Extended Data Fig. 9|. Arrestin-2 undergoes similar fluctuations in simulation as arrestin-1, suggesting a potential common activation mechanism.
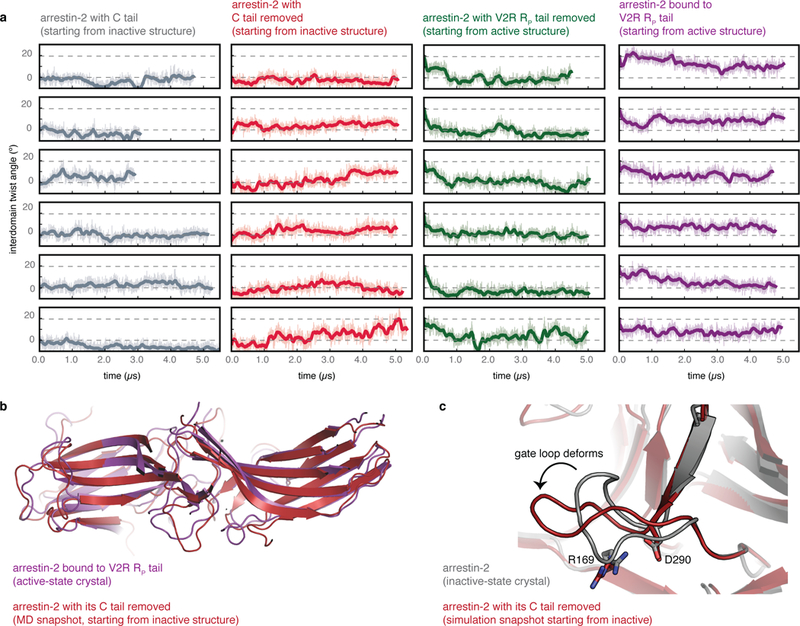
Simulations initiated from the inactive conformation but with the arrestin C tail removed reached active conformations, and simulations initiated from the active conformation but with the co-crystallized RP tail removed reached inactive conformations. a, Interdomain twist angle as a function of time for simulations of arrestin-2 performed under four conditions: active arrestin-2 bound to the V2 vasopressin receptor C-terminal phosphopeptide (PDB entry 4JQI) with the crystallographic Fab30 fragment removed; active arrestin-2 with the V2R RP tail removed; and inactive arrestin-2 with its crystallographic C tail present or absent (PDB entry 1G4M). In these simulations, arrestin-2 appears to favour more inactive-like conformations than those seen in the majority of our arrestin-1 simulations, but this may be due to the specific choice of crystal structure from which the simulations were initiated; see Extended Data Fig. 7. Dashed lines represent the inactive and active state interdomain twist angles for the arrestin-2 crystal structures. b, Snapshot of an active-like rotational state observed in simulations started from an inactive-state structure with the C tail removed (simulation 63, dark red), overlaid on the active-state structure (purple). c, Simulation snapshot from a simulation started from the inactive state with the C tail removed, in which the gate loop moves into an intermediate state (simulation 62, dark red), as seen in simulations of arrestin-1 started from the inactive state with the C tail removed. The absence of a structure of a receptor-bound β-arrestin leaves open the possibility that receptors might bind β-arrestins differently from arrestin-1, and even if the binding mode is similar, the activation mechanism might be different.
