Nucleic Acids Research, 2017, Vol. 45, No. 20, Pages 11925–11940, https://doi.org/10.1093/nar/gkx832.
The authors wish to correct an error in their article. In Figure 2, panel A was accidentally duplicated from panel B. A new Figure 2 is provided below. The article has been updated online and in print. This correction does not influence the results and overall conclusions of the article.
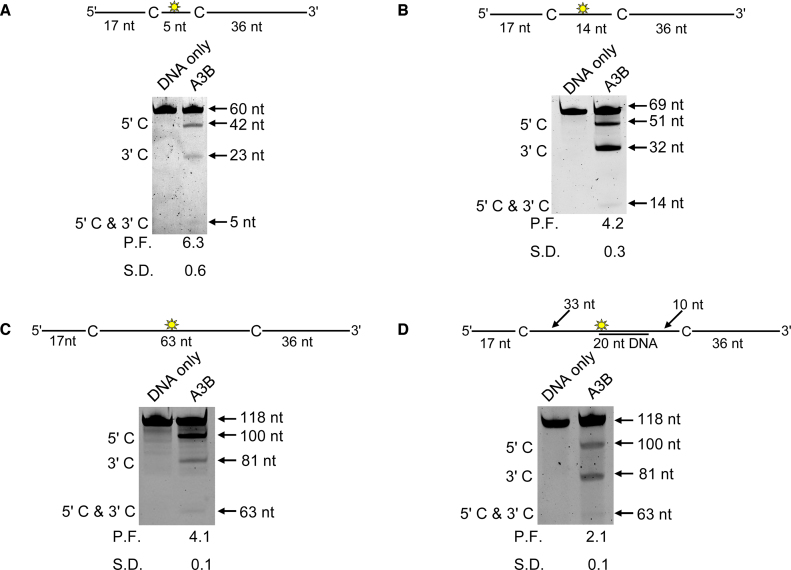
Figure 2.
A3B is a processive enzyme that both slides and moves 3-dimensionally on ssDNA. Processivity of A3B was tested on ssDNA substrates that contained a fluorescein-labeled deoxythymidine (yellow star) between two 5’ATC deamination motifs separated by different distances. (A) Deamination of a 60 nt ssDNA substrate with deamination motifs spaced 5 nt apart. Single deaminations of the 5’C and 3’C are detected as the appearance of labeled 42- and 23-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 5 nt labeled fragment. (B) Deamination of a 69 nt ssDNA substrate with deamination motifs spaced 14 nt apart. Single deaminations of the 5’C and 3’C are detected as the appearance of labeled 51- and 32-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 14 nt labeled fragment. (C) Deamination of a 118 nt ssDNA substrate with deaminated cytosines spaced 63 nt apart. Single deaminations of the 5’C and 3’C are detected as the appearance of labeled 100- and 81-nt fragments, respectively; double deamination of both C residues on the same molecule results in a 63 nt labeled fragment. (D) Deamination of a 118 nt ssDNA substrate as in (C) but with a 30 nt complementary DNA annealed between the deamination motifs. The measurements of processivity factor (P.F.) and the standard deviation (S.D.) from three independent experiments are shown below the gel.