Abstract
Progress in our understanding of the central genes, pathways, and mechanisms in the pathobiology of T-cell acute lymphoblastic leukemia (T-ALL) has identified key drivers of the disease, opening new opportunities for therapy. Drugs targeting highly prevalent genetic alterations in NOTCH1 and CDKN2A are being explored, and multiple other targets with readily available therapeutic agents, and immunotherapies are being investigated. The molecular basis of T-ALL is reviewed here and potential targets and therapeutic targets discussed.
Keywords: T-cell acute lymphoblastic leukemia, T-ALL, mutations, NOTCH1, gamma secretase inhibitor, GSI, CDKN2A, NUP214-ABL1, PTEN, JAK/STAT, NT5C2CAR, chimeric antigen receptor
Introduction
Over the last years studies dissecting the mutational landscape of T-cell acute lymphoblastic leukemia (T-ALL) have identified prominent genes that define key pathways and mechanisms essential in the pathogenesis of the disease. Among these, genetic lesions involving CDKN2A, and NOTCH1 are particularly prevalent [1]. Deletions of the CDKN2A locus encoding the P16/INK4A and P19/ARF tumor suppressors, responsible for control of cell cycle progression and P53 regulation, respectively, are present in about 70% of T-ALLs. In addition, activating mutations in NOTCH1 can be found in the majority of T-ALL cases [1,2]. However, TALL is a genetically and clinico-biologically heterogeneous disease implicating alterations in numerous transcriptional, signaling, and epigenetic factors.
Molecular basis of T-ALL
Gene expression profiling has identified major clinico-biological categories of T-ALL associated with gene expression programs related to those present during differentiation along the thymocyte development spectrum [1,3]. Early T-cell precursor T-ALL (ETP T-ALL) is characterized by activating mutations in genes regulating cytokine receptor RAS signaling, including FLT3, inactivating lesions in GATA3, ETV6, and RUNX1 disrupting hematopoietic development, and histone-modifying genes, such as EZH2, SUZ12 and EED [4]. ETP T-ALLs have a gene expression profile characteristic of very immature precursors, they are related to hematopoietic stem cells and myeloid progenitors, and they characteristically have a pattern of mutations that overlaps with that of acute myeloid leukemias [4–6]. In addition, ETP T-ALL has a lower frequency of mutations in NOTCH1 and loss of the short arm of chromosome 9 [4].
Early cortical thymocyte leukemias are primarily associated with translocations resulting in aberrant expression of TLX1, TLX3, and related homeobox transcription factor oncogenes [1,3]. They are defined as a molecular group not only by their gene expression signature and immunophenotype, which are related to those of early cortical thymic precursors, but also by the presence of associated mutations that are characteristically enriched in this group, such as mutations in BCL11B, WT1, or PHF6 or rearrangements of the ABL1 oncogene with NUP214 [1,3,7–9]. Late cortical leukemias characteristically overexpress the transcription factor oncogene TAL1 with either LMO1 or LMO2. They occur later in the pattern of gene expression programming related to T-cell development and they have the highest frequency of mutations in PTEN [1,3,10].
It is important to consider these clinical biological groups because they not only define the biology of the disease, but they are also associated with clinical outcome. ETP T-ALL has been associated with poor prognosis [1,5,11]. Early cortical thymocyte leukemias characteristically have a favorable outcome [3,12], and later cortical leukemias in the context of PTEN mutations may be associated with poor prognosis [1,3,13].
Targeting NOTCH1
The high prevalence and prominent role of NOTCH1 mutations in T-ALL make the NOTCH signaling pathway a promising target to treat the disease. NOTCH1 is activated by mutations in over 65% of T-ALL [14] and is a central driver of T-ALL cell metabolism, growth, and survival [15–18]. Hematopoietic-specific knockout of NOTCH1 results in no T-cell development, as it is the receptor that reads the instructive signals of the thymic microenvironment that commits lymphoid progenitors to become T cells [18,19]. In T-ALL this developmental circuitry is hijacked by receptor mutations that induce constitutive NOTCH1 activation or impaired degradation of active NOTCH1 [14,18]. Constitutively active NOTCH1 in T-ALL activates a broad number of cell growth and metabolism, activates MYC, and facilitates increased PI3K signaling, in support of a central disease driver role and therapeutic [15–17]. NOTCH1 is a type 1 transmembrane protein that functions as the transcription factor and requires an intramembrane proteolytic cleavage catalyzed by the γ-secretase complex for activation [18]. As result the function of NOTCH1 in the context of mutations present in T-ALL can be abrogated by blocking it at the membrane with small molecule gamma secretase inhibitors (GSIs) [14,20] (Figure 1).
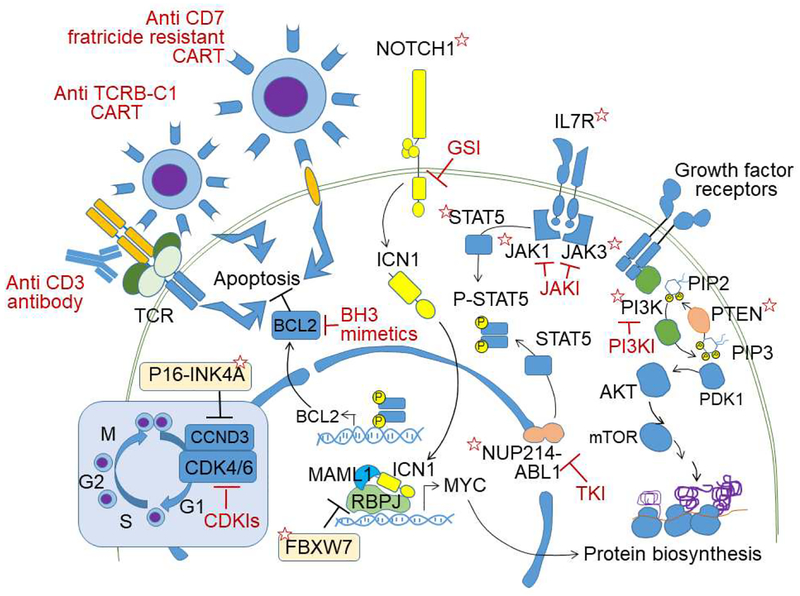
Figure 1.
Schematic representation of oncogenic driver pathways, therapeutic targets, and immunotherapy strategies in T-ALL. Proteins encoded by genes with driver mutations are indicated with red stars. Targeted therapies are indicated in red. GSI: γ-secretase inhibitors, JAKI: Janus kinase inhibitors, PI3Ki: phosphatidylinositol 3 kinase inhibitor, TKI: tyrosine kinase inhibitor, CDKi: cyclin kinase inhibitor, PIP2: Phosphatidylinositol 4,5-bisphosphate, PIP3: Phosphatidylinositol (3,4,5)-trisphosphate, ICN1: intracellular NOTCH1, TCR: T-cell receptor
Gamma secretase inhibitors
Originally developed to treat Alzheimer’s disease, GSIs have now been specifically developed in the context of oncologic applications. In early trials attempts to achieve NOTCH1 inhibition in T-ALL using this strategy had limited success, perhaps in part because these early drugs were not designed to inhibit NOTCH and had more specific activity against the production of amyloid beta peptide in the context of Alzheimer’s disease. However, biomarker analyses suggested that there was some level of inhibition of the NOTCH signaling pathway despite very limited clinical activity. Of note, exposure to the drug was limited by gastrointestinal toxicity, an on-target side effect resulting from intestinal secretory metaplasia resulting from inhibition of NOTCH1 and NOTCH2 in the intestine [18].
However, combination therapies with GSIs may offer opportunities for efficacious treatment with less toxicity in patients with mutations in NOTCH1. NOTCH1 inhibition is highly synergistic with glucocorticoids and most importantly glucocorticoids seem to be able to prevent the development of gastrointestinal toxicity associated with GSI therapy [21,22]. The synergistic interaction of GSIs and glucocorticoids in T-ALL was originally demonstrated in a xenograft model generated from a highly glucocorticoid-resistant leukemia cell line derived from a refractory patient with relapsed T-cell lymphoblastic lymphoma harboring an activating translocation driving constitutively active NOTCH1 signaling [22]. In vivo synergistic activity resulted in complete durable responses with leukemia-bearing mice treated with combination therapy surviving after 2 months of follow-up and remaining minimal residual disease-free at endpoint. Treatment with dexamethasone plus a GSI was well tolerated; however, animals that were treated with GSI alone showed accelerated mortality due to toxicity [22]. Of note, this effect was not the result of a pharmacokinetic interaction, as it could be recapitulated with genetic models of NOTCH inhibition exposed to glucocorticoids [22].
In addition, NOTCH is a central node that controls multiple aspects of biology of T-ALL, and multiple therapeutics also synergize with NOTCH inhibition in cellular and animal models [23]. Thus, inhibition with NF-kappaB with bortezomib [24], inhibition of mTOR with rapamycin [23,25], and interference with protein biosynthesis [23] are strongly synergistic with GSIs and may offer the possibility of delivering stronger cytotoxic activity and less toxicity.
Finally, NOTCH1 is also central for the control of leukemia cell metabolism [16,26]. This can be exploited therapeutically, as the inhibition of NOTCH1 results in a starvation response suppressing anabolic pathways and upregulating cellular catabolism and autophagy in leukemia [16]. Drugs that inhibit glutaminolysis are now in clinical trials in solid tumors and hematologic malignancies and downregulation of glycolysis following NOTCH inhibition with GSIs makes the leukemia cells more dependent on glutaminolysis [16]. As result, genetic inactivation of glutaminase enhances the therapeutic effects of GSIs and pharmacologic inhibition of glutaminolysis with BPTES, a potent and specific small molecule glutaminase inhibitor, is highly synergistic with inhibition of NOTCH in preclinical animal models [16].
Targeting the cell cycle
A second strategy targeting a central genetic driver of T-ALL is to block deregulated cell cycle progression resulting from loss of the P16/INK4A tumor suppressor gene [1]. As mentioned before, the loss of the P19/ARF and P16/INK4A tumor suppressors in the short arm of chromosome 9 is present in about 70% of T-ALL cases [1]. P16/INK4A functions as a negative regulator of cyclin-CDK complexes and its activity can be recapitulated via pharmacologic inhibition of CDK4/CDK6 [27] (Figure 1). Importantly, loss of the retinoblastoma tumor suppressor gene can be recurrently found in adult T-ALL [28] and loss of RB can abrogate the antitumor effects of CDK4/CDK6 inhibitors. In addition, it should be noted that CDK4/CDK6 inhibition can have an antagonistic effect with chemotherapy agents with cycle dependent activity as a result of decreased cell proliferation [29].
Targeting the PI3K pathway
The PTEN tumor suppressor gene is mutated and deleted in 10% of T-ALLs, and an additional 10% of leukemias show loss of PTEN protein expression [30]. PTEN loss results in constitutive activation of the PI3 kinase pathway, which drives cell growth, metabolism, proliferation, and survival pathway in T-ALL [31]. PI3K and NOTCH1 signaling closely interact in the regulation of cell metabolism in normal thymocyte development and in T-ALL [16,30,32]. PTEN loss in T-ALL has been associated with poor prognosis in some series, particularly if associated with RAS mutations [13,33]. Therapeutically, the PI3K pathway can be effectively blocked pharmacologically and dual inhibition of PI3 kinase gamma and PI3 kinase delta show strong antileukemic effects in preclinical models of PTEN deficient T-ALL [34] (Figure 1).
Of note, constitutive activation of PI3K can impair glucocorticoid response [35,36] and targeting PI3K, AKT, and mTOR can enhance the antileukemic effects of glucocorticoids [34–36].
Targeting the JAK/STAT pathway
Cytokine signaling provides important cues promoting proliferation and survival of lymphoid cells and leukemia lymphoblasts. In addition, activating mutations in IL-7 receptor, JAK1, JAK3, and STAT5 can be found in T-ALL, resulting in activation of JAK-STAT signaling [1,37]. Of note, IL-7 receptor mutations tend to be strongly activating alleles [38,39]; however multiple hits involving JAK1 and JAK3 mutations can be found in the same patient showing cooperative activity in the disease transformation [40]. In this context, inhibition of JAK-STAT can result in antitumor effects in preclinical models [40,41] (Figure 1). Importantly, the antileukemic effects of JAK-STAT inhibition do not seem restricted to leukemias harboring activating mutations in the pathway. In primary xenograft models of ETP, activation of the JAKSTAT signaling pathway was independent of the presence of JAK/STAT mutations and showed to be hypersensitive to stimulation with IL-7. Moreover, ruxolitinib inhibited JAK-STAT signaling and abrogated the hyperactivation effect of IL-7 and was highly effective in these preclinical models [41]. Finally, signaling pathways with altered phosphorylation after JAK inhibition (MEK, PI3K) and BCL2 can be pharmacologically inhibited, which results in synergistic antitumor effects in combination with JAK kinase inhibitors in primary T-ALL samples with JAK3 mutations [42].
Targeting NUP214-ABL1
About 5% of T-ALLS are driven by tyrosine kinase oncogene fusions, the most frequent being the NUP214-ABL1 rearrangement [7]. NUP214-ABL1 is frequently associated with the TLX1 and TLX3 group of T-ALLs and although it can be found as a subclonal alteration and does not appear to be linked with poor prognosis, it can be effectively blocked with tyrosine kinase inhibitors [43]. To date only a handful of NUP214-ABL1 positive patients have been treated with a tyrosine kinase inhibitor, yet these drugs seem to be active, demonstrating preclinical biomarkers of activity and could provide clinical benefit in some cases [44–46] (Figure 1).
Relapsed T-ALL
Relapse T-ALL is associated with high rates of secondary chemotherapy resistance and represents a particularly challenging therapeutic scenario with limited therapeutic opportunities. Genomic studies of matched diagnosis and relapsed leukemia demonstrate a frequent branched pattern of clonal evolution with relapsed tumors containing some common mutations with the main clone present at diagnosis and specific alterations not detected in the major population at diagnosis [8,47]. These results support the idea that relapse frequently emerges as a result of expansion of a preexisting ancestral clone related to but distinct from the major diagnostic population. Relapsed T-ALL is genetically heterogeneous. Among the genes altered in relapse, mutations in the cytosolic nucleotidase 2 gene (NT5C2) are particularly prevalent and can be detected in about 45% of early relapse T-ALLs and 20% of T-ALL relapses overall [47,48]. NT5C2 mutations are characteristically heterozygous and result in specific single amino-acid substitutions in characteristic hotspots, with one allele NT5C2 p.R367Q accounting for almost 90% of cases [47–50]. NT5C2 encodes a cytosolic enzyme normally involved in the degradation of purine monophosphate nucleotides. Structural and functional analyses reveal that relapse-associated NT5C2 mutations are gain of function alleles with increased nucleotidase activity [48–50].
NT5C2 activity is tightly regulated by a series of intramolecular interactions that trigger activating conformational changes in response to allosteric activation and then return the enzyme to its basal inactive state [50]. NT5C2 is a tetrameric protein composed of a dimer of dimers with a closed inactive configuration in basal conditions that transitions to an active open state upon interaction with allosteric regulators [50,51]. Most common NT5C2 mutations target an intramolecular switch off mechanism responsible for returning the enzyme back to its inactive configuration, while others lower the threshold for allosteric activation or directly activate the catalytic center resulting in allosteric-independent activation [50].
As a result, leukemia lymphoblasts harboring NT5C2 mutations have increased nucleotidase activity that not only metabolizes the normal nucleotide monophosphate intermediates in the purine biosynthesis pathway (IMP, XMP, GMP), but facilitates the clearance of Thio-IMP, Thio-XMP, and Thio-GMP, intermediate metabolites generated by the incorporation of 6-mercaptopurine (6-MP) and 6 tioguanine (6-TG) by the salvage pathway of purine biosynthesis [48–50]. Dephosphorylation and secretion out of the cell of thiopurine nucleotides decreases the effective intracellular concentration of the cytotoxic products of 6-MP and 6-TG. Consequently, expression of relapse-associated NT5C2 mutations in ALL cell lines induces resistance to thiopurine chemotherapy [48,49]. This resistance phenotype was formally verified in patient-derived xenografts and in vivo in a mouse conditional-and-inducible knock-in leukemia model with expression of the highly prevalent Nt5c2 p.R367Q allele [52]. Clonal evolution dynamic analyses in this model revealed positive selection of the Nt5c2 mutation under 6-MP chemotherapy and progression under therapy [52]. However, 6-MP resistance comes at the cost of impaired leukemia cell growth and leukemia-initiating cell activity as result of mutation-driven enhancement of purine degradation and excess export of purines [52]. In addition, the leak of purines to the media results in increased dependence on purine biosynthesis and cells harboring NT5C2 mutations show increased sensitivity to mizoribine, an inhibitor of IMPDH, a central enzyme responsible for the production of IMP downstream of both the de novo and salvage purine biosynthetic pathways [52].
Emerging immunotherapy opportunities in T-ALL
The promising results of cellular and antibody-based immunotherapies in the treatment of B-cell malignancies have generated much interest in the development of targeted immunotherapy strategies for the treatment of T-ALL. However, the development of chimeric antigen receptor (CAR) T cells with killing specificity against surface T-cell antigens is impaired by the fratricide effect of such cells and the frequent low yields of T-cell harvesting. A potential strategy to overcome these obstacles is the development via CRISPR-CAS9 of fratricide-resistant T cells devoid of CD7 and TCRA that express a CAR directed against CD7. These “off the shelf” CAR T cells show efficacy against human T-ALL without xenogeneic graft-versus-host disease [53]. Yet, even if successful in controlling disease, CAR T cell pan T-cell killing would result in long-term T-cell immunosuppression and high risk of life-threatening opportunistic infections.
In this context, allele specific CAR T cells targeting the monoallelically expressed TCRB constant chain may deliver effective antileukemic effects with preservation of T-cell immune function [54,55]. During T cell receptor β (TCRB) rearrangement, developing T cells can engage two different modules in the TCR locus both encoding the constant region of this receptor chain. These TCRB constant domains 1 and 2 (TCRBC1 and TCRBC2) are functionally equivalent but show some differences in their amino acid sequence which can be exploited for the development of TCRB1 or TCRB2-specific antibodies and CAR T cells [54,55]. Importantly, the normal T-cell pool is composed of cells expressing TCRs containing the TCRBC1 and cells with TCRs containing a β chain using instead TCRBC2. In contrast monoclonal T-cell malignancies express only one TCR that will contain a TCRB chain with either TCRBC1 or TCRBC2. By engineering anti-TCRBC CAR T cells that specifically recognize a TCRBC1-specific epitope not present in the TCRBC2, a CAR T product can be generated, as CAR expression results in fratricidal killing of TCRBC1 positive normal T cells but preserves TCRBC2+ lymphocytes [54]. The resulting TCRBC2+ anti-TCRBC1 CAR T cells can recognize and kill normal and malignant TCRBC1+ cells, but not TCRBC2+ lymphocytes. They would thus deliver effective antileukemic effects towards T-ALLs with surface expression of a TCRB1 containing TCR, while preserving much of the normal T-cell immune repertoire composed of normal T cells with TCRs containing a TCRBC2 β chain for maintenance of cellular immunity [54,55] (Figure 1).
Finally, an antibody-based immunotherapy based on the activation of the T-cell receptor using antibodies directed against the CD3 receptor similar to OKT3 may be possible, as antibody engagement of surface CD3 can induce strong TCR signaling and activate a programmed cell death response similar to that engaged during negative selection of normal autoreactive developing T cells in the thymus [56] (Figure 1).
Conclusion
Much progress has been made in the identification of oncogenic drivers and therapeutic targets in T-ALL, opening numerous new opportunities for the development of improved, highly active and less toxic therapies. Systematic efforts to identify synergistic and easily deployable new drugs and drug combinations vetted using state of the art preclinical models and the development of a path towards the clinical testing of emerging immunotherapies may guide the development of the next generation of clinical trials aiming to incorporate targeted therapies and immunotherapeutic tools for the treatment of this disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure:
No relevant financial relationship with any commercial interest.
References
- [1].Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat Rev Cancer 2016;16:494–507. [DOI] [PubMed] [Google Scholar]
- [2].Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017;49:1211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell 2002;1:75–87. [DOI] [PubMed] [Google Scholar]
- [4].Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012;481:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Haydu JE, Ferrando AA. Early T-cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol 2013;20:369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med 2011;208:2571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet 2004;36:1084–9. [DOI] [PubMed] [Google Scholar]
- [8].Tosello V, Mansour MR, Barnes K, Paganin M, Sulis ML, Jenkinson S, et al. WT1 mutations in T-ALL. Blood 2009;114:1038–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].De Keersmaecker K, Real PJ, Gatta GD, Palomero T, Sulis ML, Tosello V, et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med. 2010;16:1321–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zuurbier L, Petricoin EF 3rd, Vuerhard MJ, Calvert V, Kooi C, Buijs-Gladdines JG, et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica 2012;97:1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol 2009;10:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ferrando AA, Neuberg DS, Dodge RK, Paietta E, Larson RA, Wiernik PH, et al. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet 2004;363:535–6. [DOI] [PubMed] [Google Scholar]
- [13].Paganin M, Grillo MF, Silvestri D, Scapinello G, Buldini B, Cazzaniga G, et al. The presence of mutated and deleted PTEN is associated with an increased risk of relapse in childhood T cell acute lymphoblastic leukaemia treated with AIEOP-BFM ALL protocols. Br J Haematol 2018;182:705–11. [DOI] [PubMed] [Google Scholar]
- [14].Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004;306:269–71. [DOI] [PubMed] [Google Scholar]
- [15].Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A 2006;103:18261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Herranz D, Ambesi-Impiombato A, Sudderth J, Sanchez-Martin M, Belver L, Tosello V, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med 2015;21:1182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sanchez-Martin M, Ferrando A. The NOTCH1-MYC highway toward T-cell acute lymphoblastic leukemia. Blood 2017;129:1124–33. [DOI] [PubMed] [Google Scholar]
- [18].Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematology American Society of Hematology Education Program 2009;353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 1999;10:547–58. [DOI] [PubMed] [Google Scholar]
- [20].Palomero T, Ferrando A. Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia. Clin Lymphoma Myeloma 2009;9 Suppl 3:S205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Samon JB, Castillo-Martin M, Hadler M, Ambesi-Impiobato A, Paietta E, Racevskis J, et al. Preclinical analysis of the gamma-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol Cancer Ther 2012;11:1565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med 2009;15:50–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sanchez-Martin M, Ambesi-Impiombato A, Qin Y, Herranz D, Bansal M, Girardi T, et al. Synergistic antileukemic therapies in NOTCH1-induced T-ALL. Proc Natl Acad Sci U S A 2017;114:2006–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med 2007;13:70–7. [DOI] [PubMed] [Google Scholar]
- [25].Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood 2009;113:6172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Herranz D, Ferrando AA. Targeting NOTCH1 in T-ALL: Starving the dragon. Cell Cycle 2016;15:483–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sawai CM, Freund J, Oh P, Ndiaye-Lobry D, Bretz JC, Strikoudis A, et al. Therapeutic targeting of the cyclin D3:CDK4/6 complex in T cell leukemia. Cancer Cell 2012;22:452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, Hadler M, Paietta E, Tallman MS, et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood 2013;122:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pikman Y, Alexe G, Roti G, Conway AS, Furman A, Lee ES, et al. Synergistic Drug Combinations with a CDK4/6 Inhibitor in T-cell Acute Lymphoblastic Leukemia. Clin Cancer Research 2017;23:1012–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 2007;13:1203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Palomero T, Dominguez M, Ferrando AA. The role of the PTEN/AKT Pathway in NOTCH1-induced leukemia. Cell Cycle 2008;7:965–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol 2005;6:881–8. [DOI] [PubMed] [Google Scholar]
- [33].Trinquand A, Tanguy-Schmidt A, Ben Abdelali R, Lambert J, Beldjord K, Lengline E, et al. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol 2013;31:4333–42. [DOI] [PubMed] [Google Scholar]
- [34].Subramaniam PS, Whye DW, Efimenko E, Chen J, Tosello V, De Keersmaecker K, et al. Targeting nonclassical oncogenes for therapy in T-ALL. Cancer Cell 2012;21:459–72. [DOI] [PubMed] [Google Scholar]
- [35].Piovan E, Yu J, Tosello V, Herranz D, Ambesi-Impiombato A, Da Silva AC, et al. Direct reversal of glucocorticoid resistance by AKT inhibition in acute lymphoblastic leukemia. Cancer Cell 2013;24:766–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wei G, Twomey D, Lamb J, Schlis K, Agarwal J, Stam RW, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell 2006;10:331–42. [DOI] [PubMed] [Google Scholar]
- [37].Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of TALL. Blood 2017;129:1113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med 2011;208:901–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat genet 2011;43:932–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Degryse S, Bornschein S, de Bock CE, Leroy E, Vanden Bempt M, Demeyer S, et al. Mutant JAK3 signaling is increased by loss of wild-type JAK3 or by acquisition of secondary JAK3 mutations in T-ALL. Blood 2018;131:421–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maude SL, Dolai S, Delgado-Martin C, Vincent T, Robbins A, Selvanathan A, et al. Efficacy of JAK/STAT pathway inhibition in murine xenograft models of early T-cell precursor (ETP) acute lymphoblastic leukemia. Blood 2015;125:1759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Degryse S, de Bock CE, Demeyer S, Govaerts I, Bornschein S, Verbeke D, et al. Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia 2018;32:788–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Quintas-Cardama A, Tong W, Manshouri T, Vega F, Lennon PA, Cools J, et al. Activity of tyrosine kinase inhibitors against human NUP214-ABL1-positive T cell malignancies. Leukemia 2008;22:1117–24. [DOI] [PubMed] [Google Scholar]
- [44].Deenik W, Beverloo HB, van der Poel-van de Luytgaarde SC, Wattel MM, van Esser JW, Valk PJ, et al. Rapid complete cytogenetic remission after upfront dasatinib monotherapy in a patient with a NUP214-ABL1-positive T-cell acute lymphoblastic leukemia. Leukemia 2009;23:627–9. [DOI] [PubMed] [Google Scholar]
- [45].Clarke S, O’Reilly J, Romeo G, Cooney J. NUP214-ABL1 positive T-cell acute lymphoblastic leukemia patient shows an initial favorable response to imatinib therapy post relapse. Leuk Res 2011;35:e131–3. [DOI] [PubMed] [Google Scholar]
- [46].Chen Y, Zhang L, Huang J, Hong X, Zhao J, Wang Z, et al. Dasatinib and chemotherapy in a patient with early T-cell precursor acute lymphoblastic leukemia and NUP214-ABL1 fusion: A case report. Exp Ther Med 2017;14:3979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Oshima K, Khiabanian H, da Silva-Almeida AC, Tzoneva G, Abate F, Ambesi-Impiombato A, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2016;113:11306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med 2013;19:368–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 2013;45:290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Dieck CL, Tzoneva G, Forouhar F, Carpenter Z, Ambesi-Impiombato A, Sanchez-Martin M, et al. Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia. Cancer Cell 2018;34:136–47.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wallden K, Stenmark P, Nyman T, Flodin S, Graslund S, Loppnau P, et al. Crystal structure of human cytosolic 5’-nucleotidase II: insights into allosteric regulation and substrate recognition. J Biol Chem 2007;282:17828–36. [DOI] [PubMed] [Google Scholar]
- [52].Tzoneva G, Dieck CL, Oshima K, Ambesi-Impiombato A, Sanchez-Martin M, Madubata CJ, et al. Clonal evolution mechanisms in NT5C2 mutant-relapsed acute lymphoblastic leukaemia. Nature 2018;553:511–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cooper ML, Choi J, Staser K, Ritchey JK, Devenport JM, Eckardt K, et al. An “off-the-shelf” fratricide-resistant CAR-T for the treatment of T cell hematologic malignancies. Leukemia 2018;32:1970–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Maciocia PM, Wawrzyniecka PA, Philip B, Ricciardelli I, Akarca AU, Onuoha SC, et al. Targeting the T cell receptor beta-chain constant region for immunotherapy of T cell malignancies. Nat Med 2017;23:1416–23. [DOI] [PubMed] [Google Scholar]
- [55].Palomero T, Ferrando A. Targeted cellular immunotherapy for T cell malignancies. Nat Med 2017;23:1402–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Trinquand A, Dos Santos NR, Tran Quang C, Rocchetti F, Zaniboni B, Belhocine M, et al. Triggering the TCR Developmental Checkpoint Activates a Therapeutically Targetable Tumor Suppressive Pathway in T-cell Leukemia. Cancer Discov 2016;6:972–85. [DOI] [PubMed] [Google Scholar]