micheloyannis Sifis, Pachou Ellie, Stam Cornelis Jan, Breakspear Michael, Bitsios Panagiotis, Vourkas Michael, Erimaki Sophia, Zervakis Michael. Small-world networks and disturbed functional connectivity in schizophrenia. Schizophrenia research. 2006;87(1):60–66. doi: 10.1016/j.schres.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 142.Moretti Paolo, Muñoz Miguel A. Griffiths phases and the stretching of criticality in brain networks. arXiv preprint arXiv:1308.6661. 2013 doi: 10.1038/ncomms3521. [DOI] [PubMed] [Google Scholar]
- 143.Mori Susumu, Zijl Peter van. Fiber tracking: principles and strategies–a technical review. NMR in Biomedicine. 2002;15(7–8):468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- 144.Mucha Peter J, Richardson Thomas, Macon Kevin, Porter Mason A, Onnela Jukka-Pekka. Community structure in time-dependent, multiscale, and multiplex networks. Science. 2010;328(5980):876–878. doi: 10.1126/science.1184819. [DOI] [PubMed] [Google Scholar]
- 145.Muldoon SF, Bassett DS. Network and multilayer network approaches to understanding human brain dynamics. Philosophy of Science. 2016 Epub Ahead of Print. [Google Scholar]
- 146.Muldoon Sarah Feldt, Pasqualetti Fabio, Gu Shi, Cieslak Matthew, Grafton Scott T, Vettel Jean M, Bassett Danielle S. Stimulation-based control of dynamic brain networks. PLoS computational biology. 2016;12(9):e1005076. doi: 10.1371/journal.pcbi.1005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Muraskin Jordan, Dodhia Sonam, Lieberman Gregory, Garcia Javier O, Verstynen Timothy, Vettel Jean M, Sherwin Jason, Sajda Paul. Brain dynamics of post-task resting state are influenced by expertise: Insights from baseball players. Human brain mapping. 2016;37(12):4454–4471. doi: 10.1002/hbm.23321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Muraskin Jordan, Sherwin Jason, Lieberman Gregory, Garcia Javier O, Verstynen Timothy, Vettel Jean M, Sajda Paul. Fusing multiple neuroimaging modalities to assess group differences in perception–action coupling. Proceedings of the IEEE. 2017;105(1):83–100. doi: 10.1109/JPROC.2016.2574702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Murphy AC, Gu S, Khambhati AN, Wymbs NF, Grafton ST, Satterthwaite TD, Bassett DS. Explicitly linking regional activation and function connectivity: Community structure of weighted networks with continuous annotation. arXiv. 2016;1611:07962. [Google Scholar]
- 150.Nadakuditi RR, Newman ME. Graph spectra and the detectability of community structure in networks. Phys Rev Lett. 2012;108(18):188701. doi: 10.1103/PhysRevLett.108.188701. [DOI] [PubMed] [Google Scholar]
- 151.Nelson Steven M, Cohen Alexander L, Power Jonathan D, Wig Gagan S, Miezin Francis M, Wheeler Mark E, Velanova Katerina, Donaldson David I, Phillips Jeffrey S, Schlaggar Bradley L, et al. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67(1):156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nematzadeh Azadeh, Ferrara Emilio, Flammini Alessandro, Ahn Yong-Yeol. Optimal network modularity for information diffusion. Physical review letters. 2014;113(8):088701. doi: 10.1103/PhysRevLett.113.088701. [DOI] [PubMed] [Google Scholar]
- 153.Newman ME. Finding community structure in networks using the eigenvectors of matrices. Phys Rev E. 2006;74(3 Pt 2):036104. doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- 154.Newman ME. Spectral methods for community detection and graph partitioning. Phys Rev E. 2013;88(4):042822. doi: 10.1103/PhysRevE.88.042822. [DOI] [PubMed] [Google Scholar]
- 155.Newman MEJ. Networks: An Introduction. Oxford University Press; 2010. [Google Scholar]
- 156.Newman MEJ, Clauset A. Structure and inference in annotated networks. Nature Communications. 2016;7(11863) doi: 10.1038/ncomms11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Phys. Rev. E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- 158.Newman Mark EJ. Modularity and community structure in networks. Proceedings of the national academy of sciences. 2006;103(23):8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nichols Thomas E, Holmes Andrew P. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human brain mapping. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Noack A, Rotta R. Multi-level algorithms for modularity clustering. Experimental Algorithms. 2009:257–268. [Google Scholar]
- 161.Palla G, Derenyi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 162.Papadopoulos L, Puckett JG, Daniels KE, Bassett DS. Evolution of network architecture in a granular material under compression. Phys Rev E. 2016;94(3-1):032908. doi: 10.1103/PhysRevE.94.032908. [DOI] [PubMed] [Google Scholar]
- 163.Passaro Antony D, Vettel Jean M, McDaniel Jonathan, Lawhern Vernon, Franaszczuk Piotr J, Gordon Stephen M. A novel method linking neural connectivity to behavioral fluctuations: Behavior-regressed connectivity. Journal of neuroscience methods. 2017;279:60–71. doi: 10.1016/j.jneumeth.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 164.Pavlovic DM, Vertes PE, Bullmore ET, Schafer WR, Nichols TE. Stochastic blockmodeling of the modules and core of the Caenorhabditis elegans connectome. PLoS One. 2014;9(7):e97584. doi: 10.1371/journal.pone.0097584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Piccardi Carlo. Finding and testing network communities by lumped markov chains. PloS one. 2011;6(11):e27028. doi: 10.1371/journal.pone.0027028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pirondini Elvira, Vybornova Anna, Coscia Martina, De Ville Dimitri Van. A spectral method for generating surrogate graph signals. IEEE signal processing letters. 2016;23(9):1275–1278. [Google Scholar]
- 167.Poldrack Russell A. Region of interest analysis for fmri. Social cognitive and affective neuroscience. 2007;2(1):67–70. doi: 10.1093/scan/nsm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Poldrack Russell A. Mapping mental function to brain structure: how can cognitive neuroimaging succeed? Perspectives on Psychological Science. 2010;5(6):753–761. doi: 10.1177/1745691610388777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Porter MA, Mucha PJ, Newman ME, Warmbrand CM. A network analysis of committees in the U.S. House of Representatives. Proc Natl Acad Sci U S A. 2005;102(20):7057–7062. doi: 10.1073/pnas.0500191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Porter MA, Onnela J-P, Mucha PJ. Communities in networks. Notices of the American Mathematical Society. 2009;56(9):1082–1097. 1164–1166. [Google Scholar]
- 171.Power Jonathan D, Cohen Alexander L, Nelson Steven M, Wig Gagan S, Barnes Kelly Anne, Church Jessica A, Vogel Alecia C, Laumann Timothy O, Miezin Fran M, Schlaggar Bradley L, et al. Functional network organization of the human brain. Neuron. 2011;72(4):665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Price Cathy J, Friston Karl J. Functional ontologies for cognition: The systematic definition of structure and function. Cognitive Neuropsychology. 2005;22(3–4):262–275. doi: 10.1080/02643290442000095. [DOI] [PubMed] [Google Scholar]
- 173.Raj Ashish, Chen Yu-hsien. The wiring economy principle: connectivity determines anatomy in the human brain. PloS one. 2011;6(9):e14832. doi: 10.1371/journal.pone.0014832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Reichardt J, Bornholdt S. Detecting fuzzy community structures in complex networks with a Potts model. Phys Rev Lett. 2004;93(21):218701. doi: 10.1103/PhysRevLett.93.218701. [DOI] [PubMed] [Google Scholar]
- 175.Reichardt Jörg, Bornholdt Stefan. Statistical mechanics of community detection. Physical Review E. 2006;74(1):016110. doi: 10.1103/PhysRevE.74.016110. [DOI] [PubMed] [Google Scholar]
- 176.Rubinov Mikail, Knock Stuart A, Stam Cornelis J, Micheloyannis Sifis, Harris Anthony WF, Williams Leanne M, Breakspear Michael. Small-world properties of nonlinear brain activity in schizophrenia. Human brain mapping. 2009;30(2):403–416. doi: 10.1002/hbm.20517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sakkalis Vangelis. Review of advanced techniques for the estimation of brain connectivity measured with eeg/meg. Computers in biology and medicine. 2011;41(12):1110–1117. doi: 10.1016/j.compbiomed.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 178.Saramaaki J, Kivela M, Onnela J-P, Kaski K, Kertesz J. Generalizations of the clustering coefficient to weighted complex networks. Physical Review E. 2007;75:027105. doi: 10.1103/PhysRevE.75.027105. [DOI] [PubMed] [Google Scholar]
- 179.Satterthwaite TD, Kable JW, Vandekar L, Katchmar N, Bassett DS, Baldassano CF, Ruparel K, Elliott MA, Sheline YI, Gur RC, Gur RE, Davatzikos C, Leibenluft E, Thase ME, Wolf DH. Common and dissociable dysfunction of the reward system in bipolar and unipolar depression. Neuropsychopharmacology. 2015;40(9):2258–2268. doi: 10.1038/npp.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Satterthwaite TD, Vandekar SN, Wolf DH, Bassett DS, Ruparel K, Shehzad Z, Craddock RC, Shinohara RT, Moore TM, Gennatas ED, Jackson C, Roalf DR, Milham MP, Calkins ME, Hakonarson H, Gur RC, Gur RE. Connectome-wide network analysis of youth with Psychosis-Spectrum symptoms. Mol Psychiatry. 2015;20(12):1508–1515. doi: 10.1038/mp.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Schreiber T, Schmitz A. Improved surrogate data for nonlinearity tests. Phys. Rev. Lett. 1996;77(4):635–638. doi: 10.1103/PhysRevLett.77.635. [DOI] [PubMed] [Google Scholar]
- 182.Schreiber T, Schmitz A. Surrogate time series. Physica D. 2000;142:346–382. [Google Scholar]
- 183.Sharma A, Wolf DH, Ciric R, Kable JW, Moore TM, Vandekar SN, Katchmar N, Daldal A, Ruparel K, Davatzikos C, Elliott MA, Calkins ME, Shinohara RT, Bassett DS, Satterthwaite TD. Common dimensional reward deficits across mood and psychotic disorders: A connectome-wide association study. Am J Psychiatry. 2017;174(7):657–666. doi: 10.1176/appi.ajp.2016.16070774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shine James M, Poldrack Russell A. Principles of dynamic network reconfiguration across diverse brain states. NeuroImage. 2017 doi: 10.1016/j.neuroimage.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 185.Shine James M, Bissett Patrick G, Bell Peter T, Koyejo Oluwasanmi, Balsters Joshua H, Gorgolewski Krzysztof J, Moodie Craig A, Poldrack Russell A. The dynamics of functional brain networks: Integrated network states during cognitive task performance. Neuron. 2016;92(2):544–554. doi: 10.1016/j.neuron.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shine James M, Koyejo Oluwasanmi, Poldrack Russell A. Temporal metastates are associated with differential patterns of time-resolved connectivity, network topology, and attention. Proceedings of the National Academy of Sciences. 2016:201604898. doi: 10.1073/pnas.1604898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Simon Herbert A. The architecture of complexity. Proceedings of the American Philosophical Society. 1962;106(6):467–482. [Google Scholar]
- 188.Sizemore AE, Bassett DS. Dynamic graph metrics: Tutorial, toolbox, and tale. Neuroimage. 2017;S1053–8119(17):30564–5. doi: 10.1016/j.neuroimage.2017.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sizemore AE, Giusti C, Bassett DS. Classification of weighted networks through mesoscale homological features. Journal of Complex Networks. 2017;5(2):245–273. [Google Scholar]
- 190.Sizemore AE, Giusti C, Kahn A, Vettel JM, Betzel RF, Bassett DS. Cliques and cavities in the human connectome. J Comput Neurosci. 2017 Nov 16; doi: 10.1007/s10827-017-0672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Smit Dirk JA, Stam Cornelis J, Posthuma Danielle, Boomsma Dorret I, De Geus Eco JC. Heritability of small-world networks in the brain: A graph theoretical analysis of resting-state eeg functional connectivity. Human brain mapping. 2008;29(12):1368–1378. doi: 10.1002/hbm.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Smith Stephen M, Miller Karla L, Salimi-Khorshidi Gholamreza, Webster Matthew, Beckmann Christian F, Nichols Thomas E, Ramsey Joseph D, Woolrich Mark W. Network modelling methods for fmri. Neuroimage. 2011;54(2):875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 193.Soto Fabian A, Bassett Danielle S, Ashby F Gregory. Dissociable changes in functional network topology underlie early category learning and development of automaticity. NeuroImage. 2016;141:220–241. doi: 10.1016/j.neuroimage.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sporns O. Small-world connectivity, motif composition, and complexity of fractal neuronal connections. Biosystems. 2006;85(1):55–64. doi: 10.1016/j.biosystems.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 195.Sporns O, Betzel RF. Modular brain networks. Annu Rev Psychol. 2016;67 doi: 10.1146/annurev-psych-122414-033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sporns Olaf, Tononi Giulio, Edelman Gerald M. Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cerebral cortex. 2000;10(2):127–141. doi: 10.1093/cercor/10.2.127. [DOI] [PubMed] [Google Scholar]
- 197.Stam Cornelius J. Functional connectivity patterns of human magnetoencephalographic recordings: a small-world network? Neuroscience letters. 2004;355(1):25–28. doi: 10.1016/j.neulet.2003.10.063. [DOI] [PubMed] [Google Scholar]
- 198.Stanley Matthew L, Moussa Malaak N, Paolini Brielle M, Lyday Robert G, Burdette Jonathan H, Laurienti Paul J. Defining nodes in complex brain networks. Frontiers in computational neuroscience. 2013;7 doi: 10.3389/fncom.2013.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Stanley Matthew L, Dagenbach Dale, Lyday Robert G, Burdette Jonathan H, Laurienti Paul J. Changes in global and regional modularity associated with increasing working memory load. Frontiers in Human Neuroscience. 2014;8 doi: 10.3389/fnhum.2014.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Stanley N, Shai S, Taylor D, Mucha PJ. Clustering network layers with the strata multilayer stochastic block model. IEEE Trans Netw Sci Eng. 2016;3(2):95–105. doi: 10.1109/TNSE.2016.2537545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Stevens Alexander A, Tappon Sarah C, Garg Arun, Fair Damien A. Functional brain network modularity captures inter-and intra-individual variation in working memory capacity. PloS one. 2012;7(1):e30468. doi: 10.1371/journal.pone.0030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Telesford QK, Lynall ME, Vettel J, Miller MB, Grafton ST, Bassett DS. Detection of functional brain network reconfiguration during task-driven cognitive states. Neuroimage. 2016;142:198–210. doi: 10.1016/j.neuroimage.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Telesford QK, Ashourvan A, Wymbs NF, Grafton ST, Vettel JM, Bassett DS. Cohesive network reconfiguration accompanies extended training. Hum Brain Mapp. 2017;38(9):4744–4759. doi: 10.1002/hbm.23699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Theiler J, Prichard D. Constrained-realization Monte-Carlo method for hypothesis testing. Physica D. 1996;94(4):221–235. [Google Scholar]
- 205.Theiler J, Eubank S, Longtin A, Galdrikian B, Farmer JD. Testing for nonlinearity in time series: the method of surrogate data. Physica D. 1992;58:77–94. [Google Scholar]
- 206.Tremblay Nicolas, Borgnat Pierre. Graph wavelets for multiscale community mining. IEEE Transactions on Signal Processing. 2014;62(20):5227–5239. [Google Scholar]
- 207.Tzourio-Mazoyer Nathalie, Landeau Brigitte, Papathanassiou Dimitri, Crivello Fabrice, Etard Olivier, Delcroix Nicolas, Mazoyer Bernard, Joliot Marc. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 208.Vaiana M, Muldoon SEF. Multilayer brain networks. arXiv. 2017;1709:02325. [Google Scholar]
- 209.Vatansever Deniz, Menon David K, Manktelow Anne E, Sahakian Barbara J, Stamatakis Emmanuel A. Default mode dynamics for global functional integration. Journal of Neuroscience. 2015;35(46):15254–15262. doi: 10.1523/JNEUROSCI.2135-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Vettel Jean M, Cooper Nicole, Garcia Javier O, Yeh Frank, Verstynen Tim. White matter tractography and diffusion weighted imaging. eLS. in press. [Google Scholar]
- 211.Vindiola MM, Vettel JM, Gordon SM, Franaszczuk PJ, McDowell K. Applying eeg phase synchronization measures to non-linearly coupled neural mass models. Journal of neuroscience methods. 2014;226:1–14. doi: 10.1016/j.jneumeth.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 212.Von Economo CB, Koskinas GN. The cytoarchitectonics of the adult human cortex. Vienna and Berlin: Julius Springer Verlag; 1925. [Google Scholar]
- 213.Westphal Andrew J, Wang Siliang, Rissman Jesse. Episodic memory retrieval benefits from a less modular brain network organization. Journal of Neuroscience. 2017;37(13):3523–3531. doi: 10.1523/JNEUROSCI.2509-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wiegell Mette R, Tuch David S, Larsson Henrik BW, Wedeen Van J. Automatic segmentation of thalamic nuclei from diffusion tensor magnetic resonance imaging. NeuroImage. 2003;19(2):391–401. doi: 10.1016/s1053-8119(03)00044-2. [DOI] [PubMed] [Google Scholar]
- 215.Wig Gagan S, Schlaggar Bradley L, Petersen Steven E. Concepts and principles in the analysis of brain networks. Annals of the New York Academy of Sciences. 2011;1224(1):126–146. doi: 10.1111/j.1749-6632.2010.05947.x. [DOI] [PubMed] [Google Scholar]
- 216.Wildie Mark, Shanahan Murray. Metastability and chimera states in modular delay and pulse-coupled oscillator networks. Chaos: An Interdisciplinary Journal of Nonlinear Science. 2012;22(4):043131. doi: 10.1063/1.4766592. [DOI] [PubMed] [Google Scholar]
- 217.Ye Ming, Yang Tianliang, Qing Peng, Lei Xu, Qiu Jiang, Liu Guangyuan. Changes of functional brain networks in major depressive disorder: a graph theoretical analysis of resting-state fmri. PloS one. 2015;10(9):e0133775. doi: 10.1371/journal.pone.0133775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yeh Fang-Cheng, Vettel Jean M, Singh Aarti, Poczos Barnabas, Grafton Scott T, Erickson Kirk I, Tseng Wen-Yih I, Verstynen Timothy D. Quantifying differences and similarities in whole-brain white matter architecture using local connectome fingerprints. PLoS computational biology. 2016;12(11):e1005203. doi: 10.1371/journal.pcbi.1005203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yeo BT Thomas, Krienen Fenna M, Sepulcre Jorge, Sabuncu Mert R, Lashkari Danial, Hollinshead Marisa, Roffman Joshua L, Smoller Jordan W, Zöllei Lilla, Polimeni Jonathan R, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of neurophysiology. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yerys BE, Herrington JD, Satterthwaite TD, Guy L, Schultz RT, Bassett DS. Globally weaker and topologically different: resting-state connectivity in youth with autism. Mol Autism. 2017;8:39. doi: 10.1186/s13229-017-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yu Qingbao, Plis Sergey M, Erhardt Erik B, Allen Elena A, Sui Jing, Kiehl Kent A, Pearlson Godfrey, Calhoun Vince D. Modular organization of functional network connectivity in healthy controls and patients with schizophrenia during the resting state. Frontiers in systems neuroscience. 2011;5 doi: 10.3389/fnsys.2011.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yu Qingbao, Sui Jing, Rachakonda Srinivas, He Hao, Gruner William, Pearlson Godfrey, Kiehl Kent A, Calhoun Vince D. Altered topological properties of functional network connectivity in schizophrenia during resting state: a small-world brain network study. PloS one. 2011;6(9):e25423. doi: 10.1371/journal.pone.0025423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yu Qingbao, Du Yuhui, Chen Jiayu, He Hao, Sui Jing, Pearlson Godfrey, Calhoun Vince D. Comparing brain graphs in which nodes are regions of interest or independent components: A simulation study. Journal of Neuroscience Methods. 2017;291:61–68. doi: 10.1016/j.jneumeth.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang X, Newman ME. Multiway spectral community detection in networks. Phys Rev E. 2015;92(5):052808. doi: 10.1103/PhysRevE.92.052808. [DOI] [PubMed] [Google Scholar]
- 225.Zhou H, Lipowsky R. Network brownian motion: A new method to measure vertex-vertex proximity and to identify communities and subcommunities. International Conference on Computational Science. 2004:1062–1069. [Google Scholar]
- 226.Zippo Antonio Giuliano, Della Rosa Pasquale Anthony, Castiglioni Isabella, Mario Biella Gabriele Eliseo. Alternating dynamics of segregation and integration in human brain functional networks during working-memory task. bioRxiv. 2016:082438. doi: 10.1016/j.neuroscience.2017.12.004. [DOI] [PubMed] [Google Scholar]