Summary
Communication between the 5′ and 3′ ends of mature eukaryotic mRNAs lies at the heart of gene regulation, likely arising at the same time as the eukaryotic lineage itself. Our view of how and why it occurs has been shaped by elegant experiments that led to nearly universal acceptance of the ‘closed loop model’. However, new observations suggest that this classic model needs to be reexamined, revised, and expanded. Here, we address fundamental questions about the closed loop model, and we discuss how a growing understanding of mRNA structure, dynamics, and intermolecular interactions presents new experimental opportunities. We anticipate that the application of emerging methods will lead to expanded models that include the role of intrinsic mRNA structure and quantitative dynamic descriptions of 5′–3′ proximity linked to the functional status of an mRNA and will better reflect the messy realities of the crowded and rapidly changing cellular environment.
In brief
Communication between the two ends of mRNA is key to its fate. But how does it occur? Is it mediated by the so-called ‘closed-loop’, or is the inherent structure of RNA sufficient for this process? Vicens et al. re-examine current views, highlight new evidence, and suggest opportunities for future research.
Introduction
A common image in introductory biology textbooks is one of an extended eukaryotic messenger RNA (mRNA) circularized by a series of RNA–protein and protein–protein interactions at the 5′ and 3′ ends and with the intervening RNA ‘looped out’. By bringing the 5′ and 3′ ends into proximity, this ‘closed loop’ architecture is thought to govern the fate of a mature mRNA in the cytoplasm, controlling its translation efficiency and its eventual decay (Amrani et al., 2008; Amrani et al., 2006; Chen and Shyu, 2011; Gallie, 1991; Kahvejian et al., 2005; Wells et al., 1998). Despite near ubiquitous invocation, the importance and many characteristics of the closed loop model have recently been called into question. Specifically, as we discuss below, unanswered questions and new observations demand that this central idea of mRNA-centered post-transcriptional regulation be re-examined both intellectually and experimentally. Here, we briefly summarize evidence for, and issues with, the generally accepted version of the mRNA closed loop model. We discuss some new ways we might think about the nature of 5′–3′ proximity and propose possible avenues for exploration. We do not attempt a detailed comprehensive review of the literature or past experiments, but rather present an overview of key concepts with the goal of spurring discussion and encouraging creative experimentation.
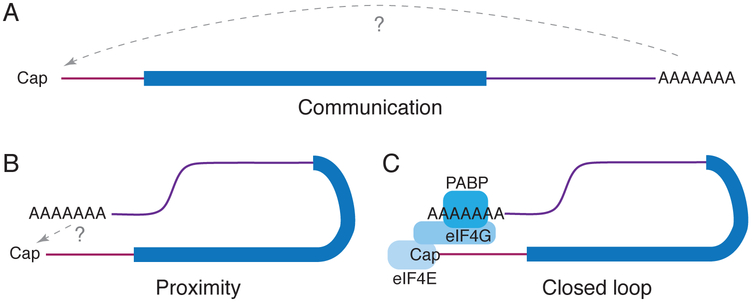
The term ‘closed loop model’ is often used interchangeably with 5′–3′ proximity and 5′–3′ communication, but these three ideas are best kept distinct. Here, we use ‘5′–3′ communication’ to refer to the conversion of regulatory information at the 3′ end of the mRNA (within the 3′ untranslated region [UTR] and poly(A) tail) into regulatory effects at the 5′ mRNA end, such as translation initiation or decapping, regardless of the molecular mechanism (Figure 1A). Formally, a signal could be transmitted from the 3′ to 5′ end along the length of an mRNA, but no signaling molecule with such property has been identified. Furthermore, 3′–5′ signal transmission along the mRNA is hard to reconcile with the presence of processive ribosomes moving in the opposite direction. Instead, when 5′–3′ communication exists, the most straightforward interpretation is that the two ends communicate by physical ‘5′–3′ proximity,’ which itself could theoretically be achieved in different ways (Figure 1B). One such way is through the ‘closed loop model,’ which refers to 5′–3′ proximity induced by four specific interactions: the 5′ cap bound by the cap binding protein eIF4E; eIF4E interacting with the translation initiation factor eIF4G; eIF4G in turn binding the poly(A) binding protein (PABP); and PABP recognizing the 3′ poly(A) tail (Figure 1C). Inherent in the closed loop model is the idea that the proximity of the 5′ and 3′ ends depends on these interactions; in other words, without those interactions, the ends are separated and so do not communicate. Stated simply, the cap–eIF4E–eIF4G–PABP–poly(A) interaction network that defines the closed loop model is an explanation for how the two ends of an mRNA are brought close together in order for 5′–3′ communication to occur, but it need not necessarily be the explanation.
Figure 1. Definitions used in this manuscript.
(A) 5′–3′ communication is the transfer of information from the 3′ to the 5′ end regardless of the mechanism. (B) 5′–3′ proximity relates to how information is generally assumed to be transferred but does not dictate how that proximity is achieved. (C) The closed loop model proposes that initiation factors eIF4E, eIF4G, and PABP are required to induce 5′–3′ proximity, while the intervening RNA loops out and the ends would be distant if not for these interactions. Maroon, 5′ UTR; dark blue, coding region; purple, 3′ UTR.
Evidence for 5′–3′ communication in eukaryotic mRNAs
Ample evidence supports the existence and importance of 5′–3′ communication in regulating the function and fate of eukaryotic mRNAs. As summarized below, this evidence is largely derived from functional studies in which perturbation of a sequence, structure, or interaction in the 3′ end of an mRNA alters events occurring at the 5′ end of that same mRNA. As stated above, we feel it is safe to assume that mRNAs achieve this through 5′–3′ proximity.
The classic (and most cited) example of 5′–3′ communication is the functional role of the poly(A) tail in mRNA translation and decay. Specifically, lengthening of the 3′ poly(A) tail is associated with an increase in cap-dependent translation initiation. A prominent example occurs during early embryonic development: maternally derived mRNAs have shortened poly(A) tails that are then lengthened upon fertilization, leading to translational activation (Barkoff et al., 1998; Gebauer et al., 1994). Shortening of the poly(A) tail not only decreases translation efficiency, but also targets the mRNA for decay via a pathway in which the 5′ end is decapped to allow 5′ to 3′ exonucleolytic degradation (Muhlrad et al., 1994; Yamashita et al., 2005). Overall, the evidence is strong enough to establish causation: changes in poly(A)-tail length are sufficient to change 5′ end activities. Thus, it is clear that the 5′ and 3′ ends of mRNAs communicate in a functionally important way and that the poly(A) tail is often a key determinant.
In addition to the poly(A) tail, elements in the encoded 3′ UTRs of both cellular and viral RNAs can alter events at the 5′ end, such as translation initiation or decapping. The 3′ UTRs of eukaryotic mRNAs are often quite long and contain a variety of regulatory RNA sequences and structural elements that are targets for interacting proteins and small regulatory RNAs (Bartel, 2018; Lai et al., 1999; Park and Maquat, 2013; Van Etten et al., 2012). These interactions can influence the fate of the mRNA, again often through a 5′ to 3′ decay process that must involve transfer of information between the two ends.
Finally, while they are not cellular mRNAs, eukaryote-infecting viral RNAs can be templates for translation and decay and thus effectively function in the cytoplasm as exogenous mRNAs. The fact that many viral RNAs use 5′–3′ communication extensively to regulate diverse processes, including translation, provides additional evidence for the importance of such communication. With viruses that use the same RNA template for both replication and translation (e.g. single-stranded positive-sense RNA viruses), 5′–3′ communication is considered critical for coordinating these two processes (Brinton and Miller, 2015). Importantly, many viruses (and some mRNAs) achieve 5′–3′ proximity without using the canonical cap, poly(A), and proteins associated with the closed loop model; they can use RNA-RNA interactions, diverse RNA protein interactions, or in some cases the mechanism is unknown (Chkuaseli and White, 2018; Filbin and Kieft, 2016; Nicholson and White, 2014). Some viruses even recruit translation initiation factors to their 3′ ends that are then used to initiate translation at the 5′ end, a process that requires 5′–3′ proximity (Simon and Miller, 2013; Truniger et al., 2017). The fact that biology has evolved many ways to achieve 5′–3′ proximity and subsequent communication underscores its power in regulation, but it also urges caution in oversimplifying how and why such communication occurs.
Evidence supporting the closed loop model
The overwhelming majority of eukaryotic mRNAs are capped and poly(A) tailed, and thus the closed loop model as depicted in Figure 1C has been accepted as a general regulatory mechanism for these mRNAs. The evidence supporting the importance of the cap–eIF4E–eIF4G–PABP–poly(A) interactions that define the closed loop model comes from biochemical, genetic and structural experiments, as well as evolution. First, the relevant RNA elements and proteins have been shown to specifically bind to each other, and disrupting these interactions leads to functional effects consistent with the closed loop model (Borman et al., 2000; Imataka et al., 1998; Kahvejian et al., 2005; Tarun and Sachs, 1996). For example, the binding of PABP to poly(A) increases the affinity of PABP for eIF4G and the affinity of eIF4E for the cap, which in turn should increase translation initiation (Borman et al., 2000). Consistent with this, mutations that abrogate or alter the PABP–eIF4G interaction result in a decrease in translation levels (Kahvejian et al., 2005). In terms of decay, shortening the 3′ poly(A) tail is an initial step in promoting decapping at the 5′ end and subsequent 5′ to 3′ decay (Muhlrad et al., 1994; Yamashita et al., 2005), which is generally assumed to be due to weakening of the closed loop interactions. Consistent with this model, yeast cells lacking PABP undergo deadenylation-independent decapping, and tethering of PABP in yeast to the 3′UTR inhibits decapping (Caponigro and Parker, 1995; Coller et al., 1998).
Structurally, the earliest evidence for circular mRNAs came from electron micrographs of polysomes on the ER (Christensen et al., 1987), and the closed loop architecture was subsequently visualized by atomic force microscopy (Wells et al., 1998). This second study remains the only direct structural evidence for the model. Here, circularized RNAs were formed only on capped and poly(A)-tailed model RNAs when all of the protein components were present and the eIF4E-eIF4G-PABP interactions were all possible (i.e., not perturbed by mutation). Importantly, the intervening RNA appeared looped out and unstructured, an observation supporting the closed loop model in which circularization of the mRNA depends on the aforementioned interactions. However, one crucial caveat in this work was that the RNA was hybridized to a complementary DNA molecule, which artificially held the RNA in this looped-out and seemingly unstructured state and may have amplified the need for cap–eIF4E–eIF4G–PABP–poly(A) interactions. Finally, the interactions between the RNA and proteins at the heart of the closed loop model have been maintained in most eukaryotes (Bannerman et al., 2018). The results of evolution provide some of the strongest evidence for the importance of the closed-loop interactions. Put together, there is clear evidence for 5′–3′ communication and for an important biological role for the cap–eIF4E–eIF4G–PABP–poly(A) interactions. Biochemical, structural and evolutionary analysis of these interactions have led to near universal acceptance of the ‘textbook’ closed loop model as a requirement for communication to occur on the vast majority of mRNAs.
Limitations of the closed loop model
Despite the evidence supporting it, several weaknesses of the closed loop model remain unaddressed. First, a prediction of the model is that disruption of any of the four core interactions will alter 5′–3′ proximity and communication, impair regulation of translation and mRNA stability, and, if on a global scale, broadly corrupt gene expression. However, no evidence currently exists to support this key prediction. Consistent with their central importance for gene expression, the loss of eIF4E, eIF4G, and PABP proteins is lethal in yeast and mammals (Caponigro and Parker, 1995; Hart et al., 2015). In contrast, perturbing the PABP–eIF4G interaction in S. cerevisiae does not cause lethality (Kessler and Sachs, 1998; Park et al., 2011). In the case of eIF4G mutants lacking the ability to bind PABP, this result has been rationalized by the realization that eIF4G has an RNA recognition motif that may bind 3′ UTRs and help circularize transcripts (Park et al., 2011). Importantly, such a model supports the importance of 5′–3′ communication without necessarily relying on the four closed loop interactions and thus supports the notion that other protein–RNA, protein–protein or RNA–RNA interactions may be involved in modulating 5′–3′ communication.
Similarly, the closed loop model predicts that disrupting interactions at the 5′ end should also impair post-transcriptional regulation. Cap-dependent translation initiation requires both the cap–eIF4E and eIF4E–eIF4G interactions outside of their roles in forming the closed loop (Jackson et al., 2010). Hence, if one of these interactions is disrupted, it is difficult to conclude whether an observed effect is due to altering the closed loop or due to altering basic steps of translation initiation. In the case of mRNA stability, disrupting the eIF4E–eIF4G interaction should provide a key test of the closed loop model. (Note that because dissociation of eIF4E from the cap is a key step in mRNA decay (Schwartz and Parker, 2000), affecting the eIF4E-cap interaction will likely have unintended consequences.) The prediction of the closed loop model is that inhibiting the interaction between eIF4E and eIF4G will decouple the ability of the 3′ end to regulate mRNA decapping. Once performed, such experiments will likely further influence the current view of the closed loop model.
Another critical unresolved issue with the closed loop model is what promotes these interactions occurring in cis, rather than in trans. A typical human cell contains hundreds of thousands of cytoplasmic mRNAs, and yet each gene may only express a handful of transcripts (Velculescu et al., 1999). While in cis interactions may be anticipated to be favored over in trans interactions, we assert this is an untested assumption given the effects of molecular crowding, localization of mRNAs in subcellular compartments, and the unknowns of high-resolution cellular organization. Thus, that trans 5′–3′ communication could formally also occur and thus a given mRNA could be regulated by 3′UTRs and poly(A) tails from other unrelated mRNAs. However, decades of research studying 5′–3′ communication have not produced evidence supporting trans regulation of this type; in fact, widespread trans effects between the 5′ and 3′ ends of different mRNAs would likely be random and incompatible with precise and accurate regulation of gene expression. In other words, cellular mRNAs must ensure that (a) 5′–3′ proximity occurs in cis and (b) it is robust enough to re-form in cis after any disruptions. However, as stated, the closed loop model does not provide an explanation for these two key issues without additional assumptions, suggesting that there must be more to 5′–3′ proximity and communication than the four interactions that define the closed loop.
5′–3′ proximity as an intrinsic property of RNA
The closed loop architecture is usually depicted with the RNA as an extended, unstructured molecule whose 5′ and 3′ ends are brought together only through the cap–eIF4E–eIF4G–PABP–poly(A) interaction (Figure 1C). This depiction, and the practice of generally drawing RNAs (mRNAs in particular) as horizontal lines with inherently distant ends, impacts how we think about the role of the closed loop interactions and the physical characteristics and role of the mRNA. In fact, the ability of every nucleobase to pair with every other nucleobase, the inherent flexibility of the RNA backbone, the power of base stacking, and multiple possible tertiary interactions mean that RNA is an inherently structured molecule regardless of whether or not it encodes a protein. In short, because of these properties, RNAs do not necessarily need to rely on external factors to achieve 5′–3′ communication.
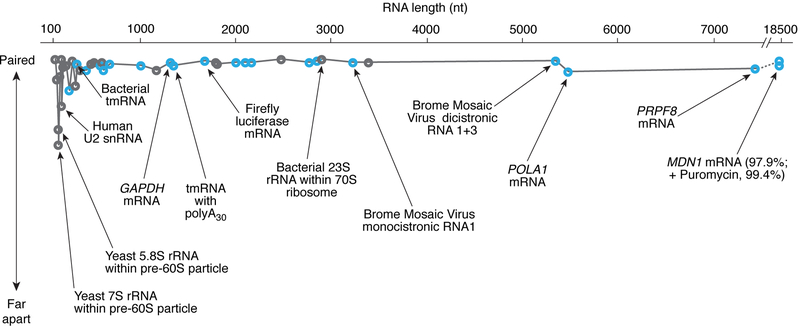
Regardless of their length, type, species, or complexity, the ends of diverse RNAs are surprisingly close (Figure 2, Table S1). As expected, the 5′ and 3′ ends are closest in tightly packed RNAs such as rRNAs and ribozymes, and farther apart in less tightly packed RNAs. But, as we argue below, there is evidence that even within large mRNAs, the 5′ and 3′ ends are often relatively close in comparison to what would be expected for fully extended transcripts (Figure 2; Table S1). These observations suggest that most RNAs likely adopt global structures that bring their 5′ and 3′ ends together. This nonintuitive idea may not be far-fetched, as global mapping in various living cells revealed that 30-40% of RNA duplexes bridge sequence distances of more than 200 nucleotides (Lu et al., 2016). On their own, the intrinsic architectural properties of RNA are sufficient to account for such long-distance juxtaposition.
Figure 2. The length of an RNA has no significant impact on how close the 5’ and 3’ ends are.
Relative 5′-3′ proximity —i.e., how close the ends are compared to the same RNA in an extended conformation— is plotted as a function of RNA length. Specifically, 5′–3′ proximity is defined as [1 – (dactual/dextended)] × 100, where dactual is an experimentally determined 5’–3’ distance, and dextended is the distance that would separate the 5′ and 3′ ends if the RNA adopted an extended conformation, with each base separated from its neighbor by 3.5 Å (i.e., dextended = # of residues × 0.35; see complete list in Table S1). 100% corresponds to base pairing between the 5′-most and the 3′-most nucleotides. Many 1000– 6000-nt long mRNAs from viruses and eukaryotes have a relative 5′–3′ proximity >98.5% (Lai et al., 2018; Leija-Martinez et al., 2014). A striking example is the 18,431 nt long MDN1 mRNA, for which a 136 nm median distance between the 5′ and 3′ ends (only 7 times larger than the diameter of a eukaryotic ribosome) is ~45-fold smaller than the distance within an extended coil conformation (>6 μm); upon removal of ribosomes, the MDN1 mRNA further compacts so that the median distance is 36 nm (Adivarahan et al., 2018). Coding RNAs are highlighted in blue; noncoding RNAs in grey.
By definition, simple antiparallel secondary structures such as hairpins bring 5′ and 3′ ends together. Because RNA architecture is modular (Westhof et al., 1996), large assemblies of RNA secondary structures likely retain this property. Certainly, for RNAs such as group I introns (Adams et al., 2004; Golden et al., 2005; Guo et al., 2004), ribosomal RNA (Garreau de Loubresse et al., 2014; Noeske et al., 2015), telomerase RNA (Nguyen et al., 2018) or snRNAs within the spliceosome (Plaschka et al., 2017; Sun et al., 2017), the ends are very close (~3 nm apart on average) (Figure 2; Table S1). While these examples could represent cases in which the natural tendency of RNA molecules to bring their ends together has been maximally harnessed, there is no obvious reason why lncRNAs, most of which are thought to consist of sequences of loosely connected structured domains (Mercer and Mattick, 2013; Novikova et al., 2012), and even mRNAs, would not adhere to this inherent property of RNA (Ding et al., 2014; Martin et al., 2011; Memczak et al., 2013; Metkar et al, 2018; Rouskin et al., 2014; von Moeller et al., 2013; Wan et al., 2014). In fact, there is no direct evidence that any RNA ever exists in a fully extended state, irrespective of the usual depictions. Interestingly, similar observations have been made about the spatial proximity of N-terminal and C-terminal ends in proteins, for which protein folding was proposed to be the main driver (Krishna and Englander, 2005).
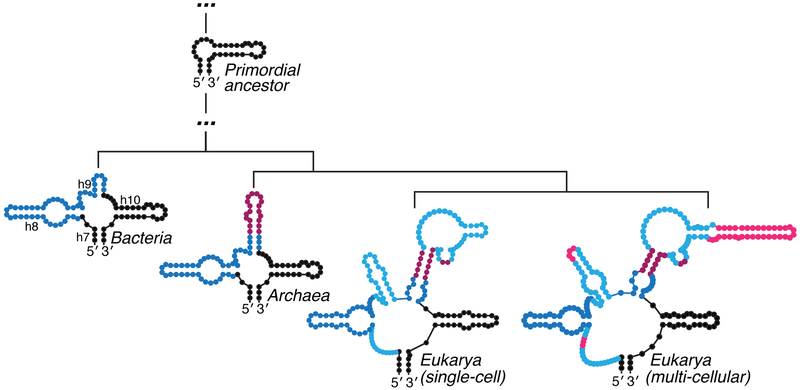
Evolutionary and functional properties of RNA, in general, are also in accord with maintaining intrinsic 5′–3′ proximity in mRNAs. A comparative analysis of ribosomes from various species (Bokov and Steinberg, 2009; Petrov et al., 2014; Petrov et al., 2015) supports earlier proposals that modern ribosomes evolved from RNAs that were, at most, a few hundred nucleotides long (Noller, 1999). Over time, expansions of rRNA occurred within loops and junctions, but the original 5′–3′ proximity was not disrupted (Figure 3). Similar additions have been described in RNase P (Kachouri et al., 2005). This model for evolution of RNA by internal accretion also accounts for what has been described as “the fractal nature of RNA secondary structure” (Purugganan, 1989). In fact, one would expect evolution to maintain intrinsic long-range contacts in RNA as it is required for many RNA-centric processes in addition to mRNA translation and decay. For example, RNA elements distant in sequence must be proximal during splicing, and this aspect can also be exploited to form circular regulatory RNAs in eukaryotes (Memczak et al., 2013).
Figure 3. The addition of new RNA during evolution does not disrupt 5′–3′ proximity.
The figure shows the evolution by accretion of SSU rRNA helices 7-10/expansion segment 3 for species of increasing complexity (adapted from (Petrov et al., 2014; Petrov et al., 2015)). Two-dimensional structures for bacteria, archaea and eukarya RNA are shown to be preserved upon serial insertion of new RNA (color coded from more ancient to recent addition as follows: dark blue, maroon, light blue, pink). 2D diagrams were drawn from PDB IDs 4YBB (E. coli) (Noeske et al., 2015), 4U3M (S. cerevisiae) (Garreau de Loubresse et al., 2014) and 4V6X (H. sapiens) (Anger et al., 2013) using RNApdbee v. 2.0 (Antczak et al., 2014) and VARNA v. 3.9 (Darty et al., 2009).
There is sufficient evidence to seriously consider the idea that inherent properties of RNA promote and conserve end-to-end spatial proximity in mRNAs and thus the cap–eIF4E–eIF4G–PABP–poly(A) interactions are not required for the ends of an mRNA to be proximal. Thus, an adjusted way to think about the closed loop interactions is that they exploit, reinforce, or sense the inherent proximity of the 5′ and 3′ ends, rather than induce that proximity. As a corollary, under some circumstances, proteins other than initiation factors and PABP may bind to mRNAs to keep the 5′ and 3′ ends apart, thereby regulating its fate.
Emerging questions and opportunities
Despite being a major focus of research, there is much about 5′–3′ communication that we do not understand. Here, we outline a few questions that we find pressing and some ideas for future research; we are excited to note that inquiry into some has already begun.
How close is close?
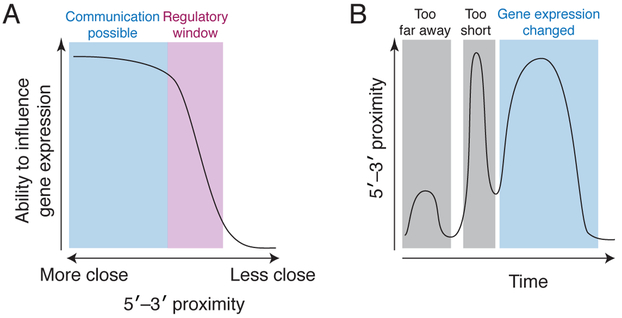
Embedded in every model of post-transcriptional regulation is the idea that the ends of an mRNA must be ‘close’ for 5′–3′ communication to occur. But ‘close’ or ‘proximal’ are non-quantitative terms. Surprisingly, more than two decades after the first observations of cap–poly(A)–tail synergy (Gallie, 1991), we still do not know whether ‘close’ means 1, 10, or 100 nm. Furthermore, functional ‘closeness’ may be different for different types of regulation (e.g., translation initiation, ribosome recycling, and mRNA decapping), and thus different regulatory mechanisms may not respond in the same way to a 2 or 5-fold change in proximity (Figure 4A). Developing a quantitative framework for the relationship between gene regulation and proximity in absolute terms is essential, although challenging; these studies ultimately will likely require single-molecule resolution to correlate a specific measurement of proximity on individual mRNAs to their functional status.
Figure 4. Linking quantitative and temporally resolved studies of 5′–3′ proximity to function.
(A) A hypothetical idealized curve of how the spatial proximity of the 5′–3′ ends of an mRNA results in changes in gene expression. The absolute values associated with communication and the regulatory window remain unknown. In addition, this curve could be different in terms of shape and position for different processes (e.g., translation initiation versus decay), for different mRNAs, or in the presence of different regulatory factors. These effects, combined with dynamic changes in the distance between the 5′–3′ ends, could provide process control of gene expression. (B) This graph depicts a hypothetical example of the 5′–3′ proximity (y-axis) as a function of time (x-axis) on a single mRNA, with the resulting changes in gene expression stated above the graph. In this example, the 5′–3′ ends are initially too far apart to allow communication (left gray box). Then the ends move closer together, but the period of time that they spend close is too short to induce changes in gene expression (middle grey box). It is only when the ends are close enough for a long enough period of time that gene expression changes (cyan box). We lack any quantitative understanding of this relationship, which may vary between different mRNAs, processes, and cellular contexts.
What are the conformational and temporal dynamics of 5′–3′ communication?
Everything in biology is in motion, and thus 5′–3′ proximity and communication must be viewed through the lens of dynamics and kinetics (Figure 4B). The closed loop model is usually depicted as a static interaction, but the 5′ and 3′ ends may not be close to one another all the time. As a point of reference, the elongating ribosome adds 6–10 amino acids per second (Morisaki et al., 2016), but there are no such measurements for the interactions and structures that govern 5′–3′ proximity. Are the 5′ and 3′ ends always the same distance apart? Or is it a transient interaction? If transient, how long is ‘long enough’ for downstream events to occur (a millisecond, one second, ten seconds, 100 seconds or longer?). These questions intertwine with those in the previous paragraph, as a full understanding of ‘how close is close’ requires knowing how long the ends spend at specific distances. Our lack of understanding of in vivo kinetics for various binding events and 5′–3′ proximity represents a major gap.
What inputs or contexts induce changes in 5′–3′proximity and communication?
In its most simple form, the closed loop model implies that 5′–3′ communication is either ‘on’ or ‘off’ and is wholly dependent on the cap-eIF4E-eIF4G-PABP-poly(A) interactions. But in fact, a more dynamic view of changing absolute proximity presents regulatory opportunities more consistent with biology. Different RNAs may have different inherent propensities for 5′–3′ proximity, and the conformation of a given RNA could be substantially impacted by the proteins bound to it at different times. In other words, there may be regulatory processes that purposedly maintain separation of the two RNA ends. We know little about how proteins other than those involved in the closed loop model affect the separation between the two ends. Things get even more complicated when one considers that different cell types and organisms have a different set of proteins that might bind mRNAs and alter their global structure. For example, the possibility exists that for a given mRNA, the time at which the 5′ and 3′ ends are in proper spatial proximity for a given function differs in different cell types based on changing protein populations.
How does translation itself alter 5′–3′ proximity?
In the standard depiction of the closed loop model, ribosomes initiate and then elongate around the looped out RNA with little effect on the mRNAs global structure because the circle is held closed only at its ends. If we think of mRNAs as inherently structured, then the processive unwinding activity of a series of ribosomes may be a major force in modulating inherent 5′–3′ proximity. Indeed, recent studies using single-molecule fluorescent in situ hybridization (smFISH) showed that 5′ and 3′ ends are > 90 nm apart for mRNAs > 5000 nt long that are being translated, and that removing ribosomes resulted in the ends being closer (~30 nm) (Table S1) (Adivarahan et al., 2018; Khong and Parker, 2018). In addition, electron microscopy studies show polysomes in a variety of superstructures that appear to change depending on the type of the mRNA being translated, the translation status, and constraints such as ribosome binding to the endoplasmic reticulum (Afonina et al., 2014, 2015; Afonina Zh et al., 2013; Brandt et al., 2010; Brandt et al., 2009; Kopeina et al., 2008; Myasnikov et al., 2014). One intriguing possibility is that the longstanding relationship between translational inhibition and mRNA decay may be explained in terms of changing the dynamics of 5′–3′ proximity. That is, inhibiting initiation gives the decay machinery, which assembles on the 3′ end, enough time to act on the 5′ end. Again, this can probably only be understood by correlating absolute values of 5′–3′ distance and dynamic changes in that distance with the translation/decay status of individual mRNAs.
The questions stated above are by no means an exhaustive list, but they illustrate the type of thorny issues with which the field must —and is starting to— wrestle. Emerging technologies, like smFISH, sophisticated applications of high-throughput sequencing, and improvements in electron and light microscopy have provided new entry points and promise to continue to give insight. However, continued creative application of these tools must be combined with development of new methods. First, given the speed of translation elongation and critical range of distances, we suspect that moving forward will require tools with even higher temporal and spatial resolution. It will also be essential to use approaches that are nondisruptive and can be used in living cells. In addition, there is a need for better ways to directly assess the global conformation and 5′–3′ proximity on a given mRNA. smFISH readily monitors 5′–3′ proximity in the cell but is less adept at identifying 5′ and 3′ ends in extended conformations. Furthermore, overall fluorescence methods inform only indirectly on the global structure of an mRNA, and so we can imagine combining time-resolved smFISH with chemical probing analysis using emerging in-cell probing (Spitale et al., 2015) (Feng et al., 2018) and spectroscopic methods (Giassa et al., 2018). Finally, although past experiments have been informative, no approach has provided a way to modulate 5′–3′ proximity in a controlled way. Given the inherent properties of RNA, developing this and other tools constitutes a nontrivial task, which will nonetheless be critical for moving beyond correlative observations.
Supplementary Material
Acknowledgements
We thank members of the Rissland and Kieft labs for stimulating and thoughtful discussions, as well as Roy Parker, Anthony Khong, and Rachel Green for useful comments on the manuscript. Support was provided from NIH grants R35GM118070 (JSK), R01AI133348 (JSK), and R35GM128680 (OSR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Adams PL, Stahley MR, Kosek AB, Wang J, and Strobel SA (2004). Crystal structure of a self-splicing group I intron with both exons. Nature 430, 45–50. [DOI] [PubMed] [Google Scholar]
- Adivarahan S, Livingston N, Nicholson B, Rahman S, Wu B, Rissland O, and Zenklusen D (2018). Spatial organization of single mRNPs at different stages of the gene expression pathway. Mol Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonina ZA, Myasnikov AG, Shirokov VA, Klaholz BP, and Spirin AS (2014). Formation of circular polyribosomes on eukaryotic mRNA without cap-structure and poly(A)-tail: a cryo electron tomography study. Nucleic Acids Res 42, 9461–9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonina ZA, Myasnikov AG, Shirokov VA, Klaholz BP, and Spirin AS (2015). Conformation transitions of eukaryotic polyribosomes during multi-round translation. Nucleic Acids Res 43, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonina Zh A, Myasnikov AG, Khabibullina NF, Belorusova AY, Menetret JF, Vasiliev VD, Klaholz BP, Shirokov VA, and Spirin AS (2013). Topology of mRNA chain in isolated eukaryotic double-row polyribosomes. Biochemistry (Mosc) 78, 445–454. [DOI] [PubMed] [Google Scholar]
- Amrani N, Ghosh S, Mangus DA, and Jacobson A (2008). Translation factors promote the formation of two states of the closed-loop mRNP. Nature 453, 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Sachs MS, and Jacobson A (2006). Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol 7, 415–425. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, and Beckmann R (2013). Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85. [DOI] [PubMed] [Google Scholar]
- Antczak M, Zok T, Popenda M, Lukasiak P, Adamiak RW, Blazewicz J, and Szachniuk M (2014). RNApdbee--a webserver to derive secondary structures from pdb files of knotted and unknotted RNAs. Nucleic Acids Res 42, W368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannerman BP, Kramer S, Dorrell RG, and Carrington M (2018). Multispecies reconstructions uncover widespread conservation, and lineage-specific elaborations in eukaryotic mRNA metabolism. PLoS One 13, e0192633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoff A, Ballantyne S, and Wickens M (1998). Meiotic maturation in Xenopus requires polyadenylation of multiple mRNAs. EMBO J 17, 3168–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokov K, and Steinberg SV (2009). A hierarchical model for evolution of 23S ribosomal RNA. Nature 457, 977–980. [DOI] [PubMed] [Google Scholar]
- Borman AM, Michel YM, and Kean KM (2000). Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5'-end. Nucleic Acids Res 28, 4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt F, Carlson LA, Hartl FU, Baumeister W, and Grunewald K (2010). The three-dimensional organization of polyribosomes in intact human cells. Mol Cell 39, 560–569. [DOI] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, and Baumeister W (2009). The native 3D organization of bacterial polysomes. Cell 136, 261–271. [DOI] [PubMed] [Google Scholar]
- Brinton MA, and Miller WA (2015). Positive strand RNA virus replication: It depends on the ends. Virus Res 206, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, and Parker R (1995). Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev 9, 2421–2432. [DOI] [PubMed] [Google Scholar]
- Chen CY, and Shyu AB (2011). Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2, 167–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chkuaseli T, and White KA (2018). Intragenomic Long-Distance RNA-RNA Interactions in Plus-Strand RNA Plant Viruses. Front Microbiol 9, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AK, Kahn LE, and Bourne CM (1987). Circular polysomes predominate on the rough endoplasmic reticulum of somatotropes and mammotropes in the rat anterior pituitary. Am J Anat 178, 1–10. [DOI] [PubMed] [Google Scholar]
- Coller JM, Gray NK, and Wickens MP (1998). mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev 12, 3226–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, and Ponty Y (2009). VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, and Assmann SM (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700. [DOI] [PubMed] [Google Scholar]
- Feng C, Chan D, Joseph J, Muuronen M, Coldren WH, Dai N, Correa IR Jr., Furche F, Hadad CM, and Spitale RC (2018). Light-activated chemical probing of nucleobase solvent accessibility inside cells. Nat Chem Biol 14, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin ME, and Kieft JS (2016). Linking Alpha to Omega: diverse and dynamic RNA-based mechanisms to regulate gene expression by 5'-to-3' communication. F1000Res 5 (F1000 Faculty Rev): 2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR (1991). The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, and Yusupov M (2014). Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Xu W, Cooper GM, and Richter JD (1994). Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J 13, 5712–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giassa IC, Rynes J, Fessl T, Foldynova-Trantirkova S, and Trantirek L (2018). Advances in the cellular structural biology of nucleic acids. FEBS Lett 592, 1997–2011. [DOI] [PubMed] [Google Scholar]
- Golden BL, Kim H, and Chase E (2005). Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol 12, 82–89. [DOI] [PubMed] [Google Scholar]
- Guo F, Gooding AR, and Cech TR (2004). Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell 16, 351–362. [DOI] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. (2015). High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Imataka H, Gradi A, and Sonenberg N (1998). A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, and Pestova TV (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachouri R, Stribinskis V, Zhu Y, Ramos KS, Westhof E, and Li Y (2005). A surprisingly large RNase P RNA in Candida glabrata. RNA 11, 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, and Sonenberg N (2005). Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler SH, and Sachs AB (1998). RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol Cell Biol 18, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, and Parker R (2018). mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. bioRxiv 366690; doi: https://doi.org/10.1101/366690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopeina GS, Afonina ZA, Gromova KV, Shirokov VA, Vasiliev VD, and Spirin AS (2008). Step-wise formation of eukaryotic double-row polyribosomes and circular translation of polysomal mRNA. Nucleic Acids Res 36, 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna MM, and Englander SW (2005). The N-terminal to C-terminal motif in protein folding and function. Proc Natl Acad Sci U S A 102, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W-JC, Kayedkhordeh M, Cornell EV, Farah E, Bellaousov S, Rietmeijer R, Mathews DH, and Ermolenko DN (2018). The formation of intramolecular secondary structure brings mRNA ends in close proximity. bioRxiv 289496; doi: https://doi.org/10.1101/289496. [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, and Blackshear PJ (1999). Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leija-Martinez N, Casas-Flores S, Cadena-Nava RD, Roca JA, Mendez-Cabanas JA, Gomez E, and Ruiz-Garcia J (2014). The separation between the 5'-3' ends in long RNA molecules is short and nearly constant. Nucleic Acids Res 42, 13963–13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. (2016). RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 165, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Barends S, Jaeger S, Schaeffer L, Prongidi-Fix L, and Eriani G (2011). Cap-assisted internal initiation of translation of histone H4. Mol Cell 41, 197–209. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- Mercer TR, and Mattick JS (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20, 300–307. [DOI] [PubMed] [Google Scholar]
- Metkar M, Ozadam H, Lajoie BR, Imakaev M, Mirny LA, Dekker J & Moore MJ (2018) Higher-order organization principles of pre-translational mRNPs. Mol Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL, et al. (2016). Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, and Parker R (1994). Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes Dev 8, 855–866. [DOI] [PubMed] [Google Scholar]
- Myasnikov AG, Afonina ZA, Menetret JF, Shirokov VA, Spirin AS, and Klaholz BP (2014). The molecular structure of the left-handed supra-molecular helix of eukaryotic polyribosomes. Nat Commun 5, 5294. [DOI] [PubMed] [Google Scholar]
- Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, and Collins K (2018). Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, and White KA (2014). Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol 12, 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, and Cate JH (2015). High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol 22, 336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF (1999). On the Origin of the Ribosome: Coevolution of Subdomains of tRNA and rRNA In The RNA World, 2nd Ed., Gesteland RF, Cech TR, and Atkins JF, eds. (Cold Spring Harbor Laboratory Press; ), pp. 197–219. [Google Scholar]
- Novikova IV, Hennelly SP, and Sanbonmatsu KY (2012). Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res 40, 5034–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, and Maquat LE (2013). Staufen-mediated mRNA decay. Wiley Interdiscip Rev RNA 4, 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, and Hinnebusch AG (2011). Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1*PABP mRNPs in vivo. EMBO J 30, 302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, et al. (2014). Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci U S A 111, 10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, et al. (2015). History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A 112, 15396–15401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin PC, and Nagai K (2017). Structure of a pre-catalytic spliceosome. Nature 546, 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD (1989). The fractal nature of RNA secondary structure. Naturwissenschaften 76, 471–473. [DOI] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, and Weissman JS (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, and Parker R (2000). mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20, 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AE, and Miller WA (2013). 3' cap-independent translation enhancers of plant viruses. Annu Rev Microbiol 67, 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C, et al. (2017). Molecular architecture of the 90S small subunit pre-ribosome. Elife 6 DOI:10.7554/eLife.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ Jr., and Sachs AB (1996). Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Truniger V, Miras M, and Aranda MA (2017). Structural and Functional Diversity of Plant Virus 3'-Cap-Independent Translation Enhancers (3'-CITEs). Front Plant Sci 8, 2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, and Goldstrohm AC (2012). Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem 287, 36370–36383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, et al. (1999). Analysis of human transcriptomes. Nat Genet 23, 387–388. [DOI] [PubMed] [Google Scholar]
- von Moeller H, Lerner R, Ricciardi A, Basquin C, Marzluff WF, and Conti E (2013). Structural and biochemical studies of SLIP1-SLBP identify DBP5 and eIF3g as SLIP1-binding proteins. Nucleic Acids Res 41, 7960–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, et al. (2014). Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, and Sachs AB (1998). Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2, 135–140. [DOI] [PubMed] [Google Scholar]
- Westhof E, Masquida B, and Jaeger L (1996). RNA tectonics: towards RNA design. Fold Des 1, R78–88. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, and Shyu AB (2005). Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12, 1054–1063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.