Summary
Predicting developmental potency and risk of posttransplantation tumor formation by human pluripotent stem cells (hPSCs) and their derivatives largely rely on classical histological analysis of teratomas. Here, we investigated whether an assay based on microRNAs (miRNA) in blood plasma is able to detect potentially malignant elements. Several hPSCs and human malignant germ cell tumor (hGCT) lines were investigated in vitro and in vivo after mouse xenografting. The multiple conventional hPSC lines generated mature teratomas, while xenografts from induced hPSCs (hiPSCs) with reactivated reprogramming transgenes and hGCT lines contained undifferentiated and potentially malignant components. The presence of these elements was reflected in the mRNA and miRNA profiles of the xenografts with OCT3/4 mRNA and the miR-371 and miR-302 families readily detectable. miR-371 family members were also identified in mouse plasma faithfully reporting undifferentiated elements in the xenografts. This study demonstrated that undifferentiated and potentially malignant cells could be detected in vivo.
Keywords: teratoma, human pluripotent stem cells, malignant elements, embryonal carcinoma, human germ cell tumors, microRNA
Highlights
-
•
Side-by-side comparison of cells and xenografts/teratomas derived from hPSCs and hGCTs
-
•
Xenografts from hPSCs and hGCTs showed the same histogenesis, mRNA and microRNA profile
-
•
miR-371 and -302 diagnosed undifferentiated/malignant cells in hPSC-derived xenografts
-
•
miR-371 diagnosed undifferentiated/malignant cells in blood of hPSCs injected mice
Salvatori et al. showed histological, mRNA, microRNA similarities between human germ cell tumors and xenografts derived from human pluripotent stem cells (hPSCs). miR-371, -302, and C19MC families were present in the blood plasma of mice after injection of hPSCs (teratoma assay) and were indicative of undifferentiated and potentially malignant cells in the growing xenograft.
Introduction
The teratoma assay is still the most widely accepted method for defining pluripotency of human stem cell lines. Putative human pluripotent stem cells (hPSCs) are injected into extra-uterine sites of immune-deficient mice where they form xenografts. These tumors are often highly heterogeneous, composed of derivatives of all three embryonic germ layers, ectoderm, endoderm, and mesoderm. They are then referred to as teratomas, while teratocarcinomas contain also malignant embryonal carcinoma (EC) cells (Buta et al., 2013, Damjanov and Andrews, 2016). Yolk sac (YS), an extraembryonic endodermal tissue, has also been described in teratomas and teratocarcinomas. The teratoma assay is laborious, animal-based, essentially qualitative, and rarely standardized, with inconsistencies reported in methodology and histopathological evaluation (Muller et al., 2010). To determine the clinical safety of hPSC-based therapies, the ability to detect undifferentiated PSCs and potentially malignant cells is extremely important (Allison et al., 2018, Damjanov and Andrews, 2016). In vitro tests such as PluriTest or ScoreCard, that describe the expression of pluripotency or lineage markers have been proposed as alternatives to the teratoma assay, but these in vitro assays may miss detecting the malignant potential hPSCs (Allison et al., 2018, Bouma et al., 2017, Muller et al., 2011, Tsankov et al., 2015). Another test, called TeratoScore, based on quantifying gene expression in xenografts, has shown good agreement with the teratoma assay with respect to identification of tissues derived from the three germ layers, but ultimately only the teratoma assay has been shown to detect both pluripotency and malignancy (Allison et al., 2018, Avior et al., 2015).
In human pathology, teratomas belong to a class of complex neoplasms called germ cell tumors (human germ cell tumor [hGCT]). During histopathological evaluation of hGCTs, the presence of undifferentiated cells and YS elements are a clear indication of malignancy. The biological significance of these elements is still debated in the context of mouse xenografts derived from injection of hPSCs. In the WHO pathology classification of tumors, various hGCT types are distinguished, of which postpubertal (type II) malignant hGCTs are of particular interest. Histologically, these are referred to as either seminoma (SE) or non-seminoma (NS). NS can represent all cell lineages present during early embryogenesis, including somatic as well as extra-somatic tissues such as YS tumor and choriocarcinoma. They all originate from EC cells, the malignant counterpart of embryonic stem cells (ESCs) (Andrews et al., 1980, Andrews et al., 1984, Pera et al., 1989). EC cells, such as ESCs, are characterized by expression of OCT3/4 and SOX2 (de Jong et al., 2008, Looijenga et al., 2003), two transcription factors also used to generate induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka, 2006). This demonstrates the similarities between hGCTs and hiPSCs (Cunningham et al., 2012). All malignant hGCTs, with the exception of teratomas (TE), are characterized by high expression of a defined set of miRNAs (Gillis et al., 2007, Looijenga et al., 2007). These include miR-371a-3p, -372-3p, -373-3p, and -367-3p, which are also highly expressed in hESCs and hiPSCs (Barroso-del Jesus et al., 2009, Lipchina et al., 2012, Parr et al., 2016, Zhang et al., 2013). These miRNAs have been shown to be specific and sensitive (liquid) biomarkers for malignant hGCTs in serum, plasma, as well as cerebrospinal fluid of patients (Belge et al., 2012, Dieckmann et al., 2012, Gillis et al., 2013, Leão et al., 2018, Murray et al., 2011, Syring et al., 2015, van Agthoven et al., 2017).
In this study, we investigated whether plasma miRNAs could serve as facile biomarkers to detect undifferentiated and potentially malignant cell types in xenografts and in plasma samples derived from mice injected with hPSC lines and hGCT cell lines. A set of hPSC lines and hGCT cell lines was investigated after in vitro culture and in vivo after being xenografted into mice. Each cell line and corresponding xenografts were examined for mRNA and miRNA expression. Patient-derived hGCTs were included for comparison. In addition, mouse plasma miRNA profiles were monitored over time during xenograft formation. The results demonstrated that the histological detection of undifferentiated and malignant components in the xenografts could be predicted with high accuracy on the basis of plasma miR-371a-3p levels. We hypothesize that this could be of value in detecting later malignancy should it arise in patients who have undergone transplantation of differentiated hPSCs as therapy.
Results
Design of the Study
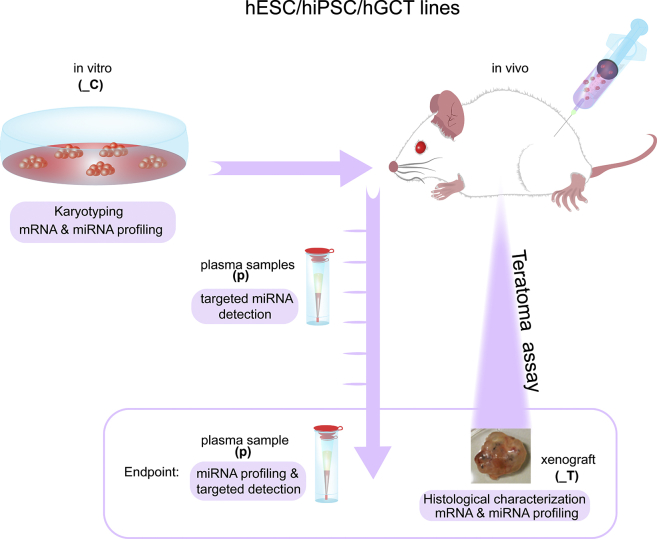
The study design is shown schematically in Figure 1. Multiple hESC and hiPSC lines and (malignant) hGCT cell lines were investigated (Table S1B). Each was examined for mRNA and miRNA expression after culture in vitro and in vivo after xenografting to mice, to investigate the extent of similarity. The xenografts were also examined (immuno)histologically by experienced pathologists. Matched serial plasma samples were profiled for miRNA expression during xenograft development and at the endpoint of the experiment when mice were euthanized. A list of all abbreviations used is given in Table S1A.
Figure 1.
Schematic Representation of the Work Flow Performed
hPSC (iPSCs and hESCs) and hGCT cell lines were injected subcutaneously into immunodeficient mice (teratoma assay). Cell lines were karyotyped before injection and collected for mRNA and miRNA analysis. During the teratoma assay, blood samples for plasma preparation were timely collected until the tumors reached the maximum allowed size of 2 cm3 (endpoint). At the endpoint, xenografts were collected for histological characterization, and for mRNA and miRNA isolation, and blood samples for plasma isolation (_C, cell lines; _T, xenografts; and p, plasma samples). The mRNA expression profile of the xenograft derived from hPSC and hGCT lines was compared with a set of hGCTs from patients (Table S2). A list of all abbreviations used in the study is given in Table S1A.
Histological Characterization of the Xenografts
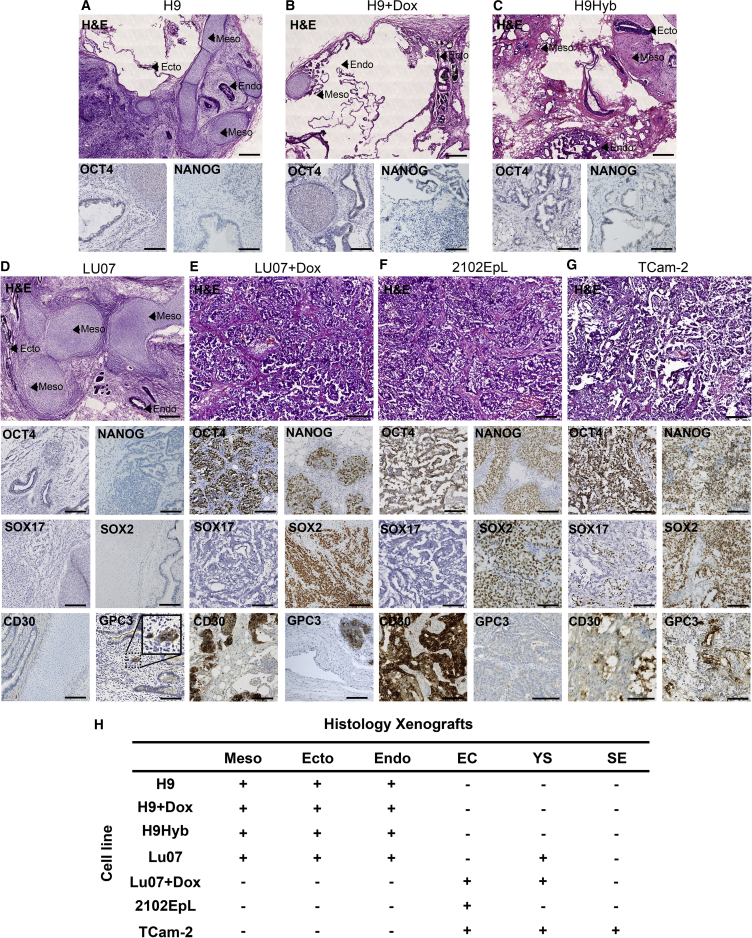
Xenografts were morphologically characterized in H&E-stained sections and by immunohistochemical detection of OCT3/4, NANOG, CD30, SOX2, SOX17, and GPC3: OCT3/4 and NANOG (Figure 2) to detect undifferentiated hPSCs and potentially malignant elements (Ulbright et al., 2014). CD30 is a survival factor and marker of transformed human EC cells (Herszfeld et al., 2006). SOX17 is informative for SE cells, SOX2 for EC, both being co-expressed with OCT3/4 and NANOG (Cheng et al., 2007, Eini et al., 2014, Hart et al., 2005, Looijenga, 2009, Looijenga et al., 2003, Rijlaarsdam et al., 2011). Glypican-3 (GPC3) identifies YS elements, associated with loss of SOX2 and gain of SOX17 (in the absence of OCT3/4 and NANOG) (Looijenga, 2009, Looijenga et al., 2003, Ulbright et al., 2014). Representative H&E-stained sections are shown in Figure 2. Xenografts derived from H9 hESCs and the H9Hyb hESC line consisted only of fully differentiated teratomas, confirming the immunohistochemical findings. The Lu07 hiPSC line also consisted predominantly of differentiated derivatives of the three germ layers but had some YS elements (see below). Xenografts of all of these cell lines contained somatic tissue structures representing all three germ layers (neural rosettes and retinal pigmented epithelium [ectoderm], intestinal epithelium [endoderm], cartilage, bone, fat, and muscle [mesoderm]) and they were negative for the pluripotency markers OCT3/4 and NANOG. This is in line with previous immunofluorescent staining for βIII-tubulin(ectoderm), human α-fetoprotein (AFP, endoderm) and human PECAM-1 (CD31, mesoderm) (Bouma et al., 2017). By contrast, the 2102Ep xenografts consisted of EC, i.e., they had not undergone somatic differentiation, as expected from their tumor origin. Xenografts from the Lu07 + Dox hiPSC line, in which the reprogramming genes had been reactivated by giving the mice doxycycline (Dox) in their drinking water, were mostly composed of EC components resembling the 2102EpL xenografts, corresponding with their immunohistochemical profiles (positive for OCT3/4, SOX2, and CD30), with small scattered areas positive for GPC3 (i.e., YS), as also found in the Lu07 hiPSC line without Dox. The TCam-2-derived xenograft showed mixed pattern of SE, EC, and YS elements, again corresponding to the immunohistochemical profile showing heterogeneous staining for OCT3/4/SOX2/CD30 (for EC), GPC3 (for YS), and OCT3/4 and SOX17 (SE). In summary, H9 and H9Hyb hESC lines gave rise to xenografts that were composed entirely of differentiated derivatives of the three germ layers; they were therefore classified as TE (Figure 2). The Lu07 hiPSC xenografts were predominantly composed of tissues derived from the three germ layers representing TE, but they also showed small scattered groups of GPC3-positive cells, i.e., YS differentiation. These YS cells were also found in the Lu07 + Dox xenografts with reactivated reprogramming factors, although these were otherwise predominantly composed of undifferentiated cells identified as EC cells.
Figure 2.
Histopathological Characterization of Xenografts Derived from Different hPSC and hGCT Cell Lines
(A–G) Representative pictures of H&E and immunohistochemistry of xenografts derived from hPSCs (A) H9, (B) H9 + Dox, (C) H9Hyb, (D) Lu07, (E) Lu07 + Dox and hGCT-derived cell lines, (F) 2102EpL, and (G) TCam-2. H&E-stained xenografts (A–D) show differentiated tissue areas representing derivatives of mesoderm (Meso), ectoderm (Ecto), and endoderm (Endo). Scale bars, 200 μm. Representative images of immunohistochemistry for the pluripotency markers OCT4 and specific markers for hGCTs SOX17, SOX2, CD30, and GPC3 are shown. Nuclei were counterstained with hematoxylin only. Scale bars, 100 μm (for the immunohistochemistry images) (see Table S3 for antibody list).
(H) Summary of presence of tissues derived from the three embryonic germ layers, undifferentiated embryonal carcinoma (EC) cells, yolk sac (YS) elements, and seminoma (SE) components in the xenograft. Notice that Lu07, although negative for all other markers, showed small groups of cells positive for GPC3 (i.e., YS differentiation).
mRNA Profiling
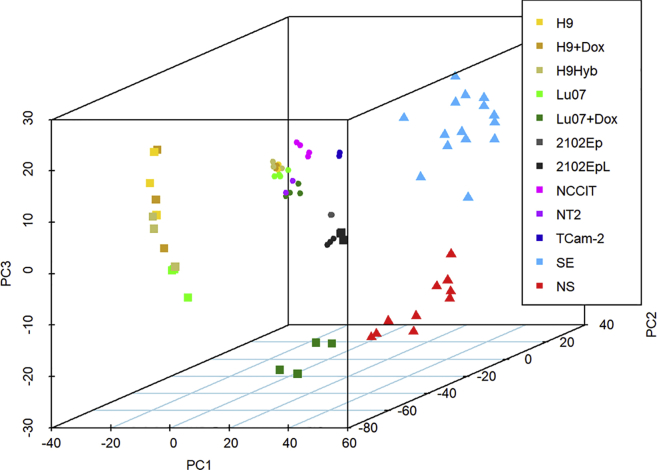
Previously, we reported the mRNA profiles of xenografts from the Lu07 + Dox hiPSC line with reactivated reprogramming transgenes and 2102EpL which completely lacked differentiated elements in histology (Bouma et al., 2017). To extend these datasets and to highlight the similarities between these and hGCTs, we included an additional series of hGCT-derived cell lines and primary testicular hGCTs, representing the various histological elements and types of hGCTs, i.e., EC, YS tumors and SEs (Table S2). To capture the major similarities between these 74 samples, principal-component analysis (PCA) of the top differentiating genes (4% of total) was performed. The 3D PCA plot clearly resolved the cell cultures, xenografts, and hGCT groups (Figure 3). The Lu07 + Dox hiPSC xenografts were separated from the xenografts derived from H9 and H9Hyb hESCs and Lu07 hiPSCs without Dox. Clustering using the top 99 differential probes confirmed the overall PCA patterns (Figure S1) and also showed the extensive overlap in expression between hGCTs and the Lu07 + Dox hiPSC xenografts. Multiple genes associated with “stem cell properties” and “malignancy” were expressed in the Lu07 + Dox hiPSC xenografts. These mRNA profiles indicated that the majority of the stem cell markers were not present in xenografts in which there was full differentiation into mature TE, while they remained high in Lu07 + Dox hiPSC xenografts, reflecting retention of their undifferentiated histology. The Lu07 + Dox hiPSC tumors therefore resembled hGCTs and the 2102EpL xenografts which also continued to express many pluripotency markers. Constitutive expression of pluripotency genes thus confers classic features of hGCTs in these hiPSC lines.
Figure 3.
Principal-Component Analysis of mRNA Expression of hPSCs, hPSC-Derived Xenografts, hGCT-Derived Cell Lines and Xenografts and Primary Testicular hGCTs
The top 1,924 differentiating mRNAs (SD > 0.75) were used for principal-component analysis. The first three components (explaining 72.7% of the variance) were plotted. The color code for the different models (and specific culture conditions) is shown on the right. The sample type is indicated by a specific symbol (culture, dot; xenograft as square; hGCT as triangle). See also Figure S1.
miRNA Profiling
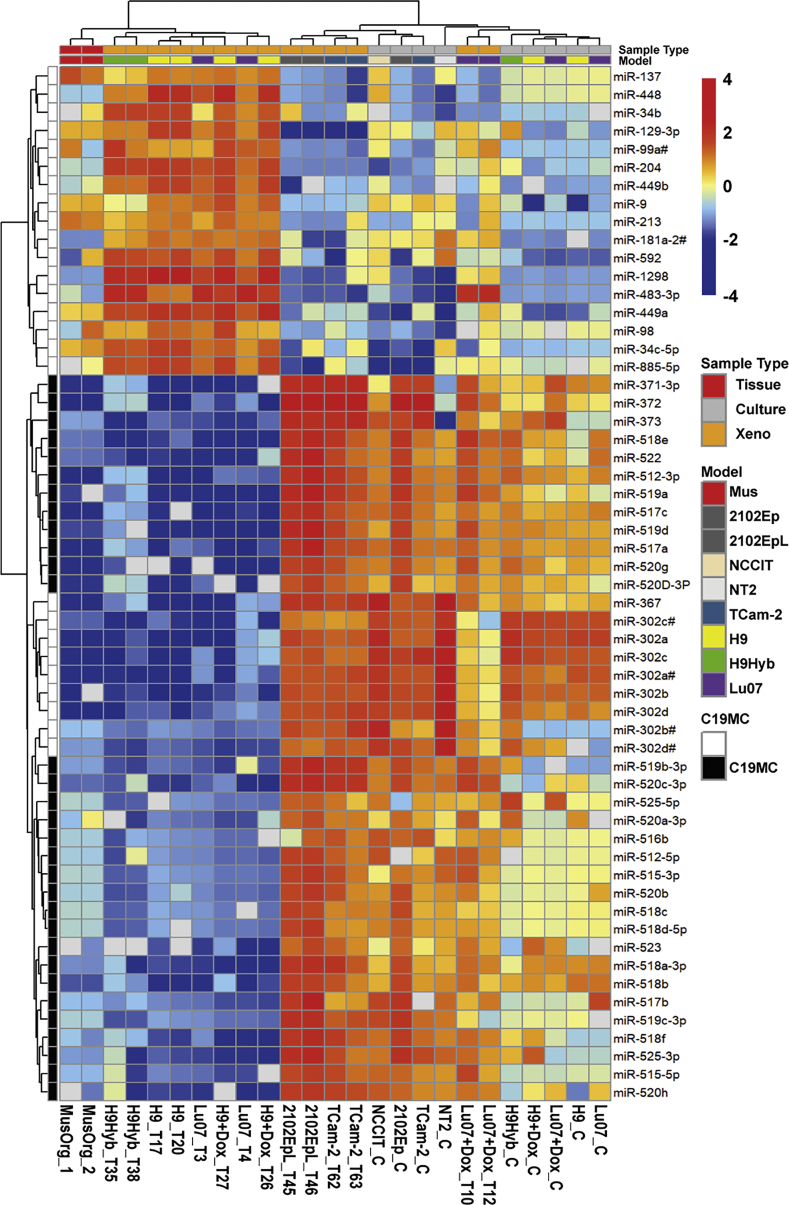
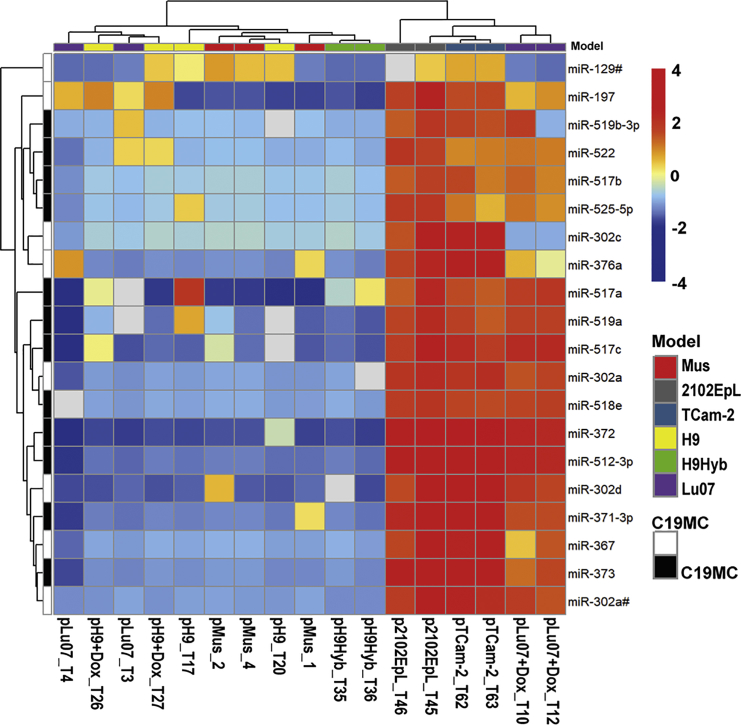
The miRNA expression profile of the xenografts was determined using TaqMan low-density arrays (TLDA cards A&B). In total, 768 individual miRNAs were assayed in the xenografts and compared with the profiles of a set of control mouse organs, human EC cell lines (NCCIT, 2102Ep, NT2), the SE cell line TCam-2, as well as the undifferentiated hPSCs (Lu07 + Dox, Lu07, H9, H9+Dox, and H9Hyb) cultured in vitro. The resulting heatmap (Figure 4) showed the relative levels of the top differentiating miRNAs (n = 57) in these samples and indicated higher levels of the different members of the miR-371, miR-302, and miR-518 families in the undifferentiated Lu07 hiPSCs, with or without Dox, Lu07 + Dox_T-derived xenografts, and the NCCIT, 2102Ep, NT2, and TCam-2 cells and xenografts. Xenografts consisting only of mature TE completely lacked expression of these miRNAs. Many of these miRNA families are located in the primate-specific chromosome 19 miRNA cluster (C19MC, Figure 4), which has been implicated in ESC function and specifically associated with malignant hGCTs (Barroso-del Jesus et al., 2009, Lipchina et al., 2012, Parr et al., 2016, Zhang et al., 2013) but not with TE (Flor et al., 2016, Gillis et al., 2007, Looijenga et al., 2007, Shen et al., 2018). A number of downregulated miRNAs was also identified, including members of the miR-34 cluster.
Figure 4.
miRNA Expression Profile Comparison between hPSCs and hGCT-Derived Cell Lines and Respective Xenografts
The miRNA expression of hPSC cell lines (Lu07 + Dox_C, Lu07_C, H9_C, H9 + Dox_C, and H9Hyb_C), hGCT-derived cell lines (NCCIT_C, 2102Ep_C, NT2_C, and TCam-2_C) and xenografts derived from the respective hPSC (Lu07 + Dox_T, Lu07_T, H9_T, H9 + Dox_T, and H9Hyb_T) and hGCT (2102EpL_T and TCam-2_T) cell lines was determined and compared. miRNA profiles of NSG mouse organs were included as internal control (MusOrg). The heatmap shows 57 of the 768 examined miRNAs of which the levels were detectable in the majority of the samples (average Cq < 36). The color key shows the relative levels of the miRNAs. Color codes for specific Sample Type and Model are indicated on the right. miRNAs located in the primate specific chromosome 19 miRNA cluster (C19MC) are indicated. Raw data are available in Table S4.
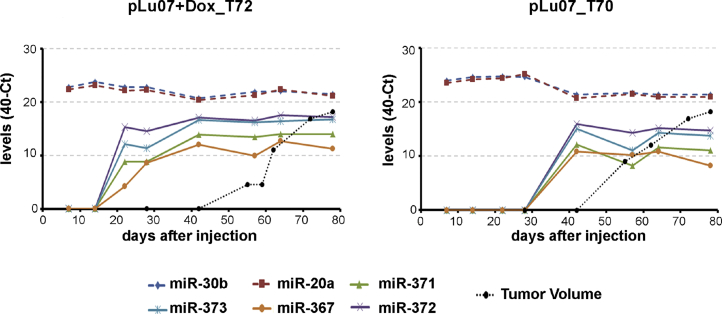
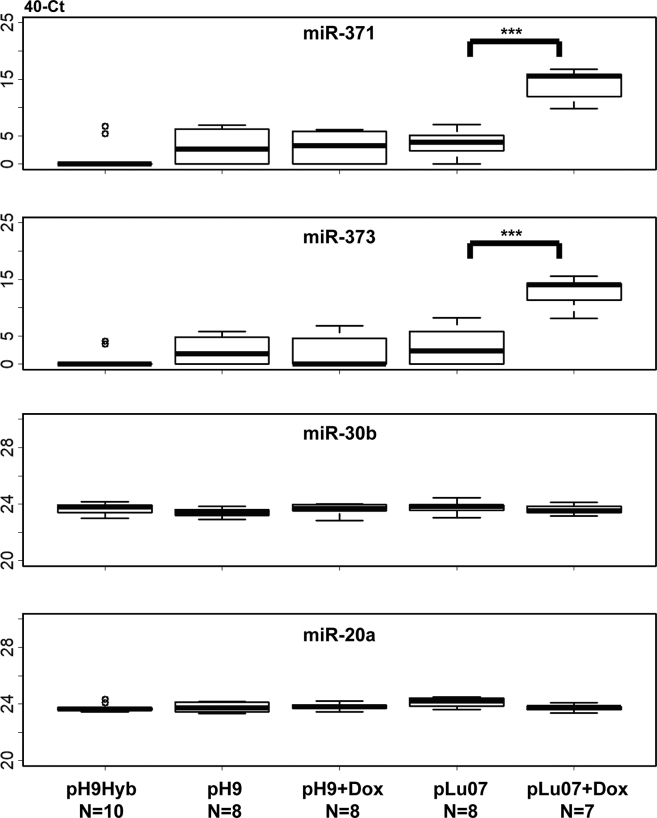
In light of these patterns of miRNAs and their apparent reflection of the histological constitution of cell lines in vitro and in vivo, mouse plasma miRNA levels were examined at the endpoint of the xenograft, using the TLDA card assays, and compared with t = 0 (before injection) as control. The same miRNA families (miR-371, miR-302, and miR-518) distinguished plasma samples of mice injected with the Lu07, H9, H9 + Dox (dox control), and H9Hyb cell lines from mice injected with the (malignant) Lu07 + Dox, 2102EpL, and TCam-2 cell lines (Figure 5), being significantly expressed in the latter group. In light of the high levels of significance of these distinguishing markers, we then examined plasma samples collected during xenograft development to determine when the onset of potential malignant growth was detectable. For each mouse investigated, 40 Ct values of miRNA levels in all plasma samples and the calculated tumor volume (mm3) were evaluated (Figures 6 and S2). In mice transplanted with Lu07 (ST70) or Lu07 + Dox (ST72) cells, plasma levels of the control miRNA (miR-20a and −30b, both cross-reactive with mouse miRNAs) remained stable throughout the experiment, while the miR-371/2/3 and -367 accumulated in the plasma during time. In the case of Lu07 + Dox mice, the time of detection was earlier and the levels were much higher than for the condition without Dox. In both cases, detection of circulating miRNAs in plasma preceded the detection of the actual tumor by physical palpation. Additional time series using mice xenografted with Lu07 hiPSCs, 2102EpL, and TCam-2 cells, confirmed these findings (Figure S2). In all cases, the selected target miRNAs were detected before the tumor became physically manifest and the levels compared with control plasma were generally >10 Ct (i.e., over 1,000-fold difference). The finding of late plasma accumulation of miR-371/2/3 and -367 in mice transplanted with Lu07 cells without Dox in the drinking water could be associated with the histological finding of a minor YS element in the corresponding xenograft (Figure 2). To resolve this further, all available additional endpoint plasma samples of mice with xenografts of H9 and H9Hyb hESCs and Lu07 hiPSCs were evaluated for miR-371/373 and the control miR-20a and -30b (Figure 7). The results showed that mice with Lu07 hiPSCs without the addition of Dox have low plasma levels of miR-371 and -373, while addition of Dox strongly enhanced these levels only in the Lu07-transplanted mice and not in any without the Dox-inducible reprogramming element in the genome. The H9 and H9hybrid hESC lines also showed very low plasma levels of these miRNAs at the endpoint of the xenograft.
Figure 5.
MicroRNA Expression Profile of the Xenograft Endpoint Plasma
Plasma samples (p) of mice xenografted with Lu07 + Dox, Lu07, H9, H9 + Dox, H9Hyb, 2102EpL, and TCam-2 cells were profiled for miRNA levels. Plasma samples of strain-, age-, and sex-matched control mice were included (n = 3, pMus). The heatmap of 20 targets, with average Cq < 36 is shown. The color key shows the relative levels of the miRNAs. Color codes for specific Sample Type and Cell lines are indicated on the right. miRNAs located in the primate specific chromosome 19 miRNA cluster (C19MC) are indicated. Spike-in miRNA levels were stable in all samples (not shown). Raw data are available in Table S4.
Figure 6.
Time Course of miR-371, -372, -373, and -367 Accumulation in the Mouse Plasma Samples of Two Representative Mice Injected with Lu07 (Right Panel) and Lu07 + Dox Cells (Left Panel)
Relative levels (40 Ct) of circulating miR-371, -372, -373, and -367 in the plasma of mice xenografted with Lu07 and Lu07 + Dox cells. Plasma samples (p) were periodically collected until the endpoint of the experiment (max day 80). Tumor size is depicted in volume (100 × levels = mm3). miR-20a and miR-30b were used as internal references.
Figure 7.
Levels of miR-371 and miR-373 for Endpoint Plasma Samples of Mice Injected with H9, H9Hyb, Lu07, and Lu07 + Dox Cell Lines
Relative levels (40 Ct) of circulating miR-371 and miR-373 in the plasma (p) of mice xenografted with H9 (+ Dox and – Dox), H9Hyb, Lu07 (+ Dox and – Dox). miR-20a and miR-30b were used as internal references. The 40-Ct values are shown on the y axis. Endpoint plasma samples derived from Lu07 + Dox injected mice have 100- to 1,000-fold higher levels of miR-371 and -373 compared with the other lines. Mann-Whitney U test, independent samples; ∗∗∗p < 0.005. Number of samples is indicated for each group.
Discussion
In this study we determined mRNA and miRNA profiles of cell lines, xenografts, and circulating miRNAs in matched plasma of mice injected with multiple hPSCs and hGCTs with various histological composition, and related these to the expected teratoma and teratocarcinoma characteristics. We demonstrated that expression levels of the miR-371, -302, and C19MC families revealed the presence of undifferentiated and malignant features in the Lu07 + Dox hiPSCs, 2102EpL and TCam-2, both in cell cultures and derivative tumors. Histologically, 2102EpL and Lu07 + Dox hiPSC tumors were similar to each other, being mainly composed of EC-like cells expressing the pluripotency markers OCT3/4 and SOX2. TCam-2 xenografts were diagnosed as mixed tumors containing SE, EC, and YS elements. The designation “mixed tumor” for the TCam-2 xenografts is in line with previously published data showing that subcutaneous TCam-2 xenotransplantation initiates reprogramming into an EC-like fate (Nettersheim et al., 2016). The H9 and H9Hyb, hESCs, and Lu07 hiPSCs, formed TE containing differentiated cells of the three germ layers and consistently lacked undifferentiated cells (shown by histology and confirmed by immunohistochemistry by the absence of OCT3/4 and NANOG expression); the Lu07 hiPSC line in the absence of Dox somewhat unexpectedly showed YS areas positive for GPC3. In this set of xenografts and terminal plasma samples, levels of the miR-371, -302, and C19MC families were below detection. These findings overlapped with previously reported human clinical pathology in which miR-371-3, -302, and C19MC family were found expressed in all testicular hGCTs except TE (Flor et al., 2016, Gillis et al., 2007, Looijenga et al., 2007, Shen et al., 2018). In particular, the miR-371 family has already proven to be a reliable serum biomarker for testicular hGCTs in patients (Belge et al., 2012, Dieckmann et al., 2012, Gillis et al., 2013, Leão et al., 2018, Murray et al., 2011, Syring et al., 2015, van Agthoven et al., 2017), and we showed here that it can be used to monitor tumor development with minimum invasiveness without the need to terminate the experiment. hGCTs much like as those derived here from the injection of hPSCs, can present highly heterogeneous histology. Although TE are assumed to be non-malignant, it is recognized that they may contain a variable number of residual undifferentiated cells, creating risk of malignancy (Damjanov and Andrews, 2016, Terenziani et al., 2015). However, malignancy features are rarely discussed when interpreting results from the teratoma assay. Teratomas are widely described in the stem cell field as benign tumors composed of mature somatic tissue in contrast to teratocarcinomas, the malignant counterparts that include EC (Damjanov and Andrews, 2007, Muller et al., 2010). This terminology contrasts with that used by clinical pathologists because the description and analysis of the malignant component is too limited; indeed EC is not the only possible malignant component of hPSC-derived tumors (Oosterhuis and Looijenga, 2005) and this should be taken into account in considering potential safety issues with transplantation of hPSC derivatives in regenerative medicine. The initiating hPSCs have the intrinsic capacity to differentiate into all three germ layers of the embryo proper, but also into extraembryonic elements such as YS. Presence of YS is mostly neglected during analysis of teratomas. It is noteworthy that this extraembryonic endodermal tissue is associated with malignant progression in pediatric TE (Gonzalez-Crussi et al., 1978, Heiftz, 1998, O'Connor and Norris, 1994), and it shows similarities with the behavior of ESCs (Oosterhuis and Looijenga, 2005), including expression of the miR-371 and -302 families (Murray et al., 2011).
Following cell transplantation, plasma samples collected from mice injected with Lu07, Lu07 + Dox, 2102EpL, and TCam-2, showed increasing levels of miR-371/2/3 and -367 targets with time. The observation that the levels plateaued in most xenografted mice prior to the maximum measured tumor size, suggests that production of miRNAs is balanced by clearing from the circulation as was also observed in patients after surgical removal of clinically manifest hGCTs (Dieckmann et al., 2017, van Agthoven et al., 2017). However, an alternative explanation might be that larger tumors may be less effective in releasing miRNAs in the circulation, possibly due to reduced growth and/or viability. This is, however, unlikely based on the correlation found between the miRNA levels and the overall tumor load (Dieckmann et al., 2012, van Agthoven and Looijenga, 2017). Interestingly, production of these miRNAs was, at the end of the experiment, detected in mice transplanted with Lu07 hiPSCs without Dox. Although this was initially unexpected based on the available results derived from tissue and plasma miRNA profiling (Figures 4 and 5), it is likely explained by the presence of a minor YS component in the corresponding xenografts. We showed in earlier work that a fraction of Lu07 and Lu07 + Dox hiPSCs contained an additional short arm of chromosome 12 (in Figure S1) (Bouma et al., 2017). The correlation between karyotypic abnormalities and their influence to malignant potential is still under discussion (Allison et al., 2018).
In the Lu07 + Dox, 2102EpL, and TCam-2 malignant xenografts, downregulation of the miR-34 cluster was identified (Figure 4). Members of miR-34 family have been shown to be p53 induced, and they act as tumor suppressors regulating key cancer pathways (Rokavec et al., 2014). This observation is therefore in line with the initial model in which the miR-371 family was found to be an alternative mechanism for inactivation of the P53 pathway in hGCTs (Voorhoeve et al., 2006).
In conclusion, this study demonstrated the similarities between hESCs, hiPSCs, and (malignant) hGCTs regarding their histological composition, mRNA, miRNA, and protein expression profiles. This combined approach resulted in an informative pre-clinical model, allowing investigation of various aspects of pluripotency regulation and tumor risk prediction. Hence, it shows that evaluation of circulating miRNAs in the plasma represents a real-time refinement of the histological analysis of the teratoma assay detecting undifferentiated, and possibly malignant elements. Specifically, the elevated levels in liquid biopsies of a defined set of embryonic miR, being miR-371, -302, and C19MC family members, is of ultimate interest because of their link to the presence of undifferentiated and potentially malignant cell populations. The patterns found in the mouse xenograft model mimic those in hGCT patients. This observation may be expected to have clinical value in development of a minimally invasive method for monitoring development of malignancy in the context of hESC and hiPSC applications in regenerative medicine.
Experimental Procedures
Cell Lines and Patient Samples
Cell lines used in this study are summarized in Table S1B. hPSC lines: hESC lines H9 and H9Hyb (H9 clone Hybrid_6), and hiPSC LUMC007iCTRL01 (Lu07) (Bouma et al., 2017) were kindly supplied by C. Freund, LUMC. The hiPSC Lu07 was generated from skin fibroblasts using a Dox-inducible lentivirus encoding mouse cDNAs for OCT3/4, SOX2, KLF4, and MYC separated by three different 2A peptides (TetO-FUW-OSKM) and a lentivirus carrying the tetracycline-controllable transactivator (FUW-M2rtTA, plasmids from Addgene) as already reported (Bouma et al., 2017). Lu07 was cultured in the absence (Lu07) or presence (Lu07 + Dox) of 1 μg/mL Dox (Sigma). As a control, H9 cells were also cultured in the absence (H9) or presence of Dox (H9 + Dox).
Five aneuploid (malignant) hGCT cell lines were used: TCam-2 representing SE (Mizuno et al., 1993), and four cell lines representing EC (NT2 [NTera-2]; Andrews et al., 1984), NCCIT (Teshima et al., 1988), and two batches of 2102Ep (Andrews et al., 1980, Josephson et al., 2007, Wang et al., 1980) [kindly supplied by P. Andrews, Sheffield, UK]). 2102Ep was maintained at the Erasmus Medical Center and the 2102EpL cultured at the LUMC, and used for the xenograft experiments.
Cell lines were characterized for genomic constitution (karyotyping) as described previously (Bouma et al., 2017) or verified for identity by STR genotyping against the DSMZ (https://www.dsmz.de/) database (hGCT cell lines).
In addition, for the human patient samples, a total of 25 primary testicular hGCTs were included in the study. This series included: 15 pure SE and 10 NS of which the histological composition (EC, TE, YS tumor, or SE) is indicated in the sample name (Table S2). Histological diagnosis was performed by expert pathologists. Use of patient tissue samples remaining after diagnosis was approved for research by the Medical Ethical Committee of the EMC (the Netherlands), permit no. 02.981. This included permission to use the secondary tissue without further consent. Samples were used according to the “Code for Proper Secondary Use of Human Tissue in The Netherlands” developed by the Dutch Federation of Medical Scientific Societies (FMWV, version, 2002; update 2011).
Teratoma Assay
Cells (1 × 106) were injected subcutaneously in the flank region of 8- to 10-week-old male NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Charles River). Tumor growth was monitored weekly by palpation, and mice were euthanized when tumors reached a volume of ≤2 cm3. Tumor size was measured using a digital caliper. Tumor volume was calculated according to the method reported before (Feldman et al., 2009). Animal experiments were approved by the Dutch Central Commission for Animal experimentation (Centrale Commissie voor Dierproeven).
When indicated, Dox (2 mg/mL, Sigma-Aldrich) was provided in the drinking water of the mice together with sucrose (10 mg/mL, Sigma-Aldrich) 1 week prior to injections and was renewed every 2 days for the duration of the experiment. Drinking water without Dox contained sucrose only. Blood samples were obtained from the animals before injections and every 7–14 days via tail bleeding. In some instances due to animal welfare considerations the bleeding interval was minimally 7 days but did not go beyond 14 days; for each bleeding, the maximum amount of blood did not exceed 7.5% of the circulating blood volume. Blood was collected in a 1.5-mL Eppendorf tube (containing 7.5 μL of heparin 1,000 μ/mL), centrifuged at 14,000 rpm for 5 min at 4°C after which plasma was stored at −80°C until analysis.
Morphological and Immunohistochemical Examination of Xenografts
Macroscopically representative parts of the tumors were snap frozen (liquid nitrogen) as well as fixed in formalin and embedded in paraffin (FFPE). H&E and various immunohistochemical stainings (antibodies specified in Table S3) were performed (frozen and FFPE) for evaluation as described previously (Bouma et al., 2017, van Casteren et al., 2009). Evaluation was performed independently by an experienced human pathologist in hGCTs (J.W.O.) and a scientist (L.H.J.L.), as well as one European board-certified veterinary pathologist (D.C.F.S.). Histological classification was based on the most recent WHO classification (Ulbright et al., 2016).
mRNA and miRNA Analysis
RNA from xenografts (_T) was isolated, labeled, and hybridized onto the Illumina Human HT-12 v4 array (Illumina, San Diego, CA, USA) and processed as described previously (Bouma et al., 2017). In addition, total mRNA from cell lines (_C), and the hGCT was profiled using Illumina HumanHT-12 v4, as described previously (Van Der Zwan et al., 2013). Sample description is provided in Table S2. Array data are available from ArrayExpress (accession E-MTAB-6312). For miRNA profiling, total RNA was extracted from cells and xenografts using the TRIzol reagent (Invitrogen) and miRVana miRNA Isolation Kit (Thermo Fischer Scientific), respectively. All reagents for miRNA isolation and quantification were obtained from Thermo Fisher Scientific (www.thermofisher.com/). Total RNA was also prepared from a mix of organs (brain, heart, kidney, testis, lungs, striated muscles, lymph nodes, small, and large intestine) of an age-, sex-, and strain-matching mouse following the same procedure adopted for the xenografts. Plasma (p) of xenografted or control mice (10–50 μL) was processed using the TaqMan miRNA ABC purification Bead Kit human panel A or B (PN: 4473087/4473088, each capturing 384 miRNAs) using the supplied protocol (PN: 4473439) and as described previously for human serum (Gillis et al., 2013). miRNAs were converted into cDNA using the specific megaplex primers (PN: 4399966/4399967) and the reverse transcription kit (PN; 3466596), pre-amplified (plasma) and quantitated on TaqMan low-density arrays (two 384-well Microfluids TLDA cards, PN: 4398965/4398966) on a TaqMan 7900 thermocycler using the supplier protocols (PN: 4399721). TaqMan miRNA array output data (sds files) were uploaded in the Thermo Fisher Cloud App (https://www.thermofisher.com/mysso/loginDisplay) and analyzed using defined threshold settings for each individual miRNA. Cq values were exported (Table S4), and globally normalized using QbasePlus (Biogazelle, Zwijnaarde, Belgium). Individual assays for specific miRNAs (hsa-miR-371a-3p, hsa-miR-372-3p, hsa-miR-373-3p, hsa-miR-367-3p, hsa-miR-20a-5p, and hsa-miR-30b-5p; Thermo Fisher Scientific) were run on a TaqMan 7500 fast thermocycler as described previously (van Agthoven et al., 2017). A non-human miRNA spike-in ath-miR-159a was added to the plasma samples for quality control of miRNA isolation and calibration (van Agthoven and Looijenga, 2017).
Analysis of mRNA and miRNA Data
Heatmaps of mRNA and miRNA data were generated in R using the “pheatmap” clustering software package using default settings. For mRNA clustering, “correlation” was used as the distance measure for columns. PCA was performed using prcomp from the R Stats package.
Author Contributions
D.C.F.S., L.C.J.D., and L.H.J.L. designed and analyzed the experiments and wrote the manuscript in collaboration with the other authors. A.J.M.G., G.P., T.v.A., M.G.F., H.S., and J.B.P. carried out the experiments. J.W.O., D.C.F.S., and L.H.J.L. reviewed the pathology. C.M. and L.H.J.L. supervised and provided the facilities. All authors read and approved the final manuscript.
Acknowledgments
This work is part of the research program “Meer Kennis met Minder Dieren” (More Knowledge with Less Animals) with project number 2017104947IZONMW, financed by the Netherlands Organisation for Scientific Research (NWO). Ton van Agthoven is supported by the Dutch Cancer Society (grant number 13-6001). Special thanks to the LUMC hiPSC core facility staff for support. We also thank Saskia Maas and Sophie Geerhardt for daily care of the mice and the histopathology work. We thank Prof Bart Janssen (GenomeScan, the Netherlands) for in kind support of the microarray assays.
Published: November 29, 2018
Footnotes
Supplemental Information includes two figures and four tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.11.002.
Contributor Information
Daniela C.F. Salvatori, Email: d.c.f.salvatori@lumc.nl.
Leendert H.J. Looijenga, Email: l.looijenga@erasmusmc.nl.
Accession Numbers
Deposited mRNA profiling data can be found in the ArrayExpress database (www.ebi.ac.uk/arrayexpress), under accession number ArrayExpress: E-MTAB-6312.
Supplemental Information
References
- Allison T.F., Andrews P.W., Avior Y., Barbaric I., Benvenisty N., Bock C., Brehm J., Brüstle O., Damjanov I., Elefanty A. Assessment of established techniques to determine developmental and malignant potential of human pluripotent stem cells. Nat. Commun. 2018;9:1925. doi: 10.1038/s41467-018-04011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews P., Damjanov I., Simon D., Banting G., Carlin C., Dracopoli N., Føgh J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- Andrews P.W., Bronson D.L., Benham F., Strickland S., Knowles B.B. A comparative study of eight cell lines derived from human testicular teratocarcinoma. Int. J. Cancer. 1980;26:269–280. doi: 10.1002/ijc.2910260304. [DOI] [PubMed] [Google Scholar]
- Avior Y., Biancotti J.C., Benvenisty N. TeratoScore: assessing the differentiation potential of human pluripotent stem cells by quantitative expression analysis of teratomas. Stem Cell Reports. 2015;4:967–974. doi: 10.1016/j.stemcr.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-del Jesus A., Lucena-Aguilar G., Menendez P. The miR-302-367 cluster as a potential stemness regulator in ESCs. Cell Cycle. 2009;8:394–398. doi: 10.4161/cc.8.3.7554. [DOI] [PubMed] [Google Scholar]
- Belge G., Dieckmann K.P., Spiekermann M., Balks T., Bullerdiek J. Serum levels of microRNAs miR-371-3: a novel class of serum biomarkers for testicular germ cell tumors? Eur. Urol. 2012;61:1068–1069. doi: 10.1016/j.eururo.2012.02.037. [DOI] [PubMed] [Google Scholar]
- Bouma M.J., van Iterson M., Janssen B., Mummery C.L., Salvatori D.C.F., Freund C. Differentiation-defective human induced pluripotent stem cells reveal strengths and limitations of the teratoma assay and in vitro pluripotency assays. Stem Cell Reports. 2017;8:1340–1353. doi: 10.1016/j.stemcr.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buta C., David R., Dressel R., Emgard M., Fuchs C., Gross U., Healy L., Hescheler J., Kolar R., Martin U. Reconsidering pluripotency tests: do we still need teratoma assays? Stem Cell Res. 2013;11:552–562. doi: 10.1016/j.scr.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Sung M.T., Cossu-Rocca P., Jones T., MacLennan G., Jong J.D., Lopez-Beltran A., Montironi R., Looijenga L. OCT4: biological functions and clinical applications as a marker of germ cell neoplasia. J. Pathol. 2007;211:1–9. doi: 10.1002/path.2105. [DOI] [PubMed] [Google Scholar]
- Cunningham J.J., Ulbright T.M., Pera M.F., Looijenga L.H.J. Lessons from human teratomas to guide development of safe stem cell therapies. Nat. Biotechnol. 2012;30:849. doi: 10.1038/nbt.2329. [DOI] [PubMed] [Google Scholar]
- Damjanov I., Andrews P.W. The terminology of teratocarcinomas and teratomas. Nat. Biotechnol. 2007;25:1212. doi: 10.1038/nbt1107-1212a. discussion 1212. [DOI] [PubMed] [Google Scholar]
- Damjanov I., Andrews P.W. Teratomas produced from human pluripotent stem cells xenografted into immunodeficient mice - a histopathology atlas. Int. J. Dev. Biol. 2016;60:337–419. doi: 10.1387/ijdb.160274id. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong J., Stoop H., Gillis A.J., Hersmus R., van Gurp R.J., van de Geijn G.J., van Drunen E., Beverloo H.B., Schneider D.T., Sherlock J.K. Further characterization of the first seminoma cell line TCam-2. Genes Chromosomes Cancer. 2008;47:185–196. doi: 10.1002/gcc.20520. [DOI] [PubMed] [Google Scholar]
- Dieckmann K.P., Radtke A., Spiekermann M., Balks T., Matthies C., Becker P., Ruf C., Oing C., Oechsle K., Bokemeyer C. Serum levels of microRNA miR-371a-3p: a sensitive and specific new biomarker for germ cell tumours. Eur. Urol. 2017;71:213–220. doi: 10.1016/j.eururo.2016.07.029. [DOI] [PubMed] [Google Scholar]
- Dieckmann K.P., Spiekermann M., Balks T., Flor I., Loning T., Bullerdiek J., Belge G. MicroRNAs miR-371-3 in serum as diagnostic tools in the management of testicular germ cell tumours. Br. J. Cancer. 2012;107:1754–1760. doi: 10.1038/bjc.2012.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eini R., Stoop H., Gillis A.J., Biermann K., Dorssers L.C., Looijenga L.H. Role of SOX2 in the etiology of embryonal carcinoma, based on analysis of the NCCIT and NT2 cell lines. PLoS One. 2014;9:e83585. doi: 10.1371/journal.pone.0083585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman J.P., Goldwawwer R., Mark S., Schwartz J., Orion I. A mathematical model for tumor volume evaluation using two-dimensions. J. Appl. Quantitative Methods. 2009;4:455–462. [Google Scholar]
- Flor I., Spiekermann M., Loning T., Dieckmann K.P., Belge G., Bullerdiek J. Expression of microRNAs of C19MC in different histological types of testicular germ cell tumour. Cancer Genomics Proteomics. 2016;13:281–289. [PubMed] [Google Scholar]
- Gillis A.J., Rijlaarsdam M.A., Eini R., Dorssers L.C., Biermann K., Murray M.J., Nicholson J.C., Coleman N., Dieckmann K.P., Belge G. Targeted serum miRNA (TSmiR) test for diagnosis and follow-up of (testicular) germ cell cancer patients: a proof of principle. Mol. Oncol. 2013;7:1083–1092. doi: 10.1016/j.molonc.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis A.J., Stoop H.J., Hersmus R., Oosterhuis J.W., Sun Y., Chen C., Guenther S., Sherlock J., Veltman I., Baeten J. High-throughput microRNAome analysis in human germ cell tumours. J. Pathol. 2007;213:319–328. doi: 10.1002/path.2230. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Crussi F., Winkler R.F., Mirkin D.L. Sacrococcygeal teratomas in infants and children: relationship of histology and prognosis in 40 cases. Arch. Pathol. Lab. Med. 1978;102:420–425. [PubMed] [Google Scholar]
- Hart A.H., Hartley L., Parker K., Ibrahim M., Looijenga L.H.J., Pauchnik M., Chung W.C., Robb L. The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer. 2005;104:2092–2098. doi: 10.1002/cncr.21435. [DOI] [PubMed] [Google Scholar]
- Heiftz S.A.e.a. Immature teratomas in children: pathologic considerations: a report from the Combined Pediatric Oncology Group/Children's Cancer Group. Am. J. Surg. Pathol. 1998;22:1115–1124. doi: 10.1097/00000478-199809000-00011. [DOI] [PubMed] [Google Scholar]
- Herszfeld D., Wolvetang E., Langton-Bunker E., Chung T.L., Filipczyk A.A., Houssami S., Jamshidi P., Koh K., Laslett A.L., Michalska A. CD30 is a survival factor and a biomarker for transformed human pluripotent stem cells. Nat. Biotechnol. 2006;24:351–357. doi: 10.1038/nbt1197. [DOI] [PubMed] [Google Scholar]
- Josephson R., Ording C.J., Liu Y., Shin S., Lakshmipathy U., Toumadje A., Love B., Chesnut J.D., Andrews P.W., Rao M.S. Qualification of embryonal carcinoma 2102Ep as a reference for human embryonic stem cell research. Stem Cells. 2007;25:437–446. doi: 10.1634/stemcells.2006-0236. [DOI] [PubMed] [Google Scholar]
- Leão R., van Agthoven T., Figueiredo A., Jewett M.A.S., Fadaak K., Sweet J., Ahmad A.E., Anson-Cartwright L., Chung P., Hansen A. Serum miRNA predicts viable disease after chemotherapy in patients with testicular nonseminoma germ cell tumor. J. Urol. 2018;200:126–135. doi: 10.1016/j.juro.2018.02.068. [DOI] [PubMed] [Google Scholar]
- Lipchina I., Studer L., Betel D. The expanding role of miR-302–367 in pluripotency and reprogramming. Cell Cycle. 2012;11:1517–1523. doi: 10.4161/cc.19846. [DOI] [PubMed] [Google Scholar]
- Looijenga L. Human testicular (non)seminomatous germ cell tumours: the clinical implications of recent pathobiological insights. J. Pathol. 2009;218:146–162. doi: 10.1002/path.2522. [DOI] [PubMed] [Google Scholar]
- Looijenga L.H., Stoop H., de Leeuw H.P., de Gouveia Brazao C.A., Gillis A.J., van Roozendaal K.E., van Zoelen E.J., Weber R.F., Wolffenbuttel K.P., van Dekken H. POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res. 2003;63:2244–2250. [PubMed] [Google Scholar]
- Looijenga L.H.J., Gillis A.J.M., Stoop H., Hersmus R., Oosterhuis J.W. Relevance of microRNAs in normal and malignant development, including human testicular germ cell tumours. Int. J. Androl. 2007;30:304–315. doi: 10.1111/j.1365-2605.2007.00765.x. [DOI] [PubMed] [Google Scholar]
- Mizuno Y., Gotoh A., Kamidono S. Establishment and characterization of a new human testicular germ cell tumor cell line (TCam-2) Nihon Hinyokika Gakkai Zasshi. 1993;84:1211–1218. doi: 10.5980/jpnjurol1989.84.1211. [DOI] [PubMed] [Google Scholar]
- Muller F.J., Goldmann J., Loser P., Loring J.F. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6:412–414. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Muller F.J., Schuldt B.M., Williams R., Mason D., Altun G., Papapetrou E.P., Danner S., Goldmann J.E., Herbst A., Schmidt N.O. A bioinformatic assay for pluripotency in human cells. Nat. Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.J., Halsall D.J., Hook C.E., Williams D.M., Nicholson J.C., Coleman N. Identification of microRNAs from the miR-371∼373 and miR-302 clusters as potential serum biomarkers of malignant germ cell tumors. Am. J. Clin. Pathol. 2011;135:119–125. doi: 10.1309/AJCPOE11KEYZCJHT. [DOI] [PubMed] [Google Scholar]
- Nettersheim D., Heimsoeth A., Jostes S., Schneider S., Fellermeyer M., Hofmann A., Schorle H. SOX2 is essential for in vivo reprogramming of seminoma-like TCam-2 cells to an embryonal carcinoma-like fate. Oncotarget. 2016;7:47095–47110. doi: 10.18632/oncotarget.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor D.M., Norris H.J. The influence of grade on the outcome of stage I ovarian immature (malignant) teratomas and the reproducibility of grading. Int. J. Gynecol. Pathol. 1994;13:283–289. doi: 10.1097/00004347-199410000-00001. [DOI] [PubMed] [Google Scholar]
- Oosterhuis J.W., Looijenga L.H.J. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer. 2005;5:210. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- Parr C.J.C., Katayama S., Miki K., Kuang Y., Yoshida Y., Morizane A., Takahashi J., Yamanaka S., Saito H. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci. Rep. 2016;6:32532. doi: 10.1038/srep32532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera M.F., Cooper S., Mills J., Parrington J.M. Isolation and characterization of a multipotent clone of human embryonal carcinoma cells. Differentiation. 1989;42:10–23. doi: 10.1111/j.1432-0436.1989.tb00602.x. [DOI] [PubMed] [Google Scholar]
- Rijlaarsdam M.A., van Herk H.A.D.M., Gillis A.J.M., Stoop H., Jenster G., Martens J., van Leenders G.J.L.H., Dinjens W., Hoogland A.M., Timmermans M. Specific detection of OCT3/4 isoform A/B/B1 expression in solid (germ cell) tumours and cell lines: confirmation of OCT3/4 specificity for germ cell tumours. Br. J. Cancer. 2011;105:854. doi: 10.1038/bjc.2011.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokavec M., Li H., Jiang L., Hermeking H. The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 2014;6:214–230. doi: 10.1093/jmcb/mju003. [DOI] [PubMed] [Google Scholar]
- Shen H., Shih J., Hollern D.P., Wang L., Bowlby R., Tickoo S.K., Thorsson V., Mungall A.J., Newton Y., Hegde A.M. Integrated molecular characterization of testicular germ cell tumors. Cell Rep. 2018;23:3392–3406. doi: 10.1016/j.celrep.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syring I., Bartels J., Holdenrieder S., Kristiansen G., Muller S.C., Ellinger J. Circulating serum miRNA (miR-367-3p, miR-371a-3p, miR-372-3p and miR-373-3p) as biomarkers in patients with testicular germ cell cancer. J. Urol. 2015;193:331–337. doi: 10.1016/j.juro.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Terenziani M., D'Angelo P., Inserra A., Boldrini R., Bisogno G., Babbo G.L., Conte M., Dall' Igna P., De Pasquale M.D., Indolfi P. Mature and immature teratoma: a report from the second Italian pediatric study. Pediatr. Blood Cancer. 2015;62:1202–1208. doi: 10.1002/pbc.25423. [DOI] [PubMed] [Google Scholar]
- Teshima S., Shimosato Y., Hirohashi S., Tome Y., Hayashi I., Kanazawa H., Kakizoe T. Four new human germ cell lines. Lab. Invest. 1988;59:328–336. [PubMed] [Google Scholar]
- Tsankov A.M., Akopian V., Pop R., Chetty S., Gifford C.A., Daheron L., Tsankova N.M., Meissner A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat. Biotechnol. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbright M.B., Balzer A.B., Berney D.M., Epstein J.I., Guo C. Germ cell tumours. In: Moch H., Humphrey P.A., Ulbright M.T., Reuter V.E., editors. WHO Classification of Tumours of the Urinary System and Male Genital Organs. IARC; 2016. pp. 189–226. [Google Scholar]
- Ulbright T.M., Tickoo S.K., Berney D.M., Srigley J.R. Best practices recommendations in the application of immunohistochemistry in testicular tumors: report from the International Society of Urological Pathology consensus conference. Am. J. Surg. Pathol. 2014;38:e50–e59. doi: 10.1097/PAS.0000000000000233. [DOI] [PubMed] [Google Scholar]
- van Agthoven T., Eijkenboom W.M.H., Looijenga L.H.J. microRNA-371a-3p as informative biomarker for the follow-up of testicular germ cell cancer patients. Cell Oncol. (Dordrecht) 2017;40:379–388. doi: 10.1007/s13402-017-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Agthoven T., Looijenga L.H.J. Accurate primary germ cell cancer diagnosis using serum based microRNA detection (ampTSmiR test) Oncotarget. 2017;8:58037–58049. doi: 10.18632/oncotarget.10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Casteren N.J., de Jong J., Stoop H., Steyerberg E.W., de Bekker-Grob E.W., Dohle G.R., Oosterhuis J.W., Looijenga L.H. Evaluation of testicular biopsies for carcinoma in situ: immunohistochemistry is mandatory. Int. J. Androl. 2009;32:666–674. doi: 10.1111/j.1365-2605.2008.00923.x. [DOI] [PubMed] [Google Scholar]
- Van Der Zwan Y.G., Stoop H., Rossello F., White S.J., Looijenga L.H. Role of epigenetics in the etiology of germ cell cancer. Int. J. Dev. Biol. 2013;57:299–308. doi: 10.1387/ijdb.130017ll. [DOI] [PubMed] [Google Scholar]
- Voorhoeve P.M., le Sage C., Schrier M., Gillis A.J., Stoop H., Nagel R., Liu Y.P., van Duijse J., Drost J., Griekspoor A. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- Wang N., Trend B., Bronson D.L., Fraley E.E. Nonrandom abnormalities in chromosome 1 in human testicular cancers. Cancer Res. 1980;40:796–802. [PubMed] [Google Scholar]
- Zhang Z., Xiang D., Heriyanto F., Gao Y., Qian Z., Wu W.-S. Dissecting the roles of miR-302/367 cluster in cellular reprogramming using TALE-based repressor and TALEN. Stem Cell Reports. 2013;1:218–225. doi: 10.1016/j.stemcr.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.