SUMMARY
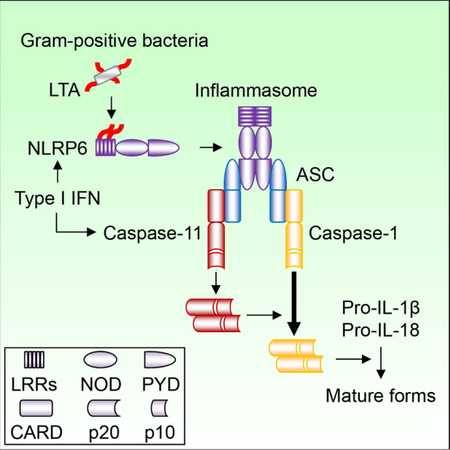
The activator and composition of the NLRP6 inflammasome remain poorly understood. We find that lipoteichoic acid (LTA), a molecule produced by Gram-positive bacteria, binds and activates NLRP6. In response to cytosolic LTA or infection with Listeria monocytogenes, NLRP6 recruited caspase-11 and caspase-1 via the adaptor ASC. NLRP6 activation by LTA induced processing of caspase-11, which promoted caspase-1 activation and IL-1β/IL-18 maturation in macrophages. Nlrp6−/− and Casp11−/− mice were less susceptible to Listeria monocytogenes infection, which was associated with reduced pathogen loads and impaired IL-18 production. Administration of IL-18 to Nlrp6−/− or Casp11−/− mice restored the susceptibility of mutant mice to Listeria monocytogenes infection. These results reveal a previously unrecognized innate immunity pathway triggered by cytosolic LTA that is sensed by NLRP6 and exacerbates systemic Gram-positive pathogen infection via the production of IL-18.
Keywords: Inflammasome, NLRP6, Lipoteichoic acid, Caspase-11, Caspase-1, Gram-positive bacteria, Listeria monocytogenes, IL-18
Graphical Abstract
ETOC BLURB
Lipoteichoid acid produced by Gram-negative bacteria is sensed by the NLRP6 inflammasome and leads to the activation of both caspase-1 and caspase-11, exacerbating infection
INTRODUCTION
Inflammasomes are genetically encoded signaling complexes that drive the activation of inflammatory caspases and induction of immune responses and pyroptosis, a proinflammatory form of cell death (Martinon and Tschopp, 2007). In response to diverse microbial and endogenous stimuli, phagocytes induce the activation of canonical inflammasomes including NLRP3, NLRC4 and AIM2 that recruits the common adaptor ASC to activate caspase-1, leading to the secretion of mature interleukin-1β (IL-1β) and IL-18 as well as the induction of pyroptosis (Schroder and Tschopp, 2010). In contrast, the noncanonical inflammasome is triggered by the binding of cytosolic lipopolysaccharide (LPS) to another class of inflammatory caspases that includes murine caspase-11 (Kayagaki et al., 2011; Kayagaki et al., 2013; Shi et al., 2014). Cytosolic LPS induces oligomerization of caspase-11 that targets gasdermin D (GSDMD) to induce pore formation in the absence of pro-caspase-11 processing (Hagar et al., 2013; Shi et al., 2014). However, recent studies showed that caspase-11 is self-cleaved after LPS stimulation (Lee et al., 2018). In addition, endogenous oxidized phospholipids bind to caspase-11 directly to induce IL-1β secretion without inducing pyroptosis in dendritic cells (Zanoni et al., 2016).
NLRP6, a member of the NOD-like receptor family, can form an inflammasome that is involved in the recognition of microbes and intestinal homeostasis (Elinav et al., 2011; Henao-Mejia et al., 2012; Levy et al., 2015; Normand et al., 2011; Wang et al., 2015; Wlodarska et al., 2014). In the intestine, NLRP6 is expressed in intestinal epithelial cells and promotes IL-18 production and epithelial cell repair in response to chemically-induced intestinal injury (Chen et al., 2011; Elinav et al., 2011; Huber et al., 2012). NLRP6 also acts within intestinal goblet cells to regulate mucus production (Birchenough et al., 2016; Wlodarska et al., 2014). In addition, NLRP6 functions within macrophages to limit commensal bacteria-driven inflammation in the intestine and to regulate systemic infection by bacterial pathogens (Anand et al., 2012; Seregin et al., 2016). Although NLRP6 can associate with caspase-1 and the adaptor ASC in overexpression studies (Levy et al., 2015), the nature of the NLRP6 inflammasome and its agonist remain unclear. In this study, we found that LTA, a molecule produced by Gram-positive bacteria, binds and activates NLRP6 leading to the recruitment and processing of caspase-11 via the adaptor ASC. Upon Listeria monocytogenes infection or the presence of cytosolic LTA, NLRP6 formed a protein complex containing caspase-11 and caspase-1 that regulates IL-18 secretion in macrophages. Our results provide new insights into the mechanism by which NLRP6 inflammasome is activated and recruits proinflammatory caspases in macrophages after microbial infection as well as the function of the NLRP6-caspase-11 pathway in systemic infection by Gram-positive bacterial pathogens.
RESULTS
Caspase-11 is processed in macrophages infected with Listeria monocytogenes and Staphylococcus aureus through NLRP6 to promote caspase-1 activation
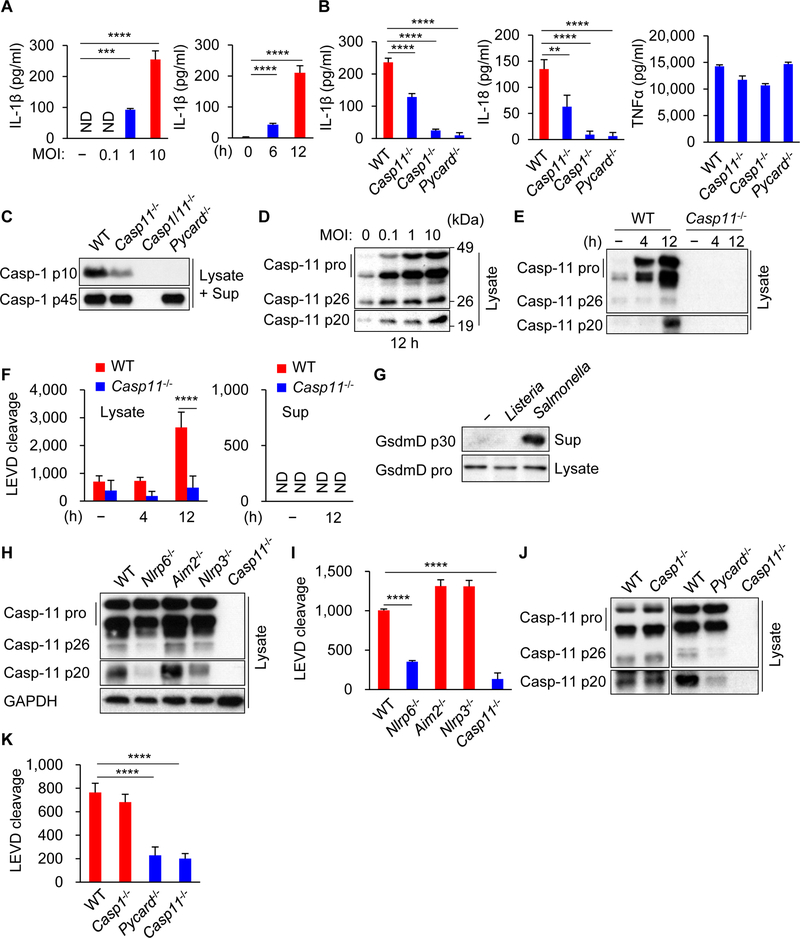
To assess whether caspase-11 can be activated by Gram-positive bacteria, bone marrow-derived macrophages (BMDMs) from wild-type (WT) and mutant mice were infected with the intracytosolic pathogen Listeria monocytogenes (Listeria). The release of IL-1β and IL-18, but not TNFα, was impaired in infected Casp11−/− BMDMs compared with WT cells (Figure 1A and 1B). Furthermore, IL-1β and IL-18 secretion induced by Listeria infection were abolished in Casp1−/−Casp11+/+ and Pycard−/− BMDMs (Figure 1B). Consistently, activation of caspase-1 induced by Listeria was reduced in Casp11−/− BMDMs and abolished in BMDMs deficient in the adaptor ASC (Figure 1C). Listeria infection induced caspase-11 expression and the cleavage of pro-caspase-11 as determined by immunoblotting with an antibody that recognizes the cleaved p20 form of caspase-11 (Figure 1D, 1E, S1A and S1B). To verify these results, we assessed the ability of cell extracts to cleave Ac-Leu-Glu-Val-Asp-7-Amino-4-methylcoumarin (Ac-LEVD-AMC), a fluorogenic substrate of mouse caspase-11 and its human caspase-4 and −5 orthologs (Martinon and Tschopp, 2007). Cell extracts from Listeria-infected BMDMs cleaved the Ac-LEVD-AMC substrate that required caspase-11, but not the caspase-8 substrate Ac-IETD-AMC or caspase-3 substrate Ac-DMQD-AMC (Figure 1F, S2A and S2B). Unlike Salmonella enterica serovar Typhimurium (Salmonella) (He et al., 2015), Listeria induced no or undetectable cleavage of pore-forming GSDMD which was associated with little cell death when compared to Salmonella (Figure 1G and S1C). To identify innate immune factors that may be required for caspase-11 cleavage, BMDMs from WT mice and mice deficient for AIM2, NLRP3 or NLRP6 as well as mutant mice lacking only caspase-1 or the adaptor ASC were infected with Listeria. The analysis revealed that NLRP6, but not AIM2 or NLRP3, was required for caspase-11 cleavage in response to Listeria infection (Figure 1H). Consistent with these results, cell extracts from Listeria-infected WT BMDMs cleaved the Ac-LEVD-AMC substrate, which was impaired in Nlrp6−/− and Casp11−/− cells, but not in Nlrp3−/− or Aim2−/− cells (Figure 1I). Additionally, Nlrp6−/− BMDMs showed reduced IL-1β and IL-18 secretion and caspase-1 activation (Figure S1D−F). Furthermore, caspase-11 cleavage induced by Listeria was significantly decreased in ASC-deficient BMDMs, but not in Casp1−/− cells which correlated with Ac-LEVD-AMC cleavage activity (Figure 1J and 1K). In addition to mouse macrophages, human THP-1 cells deficient in caspase-4 and −5 released less IL-1β than WT cells in responce to Listeria infection (Figure S1G). Another Gram-positive pathogen, Staphylococcus aureus, also induced the processing of caspase-11 and cleavage of Ac-LEVD-AMC in an NLRP6-dependent manner (Figure S1H and S1I). These results indicate that NLRP6 and ASC act upstream of caspase-11 cleavage to promote caspase-1-dependent IL-1β and IL-18 secretion in macrophages infected with Listeria.
Figure 1. Caspase-11 is processed in macrophages infected with Listeria through NLRP6 and ASC to promote caspase-1 activation.
Primary BMDMs were left uninfected or infected with Listeria at MOI = 10 or indicated MOI for 12 h or indicated times. (A, B) The supernatants were subjected to ELISA. (C−E, G, H, J) The cell lysates (Lysate) and supernatants (Sup) were subjected to immunoblotting, or (F, I, K) the lysates and (F) supernatants were subjected to caspase substrate cleavage assay. Blots of caspase-11 were cropped to reveal protein bands at different exposures. Results are representative of at least three independent experiments, and error bars denote s.d. of triplicate wells. ND, not detected. **P < 0.01, ***P < 0.001, ****P < 0.0001. See also Figures S1 and S2.
Cytosolic LTA is sensed by NLRP6 to trigger caspase-11 cleavage
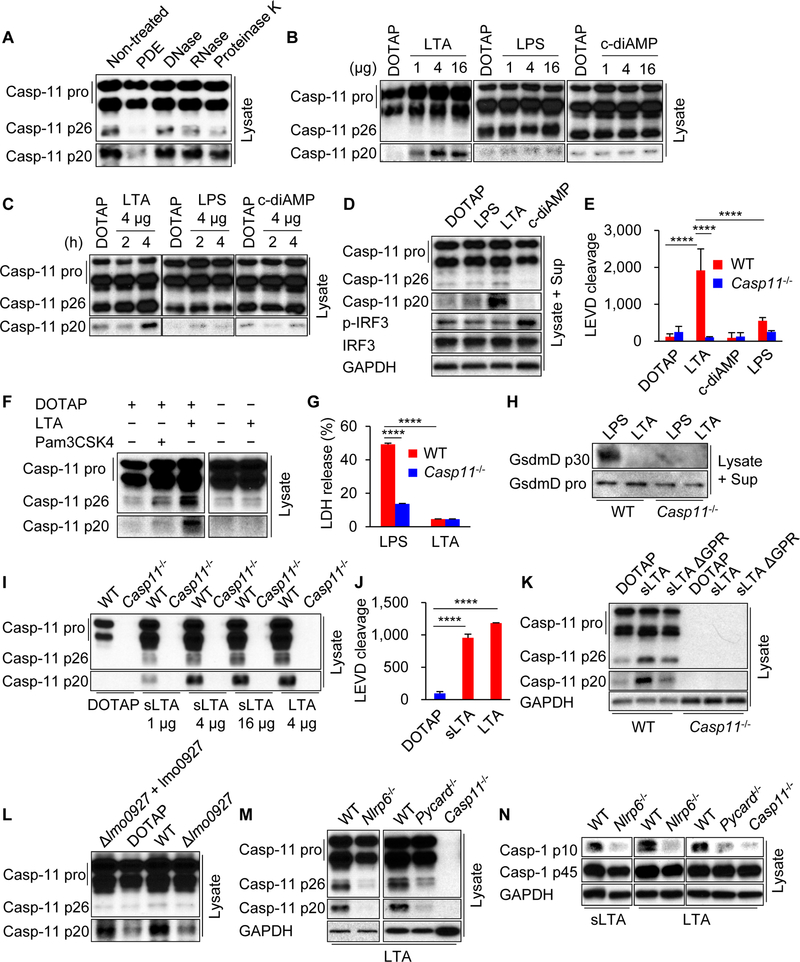
We next sought to identify the bacterial component that triggers caspase-11 cleavage during Listeria infection. Transfection of Listeria extracts into the cytosol of macrophages induced cleavage of caspase-11 (Figure 2A). The caspase-11 p20 cleavage product was markedly decreased after pre-treatment of bacterial lysates with phosphodiesterase (PDE), but not with DNase, RNase or Proteinase K (Figure 2A). Listeria produces LTA and cyclic-diAMP, which are sensitive to PDE due to the presence of phosphodiester bonds (Kolb-Maurer et al., 2003; Woodward et al., 2010). The transfection of LTA, but not cyclic-diAMP, into BMDMs resulted in the appearance of the caspase-11 p20 band associated with Ac-LEVD-AMC cleavage (Figure 2B−E and S3A). However, we did not observe the production of the caspase-11 p20 form or LEVD cleavage after LPS transfection, stimulation with nigericin or poly(dA:dT), or infection with Francisella tularensis or Salmonella (Figure 2B−E, S2C−N and S3A), which is consistent with a previous report (Hagar et al., 2013). Cytosolic delivery of LTA was required for caspase-11 cleavage as this did not occur in the absence of the liposomal transfection reagent DOTAP (Figure 2F). Unlike LTA, transfection of synthetic triacylated lipoprotein Pam3CSK4, which is also a Toll-like receptor (TLR) 2 ligand, did not result in the production of the caspase-11 p20 form (Figure 2F). LPS-mediated caspase-11 activation induces GSDMD-dependent cell death (Kayagaki et al., 2015; Shi et al., 2015). In contrast to LPS, cytosolic LTA induced neither LDH release nor detectable GSDMD cleavage after 4 h of stimulation, although LDH release increased by 16 h (Figure 2G, 2H and S3B). To rule out the possibility of an active contaminant in the purified LTA preparation responsible for caspase-11 cleavage, we assessed the ability of synthetic LTA (sLTA) to cleave caspase-11 (Figure S3C). Transfection of sLTA induced cleavage of caspase-11 and Ac-LEVD-AMC substrate in WT BMDMs which was comparable to LTA (Figure 2I and 2J). Furthermore, sLTA lacking the glycerophosphate repeat (GPR) was impaired in triggering caspase-11 cleavage (Figure 2K), suggesting that the GPR of LTA is important for the induction of caspase-11 cleavage. To further verify the role of LTA in caspase-11 cleavage, we used WT Listeria and an isogenic mutant deficient in lmo0927, a LTA synthase that is critical for the synthesis of the LTA polyglycerophosphate backbone (Webb et al., 2009). Because the LTA mutant exhibited growth defects in vitro (Webb et al., 2009), we assessed the ability of equal amounts of extracts from WT and mutant Listeria to cleave caspase-11 upon transfection into BMDMs. Extracts from the WT Listeria strain induced robust cleavage of caspase-11 unlike the lmo0927 mutant strain (Figure 2L). Reconstitution of the mutant strain with a lmo0927-expressing plasmid restored the ability of the bacterial extracts to cleave caspase-11 (Figure 2L). Consistent with the bacteria infection studies, LTA-induced caspase-11 cleavage into caspase-11 p20 and LEVD cleavage were impaired in Nlrp6−/− and Pycard−/− BMDMs (Figure 2M and S3D−F). Furthermore, caspase-1 activation induced by LTA transfection was impaired in Nlrp6−/−, Pycard−/− and Casp11−/− BMDMs (Figure 2N). Consistently, cytosolic LTA stimulation resulted in the maturation of IL-1β, which required NLRP6, caspase-11 and caspase-1, but not GSDMD which was important for the secretion of IL-1β (Figure S3G−J). Collectively, these results indicate that cytosolic LTA triggers caspase-11 processing through NLRP6 and ASC.
Figure 2. Cytosolic LTA triggers caspase-11 cleavage.
Primary BMDMs were primed with poly(I:C) for 4 h, then transfected with (A and L) Listeria extracts, (B−K, M, N) indicated ligands for 4 h. (A−D, F, H, I, K−N) The cell lysates (Lysate) and supernatants (Sup) were subjected to immunoblotting, or (E and J) the lysates were subjected to caspase substrate cleavage assay. Blots of caspase-11 were cropped to reveal protein bands at 36 different exposures. (G) The supernatants were subjected to LDH assay. Results are representative of at least (A−M) three or (N) two independent experiments, and error bars denote s.d. of triplicate wells. PDE, phosphodiesterase; sLTA, synthetic LTA; GPR, glycerophosphate repeat; lmo0927 + lmo0927, lmo0927 strain reconstituted with lmo0927-expressing plasmid. ****P < 0.0001. See also Figures S2 and S3.
NLRP6 binds LTA via the LRR domain
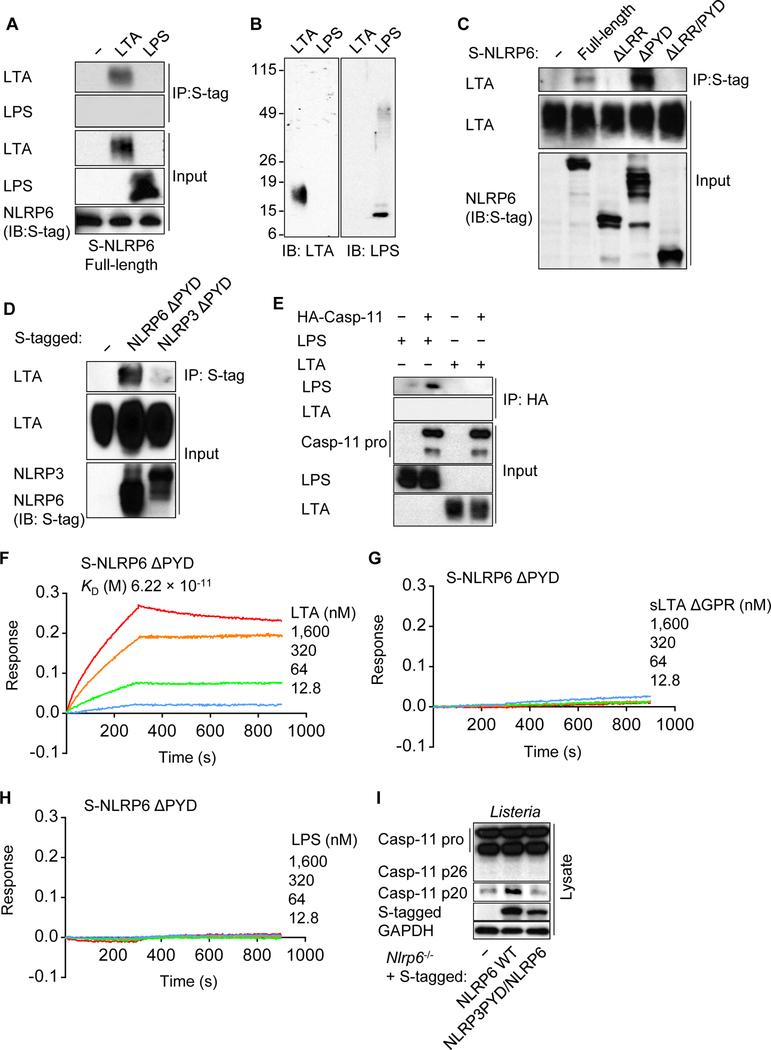
To examine whether LTA associates with NLRP6, we expressed S-tagged NLRP6 in HEK293T cells and assessed its interaction with LTA. Using specific antibodies for LTA or LPS, we observed that NLRP6 interacts with LTA, but not with LPS (Figure 3A and 3B). Although LTA did not associate with NLRP6 lacking the C-terminal leucine-rich repeats (LRRs), the interaction of LTA with NLRP6 was enhanced by deletion of the N-terminal pyrin domain (PYD) (Figure 3C). In contrast, LTA did not associate with NLRP3, AIM2, or caspase-11 (Figure 3D, 3E and S4A). LTA displayed a dose-dependent binding to immobilized purified NLRP6 ΔPYD, but not to human NOD1 or rat IgG, by Bio-layer interferometry (BLI) analysis (Figure 3F and S4B−F) . The equilibrium dissociation constant (KD) between NLRP6 ΔPYD and LTA was calculated as 6.22 × 10 −11 M, whereas there was little or no interaction of NLRP6 ΔPYD with sLTA ΔGPR, LPS or MDP under comparable conditions (Figure 3G, 3H and S4G). These results indicate that LTA directly binds NLRP6 at high affinity via the LRR domain.
Figure 3. LTA binds to the LRR domain of NLRP6.
(A, C−E) Indicated tagged NLRP6, NLRP3 or caspase-11 constructs were expressed in HEK293T cells, lysed and incubated with LTA or LPS. The lysates were immunoprecipitated (IP) and analyzed by immunoblotting (IB). (B) Full Western blots of LTA and LPS are shown as a reference. (F−H) BLI analysis of the interaction between biosensor-immobilized purified S-NLRP6 PYD and LTA, sLTA GPR or LPS. (I) Immortalized Nlrp6−/− BMDMs reconstituted with WT NLRP6 or chimeric NLRP6 with NLRP3 PYD were infected with Listeria for 9 h. The cell lysates (Lysate) were subjected to immunoblotting. Blots of caspase-11 were cropped to reveal protein bands at different exposures. Results are representative of at least (A−D, F−I) three or (E) two independent experiments. LRR, leucine-rich repeat; PYD, pyrin domain; KD, the equilibrium dissociation constant; sLTA, synthetic LTA; GPR, glycerophosphate repeat. See also Figure S4.
Amino acid alignment analysis of several PYDs revealed that the PYD of NLRP6 was unique in that it contains two gap regions in the N-terminus and a long unmatched region in the C-terminus, compared with the PYDs of NLRP3, AIM2 and Pyrin (Figure S4H). To investigate the function of NLRP6 PYD, we swapped the PYD of NLRP6 for that of NLRP3. After Listeria infection or LTA transfection, we observed production of the caspase-11 p20 in Nlrp6−/− macrophages reconstituted with WT NLRP6, but not with chimeric NLRP6 containing the PYD of NLRP3 (Figure 3I and S4I). Furthermore, caspase-11 cleavage was not observed after nigericin stimulation of Nlrp3−/− macrophages reconstituted with chimeric NLRP3 containing the NLRP6 PYD or NLRP6 containing the NLRP3 LRR (Figure S4J−L). These results suggest that the PYD of NLRP6 is essential, but not sufficient, for the induction of caspase-11 cleavage.
Listeria and cytosolic LTA induce the recruitment of caspase-1 and caspase-11 to the NLRP6 inflammasome
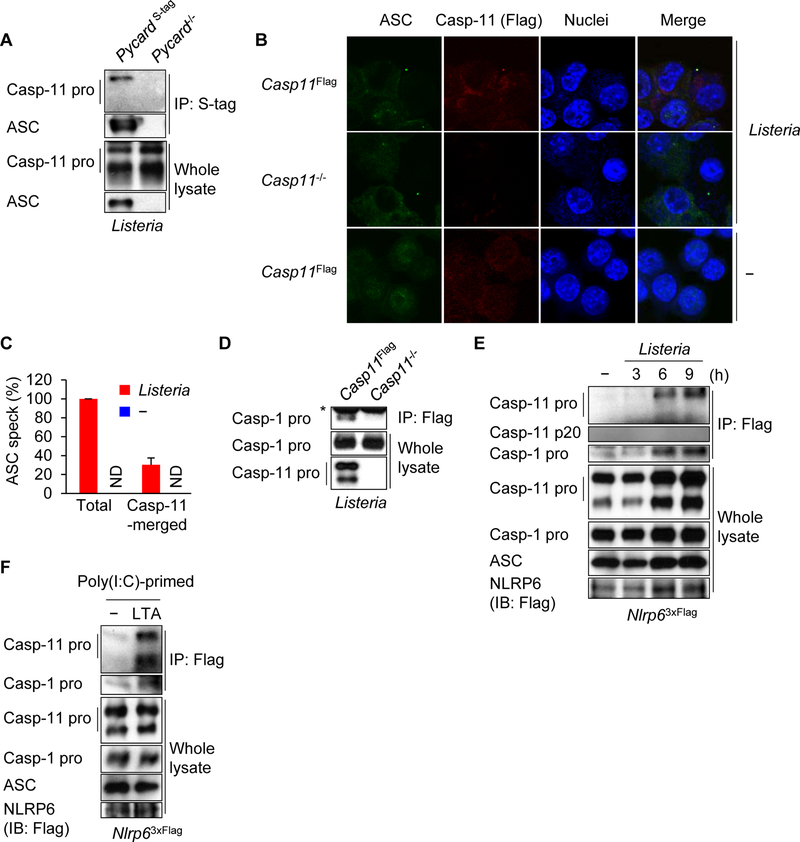
NLRP6 associates with caspase-1 through the adaptor ASC in overexpression studies (Levy et al., 2015). To determine whether caspase-11 interacts with ASC, we reconstituted Pycard−/− immortalized BMDMs with tagged ASC (Figure S5A). The interaction between caspase-11 and ASC was observed after Listeria infection in macrophages (Figure 4A). Additionally, caspase-11 co-localized with ASC “spe cks” induced by Listeria or LTA, but not by stimulation with nigericin, poly(dA:dT), LPS, Salmonella or Yersinia pseudotuberculosis yopM (Figure 4B, 4C and S5B). Furthermore, we observed that caspase-1 associated with caspase-11 in response to Listeria infection as determined by immunoprecipitation of caspase-11 followed by immunoblotting for caspase-1 (Figure 4D). In addition, caspase-11 co-localized with caspase-1 after Listeria infection (Figure S5C), suggesting that caspase-11 and caspase-1 are recruited to the NLRP6 inflammasome complex. Next, we examined the interaction between NLRP6 and inflammasome components. To assess the endogenous interaction of NLRP6 with ASC in macrophages, we used BMDMs from a knockin mouse in which 3 copies of the Flag tag were inserted at the C-terminus of NLRP6 by homologous recombination. NLRP6 co-localized with ASC in the macrophages after Listeria infection or LTA transfection (Figure S5D). Interaction between NLRP6 and caspase-11 was observed in HEK293T cells and it was enhanced by LTA transfection (Figure S5E and S5F). Immunoprecipitation of NLRP6 using an antibody against Flag revealed that NLRP6 interacts with caspase-11 as well as caspase-1 in macrophages infected with Listeria or transfected with LTA (Figure 4E and 4F). These data indicate that caspase-1 and caspase-11 are recruited to the NLRP6 inflammasome after Listeria infection or LTA transfection.
Figure 4. Caspase-11 and caspase-1 are recruited to the NLRP6 inflammasome through ASC.
(A−E) Immortalized BMDMs were left uninfected or infected with Listeria for 12 h or indicated times. (F) Poly(I:C)-primed immortalized BMDMs were incubated with LTA in Opti-MEM supplemented with 0.005% saponin for 4 h. (A, D−F) The cells were lysed, immunoprecipitated (IP) and analyzed by immunoblotting (IB). (D−F) Cells were treated with DSP and DTBP before IP. Whole cell lysates are shown as the input. Asterisk, IgG. Blots of caspase-11 were cropped to reveal protein bands at different exposures. (B and C) The cells were fixed, immunostained and ASC specks (arrowheads) were counted. ASC, green; caspase-11 (Flag), red; and nuclei, blue. Scale bars, 10 µm. ND, not detected. Results are representative of at least (A, D−F) three or (B and C) two independent experiments, and error bars denote s.d. of each group. See also Figure S5.
Caspase-11 processing is important for caspase-1-mediated IL-18 secretion
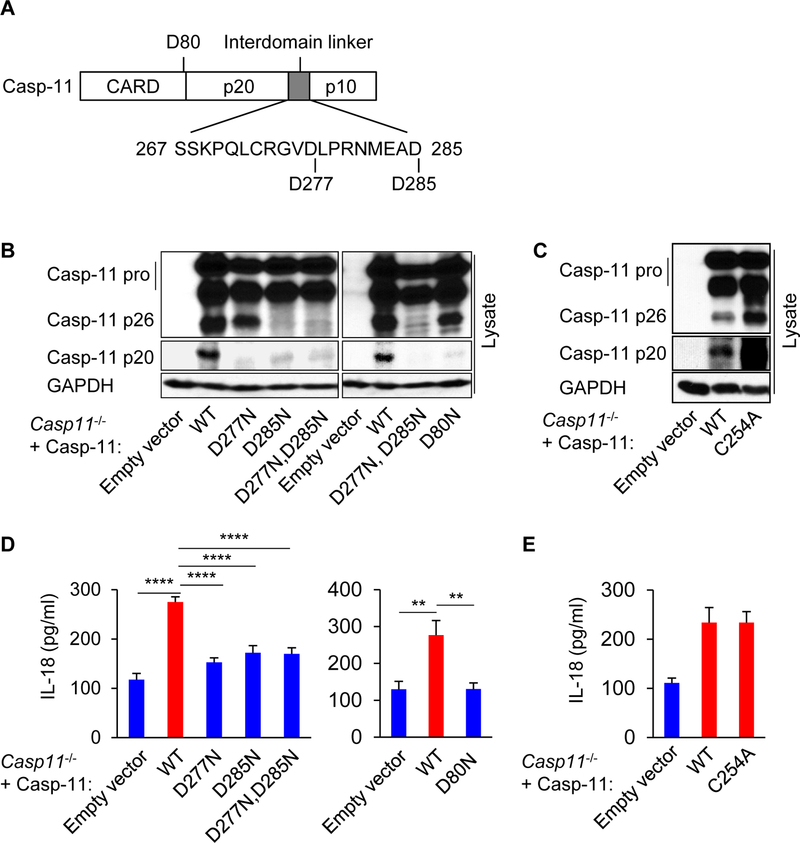
Caspase-11 does not directly process pro-IL-1β, but caspase-11 promotes caspase-1-dependent maturation of pro-IL-1β when it is co-transfected with caspase-1 (Lin et al., 2000; Wang et al., 1996). To determine whether processing of caspase-11 is required for caspase-1-dependent IL-18 secretion, we expressed WT or mutant caspase-11 that contain asparagine instead of aspartic acid at positions 277 (D277N) and 285 (D285N), which are the predicted caspase-11 cleavage sites (Broz et al., 2010), in Casp11−/− macrophages by lentiviral-mediated transduction (Figure 5A). Listeria infection induced the processing of WT pro-caspase-11 into the p20 fragment, but not of the caspase-11 mutants (Figure 5B), indicating that D277 and D285 are involved in caspase-11 cleavage. These D277N and D285N mutations did not enhance LDH release induced by LTA transfection (Figure S5G). To identify another cleavage site, we also prepared macrophages expressing caspase-11 D80N, a predicted cleavage site located between CARD and the p20 domain (Figure 5A). The formation of the p20 fragment was not observed in the D80N mutant in response to Listeria infection (Figure 5B), suggesting that D80 participates in the production of the caspase-11 p20 fragment in addition to D277 and D285. It has been reported that C254 of caspase-11 is the putative catalytic cysteine for self-cleavage (Lee et al., 2018; Wang et al., 1996). Notably, the production of caspase-11 p20 fragment was not impaired by the replacement of C254 to alanine or serine (Figure 5C and S5H), suggesting that the catalytic C254 is not essential for caspase-11 processing in response to Listeria. Consistently, IL-18 release was impaired by D277N, D285N and D80N mutations in caspase-11, but not by the C254A mutation (Figure 5D and 5E). In addition, caspase-1 cleavage was abolished by D277N and D285N mutations in caspase-11 (Figure S5I). Collectively, these results indicate that caspase-11 processing promotes caspase-1-dependent IL-18 secretion.
Figure 5. Processed caspase-11 promotes caspase-1-mediated IL-18 secretion.
(A) Schematic representation of predictive cleavage sites in pro-caspase-11. (B−E) Immortalized Casp11−/− BMDMs reconstituted with WT or mutants of caspase-11 were infected with Listeria for 12 h. (B and C) The cell lysates (Lysate) were subjected to immunoblotting, or (D and E) the supernatants were subjected to ELISA. Blots of caspase-11 were cropped to reveal protein bands at different exposures. Results are representative of at least three independent experiments, and error bars denote s.d. of triplicate wells. **P < 0.01, ****P < 0.0001. See also Figure S5.
Type I IFN signaling regulates caspase-1 activation by enhancing NLRP6 and caspase-11 expression
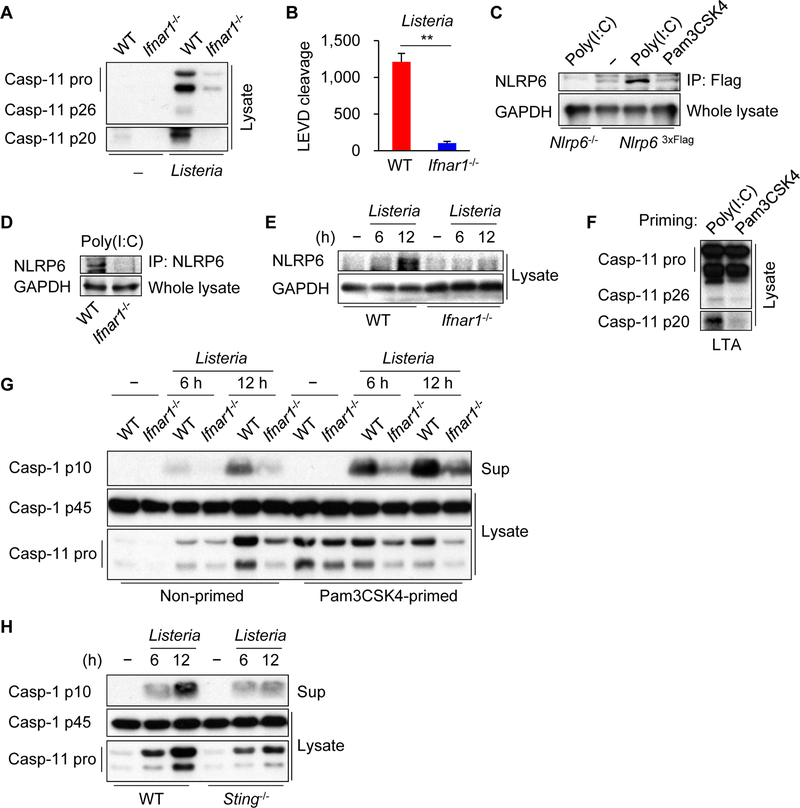
Gram-negative bacteria require type I interferon (IFN) signaling to activate the caspase-11 pathway induced by LPS (Broz et al., 2012; Gurung et al., 2012; Rathinam et al., 2012). To determine whether type I IFN signaling is important for caspase-11 processing in response to Listeria infection, we infected BMDMs from WT or Ifnar1−/− mice with the bacterium and assessed caspase-11 cleavage. Listeria infection induced the cleavage of caspase-11, which was reduced in Ifnar1−/− BMDMs (Figure 6A). Consistently, the cleavage of Ac-LEVD-AMC induced by Listeria infection was impaired in Ifnar1−/− BMDMs (Figure 6B). The defective caspase-11 processing could be explained, at least in part, by reduced induction of pro-caspase-11 in Listeria-infected Ifnar1−/− BMDMs (Figure 6A). In accord with a role for type I IFN in the regulation of NLRP6 expression in mouse embryonic fibroblasts (Wang et al., 2015), priming of BMDMs with poly(I:C) that induces type I IFN production, but not with Pam3CSK4, increased the expression of endogenous NLRP6 in macrophages, which was abolished in Ifnar1−/− BMDMs (Figure 6C and 6D). Consistent with these results, NLRP6 expression was induced in WT macrophages infected with Listeria, but not in Ifnar1−/− macrophages (Figure 6E). Furthermore, caspase-11 processing induced by LTA transfection was observed in macrophages primed with poly(I:C), but not with Pam3CSK4 that does not induce type I IFN (Figure 6F). Thus, type I IFN signaling regulates both NLRP6 and caspase-11 expression in macrophages. Consistent with a role of NLRP6 and caspase-11 in caspase-1 activation, the processing of caspase-1 was impaired in Ifnar1−/− BMDMs infected with Listeria in unprimed cells (Figure 6G). To dissociate the role of NLRP6 and caspase-11, we primed BMDMs with Pam3CSK4 prior to Listeria infection which induces pro-caspase-11 but not NLRP6 expression (Figures 6C and 6G). Caspase-1 processing induced by Listeria was impaired in Ifnar1−/− BMDMs despite adequate induction of pro-caspase-11 (Figure 6G), suggesting that impaired expression of NLRP6 via type I IFN signaling results in reduced activation of caspase-1. Furthermore, Listeria-induced caspase-11 expression and caspase-1 activation were impaired in macrophages deficient in STING, a critical adaptor regulating type I IFN production in response to Listeria infection (Figure 6H) (Ishikawa et al., 2009). Collectively, these results indicate that type I IFN signaling regulates and caspase-1 activation by enhancing NLRP6 and pro-caspase-11 expression.
Figure 6. Type I IFN signaling is required for NLRP6 and caspase-11 expression.
(A, B, E, G, H) Primary BMDMs were left uninfected or infected with Listeria for 12 h or indicated times. (C) Immortalized or (D) primary BMDMs were stimulated with indicated ligands for 22 h, immunoprecipitated (IP), and analyzed by immunoblotting. (F) Primary WT BMDMs were primed with indicated ligands for 4 h, then transfected with LTA for 4 h. The cell lysates (Lysate) and supernatants (Sup) were subjected to (A, C−H) immunoblotting or (B) caspase substrate cleavage assay. Blots of caspase-11 were cropped to reveal protein bands at different exposures. Results are representative of at least (A−C, F−H) three or (D, E) two independent experiments, and error bars denote s.d. of triplicate wells. **P < 0.01.
NLRP6-caspase-11 axis exacerbates Listeria infection through the production of IL-18
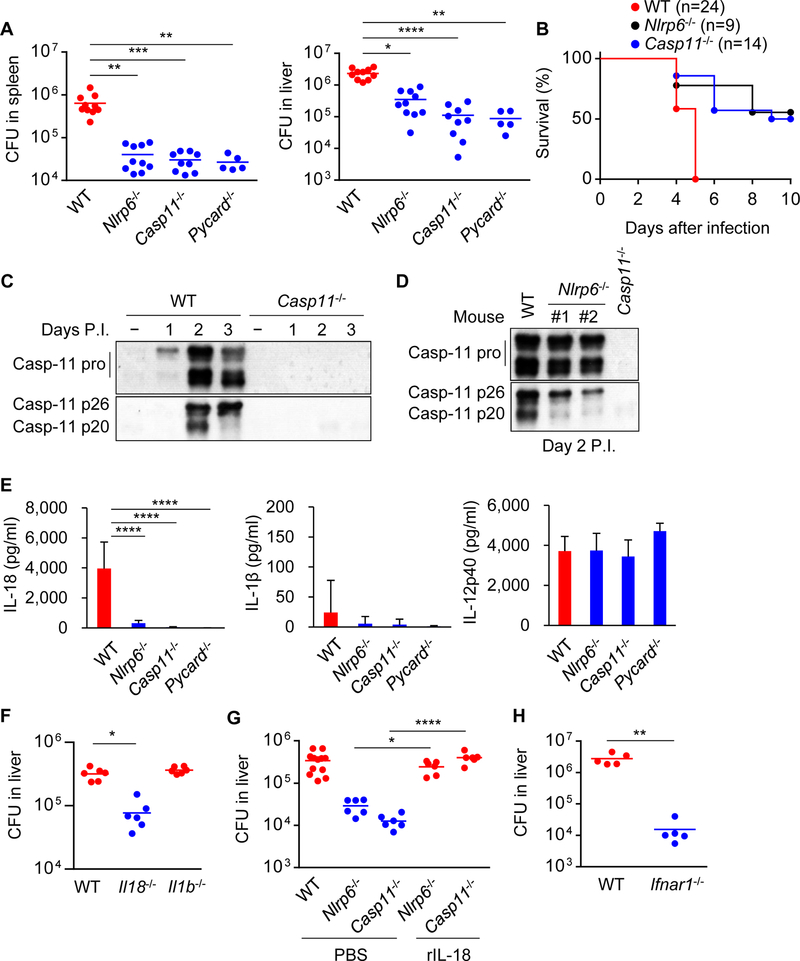
We next examined the role of NLRP6 and caspase-11 in the regulation of Listeria infection in vivo. Nlrp6−/− mice harbored reduced pathogen loads when compared with WT mice on day 4, but not on day 2, after intravenous infection with a sublethal dose of 104 cfu of Listeria (Figure 7A, S6A−C). WT mice succumbed to an infect ion with a higher dose of 105 cfu of Listeria whereas ~50% of Nlrp6−/− mice survived the infection (Figure 7B). Casp11−/− mice also harbored lower pathogen loads than WT mice after infection with WT Listeria, but not after infection with LTA-deficient Listeria or WT Salmonella (Figure 7A, S6B−E). Salmonella loads were also comparable in WT and Nlrp6−/− mice (Figure S6F). Consistent with in vitro studies, Listeria infection enhanced pro-caspase-11 expression and processing in vivo (Figure 7C). There was reduced caspase-11 p20 production in spleens from infected Nlrp6−/− mice on day 2 after infection (Figure 7D). We have also assessed Listeria loads in Aim2−/− or Nlrp3−/− mice and found less bacterial loads in Aim2−/− mice (Figure S6G). In contrast, there was no significant difference in bacterial loads between WT and Nlrp3−/− mice (Figure S6G). We have assessed the Listeria loads in pure Casp1−/− mice and found increased bacterial loads in Casp1−/− mice when compared to WT mice (Figure S6H). We also tested Gsdmd−/− mice and found that their phenotype is comparable to that of pure Casp1−/− mice (Figure S6H). Thus, the phenotype of Casp1−/− mice is different from that of Casp11−/− and Casp1/11−/− mice. Consistently, a comparable phenotype was observed in Nlrp6−/− and Casp11−/− mice and this was confirmed using two additional Listeria strains (Figure S6B and S6C).
Figure 7. NLRP6-caspase-11 axis exacerbates Listeria infection.
Mice were infected with (A, C−H) 104 or (B) 105 cfu of Listeria intravenously. (A, F−H) The organs were removed on day 4 for cfu counting. (B) Mouse survival was monitored for 10 days. (C and The spleen homogenates were immunoblotted. (E) The sera were collected on day 4, and cytokine levels were determined by ELISA. (G) Recombinant IL-18 (rIL-18) was administrated on day 2 post-infection (P.I.). Blots of caspase-11 were cropped to reveal protein bands at different exposures. Results are (A, B, F, G) pooled from two independent experiments or (C−E, H) representative of three independent experiments, and error bars denote s.d. of each group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. See also Figures S6 and S7.
Listeria induced the production of various cytokines including IL-18, IL-12p40 and IFN-γ on day 4 after infection (Figure S7A). Although infection with 104 cfu of Listeria did not induce IL-18 production in the serum on day 2, robust IL-18 production was detected on day 2 after inoculation with 106 cfu of the bacterium (Figure S7B and S7C). Notably, the production of IL-18, but not IL-12p40, was greatly reduced in the serum of infected Nlrp6−/− mice when compared to WT mice (Figure 7E and S7C). Likewise, the production of IL-18 was impaired in Casp11−/− or Pycard−/− mice infected with Listeria (Figure 7E and S7C). LTA-deficient Listeria induced little IL-18 production even after infection of WT mice with 106 cfu of the bacterium (Figure S7D). In contrast to IL-18, the amounts of IL-1β were nearly undetectable in the serum from Listeria-infected mice (Figure 7E and S7A). Nlrp6−/− and Casp11−/− mice were also more resistant to intravenous infection with Staphylococcus aureus than WT mice (Figure S6I), which correlated with reduced pathogen loads in mutant mice (Figure S6J). Furthermore, reduced amounts of IL-18, but not IL-12p40, were observed in the serum of Nlrp6−/− and Casp11−/− mice when compared with WT mice (Figure S7E). Using different model systems, IL-18 has been reported to promote both protective and detrimental effects during Listeria infection in vivo (Lochner et al., 2008; Maltez et al., 2015). To assess whether endogenous IL-1β or IL-18 regulates the clearance of Listeria, we infected WT, Il1b−/− and Il18−/− mice with Listeria, and measured the pathogen loads. Il18−/− mice contained lower pathogen loads than WT mice (Figure 7F), which is consistent with previous studies (Lochner et al., 2008; Tsuchiya et al., 2014). In contrast to Il18−/− mice, the pathogen loads of Il1b−/− mice were comparable to that of WT mice (Figure 7F). To determine whether the impairment of IL-18 production in Nlrp6−/− and Casp11−/− mice was responsible for increased pathogen clearance, we administrated recombinant IL-18 and measured pathogen loads after infection. IL-18 administration restored pathogen loads to levels observed in WT mice (Figure 7G). In early studies NLRP6 was reported to regulate the composition of the intestinal microbiota (Elinav et al., 2011), although recent studies have challenged the initial observations (Lemire et al., 2017; Mamantopoulos et al., 2017). Therefore, we co-housed WT and mutant mice for 3−4 weeks to normalize their microbiota before bacterial infection. Co-housing did not change the resistant phenotype of Nlrp6−/−, Casp11−/−, Pycard−/−, and Il18−/− mice to Listeria infection (Figure S6K). Consistent with previous studies (Auerbuch et al., 2004; Carrero et al., 2004; O’Connell et al., 2004), Ifnar1−/− mice were also more resistant to systemic infection with Listeria (Figure 7H). These results indicate that the NLRP6-caspase-11 axis exacerbates Listeria infection through the production of IL-18.
DISCUSSION
We show that LTA, a major component in the cell wall of Gram-positive bacteria, binds to NLRP6 and activates the NLRP6 inflammasome. Because NLRP6 is located intracellularly, the signaling pathway is presumably activated by Gram-positive bacteria that reach the cytosol (Berche et al., 1988; Hertzen et al., 2012; Nandi and Bishayi, 2016), by delivery of LTA into the host cytosol via the type VII secretion system of Gram-positive bacteria (Bottai et al., 2016; Cao et al., 2016; Pinheiro et al., 2016), or by bacteriolysis to liberate LTA into the cytoplasm (Man et al., 2016). Consistent with this notion, cytosolic delivery of LTA or extracts from Gram-positive bacteria was required for NLRP6-mediated caspase-11 and IL-1β processing. The molecular mechanism by which LTA activates NLRP6 upon binding remains unclear and must await the availability of atomic resolution structures of the LTA-NLRP6 complex. Structural studies of two NLRs, NOD2 and NLRC4, have revealed that these sensors are maintained in an autoinhibited monomeric state at least in part via the C-terminal LRR domain (Hu et al., 2013; Maekawa et al., 2016). Given that the LRR domain of NLRP6 mediates the interaction with LTA, the binding of LTA with NLRP6 may lead to conformation changes that overcome the autoinhibition leading to recruitment of ASC and inflammasome activation.
Based on overexpression studies, it was suggested that NLRP6 interacts with and activates caspase-1 (Levy et al., 2015). We show here that after Listeria infection or delivery of LTA to the cytosol, NLRP6 recruits and induces caspase-11 processing. Further biochemical and functional experiments show that cleavage of caspase-11 through NLRP6 requires ASC and involves homotypic ASC-ASC interactions. NLRP6 also recruited pro-caspase-1 in response to Listeria infection or delivery of LTA to the cytosol. Furthermore, caspase-11 associated with caspase-1 in response to Listeria infection. These observations suggest that the NLRP6 inflammasome can recruit both caspase-11 and caspase-1. Because NLRP6, but not NLRP3 and NLRC4, functions upstream of caspase-1 and caspase-11 in the regulation of mucus production in colonic explants (Birchenough et al., 2016), the NLRP6/caspase-11/caspase-1 signaling pathway may also operate in intestinal epithelial cells. In contrast to NLRP6, activation of the NLRP3, AIM2 and Pyrin inflammasomes did not recruit caspase-11. The molecular basis for the specific recruitment of caspase-11 by the NLRP6 inflammasome remains unclear. Analysis of chimeric NLRP3-NLRP6 proteins revealed that the PYD of NLRP6 was essential, but not sufficient, to induce caspase-11 cleavage. These results suggest that the PYD of NLRP6 acts in the context of additional NLRP6 domain(s) to mediate the specific recruitment and processing of caspase-11. However, the interpretation of NLR chimera experiments is difficult because chimeric NLR proteins may exhibit loss of critical intra-molecular interactions present in the native NLR proteins. After recruitment, caspase-11 is processed which promotes caspase-1-dependent IL-1β and IL-18 secretion in macrophages. Caspase-11 co-localized with caspase-1 in the ASC “specks” induced by Listeria infection. These results suggest a model in which the NLRP6 inflammasome recruits caspase-11 and caspase-1 leading to sequential proteolytic activation of both caspases in response to Listeria and LTA. It is also possible that caspase-1 exhibits a scaffolding function in recruiting caspase-11 to the “speck”. Consistent with the sequential activation model, processing of caspase-11 was required for IL-18 secretion in response to Listeria infection. Previous studies reported the formation of the caspase-11 p20 fragment in response to several stimuli (Lee et al., 2001; Schotte et al., 1998). In addition, the caspase-11 p20 fragment can bind to the TRPC1 substrate via amino acid residues 79 to 285 of p20 (Py et al., 2014). Consistent with these previous studies, we found that D80, D277 and D285 are required for the production of p20 fragment of caspase-11 in response to Listeria infection. Because the putative catalytic C254 residue was not required for p20 caspase-11 production, the results suggest that caspase-11 processing is mediated by another protease in response to Listeria infection.
Cryo-EM structures of the AIM2-PYD/ASC-PYD and NLRP3/ASC-PYD protein complexes have suggested that the adaptor ASC forms filamentous structures in which the PYDs localizes at the core while the CARDs forms the outer layer of the filament that recruit and nucleate caspase-1 (Cai et al., 2014; Lu et al., 2014). Thus, the CARDs of ASC in the NLRP6 inflammasome could recruit independently both caspase-11 and caspase-1 via CARD-CARD interactions. Structural studies of the NLRP6 inflammasome are needed to test whether NLRP6 can form an inflammasome containing both caspase-11 and caspase-1 through CARD-CARD interactions with ASC. In contrast to the NLRP6-caspase-11 inflammasome, cytosolic LPS induces caspase-11 activation followed by GSDMD-mediated cell death (Shi et al., 2015). Unlike cytosolic LPS stimulation, the NLRP6-caspase-11 inflammasome pathway is not associated with detectable GSDMD cleavage and robust cell death. The reason to account for the lack of or reduced GSDMD cleavage in response to LTA remains unclear. One possibility is that GSDMD is accessible to the LPS-induced caspase-11 active form, but not to cleaved caspase-11 triggered by LTA. Further work is needed to understand this question.
NLRP6 and caspase-11 exacerbate Listeria systemic infection in vivo through the secretion of IL-18. In contrast to our studies, NLRP6 has been reported to inhibit the production of inflammatory cytokine and chemokine including TNF, IL-6 and KC in macrophages which was suggested to enhance Listeria colonization in vivo (Anand et al., 2012). However, we found that the production of these cytokines induced by Listeria is not enhanced in Nlrp6−/− macrophages. We do not have an explanation for the discrepancy in results. The decreased pathogen loads in Nlrp6−/− and Casp11−/− mice may reflect adaptive evolution of Gram-positive pathogens to evade host protective mechanisms. A previous study showed comparable survival of WT and Casp11−/− mice after Listeria infection (Mueller et al., 2002), while another group showed that Casp1−/−Casp11−/− mice were more susceptible to Listeria infection (Tsuji et al., 2004). The studies by Mueller et al. and Tsuji et al. differ from ours in at least two aspects. In our model, IL-18 was robustly induced whereas the production of IL-18 in the other studies was modest which may be explained by the use of different Listeria strains (Mueller et al., 2002; Tsuji et al., 2004). We have tested three Listeria strains (EGD, 10403S and LO28) and found reduced Listeria burdens in both Nlrp6−/− and Casp11−/− mice. Although the requirement of type I IFN to activate caspase-11 in Gram-negative bacteria infection is controversial (Broz et al., 2012; Kayagaki et al., 2013; Rathinam et al., 2012), we found that it is essential for NLRP6-dependent caspase-11 processing in response to cytosolic LTA and Listeria infection. Consistent with our findings, Ifnar1−/− mice are more resistant to systemic Listeria infection (Auerbuch et al., 2004; Carrero et al., 2004; O’Connell et al., 2004), which is comparable to the phenotype observed in Nlrp6−/− and Casp11−/− mice. The role of IL-18 in Listeria infection is poorly understood and controversial. Administration of recombinant IL-18 early in infection is protective against Listeria in vivo (Maltez et al., 2015), while IL18−/− mice are more resistant than WT mice against pathogen infection (Lochner et al., 2008; Tsuchiya et al., 2014). Thus, IL-18 can exert both a protective and detrimental role in Listeria infection. Taken together, our studies reveal a novel signaling pathway induced by cytosolic LTA to activate the NLRP6 inflammasome leading to caspase-11 and caspase-1 processing that regulates Gram-positive bacterial infection.
STAR METHODS
Mice.
Nlrp3−/−, Pycard−/−, Casp11−/−, Casp1−/−Casp11−/− (He et al., 2016), Nlrp6−/− (Chen et al., 2011), Il18−/− (Seregin et al., 2017), Aim2−/−, Il1b−/− (Seo et al., 2015), Casp1−/− (Man et al., 2017), and Gsdmd−/− mice (Rauch et al., 2017) on C57BL/6 background have been reported. C57BL/6 WT and Ifnar1−/− (Muller et al., 1994) mice were purchased from Jackson Laboratories. The Nlrp6–3xFlag-IRES-eGFP mouse was generated at the Institut Clinique de la Souris (Illkirch, France) from C57BL/6N embryonic stem cells (K4711TB1–2) containing a 3xFlag-IRES-eGFP cassette. Briefly, a 3xFlag-IRES-eGFP cassette was inserted into Nlrp6 exon 8 ahead of its translation termination codon by homologous recombination. All animal studies were approved by the University of Michigan Committee on Use and Care of Animals.
Reagents.
Purified LTA (tlrl-pslta), poly(I:C) (LMW) (tlrl-picw), Pam3CSK4 (tlrl-pms), c-diAMP (tlrl-nacda), MDP (tlrl-mdp), PGN (tlrl-pgns2), LPS-SM (tlrl-smlps), nigericin (tlrl-nig), poly(dA:dT) (tlrl-patn), and LPS-EK (tlrl-eklps) were from Invivogen, DNase I from Worthington, RNase A from Omega, PDE I from Affymetrix, proteinase K from Roche, cytotoxicity detection kit from Clontech, Ac-LEVD-AMC (ALX-260–083), Ac-IETD-AMC (ALX-260–042) and Ac-DMQD-AMC (ALX-260–078) from Enzo Life Sciences, z-VAD-FMK (FMK001) from R&D systems, pHIV-EGFP (21373) from Addgene (a gift from Bryan Welm and Zena Werb) (Welm et al., 2008), recombinant human NOD1 protein (H00010392-P01) from Novus biologicals, Silver Staining kit from Thermo Fisher Scientific, and recombinant mouse IL-18 from MBL. Antibodies for NLRP6 (SAB1302240), caspase-11 (C1354), GSDMD (G7422) and Flag (F1804) were from Sigma-Aldrich, LTA (HM2048) and LPS (HM6011) from Hycult Biotech, GAPDH (MAB374) and S-tag (71549–3) from Millipore, HA (TA180128S) from Origene, caspase-1 (sc-514) from Santa Cruz, ASC (rabbit; AG-25B-0006) from Adipogen, and IL-1β (AB-401) from R&D systems. Rat anti-ASC antibody was generated in our laboratory (He et al., 2016). sLTA and sLTA GPR were described (Fukase et al., 1992). Endotoxin concentration in the synthetic LTAs was measured using Limulus amebocyte lysate chromogenic assay kit (Thermo Fisher Scientific) and found to be lower than the detection limit (< 0.1 EU/ml).
Cell culture.
BMDMs were generated by differentiating bone marrow progenitors from the tibia and femur for 5 days in RPMI 1640 (Gibco) supplemented with 10% FBS, 30% L929 cell supernatant, non-essential amino acids, sodium pyruvate and antibiotics (penicillin, streptomycin and amphotericin B). BMDMs were cultured with J2 virus to generate immortalized BMDMs (Blasi et al., 1985). Cas9-expressing parental THP-1 cells and caspase-4/5- or caspase-1-deficient THP-1 cells were a gift from Seth L. Masters (Baker et al., 2015). Immortalized BMDM, L929, HEK293T, and THP-1 cells were cultured in RPMI1640 supplemented with 10% FBS, non-essential amino acids, sodium pyruvate and gentamicin.
Stimulation of cells.
Cells were plated at a density of 5 × 10 5 cells per well in 24-well microplates. Culture medium was replaced with RPMI 1640 supplemented with 0.3% FBS, non-essential amino acids, sodium pyruvate before infection. Cells were infected with Listeria monocytogenes EGD (MOI = 10 or indicated MOI) (Hara et al., 2008), Staphylococcus aureus 8325–4 (MOI = 10) (Munoz-Planillo et al., 2009), Salmonella enterica serovar Typhimurium SL1344 (MOI = 10) (Franchi et al., 2012), Yersinia pseudotuberculosis 32777 yopM (MOI = 30) (Chung et al., 2016) or Francisella tularensis U112 (MOI = 500) (Fernandes-Alnemri et al., 2010). Gentamicin (10 µg/ml) was added to the cultures 1 h after infection when the cells were continuously cultured for more than 2 h. For transfection, cells were primed with poly(I:C) (1 µg/ml) for 4 h and culture medium was replaced with Opti-MEM (Gibco). To prepare bacterial extracts, 109 cfu of bacteria were suspended in 1 ml of PBS, incubated with 100 µg/ml lysozyme (Fisher Scientific), 5 mM EDTA and protease inhibitor cocktail (Sigma-Aldrich) for 30 min at room temperature, sonicated, and centrifuged at 21,000×g for 1 min. The supernatants were used as bacterial extracts. The extracts were treated with DNase I (10 µg/ml), RNase A (10 µg/ml), PDE (1 unit/reaction), or Proteinase K (100 µg/ml) in the presence of 14 mM MgCl2 at 37 ºC for 3 h, then heated at 100 ºC for 10 min. 1 µg of bacterial ligand or 1.25 µl of bacterial extract was suspended in 10 µl of Opti-MEM. 3750 ng of DOTAP (Roche) or 2500 ng of Lipofectamine 2000 (Thermo Fisher Scientific) was suspended in 10 µl of Opti-MEM for 5 min, and then the suspensions were mixed and incubated for 30 min at room temperature (Hagar et al., 2013). The volumes were then brought up to 300 µl with Opti-MEM and the cells were cultured for 4 h. LPS (50 ng/ml) priming was performed for 4 h and cells were stimulated with nigericin (5 µM) or transfected with 0.2 µg of poly(dA:dT) using 1 µl of Lipofectamine LTA and 0.2 µl of PLUS reagent (Thermo Fisher Scientific).
HEK293T cell transfection.
HEK293T cells were plated into 100 mm culture dish (7 × 106 cells) overnight. To examine an interaction between bacterial ligands and intracellular proteins, cells were transfected for 24 h with 10 µg of pcDNA3 expression plasmids expressing S-tagged full-length NLRP6 (amino acids 1–869), LRR (amino acids 1–459), PYD (amino acids 129–869) and LRR/PYD (amino acids 129–459), S-tagged full-length NLRP3 (amino acids 1–1033) and PYD (amino acids 92–1,033), or S-tagged full-length AIM2 (amino acids 1–354) using Lipofect amine 2000, lysed, centrifuged, then incubated with 10 µg of LTA or LPS for 4 h. After the incubation, the lysates were treated with S-protein beads (Novagen) at 4 ºC overnight. To test caspase-11 recruitment by LTA transfection, cells were transfected for 24 h with 10 µg of plasmids expressing Flag-tagged NLRP6, Myc-tagged ASC, and HA-tagged caspase-11, then transfected with 28 µg of LTA using Lipofectamine 2000 for 4 h.
Reconstitution in macrophages.
Immortalized Pycard−/− or Casp11−/− BMDMs were transduced with lentivirus containing pHIV-ASC-SFP (S-tag, Flag and streptavidin-binding tag) (He et al., 2016), pHIV-Casp11-SFP, pHIV-Casp11 WT or mutant (C254A, C254S, D80N, D277N and/or D285N), or empty vector. Immortalized Nlrp6−/− BMDMs were transduced with lentivirus containing pHIV-NLRP6-S-tag or pHIV-NLRP3 PYD (1– 91)/NLRP6 (129–869)-S-tag. Immortalized Nlrp3−/− BMDMs (Wu et al., 2010) were transduced with lentivirus containing pHIV-NLRP6 PYD (1–128)/NLRP3 (92–1033)-S-tag or pHIV-NLRP6 (1–459)/NLRP3 (739–1033)-S-tag. After 3 days, transduced cells were sorted by flow cytometry using eGFP as a marker. Expression of reconstituted proteins was determined by immunoblotting.
Immunoblotting.
Cells were lysed with SDS sample buffer and supernatants were concentrated with trichloroacetic acid. For immunoprecipitation, cells were lysed with 0.5% Nonidet P-40 in PBS with protease inhibitor cocktail. Cell lysates were clarified by centrifugation at 21,000×g for 15 min. Pre-cleared cell lysates were incubated with anti-Flag beads (Sigma-Aldrich), S-protein beads, anti-NLRP6 antibody or anti-HA antibody with protein G beads (Genscript) at 4 ºC overnight. The beads were washed 4 times with lysis buffer, and added SDS sample buffer. For crosslinking, cells were treated with 2 mM DSP (dithiobis[succinimidylpropionate]) and 16.6 mM DTBP (dimethyl 3,3’-dithiobispropionimidate·2HCl) for 30 min before cell lysis. The lysates and precipitates were subjected to SDS-PAGE and subsequently transferred to PVDF membranes by electroblotting. For LTA or LPS detection (Di Padova et al., 1993; Grundling and Schneewind, 2007; Jimenez-Dalmaroni et al., 2009; Tsuneyoshi et al., 2005), samples were heated at 100 ºC for 5 min, subjected to SDS-PAGE, and transferred to PVDF membranes. The membrane was blocked with 5% nonfat dry milk in TBS with 0.1% Tween 20 for 3 h at room temperature. After washing with TBS and 0.1% Tween 20, the membrane was incubated with mouse primary antibody (1:1000) overnight at 4 ºC, washed, then treated with HRP-anti-mouse IgG (1:5000; Jackson Immuno Research) for 90 min at room temperature. Blots were developed using ECL Western blotting substrate (Thermo Fisher Scientific).
Caspase substrate cleavage assay.
Cells were lysed with caspase assay buffer (50 mM HEPES pH7.4, 100 mM NaCl, 0.1% CHAPS, 10 mM DTT, 1 mM EDTA and 10% glycerol), centrifuged at 21,000×g for 2 min, then mixed with Ac-LEVD-AMC, Ac-IETD-AMC or Ac-DMQD-AMC (final concentration 300 µM) in 96-well black plate (Costar). Fluorescence was measured using SPECTRAmax M2 (Molecular Devices, Excitation 350 nm, Emission 450 nm). The cleavage activity was shown as an increase of the fluorescence after 1 h.
ELISA.
Levels of mouse IL-18, IL-1β, IL-12p40, TNFα, IFN-γ, IL-6, KC, and human IL-1β were measured by ELISA (R&D systems) according to the manufacturer’s manual.
Immunostaining.
Cells were fixed and permeabilized with 0.1% Triton X100 in blocking buffer (2.5% BSA in PBS) for 30 min. Cells were washed with PBS and incubated for 90 min at room temperature with the following antibodies: rabbit anti-ASC (Adipogen, 1:400), rabbit anti-caspase-1 (Santa Cruz, 1:100) and/or mouse anti-Flag (Sigma, 1:200). Cells were washed, and incubated with anti-rabbit Alexa Fluor 488 antibody (Thermo Fisher Scientific, 1:400) and anti-mouse Alexa Fluor 594 antibody (Thermo Fisher Scientific, 1:400) for 45 min. After washing, cells were mounted in ProLong Gold Antifade with DAPI (Thermo Fisher Scientific), and imaged with a FV 500 confocal microscopy (Olympus) or Nikon A1 confocal microscopy (Nikon). Three hundred cells were examined in each group. z-VAD-FMK was added to the culture 1 h post infection.
Analysis of ligand-protein interaction.
To measure an interaction between bacterial ligand and host proteins, BLI analysis was performed using Octet RED96 system (ForteBio). Recombinant S-NLRP6 ΔPYD was purified from HEK293T cells. Cells were transfected for 24 h with 10 µg of pcDNA3 plasmid expressing S-tagged NLRP6 PYD, lysed in lysis buffer containing 0.5% Nonidet P-40 in PBS with 300 mM NaCl, 5 mM 2-mercaptoethanol and protease inhibitor cocktail. After centrifugation at 21,000×g for 15 min, the clear lysates were incubated with S-protein beads at 4 ºC for 1 h and applied to spin column. The beads were washed with lysis buffer 5 times and eluted with 3 M MgCl2. The purified protein was biotinylated, dialyzed using Dialysis Cassette (Thermo Fisher Scientific; 20,000 MWCO), and immobilized to Streptavidin Biosensor at 0.025 mg/ml in PBS with or without 0.001% Tween 20. Biotin-rat IgG and human NOD1 protein (Novus biologicals) were used as negative controls. LTA, LTA GPR, LPS or MDP were diluted to indicated concentrations with PBS with or without 0.001% Tween 20. The association process was performed for 300 sec at 25°C with shaking, and the dissociation process was performed for 600 sec in PBS with or without 0.001% Tween 20 with shaking. The resulting data were analyzed after subtracting background and the equilibrium dissociation constant KD was calculated using Octet RED96 analysis software 7.0 and GraphPad Prism 6.
Infection in vivo.
Mice were infected intravenously with 104 to 106 cfu of Listeria (Yamamoto et al., 2012). To count the bacterial numbers, spleens and livers were collected at 2 or 4 days after infection, homogenized in 5 ml of PBS, serially diluted, and plated on brain heart infusion agar. Mice were infected with 2 × 10 8 cfu of Staphylococcus aureus intravenously and livers were collected and homogenized 8 h after infection and bacterial numbers were counted on LB agar plates. Mice were infected with 104 cfu of Salmonella intraperitoneally or intravenously and livers were collected and homogenized on day 3 after infection and bacterial numbers were counted on LB agar plates. Levels of cytokines in sera were determined by ELISA. For immunoblotting, spleens were homogenized in 2 ml of PBS with 0.1% Triton X-100 and protease inhibitor cocktail. Recombinant mouse IL-18 was administrated by intraperitoneal injection (1 µg/mouse) on day 2 after infection.
Statistical analysis.
For two-group comparisons by Gaussian distribution, a two-tailed unpaired t-test with Welch’s correction was used when the variances of the groups were judged to be equal by the F test. For two-group comparisons with non-Gaussian distribution, a Mann-Whitney test was used. Multigroup comparisons with Gaussian distribution, one-way ANOVA with Tukey-Kramer’s multiple-comparison test was used after the confirmation of homogeneity of variance among the groups by Bartlett’s test. For multigroup comparisons with non-Gaussian distribution, a Kruskal-Wallis test with Dunn’s test was used. P values of 0.05 or less were the threshold for statistical significance. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
HIGHLIGHTS.
LTA from Gram-positive bacteria binds and activat es NLRP6.
Listeria and cytosolic LTA induce caspase-11 processing via NLRP6 and ASC.
Processed caspase-11 promotes caspase-1 activation and IL-18 secretion.
NLRP6 and caspase-11 exacerbate Gram-positive pathogen infection in vivo.
ACKNOWLEDGMENTS
We thank Millennium Pharmaceuticals for Casp1−/−Casp11−/− mice, R. E. Vance (University of California, Berkeley) for Gsdmd−/− mice, T.-D. Kanneganti (St. Jude Children’s Research Hospital) for Casp1−/− mice, S. L. Masters (The Walter and Eliza Hall Institute of Medical Research) and M. A. Gavrilin (The Ohio State University) for THP-1 mutant cells, J. B. Bliska (Stony Brook University) for Yersinia mutant, G. N. Barber (University of Miami Miller School of Medicine) for Sting−/− BM cells, R. A. Flavell (Yale University School of Medicine) for NLRP6 plasmid, A. Gründling (Imperial College London) for Listeria lmo0927 mutant, J. Whitfield (University of Michigan) for ELISA assays, James Delproposto (University of Michigan) for BLI analysis, and L. Haynes (University of Michigan) for animal husbandry. H. H. was supported by a JSPS Postdoctoral Fellowship for Research Abroad and S. S. by Training grant F32CA200144 from the NCI. This work was supported by NIH grants R01AI063331 and R01DK091191 to G. N., R01CA166879 to G. Y. C., INCa (Plbio-2012–106) to M. C., AR055398 to E. S. A., and funds to the Michigan Comprehensive Cancer Center Immunology Monitoring Core from the University of Michigan’s Cancer Center Support Grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, Lamkanfi M, and Kanneganti TD (2012). NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 488, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, and Portnoy DA (2004). Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med 200, 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker PJ, Boucher D, Bierschenk D, Tebartz C, Whitney PG, D’Silva DB, Tanzer MC, Monteleone M, Robertson AA, Cooper MA, et al. (2015). NLRP3 inflammasome activation downstream of cytoplasmic LPS recognition by both caspase-4 and caspase-5. Eur. J. Immunol 45, 2918–2926. [DOI] [PubMed] [Google Scholar]
- Berche P, Gaillard JL, and Richard S (1988). Invasiveness and intracellular growth of Listeria monocytogenes. Infection 16 Suppl 2, S145–148. [DOI] [PubMed] [Google Scholar]
- Birchenough GM, Nystrom EE, Johansson ME, and Hansson GC (2016). A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, and Rapp UR (1985). Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature 318, 667–670. [DOI] [PubMed] [Google Scholar]
- Bottai D, Groschel MI, and Brosch R (2016). Type VII Secretion Systems in Gram-Positive Bacteria. Curr Top Microbiol. Immunol 404, 235–265. [DOI] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N, Dixit VM, and Monack DM (2012). Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490, 288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, von Moltke J, Jones JW, Vance RE, and Monack DM (2010). Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, and Chen ZJ (2014). Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Casabona MG, Kneuper H, Chalmers JD, and Palmer T (2016). The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol 2, 16183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, and Unanue ER (2004). Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med 200, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GY, Liu M, Wang F, Bertin J, and Nunez G (2011). A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol 186, 7187–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung LK, Park YH, Zheng Y, Brodsky IE, Hearing P, Kastner DL, Chae JJ, and Bliska JB (2016). The Yersinia Virulence Factor YopM Hijacks Host Kinases to Inhibit Type III Effector-Triggered Activation of the Pyrin Inflammasome. Cell Host Microbe 20, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Padova FE, Brade H, Barclay GR, Poxton IR, Liehl E, Schuetze E, Kocher HP, Ramsay G, Schreier MH, McClelland DB, et al. (1993). A broadly cross-protective monoclonal antibody binding to Escherichia coli and Salmonella lipopolysaccharides. Infect. Immun 61, 3863–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. (2011). NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145, 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. (2010). The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol 11, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, Shaw MH, Kim YG, and Nunez G (2012). NLRC4-driven production of IL-1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol 13, 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukase K, Matsumoto T, Ito N, Yoshimura T, Kotani S, and Kusumoto S (1992). Synthetic Study on Lipoteichoic Acid of Gram-Positive Bacteria .1. Synthesis of Proposed Fundamental Structure of Streptococcus-Pyogenes Lipoteichoic Acid. Bull. Chem. Soc. Jap 65, 2643–2654. [Google Scholar]
- Grundling A, and Schneewind O (2007). Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Natl. Acad. Sci. U S A 104, 8478–8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P, Malireddi RK, Anand PK, Demon D, Vande Walle L, Liu Z, Vogel P, Lamkanfi M, and Kanneganti TD (2012). Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-beta (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J. Biol. Chem 287, 34474–34483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, and Miao EA (2013). Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Tsuchiya K, Nomura T, Kawamura I, Shoma S, and Mitsuyama M (2008). Dependency of caspase-1 activation induced in macrophages by Listeria monocytogenes on cytolysin, listeriolysin O, after evasion from phagosome into the cytoplasm. J. Immunol 180, 7859–7868. [DOI] [PubMed] [Google Scholar]
- He WT, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang ZH, Zhong CQ, and Han J (2015). Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res 25, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zeng MY, Yang D, Motro B, and Nunez G (2016). NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, et al. (2012). Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzen E, Johansson L, Kansal R, Hecht A, Dahesh S, Janos M, Nizet V, Kotb M, and Norrby-Teglund A (2012). Intracellular Streptococcus pyogenes in human macrophages display an altered gene expression profile. PLoS One 7, e35218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, Wang R, Zhang Y, Martinon F, Miao D, et al. (2013). Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175. [DOI] [PubMed] [Google Scholar]
- Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, Hu B, Hedl M, Zhang W, O’Connor W Jr., Murphy AJ, et al. (2012). IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, and Barber GN (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Dalmaroni MJ, Xiao N, Corper AL, Verdino P, Ainge GD, Larsen DS, Painter GF, Rudd PM, Dwek RA, Hoebe K, et al. (2009). Soluble CD36 ectodomain binds negatively charged diacylglycerol ligands and acts as a co-receptor for TLR2. PLoS One 4, e7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. (2015). Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. (2011). Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, et al. (2013). Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249. [DOI] [PubMed] [Google Scholar]
- Kolb-Maurer A, Kammerer U, Maurer M, Gentschev I, Brocker EB, Rieckmann P, and Kampgen E (2003). Production of IL-12 and IL-18 in human dendritic cells upon infection by Listeria monocytogenes. FEMS Immunol. Med. Microbiol 35, 255–262. [DOI] [PubMed] [Google Scholar]
- Lee BL, Stowe IB, Gupta A, Kornfeld OS, Roose-Girma M, Anderson K, Warming S, Zhang J, Lee WP, and Kayagaki N (2018). Caspase-11 auto-proteolysis is crucial for noncanonical inflammasome activation. J. Exp. Med 215, 2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Hur J, Lee P, Kim JY, Cho N, Kim SY, Kim H, Lee MS, and Suk K (2001). Dual role of inflammatory stimuli in activation-induced cell death of mouse microglial cells. Initiation of two separate apoptotic pathways via induction of interferon regulatory factor-1 and caspase-11. J. Biol. Chem 276, 32956–32965. [DOI] [PubMed] [Google Scholar]
- Lemire P, Robertson SJ, Maughan H, Tattoli I, Streutker CJ, Platnich JM, Muruve DA, Philpott DJ, and Girardin SE (2017). The NLR Protein NLRP6 Does Not Impact Gut Microbiota Composition. Cell Rep 21, 3653–3661. [DOI] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. (2015). Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 163, 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XY, Choi MS, and Porter AG (2000). Expression analysis of the human caspase-1 subfamily reveals specific regulation of the CASP5 gene by lipopolysaccharide and interferon-gamma. J. Biol. Chem 275, 39920–39926. [DOI] [PubMed] [Google Scholar]
- Lochner M, Kastenmuller K, Neuenhahn M, Weighardt H, Busch DH, Reindl W, and Forster I (2008). Decreased susceptibility of mice to infection with Listeria monocytogenes in the absence of interleukin-18. Infect. Immun 76, 3881–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, and Egelman EH (2014). Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Ohto U, Shibata T, Miyake K, and Shimizu T (2016). Crystal structure of NOD2 and its implications in human disease. Nat. Commun 7, 11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltez VI, Tubbs AL, Cook KD, Aachoui Y, Falcone EL, Holland SM, Whitmire JK, and Miao EA (2015). Inflammasomes Coordinate Pyroptosis and Natural Killer Cell Cytotoxicity to Clear Infection by a Ubiquitous Environmental Bacterium. Immunity 43, 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamantopoulos M, Ronchi F, Van Hauwermeiren F, Vieira-Silva S, Yilmaz B, Martens L, Saeys Y, Drexler SK, Yazdi AS, Raes J, et al. (2017). Nlrp6- and ASC-Dependent Inflammasomes Do Not Shape the Commensal Gut Microbiota Composition. Immunity 47, 339–348 e334. [DOI] [PubMed] [Google Scholar]
- Man SM, Karki R, Briard B, Burton A, Gingras S, Pelletier S, and Kanneganti TD (2017). Differential roles of caspase-1 and caspase-11 in infection and inflammation. Sci. Rep 7, 45126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, Frase S, Zhu Q, Malireddi RK, Kuriakose T, et al. (2016). IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell 167, 382–396 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, and Tschopp J (2007). Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ 14, 10–22. [DOI] [PubMed] [Google Scholar]
- Mueller NJ, Wilkinson RA, and Fishman JA (2002). Listeria monocytogenes infection in caspase-11-deficient mice. Infect. Immun 70, 2657–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, and Aguet M (1994). Functional role of type I and type II interferons in antiviral defense. Science 264, 1918–1921. [DOI] [PubMed] [Google Scholar]
- Munoz-Planillo R, Franchi L, Miller LS, and Nunez G (2009). A critical role for hemolysins and bacterial lipoproteins in Staphylococcus aureus-induced activation of the Nlrp3 inflammasome. J. Immunol 183, 3942–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A, and Bishayi B (2016). Intracellularly survived Staphylococcus aureus after phagocytosis are more virulent in inducing cytotoxicity in fresh murine peritoneal macrophages utilizing TLR-2 as a possible target. Microb. Pathog 97, 131–147. [DOI] [PubMed] [Google Scholar]
- Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, Lemoine Y, Hot D, and Chamaillard M (2011). Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc. Natl. Acad. Sci. U S A 108, 9601–9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, et al. (2004). Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med 200, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Reis O, Vieira A, Moura IM, Zanolli Moreno L, Carvalho F, Pucciarelli MG, Garcia-Del Portillo F, Sousa S, and Cabanes D (2016). Listeria monocytogenes encodes a functional ESX-1 secretion system whose expression is detrimental to in vivo infection. Virulence 8, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py BF, Jin M, Desai BN, Penumaka A, Zhu H, Kober M, Dietrich A, Lipinski MM, Henry T, Clapham DE, et al. (2014). Caspase-11 controls interleukin-1beta release through degradation of TRPC1. Cell Rep 6, 1122–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, and Fitzgerald KA (2012). TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell 150, 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I, Deets KA, Ji DX, von Moltke J, Tenthorey JL, Lee AY, Philip NH, Ayres JS, Brodsky IE, Gronert K, et al. (2017). NAIP-NLRC4 Inflammasomes Coordinate Intestinal Epithelial Cell Expulsion with Eicosanoid and IL-18 Release via Activation of Caspase-1 and −8. Immunity 46, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte P, Van Criekinge W, Van de Craen M, Van Loo G, Desmedt M, Grooten J, Cornelissen M, De Ridder L, Vandekerckhove J, Fiers W, et al. (1998). Cathepsin B-mediated activation of the proinflammatory caspase-11. Biochem. Biophys. Res. Commun 251, 379–387. [DOI] [PubMed] [Google Scholar]
- Schroder K, and Tschopp J (2010). The inflammasomes. Cell 140, 821–832. [DOI] [PubMed] [Google Scholar]
- Seo SU, Kamada N, Munoz-Planillo R, Kim YG, Kim D, Koizumi Y, Hasegawa M, Himpsl SD, Browne HP, Lawley TD, et al. (2015). Distinct Commensals Induce Interleukin-1beta via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity 42, 744–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seregin SS, Golovchenko N, Schaf B, Chen J, Eaton KA, and Chen GY (2016). NLRP6 function in inflammatory monocytes reduces susceptibility to chemically induced intestinal injury. Mucosal Immunol 10, 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, Baxter NT, Zhao L, Schloss PD, Martens EC, et al. (2017). NLRP6 Protects Il10−/− Mice from Colitis by Limiting Colonization of Akkermansia muciniphila. Cell Rep 19, 733–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, and Shao F (2015). Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, and Shao F (2014). Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Hara H, Fang R, Hernandez-Cuellar E, Sakai S, Daim S, Chen X, Dewamitta SR, Qu H, Mitsuyama M, et al. (2014). The adaptor ASC exacerbates lethal Listeria monocytogenes infection by mediating IL-18 production in an inflammasome-dependent and -independent manner. Eur. J. Immunol 44, 3696–3707. [DOI] [PubMed] [Google Scholar]
- Tsuji NM, Tsutsui H, Seki E, Kuida K, Okamura H, Nakanishi K, and Flavell RA (2004). Roles of caspase-1 in Listeria infection in mice. Int. Immunol 16, 335–343. [DOI] [PubMed] [Google Scholar]
- Tsuneyoshi N, Fukudome K, Kohara J, Tomimasu R, Gauchat JF, Nakatake H, and Kimoto M (2005). The functional and structural properties of MD-2 required for lipopolysaccharide binding are absent in MD-1. J. Immunol 174, 340–344. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, Zheng Y, Rongvaux A, Sun Q, Yang G, et al. (2015). Nlrp6 regulates intestinal antiviral innate immunity. Science 350, 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Miura M, Jung Y, Zhu H, Gagliardini V, Shi L, Greenberg AH, and Yuan J (1996). Identification and characterization of Ich-3, a member of the interleukin-1beta converting enzyme (ICE)/Ced-3 family and an upstream regulator of ICE. J. Biol. Chem 271, 20580–20587. [DOI] [PubMed] [Google Scholar]
- Webb AJ, Karatsa-Dodgson M, and Grundling A (2009). Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol. Microbiol 74, 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm BE, Dijkgraaf GJ, Bledau AS, Welm AL, and Werb Z (2008). Lentiviral transduction of mammary stem cells for analysis of gene function during development and cancer. Cell Stem Cell 2, 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. (2014). NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156, 1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, and Portnoy DA (2010). c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Fernandes-Alnemri T, and Alnemri ES (2010). Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J. Clin. Immunol 30, 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Hara H, Tsuchiya K, Sakai S, Fang R, Matsuura M, Nomura T, Sato F, Mitsuyama M, and Kawamura I (2012). Listeria monocytogenes strain-specific impairment of the TetR regulator underlies the drastic increase in cyclic di-AMP secretion and beta interferon-inducing ability. Infect. Immun 80, 2323–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanoni I, Tan Y, Di Gioia M, Broggi A, Ruan J, Shi J, Donado CA, Shao F, Wu H, Springstead JR, et al. (2016). An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352, 1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.