Abstract
Aberrant chromatin remodeling and activation of the PI3K pathway have been identified as important mediators of pediatric high-grade glioma (pHGG) and diffuse intrinsic pontine glioma (DIPG) pathogenesis. As inhibition of these pathways are promising therapeutic avenues and radiation is the only modality to prolong survival of patients with DIPG, we sought to explore radiosensitizing functions of such inhibition and to explore mechanisms of action of such agents. Here, we demonstrate that combined treatment with radiotherapy and CUDC-907, a novel first-in-class dual inhibitor of histone deacetylases (HDAC) and PI3K, evokes a potent cytotoxic response in pHGG and DIPG models. CUDC-907 modulated DNA damage response by inhibiting radiation-induced DNA repair pathways including homologous recombination and nonhomologous end joining. The radiosensitizing effects of CUDC-907 were mediated by decreased NFκB/Forkhead box M1 (FOXM1) recruitment to promoters of genes involved in the DNA damage response; exogenous expression of NFκB/FOXM1 protected from CUDC-907-induced cytotoxicity. Together, these findings reveal CUDC-907 as a novel radiosensitizer with potent antitumor activity in pHGG and DIPG and provide a preclinical rationale for the combination of CUDC-907 with radiotherapy as a novel therapeutic strategy against pHGG and DIPG. More globally, we have identified NFκB and FOXM1 and their downstream transcriptional elements as critical targets for new treatments for pHGG and DIPG.
Significance:
These findings describe the radiosensitizing effect of a novel agent in pediatric high-grade gliomas, addressing a critical unmet need of increasing the radiation sensitivity of these highly aggressive tumors.
Introduction
Pediatric high-grade glioma (pHGG) and diffuse intrinsic pontine glioma (DIPG) are among the most recalcitrant malignancies with dismal prognoses despite aggressive multimodal treatment (1). The 5-year survival for patients with pHGG is <20% and most children with DIPG do not survive beyond one year after diagnosis (2). Radiotherapy is the standard of care for pHGG and DIPG, but currently radiation fails to provide longterm disease control or cure. Radiation dose escalation is not a viable approach due to its associated toxicities, thus there is an urgent need for effective radiosensitizers (3).
Recent large-scale genomic studies have facilitated a better understanding of molecular mechanisms underlying the pathogenesis of pHGG and DIPG, and aberrations in chromatin regulation have emerged as a central theme (4–6). Driver mutations in histone genes and histone chaperones are common in pHGG and DIPG, several of these mutations are associated with poor outcomes, and targeting chromatin regulation through HDAC inhibition shows potent tumor growth suppression in vitro and in vivo (4–9). In parallel, most pHGGs and DIPGs show activation of the PI3K pathway through PTEN silencing by DNA hypermethylation or activating mutations in the PI3K pathway (10, 11). These findings suggest alterations in chromatin remodeling and aberrant activation of the PI3K pathway as important contributors to tumorigenesis in pHGG and DIPG, and targeting these two critical oncogenic pathways may serve as a novel therapeutic approach.
Radiotherapy induces DNA breaks and resultant damaged cells rely on (i) DNA repair pathways, especially homologous recombination (HR) and nonhomologous end joining (NHEJ), to repair DNA double strand breaks (DSB); and (ii) cell-cycle checkpoints to arrest progression of damaged cells, allowing sufficient time for DNA repair. In the event that cells enter mitosis with damaged DNA, they undergo mitotic catastrophe and apoptosis. Hence, drugs that impair DNA repair or cell-cycle arrest are potential radiosensitizers. Several studies have shown that HDAC and PI3K inhibitors interfere with DNA repair pathways (12–16). HDAC inhibition decreases expression of critical HR and NHEJ regulators, such as BRCA1, BRCA2, and KU80, and G2–M checkpoint kinases WEE1 and CHK1 (12, 13, 15). Similarly, inhibition of the PI3K pathway in breast cancer leads to BRCA1 downregulation, and consequently HR deficiency (14). Inhibitors of PI3K and HDACs act synergistically in medulloblastoma in vitro and in vivo, and this effect is partially mediated by the FOXO1 pathway that participates in DNA damage response (DDR; refs. 17, 18). These complementary roles of HDACs and PI3K in regulating DDR suggest that HDAC and PI3K inhibitors may synergize in their anticancer and radiosenstizing functions.
A novel oral dual pan-HDAC and PI3K inhibitor, CUDC-907, exhibits broad antitumor activity in hematologic and solid tumor preclinical models, and demonstrates greater inhibition of tumor growth in vitro and in vivo than HDAC or PI3K inhibitors as monotherapy (19–21). Interestingly, CUDC-907 also inhibits compensatory signaling pathways, such as RAF, MEK, and MAPK, suggesting that CUDC-907 may evade innate and/or acquired tumor resistance (20). The current potential therapeutic benefit of dual HDAC and PI3K inhibition, and in particular by CUDC-907, alone or in concert with radiotherapy has not been explored in pHGG or DIPG.
Here, we demonstrate that CUDC-907 in combination with radiotherapy provides synergistic cytotoxicity in pHGG and DIPG cell lines, and extends survival in mouse models of pHGG and DIPG. Furthermore, we find that CUDC-907 reduces DDR and induces sustained DNA damage in combination with radiation treatment through a NFκB- and Forkhead boxM1 (FOXM1)- mediated pathway. Taken together, these data reveal the potential for a new approach to the treatment of pHGG and DIPG by demonstrating precise molecular elements that should be targeted in combination with radiation: NFκB and FOXM1. CUDC-907 engages this novel strategy due to its ability to transcriptionally downregulate NFκB and FOXM1, block the repair of DNA DSBs, and markedly radiosensitize pHGG and DIPG.
Materials and Methods
Drugs, plasmids, and cell culture
CUDC-907 was obtained from Curis Inc. under a material transfer agreement. Panobinostat and BKM120 were purchased from Selleck Chemicals. Plasmids for expression of NFKB1/p50 (pcDNA-cFlag p50), RELB (pcDNA-cFlag RELB), and FOXM1b (pCW57.1-HA-Flag-FOXM1b) were purchased from Addgene Inc. Cells were tested for mycoplasma contamination at the beginning of the study. Cell lines SF188, SF8628, SF9427 were obtained from the Brain Tumor Research Center (BTRC) Tissue Bank at the University of California, San Francisco (UCSF, San Francisco, CA) and authenticated by the UCSF Genomics Core using short tandem repeat (STR) profiling. SF10693 was established in S. Mueller's laboratory and authenticated by STR profiling and sequencing. BT245 and BT869 were established in K.L. Ligon's laboratory and the lines were authenticated by molecular profiling using the Agilent 1 M feature array for comparative genomic hybridization. Trp53-null mouse neural progenitor cells lacking Pten (d333) or expressing BRAFV600E/KIAA1549:BRAF were established in the laboratories of Drs. Zhou and Stiles (Dana Farber Cancer Institute, Boston, MA), respectively. The loss of Trp53 and Pten was verified by genotyping and immunoblotting and BRAFV600E/KIAA1549:BRAF insertion was confirmed by genomic sequencing. KNS42 was obtained from JCRB (Japan Cancer Research Resources) cell bank. BT245 was grown as a suspension culture in human NeuroCult (StemCell Technologies Inc.) media supplemented with 20 ng/mL EGF, 20 ng/mL FGF2, 0.0002% heparin. SF9427 was grown as an adherent culture in media as for BT245 above, with further 5% FBS supplementation. SF188, SF8628, and KNS-42 cell lines were cultured as adherent cells in DMEM supplemented with 10% FBS. SF10693 (adherent) and BT869 (suspension) were cultured in a 1:1 mix of neurobasal media and DMEM F-12 with B27 supplement, 20 ng/mL EGF, 20 ng/mL FGF2, 10 ng/mL PDGF-AA, 10 ng/mL PDGF-BB, and 0.0002% heparin. Normal human astrocytes were purchased from Lonza Inc. and cultured as per manufacturer's recommendation.
Lentiviral production and establishment of cell line with FOXM1 overexpression
For lentiviral production, 80% confluent 293 cells in 100-mm plates were transfected with 1.5 μg of pMD8.2, 3.0 μg of psPAX2, and 6 μg of either pCW57.1HA-Flag-FOXM1b, pCVL. SFFV-SceOpt.I.IRES.BFP, or pCVL.SFFV-IRES.BFP using Lipofectamine 2000 as per manufacturer's instructions; supernatant was harvested 72 hours posttransfection, debris removed by centrifugation at 3,000 rpm for 5 minutes, and filtered through 0.45-μm filter before use or storage at −80°C. Supernatant containing the lentiviral particles with the doxycycline-inducible HA-tagged FOXM1b cDNA was used to transduce SF188 cells in the presence of 8 μg/mL polybrene. Cells stably expressing FOXM1b were selected by adding puromycin at 2.5 μg/mL after 48 hours of infection and selection was performed for two weeks. The doxycycline-inducible expression of HAFOXM1b was confirmed by Western blotting. Doxycycline treatment (250 ng/mL) for 24 hours induced modest HAFOXM1b expression.
Surgical procedure, in vivo treatment of orthotopic tumorbearing mice, and IHC analyses
DFCI and UCSF Institutional Animal Care and Use Committees (IACUC) approved all animal protocols. Five-week-old athymic mice were used for orthotopic glioma tumor formation. Mice were anesthetized by intraperitoneal injection of 100 mg/kg ketamine and 10 mg/kg xylazine in 0.9% saline. The skulls of the mice were exposed and a small opening made using a 25-gauge needle at 1.5 mm to the right of midline, posterior to the lambdoid suture for injection into the pontine tegmentum for luciferized SF8628 cells while an incision was made at 2 mm to the right of the midline for d333 cells. A total of 1.0 × 105 cells in a volume of 1 μL, were injected into the pontine tegmentum or cerebrum for SF8628 (DIPG) and d333 (HGG), respectively. Tumor-bearing mice were randomized to one of the following treatment groups: (i) control; (ii) radiation (137Cs source); (iii) CUDC-907; (iv) CUDC-907 + radiation. Tumor-bearing mice were treated daily by oral gavage for 10 days (Monday–Friday, 2 weeks) and 12 days (Monday–Thursday, 3 weeks) for SF8628 and d333, respectively, using 100 mg/kg CUDC-907 (constituted fresh daily at 7.5 mg/mL in 30% Captisol). Radiation was administered at 0.5 Gy every other day (Monday–Wednesday–Friday for SF8628 and Monday–Wednesday for d333), for a total dose of 3.0 Gy. Mice in control and radiation only groups were administered vehicle (30% Captisol); in the combination arm radiation was administered 3 hours after CUDC-907. Mice were monitored daily for tumor-related signs and were euthanized for indications of abnormal neurologic function according to the IACUC protocol. Prism software was used to plot the survival Kaplan-Meier curves. As all the animals died, to determine the survival differences among the groups oneway ANOVA (after logarithmic transformation of the values) followed by the Newman-Keuls method were performed. Additional mice from each cohort were sacrificed 2 hours after completion of treatments for SF8628 and following 1 week of treatments for d333; brains were then resected and fixed in 4% paraformaldehyde overnight followed by storage in 70% ethanol before paraffin embedding and sectioning for IHC staining. The slides were dewaxed with xylene, hydrated, antigen unmasked with citrate buffer, blocked, incubated with either γH2AX or Ki-67 antibodies overnight before signal was detected using 3,3′ Diaminobenzidine (DAB) chromogen and slides were counterstained with hematoxylin prior to dehydration and mounting.
Cell viability, clonogenic survival assay, cell cycle, apoptosis, and γH2AX detection assays
To measure cell viability, 1 × 104 cells were plated in triplicates in 96-well dish and cell viability was detected by performing CellTiter-Glo assays after 72 hours of drugs treatment or silencer RNA (siRNA) transfections.
To evaluate colony-forming potential, cells were seeded into a 6-well plate in triplicates for each condition. Cells were allowed to grow and adhere overnight before CUDC-907 (2 nmol/L) was added. After 16 hours of CUDC-907 treatment, cells were irradiated at varying doses and grown for 10–20 days to allow colony formation. Colonies of ≥50 cells were counted for each condition and normalized to the plating efficiency of each line. Dose enhancement ratio (DER), described as radiation dose required to achieve 10% survival in [radiotherapy group/CUDC-907 + radiotherapy group] was calculated by fitting a liner quadratic model using PRISM software and a DER of >1 indicates that CUDC-907 acts as a radiosensitizer.
For cell-cycle analysis, 60% confluent SF188 and SF8628 cells were treated with 2.5 nmol/L and 5.0 nmol/L CUDC-907 overnight (~16 hours) and then irradiated at 4 Gy in a γ-irradiator (137Cs source). Cells were harvested by trypsinization 6 hours after irradiation, fixed in 70% ethanol overnight at 4°C before cells were washed with 1× PBS, and stained with propidium iodide using RNaseA containing propidium iodide solution (Cell Signaling Technology). Cells were analyzed by flow cytometry (Sony SP6800) and cell-cycle profiles were determined using the MODFIT software.
Apoptosis was measured using the caspase 3/7 assay kit from Promega Inc. and Annexin Vstaining kit from BD Pharmingen Inc. For caspase 3/7 assay, 1 × 104 cells were plated in 96-well plate in triplicates 24 hours prior to treatment with 1.25 nmol/L CUDC-907. Radiation (4 Gy) was administered to the cells after 16 hours of CUDC-907 exposure and caspase activity was measured 48 hours after the commencement of CUDC-907 treatment as per the manufacturer's protocol (Promega Inc.). For Annexin V detection, 60% confluent cells were treated with 100 nmol/L CUDC-907 for 16 hours before irradiation (4 Gy) where indicated. Cells were harvested together at a time point corresponding to 4 hours after irradiation and Annexin V levels were analyzed by flow cytometry (CytoFLEXS) using the Annexin V detection kit from BD Biosciences as per the recommended protocol; data analyses were performed using FlowJo software.
γ-H2AX–positive cells were determined using a flow cytometry-based assay. Cells were treated with either CUDC-907 (100 nmol/L), or radiation (4 Gy), or combination of CUDC-907 (100 nmol/L) followed by irradiation (4 Gy). Control- and radiation-treated cells were incubated with DMSO while the CUDC-907 and combination-treated cells were incubated with 100 nmol/L CUDC-907 for 16 hours prior to irradiation and cells were harvested at the indicated time points (30 minutes, 5 hours, and 24 hours after irradiation) by trypsinization. Cells were washed, fixed in 1.5% paraformaldehyde for 15 minutes, washed, refixed in cold 90% methanol and stored at −20°Cuntil all samples were collected. Following at least a 30-minute fixation with 90% methanol, cells were washed with 1× PBS and resuspended in 100 μL of staining buffer (1 mg BSA/mL of 1× PBS). To block nonspecific signal from Fc receptor, 5 μL of TruStain FcX blocker (Biolegend Inc.) was added and samples were incubated at room temperature for 10 minutes. Following the blocking, 5 μL of FITC-conjugated γH2AX antibody (Biolegend Inc.) was added and cells were incubated in the dark for 1 hour. Cells were washed once with 1× PBS, resuspended in 250 μL of staining buffer containing 2.5 μg/mL propidium iodide, and stained cells were identified by flow cytometry (CytoFlexS). Flow cytometry data were analyzed using the FlowJo software.
DNA repair assays
DR-GFP and EJ5 cells were treated with CUDC-907 (2.5 or 5.0 nmol/L) or transfected with siRNA (Supplementary Table S1) for 24 hours prior to addition of lentiviral particles expressing I-SceI endonuclease and blue fluorescent protein (BFP). Lentiviral particles were generated as described above and the minimum volume of viral supernatant that resulted in approximately 50% BFP-positive cells in 48 hours was used for transduction. After 48 hours of transduction of DR-GFP and EJ5 cells, cells were harvested and analyzed by flow cytometry (Sony SP6800) to detect BFP- and GFP-positive cells. The percentage of double positive cells represents the frequency of DNA break repair.
Reverse transcriptase-PCR and chromatin immunoprecipitation-PCR
SF188 cells were treated with DMSO, CUDC-907 (100 nmol/L) for 16 hours, radiation (4 Gy), or CUDC-907 pretreatment followed by radiation 16 hours later. Total RNA was isolated for RT-PCR analysis 3 hours after irradiation. Reverse transcription was performed on 2 μg of total RNA using Superscript IV (Invitrogen Inc.) and real-time PCR was performed on Roche Light Cycler 96 using the specific primers with SYBR Green chemistry (Supplementary Table S2). Expression of mRNA was calculated using the ΔΔCt method relative to DMSO-treated samples. For ChIP-PCR analysis, 60% confluent SF188 cells in 100-mm plates were treated as above and DNA–protein interactions were captured 1-hour postirradiation by crosslinking with 1% formaldehyde for 10 minutes. Crosslinking was terminated by adding 125 mmol/L glycine-HCl (pH 3.5) and cells were washed twice with 1× PBS; cells were collected in 2 mL of lysis buffer I (50 mmol/L HEPES-KOH, pH7.4; 140 mmol/L NaCl; 1 mmol/L EDTA; 10% glycerol; 0.5% NP-40; 0.25% Triton X-100; protease inhibitors) and incubated on rotator at 4° C for 15 minutes. Nuclei were collected by centrifugation and washed for 10 minutes at room temperature with buffer 1 (10 mmol/L Tris-HCl, pH 8.0; 200 mmol/L NaCl; 1 mmol/L EDTA; 0.5 mmol/L EGTA; protease inhibitors). Next, nuclei were resuspended in 1 mL of lysis buffer II (10 mmol/L Tris-HCl, pH 8.0; 100 mmol/L NaCl; 1 mmol/L EDTA; 0.5 mmol/L EGTA; 0.1% sodium deoxycholate; 0.5% N-lauroylsarcosine; protease inhibitors) and DNA was fragmented using a Bioruptor on a 30-second ON and 30-second OFF cycle, four times for 5 minutes each (20 minutes total) to a size range of 200–700 bp. Finally, Triton-X100 was added to a final concentration of 1% and solubilized chromatin was clarified by centrifugation. The solubilized chromatin was precleared with Protein G-dyna beads for 2 hours before it was incubated with 20 μL of protein G-dyna beads that were prebound with 2 μg of antibody (IgG isotype control, anti-FOXM1 or anti-RELB) for 16–18 hours at 4°C on a nutator. The bound nucleoprotein complexes were washed extensively before elution, followed by crosslinking reversal at 65°C for 16 hours, ProteinaseK and RNAse treatment for 2 hours, and finally DNA purification using the MinElute PCR Purification Kit (Qiagen Inc.). Eluted DNA was PCR amplified using specific primers for each locus to detect recruitment of FOXM1 and RELB (Supplementary Table S3).
Antibodies, Western blotting, nuclear–cytoplasmic fractionation, immunoprecipitation, and immunofluorescence
Anti-RAD51, 53BP1, XLF, KU70, KU80, WEE1, CDKN1A/p21, FOXM1, NFKB1/p50, RELA, and RELB were purchased from Santa Cruz Biotechnology. Anti-phospho-Ser473-AKT, γH2AX, pPRAS40, p4EBP1, CHK1, Acetyl-TUBULIN, Acetyl-Histone H3, Histone H3, TUBULIN, pCDK1/2, ACTIN, and VINCULIN were purchased from Cell Signaling Technology. FITC-conjugated γH2AX and isotype control antibodies were purchased from BioLegend.
Cells were either treated with DMSO or CUDC-907 (100 nmol/L) for 16 hours, and irradiated (4 Gy) as indicated. Western blot analysis was performed on whole-cell extracts (20 μg) prepared using RIPA buffer (Pierce Biotechnology Inc.) 5 hours after irradiation, separated by SDS-PAGE, and transferred onto polyvinylidene difluoride membrane. Western blotting was performed using the recommended condition for each antibody as per manufacturer's recommendations.
To extract nuclear and cytoplasmic fractions, cells were washed with 1× PBS and collected by centrifugation. Cell pellet was resuspended in 3× volume of hypotonic buffer [10 mmol/L HEPES-KOH (pH 7.5), 10 mmol/L KCl, 1 mmol/L EDTA, 1.5 mmol/L MgCl2, protease, and phosphatase inhibitors] and incubated on ice for 10 minutes. The cell suspension was homogenized by passing through 27 G needle, 15 times, and the supernatant representing the cytoplasmic fraction was collected by centrifugation. The pellet representing the nuclei was washed twice with 3× volumes of hypotonic buffer, resuspended in 1× volume of high salt buffer [10 mmol/L HEPES-KOH (pH 7.5), 400 mmol/L KCl, 1 mmol/L EDTA, 1.5 mmol/L MgCl2, 10% glycerol, protease, and phosphatase inhibitors] and incubated on ice for 40 minutes followed by centrifugation to clarify the nuclear extracts. Following centrifugation, equal volume of low salt buffer [10 mmol/L HEPES-KOH (pH 7.5), 1 mmol/L EDTA, 1.5 mmol/L MgCl2, 10% glycerol, protease, and phosphatase inhibitors] was added and extracts were further analyzed by immunoblotting.
For immunoprecipitation experiments, whole-cell extracts were prepared by lysis in buffer containing 25 mmol/L Tris-HCl pH 8.0, 0.5% NP-40, 1 mmol/L EDTA, 150 mmol/L NaCl; 100 μg of extract was incubated with 2 μg of antibody bound to 15 μL of protein G-dyna beads overnight. Bound protein–antibody complexes were washed extensively with lysis buffer before elution with 2× SDS-PAGE loading dye and Western blotting.
For immunofluorescence experiments, cells were plated on coverslips overnight, treated with CUDC-907 (100 nmol/L) or DMSO for 16 hours, then irradiated (4 Gy) where appropriate, before fixation with 1% paraformaldehyde containing 0.5% Triton-X100 for 30 minutes at room temperature, followed by methanol fixation for 20 minutes at −20°C. All blocking was performed with 3% goat serum in 1× PBS containing 0.1% Triton X-100,1 mmol/L EDTA, and 1 mg/mL BSA for 1 hour and primary incubations were done overnight at 4°C with antibodies diluted in blocking buffer (1:200 for anti-RAD51, 1:1,000 for anti-γH2AX, and anti-53BP1). After overnight incubations, coverslips were washed twice with blocking buffer and once with 1× PBS prior to incubation with secondary antibody solution containing Alexa Fluor488–conjugated anti-mouse and Alexa Fluor 594–conjugated anti-rabbit for 1 hour. Coverslips were washed as before and slides were mounted on slides. Nuclei were counterstained with DAPI and images were taken at ×63 magnification using the Zeiss microscope. At least 100 cells were counted for each condition and cells with more than 5 foci were considered positive.
NFκB reporter assay, plasmid, and siRNA transfection
All plasmid transfections were performed using Lipofectamine 3000 (Invitrogen Inc.) as per the manufacturer's instructions. NFκB reporter assays were performed in 96-well plates in triplicate in either SF188 cells or SF188/FOXM1b (SF188 cells stably expressing doxycycline-inducible FOXM1b) cells. The plasmid pHAGE NFκB-TA-luc-UBC-GFP-W (100 ng) was used as the luciferase-based reporter and pRenillaLuciferase (0.5 ng) was used as the transfection efficiency control. Firefly and Renilla luciferase expression was measured 48 hours after transfection using the Dual Glo luciferase assay system (Promega Inc.). To overexpress NFκB, pcDNA3.1/cFlagRELB and pcDNA3.1/cFlagp50 (1:1) were cotransfected and to overexpress FOXM1b, doxycycline (250 ng/mL) was added to the SF188/FOXM1b cells 4 hours after transfection. For reporter assays and cell viability rescue assays, CUDC-907 was added for 5 hours after transfection and cells were irradiated (2 Gy) 1 hour prior to luciferase assay measurements.
All siRNA transfections were performed using RNAiMAX (Invitrogen Inc.) with indicated siRNAs (Supplementary Table S1) according to manufacturer's protocols. For radiosensitization experiments, radiation was administered 24 hours after siRNA transfection and cell viability was measured 70 hours postirradiation.
Statistical analysis
Unless indicated, all data are represented as mean ± SEM and EC50 were calculated using the PRISM software. Whenever multiple groups were compared we used one-way ANOVA followed by post hoc tests whether significance was noted. Pairwise comparisons were performed using the Newman–Keuls test (if all groups were to be compared) or the Dunnett test whenever all groups were compared with a single reference group. Asterisks indicate statistically significant (*, P< 0.05; **, P<10−2; ***, P<10−3; ****, P <10−4) values.
Results
CUDC-907 induces cytotoxicity and synergism with radiotherapy
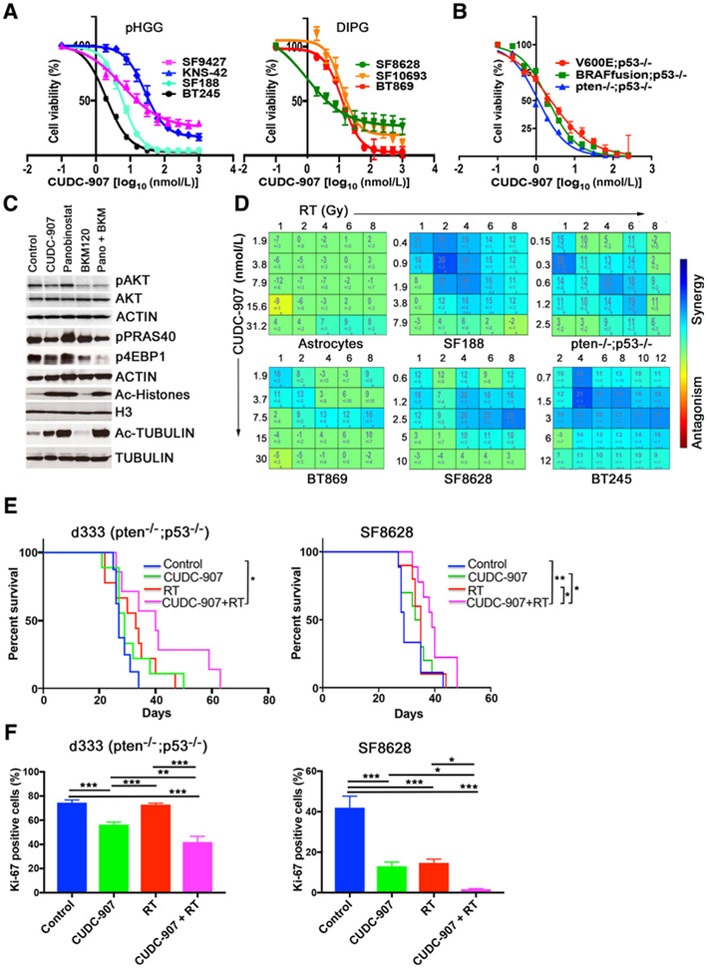
The cooccurrence of PI3K pathway activating mutations and histone mutations in pHGG and DIPG along with observed cytotoxicity of HDAC inhibition in DIPG cells led us to investigate the antineoplastic activity of CUDC-907 (dual pan-PI3K and pan-HDAC inhibitor) in pHGG and DIPG. We assessed in vitro cytotoxicity of CUDC-907 in seven human cell lines derived from pHGGs (BT245, KNS42, SF188, and SF9427) and DIPGs (BT869, SF8628, and SF10693) and in PI3K- or BRAF- activated mouse neural stem cells (mNSC) that produce gliomas in mice upon intracranial injection (22). All pediatric glioma cell lines and mNSC models were sensitive to CUDC-907 treatment with half maximal effective concentrations (EC50) at low nanomolar ranges (Fig. 1A and B). All glioma lines except KNS-42 showed 2–10 fold greater sensitivity to CUDC-907 than normal human astrocytes; in particular the c-MYC–amplified SF188 and BT245 lines exhibited marked sensitivity to CUDC-907 (Fig. 1A; Supplementary Fig. S1A).
Figure 1.
CUDC-907 induces cytotoxicity and synergizes with radiation in pHGG and DIPG models in vitro and in vivo. A and B, Cell viability of pHGG and DIPG cell lines (A) and genetically defined mNSC HGG cell lines after treatment with CUDC-907 for 72 hours (B). C, Immunoblot analyses (SF188) of downstream elements after CUDC-907 (100 nmol/L), panobinostat (100 nmol/L), or BKM120 (300 nmol/L) for either 2 hours for AKT, p-AKT, and acetylated proteins, or 16 hours for p-PRAS40 and p-4EBP1. D, Synergism–antagonism analyses using Combenefit software display marked synergy between CUDC-907 and radiation in pHGG and DIPG lines but not normal human astrocytes. Cellswere pretreated with CUDC-907 for 24 hours prior to irradiation and cell viability was determined 48 hours after irradiation (23). E, Kaplan-Meier survival curves for SF8628 and d333 (Pten−/−;Trp53−/−) orthotopic glioma models randomized into four treatment arms: (i) control, (ii) CUDC-907 (100 mg/kg), (iii) radiation (RT 0.5 Gy, Monday-Wednesday-Friday), and (iv) CUDC-907, followed by radiotherapy (3 hours later). F, Quantification of IHC analyses for Ki-67 in d333 and SF8628 orthotropic tumors using the Cell Profiler software. At least five representative tumor areas were imaged per slide and brain sections from two mice were analyzed per group. Values are mean ± SEM and only P values <0.05 are shown as *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We confirmed dual inhibition of HDAC and PI3K pathways after CUDC-907 treatment by measuring levels of acetylated proteins (histones and tubulin) as markers for HDAC inhibition (19, 20) and levels of phospho-proteins downstream of PI3K (pAKT, pPRAS40, p4EBP1) in SF188 and SF8628 cells (Fig. 1C; Supplementary Fig. S1B and S1C). CUDC-907 decreased phosphorylation of AKT within two hours and downstream targets of AKT by 16 hours, similar to BKM120, and increased acetylated histones and tubulin within two hours, similar to panobinostat (Fig. 1C; Supplementary Fig. S1B and S1C).
Given the roles of HDAC and PI3K inhibitors in DNA damage repair (12–16), we tested combinations of CUDC-907 and radiation in genetically representative pHGG and DIPG cell lines and in a mNSC model of high-grade glioma. Using Combenefit software (23), we observed significant synergy between radiation and CUDC-907 in all in vitro models of pHGG and DIPG while there was little to no synergy in normal human astrocytes (Fig. 1D; Supplementary Fig. S2A). We also performed clonogenic cell survival assay on SF188 and SF8628 cells and the dose enhancement ratio (DER>1.0) also indicated radiosensitization by CUDC-907 in both pHGG and DIPG lines (Supplementary Fig. S2B). The synergistic effects of combination treatment of CUDC-907 and radiotherapy were observed even at low concentrations of CUDC-907 (0–1 nmol/L) and clinically relevant doses of radiation (1–4 Gy; Fig. 1D). This dramatic synergistic activity of sequentially administered CUDC-907 and radiation was also seen when both CUDC-907 and irradiation were administered concurrently (Supplementary Fig. S2C).
Weasked whether the radiosensitizing functions of CUDC-907, observed in vitro, would be recapitulated in vivo, and tested this hypothesis in genetically engineered mNSC (Pten−/−;Trp53−/−, d333) and DIPG (SF8628) orthotopic glioma models. In mice bearing SF8628 brainstem tumors, combination treatment with CUDC-907 and radiation significantly increased survival compared with control or single agent [control vs. combination P = 0.005; CUDC-907 or radiotherapy vs. combination P = 0.046]. Similar results were observed in mice bearing orthotopic murine d333 intracranial tumors (control vs. combination, P = 0.04; Fig. 1E). Single modality treatment with CUDC-907 or radiation did not increase survival significantly compared with control in either orthotopic glioma model (Fig. 1E). Furthermore, Ki-67 immunostaining demonstrated that cellular proliferation was significantly reduced in d333 and SF8628 orthotopic tumors treated with CUDC-907 and radiotherapy compared with tumors treated with vehicle, radiation alone, or CUDC-907 monotherapy (Fig. 1F; Supplementary Fig. S3). Together, these data establish CUDC-907 as a novel potent radiosensitizer in pHGG and DIPG in vitro and in vivo.
CUDC-907 induces sustained DNA damage and reduces DNA damage response
We sought to understand the mechanisms underlying the antineoplastic effects of CUDC-907 as a single agent and in synergy with radiation. Because radiation induces a G2–M cell-cycle arrest to allow for DNA damage repair, we first studied the effect of CUDC-907 on cell-cycle regulation. We found that CUDC-907 monotherapy induced a G1 cell cycle arrest and pre-treatment with CUDC-907 abolished radiation-induced G2–M arrest (Fig. 2A; Supplementary Fig. S4A). Given that induction of CDKN1A/ p21 is important in regulating G1 cell cycle arrest (24) and phosphorylation of CDK1 by WEE1 and CHEK1 is important in mediating G2–M arrest (25), we examined p21 and pCDK1 levels after CUDC-907 treatment. p21 expression was markedly elevated and pCDK1 levels were reduced after treatment with CUDC-907 as monotherapy or with radiation, suggesting a mechanism for CUDC-907-induced G1 arrest (Fig. 2B; Supplementary Fig. S4B). To delve further into the mechanism of such p21 protein induction, we measured steady-state levels of p21 mRNA following treatment with CUDC-907 and observed elevated p21 mRNA (Fig. 2C). To investigate the mechanism of CUDC-907–induced p21 mRNA increases, we examined the transcriptional activation marks, H3K9 acetylation and RNA pol II recruitment, at the p21 gene. We observed high levels of histone H3K9 acetylation at the p21 promoter in the presence of CUDC-907 (Fig. 2D). In contrast, the recruitment of RNA Pol II to the p21 promoter was not altered but enrichment was seen in the gene body only in the presence of CUDC-907 (Fig. 2E). Taken together, our data indicate that although RNA Pol II is bound to the p21 promoter, hyperacetylation at H3K9 after CUDC-907 treatment leads to increased transcription of the p21 gene (Fig. 2C–E). We also sought to explore the contribution of apoptosis by measuring caspase-3/7 activity and Annexin V staining, and found that CUDC-907 induced apoptosis, an effect that was augmented by radiation, whereas radiation alone induced little to no apoptosis (Fig. 2F; Supplementary Fig. S4C).
Figure 2.
CUDC-907 induces G1 cell-cycle arrest, CDKN1A/p21 expression, and apoptosis. A, Cell-cycle distribution of SF188 (pHGG) and SF8628 (DIPG) following CUDC-907 (2.5 or 5.0 nmol/L) for 16 hours with or without irradiation (4 Gy); all samples were fixed 6 hours after irradiation. B and C, Protein levels of cell-cycle regulators p21 and p-CDK1/2 (B) and 18S rRNA normalized mRNA expression of p21 (C), 16 hours after CUDC-907 (100 nmol/L), irradiation (4 Gy) or combination (4 Gy 16 hours after CUDC-907). D and E, Enrichment of H3K9 acetylation (D) and RNA Pol II recruitment (E) to the promoter and in the body of the p21 gene in SF188 cells. F, SF188 and SF8628 assessed for apoptosis, measured by caspase-3/7 activity, 48 hours after CUDC-907 (1.25 nmol/L), irradiation (4 Gy), or combination (4 Gy administered 16 hours after CUDC-907). Values are mean ± SEM, normalized to control (DMSO) and only P < 0.05 are shown (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
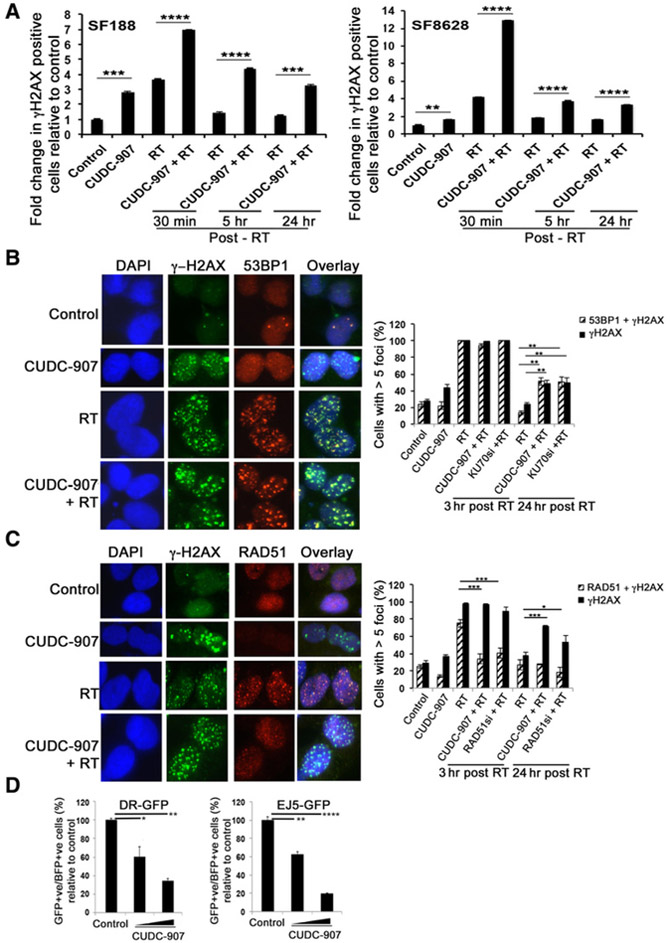
We hypothesized that, in addition to abrogating the radiation-induced G2–M cell-cycle arrest, CUDC-907 radiosensitizes pediatric gliomas by an additional mechanism of directly blocking repair of radiation-induced DNA damage (26). We tested whether CUDC-907 augments and sustains levels of radiation-induced γH2AX, an indicator of DNA damage. Using flow cytometry and immunostaining, we quantified γH2AX-positive cells at multiple time points after CUDC-907 and irradiation. CUDC-907 or radiation significantly increased the number of γH2AX-positive cells, and combination therapy elevated levels of γH2AX significantly more than either single treatment modality (Fig. 3A; Supplementary Fig. S5A and S5B). Moreover, combination therapy led to sustained elevations of γH2AX-positive cells for longer duration compared with radiation alone (Fig. 3A; Supplementary Fig. S5A and S5B).
Figure 3.
CUDC-907 impairs DDR pathways. A, DNA damage measured by γH2AX flow cytometry at specific time points after CUDC-907 (100 nmol/L), irradiation (4 Gy), or combination (4 Gy administered 16 hours after CUDC-907). B and C, Representative images and quantification of immunofluorescence staining for γH2AX and 53BP1 or γH2AX and RAD51 in SF188 cells treated with radiotherapy (4 Gy), CUDC-907 (100 nmol/L), or combination treatment in which radiotherapy was combined with KU70 siRNA, RAD51 siRNA (48 hours after siRNA transfection), or CUDC-907 (16 hours prior to 4 Gy). For staining, quantitated 3 or 24 hours after the last treatment, a minimum of 100 cells were counted per condition and nuclei containing ≥5 foci of γH2AX or colocalized γH2AX and 53BP1 or γH2AX and RAD51 were considered positive. D, Functional assays for HR (DR-GFP) and end-joining (EJ5-GFP) demonstrate that CUDC-907 impairs both DNA repair pathways in a dose-dependent fashion. Cells were treated with CUDC-907 (2.5 and 5.0 nmol/L) for 16 hours prior to introduction of dsDNA breaks with lentivirus expressing ISceI restriction enzyme and BFP tracker. GFP- and BFP-positive cells, reflecting proficient DNA repair, were identified by flow cytometry 48 hours after lentiviral infection. All values are mean ± SEM and only P < 0.05 are indicated. n = 3 independent samples; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Statistical analysis for A, compared radiotherapy vs. CUDC-907 + radiotherapy sample at each time point. Detailed P values for panel B and C are provided in Supplementary Table S4.
In concordance with these in vitro results, IHC staining of orthotopic tumors treated in vivo revealed that combination of CUDC-907 and radiotherapy significantly increased γH2AX levels in d333 orthotopic tumors compared with controls (P = 0.00014) or CUDC-907 monotherapy (P = 0.00011) or irradiation alone (P = 0.0001; Supplementary Fig. S6A and S6B). Similar results were seen for the SF8628 orthotopic tumors (Supplementary Fig. S6A and S6B). These findings suggest that CUDC-907 induces DNA damage and inhibits efficient DNA repair.
As CUDC-907 treatment led to persistent DNA damage and the most lethal lesions (double strand breaks) are primarily repaired by NHEJ and HR, we examined effects of CUDC-907 on these particular DNA repair pathways. To study HR, we measured RAD51 focus formation, while 53BP1 focus formation was used as an indicator for NHEJ pathway choice (Fig. 3B and C; refs. 27–29). As expected, irradiation of SF188 cells induced formation of 53BP1 and RAD51 foci in γH2AX-positive cells and dsDNA breaks were repaired efficiently by 24 hours, as indicated by return of γH2AX, 53BP1 and RAD51 foci to basal, pretreatment levels (Fig. 3B and C). When CUDC-907 was combined with irradiation, CUDC-907 pretreatment blunted radiation-induced RAD51 focus formation to levels comparable with that of siRNA knockdown of RAD51 prior to irradiation, consistent with CUDC-907's function of blocking HR (Fig. 3C). Evaluation of NHEJ revealed that CUDC-907 prevented resolution of radiation-induced 53BP1 foci with levels comparable with that of siRNA knockdown of KU70 prior to irradiation (Fig. 3B). CUDC-907 treatment did not reduce radiation-induced 53BP1 focus formation (Fig. 3B). Loss of RAD51 focus formation and sustained 53BP1 foci in combination therapy samples, accompanied by increased γH2AX foci, indicate defective HR and NHEJ resulting in augmented DNA damage. siRNA-mediated knockdown of core NHEJ and HR components mimicked these effects of CUDC-907.
To confirm that CUDC-907 causes functional defects in NHEJ and HR, we used established HeLa-based EJ5-GFP and U20Sbased DR-GFP reporter assays, respectively (Fig. 3D; ref. 30). In both systems, I-SceI induces double-strand breaks; DNA repair by either HR (in DR-GFP cells) or end joining (classical NHEJ or Alt-EJ in EJ5-GFP cells) restores GFP expression (30). These functional assays demonstrated that CUDC-907 significantly inhibited both end joining (40%–80% inhibition) and HR (40%–65% inhibition) in dose-dependent manners (Fig. 3D).
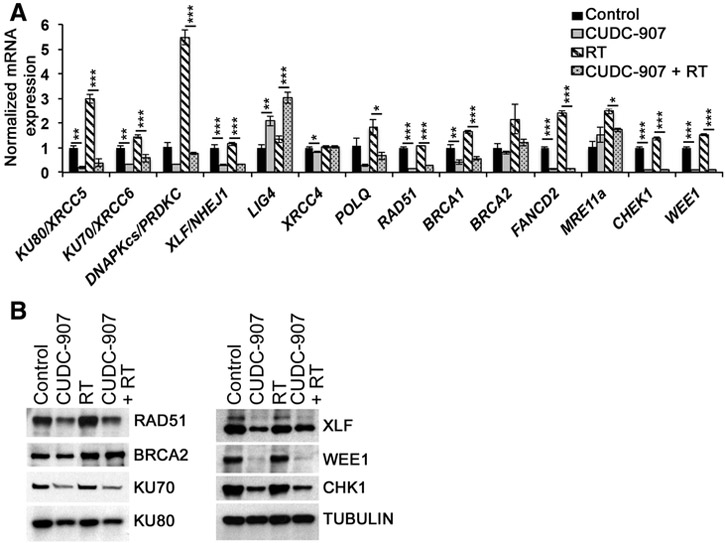
To elucidate the detailed mechanisms by which CUDC-907 affects DDR, we next asked if CUDC-907 reduces expression of key proteins in the DNA repair machinery. We examined the expression of important HR (RAD51, BRCA1, BRCA2, FANCD2,) NHEJ (KU80/XRCC5, KU70/XRCC6, DNAPKcs/PRDKC, XRCC4, LIG4, NHEJ1/XLF) and Alt-EJ (POLQ) pathway genes, and G2–M checkpoint kinases (CHEK1 and WEE1) in SF188 cells. Differential expression of these genes has been previously described as mechanisms of radiosensitization (15, 16, 26, 31). As assessed by quantitative real-time PCR, expression of RAD51, BRCA1, FANCD2, KU80, KU70, DNAPKcs, XLF, CHEK1, WEE1, and POLQ was significantly reduced after treatment with CUDC-907 (Fig. 4A). Furthermore, radiation-induced upregulation of BRCA1, BRCA2, FANCD2, KU80, KU70, DNAPKcs, CHEK1, WEE1, and POLQ was ablated by CUDC-907 pretreatment (Fig. 4A). The effects of CUDC-907 on these particular DDR genes were specific, as other DDR genes, such as MRE11a and LIG4, were upregulated and XRCC4 was unaffected by CUDC-907 (Fig. 4A). We confirmed that these quantitative real-time PCR results are reflected in protein expression by utilizing Western blot analyses in SF188 and SF8628 cells. Indeed, protein levels of RAD51, KU70, KU80, XLF, WEE1, and CHEK1, but not BRCA2, were significantly reduced after CUDC-907 monotherapy and in combination with radiation (Fig. 4B; Supplementary Fig. S7A and S7B).
Figure 4.
CUDC-907 downregulates DDR pathway genes. mRNA (A) and protein expression of genes involved in end joining, HR, and cell-cycle checkpoints in SF188 cells treated with CUDC-907 (100 nmol/L), radiation (4 Gy), and combination (CUDC-907 pretreatment for 16 hours, followed by 4 Gy; B). mRNA expression is normalized to 18S rRNA; Graph represents mean ± SEM and ANOVA P value for each gene is significant (P < 0.05) except for BRCA2; P values of pairwise comparisons are shown in Supplementary Table S5.
Together, these data reveal CUDC-907 as a regulator of DDR, inhibiting end joining (NHEJ and Alt-EJ), HR, and checkpoint kinases. CUDC-907 not only decreases expression of DDR proteins when used as a single agent, but also prevents induction of these DDR proteins after irradiation, thus leading to sustained DNA damage and radiosensitization.
CUDC-907 inhibits NFκB and FOXM1 expression and transcriptional activity
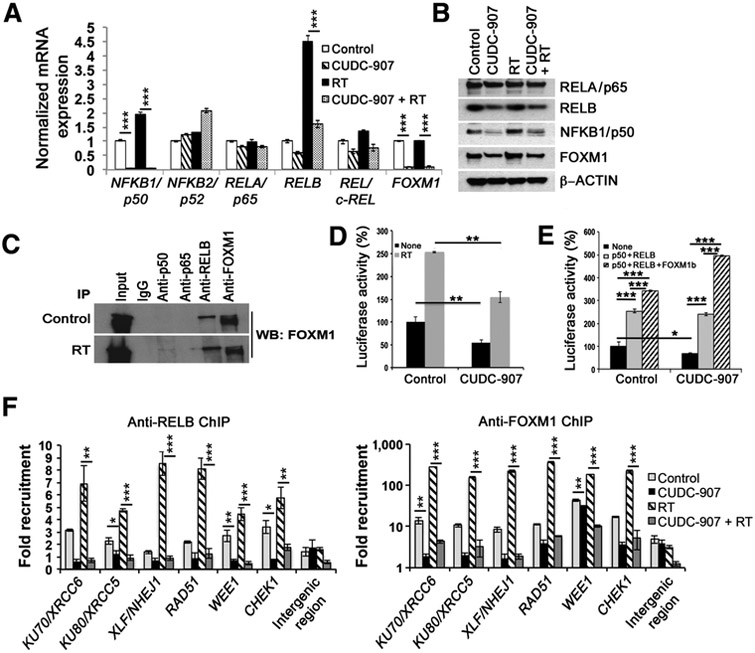
To further elucidate mechanisms by which CUDC-907 confers its synergistic antitumor effects with radiation, we explored the biochemical machineries by which dual HDAC- and PI3K inhibition modulate expression of DDR genes. We first conducted in silico analysis on oPOSSUM (http://opossum.cisreg.ca/oPOS SUM3/) to identify transcription factor families with binding sites in the promoter regions of DDR genes including those specifically regulated by CUDC-907 (Fig. 4A; ref. 32). Binding sites for NFκB and forkhead proteins were significantly enriched in promoter regions of the examined DDR genes (z-score = −10.19 and −2.87, respectively). We focused on NFκB and FOXM1, among the forkhead proteins, because NFκB and FOXM1 have been linked to radio- and chemoresistance in diverse cancers and were recently identified as transcriptional binding partners (33–36). Interestingly, NFκB and FOXM1 have been previously described as targets downregulated by HDAC inhibitors (17). To characterize effects of CUDC-907 on NFκB and FOXM1, we examined mRNA and protein expression of FOXM1 and NFκB subunits (RELA/p65, RELB, REL/c-REL, NFKB1/p50, NFKB2/p52.) Both mRNA and protein expression of NFKB1/ p50, RELB, and FOXM1 were significantly reduced after treatment with CUDC-907 alone or in combination with radiation (Fig. 5A and B; Supplementary Fig. S8A and S8B). Of particular note, CUDC-907 prevented radiation-induced upregulation of NFKB1/p50, RELB, and FOXM1 (Fig. 5A and B). These effects were specific, as expression of NFKB2/p52 and RELA/p65 was not substantially affected by CUDC-907 (Fig. 5A and B) and expression of forkhead transcription factors FOXP3 and FOXO1, both implicated in DDR response (18, 37), was upregulated by CUDC-907 (Supplementary Fig. S9A). Furthermore, as transcriptional activity of NFκB is regulated through its nuclear-cytoplasmic translocation, a process that is modulated by PI3K (38, 39), we investigated whether CUDC-907 interferes with NFKB1/p50 and RELB cytoplasmic-nuclear translocation. Western blot and immunostaining demonstrated that radiation increases nuclear localization of NFKB1/p50 and RELB, but CUDC-907 pretreatment blocks radiation-induced nuclear localization, resulting in continued diffuse cytoplasmic staining of NFKB1/p50 and RELB (Supplementary Fig. S9B and S9C). These findings indicate that CUDC-907 inhibits expression of FOXM1 and subunits of the NFκB pathway, NFKB1/p50 and RELB, and impairs nuclear localization of NFKB1/p50 and RELB, thus preventing transcriptional activity of these important DDR regulators.
Figure 5.
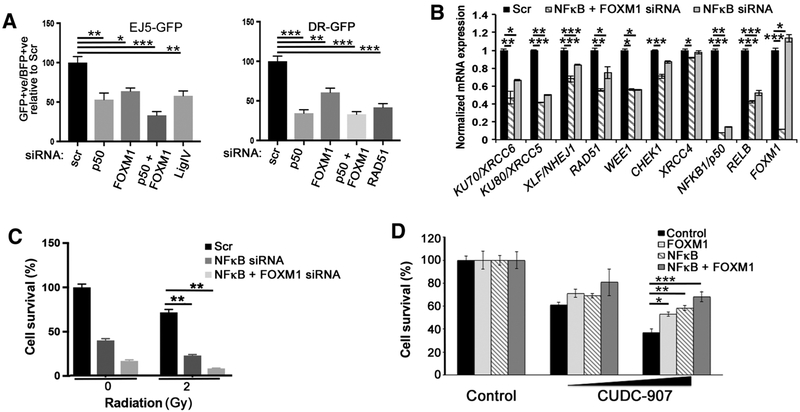
CUDC-907 impedes the recruitment and transcriptional activity of NFκB and FOXM1. mRNA (A) and protein expression of NFκB pathway genes and FOXM1 in SF188 cells after CUDC-907 (100 nmol/L), radiation (4 Gy) or combination (4 Gy administered 16 hours after CUDC-907; B). C, Coimmunoprecipitation of SF188 cells with or without radiation (4 Gy) using indicated antibodies and FOXM1 immunoblot. Input represents 10% of protein extract used in immunoprecipitation analysis. D and E, NFκB-driven luciferase activity in SF188 cells treated with CUDC-907 plus radiation (4 Gy; D) or overexpression of NFκB (NFKB1/p50 and RELB; E) and FOXM1b. F, Chromatin immunoprecipitation followed by PCR (ChIP-PCR) using anti-RELB and anti-FOXM1 antibodies on cross-linked chromatin from SF188 cells following indicated treatments. Graphs show the average ± SEM, and significant P values are indicated as *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For A and F, P value summary of pairwise comparisons are provided in Supplementary Tables S6 and S7, respectively.
Recent findings in lymphoblastoid cells identified FOXM1 as a transcriptional partner for NFκB based on cooccupancy at NFκB DNA-binding sites; therefore, we sought to confirm the interaction between FOXM1 and NFκB by coimmunoprecipitation (40). Wefound that FOXM1 strongly bound to RELB, but not RELA/p65 in pediatric glioma cells under normal and irradiated conditions, and FOXM1 weakly interacted with NFKB1/p50 after irradiation (Fig. 5C). Next, using an NFκB-driven luciferase reporter assay, we explored the transcriptional significance of the RELB-FOXM1 interaction, and the reduced NFκB and FOXM1 expression after CUDC-907 treatment. Radiation induced a marked increase in NFκB-driven luciferase activity; however, CUDC-907 inhibited NFκB-driven luciferase activity and significantly diminished radiation-induced increase in NFκB transcriptional activity (Fig. 5D). We next sought to confirm that NFKB1/p50, RELB, and FOXM1 mediate the observed alterations in NFκB transcriptional activity by investigating the effect of exogenous overexpression of NFKB1/ p50 and RELB with or without FOXM1 before CUDC-907 treatment. Reduction of luciferase activity by CUDC-907 was rescued by exogenous overexpression of NFKB1/p50 and RELB, and this effect was further augmented by the exogenous overexpression of FOXM1b in combination with NFKB1/p50 and RELB (Fig. 5E). Thus, we found that CUDC-907 impairs NFκB-driven gene expression, a novel pathway by which dual HDAC and PI3K inhibition causes transcriptional dysregulation.
CUDC-907 inhibits expression of critical DDR genes through a NFκB- and FOXM1-mediated pathway
Given that CUDC-907 downregulates expression of key DDR genes, inhibits NFκB and FOXM1 transcriptional activity, and NFκB- and FOXM1-binding sites are enriched in promoter regions of DDR genes, we hypothesized that NFκB and FOXM1 directly bind to the promoter regions of DDR genes to modulate their expression. We first performed chromatin immunoprecipitation with anti-RELB and anti-FOXM1 antibodies followed by PCR analyses to identify recruitment of NFκB and FOXM1 to the promoter regions of specific DDR genes. We found significantly enriched binding of RELB and FOXM1 to the promoter regions of RAD51, KU70, KU80, XLF, RAD51, WEE1, CHEK1, but not to a gene desert genomic locus in control cells (Fig. 5F). Furthermore, recruitment of RELB and FOXM1 to these promoter regions was enhanced by irradiation, consistent with our described radiation-induced increase in mRNA expression (Fig. 5F). These findings suggest important roles for NFκB and FOXM1 in driving basal transcription of critical DDR genes and regulating induction of these genes by radiation (Fig. 5F). CUDC-907 significantly abrogated binding of RELB and FOXM1 to promoter regions of all six DDR genes with and without radiation (Fig. 5F). These results are consistent with the hypothesis that dual HDAC and PI3K inhibition by CUDC-907 impairs DDR by reducing NFκB and FOXM1 transcriptional activities, which are critical in regulating appropriate DDR gene expression.
To further confirm these findings, we used siRNAs to knockdown NFκB and FOXM1 and evaluated DNA damage, DDR gene expression, and functional DDR. Knockdown of NFκB1/p50 and FOXM1 increased γH2AX and reduced cell viability, whereas cell-cycle profile was only altered by NFκB knockdown (Supplementary Fig. S10A-S10C). To understand the mechanisms by which loss of NFκB1/p50 and FOXM1 augmented γH2AX levels, we examined HR- and NHEJ-mediated repair using DR-GFP and EJ5- GFP systems after NFκB1/p50 and FOXM1 knockdown. In both systems, we observed significantly reduced GFP expression; a phenotype recapitulated by downregulation of key HR and NHEJ proteins, RAD51 and LIG4, respectively. These results indicate that loss of NFκB and FOXM1 inhibits HR- and NHEJ-mediated dsDNA break repair (Fig. 6A). We also demonstrated that HR and end-joining defects were associated with reduced mRNA and protein expression of NFκB- and FOXM1-mediated DDR genes, RAD51, KU70, KU80, XLF, WEE1, and CHEK1 (Fig. 6B; Supplementary Fig. S10D). Expression of XRCC4, a gene whose expression was not altered by CUDC-907, was similarly not affected by siRNA knockdown of NFκB and FOXM1 (Fig. 6B).
Figure 6.
NFκB- and FOXM1-driven transcription regulates the DNA-damage response and CUDC-907 mediates cytotoxicity through NFκB and FOXM1. A, Functional assays for HR (DR-GFP) and end-joining (EJ5-GFP), after siRNA knockdown of indicated genes, demonstrate that loss of NFκB and/or FOXM1 inhibits HR- and NHEJ-mediated dsDNA break repair. B, mRNA levels of indicated genes after siRNA knockdown of NFκB (NFKB1/p50 and RELB) and FOXM1 (for 72 hours in SF188 cells). C, NFKB1/p50 and FOXM1 siRNA knockdown enhance radiation-induced cytotoxicity (SF188 cells, 2 Gy). D, Cell viability of SF188 after CUDC-907 treatment (1.25 or 2.5 nmol/L) with or without exogenous overexpression of NFκB (p50 and RELB) or NFκB and FOXM1b, demonstrating that exogenous overexpression of NFκB and FOXM1 significantly decreases CUDC-907-induced cell death. All graphs represent values as mean ± SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant). The P values for all pairwise comparisons of B are provided in Supplementary Table S8.
Having demonstrated that NFκB and FOXM1 are important transcriptional regulators through which CUDC-907 mediates its effects on DDR gene expression, we asked whether NFκB and FOXM1 knockdown recapitulates the radiosensitizing effects of CUDC-907 and whether overexpression of NFκB and FOXM1 rescues cytotoxic effects of CUDC-907. NFκB and FOXM1 knockdown markedly enhanced radiation-induced cytotoxicity in SF188 cells (Fig. 6C). Interestingly, exogenous overexpression of NFκB and FOXM1 significantly decreased CUDC-907-induced apoptosis (Fig. 6D; Supplementary Fig. S10E).Taken together, our findings further demonstrate that CUDC-907's antitumor and radiosensitizing properties are dependent on NFκB- and FOXM1- mediated pathways.
Discussion
Here, we establish CUDC-907, a potent dual PI3K and HDAC inhibitor, as a novel radiosensitizer with therapeutic potential in models of pHGG and DIPG, pediatric brain tumors with dismal survival rates and limited treatment options. In recent years, HDAC inhibitors have emerged as a novel therapeutic strategy for DIPG (8, 41) and the combination of radiation and CUDC-907 is a novel treatment concept in pHGGs and DIPG that capitalizes on known effects of dual PI3K and HDAC inhibition on DNA repair pathways (4–7, 12–15).
We chose to investigate the dual inhibitor, CUDC-907, rather than using combinations of single agents targeting the PI3K and HDAC pathways, because such a single dual inhibitor simplifies pharmacokinetic approaches, avoids undesirable drug-drug interactions that may lead to dose-limiting toxicities, and allows us to build on the CUDC-907 backbone for further combination studies. It is therefore not surprising that this dual inhibitor, that provides a simple treatment regimen, is already quite far along in pediatric clinical development, with a phase II pediatric dose already established. We demonstrate that CUDC-907 has potent cytotoxic effects in the investigated pHGG and DIPG cell lines, and in combination with radiation, synergistic cell death is observed in in vitro glioma models harboring a wide range of genetic aberrations with limited effects on wild-type astrocytes (Fig. 1). In in vivo HGG and DIPG models, despite the limited blood–brain barrier (BBB) penetration of orally delivered CUDC-907 (<10% distribution in normal brain relative to plasma, personal communication, Curis Inc.), CUDC-907 and radiation significantly prolonged survival, indicating a potential therapeutic advantage for the combination (Fig. 1).
We elucidated novel mechanisms by which CUDC-907 confers its cytotoxicity and synergistic activity with radiation in vitro, finding that CUDC-907 regulates cell-cycle progression and impairs DDR. We observed that CUDC-907 treatment induced G1 cell-cycle arrest likely through CDKN1A/p21 promoter hyperacetylation-driven transcriptional activation, downregulation of CDK1 phosphorylation, and abrogation of the G2–M block induced by radiation treatment (Fig. 2). The G2–M arrest upon radiation treatment is crucial for the repair of DSBs through HR and end-joining (NHEJ and Alt-EJ) processes. In the presence of CUDC-907, the G1 arrest excludes the error-free HR pathway and cells depend on error-prone end-joining pathways to repair DNA and maintain genomic integrity. We show that CUDC-907 by itself induces DNA damage as measured by γH2AX and levels of DNA damage are significantly heightened by the addition of radiation treatment (Fig. 3). Furthermore, we observed a significant delay in the resolution of γH2AX in cells treated with CUDC-907 and radiation. Both PI3K and HDAC inhibition have been shown to block DNA repair pathways, and HDAC inhibitors alter expression of critical DNA repair genes (4–7, 12–16, 26, 31). We explored the effects of CUDC-907 on DDR pathways by employing functional assays and immunofluorescence-based foci formation, and found that both HR and end-joining DNA repair efficiencies were reduced in the presence of CUDC-907 (Fig. 3). Specifically, this loss of DDR is a result of transcriptional down-regulation of the critical players of HR (RAD51, BRCA1, FANCD2) and end-joining pathways (KU70, KU80, XLF, DNAPKcs, POLQ; Fig. 4). Interestingly, CUDC-907 ablated the radiation-induced upregulation of DDR pathways. As HR is known to function only during S and G2–M phase (42), CUDC-907–mediated G1 cell-cycle arrest also contributes to the reduction in HR efficiency. Together, these data demonstrate that CUDC-907 functions as a novel radiosensitizer by inducing a G1 cell-cycle arrest and inhibiting important DNA damage repair pathways, leading to sustained radiation-induced DNA damage and eventually apoptosis.
The effects of CUDC-907 on diverse DDR pathways led us to investigate transcriptional networks downstream of CUDC-907 and we demonstrated that CUDC-907 inhibits a transcriptional program driven by RELB-NFKB1/p50 axis of NFκB and FOXM1. NFκB and FOXM1 participate in oncogenesis of many cancers and contribute to radio- and chemoresistance; however, the exact mechanisms by which NFκB, particularly RELB-NFKB1/p50, and FOXM1 regulate DDR are not well understood (33–36). CUDC-907 decreased expression of NFKB1/p50 and RELB components of the NFκB pathway, and FOXM1, a transcriptional partner for NFκB in lymphoblastoid cells. CUDC-907 abolished radiation-induced upregulation of NFKB1/p50, RELB, and FOXM1 and their transcriptional activities (Fig. 5). Similar to our findings, decreased expression of FOXM1 was observed after treatment with trichostatin A, an HDAC inhibitor, and LY294002, a PI3K inhibitor (35, 43, 44). Although HDAC inhibitors can activate or inhibit the NFκB pathway depending on cellular context (45, 46), PI3K-Akt pathway activation upregulates NFκB transcriptional activity through nuclear translocation (38, 39). We show that FOXM1 interacts with RELB but not RELA in glioma cells, and augments RELB-NFKB1/p50–driven transcriptional activity (Fig. 5C–E). In agreement with above observations, CUDC-907 treatment decreased recruitment of RELB on target genes like RAD51, KU70, KU80, XLF, WEE1, CHEK1, leading to reduced expression of these key DDR genes and HR and end-joining mediated repair; an effect recapitulated by siRNA-mediated knockdown of NFKB1/p50, RELB, and FOXM1 (Figs. 5F and6B; Supplementary Fig. S10D). In a manner similar to CUDC-907 treatment, RELB-NFKB1/p50 and FOXM1 knockdown radiosensitizes glioma cells and the exogenous overexpression of NFKB1/ p50, RELB, and FOXM1 partially rescues CUDC-907–induced cell death, in keeping with the hypothesis that cytotoxicity of CUDC-907 in glioma models depends on the DDR pathways regulated by NFKB1/p50, RELB, and FOXM1 (Fig. 6C and D). Given the partial rescue, CUDC-907 likely also affects other transcriptional regulators of DDR. For example, in silico analysis also revealed high confidence predictions for c-MYC binding site (E-box) upstream of many DDR genes (Z score = 5.82), and c-MYC is a known target of HDAC and PI3K inhibition (47, 48). Future characterization of additional downstream targets of CUDC-907 will further illuminate the diverse antitumor and radiosensitizing effects of CUDC-907.
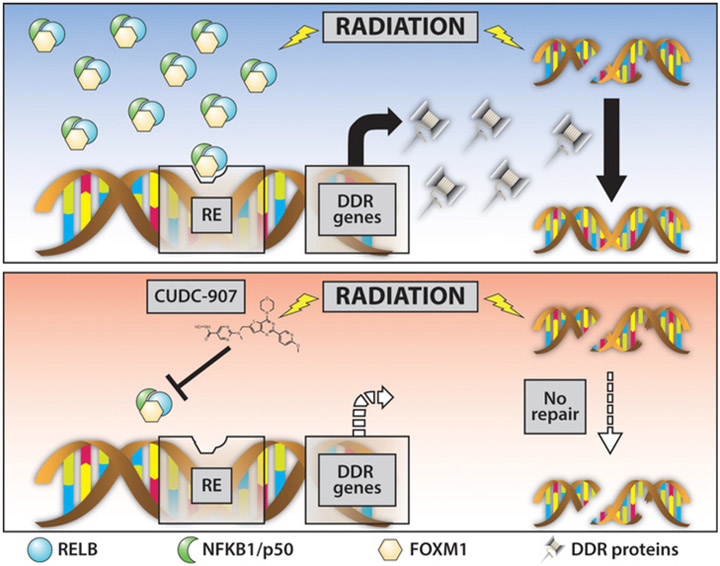
Radiosensitization can be accomplished by targeting different participants in DDR pathways, including DNA damage sensors, signaling kinases, cell-cycle effectors, and transcription factors (49). Here, we demonstrate that CUDC-907 synergizes with radiation in pHGG and DIPG models by inducing G1 cell-cycle arrest and affecting critical DDR pathways involved in dsDNA break repair and cell-cycle arrest by decreasing transcriptional regulators, NFκB and FOXM1. DDR is critical for repairing DNA damage after radiation, and combination treatment of CUDC-907 and radiotherapy leads to loss of DNA repair capabilities and hence sustained DNA damage after radiation, resulting in greater apoptosis (Fig. 7). Our findings also support targeting NFκB and FOXM1, and their downstream transcriptional programs as a novel therapeutic strategy in pHGG and DIPG. Wethus contribute to an expanding literature in which investigations of cellular responses to radiation disclose specific molecular elements that warrant further investigation as anticancer targets in precision medicine. Importantly, initial clinical trials of CUDC-907 have shown a favorable toxicity profile in adults (50) and a pediatric phase I trial is ongoing (NCT02909777). Our preclinical data presented here suggest that pHGG and DIPG can benefit from dual PI3K and HDAC inhibition, particularly by CUDC-907, especially in concert with radiation, and this novel promising therapeutic avenue warrants further clinical investigation.
Figure 7.
Novel NFκB- and FOXM1-dependent mechanism of cytotoxicity and radiosensitization in glioma cells. CUDC-907 augments unrepaired DNA damage by simultaneously directly inducing DNA damage and reducing DNA repair capabilities through reduced NFκB and FOXM1 activity, thus leading to apoptotic cell death as a single agent and synergistically with radiation.
Supplementary Material
Acknowledgments
We are thankful to Dr. Ramana Davuluri for helping with TF binding site analysis, and Dr. Alan D'Andrea and Dr. Geoffrey Shapiro for helpful critical comments. This research was supported in part by the NIH Brain Tumor SPORE grant P50 CA097257 (to S. Mueller), NINDS 1R01NS091620 (to D. Haas- Kogan, W. Weiss, A. Resnick), the Grand Philanthropic Fund (to D. Haas- Kogan), and the William M. Wood foundation (to D. Haas-Kogan). In addition, S. Mueller was supported by the National Center for Advancing Translational Sciences, NIH, through UCSF-CTSI grant number KL2TR000143 and D. Kozono was supported by NCI K08CA172354.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
W.A. Weiss has ownership interest (including patents) in StemSynergy Therapeutics and has provided expert testimony. S.G. Dubois is a consultant/advisory board member and has provided expert testimony for Loxo Oncology and Roche Genentech. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Jones C, Karajannis MA, Jones DTW, Kieran MW, Monje M, Baker SJ, et al. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro Oncol 2017;19:153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dasgupta T, Haas-Kogan DA. The combination of novel targeted molecular agents and radiation in the treatment of pediatric gliomas. Front Oncol 2013;3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22:425–37. [DOI] [PubMed] [Google Scholar]
- 5.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 2014;46:444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–31. [DOI] [PubMed] [Google Scholar]
- 7.Gajjar A, Bowers DC, Karajannis MA, Leary S, Witt H, Gottardo NG. Pediatric brain tumors: innovative genomic information is transforming the diagnostic and clinical landscape. J Clin Oncol 2015;33:2986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 2015;21:827. [DOI] [PubMed] [Google Scholar]
- 9.Lulla RR, Saratsis AM, Hashizume R. Mutations in chromatin machinery and pediatric high-grade glioma. Sci Adv 2016;2:e1501354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mueller S, Phillips J, Onar-Thomas A, Romero E, Zheng S, Wiencke JK, et al. PTEN promoter methylation and activation of the PI3K/Akt/mTOR pathway in pediatric gliomas and influence on clinical outcome. Neuro Oncol 2012;14:1146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun 2016;7:11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol Cancer Ther 2003;2:151–63. [PubMed] [Google Scholar]
- 13.Bangert A, Cristofanon S, Eckhardt I,Abhari BA, Kolodziej S, Hacker S, et al. Histone deacetylase inhibitors sensitize glioblastoma cells to TRAIL-induced apoptosis by c-myc-mediated downregulation of cFLIP. Oncogene 2012;31:4677–88. [DOI] [PubMed] [Google Scholar]
- 14.Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov 2012;2:1036–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornago M, Garcia-Alberich C, Blasco-Angulo N, Vall-Llaura N, Nager M, Herreros J, et al. Histone deacetylase inhibitors promote glioma cell death by G2 checkpoint abrogation leading to mitotic catastrophe. Cell Death Dis 2014;5:e1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roos WP, Krumm A. The multifaceted influence of histone deacetylases on DNA damage signalling and DNA repair. Nucleic Acids Res 2016;44: 10017–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei Y, Liu KW, Wang J, Garancher A, Tao R, Esparza LA, et al. HDAC and PI3K antagonists cooperate to inhibit growth of MYC-driven medulloblastoma. Cancer Cell 2016;29:311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science 2006;314:294–7. [DOI] [PubMed] [Google Scholar]
- 19.Mondello P, Derenzini E, Asgari Z, Philip J, Brea EJ, Seshan V, et al. Dual inhibition of histone deacetylases and phosphoinositide 3-kinase enhances therapeutic activity against B cell lymphoma. Oncotarget 2017;8: 14017–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian C, Lai CJ, Bao R, Wang DG, Wang J, Xu GX, et al. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clin Cancer Res 2012;18:4104–13. [DOI] [PubMed] [Google Scholar]
- 21.Sun K, Atoyan R, Borek MA, Dellarocca S, Samson ME, Ma AW, et al. Dual HDAC and PI3K inhibitor CUDC-907 downregulates MYC and suppresses growth of MYC-dependent cancers. Mol Cancer Ther 2017;16:285–99. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Alberta JA, Pilarz C, Calligaris D, Chadwick EJ, Ramkissoon SH, et al. A brain-penetrant RAF dimer antagonist for the noncanonical BRAF oncoprotein of pediatric low-grade astrocytomas. Neuro Oncol 2017;19:774–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Veroli GY, Fornari C, Wang D, Mollard S, Bramhall JL, Richards FM, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016;32:2866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53- mediated G1 arrest in human cancer cells. Cancer Res 1995;55:5187–90. [PubMed] [Google Scholar]
- 25.Den Haese GJ, Walworth N, Carr AM, Gould KL. The Wee1 protein kinase regulates T14 phosphorylation of fission yeast Cdc2. Mol Biol Cell 1995;6:371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller S, Hashizume R, Yang X, Kolkowitz I, Olow AK, Phillips J, et al. Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro Oncol 2014;16:352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol 2014;15:7–18. [DOI] [PubMed] [Google Scholar]
- 28.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci 1998;23:247–51. [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann M, de Lange T.53BP1: pro choice in DNA repair. Trends Cell Biol 2014;24:108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 2008;4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins GS, Prevo R, Lee YF, Helleday T, Muschel RJ,Taylor S, et al. A small interfering RNA screen of genes involved in DNA repair identifies tumorspecific radiosensitization by POLQ knockdown. Cancer Res 2010;70: 2984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009;138:1019–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding GR, Honda N, Nakahara T, Tian F, Yoshida M, Hirose H, et al. Radiosensitization by inhibition of IkappaB-alpha phosphorylation in human glioma cells. Radiat Res 2003;160:232–7. [DOI] [PubMed] [Google Scholar]
- 34.Wierstra I FOXM1 (Forkhead box M1)in tumorigenesis: overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumorsuppressive properties, and target of anticancer therapy. Adv Cancer Res 2013;119:191–419. [DOI] [PubMed] [Google Scholar]
- 35.Li L, Fan B, Zhang LH, Xing XF, Cheng XJ, Wang XH, et al. Trichostatin A potentiates TRAIL-induced antitumor effects via inhibition of ERK/FOXM1 pathway in gastric cancer. Tumour Biol 2016;37:10269–78. [DOI] [PubMed] [Google Scholar]
- 36.Zona S, Bella L, Burton MJ, Nestal de Moraes G, Lam EW. FOXM1: an emerging master regulator of DNA damage response and genotoxic agent resistance. Biochim Biophys Acta 2014;1839:1316–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung DJ, Jin DH, Hong SW, Kim JE, Shin JS, Kim D, et al. Foxp3 expression in p53-dependent DNA damage responses. J Biol Chem 2010;285:7995–8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karin M Nuclear factor-kappaB in cancer development and progression. Nature 2006;441:431–6. [DOI] [PubMed] [Google Scholar]
- 39.Bai D, Ueno L, Vogt PK. Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int J Cancer 2009;125:2863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SC, Greenfeld H, et al. The NF-kappaB genomic landscape in lymphoblastoid B cells. Cell Rep 2014;8:1595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 2017;31:635–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Z, Bozzella M, Seluanov A, Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008;7:2902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer 2013;13:482–95. [DOI] [PubMed] [Google Scholar]
- 44.Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol 2004;24:2649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Place RF, Noonan EJ, Giardina C. HDAC inhibition prevents NF-kappa B activation by suppressing proteasome activity: down-regulation of proteasome subunit expression stabilizes I kappa B alpha. Biochem Pharmacol 2005;70:394–406. [DOI] [PubMed] [Google Scholar]
- 46.Dai Y, Rahmani M, Dent P, Grant S. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol 2005;25:5429–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schild C, Wirth M, Reichert M, Schmid RM, Saur D, Schneider G. PI3K signaling maintains c-myc expression to regulate transcription of E2F1 in pancreatic cancer cells. Mol Carcinog 2009;48:1149–58. [DOI] [PubMed] [Google Scholar]
- 48.Nebbioso A, Carafa V, Conte M, Tambaro FP, Abbondanza C, Martens J, et al. c-Myc modulation and acetylation is a key HDAC inhibitor target in cancer. Clin Cancer Res 2017;23:2542–55. [DOI] [PubMed] [Google Scholar]
- 49.Raleigh DR, Haas-Kogan DA. Molecular targets and mechanisms of radiosensitization using DNA damage response pathways. Future Oncol 2013; 9:219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Younes A, Berdeja JG, Patel MR, Flinn I, Gerecitano JF, Neelapu SS, et al. Safety, tolerability, and preliminary activity of CUDC-907, a first-in-class, oral, dual inhibitor of HDAC and PI3K, in patients with relapsed or refractory lymphoma or multiple myeloma: an open-label, dose-escalation, phase 1 trial. Lancet Oncol 2016;17:622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.