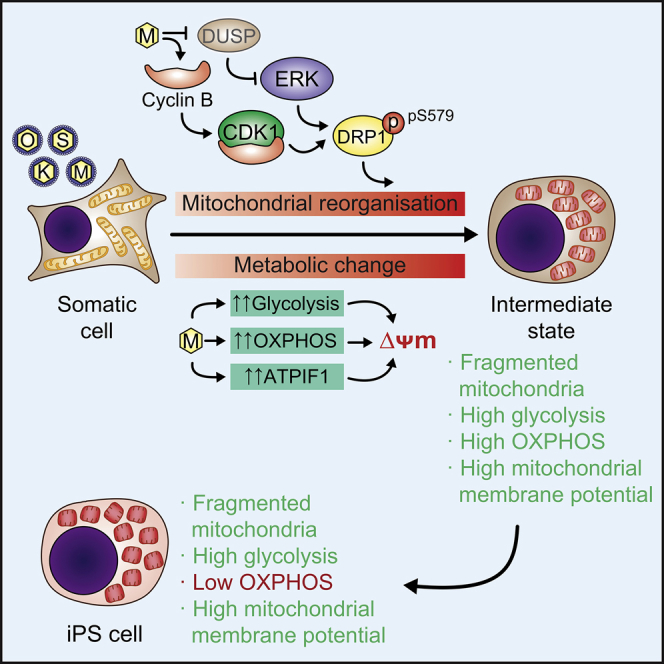
Summary
Cell reprogramming is thought to be associated with a full metabolic switch from an oxidative- to a glycolytic-based metabolism. However, neither the dynamics nor the factors controlling this metabolic switch are fully understood. By using cellular, biochemical, protein array, metabolomic, and respirometry analyses, we found that c-MYC establishes a robust bivalent energetics program early in cell reprogramming. Cells prone to undergo reprogramming exhibit high mitochondrial membrane potential and display a hybrid metabolism. We conclude that MYC proteins orchestrate a rewiring of somatic cell metabolism early in cell reprogramming, whereby somatic cells acquire the phenotypic plasticity necessary for their transition to pluripotency in response to either intrinsic or external cues.
Keywords: cell reprogramming, metabolism, mitochondrial dynamics, cell signaling
Graphical Abstract
Highlights
-
•
Endogenous MYC biological activity is necessary for cell reprogramming
-
•
MYC drives mitochondrial fission early in cell reprogramming
-
•
MYC establishes a bivalent energetics state necessary for cell reprogramming
-
•
MYC polarizes mitochondria, poising cells for reprogramming
Torres and colleagues describe an MYC-dependent remodeling of mitochondrial dynamics and metabolism during the first stage of cell reprogramming. Endogenous MYC activity was found to be necessary for cell reprogramming, likely by establishing a hybrid metabolic state characterized by elevated glycolytic flux and a somatic oxidative metabolic rate. MYC polarized mitochondria by increasing ATPIF1 and labeled cells prone to cell reprogramming.
Introduction
Somatic cells can be reprogrammed to pluripotency by forced expression of OCT4, SOX2, KLF4, and c-MYC (OSKM hereafter) (Takahashi and Yamanaka, 2006). Genome-wide and proteomic studies have shown that OSKM-induced reprogramming is a multi-step process characterized by two waves of gene expression and proteome resetting (Buganim et al., 2012, Hansson et al., 2012, Polo et al., 2012). During the first wave, there is a stochastic activation of genes controlling cell proliferation, metabolism, and cytoskeletal reorganization, and a downregulation of somatic gene expression signatures. A transient phase then follows, whereby a reduced group of cells upregulates early stem cell markers. In the second wave, the robust upregulation of core pluripotency circuitry genes, silencing of exogenous reprogramming factors, and the complete resetting of somatic epigenetic marks takes place, leading to the definitive stabilization of the pluripotent state.
MYC family members of transcription factors play central roles in the regulation of a wide range of cellular processes by controlling the expression of discrete groups of genes directly or in a secondary response fashion (Dang, 2016, Kress et al., 2015). Although its overexpression is dispensable for the process, the presence of c-MYC in the cocktail is favorable for cell reprogramming (Nakagawa et al., 2008, Wernig et al., 2008). Paradoxically, while sustained high levels of c-MYC are beneficial at early stages, elevated expression of this proto-oncogene at later stages is detrimental for cell reprogramming (Sridharan et al., 2009, Zhuang et al., 2018).
A hallmark of cell reprogramming is the metabolic switch undertaken by somatic cells from an oxidative phosphorylation (OXPHOS)-based metabolism to a metabolic state substantiated on aerobic glycolysis (Folmes et al., 2011). As this postulation is based on the comparison between the somatic and pluripotent endpoint metabolic profiles, neither the dynamics nor the factors orchestrating this metabolic switch during the reprogramming process are completely understood. Here we show that c-MYC plays a central role in modulating metabolic pathways to favor somatic cell fate change.
Results
Cell Reprogramming-Induced Mitochondrial Fission Is c-MYC Dependent
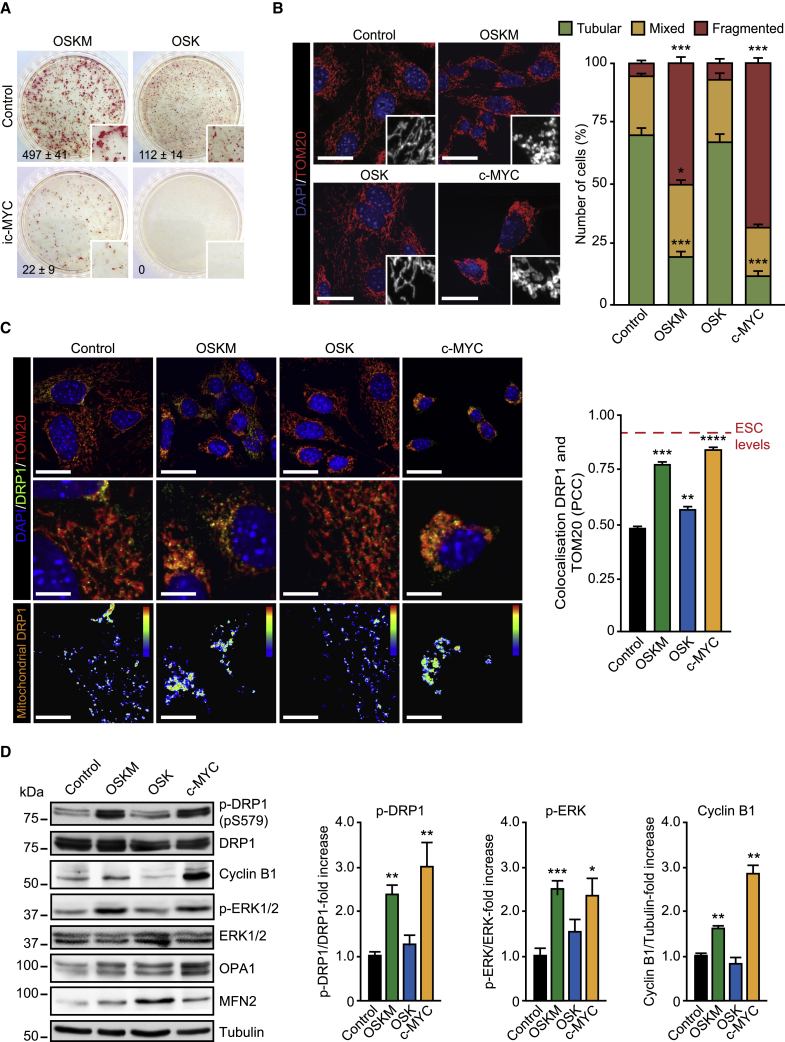
Cell reprogramming experiments in the presence or absence of exogenous c-MYC (Nakagawa et al., 2008, Wernig et al., 2008) have nourished the perception of a non-essential role in the reprogramming process for the MYC gene family. As these experiments have only tested the overexpression effect of MYC genes, an important role for this gene family in the reprogramming process may have been overlooked. We therefore sought to investigate the role of endogenous MYC activity in somatic cell reprogramming. For this, we conducted cell reprogramming experiments in the presence or absence of the MYC inhibitor 10058-F4 (ic-MYC), known to impair endogenous MYC biological activity (Scognamiglio et al., 2016). Cell reprogramming, assessed by scoring the number of alkaline phosphatase (AP)-positive colonies, induced by overexpression of OSKM in mouse embryonic fibroblasts (MEFs) was greatly impaired in the presence of the MYC inhibitor (Figure 1A). Remarkably, cell reprogramming in the absence of exogenous c-MYC, induced by ectopic expression of OCT4, SOX2, and KLF4 (OSK hereafter), was completely abolished by treatment of the cells with the MYC inhibitor and no AP-positive colonies were detected (Figure 1A). These results indicate that endogenous MYC activity is necessary for somatic cell reprogramming.
Figure 1.
Role of c-MYC in Cell Reprogramming-Induced Mitochondrial Fission
(A) Representative bright-field images after alkaline phosphatase (AP) staining of plates containing MEFs after 25 days of either OSK (right panels) or OSKM (left panels) retroviral delivery in the presence of DMSO (control) or the MYC inhibitor 10058-F4 (ic-MYC, 10 μM). Inset shows a magnification of a selected area from the AP-stained plates. Data on the bottom left-hand side of the pictures represent the mean ± SEM of three independent experiments.
(B) MEFs were mock-infected (control) or transduced with the indicated factors. At day 4 post transduction, cells were fixed and mitochondrial morphology assessed by immunofluorescence. Left panels: representative confocal images of MEFs stained with anti-TOM20 antibodies (red) before (control) or after expressing the indicated factors. Inset shows a black-and-white magnification of the pictures. DAPI (blue) was used as a nuclear counterstaining. Graph on the right shows the quantification of the different mitochondrial morphologies.
(C) Representative confocal images of MEFs before (Control) or 4 days after OSKM, OSK, or c-MYC expression stained with anti-DRP1 (green) or anti-TOM20 (red) antibodies. DAPI (blue) was used as a nuclear counterstaining. Middle panels show a magnification of the pictures displayed in the upper panels. Bottom images are color map representations of the pictures in the middle panels to display co-localized pixels between both fluorophores according to the color bar shown on the upper-right corner of the pictures. Warm colors depict pixels with highly correlated intensity and spatial overlap while cold colors are indicative of random or anti-correlation. Graph on the right shows the quantification of the Pearson's correlation coefficient (PCC) to display the degree of co-localization between DRP1 and TOM20 in cells transduced with the indicated factors. Red dashed line indicates the levels of DRP1 and TOM20 co-localization found in ESCs.
(D) Lysates of MEFs control or expressing OSKM, OSK, or c-MYC for 4 days were analyzed by immunoblotting using the indicated antibodies. Graphs on the right show the quantification of the data.
Data represent mean ± SEM, one-tailed unpaired t test (n = 3): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Scale bars, 24 μm in (B) and upper panels of (C); 12 μm in middle and bottom panels of (C). See also Figure S1.
ERK1/2-mediated mitochondrial fission is a necessary event for OSKM-induced cell reprogramming (Prieto et al., 2016a, Prieto et al., 2016b). We next investigated the role of MYC in OSKM-induced mitochondrial fission early in cell reprogramming. OSK cells transduced for 4 days showed identical mitochondrial morphology to that of controls whereas ∼50% of OSKM-transduced cells displayed fragmented mitochondria (Figure 1B). Remarkably, ∼70% of the cells presented fragmented mitochondria in c-MYC-expressing cells (Figure 1B). OSKM or c-MYC induced a robust recruitment of dynamin-related protein 1 (DRP1) to mitochondria, whereas the association of DRP1 with these organelles augmented only slightly by OSK (Figure 1C). Accordingly, and compared with control and OSK-expressing MEFs, ERK1/2 and DRP1-S579 phosphorylation were increased about 3-fold by OSKM- or c-MYC (Figure 1D), indicating that OSKM-induced mitochondrial fission is c-MYC dependent. Also, we observed an increase in cyclin B1 protein in OSKM- and c-MYC-expressing cells (Figure 1D and see below).
Treatment of c-MYC-expressing cells with a MEK1/2 inhibitor decreased both DRP1-S579 phosphorylation and mitochondrial fission (Figures S1A and S1B, respectively). Interestingly, reduction of c-MYC-induced mitochondrial fission by the MEK1/2 inhibitor was rescued by co-expression of DRP1-S579D phosphomimetic mutation, but not by the wild-type form of the dynamin (Figure S1C). Activation of ERK signaling early in reprogramming is associated with a decrease in Dusp6 gene expression (Prieto et al., 2016a), the major cytosolic ERK1/2-phosphatase (Kidger and Keyse, 2016, Ríos et al., 2014). While expression of this phosphatase decreased by ∼20% in c-MYC- and OSK-expressing cells, OSKM expression led to ∼40% reduction in Dusp6 mRNA levels (Figure S1D), suggesting a cooperation between the four factors in reducing the expression of this phosphatase. In addition to Dusp6, other phosphatases known to target ERK1/2 (Ríos et al., 2014) were also downregulated in response to OSKM, OSK, or c-MYC expression (Figure S1D). Altogether, our results indicate that OSKM-induced DRP1 activation and mitochondrial fission are c-MYC dependent.
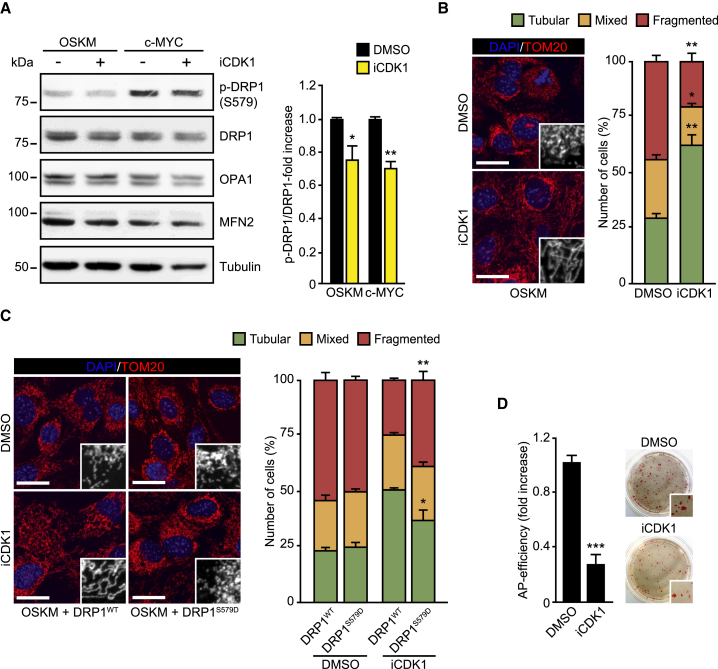
Overexpression of c-MYC triggers cell-cycle progression (Figure S2A), a cell reprogramming hallmark. We found that this rise in cell proliferation by either OSKM or c-MYC was associated with an increase in Ccnb1 and Ccnb2 gene transcripts (Figure S2B), cyclin B1 protein, and DRP1-S579 phosphorylation (Figures 1D and S2C). Interestingly, cyclin B1 is a key component of the CDK1 complex (Bretones et al., 2015). CDK1 phosphorylates DRP1 and induces mitochondrial fission during cell division (Taguchi et al., 2007). In this regard, CDK1 inhibition (iCDK1) decreased DRP1-S579 phosphorylation in OSKM- or c-MYC-transduced cells (Figures 2A and 2B) and impaired OSKM- or c-MYC-induced mitochondrial fission (Figures 2B and S2D, respectively). Interestingly, the inhibition of mitochondrial fission by the CDK1 inhibitor in OSKM or c-MYC-expressing cells could be rescued by co-expression of DRP1-S579D phosphomimetic mutation but not by the wild-type form of the dynamin (Figures 2D and S2E). Furthermore, inhibition of CDK1 impaired OSKM-mediated cell reprogramming (Figure 2D). Our results therefore suggest that CDK1 collaborate with ERK1/2 in the activation of DRP1 during early cell reprogramming to induce mitochondrial fragmentation in a c-MYC-dependent manner.
Figure 2.
Role of CDK1 in c-MYC-Induced DRP1-S579 Phosphorylation
(A) Left panels: lysates from OSKM or c-MYC-transduced MEFs for 3.5 days that were incubated during 10 hr with DMSO (as vehicle control) or the CDK1 inhibitor RO-3306 (1 μM) were analyzed by immunoblotting using the indicated antibodies. Right graph shows the quantification of ERK1/2 or DRP1 phosphorylation ratios in cells treated with either DMSO (black bars) or RO-3306 (yellow bars).
(B) OSKM-transduced cells for 3.5 days were incubated with DMSO (as vehicle control) or the CDK1 inhibitor RO-3306 (1 μM) for 10 hr (iCDK1). Cells were then fixed and mitochondrial morphology assessed by immunofluorescence. Left panels: representative confocal images of MEFs stained with anti-TOM20 antibodies (red). Inset shows a black-and-white magnification of the pictures. DAPI (blue) was used as a nuclear counterstaining. Graph on the right shows the quantification of the different mitochondrial morphologies observed.
(C) Left panels show representative confocal images of MEFs expressing OSKM, together with DRP1 wild-type (DRP1WT) or the phosphomimetic S579D mutation (DRP1S579D), during 3.5 days. Cells were then treated, fixed, and stained as in (B). Graph on the right shows the quantification of the indicated mitochondrial morphologies observed in the cells transduced and treated as indicated.
(D) Graphs showing the number of AP-positive colonies scored in MEFs after 25 days of OSKM retroviral delivery in the presence of DMSO (as vehicle control) or the CDK1 inhibitor RO-3306 (1 μM) (iCDK1) (panels on the right). Right panels: representative bright-field images from the plates of the indicated cultures after AP staining. Inset shows a magnification of a selected area from the AP-stained plates.
Data represent mean ± SEM, one-tailed unpaired t test (n = 3): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Scale bars, 24 μm in (B) and (C). See also Figure S2.
A Hybrid Energetics Program Is Induced by c-MYC during Cell Reprogramming
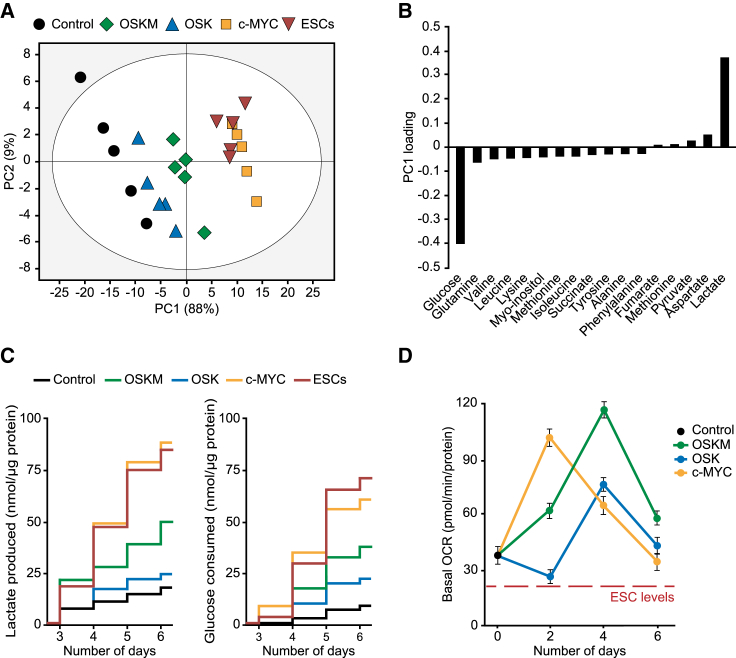
Somatic cell metabolism is switched from oxidative- to glycolytic-based energetics during the reprogramming process (Folmes et al., 2011). We next investigated whether this metabolic change could be driven by c-MYC. For this, we first carried out a metabolomics study, based on nuclear magnetic resonance (NMR), to analyze extracellular metabolite variations in either control or OSKM-, OSK-, or c-MYC-expressing MEFs at day 4 post transduction, or in mouse embryonic stem cells (ESCs) as the endpoint control of the reprogramming process (Figures 3 and S3). Interestingly, principal component analysis (PCA) of the data showed the clustering of c-MYC-expressing MEFs with ESCs (Figure 3A). Plotting of the extracellular metabolite changes along PC1 variable revealed a deep increase in glucose depletion and a robust accumulation of lactate in ES- and c-MYC-expressing cells (Figure 3B). Time-series measurements of lactate and glucose showed a high parallelism between c-MYC-transduced MEFs and ESCs (Figure 3C). The variation of these metabolites over time in control and OSK-transduced cells was similar, and OSKM-expressing cells showed intermediate dynamics between that of ESCs and control cells (Figure 3C). Similar trends, and c-MYC dependency, were observed when measuring the kinetics of lactate production, glucose consumption, acetate production, or glutamine consumption at day 4 of the process (Figure S3B).
Figure 3.
Glycolysis and OXPHOS Are Induced by c-MYC Early in Cell Reprogramming
(A) Principal Component 1 and 2 (PC1, PC2) projections of extracellular metabolite variations in conditioned medium taken from cells at day 4 post transduction with the indicated factors. PC1: R2 = 0.88, Q2 = 0.84; PC2: R2 = 0.08, Q2 = 0.07.
(B) Graph displaying the loadings of metabolite variations along PC1 axis.
(C) Time-course assessment of lactate accumulation (right graph) or glucose dissipation (left graph) in culture medium from ESCs or MEFs transduced with the indicated factors.
(D) Graph displaying the basal oxygen consumption rate (OCR) in MEF control or expressing the indicated factors for the days shown. Red dashed line indicates basal OCR values in ESCs.
Data represent mean ± SEM, one-tailed unpaired t test (n = 5 in A–C; n = 3 in D); all results were statistically significant (p < 0.05). See also Figures S3 and S4.
These findings suggest that expression of c-MYC, alone or in combination with OSK, activates the glycolytic pathway. In fact, glycolytic flux and capacity were upregulated during the first days of OSKM expression (Figure S4A) in a c-MYC-dependent manner (Figure S4B). Our results therefore suggest that c-MYC articulates the metabolic switch associated with cell reprogramming, which has been shown to be characterized by both upregulation of glycolysis and inhibition of OXPHOS (Folmes et al., 2011).
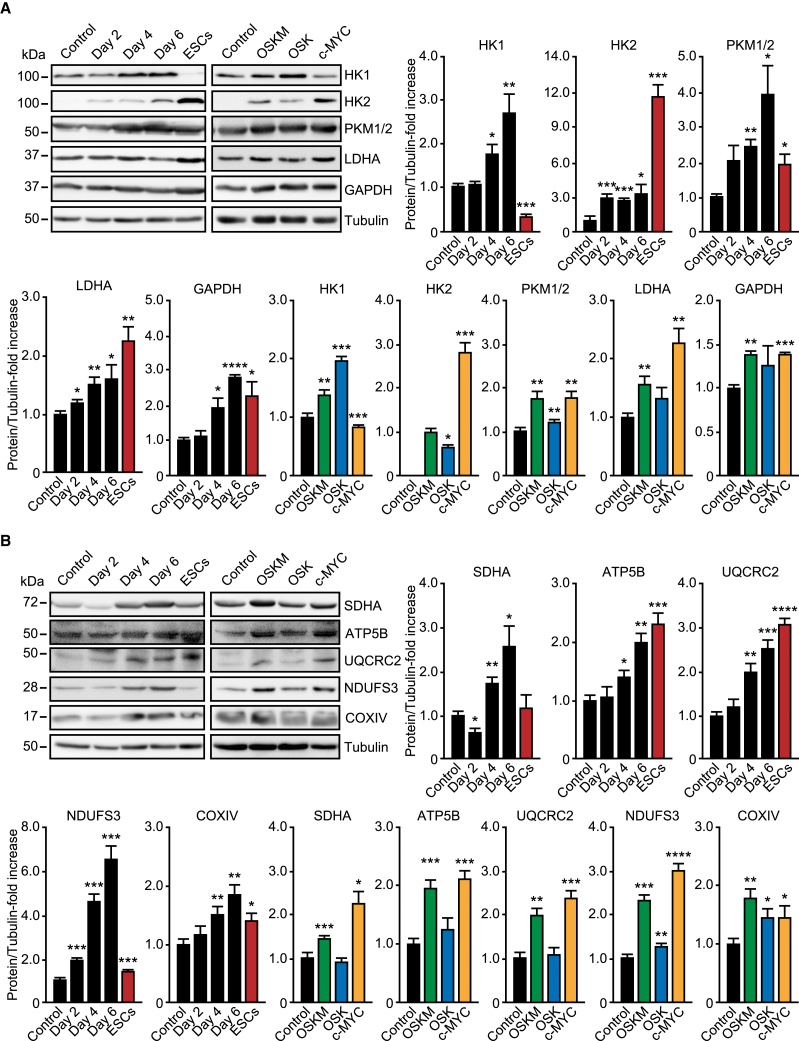
Time-course measurements of oxygen consumption rates (OCRs) revealed the induction of a transient OXPHOS peak following expression of OSKM, OSK, or c-MYC, albeit at different times and with distinctive intensities (Figure 3D). As previously reported (Kida et al., 2015), OSKM induced a robust OXPHOS peak at day 4 of reprogramming (Figures 3D and S4C). Interestingly, OSK expression also produced an OXPHOS peak at day 4, although to a much lesser extent than OSKM. Overexpression of c-MYC alone also led to an OXPHOS peak at day 2 post transduction, which was similar in intensity to that of OSKM (Figure 3D). Surprisingly, our respirometry experiments also showed that OCRs decreased at day 6 to values found at day zero, but these were not yet diminished to levels displayed by ESCs (Figure 3D). Thus, and in agreement to previously published data, our results showed that cell reprogramming induces a steady upregulation of glycolysis. However, and conversely to what was previously thought, cell reprogramming does not involve an inhibition of OXPHOS below somatic levels following the early metabolic burst. In agreement with these findings, combined immunoblotting analyses (Figure 4) and quantitative protein microarrays (Figure S5) showed that both OXPHOS and glycolysis enzymatic machineries were upregulated during early cell reprogramming in a c-MYC-dependent manner.
Figure 4.
Induction of Glycolytic and OXPHOS Enzymes by c-MYC
(A and B) Lysates of ESCs, MEFs control, or expressing OSKM for the specified days (left panels, black and red bars), or the indicated factors for 4 days (right panels, colored bars), were analyzed by immunoblotting using antibodies against the glycolytic (A) or OXPHOS (B) enzymes shown. Graphs show the quantification of the data. Data represent mean ± SEM, one-tailed unpaired t test (n = 3): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S5.
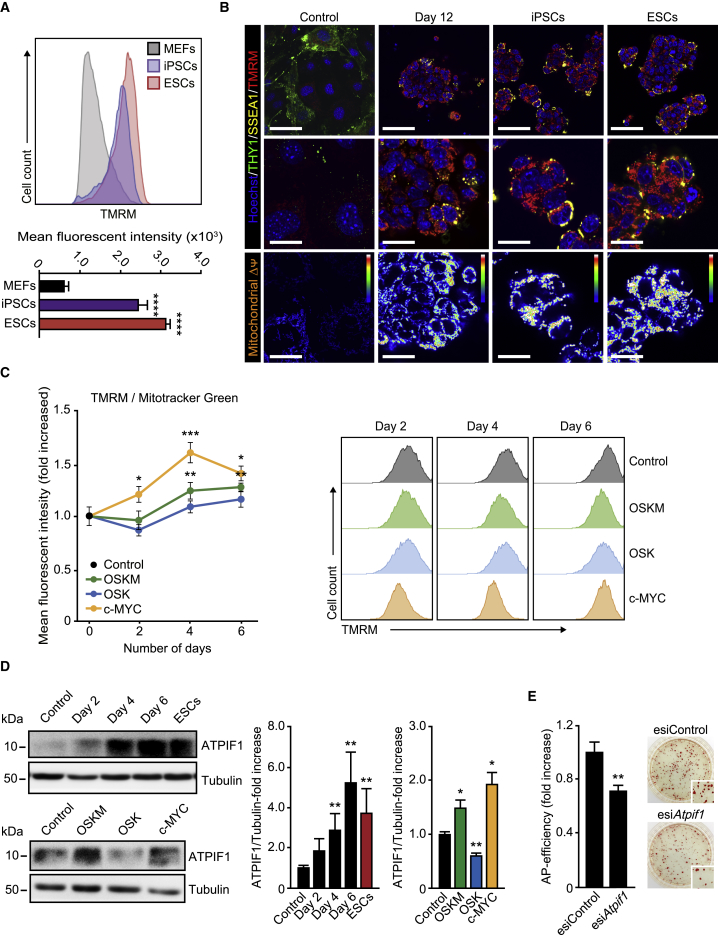
Mitochondrial Membrane Potential Marks Cells Prone to Cell Reprogramming
We next sought to investigate the importance of maintaining somatic OXPHOS rates for cell reprogramming. First, we wanted to assess mitochondrial membrane potential (MMP), as an indicator of mitochondrial function, using the TMRM probe. Flow-cytometry analyses revealed elevated MMP levels in pluripotent cells compared with controls (Figure 5A). Immunofluorescence examination of MMP also showed that ESCs displayed high TMRM levels compared with controls (Figure 5B). As previously reported (Folmes et al., 2011), we observed colonies with intense TMRM staining in OSKM-expressing cells at day 12 post transduction (Figure 5B). These TMRM-positive colonies were also positive for SSEA-1 staining, suggesting that functional mitochondria are important for cells undergoing reprogramming. As we have recently described (Prieto et al., 2016a), total mitochondrial content, measured by flow cytometry using the MMP-independent probe MitoTracker green, was reduced in pluripotent cells compared with control cells (Figure S6A) and decreased with time in either OSKM-, OSK-, or c-MYC-transduced cells (Figures S6B and S6C). These observations led us to take into consideration this feature when analyzing MMP by this technique (see below). Time-course examination of TMRM/MitoTracker green ratios in OSK- or OSKM-expressing cells showed a gradual increase in the relative MMP that reached a plateau at day 6, OSKM inducing a higher increase in these ratios than OSK (Figure 5C). Interestingly, the relative MMP values observed in OSKM-expressing cells were higher than those found in either somatic or OSK-expressing cells. Relative MMP in cells expressing c-MYC alone displayed a steep increase, which peaked at day 4 and decreased at day 6 (Figure 5C). Interestingly, we observed an upregulation of the ATPase inhibitor factor 1 (ATPIF1), a physiological inhibitor of the H+-ATP synthase (Gledhill et al., 2007), during OSKM-induced cell reprogramming (Figure 5D, upper panels). Expression of ATPIF1 protein was reduced by OSK and highly increased by either OSKM or c-MYC (Figure 5D, lower panels). Furthermore, knockdown of ATPIF1 protein (Figure S6D) reduced the emergence of AP-positive colonies in OSKM-transduced cells by ∼30% (Figure 5E). Taken together, these results suggest that elevated MMP, induced by c-MYC and likely upheld by ATPIF1-mediated inhibition of the ATPase, could mark cells prone to cell reprogramming.
Figure 5.
Increased Mitochondria Polarization during Cell Reprogramming
(A) Representative flow-cytometry histograms of MEFs, induced pluripotent stem cells (iPSCs), and ESCs stained with TMRM to assess mitochondrial membrane potential. Graph underneath shows the quantification of the mean fluorescence intensity of the histograms shown above.
(B) Representative confocal images of live MEFs before (Control) or 12 days after OSKM expression (Day 12), iPSCs, and ESCs stained with anti-THY1 (green) or anti-SSEA1 (yellow) antibodies, combined with the cell-permeable TMRM dye (red). Hoechst 33342 (blue) was used as a nuclear counterstaining. Lower pictures are color map representations of the pictures in the middle panels showing TMRM intensity according to the displayed color bar. Scale bars, 24 μm (upper images) and 12 μm (middle and bottom images).
(C) Right: histograms of TMRM staining in MEFs expressing the indicated factors for the days shown. Left graph represents the TMRM/MitoTracker green ratio dynamics along the indicated days.
(D) Lysates of MEF control, expressing OSKM for the specified days or ESCs (upper panels, black and red bars in the right graph), or the indicated factors for 4 days (lower panels, colored bars in the right graph) were analyzed by immunoblotting using the shown antibodies. Graphs on the right show the quantification of the data.
(E) MEFs transduced with OSKM factors were transfected with esiRNAs targeting either eGFP (Control) or Atpif1 at day 1 post transduction. Graph shows the number of AP-positive colonies obtained after 25 days of retroviral delivery. Panels on the right show representative bright-field images from the plates of the indicated cultures after AP staining; inset shows magnification of a selected area from the AP-stained plates.
Data represent mean ± SEM, one-tailed unpaired t test (n = 3): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S6.
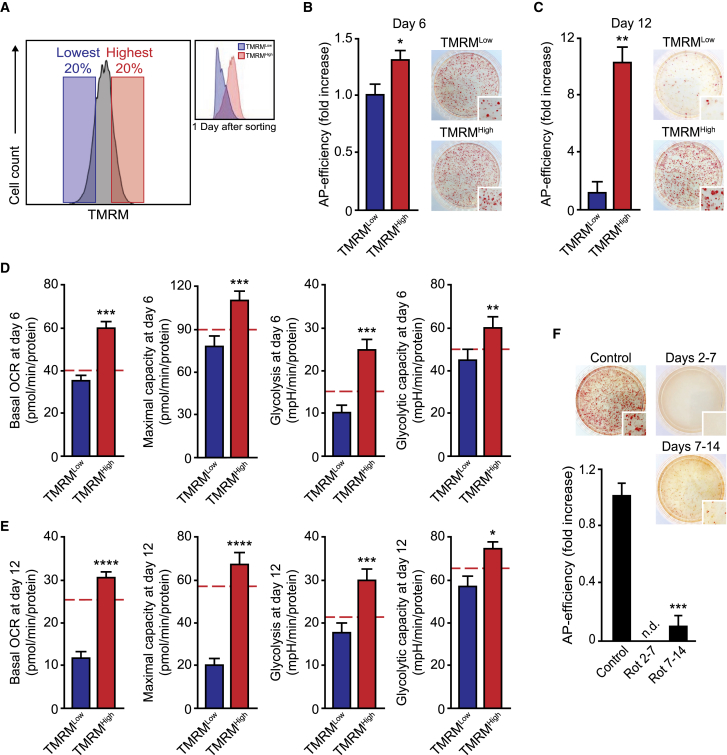
To investigate this possibility, we sorted OSKM-expressing cells based on their TMRM intensity at either day 6 or day 12 post transduction (Figure 6A) and compared their ability to generate AP-positive colonies as an estimation of reprogramming efficiency. OSKM-expressing TMRM-high cell population sorted at day 6 post transduction showed ∼25% increase in the number of AP-positive colonies when compared with TMRM-low counterparts (Figure 6B). Remarkably, this increase in the reprogramming efficiency shown by TMRM-high cells was clearly evident when OSKM-expressing cells were sorted at day 12 post transduction, and TRMR-high cells displayed a >10-fold increase in the number of AP-positive colonies (Figure 6C). These results suggest that high MMP marks cells prone to reprogramming.
Figure 6.
Cells Prone to Reprogramming Display a Hybrid Metabolism
(A) Flow-cytometry histograms illustrating the windows used for sorting OSKM-expressing cells based on their TMRM staining intensity (left histograms) and the TMRM profiles following overnight culture of the sorted cells at the indicated days post transduction (right histograms).
(B and C) Graphs showing the number of AP-positive colonies scored in MEFs after 25 days of OSKM retroviral delivery that were sorted based on their TMRM loading intensity (TMRMLow or TMRMHigh) at day 6 (B) or day 12 (C) post transduction. Right panels: representative bright-field images from the plates of the indicated cultures after AP staining. Inset shows magnification of a selected area from the AP-stained plates.
(D and E) Graphs displaying basal oxygen rate (OCR), maximal respiratory capacity, glycolytic flux (Glycolysis), or the glycolytic capacity of OSKM-transduced cells sorted as above at day 6 (D) or day 12 (E) post transduction. Dashed red lines show the values of the mean, corresponding to non-transduced MEF controls in each assay.
(F) Graph showing the scoring of AP-positive colonies obtained in MEFs after 25 days of OSKM retroviral delivery incubated with DMSO (as vehicle control), or the ETC complex I inhibitor rotenone (0.5 μM) for the indicated days. Right panels: representative bright-field images from the plates of the indicated cultures after AP staining; inset shows magnification of a selected area from the AP-stained plates.
Data represent mean ± SEM, one-tailed unpaired t test (n = 3): ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; n.d., not detected. See also Figure S6.
We next examined the metabolic profiles of these OSKM-expressing cell populations displaying different MMPs. Cell sorting was performed at day 6 (Figures 6D and S6E) or day 12 (Figures 6E and S6F) post transduction into TMRM-low (blue bars and lines) and TMRM-high (red bars and lines) cell populations. TMRM-low cells sorted at either day 6 or day 12 post transduction displayed similar (day 6) or reduced (day 12) OCRs and glycolytic rates compared with controls. Remarkably, TMRM-high cell populations sorted at either day 6 or day 12 post transduction showed increased OCRs and glycolytic rates. Furthermore, transient inhibition of ETC complex I by rotenone at different days of the process blunted cell reprogramming (Figure 6F), underscoring the importance of OXPHOS for cell reprogramming. Taken together, our results indicate that both glycolysis and OXPHOS play central roles in the phenotypic transition of somatic cells to pluripotency.
Discussion
It has been reported that overexpression of exogenous c-MYC is beneficial but not necessary for cell reprogramming (Nakagawa et al., 2008, Wernig et al., 2008). Moreover, it has been shown that MYC family members do not directly regulate pluripotency in ESCs or mouse embryos (Scognamiglio et al., 2016). These findings have nourished the idea that MYC does not play a major role in cell reprogramming. Here we present evidence showing that MYC is necessary for cell reprogramming, likely by inducing a hybrid energetics program to favor the phenotypic transition to pluripotency.
Somatic cells undergo a dramatic remodeling in their metabolic profiles during their transit to pluripotency (Folmes et al., 2011). It has been suggested that this metabolic transition is a synchronous phenomenon in which a gradual increase in glycolytic flux parallels a reduction in cellular respiration during the reprogramming process (Folmes et al., 2011, Panopoulos et al., 2012, Varum et al., 2011). However, we found that a hybrid OXPHOS/glycolysis metabolic phenotype, similar to that found in naive pluripotent stem cells (Takashima et al., 2014, Zhou et al., 2012), is generated early during the process by c-MYC and marks cells prone to cell reprogramming.
The upregulation of both glycolytic and OXPHOS enzymes by c-MYC (Sridharan et al., 2009, Hansson et al., 2012, Cao et al., 2015, Sone et al., 2017; and this study) may account for the elevated OXPHOS and glycolytic activities observed during early cell reprogramming. OSKM expression causes an early and transient activation peak of OXPHOS (Kida et al., 2015, Sone et al., 2017; and this study), which we found is induced by the combined activity of OSK and c-MYC. The steep OXPHOS downregulation to near somatic levels observed in these studies coincided with the upregulation of glycolytic fluxes. The opposite dynamics in the activity of these two pathways could suggest the negative regulation of OXPHOS by high glycolytic metabolic rates, which may limit pyruvate availability in mitochondria to maximize NAD+ recycling. However, an orchestrated regulation of both pathways by the crosstalk of different gene expression programs induced by the reprogramming factors could not be ruled out. In this regard, the upregulation of ATPIF1 protein by c-MYC may play a role in modulating OXPHOS activity, as it does in cancer cells (Santacatterina et al., 2016). Although necessary for cell reprogramming (Kida et al., 2015 and this study), whether this OXPHOS peak is solely a by-product of the forced expression of the reprogramming factors or plays a specific role in the process remains unknown. In this regard, it is possible that the surge in OXPHOS could induce a burst in reactive oxygen species (ROS) during cell reprogramming (Esteban et al., 2010). As ROS can activate pro-proliferative signaling pathways (Hawkins et al., 2016, Reczek and Chandel, 2015), an early and transitory increase in ROS levels could stimulate cell proliferation and therefore be favorable for cell reprogramming. In keeping with this, optimal ROS signaling has been found to be critical for nuclear reprogramming (Zhou et al., 2016). Remarkably, our observations showing that cells displaying high MMP are prone to cell reprogramming underscore the role of mitochondria in this process. These high MMP cells displayed both increased glycolysis and somatic OXPHOS rates, which indicates that a hybrid metabolism is readily established during the first stages of cell reprogramming, likely by c-MYC.
Overall, our work showing the early events driving the metabolic rewiring by MYC reinforce the existent parallels between cell reprogramming and tumorigenesis (Abad et al., 2013, Apostolou and Hochedlinger, 2013, Hochedlinger et al., 2005, Mosteiro et al., 2016, Ohnishi et al., 2014, Prieto and Torres, 2017). Also, our findings suggest that variation of endogenous MYC levels is a determining factor for cell reprogramming mediated by either forced OSK expression or chemical cocktails, in line with studies in other biological contexts (Clavería et al., 2013, Sancho et al., 2013).
Experimental Procedures
Cell Culture, Reprogramming Assays, Reagents, and Plasmids
The induced pluripotent stem cell lines used in this study have been described elsewhere (Prieto et al., 2016a, Prieto et al., 2016b) and were grown in gelatinized plates in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), 1× non-essential amino acids, 1× sodium pyruvate, 1× penicillin/streptomycin (all from Biowest), 0.1 mM 2-mercaptoethanol (Sigma), in the presence of human leukemia inhibitory factor (prepared in-house). Wild-type MEFs (homogeneous C57BL/6 background) were prepared from E13.5 pooled embryos and cultured in high-glucose DMEM supplemented with 10% FBS and 1× penicillin/streptomycin. Early-passage MEFs (third passage at most) were used in all experiments. All procedures were carried out in accordance with the guidelines of the Ethics committee at the University of Valencia. Mice were crossed and maintained at the University of Valencia animal core facility in accordance with Spanish regulations (RD53/2013). The experimental protocol (no. 2015/VSC/PEA/00,079) was approved by the Animal Experimentation Ethics Committee of the University of Valencia and the Generalitat Valenciana government (Spain). Retroviral constructs pMX-Oct4, pMX-Sox2, pMX-Klf4, and pMX-c-Myc (Takahashi and Yamanaka, 2006) were from Addgene. All cells have been routinely tested for mycoplasma contamination using the Lookout MYCoplasma PCR detection kit (Sigma-Aldrich).
Reprogramming was carried out by transduction of MEFs with retroviruses encoding OCT4, SOX2, KLF4, and c-MYC as previously described (Prieto et al., 2016a, Prieto et al., 2016b, Takahashi and Yamanaka, 2006). Reprogramming was assessed 25 days after transduction of MEFs with OSKM or OSK-encoding retroviruses by scoring all the AP-positive colonies per 60 mm. AP staining was performed according to the manufacturer's instructions (Alkaline Phosphatase Detection Kit, Millipore). See Supplemental Experimental Procedures.
Immunofluorescence and Flow Cytometry
For live immunofluorescence analysis, cells were plated on gelatin-coated tissue culture dishes. Cells, untreated or transduced with the indicated viruses, were plated at 1.5 × 104 cells/cm2 the day before processing. The cells were incubated with the indicated antibodies diluted in fresh culture medium for 45 min at 37°C in a CO2 incubator. Cells were then washed three times and medium replaced with fresh culture medium containing 100 nM TMRM (tetramethylrhodamine methyl ester) and 2 μg/mL of cell-permeable Hoechst 33,342 (both from Thermo Fisher) for 30 min in a CO2 incubator at 37°C. Cells were washed three times with fresh medium and analyzed immediately by confocal microscopy.
Confocal immunofluorescence images were taken using a Fluoview FV10i confocal microscope equipped with 405-, 488-, and 633-nm lasers and a live cell environmental module (Olympus). Three-dimensional reconstructions of z stacks and color map representations of the images were performed using FV10-ASW 2.1 viewer software. Co-localization of TOM20 and DRP1 staining was evaluated by calculating the Pearson’s correlation coefficient using the freely available JACoP plug-in (http://rsb.info.nih.gov/ij/plugins/track/jacop.html) for ImageJ analysis software. All images were compiled using Adobe Illustrator CS5.
For assessing or sorting cells based on their MMP by flow cytometry, cells treated as indicated in the text were trypsinized, resuspended in culture medium containing 1% FBS and 100 nM TRMR, and incubated at 37°C in an incubator with CO2 supply for 15 min. Cells were then processed for analysis or cell sorting by flow cytometry. Analytical flow-cytometry measurements were taken using a FACSVerse flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star). At least 10,000 events from each sample were recorded. Fluorescence-activated cell sorting based on TMRM intensity was carried out using a FACSAria III (Becton Dickinson). Details about the antibodies used in this study are provided in Tables S1 and S2.
Western Blot
Cells transduced as indicated in the main text were lysed in RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 0.5% sodium deoxycholate, 100 mM NaF, 2 mM Na3VO4, 20 mM Na4P2O7, and 1× complete proteinase inhibitor cocktail from Roche). Cellular lysates were used for immunoblotting with the indicated antibodies using standard procedures (see Supplemental Experimental Procedures). Details about the antibodies used in this study are provided in Tables S1 and S2.
Printing and Processing of Reverse-Phase Protein Arrays
The protein concentration of the samples was determined with the Bradford reagent (Bio-Rad) using BSA as standard, and diluted in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4 [pH 7.4]) to a final protein concentration of 0.5 μg/μL before printing. After printing, arrays were allowed to dry and further blocked in PBS with Tween 20 containing 5% skimmed milk. Thereafter, each pad in the array was incubated overnight at 4°C with the indicated concentrations of highly specific primary monoclonal or polyclonal antibodies. Microarrays were scanned using a Typhoon 9410 scanner (GE Healthcare). Details of the antibodies used can be found in Tables S1 and S2.
Nucleic Acid Purification and qPCR Analysis
Total RNA was extracted using TRIzol reagent and cDNA synthesized using a SuperScript III reverse transcriptase kit (both from Invitrogen). cDNA products were amplified using an Applied Biosystems StepOne plus Fast Real-Time PCR System. Sequences of the oligonucleotides used in this study are listed in Table S3 and correspond to predesigned KiCqStart SYBR Green primers (Sigma-Aldrich).
Metabolomic Footprinting
Metabolic profiling of cellular medium was carried out with NMR spectroscopy. Samples of culture medium were collected 10, 24, 48, and 72 hr after initial cell incubation with fresh medium at day 2.5 post transduction. Metabolite assignment was performed with the help of the Human Metabolome database and published literature. Metabolite concentrations in basal media were subtracted from concentrations of metabolites in 10- to 72-hr conditioned media for the calculation of net fluxes. The resulting rates were normalized to total cellular protein content determined by a BCA protein assay (Pierce, Thermo-Scientific). See Supplemental Experimental Procedures for details.
Extracellular Metabolic Flux Analysis
Basal and uncoupled OCR, or extracellular acidification rate (ECAR), were measured using a Seahorse bioanalyzer (XF96) and the Mito or Glycolysis stress test kits (both from Seahorse Bioscience, Millipore), respectively, using 20,000–30,000 cells per well. Each experiment was conducted in triplicate, repeated at least three times, and normalized to total protein contents. See Supplemental Experimental Procedures for details.
Statistical Methods
Principal component analysis was performed on Pareto scaled and mean centered data using SIMCA-p+ 12.0 PCA (Umetrics). The model quality was assessed by R2 (goodness of fit) and Q2 (goodness of prediction).
Where indicated, Student's t test was used to estimate statistical significance between categories. Relative values (percentages) were normalized using arcsine transformation before carrying out their statistical comparison. Results are presented as mean ± SEM or mean ± SD as indicated. The number of independent experiments used to estimate statistical significance is denoted as “n” in every figure legend.
Author Contributions
All of the authors designed and discussed the experiments. J.P. conducted most of the experimental work. M.L., K.G., A.V.-S., and X.P. contributed to reprogramming experiments. F.S., L.T., and J.M.C. carried out RPA. A.Y.S. and J.L.-S. contributed to OCR and ECAR measurements. M.P.-S. and A.P.-L. carried out NMR measurements. J.P. and J.T. conceived the project. J.T. provided most of the funding, supervised the experiments, and wrote the manuscript.
Acknowledgments
We are indebted to Lisa M. Sevilla for critically reading the manuscript. We thank F. Pallardó and J. Viña teams for their help with Seahorse Analyzer. This work was supported by grant BFU2015-68366-R MINECO/FEDER, UE (J.T.); grant SAF2013-41945-R MINECO and Fundación Ramón Areces (J.M.C.), and grant SAF2014-53977-R MINECO (A.P.-L.). J.P. was supported by a VALi + d predoctoral fellowship from Generalitat Valenciana and a Journal of Cell Science Traveler fellowship.
Published: November 21, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.10.018.
Supplemental Information
References
- Abad M., Mosteiro L., Pantoja C., Cañamero M., Rayon T., Ors I., Graña O., Megías D., Domínguez O., Martínez D. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. doi: 10.1038/nature12586. [DOI] [PubMed] [Google Scholar]
- Apostolou E., Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretones G., Delgado M.D., León J. MYC and cell cycle control. Biochim. Biophys. Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Guo W.T., Tian S., He X., Wang X.W., Liu X., Gu K.L., Ma X., Huang D., Hu L. miR-290/371-Mbd2-MYC circuit regulates glycolytic metabolism to promote pluripotency. EMBO J. 2015;34:609–623. doi: 10.15252/embj.201490441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavería C., Giovinazzo G., Sierra R., Torres M. MYC-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500:39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- Dang C.V. A time for MYC: metabolism and therapy. Cold Spring Harb. Symp. Quant. Biol. 2016;81:79–83. doi: 10.1101/sqb.2016.81.031153. [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Folmes C.D., Nelson T.J., Martinez-Fernandez A., Arrell D.K., Lindor J.Z., Dzeja P.P., Ikeda Y., Perez-Terzic C., Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gledhill J.R., Montgomery M.G., Leslie A.G., Walker J.E. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc. Natl. Acad. Sci. U S A. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J., Rafiee M.R., Reiland S., Polo J.M., Gehring J., Okawa S., Huber W., Hochedlinger K., Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins K.E., Joy S., Delhove J.M., Kotiadis V.N., Fernandez E., Fitzpatrick L.M., Whiteford J.R., King P.J., Bolanos J.P., Duchen M.R. NRF2 orchestrates the metabolic shift during induced pluripotent stem cell reprogramming. Cell Rep. 2016;14:1883–1891. doi: 10.1016/j.celrep.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K., Yamada Y., Beard C., Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Kida Y.S., Kawamura T., Wei Z., Sogo T., Jacinto S., Shigeno A., Kushige H., Yoshihara E., Liddle C., Ecker J.R. ERRs mediate a metabolic switch required for somatic cell reprogramming to pluripotency. Cell Stem Cell. 2015;16:547–555. doi: 10.1016/j.stem.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidger A.M., Keyse S.M. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs) Semin. Cell Dev. Biol. 2016;50:125–132. doi: 10.1016/j.semcdb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress T.R., Sabò A., Amati B. MYC: connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer. 2015;15:593–607. doi: 10.1038/nrc3984. [DOI] [PubMed] [Google Scholar]
- Mosteiro L., Pantoja C., Alcazar N., Marión R.M., Chondronasiou D., Rovira M., Fernandez-Marcos P.J., Muñoz-Martin M., Blanco-Aparicio C., Pastor J. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354 doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without MYC from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Semi K., Yamamoto T., Shimizu M., Tanaka A., Mitsunaga K., Okita K., Osafune K., Arioka Y., Maeda T. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Panopoulos A.D., Yanes O., Ruiz S., Kida Y.S., Diep D., Tautenhahn R., Herrerías A., Batchelder E.M., Plongthongkum N., Lutz M. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J., Torres J. Mitochondrial dynamics: in cell reprogramming as it is in cancer. Stem Cells Int. 2017;2017:8073721. doi: 10.1155/2017/8073721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J., León M., Ponsoda X., Sendra R., Bort R., Ferrer-Lorente R., Raya A., López-García C., Torres J. Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming. Nat. Commun. 2016;7:11124. doi: 10.1038/ncomms11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J., León M., Ponsoda X., García-García F., Bort R., Serna E., Barneo-Muñoz M., Palau F., Dopazo J., López-García C. Dysfunctional mitochondrial fission impairs cell reprogramming. Cell Cycle. 2016;15:3240–3250. doi: 10.1080/15384101.2016.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek C.R., Chandel N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015;33:8–13. doi: 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos P., Nunes-Xavier C.E., Tabernero L., Köhn M., Pulido R. Dual-specificity phosphatases as molecular targets for inhibition in human disease. Antioxid. Redox Signal. 2014;20:2251–2273. doi: 10.1089/ars.2013.5709. [DOI] [PubMed] [Google Scholar]
- Sancho M., Di-Gregorio A., George N., Pozzi S., Sánchez J.M., Pernaute B., Rodríguez T.A. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev. Cell. 2013;26:19–30. doi: 10.1016/j.devcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacatterina F.L., Sánchez-Cenizo L., Formentini M.A., Mobasher E., Casas C.B., Rueda I., Martínez-Reyes I., Núñez de Arenas C., García-Bermúdez J., Zapata J.M. Down-regulation of oxidative phosphorylation in the liver by expression of the ATPase inhibitory factor 1 induces a tumor-promoter metabolic state. Oncotarget. 2016;7:490–508. doi: 10.18632/oncotarget.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scognamiglio R., Cabezas-Wallscheid N., Thier M.C., Altamura S., Reyes A., Prendergast Á.M., Baumgärtner D., Carnevalli L.S., Atzberger A., Haas S. MYC depletion induces a pluripotent dormant state mimicking diapause. Cell. 2016;164:668–680. doi: 10.1016/j.cell.2015.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Morone N., Nakamura T., Tanaka A., Okita K., Woltjen K., Nakagawa M., Heuser J.E., Yamada Y., Yamanaka S. Hybrid cellular metabolism coordinated by Zic3 and Esrrb synergistically enhances induction of naive pluripotency. Cell Metab. 2017;25:1103–1117.e6. doi: 10.1016/j.cmet.2017.04.017. [DOI] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. Mitotic phosphorylation of dynamin-related GTPase DRP1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S., Rodrigues A.S., Moura M.B., Momcilovic O., Easley C.A., 4th, Ramalho-Santos J., Van Houten B., Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-MYC is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Meng S., Li Y., Ghebre Y.T., Cooke J.P. Optimal ROS signaling is critical for nuclear reprogramming. Cell Rep. 2016;15:919–925. doi: 10.1016/j.celrep.2016.03.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Q., Li W., Benda C., Huang Z., Ahmed T., Liu P., Guo X., Ibañez D.P., Luo Z., Zhang M. NCoR/SMRT co-repressors cooperate with c-MYC to create an epigenetic barrier to somatic cell reprogramming. Nat. Cell Biol. 2018;20:400–412. doi: 10.1038/s41556-018-0047-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.