Abstract
CREBBP, encoding an acetyltransferase, is among the most frequently mutated genes in small cell lung cancer (SCLC), a deadly neuroendocrine tumor type. We report acceleration of SCLC upon Crebbp inactivation in an autochthonous mouse model. Extending these observations beyond the lung, broad Crebbp deletion in mouse neuroendocrine cells cooperated with Rb1/Trp53 loss to promote neuroendocrine thyroid and pituitary carcinomas. Gene expression analyses showed that Crebbp loss results in reduced expression of tight junction and cell adhesion genes, including Cdh1, across neuroendocrine tumor types, while suppression of Cdh1 promoted transformation in SCLC. CDH1 and other adhesion genes exhibited reduced histone acetylation with Crebbp inactivation. Treatment with the histone deacetylase inhibitor Pracinostat increased histone acetylation and restored CDH1 expression. Additionally, a subset of Rb1/Trp53/Crebbp-deficient SCLC exhibited exceptional responses to Pracinostat in vivo. Thus, CREBBP acts as a potent tumor suppressor in SCLC and inactivation of CREBBP enhances responses to a targeted therapy.
Keywords: CREBBP, CBP, SCLC, CDH1, E-Cadherin
Introduction
Recent identification of the genomic alterations in small cell lung cancer (SCLC), a deadly type of lung cancer may provide new opportunities for therapeutic intervention (1–3). Critical challenges remain, however, as few of these SCLC alterations are readily actionable and a majority of them have not been validated for their roles in disease initiation and progression (4). Along with RB1 and TP53 inactivation, mutations in the CREBBP and EP300 acetyltransferases are among the most frequent in SCLC, appearing in 15–17% and 5–13% of SCLC patient tumors, respectively (1,2,5,6). In SCLC, deletions and truncating mutations in CREBBP and EP300 genes along with missense mutations in the histone acetyltransferase (HAT) domain are frequent, and these occur in a mutually exclusive manner. For CREBBP, HAT domain mutations observed in SCLC samples have been shown to abrogate CREBBP-mediated histone acetylation (6). CREBBP and EP300 acetylation of lysine residues on histone tails neutralizes their positive charge and can increase chromatin accessibility. Acetylation of a specific histone residue, histone H3K27 by CREBBP/EP300 can promote transcriptional enhancer function (7) and deletion of Crebbp/Ep300 in mouse fibroblasts eliminates the vast majority of H3K27 acetylation (8). CREBBP/EP300 also acetylate non-histone proteins, such as p53 and BCL6 (9,10). CREBBP is mutated in lymphomas, urothelial carcinoma, and other human tumor types (11–13). Studies employing mouse models have demonstrated that Crebbp functions as a tumor suppressor in leukemia and lymphoma (14–17). However, in vivo evidence that Crebbp functions as a tumor suppressor in solid tumors is lacking. In lymphoma, it has been posited that loss of CREBBP-mediated acetylation and activation of p53 drives tumorigenesis (13,17). p53-dependent mechanisms of tumor suppression mediated by CREBBP are likely not relevant to tumors such as SCLC that almost invariably harbor TP53 mutations (1). Thus, elucidating roles for p53-independent tumor-suppressive activities of CREBBP in SCLC is important. In this study, we demonstrate p53-independent Crebbp tumor suppressor function not only in SCLC but across multiple neuroendocrine tumor types. We report CREBBP-control of adhesion related transcript expression, including CDH1, encoding E-CADHERIN, as contributing to tumor suppression and we identify a potential therapeutic approach for treating CREBBP-deficient SCLC.
Results
Crebbp mutation promotes tumorigenesis of pre-neoplastic neuroendocrine cells
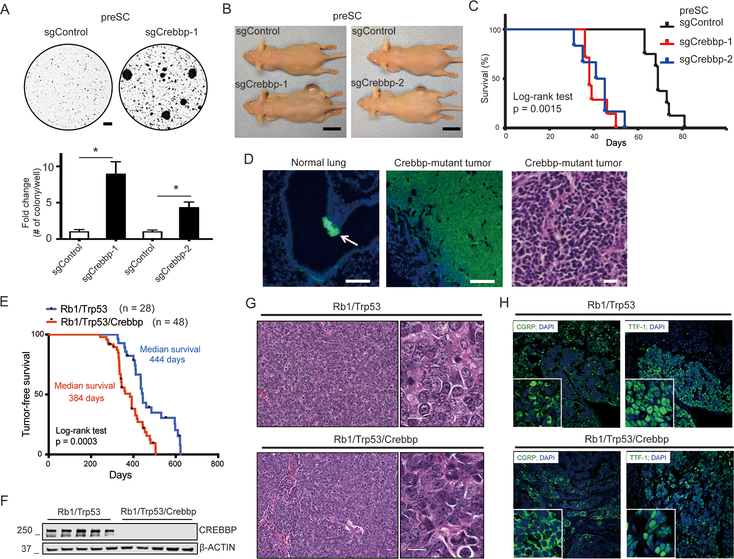
To study the potential role of Crebbp in SCLC tumor suppression, we mutated Crebbp in a cell-based model of early-stage SCLC that we previously described (18). Rb1/Trp53/Rbl2-deficient “preSC” cells, derived from a mouse SCLC model at an early stage in tumorigenesis become fully transformed with ectopic expression of SCLC oncogenes such as MYCL (18). Here we expressed Cas9 and two sgRNAs targeting DNA sequences encoding the HAT domain of the murine Crebbp gene and validated loss of CREBBP protein (Supplementary Figure S1A,B). We found that Crebbp-mutant preSC cells formed more and individually larger colonies in soft agar compared to control preSC cells (Figure 1A). When preSC cells were injected into the flanks of immune-compromised mice, tumors emerged at 30–50 days in the sites injected with the Crebbp-mutant preSC cells but not in those injected with control cells (Figure 1B). Further aging of the mice injected with control preSC cells showed that these cells also formed tumors, with delayed kinetics (Figure 1C). Hematoxylin-eosin staining showing typical SCLC morphology and immunostaining showed the expression of UCHL1 and CGRP, markers of both SCLC as well as normal pulmonary neuroendocrine cells (Figure 1D and Supplemental Figure S1C). These data support a role for CREBBP in SCLC tumor suppression.
Figure 1. Inactivation of Crebbp accelerates SCLC in mouse models.
A, Representative images of control and Crebbp-targeted preSC cells in soft agar 3 weeks after seeding of 1×104 cells. Two independent sgRNAs were employed (sgCrebbp-1, sgCrebbp-2). Bottom: quantification of colonies > 0.1 mm in diameter (n=4). Scale bar = 0.5 mm.
B, Images of preSC-derived allografts, 40 days after subcutaneous injection of cells. Scale bar = 1cm.
C, Kaplan-Meier overall survival curves of mice injected with control-preSC cells (Control, n= 6) and mice injected with Crebbp-knockout preSC cells (n= 5 each). Statistical significance was calculated using log-rank (Mantel-Cox) test.
D, Images of UCHL1-stained sections of Crebbp-mutant tumor and normal lung. Arrow points to neuroepithelial body in the airway. Scale bar = 100μm. Representative section of Crebbp-mutant tumors stained with hematoxylin and eosin (H+E). Scale bar = 20μm.
E, Kaplan-Meier tumor-free survival curves of Rb1/Trp53 mutant (blue, n=28) and Rb1/Trp53/Crebbp mutant (red, n=48) mice from autochthonous model infected with Ad-CGRP-Cre (Day 0). Statistical significance was calculated using log-rank (Mantel-Cox) test.
F, Representative immunoblotting results of CREBBP protein levels in 5 lung tumor tissues from each cohort (Rb1/Trp53 vs. Rb1/Trp53/Crebbp). Beta-ACTIN was used as a loading control.
G, Representative H&E stained section of SCLC in each cohort (Rb1/Trp53 vs. Rb1/Trp53/Crebbp). Scale bar, 20μm.
H, Representative immunofluorescence for SCLC markers TTF-1 and CGRP in each cohort (Rb1/Trp53 vs. Rb1/Trp53/Crebbp). DAPI was used as a nuclear stain. Original magnification, 40X.
Crebbp inactivation accelerates SCLC in an autochthonous mouse model
To further investigate the contribution of Crebbp inactivation to SCLC development in vivo we employed an autochthonous model. We performed a genetic cross to incorporate a floxed Crebbp allele (14) into a Rb1/Trp53 deleted model of SCLC that develops lung tumors with histopathological and molecular features of human SCLC (19,20). Via intratracheal instillation, we infected Rb1lox/lox;Trp53lox/lox (herein Rb1/Trp53) and Rb1lox/lox;Trp53lox/lox;Crebbplox/lox (herein Rb1/Trp53/Crebbp) mice with adenovirus expressing Cre recombinase under control of a neuroendocrine-specific CGRP promoter (Ad-CGRP-Cre) (21). Following Ad-CGRP-Cre infection, we found that Rb1/Trp53/Crebbp mice developed lung tumors and became moribund significantly earlier, with 384 days of median tumor-free survival compared to 444 days in Rb1/Trp53 mice (p=0.0003, log-rank test) (Figure 1E). Immunoblot analysis verified complete loss of CREBBP protein in the tumors from Rb1/Trp53/Crebbp mice infected with Ad-CGRP-Cre (Figure 1F). Histopathology review by a lung cancer pathologist (A.G.) confirmed SCLC histology in both groups (Figure 1G) and tumors from both groups stained positive for markers of SCLC, including TTF1 and the neuroendocrine marker CGRP (Figure 1H). The Rb1/Trp53/Crebbp tumors were uniformly of “classic” SCLC histology and showed vascular invasion and liver metastases (Supplemental Figure S2A-F). No difference in proliferation between Rb1/Trp53 and Rb1/Trp53/Crebbp late-stage lung SCLC was observed, based on phospho histone H3 immunostaining of mitotic cells (Supplemental Figure S3A,B) and a panel of neuroendocrine markers associated with SCLC were expressed at similar levels (Supplemental Figure S3C). Rates of liver metastases were also similar between the Rb1/Trp53 and Rb1/Trp53/Crebbp models (Supplemental Figure S2G-I). These data definitively show that Crebbp functions as a tumor suppressor in small cell lung cancer.
Crebbp loss accelerates development of pituitary and thyroid neuroendocrine tumors
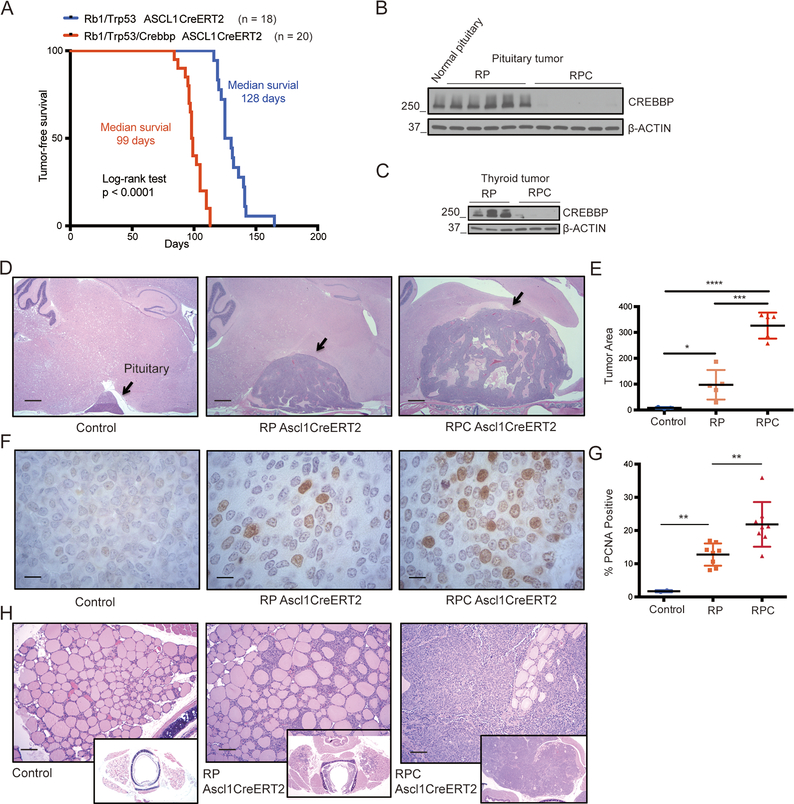
To further evaluate whether Crebbp acts as a tumor suppressor across multiple neuroendocrine tissue compartments, we employed a tamoxifen-inducible neuroendocrine expressing Cre driver strain (Ascl1Cre-ERT2 knockin allele) (22) and generated Ascl1Cre-ERT2/Rb1/Trp53 and Ascl1Cre-ERT/Rb1/Trp53/Crebbp mice. Following tamoxifen injection into the peritoneum to delete conditional alleles, we monitored mice for tumor-associated morbidity. In the control group Ascl1Cre-ERT2/Rb1/Trp53 mice became moribund at a median of 128 days post tamoxifen injection, with 100% incidence of massive pituitary tumors that distorted the brain (Figure 2A, Supplemental Figure S4A and S4B). Histological review of these tumors, performed by a veterinary pathologist (S.P.), indicated pituitary carcinoma of the intermediate lobe, a neuroendocrine tumor type previously described in Rb1- and Rb1/Trp53-mutant mouse models (23–25). The tumors exhibited multifocal invasion of the overlying neural parenchyma. We also observed invasion of the underlying sphenoid bone with islands of neoplastic cells within bone marrow (Supplemental Figure S4C). Ascl1Cre-ERT2/Rb1/Trp53 mice also exhibited thyroid neuroendocrine C-cell adenomas, characterized by discrete nodular foci composed of well-differentiated C cells that compressed adjacent follicles (Supplemental Figure S4D, S4E). Thyroid C-cell tumors have also been previously shown to be frequent in mice with Rb1 and Trp53 mutation (26). Ascl1Cre-ERT2/Rb1/Trp53/Crebbp mice became moribund significantly more rapidly than Rb1/Trp53 mutant controls (Figure 2A), as pituitary tumor burden led to euthanasia at a median of 99 days (p<0.0001, log-rank test). The pituitary carcinomas in Rb1/Trp53/Crebbp mutant mice were histologically similar to those from the Rb1/Trp53 mutant model (Supplemental Figure S4F and S4G). In contrast to small thyroid C-cell adenomas in Rb1/Trp53 mutants, the Rb1/Trp53/Crebbp mutant animals invariably exhibited large, bilateral and invasive thyroid medullary C-cell carcinomas with multifocal necrosis, marked nuclear atypia and high mitotic rates (Supplemental Figure S4H,I). The tumor cells also showed vascular invasion (Supplemental Figure S4J). We confirmed complete loss of CREBBP protein expression in the pituitary and thyroid tumors from Ascl1Cre-ERT2/Rb1/Trp53/Crebbp mice (Figure 2B,C).
Figure 2. Crebbp inactivation accelerates pituitary and thyroid neuroendocrine tumors.
A, Kaplan-Meier tumor-free survival curves of double knockout mice (Rb1lox/lox;Trp53lox/lox;ASCL1CreERT2, green, n=18) and triple knockout mice (Rb1lox/lox;Trp53lox/lox;Crebbplox/lox;ASCL1CreERT2 red, n=20). Log-rank (Mantel-Cox) test was used to determine the significance of tumor-free survival between the cohorts.
B, Representative immunoblotting results of CREBBP protein levels in pituitary tumors from 5 mice in each cohort. Normal mouse pituitary was used as a control. RPC represents Rb1/Trp53/Crebbp; RP represents Rb1/Trp53.
C, Representative immunoblotting showing CREBBP protein levels in thyroid tumors from 3 mice in each cohort.
D-E, Mice with the indicated genotypes were euthanized 3 months after tamoxifen (TAM) injection. (D) Representative H&E stained sections showing normal pituitary gland and pituitary tumors, scale bar = 500μm. (E) Average area of pituitary and pituitary carcinomas in each cohort quantified with n= 5 mice in each cohort. Statistical significance was determined by two-tailed unpaired Student’s t- test; *, p<0.05; ***, p<0.001; ****, p<0.0001.
F-G, Representative IHC of PCNA in normal pituitary and in pituitary tumors from each cohort. Scale bar = 20μm. (G) Quantification of positive PCNA staining in normal pituitary tissues and pituitary tumors in each cohort. **, p<0.01.
H, Representative H&E stained sections of normal thyroid and thyroid tumors from mice in each cohort, 3 months after TAM injection. Scale bar = 500μm. Also see related Figure S4 for images of tumor histology at time of animal morbidity.
To determine how Crebbp inactivation accelerated tumor development in these tissues, we examined affected pituitary at 3-months post tamoxifen and found significantly larger pituitary tumor size in the triple mutant compared to the double mutant mice (Figure 2D and 2E). Immunohistochemistry for PCNA, a marker of proliferating cells, showed a higher rate of proliferation in the Ascl1Cre-ERT2/Rb1/Trp53/Crebbp compared to Ascl1Cre-ERT2/Rb1/Trp53 pituitary tumors (Figure 2F and 2G), explaining the faster tumor growth upon Crebbp deletion. While Ascl1Cre-ERT2/Rb1/Trp53 mice exhibited hyperplastic medullary thyroid lesions or small early adenomas at 3-months post tamoxifen, the Ascl1Cre-ERT2/Rb1/Trp53/Crebbp group exhibited large bilateral thyroid C-cell carcinomas (Figure 2H). These data indicate that Crebbp cooperates with Rb1 and Trp53 to suppress tumorigenesis not only in SCLC, but across multiple neuroendocrine cell types.
Crebbp-regulated transcriptional changes in neuroendocrine tumors
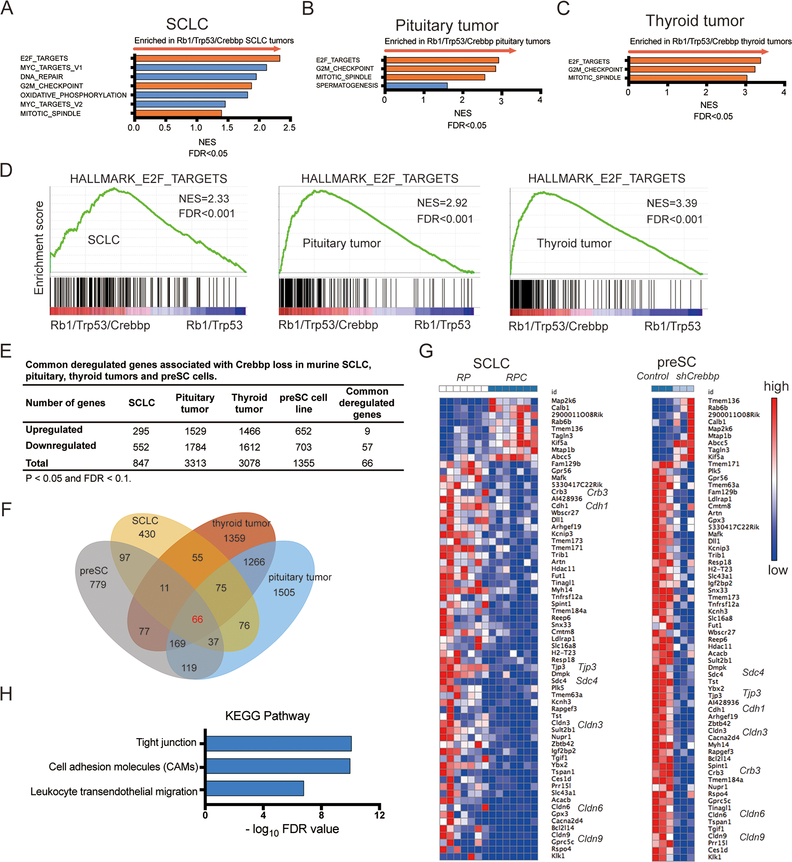
We hypothesized that Crebbp loss cooperates with Rb1/Trp53 deletion to promote SCLC, thyroid medullary C-cell and pituitary carcinomas (all neuroendocrine tumors) through control of gene expression. We performed RNAseq analyses to compare Rb1;Trp53 vs. Rb1;Trp53;Crebbp tumors, initially examining each of these 3 tumor types individually. To identify pathways and gene sets enriched in the Crebbp-mutant tumors, we performed Gene Set Enrichment Analyses (GSEA) (27). We queried the 50 gene-set “Hallmark” gene signatures from MSigDB (28). For SCLC, thyroid and pituitary tumor types, gene sets related to E2F_TARGETS, G2/M_CHECKPOINT and MITOTIC_SPINDLE were commonly enriched in the Rb1/Trp53/Crebbp compared to the Rb1/Trp53 group (Figure 3A-D and Supplemental Figure S5). This result is consistent with the increased proliferation observed in vivo with Crebbp inactivation in pituitary carcinomas (Figure 2F and 2G). Next, to identify genes significantly deregulated upon Crebbp inactivation we used edgeR (29) and found 847 differentially expressed genes between the double mutant and triple mutant SCLC and 3313 and 3078 genes similarly altered in pituitary and thyroid tumors, respectively, with FDR<0.05 (Figure 3E, Supplemental Tables S1–3). The large number of differentially expressed genes in any given tumor type complicated determination of which genes were likely to be functionally important. We hypothesized that key mediators of Crebbp tumor suppressive activity are likely to be commonly deregulated across the three different Rb1/Trp53-deleted neuroendocrine tumor types that were each accelerated upon Crebbp genetic inactivation. We identified a set of 141 genes consistently regulated in a Crebbp-dependent manner across the three tumor types (Figure 3E,F). To further define a core set of Crebbp-regulated transcripts relevant to SCLC, we knocked down Crebbp expression in murine preSC cells using lentiviral shRNA vectors and identified 1355 differentially expressed genes (Supplemental Table S4), with 66 genes commonly deregulated across each of the 4 Crebbp-perturbed comparisons (mouse SCLC, pituitary tumors, thyroid tumors and preSC cells, Figure 3E and 3F). A heat map of this core set of 66 Crebbp-dependent genes in the mouse SCLC samples and in preSC cells is shown in Figure 3G. Interestingly, there were 6-fold more core genes downregulated upon Crebbp deletion than upregulated, consistent with CREBBP acting, in general, as a positive regulator of gene expression. KEGG (Kyoto Encyclopedia of Genes and Genomes) analyses of these 66 genes indicated significant changes in tight junction and cell adhesion genes, reflecting decreased expression of functionally related genes such as Cdh1, Cldn3, Cldn6, Cldn9 and Tjp3 (Figure 3H). Data from cross-tumor analyses indicate that CREBBP expression in neuroendocrine tumor cells promotes a cellular adhesion related transcriptional program.
Figure 3. Gene expression analyses of CREBBP-perturbed neuroendocrine tumors.
A-C, Gene set enrichment analysis (GSEA) identifies biological processes and pathways enriched in Crebbp-deleted tumors across 3 murine neuroendocrine tumor types (A) SCLC (n=7 Rb1/Trp53/Crebbp vs. 7 Rb1/Trp53), (B) pituitary carcinomas (n=8 Rb1/Trp53/Crebbp vs. 9 Rb1/Trp53), and (C) thyroid c-cell tumors (n= 5 Rb1/Trp53/Crebbp vs. 7 Rb1/Trp53). Red bars show 3 gene sets shared among the three murine neuroendocrine tumor types with FDR<0.05.
D, GSEA plots showing the top gene set enriched in the Crebbp-deficient neuroendocrine tumors, “E2F_Targets.”
E, Summary of commonly dysregulated genes upon Crebbp deletion in three neuroendocrine tumor types (SCLC, pituitary and thyroid tumors) and Crebbp knockdown preSC cells. p< 0.05 and FDR< 0.1.
F, Venn diagram showing 66 genes commonly deregulated in 3 Crebbp-deficient neuroendocrine tumor types compared to Crebbp-wild type controls as well as in preSC cells with lentiviral Crebbp shRNA expression (3 different shRNA sequences included in analysis).
G, Heat map of the 66 commonly dysregulated genes with Crebbp suppression in SCLC primary tumors and preSC cells. Red denotes high expression, blue denotes low expression. RPC represents Rb1/Trp53/Crebbp; RP represents Rb1/Trp53.
H, KEGG pathway enrichment analysis of 66 differentially expressed genes identified significantly enriched biological pathways such as tight junctions and cell adhesion molecules. Top enriched pathways shown.
Features of epithelial mesenchymal transition in Crebbp-deleted SCLC
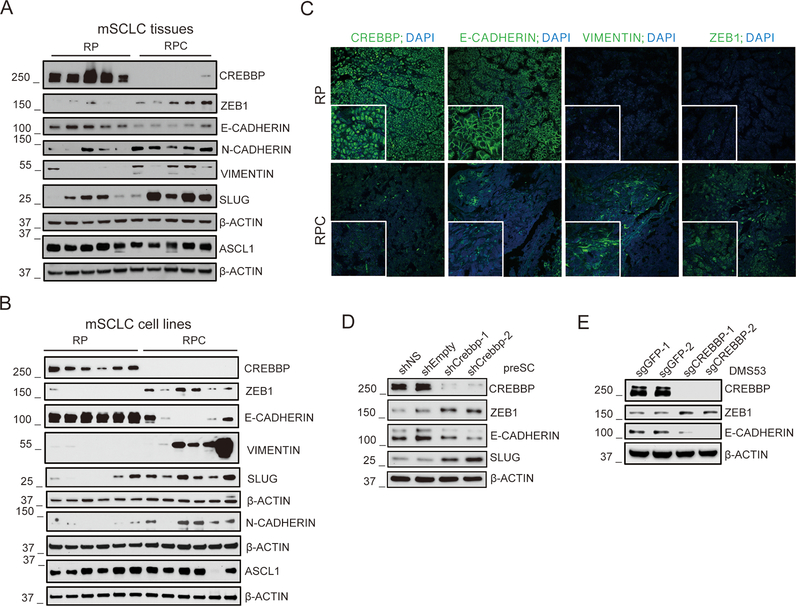
CREBBP-dependent expression of CDH1 was of particular interest, as CDH1 is a tumor suppressor gene that controls epithelial to mesenchymal transitions (EMT) (30). E-CADHERIN is normally expressed in lung neuroendocrine cells, the cells of origin for a majority of SCLC (21), but transient suppression of E-CADHERIN and concomitant upregulation of mesenchymal proteins occur when neuroendocrine cells migrate towards developing neuroendocrine bodies (31). We wanted to determine whether Crebbp loss in SCLC was associated with additional features of EMT. Immunoblot analysis confirmed reduced E-CADHERIN level in Rb1/Trp53/Crebbp primary tumors relative to Rb1/Trp53 SCLC (Figure 4A). Furthermore, in the Rb1/Trp53/Crebbp-null mouse SCLC tumors, we observed increased expression of proteins associated with EMT, including ZEB1, N-CADHERIN, VIMENTIN and SLUG (Figure 4A). Cell lines derived from Rb1/Trp53/Crebbp-deficient mouse tumors also exhibited decreased E-CADHERIN and increased expression of EMT-associated proteins while maintaining ASCL1 expression, an indication that neuroendocrine features were still maintained (Figure 4B). We noted that the magnitude of EMT marker changes upon Crebbp deletion was enhanced in the cell lines compared to primary tumors. Reduction in E-CADHERIN and increased expression of VIMENTIN, and ZEB1, two mesenchymal markers, were confirmed using immunofluorescence in SCLC tumors from the Rb1/Trp53/Crebbp model (Figure 4C and Supplemental Figure S6). We noticed non-uniform E-CADHERIN distribution in the Rb1/Trp53/Crebbp model and the E-CADHERIN low cells maintained CGRP expression (Supplemental Fig S6). Mining data from human SCLC in which CREBBP mutation status was annotated, 11/81 tumors with associated RNAseq data exhibited CREBBP mutation (1). GSEA analyses revealed a trend (FDR =0.19) towards enrichment in the Hallmark “EMT” gene set in the CREBBP-mutant samples (Supplemental Fig S7). Knockdown of CREBBP in preSC cells using two different shRNAs led to reduction in E-CADHERIN levels and increased expression of ZEB1 (Figure 4D). Similarly, knockout of CREBBP in the human SCLC cell line DMS53 also led to reduced E-CADHERIN and increased ZEB1 expression (Figure 4E). Importantly, knockout of CREBBP in DMS53 was associated with increased growth while overexpression of CDH1 had the opposing effect (Supplementary Figure S8). Collectively, these findings indicate that CREBBP inactivation can induce a partial EMT program with reduced expression of CDH1.
Figure 4. Loss of CREBBP induces a partial EMT in SCLC.
A, Immunoblotting of CREBBP, EMT markers (ZEB1, E-CADHERIN, N-CADHERIN, VIMENTIN and SLUG) and neuroendocrine marker ASCL1 in 5 Crebbp wide-type and 5 Crebbp-deficient mouse SCLC tumor tissues. Beta-ACTIN was used as a loading control.
B, Immunoblotting of CREBBP, EMT markers (ZEB1, E-CADHERIN, N-CADHERIN, VIMENTIN and SLUG) and neuroendocrine marker ASCL1 in 6 Crebbp wide-type and 6 Crebbp-deleted murine SCLC cell lines.
C, Representative images of immunofluorescence staining of CREBBP and EMT markers (E-CADHERIN, VIMENTIN and ZEB1) in Crebbp wide-type and Crebbp-deleted mouse SCLC tumors. Nuclear DNA stained using DAPI. Original magnification, 40×.
D, Immunoblotting of CREBBP, ZEB1, E-CADHERIN and SLUG protein levels in murine preSC cells with or without Crebbp knockdown using shRNAs. Beta-ACTIN was used as loading control.
E, Immunoblotting of CREBBP, ZEB1 and E-CADHERIN protein levels in human SCLC cell line DMS53 with or without CREBBP knockout using sgRNAs. Beta-ACTIN was used as loading control.
CREBBP restoration results in increased CDH1 expression and impaired proliferation
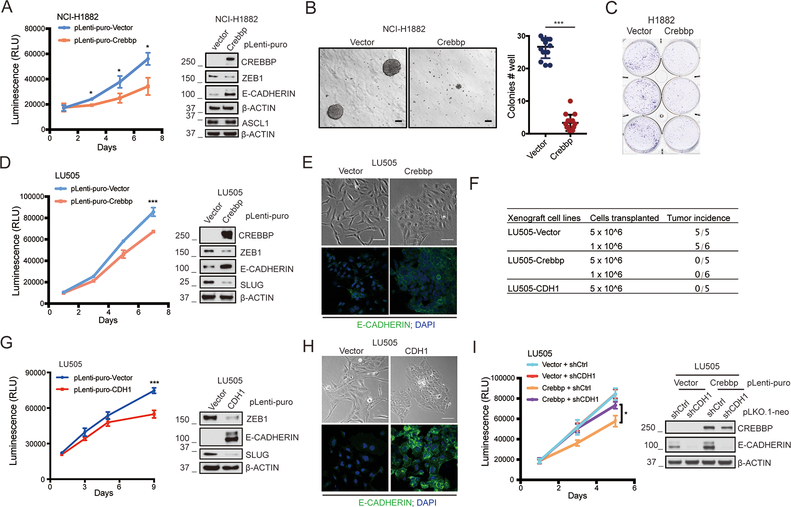
To better understand the functional effects of CREBBP inactivation in SCLC we performed a rescue experiment using lentiviral expression of CREBBP in NCI-H1882, a human SCLC cell line that lacks expression of CREBBP owing to a large deletion (Supplemental Figure S9A). Restoration of CREBBP expression in NCI-H1882 resulted in reduced cell proliferation, diminished growth capacity in soft agar, reduced colonies upon low density plating and increased E-CADHERIN expression (Figure 5A-C). CREBBP re-expression also led to increased E-CADHERIN and reduction in ZEB1 (Figure 5A). We extended this experiment to a cell line that we derived from an SCLC PDX model, LU505. Targeted sequencing and copy number variation analysis indicated that LU505 harbors a genomic deletion encompassing CREBBP exon 1 which leads to lack of CREBBP expression (Figure 5D, Supplemental Figure S9B and S10A). Features of LU505 include high expression of NEUROD1 and of mesenchymal markers ZEB1 and SLUG along with low expression of ASCL1 and MYCL amplification/overexpression (Supplemental Figure S10). This PDX model has classic SCLC morphology in vivo, although cells derived from LU505 and expanded in cell culture exhibit mesenchymal features (Supplemental Figure S10B,C). Overall, this PDX model has some features of a “variant” form of SCLC (NEUROD1 expression, low levels of ASCL1 and adherent growth in culture) while also exhibiting features of more typical SCLC (“classic” histology and MYCL amplification/overexpression). Lentiviral expression of CREBBP in LU505-derived cells expanded in culture also led to increased E-CADHERIN and reduced levels of ZEB1 and SLUG (Figure 5D). These molecular changes coincided with reduced proliferation and a switch from an elongated and fibroblast-like morphology, to polygonal morphology with epithelial cell features and patchy growth (Figure 5E). Restoration of CREBBP also completely eliminated growth of these cells in immunocompromised mice, compared to control vector infected LU505 cells that readily formed tumors (Figure 5F, Supplemental Figure S11).
Figure 5. CREBBP re-expression in CREBBP-deleted human SCLC cells inhibits transformation.
A, CellTiter-Glo viability assay of ectopic expression of CREBBP in human CREBBP-deficient NCI-H1882 cells (n= 3 independent experiments). *, p<0.05. Immunoblotting of CREBBP, ZEB1, ASCL1 and E-CADHERIN were performed in these cells.
B, Anchorage-independent assay to test impact of CREBBP overexpression on growth of NCI-H1882 cells in soft agar. Cells were seeded at 1.0×105 cells/well (6 well plate). n = 3 independent experiments. ***, p<0.001.
C, Colony formation assay to test impact of CREBBP overexpression on ability of NCI-H1882 cells to grow when plated at low density (n= 3 independent experiments). 3 representative technical replicates per condition were shown.
D, CellTiter-Glo viability assay of CREBBP overexpression in human CREBBP-deficient LU505 cells (derived from a PDX tumor). n= 3 independent experiments. ***, p<0.001. Immunoblotting of CREBBP, ZEB1, E-CADHERIN and SLUG were performed in these cells.
E, Representative phase contrast microscopic photos of LU505 cells with or without CREBBP overexpression (Upper). Scale bar = 100μm. Representative immunofluorescence images of E-CADHERIN staining in LU505 cells with or without CREBBP overexpression (Lower). DAPI was used as a nuclear stain. Original magnification, 40×.
F, Summary of differences in tumor-initiating ability of LU505-vector, LU505-CREBBP and LU505-CDH1 cells upon transplantation of 5 × 106 cells or 1 × 106 cells into immunocompromised NSG mice.
G, CellTiter-Glo viability assay of CDH1 overexpression in LU505 cells (n= 3 independent experiments). ***, p<0.001. Immunoblotting of ZEB1, E-CADHERIN and SLUG were performed in these cells.
H, Representative phase contrast microscopic photos of LU505 cells with or without CDH1 overexpression (Upper). Scale bar = 100μm. Representative immunofluorescence images of E-CADHERIN staining in LU505 cells with or without CDH1 overexpression. DAPI was used to stain nuclear. Original magnification, 40×.
I, CellTiter-Glo viability assay of shRNA-mediated CDH1 knockdown on the proliferation suppression induced by CREBBP overexpression in LU505 cells (n= 3 independent experiments). *, p<0.05. Re-expression of CREBBP and knockdown of CDH1 in this cell line were validated by immunoblotting.
Interestingly, ectopic CDH1 overexpression was sufficient to reduce proliferation and decreased the levels of ZEB1 and SLUG in LU505 (Figure 5G). Overexpression of CDH1, like CREBBP, also eliminated tumorigenesis of LU505 cells in immunocompromised mice (Figure 5F). CDH1 overexpression also phenocopied the acquisition of epithelial cell morphology seen in the CREBBP-restored cells (Figure 5H). To determine whether this axis of CDH1 expression downstream of CREBBP is functionally important, we re-expressed CREBBP and simultaneously suppressed CDH1 using lentiviral shRNAs. CDH1 knockdown partially blocked the decrease in LU505 proliferation mediated by CREBBP restoration (Figure 5I). Consistent with the notion that changes in the EMT pathway can enable tumor invasive abilities, overexpression of either CREBBP or CDH1 decreased the migratory capacity of LU505 cells in a transwell migration assay (Supplemental Figure S12A,B). Taken together, our findings suggest that CREBBP suppresses tumorigenesis and restrains EMT in part through positive regulation of CDH1.
Crebbp loss leads to epigenetic suppression of CDH1 and other cell adhesion genes
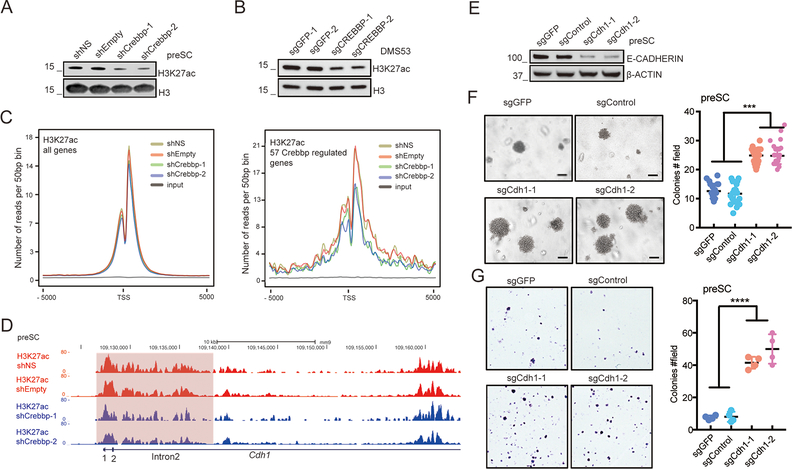
CREBBP acetylates multiple histone residues, including histone H3 lysine 27 (H3K27), a key site for transcriptional enhancer activation (7). We hypothesized that the reduced histone acetylation following Crebbp deletion might contribute to the reduced expression of CDH1. Crebbp knockdown in preSC cells resulted in a global decrease in acetylated H3K27 (H3K27Ac) (Figure 6A, also see Figure 4D). A similar result was seen with CREBBP deletion in the human DMS53 cell line (Figure 6B). We performed ChIP-Seq analyses to examine H3K27Ac levels surrounding the transcriptional start sites (TSS) across the genome of preSC cells and found Crebbp knockdown did not correlate significantly with a reduction in H3K27Ac in these regions (Figure 6C). However, limiting our analysis to the 57 genes consistently downregulated upon Crebbp knockdown in preSC cells and in the neuroendocrine tumors previously described (Figure 3), we found that Crebbp knockdown in preSC cells exhibited reduced H3K27Ac associated with these core CREBBP regulated genes (Figure 6C). Further examining the pattern of H3K27Ac at the Cdh1 gene, Crebbp knockdown in preSC cells resulted in reduced H3K27Ac levels in potential enhancer regions extending from intron 1 through intron 2 (Figure 6D). Other core Crebbp-regulated adhesion genes such as Cldn6, Cldn3, Cldn9, Tjp3 and Sdc4 also exhibited reduced H3K27 acetylation in response to Crebbp knockdown (Supplemental Figure S13). These results suggest a role for CREBBP-mediated H3K27 acetylation in regulating the expression of CDH1 and other CREBBP regulated cellular adhesion genes.
Figure 6.
Epigenetic control of cell adhesion molecule expression by CREBBP contributes to transformation of SCLC.
A, Knockdown of Crebbp decreases global H3K27ac levels in preSC cells. Histone H3 was used as loading control.
B, Knockout of CREBBP decreases global H3K27ac level in human SCLC cell line DMS53. Histone H3 was used as loading control.
C, Metaplots of the H3K27ac distribution across all transcripts and across the core 57 genes consistently downregulated upon Crebbp suppression in preSC cells and in the 3 neuroendocrine tumor types. Data from two controls (non-silencing shRNA and empty vector) and two Crebbp shRNAs (shCrebbp-1 and shCrebbp-2) along with input are shown. TSS, transcriptional start site. −5000 and 5000 represent base pairs upstream and downstream of the TSS.
D, ChIP-seq read density plots shows decreased H3K27ac levels in introns 1 and 2 of Cdh1 in two Crebbp knockdown preSC cells (shCrebbp-1 and shCrebbp-2; blue) compared to two control preSC cells (shNS and shEmpty; red).
E, Immunoblotting of E-CADHERIN protein in 2 control preSC cells and 2 sgRNA-mediated Cdh1 knockout preSC cells. Beta-ACTIN was used as loading control.
F, Anchorage-independent assay of sgRNA-mediated Cdh1 knockout on the growth ability of preSC cells in soft agar. Representative images of colonies in soft agar are shown. Cells were seeded at 2.5×105 cells/well (6 well plate). The number of colonies from 15 fields was counted. ***, p<0.001. n= 3 independent experiments. Scale bar = 100μm.
G, Colony formation assay of Cdh1 knockout on the growth ability of preSC cells. Cells were seeded at 6×103 cells/well (6 well plate). Representative images are shown. The number of colonies from 4 fields representing the entire well was counted. ****, p<0.0001. n= 3 biological replicates.
CDH1 suppression increases transformation
To evaluate importance of Cdh1 downregulation for cellular transformation, we deleted Cdh1 in preSC cells using lentiviral CRISPR-Cas9 vectors and verified that this led to decreased E-CADHERIN expression (Figure 6E). Cdh1 deletion promoted anchorage-independent growth of targeted preSCs in soft agar and increased their colony-forming capacity upon low-density plating (Figure 6F and 6G). These results indicate that the inactivation of Cdh1 promotes cell transformation, a necessary step toward full-blown tumor development. On the other hand, re-expression of CDH1 inhibited proliferation and eliminated tumor-initiating ability of CREBBP-deleted LU505 cells (Figure 5F and 5G). Thus, Cdh1 itself exhibits properties of a tumor suppressor gene in SCLC.
Histone deacetylase inhibition suppresses effects of Crebbp deletion
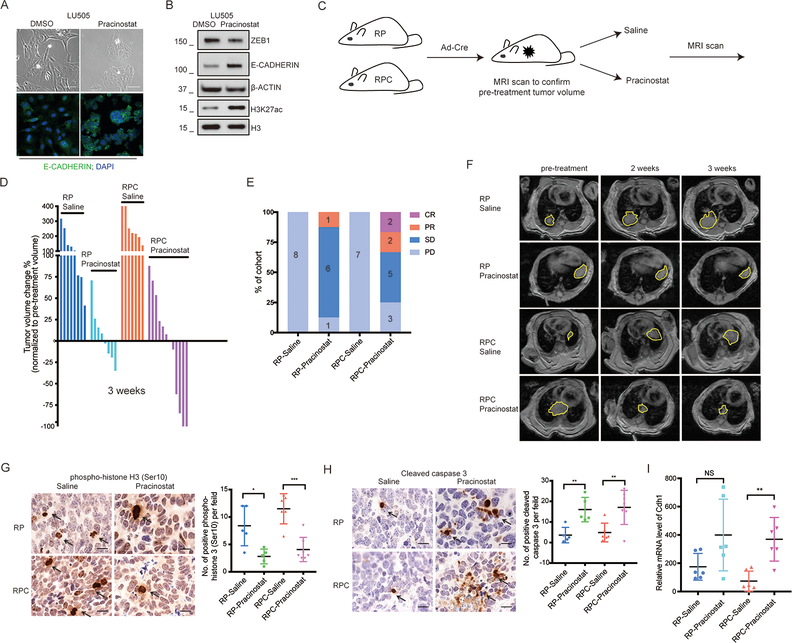
The balance of histone acetylation and deacetylation play critical roles in the regulation of gene expression. Effects of histone acetyltransferases such as CREBBP are opposed by histone deacetylases (HDACs). We show that CREBBP inactivation leads to reduced acetylation of H3K27 and transcriptional repression of CDH1. If reductions in acetylation upon CREBBP deletion contribute to reduced gene expression, then the effects could potentially be reversed with HDAC inhibition. We employed an inhibitor of class I,II and IV HDACs, Pracinostat (32), currently being tested in advanced clinical trials outside of SCLC, including a Phase III trial in acute myeloid leukemia (Clinical trials ID: NCT03151408). In a cell line that we derived from the human CREBBP-deleted PDX model LU505, we found that Pracinostat treatment resulted in increased E-CADHERIN (Figure 7A,B), which correlated with increased global H3K27Ac (Figure 7B). Pracinostat treatment also led to a global increase in H3K27Ac, H3K18Ac and increased E-CADHERIN expression in DMS53 human SCLC cells with CRISPR-generated CREBBP-deletion (Supplementary Figure S14). These findings suggest that Pracinostat treatment can partially reverse effects of CREBBP loss on CDH1 expression. We mined human SCLC cell line data from a study that tested response of 66 human SCLC cell lines to a large panel of small molecule inhibitors (33). We found a significant inverse correlation between activity of Pracinostat and expression of CREBBP (Supplemental Figure S15) suggesting that loss of CREBBP may increase responsiveness to this therapeutic approach.
Figure 7. Enhanced efficacy of HDAC inhibition with Crebbp mutation in SCLC.
A, Representative phase contrast microscopic photos (top) and E-CADHERIN immunofluorescence (bottom) in LU505 cells treated with DMSO or Pracinostat (125nM) for 21 days (Upper). Scale bar = 100μm. DAPI was used to stain nucleus. Original magnification, 40×.
B, LU505 SCLC cells treated with DMSO or Pracinostat for 21 days were probed for ZEB1, E-CADHERIN and SLUG (whole cell lysates) and H3K27ac and H3 (acid extraction of histones).
C, Schematic of treatment trial with saline (control) and Pracinostat in RP (Rb1/Trp53 mutant) and RPC (Rb1/Trp53/Crebbp mutant) SCLC autochthonous models with lung tumor burden detected by MRI. Pracinostat dosed at 100mg/kg orally, 5 times per week.
D, Tumor volume changes (%) based on MRI scan quantification in RP and RPC mice treated with saline and Pracinostat for 3 weeks, normalized to pre-treatment tumor volume.
E, Treatment response divided into PD (progressive disease, >30% tumor volume increase), SD (stable disease, < 30% change in tumor volume in either direction), PR (partial regression >30% regression) and CR (>90% regression) in RP and RPC mice treated with saline and Pracinostat. Data are presented as percentage of mice in each treatment group.
F, Representative MRI images of the thorax regions of mice treated with saline or Pracinostat at pre-treatment compared to 2- and 3-weeks of treatment in each group.
G-H, Representative IHC staining of phospho-histone H3 (Ser10) and cleaved caspase 3 in primary SCLC tumors of mice treated with saline vs. Pracinostat at 3 weeks (end of treatment). Scale bar = 20μm. Data are presented as number of positive phospho-histone H3 (Ser10) cells per field. The p value between different groups was calculated using unpaired student’s t-test. RP-saline, n= 5 mice, RP-Pracinostat, n = 5 mice, RPC-saline, n = 7 mice and RPC-Pracinostat, n = 6 mice. *, p<0.05. **, p<0.01. ***, p<0.001.
I, Quantitative reverse-transcription PCR analysis of Cdh1 mRNA levels in primary SCLC tissues derived from RP and RPC mice treated with saline or Pracinostat at 3 weeks. n = 6 mice in each treatment group. NS, not significant. **, p<0.01, unpaired student’s t-test.
To determine whether Pracinostat exhibits therapeutic efficacy in SCLC, we employed the autochthonous Rb1/Trp53 and Rb1/Trp53/Crebbp models. We screened mice by MRI for lung tumor burden and entered mice into treatment groups upon detection of adequately sized tumors (see methods). Mice underwent subsequent MRI at 2- and 3-weeks of treatment followed by animal euthanasia and tumor analyses (Figure 7 C-F and Supplemental Figure S16). Focusing on the 3-week time point, in saline treated Rb1/Trp53 and Rb1/Trp53/Crebbp models, all mice exhibited progressive disease (PD) (> 30% increase in tumor volume) (Figure 7D-F). In the Rb1/Trp53 model treated with Pracinostat, only 1/8 animals exhibited PD, while 6/8 exhibited stable disease and 1/8 a partial response (PR) (>30% decrease in tumor volume). Thus, Pracinostat exhibits efficacy as monotherapy in the Rb1/Trp53-deleted autochthonous model. We also tested Pracinostat in the Rb1/Trp53/Crebbp-deleted model. Of 12 Pracinostat treated Rb1/Trp53/Crebbp mice, 4 exhibited partial or complete responses, which included 2 complete regressions, and 2 strong regressions (84% and 62% reductions in tumor volume). Stable disease was seen in an additional 5 mice (Figure 7D,E). We could not study molecular properties of the exceptional responders, owing to a lack of tumor material available; however, of the tumors we could analyze, Pracinostat treatment in both Rb1/Trp53 and Rb1/Trp53/Crebbp models resulted in reduced proliferation, as shown by reduced phospho Ser-10 histone H3 positive cells, and increased apoptosis, as shown by cleaved caspase-3 immunostaining (Figure 7G, H). In the Pracinostat-treated Rb1/Trp53/Crebbp SCLC tumors, we also observed increased expression of CDH1 (Figure 7I). Thus, although Pracinostat exhibits efficacy as monotherapy regardless of Crebbp status, we found that Crebbp deletion in SCLC results in the potential to elicit striking responses, including complete regressions.
Discussion
CREBBP is a tumor suppressor gene in SCLC
CREBBP is one of the most frequently mutated genes in human SCLC yet the functional contribution of CREBBP inactivation to SCLC has been largely uncharacterized. This study definitively demonstrates that CREBBP functions as a tumor suppressor not only in SCLC but also across other neuroendocrine tumor types. CREBBP tumor suppressor activity has been best described in lymphoma and leukemia, where Crebbp inactivation in mouse models led to tumor acceleration (13,15–17). This was in part mediated by impaired acetylation and reduced activity of another tumor suppressor, p53 (13,17). In contrast, CREBBP perturbation in neuroendocrine cells/tumors consistently leads to p53-independent changes in the expression of genes related to tight junctions and cell adhesion, including CDH1, and we show that CDH1 itself exhibits properties of a tumor suppressor in SCLC. Our observation that CREBBP controls adhesion molecule expression in driving SCLC is a novel finding that has not been described in CREBBP-mutant leukemia/lymphoma. These findings illustrate the importance of cellular context for CREBBP function as a tumor suppressor.
CREBBP regulation of CDH1 and cellular adhesion genes in SCLC
Neuroendocrine tumors have common features and we leveraged the generation of three different Rb/p53-deleted neuroendocrine tumor types that were each accelerated by Crebbp loss to hone in on consistently regulated CREBBP targets. We further refined our identification of core CREBBP-regulated transcripts by suppressing CREBBP expression in preSC cells. Among the 66 genes commonly deregulated with Crebbp suppression across each of these 4 comparisons, we found that tight junction and adhesion genes were strongly enriched (Figure 3H). We were particularly interested in the reduced expression of Cdh1 upon Crebbp loss, as we initially hypothesized that CREBBP positively controls tumor suppressor pathways relevant to SCLC and CDH1 is a tumor suppressor gene, mutated in gastric, breast, bladder and other cancers (34–36). While CDH1 itself is not a target of inactivating mutations in SCLC, loss of CDH1 expression has been implicated in SCLC chemoresistance (37) and in advanced murine SCLC (38). Moreover, downregulation of CDH1, a key regulator of EMT, and upregulation of EMT-associated genes occurs in normal neuroendocrine cell development as ASCL1-positive lung neuroendocrine cells migrate to form neuroendocrine cell bodies (31). Crebbp suppression in SCLC may contribute to epigenetic reactivation of a pathway active in developing neuroendocrine cells, which are likely major cells of origin for SCLC (21). CREBBP suppression resulted in increased expression of EMT markers such as VIMENTIN and ZEB1 (Figure 4) with the converse effects were observed upon CREBBP reintroduction to CREBBP-null SCLC cells (Figure 5). It has increasingly become appreciated that functionally important EMT-like programs in cancer are often only partially activated, without overt mesenchymal transformation (39,40). Importantly, ASCL1 and other neuroendocrine markers were found to be co-expressed with EMT markers in Crebbp-deleted SCLC. These data suggest that CREBBP loss drives partial acquisition of mesenchymal programs, while still maintaining neuroendocrine features. This is consistent with the “classic” SCLC histology observed in the Rb1/Trp53/Crebbp Ad-CGRP Cre mouse model that was similar to Rb1/Trp53 controls.
Our finding that CREBBP inactivation leads to a reduction in CDH1 expression and partial acquisition of mesenchymal features has important clinical relevance. In clinical samples, positive expression of E-CADHERIN was significantly associated with a better outcome in SCLC patient samples, while expression of mesenchymal markers, such as VIMENTIN and SNAIL1 was associated with a worse outcome (41). Factors upstream of E-CADHERIN expression in SCLC have been poorly understood, and we link a major SCLC driver gene as a key regulator of E-CADHERIN expression. The full breadth of phenotypes caused by CREBBP loss in SCLC remains to be determined. In a set of 51 SCLC cell lines, expression of E-CADHERIN was a top marker of sensitivity to cisplatin (37). It will be critical for future studies to determine whether reduced E-CADHERIN upon CREBBP loss contributes to chemoresistance in SCLC. Notably, we found a high rate of liver metastasis in both the Rb/p53/Crebbp and Rb/p53 models without clear differences between these genotypes (Supplemental Figure S2). Nonetheless, detailed examination of the consequences of Crebbp loss on each step in the metastatic cascade is warranted.
HDAC inhibitors have been employed in clinical trials without success in SCLC, but there have been no attempts to select SCLC patient subsets for clinical trial entry. Our study suggests a new avenue to target CREBBP-mutant SCLC, using inhibitors of histone deacetylases as a strategy to restore H3K27Ac and CDH1 expression following CREBBP deletion. Pracinostat, an HDAC inhibitor with excellent pharmacokinetic characteristics has yielded promising data in AML that has led to its current testing in a Phase III clinical trial for this tumor type. Our observations of efficacy in the Rb1/Trp53 autochthonous model support investigation of Pracinostat broadly in SCLC. Moreover, our observations of examples of complete regressions in the context of Crebbp-inactivated SCLC further suggest that targeting this approach to subsets of SCLC patients most likely to respond would increase potential for clinical success. We focused our study on CREBBP, but mutations in either CREBBP or the related acetyltransferase EP300, both prevalent in human SCLC (1) could be used to stratify patients to treatment in SCLC. Future studies using the Crebbp-deficient mouse model should also test Pracinostat efficacy together with cisplatin-etoposide chemotherapy, the mainstay of first-line SCLC therapy in the clinic. The preclinical mouse model we developed will also be invaluable to test other ideas to specifically target the substantial subset of SCLC that harbors CREBBP-inactivation. For example, observed synthetic lethality between CREBBP and EP300 in some contexts could lead to a specific sensitivity of CREBBP-deleted SCLC towards pharmacological inhibition of EP300 (42,43).
In summary, we have demonstrated that CREBBP plays a critical role in SCLC tumor suppression. The mouse model of SCLC that we have generated will help delineate mechanisms underlying CREBBP tumor suppression in SCLC and will be an ideal tool to test novel approaches to treating CREBBP-mutant SCLC.
Materials and Methods
Mice
We thank Drs. Tyler Jacks, Anton Berns and Jane Johnson for Rb1lox/lox, Trp53lox/lox and Ascl1-CRE-ERT2 strains, respectively. We thank Dr. Jan van Deursen (Mayo Clinic) for providing the Crebbp floxed strain (14). The Rb1lox/lox/Trp53lox/lox Adeno-Cre model of SCLC has been reported previously (19). We employed Ad-CGRP-Cre (21), which uses a neuroendocrine specific promoter to drive Cre expression; this was obtained from the University of Iowa Gene Vector Core, with permission of Dr. Anton Berns. The floxed Crebbp mice were bred to compound Rb1lox/lox;Trp53lox/lox mice to obtain Rb1lox/lox;Trp53lox/lox;Crebbplox/lox mice used for intra-tracheal infections with Ad-CGRP-Cre. The titer of adenoviral vector used in this study was 1.25 × 109 pfu per mouse. Ascl1CreERT2 knock-in mice, previously described (22) were bred to the Rb1lox/lox;Trp53lox/lox and Rb1lox/lox;Trp53lox/lox;Crebbplox/lox mice to obtain Rb1lox/lox;Trp53lox/lox;Ascl1CreERT2 and Rb1lox/lox;Trp53lox/lox;Crebbplox/lox;Ascl1CreERT2 mice. Genotypes of these mice were confirmed by PCR. Intra-tracheal infection was used to deliver adenoviral vectors expressing Cre recombinase to lungs of adult mice as described previously (44). Tamoxifen (TAM) induction of Cre recombinase under the control of Ascl1 promoter was accomplished by intraperitoneal injection of mice with 150 mg/kg/day TAM, prepared in corn oil, over five consecutive days. Mice were monitored every week after virus infection or TAM injection. Mice were euthanized when moribund with labored breathing (Ad-CGRP-Cre study) or when they exhibited poor body condition or other symptoms of advanced pituitary tumor burden (Ascl1Cre-ERT2 study). Tumor tissues were used for primary cell culture in media optimized for SCLC. Tumor fragments were also frozen for molecular and histological analyses as well as immunofluorescence experiments. After excising tumor pieces for molecular analyses, the whole lung was inflated with neutral buffered formalin (NBF) and processed for histological analyses. All animal procedures related to Figure 1B-D were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Virginia, accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). The IACUC at the Fred Hutchinson Cancer Research Center approved the remaining animal procedures described.
Subcutaneous allografts
A total of 5.0X105 of control or Crebbp-targeted preSC cells were injected into the flanks of immune-compromised mice (Hsd: Athymic Foxn1nu) purchased from Envigo. The injected mice were maintained and observed for palpable tumors according to procedures approved by IACUC and euthanized when tumor size reaches 2 cm in diameter, the end-point of allograft study under the guideline of the institutional animal policy. Kaplan-Meier curve was used to plot the time of survival (maximal tumor volume). For xenograft experiments, LU505 cells expressing control vector, Crebbp or CDH1 were re-suspended in a 100μl of 50% Matrigel/DMEM mix and injected into the flanks of NSG mice. Mice were euthanized 8 weeks after the injection of 5.0 × 106 cells or when tumors reached maximum tumor volume (2000 mm3) when 1.0 × 106 cells were injected. Gross tumors were photographed and fixed in NBF for histological analysis.
HDAC inhibitor treatments
Rb1/Trp53 and Rb1/Trp53/Crebbp mice were infected with Adeno-Cre and monitored by MRI scan (ICON small animal MRI system, Bruker Biospin) to identify mice with lung tumor burden. Respiratory gating was employed and a total of 15 slices of 1 mm thickness was acquired to cover the entire lung volume. Mice were anesthetized with isoflurane during imaging. Tumor volume per animal was quantified using 3D ImageJ Suite with manual quantification of consecutive axial image slices. Once tumor size met defined criteria (present on 3 consecutive 1mm slices), mice were treated with saline (control) or Pracinostat for 3 weeks. Pracinostat was provided by MEI Pharma. 10 mg/ml Pracinostat was prepared in 0.5% methylcellulose, 0.1% Tween 80 in sterile water. Pracinostat was given using oral gavage 100mg/kg for 3 weeks (daily, 5 days per week). Saline was also given daily for 3 weeks (daily, 5 days per week). MRI was performed to follow tumor volume after treatment start and weights were monitored daily during the course of treatment.
Histology and Immunohistochemistry
Mouse lungs, lung tumors, pituitary and thyroid tumors were fixed in NBF for 24 hours and then transferred to 70% ethanol before paraffin embedding. 4 μm-thick tissue sections were stained with hematoxylin and eosin (H&E) or used for immunohistochemistry performed using standard procedures. Anti-PCNA (1:1000; Rabbit polyclonal, ab18197, Abcam) and anti-phospho-histone H3 (1:500; 06–570, EMD Millipore) were used as markers of proliferating cells. Anti-cleaved caspase 3 (1:100; #9661, Cell Signaling Technology) was used as a marker of apoptosis. Immunofluorescence using paraffin sections was performed on preSC allografts with anti-UCHL1 (Sigma, HPA005993) primary antibody and Alexa 488 fluor-conjugated secondary antibody (Invitrogen). For immunofluorescence of mouse SCLC tumors, fresh lung tumor tissues were fixed in 4% paraformaldehyde (PFA) overnight at 4°C, then transferred to 30% sucrose at 4°C overnight and then embedded in Tissue-Tek OCT solution and stored at −80°C. 8 μm-thick serial sections were cut using a Leica CM1950 cryostat at −20°C. For immunofluorescence staining of cultured adherent cells, cells were first seeded onto 8-well chamber slides (354688, Corning). After cells adhered, slides were washed with PBS and fixed in 4% PFA for 10 minutes at room temperature. Immunofluorescence staining was performed with standard procedures. The following primary antibodies were used: anti-CGRP (1:1000, Rabbit polyclonal, Sigma), anti-CGRP (1:100, Guinea Pig, Peninsula Laboratories), anti-TTF-1 (1:100; Rabbit polyclonal, Cell Signaling Technology), anti-E-CADHERIN (1:200; Rabbit polyclonal, Cell Signaling Technology), anti-CREBBP (1:100; Rabbit polyclonal, Cell Signaling Technology), anti-VIMENTIN (1:200; Rabbit polyclonal, Cell Signaling Technology) and anti-ZEB1 (1:50; Rabbit polyclonal, NBP1–05987, Novus). Dylight 554 Phalloidin was used to stain F-actin (1:200, Cell Signaling Technology). Tissues/cells were dual-labelled using second antibodies: Alexa Fluor 488 Goat Anti-Rabbit IgG (A11034, ThermoFisher) or Alexa Fluor 594 Goat Anti-Guinea Pig (A11076, ThermoFisher). Fluorescence images were obtained using a Zeiss LSM 700 confocal microscope. For preserving fluorescence and nuclear counter staining, anti-fade reagent with DAPI (Vector Laboratories) was used. H&E and immunostained images were acquired using Nikon Eclipse microscope.
Protein extraction and western blot analyses
The T-PER Tissue protein extraction reagent (ThermoFisher) was used to extract protein from mouse tumor tissues and cultured human and murine cell lines following the manufacturer’s procedure. The immunoblot experiments were performed with standard procedures. The following antibodies were used in this study: anti-CREBBP (7389; Cell Signaling Technology), anti-E-CADHERIN (3195; Cell Signaling Technology), anti-N-CADHERIN (13116; Cell Signaling Technology), anti-ZEB1 (3396; Cell Signaling Technology), anti-SLUG (9585; Cell Signaling Technology), anti-VIMENTIN (5741; Cell Signaling Technology), anti-ASCL1 (556604, BD Biosciences) anti-NEUROD1 (ab60704; Abcam), and anti-β-ACTIN (A3854; Sigma).
Histone extraction
Total histone proteins were extracted by acid extraction as described previously (45). Briefly, cells were suspended in hypotonic buffer and lysed at 4°C for 30 minutes. Lysates were centrifuged at 10,000g for 5min and the supernatant was discarded. The pellets were re-suspended in 400μL of 0.4N H2SO4, and incubated on a rotator overnight at 4°C. Samples were centrifuged at 10,000g for 10min at 4°C, and the supernatant was transferred to a new tube. To precipitate histones, 132μL TCA was added solution mixed by inverting. The mixtures were incubated on ice for 1 hour. Total histones were pelleted by centrifugation at 10,000g for 10min at 4°C, and washed twice with ice-cold acetone. Histone pellets were dissolved in H2O. Primary antibodies against H3K27ac (8173, Cell Signaling Technology) and total Histone H3 (3638, Cell Signaling Technology) were used in the immunoblot experiments.
Plasmids and reagents
The plasmids used in this study are summarized in Supplementary Table S5. The lentiCRISPRv2 (a gift from Feng Zhang) was used to knock out the human CREBBP gene in human SCLC cells and murine Cdh1 gene in preSC cells. The pL-CRISPR.EFS.tRFP (a gift from Benjamin Ebert #57819) was used to mutate murine Crebbp. Guide RNA (sgRNA) sequences for Crebbp and Cdh1 genes were generated using the online CRISPR design tool at crispr.mit.edu and the sequences provided in Supplemental Table S5. The lentiviral vectors pLKO.1-TRC (#10878, addgene, a gift from David Root) and pLKO.1-neo (#13425, addgene, a gift from Shiela Stewart) were used to knock down mouse Crebbp and human CDH1 genes in SCLC cells, and the shRNA sequences are provided in Supplemental Table S5. The pLenti-puro lentiviral vector (#39481, addgene, a gift from le-Ming Shih) was used to subclone coding sequences of mouse Crebbp and human CDH1 genes. The cDNA of mouse Crebbp was cloned out of the MSCV-IRES-CBP vector into the pLenti-puro vector. The cDNA of human CDH1 gene was cloned from the CDH1-GFP vector (#28009, addgene, a gift from Jennifer Stow) into the pLenti-puro vector. Lentivirus production was performed by co-transfecting the transfer plasmids with 2 packaging plasmids (psPAX2 and pMD2.G, gifts from Didier Trono) into 293FT cells using Lipofectamine 2000 (Invitrogen). Supernatants containing the lentiviral particles were harvested 48 hours after transfection and filtered through 0.45μm PVDF filter (Millipore). Infections were performed with viral supernatant in the presence of 8μg/ml polybrene (H9268, Sigma-Aldrich). Puromycin (A1113803, ThermoFisher) and G418 (10131035, Fisher) were used to select stably transduced cells.
Cell lines, cell culture and generation of knockdown and CRISPR-mediated knockout cells
Human SCLC cell lines (DMS53 and NCI-H1882) and 293FT cells, obtained from ATCC, were cultured in normal DMEM media supplemented with 10% FBS and Pen/strep. Murine SCLC cell lines and the human PDX LU505 SCLC model were cultured in DMEM supplemented with 15% FBS, 1mM sodium pyruvate, 100μm beta-mercaptoethanol and 10μg/ml insulin and Pen/strep. The LU505 cells were extracted from a SCLC PDX model generated and utilized as previously described (46). preSC cells were cultured in RPMI-1640 media supplemented with 10% FBS and Pen/Strep. No Mycoplasma testing was performed. HDAC inhibitor Pracinostat (MEI pharma) dissolved in DMSO was used to treat LU505 and DMS53 cells. DMSO was used a vehicle control. To delete CREBBP in the human SCLC cell line DMS53, a single cell clone was isolated and expanded from the parental DMS53 cells. This single clone was further infected with lentiCRISPRv2-sgRNAs targeting exon9 of the human CREBBP gene (Supplemental Table S5). Puromycin was used to select stably transduced cells, and single clones with CREBBP deletion were isolated and validated by PCR based sequencing and immunoblot. Two control sgRNAs were used in this study (Supplemental Table S5). To generate Cdh1 knockout preSC cells, the parental cells were infected with the lentiCRISPRv2-sgRNAs targeting exon1 and exon9 of the mouse Cdh1 gene respectively (Supplemental Table S5). Puromycin was used to select stably transduced cells, and preSC polyclonal cells with Cdh1 knockout were validated by immunoblot. To generate Crebbp-mutant preSCs, the parental cells were transfected with CRISPR/Cas9-RFP vector carrying single sgRNAs which target the sequence encoding the HAT domain of CREBBP or empty vector (control) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction. Forty-eight hours later, the RFP-positive cells were sorted using FACS (BD Influx cell sorter).
Nucleic acid extraction, RT-qPCR, and RNA-seq experiments
Total genomic DNA was isolated from cells, and PCR was performed to amplify fragments containing target areas using specific primer sets described in Supplemental Table S5. The same primers were also used for sequencing the PCR fragments. The sequencing results were visualized using Snapgene software. Total RNA from tumors or cultured cells were isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized using the Protoscript II First Strand cDNA Synthesis Kit (New England Biolabs) or iScript reverse transcription supermix for RT-qPCR (BIO-RAD). Quantitative PCR was performed with All-in-One qPCR mix (GeneCopoeia). Quantitative expression data were acquired and analyzed with a 7900 Real-time PCR System (Applied Biosystems) using power SYBR Green PCR master mix (Thermo Fisher Scientific) and StepOnePlus System (Applied Biosystems). Primers were designed to detect targeted genes and the sequences of each primer are provided in Table S5. RNA-seq libraries were generated using the NEBNext Ultra RNA Library Prep Kit for Illumina (E7530L, New England BioLabs Inc) following the manufacturer’s protocol. Oligo dT purified mRNA from 500ng of total RNA was used as starting material. Libraries were submitted to the genomics core facility at Fred Hutchinson Cancer Research Center. 50-bp single-end sequencing was performed on an Illumina HiSeq 2500 system. Reads were mapped to the mm9 genome using TopHat (47) and the fragment per kilobase per million (FPKM) values were generated using Cuffdiff (48). Differentially expressed genes regulated by Crebbp were identified using the edgeR package (29). FPKM values of each sequenced sample were used for Gene Set Enrichment Analysis (GSEA, (http://www.broadinstitute.org/gsea/))(27). For all the mouse tumor tissues and matched preSC cell lines, gene set permutation type was used. The Hallmark gene sets in the Molecular Signatures Database collection was selected for each of the analyses (28). Genomic data has been deposited to an appropriate repository with GEO accession number GSE117552.
Chromatin immunoprecipitation and ChIP-seq library preparation and sequencing
ChIP experiments in preSC cells were performed using the ChIP-IT High Sensitivity Kit (53040, Active Motif) following the manufacturer’s manual with some modifications. Briefly, 20 million cells were fixed using the cell fixative solution provided by the kit followed by nuclear preparation and sonication (Covaris E220, Covaris) to obtain short fragment chromatin (~200 bp). 5μg H3K27ac antibody (39685, Active Motif) was added to lysate of sheared chromatin and incubated at 4°C overnight. Enriched chromatin was collected by Protein G agarose beads and then processed by reverse cross-link and DNA purification. ChIP-Seq libraries were generated using the NEBNext ChIP-Seq Library Prep Reagent Set for Illumina (E6200L, New England BioLab Inc) following the manufacturer’s protocol. 10ng of ChIP DNA or input DNA was used to generate the libraries. Libraries were submitted to the genomics core facility at Fred Hutchinson Cancer Research Center. 50-bp single-end sequencing was performed on an Illumina HiSeq 2500 system. Reads were aligned to the mm9 and hg19 versions of the mouse and human genomes, respectively, using Burrows-Wheeler Aligner (BWA, version 0.7.12) (49). Only reads with mapQ > 20 were used for subsequent analyses. Read depth normalized bigwig files for read density visualization were generated using Homer (50). H3K27Ac profiles around Transcription Start Sites were calculated with the ‘normalizeToMatrix’ function of the Bioconductor package EnrichedHeatmap [https://github.com/jokergoo/EnrichedHeatmap] using 50bp bins across 10Kbp regions.
Cell proliferation, colony formation, and anchorage-independent growth assays
Cell viability experiments were performed using the CellTiter-Glo Luminescent Cell Viability Assay following the protocol provided by the manufacturer with some modifications (Promega). Briefly, 100μl of cell suspension was seeded in white-walled 96-well plates. The CellTiter-Glo reagent was added directly to each well at a 1:1 ratio. The plate was then mixed on a plate shaker for 5 minutes and incubated at room temperature for another 5 minutes before recorded on a plate reader (BioTek). All of the experiments were performed in biological triplicates. For colony formation assay experiments in Figure 5C, control and Crebbp-overexpressed NCI-H1882 cells were seeded at 10000 cells/well in technical triplicates in 6-well plates. 3 weeks later, cells were washed twice with PBS, fixed in 4% PFA at room temperature for 10min and stained with crystal violet (0.05%). Experiments were repeated twice and representative 3 wells of each condition are shown. For colony forming assays in Figure 6G, control and Cdh1-knockout preSC cells were seeded at 6000 cells/well in triplicate in 6-well plates. 2 weeks later, cells were fixed and stained with crystal violet (0.05%). 4 fields representing the entire well were counted. Experiments were performed in biological triplicates and fields from representative wells shown. For anchorage-independent growth assays in Figure 1A, cells were plated in at least triplicate in 6 well plates in 0.5mL of growth media containing 0.3% agar and seeded on top of a 0.5mL base layer of media containing 0.5% agar. The media was regularly changed every 3 days for 3 weeks. Colonies were stained with 0.05% crystal violet after fixation in 4% PFA. For experiments in Figure 5B and 6F, cells were seeded in 0.3% low melting point SeaPlaqueTM agarose (Lonza, Catalogue no: 50101) on top of 0.6% low melting point SeaPlaqueTM agarose layer. Low melting point agarose was pre-mixed with Dulbecco’s Modified Eagle’s Media 2X (Fisher Scientific, SLM202B) complemented with 20% FBS, sodium pyruvate (2mM), 200 U ml−1 penicillin and 200 μg/ml streptomycin. Cells were allowed to grow at 37 °C with 5% CO2 for 2–4 weeks. For each well, colonies from at least 5 random fields were counted (a total of at least 15 fields/condition). Representative microscopic images are displayed.
Transwell migration assay
Cell migration assay was performed using 8μm cell culture insert (08–771-21, Falcon). Briefly, 7.5×104 cells in serum-free DMEM media were seeded per well into the inserts, and 10% FBS DMEM media was used as chemokine added into the plate wells. The inserts were washed with PBS after 17 hours incubation and cells migrated to the basal side of the membrane were fixed using 4% PFA and then stained with 0.5% crystal violet. For each well, at least 5 random fields were photographed and counted. The experiments were performed using biological triplicates.
Statistical analyses
Statistical analyses and graphical presentation were performed with GraphPad Prism 7.0. The results are presented as the mean ± s.d. or the mean ± s.e.m. and evaluated using an unpaired Student’s t-test (two-tailed; p<0.05 was considered to be significant). Kaplan-Meier curves of lung tumor-free survival were generated using Prism 7.0 (Log-rank test; p<0.05 was considered to be significant).
Supplementary Material
Significance.
Our findings demonstrate that CREBBP loss in SCLC reduces histone acetylation and transcription of cellular adhesion genes, while driving tumorigenesis. These effects can be partially restored by histone deacetylase inhibition, which exhibited enhanced effectiveness in Crebbp-deleted tumors. These data provide rationale for selectively treating CREBBP-mutant SCLC with HDAC inhibitors.
Acknowledgements
We thank Scott Dylla and Jorge Aguilar (Abbvie Stemcentrx) for providing the LU505 PDX model. We thank Ryan Basom and Qing Zhang, as well as the Fred Hutch Genomics and Bioinformatics Shared Resource for help with the generation and analysis of next generation sequencing data, and Joe Hiatt for GSEA analyses. We also acknowledge support from the Fred Hutch Histopathology and small animal imaging Shared Resources as well as the Research Histology Core and the Flow Cytometry Core at the UVA Medical Center. We are grateful to Valera Vasioukhin, Taran Gujral, Julien Sage and Slobodan Beronja for critical reading of the manuscript.
Grant Support
This work was supported by NIH (R01CA200547 to D. MacPherson and R01CA194461 and R03CA215777 to K-S. Park), American Cancer Society (RSG-15–066-01-TBG to K-S. Park), the David R. Jones Fund at University of Virginia (to K-S. Park). D. Jia was supported by a postdoctoral fellowship from the International Association for the Study of Lung Cancer. E. Eastwood was supported by NIH training grant T32CA009657. This project was supported by the NIH Support Grants for FHCRC/UW Cancer Consortium Cancer Center (P30CA015704) and for University of Virginia Cancer Center (P30CA044579).
Footnotes
The authors report no conflicts of interest.
References
- 1.George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524(7563):47–53 doi 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nature genetics 2012. doi 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Augert A, Zhang Q, Bates B, Cui M, Wang X, Wildey G, et al. Small Cell Lung Cancer Exhibits Frequent Inactivating Mutations in the Histone Methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2017;12(4):704–13 doi 10.1016/j.jtho.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nature reviews Cancer 2017;17(12):725–37 doi 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- 5.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13 doi 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics 2012. doi 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America 2010;107(50):21931–6 doi 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. The EMBO journal 2011;30(2):249–62 doi 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, et al. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J 2001;20(6):1331–40 doi 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet 2002;32(4):606–13 doi 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 11.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011;471(7337):235–9 doi 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014. doi 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 2011;471(7337):189–95 doi 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang-Decker N, Tong C, Boussouar F, Baker DJ, Xu W, Leontovich AA, et al. Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell 2004;5(2):177–89. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer discovery 2017;7(3):322–37 doi 10.1158/2159-8290.CD-16-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, et al. CREBBP Inactivation Promotes the Development of HDAC3-Dependent Lymphomas. Cancer discovery 2017;7(1):38–53 doi 10.1158/2159-8290.CD-16-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton SJ, Giotopoulos G, Yun H, Vohra S, Sheppard O, Bashford-Rogers R, et al. Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors. Nat Cell Biol 2017;19(9):1093–104 doi 10.1038/ncb3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DW, Wu N, Kim YC, Cheng PF, Basom R, Kim D, et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30(11):1289–99 doi 10.1101/gad.279307.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 2003;4(3):181–9. [DOI] [PubMed] [Google Scholar]
- 20.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2015;10(4):553–64 doi 10.1097/JTO.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 2011;19(6):754–64 doi 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 22.Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE. Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PloS one 2011;6(3):e18472 doi 10.1371/journal.pone.0018472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature 1992;359(6393):295–300. [DOI] [PubMed] [Google Scholar]
- 24.Tsai KY, MacPherson D, Rubinson DA, Nikitin AY, Bronson R, Mercer KL, et al. ARF mutation accelerates pituitary tumor development in Rb+/− mice. Proc Natl Acad Sci U S A 2002;99(26):16865–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams BO, Remington L, Albert DM, Mukai S, Bronson RT, Jacks T. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat Genet 1994;7(4):480–4. [DOI] [PubMed] [Google Scholar]
- 26.Harvey M, Vogel H, Lee EY, Bradley A, Donehower LA. Mice deficient in both p53 and Rb develop tumors primarily of endocrine origin. Cancer Res 1995;55(5):1146–51. [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1(6):417–25 doi 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40 doi 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nature reviews Clinical oncology 2017. doi 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo CS, Krasnow MA. Formation of a Neurosensory Organ by Epithelial Cell Slithering. Cell 2015;163(2):394–405 doi 10.1016/j.cell.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novotny-Diermayr V, Sausgruber N, Loh YK, Pasha MK, Jayaraman R, Hentze H, et al. Pharmacodynamic evaluation of the target efficacy of SB939, an oral HDAC inhibitor with selectivity for tumor tissue. Mol Cancer Ther 2011;10(7):1207–17 doi 10.1158/1535-7163.MCT-11-0044. [DOI] [PubMed] [Google Scholar]
- 33.Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, et al. Small Cell Lung Cancer Screen of Oncology Drugs, Investigational Agents, and Gene and microRNA Expression. J Natl Cancer Inst 2016;108(10) doi 10.1093/jnci/djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015;163(2):506–19 doi 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9 doi 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al-Ahmadie HA, Iyer G, Lee BH, Scott SN, Mehra R, Bagrodia A, et al. Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat Genet 2016;48(4):356–8 doi 10.1038/ng.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart CA, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, et al. Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 2017. doi 10.18632/oncotarget.15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Semenova EA, Kwon MC, Monkhorst K, Song JY, Bhaskaran R, Krijgsman O, et al. Transcription Factor NFIB Is a Driver of Small Cell Lung Cancer Progression in Mice and Marks Metastatic Disease in Patients. Cell reports 2016;16(3):631–43 doi 10.1016/j.celrep.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013;19(11):1438–49 doi 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell 2016;166(1):21–45 doi 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 41.Canadas I, Rojo F, Taus A, Arpi O, Arumi-Uria M, Pijuan L, et al. Targeting epithelial-to-mesenchymal transition with Met inhibitors reverts chemoresistance in small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2014;20(4):938–50 doi 10.1158/1078-0432.CCR-13-1330. [DOI] [PubMed] [Google Scholar]
- 42.Ogiwara H, Sasaki M, Mitachi T, Oike T, Higuchi S, Tominaga Y, et al. Targeting p300 addiction in CBP-deficient cancers causes synthetic lethality via apoptotic cell death due to abrogation of MYC expression. Cancer discovery 2015. doi 10.1158/2159-8290.CD-15-0754. [DOI] [PubMed] [Google Scholar]
- 43.Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, et al. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 2017;550(7674):128–32 doi 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 2009;4(7):1064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nature protocols 2007;2(6):1445–57 doi 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 46.Anderson WC, Boyd MB, Aguilar J, Pickell B, Laysang A, Pysz MA, et al. Initiation and characterization of small cell lung cancer patient-derived xenografts from ultrasound-guided transbronchial needle aspirates. PLoS One 2015;10(5):e0125255 doi 10.1371/journal.pone.0125255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25(9):1105–11 doi 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology 2013;31(1):46–53 doi 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26(5):589–95 doi 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 2010;38(4):576–89 doi 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.