Abstract
Objectives:
To determine the prognostic significance of spatial proximity of lung cancer cells and specific immune cells in the tumor microenvironment.
Materials and Methods:
We probed formalin-fixed, paraffin-embedded (FFPE) tissue microarrays using a novel tyramide signal amplification multiplexing technique labelling CD8, CD4, Foxp3, and CD68+ cells. Each multiplex stained immunohistochemistry slide was digitally processed by Vectra INFORMS software, and an X- and Y-coordinate assigned to each labeled cell type. The abundance and spatial location of each cell type and their proximity to one another was analyzed using a novel application of the G-cross spatial distance distribution method which computes the probability of finding at least one immune cell of any given type within a r µm radius of a tumor cell. Cox proportional hazards multiple regression was used for multivariate analysis of the influence of proximity of lymphocyte types.
Results:
Pathologic tumor specimens from 120 NSCLC patients with pathologic tumor stage I-III disease were analyzed. On univariate analysis, age (P=0.0007) and number of positive nodes (P=0.0014) were associated with overall survival. Greater area under the curve (AUC) of the Gcross function for tumor cell-Treg interactions was significantly associated with worse survival adjusting for age and number of positive nodes (HR 1.52 (1.11–2.07), P=0.009). Greater G-cross AUC for T-reg-CD8 was significantly associated with better survival adjusting for age and number of positive lymph nodes (HR 0.96 (0.92–0.99), P=0.042).
Conclusion:
Increased infiltration of regulatory T cells into core tumor regions is an independent predictor of worse overall survival in NSCLC. However, increased infiltration of CD8+ cytotoxic T cells among regulatory T cells seems to mitigate this effect and was significantly associated with better survival. Validation of the G-cross method of measuring spatial proximity between tumor and immune cell types and exploration of its use as a prognostic factor in lung cancer treatment is warranted.
Keywords: Non-small cell lung cancer, intratumoral T cells, immune cells, spatial distances, T regulatory cells, spatial computation
INTRODUCTION
The immune microenvironment plays a critical role in the antitumor response against lung cancer.1–3 Further characterization of T-cell mediated antitumor responses is imminently necessary to understand the heterogeneity of response to immune checkpoint inhibitors which can lead to a substantial durable response and prolong survival, but only in 10–20% of lung cancer patients.4
There have been several recent attempts to study the type and distribution of tumor infiltrating lymphocytes (TILs) and their relationship to disease prognosis.5–13 A recent study from the Lung Adjuvant Cisplatin Evaluation Biomarker group identified intense tumor lymphocytic infiltration, comprising only 9% of samples, as a favorable prognostic immunophenotype for survival in resected non-small cell lung cancer.14 With regard to specific cell types, CD8+ cytotoxic T cells are immunogenic and directly involved in tumor-directed adaptive immunity, and a high CD8+ TIL rate is associated with favorable cancer prognosis.6,15–18 Conversely, Foxp3+ regulatory T cells (Tregs) are a subclass of CD4+ T cells with immunosuppressive characteristics.19–21 However, the significance of Tregs within the tumor microenvironment has been somewhat controversial, with studies demonstrating both poor prognosis22–25 as well as better locoregional control and prognosis.26–29
Most previous attempts to study TILs have used simpler methods to analyze lymphocyte number such as a numerical count of lymphocytes per slide but lacked spatial information about precise cell location, intercellular interactions, and core tumor vs. invasive margin location. The lack of that information could account for conflicting findings regarding the prognostic significance of Tregs. Studies have shown that there is additional value and information in going beyond simple cell counts towards spatial analysis based on distances between two immune cell types.11,30
In this study, we apply a method of spatial analysis, based on the cross-type nearest neighbor distance distribution function (G-cross),31 to account for spatial relationships between tumor cells and specific types of T cells. This method of analysis is a novel application of spatial modelling previously used in ecology to reveal information on plant and animal interactions, population dynamics, and competition.32,33 Specifically, we use the G-cross method to determine whether a quantitative measure of spatial proximity between tumor and T cells as well as between different types of T cells can independently predict survival in patients with resected lung cancer.
MATERIALS AND METHODS
Patients
This retrospective, institutional review board-approved study included 120 patients with NSCLC who were diagnosed from 1998 to 2009 and treated at a single institution. All patients included had non-metastatic, pathologically confirmed diagnosis of NSCLC and underwent surgery followed by adjuvant radiation treatment with or without adjuvant chemotherapy. Clinical and pathologic characteristics including age, performance status, smoking history, tumor histology, pathologic tumor stage, tumor and lymph node size and involvement, and evidence of extracapsular extension, perineural and lymphovascular invasion were extracted.
Tissue Multiplex Immunohistochemistry
Immunofluorescent staining was performed using the opal multiplex immuno-fluorescent system (Perkin Elmer, Hopkinton, MA). Briefly, TMA slides were deparaffinized and tissues were fixed with 10% neutral buffered formalin. Heat- induced epitope retrieval (HIER) was performed in a conventional microwave oven using citrate buffer (pH 6.0). Each section was subjected to six sequential rounds of staining (pan-cytokeratin, DAPI, CD8, CD4, Foxp3, and CD68). Each round of staining included a protein block followed by staining with primary antibody and its corresponding secondary HRP- conjugated polymer. Signal amplification was achieved by using a specific TSA (Tyramide Signal amplification) solution for each antibody. Following this covalent reaction between the labeled tyramide and the tissue, the TMA slides were further subjected to HIER to remove unbound antibodies before the slides were stained with the next antibody. After all five sequential antibody staining, slides were counterstained with spectral DAPI (Perkin Elmer, Hopkinton, MA) and mounted with vectashield fluorescence mounting medium (Vector Labs, Burlingame, CA). Multiplex stained TMA slides were imaged using Vectra multispectral imaging system (Perkin Elmer, Hopkinton, MA) and the images were analyzed using inform software (Perkin Elmer, Hopkinton, MA)
Tumor and Immune Cell Localization Method
Each multiplex stained immunohistochemistry slide was digitally processed by Vectra INFORMS software, and an X- and Y-coordinate assigned to each labeled cell type. Cells that stained positive for each marker type were represented by different colors.
Spatial Analysis Computational Method: The G-cross function
The G-cross function computes the probability distribution of immune cells nearest to tumor cells within any given distance. Specifically, the G-cross is a spatial distance distribution metric that represents the probability of finding at least one given immune cell of any given type within a r µm radius of any tumor cell. These probability distributions can be applied to quantify relative proximity of any two types of cells. Therefore, the G-cross function is a quantitative surrogate of immune infiltration, as it can differentiate between immune cells that are close to but clustered away from tumor cells vs. those that are densely co-located with them (Figures 1–2).
Figure 1:
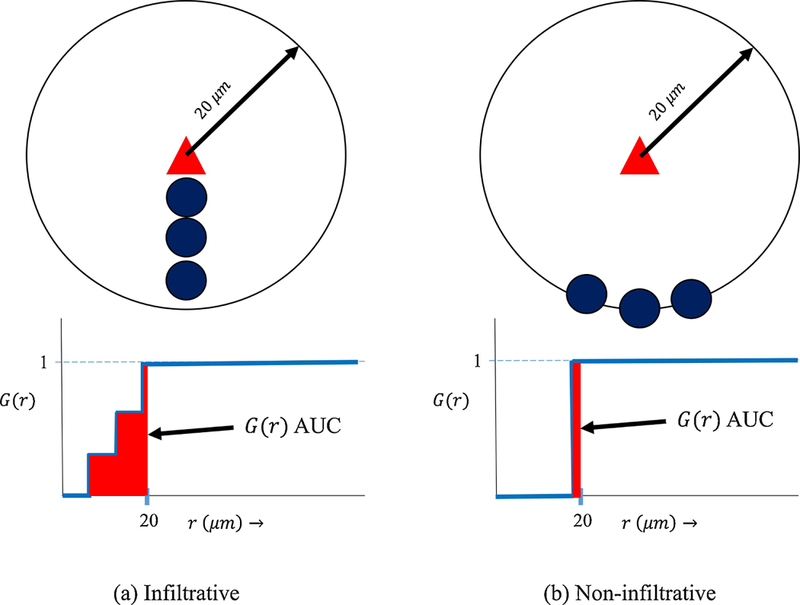
The G-cross function quantitatively differentiates between scenarios (a) and (b) representing infiltrative cells (dark blue circles) densely co-located with tumor cells (triangle) (a) vs. non-infiltrative cells close to but clustered away from tumor cells (b) within a 20 μm radius. A simple count of number of immune cells within a 20 μm radius of the tumor cell cannot distinguish between the different intercellular spatial relationships in (a) and (b). The corresponding area under the G-cross curves (AUC) for scenario (a) vs (b) is much greater in scenario (a), clearly differentiating the levels of infiltration visually observed.
Figure 2:
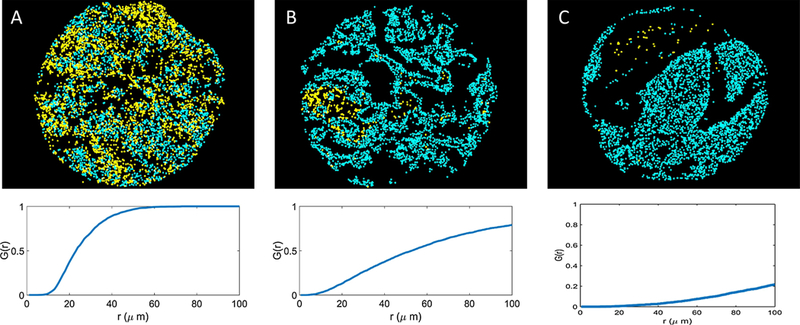
The G-cross function is a quantitative measure of qualitative immune cell infiltration. Here, by computing the area under the G-cross curves for yellow and blue cells in slides A, B, and C, we quantify the infiltration of the yellow cells into the blue cells on each respective slide. These examples demonstrate different levels of yellow: blue cell infiltration: (A) high, (B) medium, and (C) low. Corresponding G-cross curves rise sharply in (A), moderately in (B), and slowly in (C). Each infiltration type A, B, and C is associated with a unique G-cross signature.
Mathematically, the G-cross function is expressed as follows:
The subscripts ‘x’ and ‘y’ indicate that we compute the spatial distribution of cells of type ‘y’ relative to cells of type ‘x’. The term λy is the overall density of cells of type ‘y’ in the slide. The area under the curve (AUC) of the G-cross function above represents the level of infiltration of cell ‘y’ within a given distance from cells of type ‘x’.
Differentiation of Core Tumor and Invasive Margin Regions
The presence of different immune cells in the core tumor region as opposed to the tumor periphery, has been shown to impact disease prognosis differently.
We use an automated approach to estimate core tumor vs. invasive margin regions using a data-driven approach: kernel density estimation.35,36 We defined the core tumor region as the central region containing the majority, or approximately 60% of tumor cells, and the invasive margin as the scattered cells outside this perimeter (Supplementary Figure 1). The estimation technique is based on the technique previously described by Bowman & Azzalini.37 We then estimate the G-cross function separately for the core tumor and invasive margin regions. The AUC was computed over a spatial distance of r = 0 to r = 20 μm.
Statistical Analysis
Assessment of the correlation between G-cross AUC scores and other clinicopathologic factors were performed by using Spearman rank order correlation between continuous variables, Kruskal Wallis method between continuous and categorical variables, and Fisher test between categorical variables. P-values were adjusted for multiple testing using the Benjamini and Hochberg method.38 Clinical factors including patient age, sex, performance status, smoking history, tumor histology, tumor differentiation, pathologic disease stage, number of positive lymph nodes, lymphovascular space invasion, and extracapsular extension were also tested for association with overall survival using univariate Cox regression analysis. Cox proportional hazard multiple regression was used to model the influence of proximity of lymphocyte types using G-cross AUC scores on overall survival adjusting for significant clinical variables identified on univariate analysis. Survival and multivariate analyses were conducted using R (R Core Team, Vienna, Austria). Computational code for calculating G-cross AUC from G-cross curves and core tumoral-invasive margin segregation, was written and executed in MATLAB 2014a (The MathWorks, Natick, MA).
RESULTS
G-cross as a quantitative measure of qualitative cell infiltration
One hundred and twenty NSCLC patients with pathologic tumor stage I-III disease and their corresponding pathologic tumor specimens were included in the analysis. Patient demographic and tumor clinicopathological characteristics are shown in Table 1. The G-cross function was a robust quantitative surrogate of qualitative immune cell infiltration patterns. Computation of the area under the G-cross curves for slides with differing qualitative visual patterns of cell-cell infiltration were associated with unique G-cross signatures (Figure 2).
Table 1.
Baseline patient, tumor, and treatment characteristics
| Characteristic | No. of Pts (n = 120) |
|---|---|
| Median age at diagnosis, years (range) | 63 (34–89) |
| Gender | |
| Male | 52 (43%) |
| Female | 68 (57%) |
| Karnofsky performance status, median (range) | 80 (50–100) |
| Smoking history | |
| None, n (%) | 20 (17%) |
| Current or former smoker, n (%) | 100 (83%) |
| Pathologic tumor stage, n (%) | |
| Stage IA | 2 (2%) |
| Stage IB | 3 (3%) |
| Stage IIA | 2 (2%) |
| Stage IIB | 11 (9%) |
| Stage IIIA | 82 (68%) |
| Stage IIIB | 20 (17%) |
| Tumor Histology, n (%) | |
| Squamous | 35 (29%) |
| Adenocarcinoma | 72 (60%) |
| NSCLC, NOS | 12 (10%) |
| Sarcomatoid | 1 (1%) |
| Tumor differentiation, n (%) | |
| Well | 12 (10%) |
| Moderate | 48 (40%) |
| Poor | 60 (50%) |
| LVSI, n (%) | |
| Yes | 32 (27%) |
| No | 88 (73%) |
| Adjuvant chemotherapy treatment, n (%) | |
| Yes | 32 (27%) |
| No | 88 (73%) |
Abbreviations: LVSI = lymphovascular space invasion.
G-cross association with tumor pathologic characteristics
The correlation coefficients between G-cross AUC scores and tumor pathologic factors in this cohort are shown in Table 2, along with adjusted P-values. None of the G-cross AUC variables were significantly correlated with pathologic factors including tumor histology, tumor differentiation, tumor size, number of positive lymph nodes, pathologic tumor stage, perineural invasion, lymphovascular space invasion, and extracapsular extension.
Table 2.
Associations (correlation coefficients) of G-cross scores with tumor pathologic factors
| Tumor pathologic variables | ||||||||
|---|---|---|---|---|---|---|---|---|
| G-Cross AUC Variables | Tumor histology |
Tumor differentiati on |
Tumor size |
Number of positive lymph nodes |
Pathologic tumor stage |
Perineural invasion |
LVSI | ECE |
| Tumor: CD8 | 0.544 (P=0.91) |
6.804 (P=0.10) |
−0.057 (P=0.84) |
−0.090 (P=0.69) |
1.652 (P=1.0) |
0.141 (P=0.95) |
1.627 (P=0.45) |
0.290 (P=0.91) |
| Tumor: CD68 | 2.826 (P=0.81) |
2.608 (P=0.28) |
−0.137 (P=0.49) |
0.033 (P=0.89) |
4.841 (P=1.0) |
0.081 (P=0.95) |
4.335 (P=0.25) |
0.790 (P=0.91) |
| Tumor: CD4 | 0.958 (P=0.91) |
1.421 (P=0.45) |
−0.051 (P=0.86) |
0.008 (P=0.95) |
1.795 (P=1.0) |
1.077 (P=0.95) |
0.370 (P=0.76) |
1.104 (P=0.91) |
| Tumor: FoxP3 | 2.239 (P=0.88) |
2.486 (P=0.28) |
0.027 (P=0.92) |
−0.086 (P=0.69) |
8.219 (P=0.67) |
0.014 (P=0.97) |
0.177 (P=0.76) |
0.122 (P=0.91) |
| Tumor: CD8 + CD68 | 2.283 (P=0.88) |
4.776 (P=0.16) |
−0.066 (P=0.82) |
−0.007 (P=0.95) |
2.727 (P=1.0) |
0.190 (P=0.95) |
7.005 (P=0.07) |
2.116 (P=0.89) |
| Tumor: CD8 + CD4 | 0.863 (P=0.91) |
5.900 (P=0.10) |
−0.046 (P=0.86) |
−0.036 (P=0.89) |
2.017 (P=1.0) |
0.418 (P=0.95) |
2.461 (P=0.39) |
0.132 (P=0.91) |
| Tumor: CD8 + FoxP3 | 0.827 (P=0.91) |
6.409 (P=0.10) |
−0.033 (P=0.92) |
−0.086 (P=0.69) |
1.425 (P=1.0) |
0.032 (P=0.96) |
1.517 (P=0.45) |
0.197 (P=0.91) |
| Tumor: CD68 + CD4 | 1.303 (P=0.91) |
2.191 (P=0.31) |
−0.094 (P=0.71) |
0.043 (P=0.90) |
3.284 (P=1.0) |
0.684 (P=0.95) |
2.893 (P=0.37) |
0.040 (P=0.91) |
| Tumor: CD68 + FoxP3 | 1.116 (P=0.91) |
4.206 (P=0.18) |
−0.081 (P=0.74) |
0.009 (P=0.95) |
3.020 (P=1.0) |
0.044 (P=0.96) |
3.386 (P=0.35) |
0.675 (P=0.91) |
| Tumor: CD4 + Foxp3 | 1.815 (P=0.91) |
3.407 (P=0.22) |
−0.018 (P=0.92) |
−0.011 (P=0.95) |
2.509 (P=1.0) |
0.380 (P=0.95) |
0.270 (P=0.76) |
0.490 (P=0.91) |
| FoxP3: CD8 | 11.396 (P=0.09) |
0.0003 (P=1.0) |
0.140 (P=0.49) |
−0.041 (P=0.90) |
10.700 (P=0.33) |
0.190 (P=0.95) |
0.286 (P=0.76) |
0.197 (P=0.91) |
None of the above correlations reached statistical significance, adjusted P < 0.05.
Abbreviations: KPS = Karnofsky performance status, LVSI = lymphovascular space invasion.
Survival Analyses
On univariate analysis, age, number of positive nodes, and tumor differentiation were associated with overall survival (Table 3). Multivariable models adjusting for age and number of positive nodes identified Tumor: FoxP3 and CD8: FoxP3 G-cross AUC as significant predictors of overall survival (Table 4). Greater G-cross AUC for tumor cell-Treg interactions was significantly associated with worse survival adjusting for age and number of positive nodes (HR 1.52 (1.11–2.07), P=0.009). Greater G-cross AUC for T-reg-CD8 was also significantly associated with better survival adjusting for age and number of positive lymph nodes (HR 0.96 (0.92–0.99), P=0.042). Other tumor-immune cell interactions were not significantly associated with survival (Table 4).
Table 3.
Univariate Cox regression of clinicopathologic and treatment factors associated with overall survival
| Characteristic | HR | 95% CI | P value |
|---|---|---|---|
| Age | 1.04 | 1.02–1.07 | 0.0007 |
| Gender | 1.26 | 0.78–2.0 | 0.34 |
| KPS | 1.003 | 0.98–1.03 | 0.81 |
| Positive smoking history | 1.002 | 0.99–1.01 | 0.70 |
| Pathologic tumor stage I II III |
1 1.65 1.13 |
- 0.46–5.88 0.36–3.64 |
- 0.44 0.83 |
| Tumor histology Adenocarcinoma Squamous NSCLC, NOS Sarcomatoid |
1 0.9 0.86 0.64 |
- 0.53–1.52 0.4–1.84 0.09–4.73 |
- 0.68 0.70 0.67 |
| Tumor differentiation Well Moderate Poor |
1 0.55 0.36 |
- 0.27–1.12 0.18–0.75 |
- 0.10 0.006 |
| Number of positive lymph nodes | 1.13 | 1.05–1.21 | 0.0014 |
| Perineural invasion | 1.85 | 0.67–5.11 | 0.24 |
| LVSI | 0.89 | 0.51–1.53 | 0.67 |
| ECE | 1.10 | 0.50–2.42 | 0.81 |
| Adjuvant chemotherapy treatment | 0.72 | 0.46–1.15 | 0.17 |
Boldface type indicates statistical significance on univariate analysis.
Abbreviations: KPS = Karnofsky performance status, LVSI = Lymphovascular space invasion, ECE = Extracapsular extension, HR = hazard ratio, CI = confidence interval.
Table 4.
Multivariate Cox regression of G-cross AUC scores and clinical factors with overall survival
| Characteristic | HR | 95% CI | P value |
|---|---|---|---|
| Age | 1.04 | 1.01–1.06 | 0.002 |
| Number of positive lymph nodes | 1.18 | 1.08–1.29 | 0.0002 |
| Tumor: CD8 (G-cross) | 0.96 | 0.87–1.06 | 0.42 |
| Tumor: CD68 (G-cross) | 0.91 | 0.78–1.06 | 0.24 |
| Tumor: CD4 (G-cross) | 0.96 | 0.83–1.12 | 0.60 |
| Tumor: FoxP3 (G-cross) | 1.52 | 1.11–2.07 | 0.009 |
| CD8: FoxP3 (G-cross) | 0.96 | 0.92–0.99 | 0.042 |
| Tumor: CD8+CD68 (G-cross) | 0.95 | 0.88–1.03 | 0.23 |
| Tumor: CD8+CD4 (G-cross) | 0.97 | 0.89–1.05 | 0.40 |
| Tumor: CD8+FoxP3 (G-cross) | 0.99 | 0.90–1.08 | 0.77 |
| Tumor: CD4+CD68 (G-cross) | 0.95 | 0.86–1.05 | 0.33 |
| Tumor: FoxP3+CD68 (G-cross) | 0.98 | 0.86–1.11 | 0.71 |
| Tumor: FoxP3+CD4 (G-cross) | 0.99 | 0.90–1.15 | 0.81 |
Boldface type indicates statistical significance on multivariate analysis.
Abbreviations: AUC = area under the curve, HR = hazard ratio, CI = confidence interval.
G-cross analysis of core tumor and invasive margin regions
The tumor-T reg interaction was significantly associated with overall survival in core tumor (HR = 1.48, 95% CI 1.13–1.94, P = 0.003), but not in the invasive margin region (HR = 1.14, 95% CI 0.96–1.36, P = 0.09). The Treg-CD8 interaction was significantly associated with OS in the invasive margin region (HR = 0.99, 95% CI 0.98–0.99 P = 0.03), but not in the core tumor (HR = 0.99, 95% CI 0.98–1.01, P = 0.2).
DISCUSSION
A detailed understanding of the tumor immune environment is needed in order to characterize the anti-tumor immune response in lung cancer, particularly in the era of immunotherapy. Considering spatial relationships between tumor and immune cells and not only raw immune cell counts is an essential step towards our understanding and may provide insight into new prognostic indicators for treatment response. In our study, we show that the G-cross method can be used as a quantitative descriptor of tumor immune cell infiltration, accounting for the spatial distribution of specific cells of interest. Our data shows that increased infiltration of regulatory T cells into core tumor regions (as measured by the area under the G-cross curve computed for tumor: Treg interaction between r = 0 µm and r = 20 µm) is an independent predictor of worse overall survival. However, increased infiltration of CD8+ cytotoxic T cells among regulatory T cells (as measured by the G-cross AUC for T-reg:CD8 between r = 0 µm and r = 20 µm) seems to mitigate this effect and was significantly associated with better survival. Additionally, we were able to separately evaluate cell-cell interactions in the core tumor region and invasive tumor margin and found that increased tumor-T regulatory cell interaction in the core but not in the periphery was associated with worse overall survival.
Our data is consistent with previous studies indicating that regulatory T cells and CD8+ cytotoxic T cells in the tumor microenvironment are prognostic for clinical outcome in lung cancer. A meta-analysis of the prognostic value of tumor-infiltrating lymphocytes in lung cancer identified that overall, high levels of CD8+ cells within the tumor or tumor stroma was associated with improved OS.15 Peritumoral CD4+ was associated with improved OS but intratumoral counts were not.15 A high density of Foxp3+ regulatory T cells in the tumor stroma was associated with worse progression free survival and was a negative prognostic factor.15 An analysis of 129 pathologic specimens from patients with stage II/III surgically resected lung cancer identified higher CD8 cell concentration, CD45RO+ memory T cell concentration, and CD57+ effector T cell concentration in the peritumoral stroma as factors associated with improved OS.39 Elevated intratumoral CD8 cell and FOXP3 cell concentration were independently associated with favorable OS.39 However, the precise relationship between Tregs, tumor, and survival has been controversial. For example, a study of TILs in 80 patients with resected NSCLC identified a high number of tumor stroma infiltrating Foxp3+ Tregs was associated with improved OS.16
Recent studies have demonstrated that the spatial context of the tumor microenvironment may hold additional prognostic value.30,34 Most prior studies reporting on TILs use methods requiring manual demarcation of regions of interest for computing spatial statistics. Furthermore, the considered parameters are simplistic, using measures such as count, cell density, or a subjective gestalt assessment of infiltration by a pathologist.6,11,14 Specifically, the spatial context of immune cells has been shown to be critical for cancer development.5,6,9–13 For example, the count of CD8+ cells in distant stromal regions was shown to be an independent predictor of breast cancer-specific survival.6 A high density of CD3+ cells in the invasive margin has been shown to be significantly associated with disease-free survival in colorectal cancer.11 Studies have shown that there is additional value and information in going beyond simple cell counts, towards spatial analysis based on tumor-vessel distances,40 and distances between two immune cell types.30 Furthermore, Feichtenbeiner et al., showed that high intraepithelial infiltration of CD8(+) and FoxP3(+) cells was associated with the improved 10-year metastasis-free survival, indicating the potential power of spatial analysis in prognosis.30 Recent advances in the fields of image analysis, computer vision, and machine learning is transforming histopathology.41–43 We used a digital histopathology platform to identify the precise cell location and phenotype to analyze spatial proximity.
Our study shows the feasibility of using a fully automated, quantitative approach to studying spatial relationships between cells using the G-cross framework, which accounts for the heterogeneity in the spatial distribution of tumor cells and immune cells. Applying a computational framework with origins in the study of complex inter-species interactions in ecosystems, we are able to quantitatively capture cell infiltration with compelling visual agreement. Cells that are highly infiltrative among tumor cells have sharply-rising G-cross curves and greater G-cross AUC. On the other hand, cells that are dispersed and non-infiltrated among tumor cells have slow-rising G-cross curves and low G-cross AUC. We found that the area under the G-cross curve has prognostic value, computed over an interaction distance of 20 μm.
We found that close proximity of regulatory T cells with core tumor cells or highly infiltrative T regs has a significant negative association with survival and that this effect can be partially mediated increased proximity to CD8+ cells. Tregs mediate the magnitude of the adaptive immune response to foreign antigens and are specialized for immune suppression.20 Tregs inhibit anti-tumor immune surveillance in normal individuals and in cancer patients via suppression mechanisms that include secretion of inhibitory cytokines such as IL10 and TGFb and direct cytolysis of natural killer and cytotoxic T cells.20,44 Removal of Tregs can enable effective anti-tumor immune reactivity in animals with otherwise ineffective anti-tumor responses.45 Interestingly, the direct cytolytic effect of Tregs on natural killer cells and CD8+ lymphocytes via granzyme-B and perforin suggests that Tregs act in a contact-dependent manner.46 This suggests that precise analysis of very small cell-cell distances is very important in studying immune mediated responses involving Tregs. We hypothesize that increased number and proximity of CD8+ cells to Tregs in the core tumor region can somewhat overwhelm the regulatory function of Tregs and thus be associated with better prognosis.
A recent study suggests that NSCLC resected patients with high CD+ lymphocytes lacking PD-1 inhibitory receptor had longer overall survival and better response to treatment with nivolumab.47 Future directions include staining for PD-1 and subdividing the CD8+ cell population into PD-L1+ and PD-L1- components, and the tumor population into PD-L1+ and a PD-L1-components, and analyzing the difference in spatial interactions between these sub-categories. This would enable characterization of the infiltrative behavior of T lymphocytes as a function of their PD-1 expression and potentially further differentiate disease prognosis.
Several limitations apply to our study. First, while we use spatial proximity (approximating 20 µm spatial distance as cell-cell abutment) as a proxy for function, we realize that this is just one parameter and does not capture the full extent and magnitude of intercellular interactions. Second, while all patients in this study underwent resection and radiotherapy for NSCLC lung cancer, we recognize that this is a heterogeneous cohort. Validation of our findings is therefore warranted in other NSCLC patient cohorts. Lastly, while the multiplex staining herein is a routine method for research and standard within Cancer Immune Monitoring and Analysis Centers (CIMAC), adoption into routine clinical practice will take further validation and development. However, the G-cross analysis framework itself can be easily applied once staining has been performed as it uses the spatial information of different cell types as input, regardless of staining or imaging technique. Furthermore, the G-cross analysis was written using the freely available R software, making it easy to install and use.
In summary, the current study shows that increased infiltration of regulatory T cells into core tumor regions is an independent predictor of worse overall survival in non-small cell lung cancer patients. However, increased infiltration of CD8+ cytotoxic T cells among regulatory T cells seems to mitigate this effect and was significantly associated with better survival. Validation of the G-cross method of measuring spatial proximity between tumor and immune cell types and exploration of its use as a prognostic factor in lung cancer treatment with or without immunotherapy is warranted. This study offers new insights into the nature of NSCLC immunity with potential for harnessing this information towards effective immunomodulatory strategies.
Supplementary Material
Highlights.
Nearest neighbor spatial models show relationships between tumor and immune cells
Tumor and T regulatory cell spatial proximity predicts worse lung cancer survival
Spatial proximity of CD8 to T regulatory cells predicts better lung cancer survival
Tumor-immune cell spatial modeling helps our understanding of individual responses
Acknowledgements:
Funding: This work was partly funded by the Lung Cancer Priority Program at MD Anderson Cancer Center and the Mabuchi Research Fund. AR was supported by CCSG Bioinformatics Shared Resource P30 CA01667, an Institutional Research Grant from The University of Texas MD Anderson Cancer Center (MD Anderson), a Career Development Award from the MD Anderson Brain Tumor SPORE, A gift from Agilent technologies, and a Research Scholar Grant from the American Cancer Society (RSG-16–005-01).
SHL receives research grant from STCube Pharmaceuticals, Inc., Genentech Inc., Hitachi Chemical Diagnostics, Inc., and Peregrine Pharmaceuticals., and honorarium from AstraZeneca.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lewis JS, Ali S, Luo J, Thorstad WL & Madabhushi A A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am. J. Surg. Pathol 38, 128–137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D & Coussens LM Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Junttila MR & de Sauvage FJ Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med 348, 203–213 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Mahmoud SMA et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 29, 1949–1955 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Mlecnik B et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 29, 610–618 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Bremnes RM et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 6, 824–833 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Loi S et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02–98. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 31, 860–867 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Denkert C et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 28, 105–113 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Galon J et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313, 1960–1964 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Galon J et al. Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumours. J. Pathol 232, 199–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridman WH, Pagès F, Sautès-Fridman C & Galon J The immune contexture in humantumours: impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Brambilla E et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non-Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 34, 1223–1230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng Y et al. Prognostic Role of Tumor-Infiltrating Lymphocytes in Lung Cancer: a Meta-Analysis. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol 37, 1560–1571 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Jackute J et al. The prognostic influence of tumor infiltrating Foxp3(+)CD4(+), CD4(+) and CD8(+) T cells in resected non-small cell lung cancer. J. Inflamm. Lond. Engl 12, 63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye S-L, Li X-Y, Zhao K & Feng T High expression of CD8 predicts favorable prognosisin patients with lung adenocarcinoma: A cohort study. Medicine (Baltimore) 96, e6472 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruffini E et al. Clinical significance of tumor-infiltrating lymphocytes in lung neoplasms. Ann. Thorac. Surg 87, 365–371; discussion 371–372 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Seddiki N et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J. Exp. Med 203, 1693–1700 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakaguchi S, Yamaguchi T, Nomura T & Ono M Regulatory T cells and immunetolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Zheng Y et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Curiel TJ et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med 10, 942–949 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Bates GJ et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 24, 5373–5380 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Griffiths RW et al. Frequency of regulatory T cells in renal cell carcinoma patients and investigation of correlation with survival. Cancer Immunol. Immunother CII 56, 1743–1753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiraoka N, Onozato K, Kosuge T & Hirohashi S Prevalence of FOXP3+ regulatory Tcells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 12, 5423–5434 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Perrone G et al. Intratumoural FOXP3-positive regulatory T cells are associated with adverse prognosis in radically resected gastric cancer. Eur. J. Cancer Oxf. Engl. 1990 44, 1875–1882 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Badoual C et al. Prognostic value of tumor-infiltrating CD4+ T-cell subpopulations in head and neck cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res 12, 465–472 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Bron L et al. Prognostic value of arginase-II expression and regulatory T-cell infiltration in head and neck squamous cell carcinoma. Int. J. Cancer 132, E85–93 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Salama P et al. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol 27, 186–192 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Feichtenbeiner A et al. Critical role of spatial interaction between CD8+ and Foxp3+ cells in human gastric cancer: the distance matters. Cancer Immunol. Immunother CII 63, 111–119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Lieshout MNM & Baddeley AJ Indices of Dependence Between Types in Multivariate Point Patterns. Scand. J. Stat 26, 511–532 (1999). [Google Scholar]
- 32.Haase P Spatial pattern analysis in ecology based on Ripley’s K-function: Introduction and methods of edge correction. J. Veg. Sci 6, 575–582 (1995). [Google Scholar]
- 33.Fortin M-J, Dale MRT & ver Hoef J Spatial analysis in ecology. in Encyclopedia of Environmetrics 4, 2051–2058 (John Wiley & Sons, Ltd., 2002). [Google Scholar]
- 34.Carstens JL et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat. Commun 8, 15095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenblatt M Remarks on Some Nonparametric Estimates of a Density Function. Ann. Math. Stat 27, 832–837 (1956). [Google Scholar]
- 36.Parzen E On Estimation of a Probability Density Function and Mode. Ann. Math. Stat 33, 1065–1076 (1962). [Google Scholar]
- 37.Bowman AW & Azzalini A Applied Smoothing Techniques for Data Analysis: The Kernel Approach with S-Plus Illustrations (OUP; Oxford, 1997). [Google Scholar]
- 38.Benjamini Y & Hochberg Y Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol 57, 289–300 (1995). [Google Scholar]
- 39.Sepesi B et al. Tumor-Infiltrating Lymphocytes and Overall Survival in Surgically Resected Stage II and III Non-Small Cell Lung Cancer. Int. J. Radiat. Oncol. Biol. Phys 98, 223 (2017). [Google Scholar]
- 40.Balsat C et al. Improved computer-assisted analysis of the global lymphatic network in human cervical tissues. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 27, 887–898 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Kothari S, Phan JH, Stokes TH & Wang MD Pathology imaging informatics forquantitative analysis of whole-slide images. J. Am. Med. Inform. Assoc. JAMIA 20, 1099–1108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV & Fotiadis DI Machinelearning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J 13, 8–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Litjens G et al. A Survey on Deep Learning in Medical Image Analysis. ArXiv170205747 Cs (2017). [DOI] [PubMed]
- 44.Vignali DAA, Collison LW & Workman CJ How regulatory T cells work. Nat. Rev. Immunol 8, 523–532 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang HY & Wang R-F Regulatory T cells and cancer. Curr. Opin. Immunol 19, 217–223 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Grossman WJ et al. Differential expression of granzymes A and B in human cytotoxic lymphocyte subsets and T regulatory cells. Blood 104, 2840–2848 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Mazzaschi G et al. Low PD-1 Expression in Cytotoxic CD8+ Tumor-Infiltrating Lymphocytes Confers an Immune-Privileged Tissue Microenvironment in NSCLC with a Prognostic and Predictive Value. Clinical Cancer Research (2017). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


