Abstract
Cardiovascular disease is still a major cause of ill-health and mortality, heart failure and arrhythmia being among the causes of sudden cardiac death. There are few drugs available for treatment or prevention and it remains difficult to predict who will develop these conditions, even when disease-causing mutations or associated gene variants are identified in individuals or families. This is in part because widely used rodent models may not fully capture the physiology of the human heart. The advent of pluripotent stem cell technology that allows cardiovascular cells to be derived from patients and healthy individuals, in some cases genetically matched through mutation repair, is leading to paradigm shifts in how cardiovascular diseases are studied in humans. However, these cells are often only partially mature imposing some limitations in use. This Perspective reviews aspects of recent advances but also remaining challenges.
Keywords: hiPSC, cardiomyocytes, disease models, cardiomyocyte maturation, cardiovascular cells, microtissues
Christine Mummery provides a Perspective on applications of human iPSC-cardiomyocytes for disease modeling and drug testing, including recently developed advanced 3D-engineered cultures and multicellular formats enhancing the maturity of cardiomyocytes to rapidly expand their utility in drug discovery and cardiotoxicity screens beyond effects on ion channels.
Main Text
Introduction
The ability to generate induced pluripotent stem cells (iPSCs) by reprogramming somatic tissues is arguably the greatest breakthrough in biomedical science of the last decade. It means that the most inaccessible cells of the body, such as those of the heart, can now be derived repeatedly from any individual without the ethical limitations of embryonic stem cells (ESCs). While first excitement concerned the potential for allogenic transplantation in regenerative medicine, focus has now shifted to more medically relevant areas: (cardiac) disease modeling, target discovery, and drug development. These applications, however, are requiring new levels of sophistication in bioassays to monitor disease phenotypes so that the predictive value of hiPSCs can be realized for acute and chronic genetic and somatic diseases. In addition, generating cell subtypes in the heart benefits from detailed knowledge of the developmental pathways that lead to the formation of these cells in the embryo (reviewed in Birket and Mummery, 2015). As these areas have merged over the last several years, they have created unique opportunities to create new human models of organ physiology and pathology that are leading to greater understanding of mechanisms underlying abnormal human development and the cause of disease. This is now beginning to provide unprecedented opportunities to identify new markers for diagnosis and molecular targets for prevention and therapeutic intervention in multiple disease states.
Human Heart and Its Vessels
Cardiovascular disease is a major cause of morbidity and mortality in the developed world, second only to cancer in the number of individuals affected. In contrast to cancer, however, there are few new drugs in the pipeline, in part because cardiac and vascular diseases have been difficult to model in laboratory assays or experimental animals. Rodent hearts, for example, beat between 250 and 500 times per min, a human heart at rest only 60 times per min. Physiology between species is therefore different, in part due to differences in size, in part due to different levels of ion channel proteins that mediate contractile and electrical signals (Figure 1). Most notably, one important K+-channel (the HERG channel) in human cardiomyocytes that regulates heart rate and is the target of drugs and gene mutations causing arrhythmias, is negligible in mice so that abnormalities mediated by aberrant behavior of this channel are largely undetectable. As a result of these and other differences, heart failure, atherosclerosis, and cardiovascular complications of diabetes, have few treatment options that can be offered in the clinic despite huge investments by the pharmaceutical industry (Paul et al., 2010).
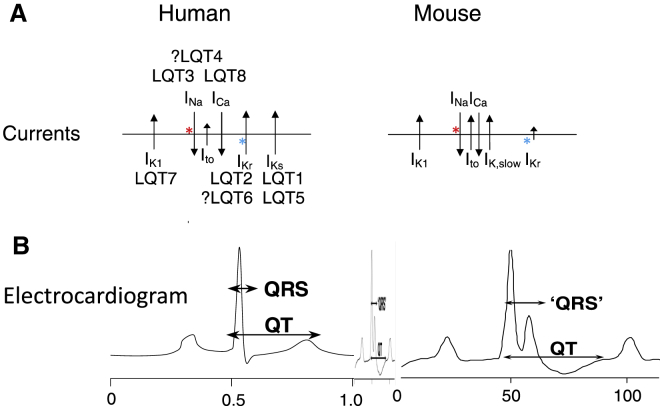
Figure 1.
Comparison of Ion Channel Use on Human and Mouse Cardiomyocytes
Mouse hearts are smaller, beat faster, and have different ion channel expression profiles that human cardiomyocytes.
(A) Ion channels present on mouse and human cardiomyocytes; the length of the arrows corresponds to the amount of current carried by the channel (INa values are similar in mice and humans so that mutations or drugs affecting these channels will show similar responses in both species; IKr [HERG channel] differs between species so that mouse hearts show lower responses than humans). LQT (long QT) refers to the syndromes associated with mutations in the ion channels indicated.
(B) Electrocardiograms of humans and mice show corresponding differences in shape. The middle electrocardiogram shows that of mice on the same scale as human.
Since human iPSCs (hiPSCs) can divide indefinitely and in principle form any cell of the body, heart, and vascular cells can be generated from individuals of any ethnic background who may be healthy or have any of a multiplicity of diseases of genetic or unknown origin. In addition, recent technical advances in gene targeting using CRISPR/Cas9 technology in human PSCs (hPSCs) mean that any mutation or gene variant of interest can be introduced either to correct gene mutations or introduce them on a healthy control background (Bellin et al., 2013). In addition, rapid targeting of reporter genes into loci of interest facilitates the lineage tracing in culture and cell subtype selection (Schwach et al., 2017). Thus, not only is there potential to use these new windows of opportunity to slow disease progression and identify factors that predetermine its degree of severity but it may also be possible to target specific disease mechanisms and develop cures. If the symptoms in the hiPSCs and their derivatives could be alleviated, the same approach may be feasible directly in the patient. If adverse drug responses were identified in specific ethnic groups, it may be possible to predict these prior to clinical disaster. Side effects of drugs are now among the leading causes of death worldwide and many of these effects are due to cardiotoxicity.
hiPSCs can now differentiate efficiently into cardiomyocytes and vascular cells (reviewed in Mummery et al., 2012) and there are numerous examples of how this approach has led to important insights in cardiac disease mechanisms (Bellin et al., 2013, Wang et al., 2014). This is despite the widely reported immaturity of differentiated derivatives of hiPSCs, including cardiomyocytes (reviewed in Veerman et al., 2015). It is now feasible to obtain certain types of cardiac and vascular cells that capture entire genetic profiles, not only of mutated genes (when the gene is known) but also all the genetic modifiers that play an important but yet largely unknown role in the pathology of cardiovascular disease and drug responses.
The concept of differentiated hiPSCs and (mutated) hESCs as “patients” has started receiving robust support in experimental data: reprogrammed cells from patients with rare genetic disorders are already experimental paradigms that are providing new clues on how diseases cause damage and how this might be alleviated or reversed.
Lessons from Development: Directing Cardiovascular Differentiation of hPSCs
In vitro differentiation of stem cells to cardiomyocytes and vascular cells mimics the sequential stages of embryonic cardiac development (for review see Birket and Mummery, 2015). The heart and associated vessels are the first identifiable tissues to develop in vertebrate embryos. The heart forms soon after gastrulation from anterior migrating mesodermal cells that intercalate between the ectoderm and endoderm cell layers in the primitive streak. The first vessels appear on the yolk sac enveloping the embryo. Multipotent cardiac progenitor cells are primarily localized in the mid-streak where signals from the adjacent endoderm, in particular, appear to have a highly conserved instructive function in cardiogenesis. Two families of protein growth factors are thought to control these early stages of mesoderm formation and cardiogenesis: nodal and bone morphogenetic proteins (BMPs), which are members of the transforming growth factor β superfamily, and the Wnt proteins. These factors, or their inhibitors, are expressed in the endoderm; genetic disruption of their signaling has dramatic effects on cardiac development. In vertebrates, BMP signaling generally promotes cardiogenesis, while Wnt proteins are involved in cardiac specification. Differentiation of mesoderm to vascular cells requires different signaling pathways, the most important being that induced by vascular endothelial growth factor (Carmeliet, 2003). Overall, it appears that the timing and relative expression of different growth factor combinations induce, then pattern, the cardiogenic mesoderm. Once mesoderm cells have received appropriate signals, they switch on a highly conserved heart-specific combination of transcription factors that establish the cardiac transcriptional program. Initially, the mesodermal precursor cells in the primitive streak express transcription factors such as the T-box factor Brachyury (T) and the homeodomain protein, Mixl1. Prior to migration from the streak, these cells transiently activate the basic helix-loop-helix transcription factor mesoderm posterior 1 (Mesp1) to enter a “precardiac” mesoderm stage of development. A subset of the Mesp1+ cell population then begins to express the homeodomain transcription factor Nkx2-5, the T-box protein Tbx5, and Isl1, a LIM homeodomain transcription factor, which are early markers of the cardiac lineage that are activated shortly after the formation of the heart fields. In mice, Nkx2-5 is essential for the interpretation of patterning signals within the primitive heart tube and is likely to act in concert with Tbx5 during formation of both the atrial and left ventricular compartments to positively regulate transcription. Nkx2-5 and Tbx5 associate with members of the GATA family of zinc-finger transcription factors and with serum response factor to activate cardiac structural genes, such as actin, myosin light chain, myosin heavy chain, troponins, and desmin. Tbx5 can also cooperate with Nkx2-5 to activate expression of ANF and the junctional protein connexin 40. Members of the myocyte enhancer factor 2 family of transcription factors also play key roles in cardiomyocte differentiation by regulating cardiac muscle structural genes. Thus, multiple complex interactions between this highly conserved gene-regulatory networks control the initial differentiation, proliferation, and maturation of cardiomyocytes. Apart from their functional role, many of these factors can be used as markers of emerging cardiomyocytes in differentiating cultures of hPSCs. In rather a similar way, a set of markers that may also be functional proteins have been described for endothelial cells and vascular smooth muscle cells. These include the vascular endothelial growth factor receptors, α-smooth muscle actin and platelet endothelial cell adhesion molecule.
hiPSC Lines Modeling Cardiac Disease
hiPSC lines have been created from many individuals with cardiac disease, predominantly with mutations in ion channel genes (reviewed in Brandão et al., 2017) or in sarcomeric proteins (reviewed in Giacomelli et al., 2017b). For the ion channel mutations that lead to arrhythmias and sometimes sudden cardiac death (SCD), hiPSC-derived cardiomyocytes have not only been shown to recapitulate the disease phenotypes but, as mentioned earlier, have also provided new insights into underlying disease mechanisms or have indicated treatment modalities. These ion channelopathies are of particular interest because ion channel use is different in the smaller hearts of rodents so that relevant animal models are unavailable. For example, mouse models with mutations in Kcnq1 are available but there are significant differences in the handling of rectifying potassium currents between mouse and human (Davis et al., 2011). Since cardiac ion channels are expressed within 2 weeks of initiating cardiomyogenic differentiation in hPSCs, cardiomyocyte immaturity has had a relatively little impact on predictive outcomes for adult humans. For this reason, hPSC models have to some extent surpassed mice as useful for determining drug sensitivities, since many of these channelopathies are autonomous to the cardiomyocytes (Blinova et al., 2018, Braam et al., 2013).
Changes in electrical properties of hPSC-cardiomyocytes as a result of disease or in response to drugs can be determined relatively reliably using electrophysiology (patch clamp with microelectrodes or microelectrode arrays) or optical methods using voltage- and calcium-sensitive dyes (reviewed in van Meer et al., 2016). By contrast, phenotypes resulting from cardiac hypertrophy or myopathy have been more difficult to capture since until recently there have been few satisfactory ways to measure the alterations in cardiomyocyte size or contractility evident in patients.
The first report of hiPSC-cardiomyocytes from a patient with hypertrophic cardiomyopathy concerned LEOPARD syndrome. Patients with this condition suffer from various symptoms, with hypertrophic cardiomyopathy the cause of mortality in some sufferers. This autosomal-dominant hereditary disorder is commonly caused by mutations in the tyrosine phosphatase gene, PTPN11, which is ubiquitously expressed throughout the body and is essential for normal development. hiPSCs, generated from two patients with one of the most recurrent mutations, T468M, were differentiated to cardiomyocytes, and assessed for hypertrophy (Carvajal-Vergara et al., 2010). The hiPSC-cardiomyocytes displayed a higher degree of sarcomeric organization and nuclear NFATC4 localization and were larger than wild-type hiPSC-cardiomyocytes, which are highly disorganized and immature. However, not all standard criteria to analyze cardiomyocyte hypertrophy could be performed due to the mixed population of cells obtained from the differentiation procedure and the interpretation of increased cardiomyocyte spreading as increased size is perhaps in retrospect somewhat over interpreted. Nevertheless, this type of study set the stage for many more robust myopathy models and there is now much greater insight into these conditions as a result of improved differentiation strategies to enrich for cardiomyocytes and better functional assays that reflect disease phenotypes in patients.
Cell-Based Screening for Drug Discovery
Although hiPSC technology could be instrumental in understanding the biology of healthy and diseased cardiomyocytes, this might not be enough for efficient drug target identification. Over the last two decades, several gene mutations causing cardiac disease have been identified through candidate gene approaches in families exhibiting hereditary cardiac abnormalities, but it has proven extremely difficult to translate this to the identification of drug targets and therapies.
Cell-based screening with a phenotypic endpoint is now proving to be a successful solution to this in some disease areas. Typically, primary human cells are used to create in vitro disease models in culture with a finite “window for screening.” These models can be used in combination with either RNAi technology to find drug targets, or chemical libraries to find novel lead molecules. As previously mentioned, primary human cardiomyocytes are difficult to obtain and cannot be maintained in culture for longer than a few days, making them unsuitable for high throughput screening. Using hPSCs and differentiation protocols that generate high yields of cardiomyocytes might solve this problem. hPSC disease models that accurately reflect the disease pathology could be developed into miniaturized high-throughput and high-content phenotypic screening assays. A range of assays to identify a response are available and can be based on fluorescent imaging, immunochemistry, and/or multiplexing (van Meer et al., 2016). This allows for a complex assay design, based on simultaneous measurements of parameters such as cell proliferation, differentiation, morphology, and viability and will be the approach taken in the present proposal.
Personalized Medicine and Stem Cell Banks
The late-stage attrition of drugs due to limited effectiveness or to unforeseen side effects is perhaps the biggest problem/risk in drug development. To some extent, this is because not all individuals receiving a drug for a specific symptom will have a beneficial response or suffer a toxic side effect. A panel of PSC lines with different genotypes and from different racial backgrounds will allow in vitro analyses, predicting both drug effectiveness and toxicity at an individual level against a specific genetic background, as well as identifying biomarkers for the development of companion diagnostic assays to predict individual therapy response and toxicity in the clinic.
The potential clinical implications of personalized hiPSC-cardiomyocytes and how they might be implemented to guide clinical decisions was recently illustrated by a study in which genome editing was combined with hiPSCs from an asymptomatic patient carrying a hypertrophic cardiomyopathy genetic variant in the sarcomeric protein MYL3 (Ma et al., 2018). This variant has been classified as “likely pathogenic,” creating a clinical dilemma on whether to place an intra cardiac device (ICD) into a patient without symptoms in case an arrhythmic event should take place. ICDs may activate spontaneously in the absence of arrhythmia, causing significant patient stress. Clinical genetic testing is emerging as the standard of care in patients but interpretation of clinical risk is often confounded by “variants of unknown significance” (VUS). VUS patient hiPSC-cardiomyocytes derived from the heterozygous carrier underwent targeting of the second (healthy) allele either with the VUS or a frameshift mutation in the same gene to create homozygous isogenic pairs. The heterozygous VUS-hiPSC-cardiomyocytes did not show the hypertrophic cardiomyopathy phenotype, but, more importantly, neither did the gene-edited homozygous lines, supporting a benign assessment of this particular MYL3 variant. This hiPSC-based assay is now being used to indicate which patients would be at risk of SCD and would benefit from placement of an ICD. This is in line with earlier studies showing that any drugs or gene variants that compromise repolarization reserve present an additional risk (Braam et al., 2013).
Challenges for the Field
While many laboratories worldwide are developing human iPSC disease models, and the overall disease phenotype of the patient is apparently retained in these cells, there are at least two general questions still to be answered. Firstly, what can we learn about the disease that cannot be gleaned from studying the patients. For example, in many electrophysiological disorders, the physiological basis for the arrhythmia is already known. While there is emerging evidence that hiPSC models for genetic ion channelopathies can help us further understand the molecular mechanism behind the disease and develop new therapeutic interventions (e.g., Braam et al., 2013, Zhang et al., 2014), it is not yet clear how closely the in vitro disease phenotype will match that of the patient from whom they were derived, particularly in terms of severity. In some familial hypertrophic cardiomyopathies, one example described a boy in which the cardiac septum was 10 times thicker than normal, yet his father with the same mutation has no apparent symptoms (Birket et al., 2015). It remains to be seen whether such obvious differences in the sensitivity of the patient's cardiomyocytes to undergo hypertrophy will be similarly reflected in the cardiomyocytes derived from their respective iPSC lines.
A potential technical issue mentioned earlier is that in-vitro-differentiated derivatives of hPSCs typically have an immature phenotype. For hPSC-cardiomyocytes this means that their gene expression profile resembles that of fetal cardiomyocytes (reviewed in Veerman et al., 2015). In addition, their resting membrane potentials and upstroke velocities are small compared with adult cardiomyocytes, and their sarcomeres and shapes are relatively disorganized and not aligned as in adult cells, potentially limiting their utility in disease modeling and drug screening. Understanding and recapitulating in vitro the physiological and biochemical triggers active in normal development will be essential to achieve this goal. One study recently reported such an approach where hPSC-cardiomyocytes were cultured as engineered heart tissues (EHTs) then subjected to “exercise” at increasing pacing rates up to 6 Hz (Ronaldson-Bouchard et al., 2018). EHTs are 3D heart muscle equivalents in which hPSC-cardiomyocytes are embedded in collagen gels such that they contract against a load provided by poles at each end of the tissue (Ulmer et al., 2018). The paced tissues showed many features of mature myocardium including increased numbers of mitochondria, reflecting altered metabolism, and t-tubules, structures evident by electron microscopy that are essential for proper calcium handling and only develop postnatally in humans.
It should also be noted that the heart is not only composed of cardiomyocytes (∼30% of the total cells present in the adult heart; ∼60% in fetal heart), but also two other principal cell types, cardiac fibroblasts or smooth muscle cells and vascular endothelial cells. While electrophysiological disorders and cardiomyopathies are cell-autonomous disorders affecting solely the cardiomyocytes, for other cardiac diseases these other cell types also play important roles, influencing the cardiomyocytes both in terms of their response to injury and to cardiac and non-cardiac drugs. A stem cell-based model that recapitulates cardiac organogenesis, essentially creating an artificial human myocardium, would be invaluable.
Aside from the genetic forms of cardiac disease, there are many cardiac conditions caused by abnormal physiological insults. In the human heart itself, these can include lack of oxygen during a myocardial infarction, high blood pressure, high glucose levels due to diabetes, and changes in cardiomyocyte shape from cardiac remodeling post myocardial infarction or heart failure. In some cases, these changes can be recapitulated in the laboratory using chemicals or drugs such as phenylephrine or in the case of shape changes, by micropatterning extracellular matrix proteins such as fibronectin onto synthetic polymers. Alternatively, cryoinjury has been used to mimic aspects of cardiac repair in vitro in cardiac organoids (Mills et al., 2017a, Mills et al., 2017b). Combined with monogenetic disease hPSC lines, this will provide an extensive collection of paradigms for downstream applications.
Toward Solutions: Implementing hPSCs in Disease Pathology and Drug Discovery
Several recent studies including from our own lab (Giacomelli et al., 2017a) have described methods for creating synthetic human myocardium and associated vasculature that show the features of mature heart tissue, but which can also show characteristics of disease. Robust methods for producing the cellular components of the heart and vasculature are being developed, and assembled into either synthetic or natural structures that can be mechanically, electrically, or biochemically challenged, and for which readouts of function are quantifiable and reproducible. This may contribute in the future to heart and vascular repair, but a more immediate outcome is the creation of authentic models of the healthy and diseased human cardiovascular system and accompanying bioassays of functionality that can be applied to drug safety and discovery, as mentioned earlier.
New diseases are continuously being added to the list of those that recapitulate not only genetic but also sporadic forms of disease. This new technology thus has great potential in bringing new insights into cardiovascular development and disease in humans far beyond the anticipated uses in cell-replacement therapy. Patient-specific iPSCs have rapidly emerging applications in disease modeling, drug testing, and drug discovery since, compared with other organs, heart tissues are particularly inaccessible as patient biopsies. Models of disease “in a dish,” are now allowing experiments that, not so long ago, would have been inconceivable. Before this potential can be fully realized, however, there are a number of important hurdles to overcome. These will not only require exceptional expertise in hPSC biology, but also exquisite knowledge of the underlying developmental biology, to drive differentiation efficiently and reproducibly to the required cell types and multidisciplinary approaches to developing sensitive and quantitative assays of deviant phenotypes in “diseased” cells.
The impact will be to provide new paradigms for understanding disease mechanisms, drug safety pharmacology, and discovering new therapeutic drugs.
Acknowledgments
I am grateful to the many talented past and present members of my lab who have contributed to some the work referred to in this Perspective, and to our funders, most recently the European Research Council (ERC-AdG 323182 STEMCARDIOVASC) and the Netherlands Organ-on-Chip Initiative (NOCI), a Gravitation project (024.003.001) funded by the Netherlands Wetenschaps Organisatie (NWO). Apologies to all of those whose work I have not cited, an intrinsic limitation of a Perspective. References to relevant reviews are provided for primary data references.
References
- Birket M.J., Mummery C.L. Pluripotent stem cell derived cardiovascular progenitors–a developmental perspective. Dev. Biol. 2015;400:169–179. doi: 10.1016/j.ydbio.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Kosmidis G., Ward D., Leitoguinho A.R., van de Pol V., Dambrot C., Devalla H.D., Davis R.P., Mastroberardino P.G. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep. 2015;13:733–745. doi: 10.1016/j.celrep.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M., Casini S., Davis R.P., D'Aniello C., Haas J., Ward-van Oostwaard D., Tertoolen L.G., Jung C.B., Elliott D.A., Welling A. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32:3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blinova K., Dang Q., Millard D., Smith G., Pierson J., Guo L., Brock M., Lu H.R., Kraushaar U., Zeng H. International multisite study of human-induced pluripotent stem cell-derived cardiomyocytes for drug proarrhythmic potential assessment. Cell Rep. 2018;24:3582–3592. doi: 10.1016/j.celrep.2018.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam S.R., Tertoolen L., Casini S., Matsa E., Lu H.R., Teisman A., Passier R., Denning C., Gallacher D.J., Towart R., Mummery C.L. Repolarization reserve determines drug responses in human pluripotent stem cell derived cardiomyocytes. Stem Cell Res. 2013;10:48–56. doi: 10.1016/j.scr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Brandão K.O., Tabel V.A., Atsma D.E., Mummery C.L., Davis R.P. Human pluripotent stem cell models of cardiac disease: from mechanisms to therapies. Dis. Model. Mech. 2017;10:1039–1059. doi: 10.1242/dmm.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. Blood vessels and nerves: common signals, pathways and diseases. Nat. Rev. Genet. 2003;4:710–720. doi: 10.1038/nrg1158. [DOI] [PubMed] [Google Scholar]
- Carvajal-Vergara X., Sevilla A., D'Souza S.L., Ang Y.S., Schaniel C., Lee D.F., Yang L., Kaplan A.D., Adler E.D., Rozov R. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.P., van den Berg C.W., Casini S., Braam S.R., Mummery C.L. Pluripotent stem cell models of cardiac disease and their implication for drug discovery and development. Trends Mol. Med. 2011;17:475–484. doi: 10.1016/j.molmed.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Giacomelli E., Bellin M., Sala L., van Meer B.J., Tertoolen L.G., Orlova V.V., Mummery C.L. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development. 2017;144:1008–1017. doi: 10.1242/dev.143438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E., Mummery C.L., Bellin M. Human heart disease: lessons from human pluripotent stem cell-derived cardiomyocytes. Cell. Mol. Life Sci. 2017;74:3711–3739. doi: 10.1007/s00018-017-2546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Zhang J., Itzhaki I., Zhang S.L., Chen H., Haddad F., Kitani T., Wilson K.D., Tian L., Shrestha R. Determining the pathogenicity of a genomic variant of uncertain significance using CRISPR/Cas9 and human-induced pluripotent stem cells. Circulation. 2018;138 doi: 10.1161/CIRCULATIONAHA.117.032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R.J., Voges H.K., Porrello E.R., Hudson J.E. Cryoinjury model for tissue injury and repair in bioengineered human striated muscle. Methods Mol. Biol. 2017;1668:209–224. doi: 10.1007/978-1-4939-7283-8_15. [DOI] [PubMed] [Google Scholar]
- Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G., Kamp T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ. Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S.M., Mytelka D.S., Dunwiddie C.T., Persinger C.C., Munos B.H., Lindborg S.R., Schacht A.L. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat. Rev. Drug Discov. 2010;9:203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K., Ma S.P., Yeager K., Chen T., Song L., Sirabella D., Morikawa K., Teles D., Yazawa M., Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018;556:239–243. doi: 10.1038/s41586-018-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach V., Verkerk A.O., Mol M., Monshouwer-Kloots J.J., Devalla H.D., Orlova V.V., Anastassiadis K., Mummery C.L., Davis R.P., Passier R. A COUP-TFII human embryonic stem cell reporter line to identify and select atrial cardiomyocytes. Stem Cell Reports. 2017;9:1765–1779. doi: 10.1016/j.stemcr.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer B.M., Stoehr A., Schulze M.L., Patel S., Gucek M., Mannhardt I., Funcke S., Murphy E., Eschenhagen T., Hansen A. Contractile work contributes to maturation of energy metabolism in hiPSC-derived cardiomyocytes. Stem Cell Reports. 2018;10:834–847. doi: 10.1016/j.stemcr.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer B.J., Tertoolen L.G., Mummery C.L. Concise review: measuring physiological responses of human pluripotent stem cell derived cardiomyocytes to drugs and disease. Stem Cells. 2016;34:2008–2015. doi: 10.1002/stem.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerman C.C., Kosmidis G., Mummery C.L., Casini S., Verkerk A.O., Bellin M. Immaturity of human stem-cell-derived cardiomyocytes in culture: fatal flaw or soluble problem? Stem Cells Dev. 2015;24:1035–1052. doi: 10.1089/scd.2014.0533. [DOI] [PubMed] [Google Scholar]
- Wang G., McCain M.L., Yang L., He A., Pasqualini F.S., Agarwal A., Yuan H., Jiang D., Zhang D., Zangi L. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., D'Aniello C., Verkerk A.O., Wrobel E., Frank S., Ward-van Oostwaard D., Piccini I., Freund C., Rao J., Seebohm G. Recessive cardiac phenotypes in induced pluripotent stem cell models of Jervell and Lange-Nielsen syndrome: disease mechanisms and pharmacological rescue. Proc. Natl. Acad. Sci. U S A. 2014;111:E5383–E5392. doi: 10.1073/pnas.1419553111. [DOI] [PMC free article] [PubMed] [Google Scholar]