Abstract
Eyeless (ey) is one of the most critical transcription factors for initiating the entire eye development in Drosophila. However, the molecular mechanisms through which Ey regulates target genes and pathways have not been characterized at the genomic level. Using ChIP-Seq, we generated an endogenous Ey-binding profile in Drosophila developing eyes. We found that Ey binding occurred more frequently at promoter compared to non-promoter regions. Ey promoter binding was correlated with the active transcription of genes involved in development and transcription regulation. An integrative analysis revealed that Ey directly regulated a broad and highly connected genetic network, including many essential patterning pathways, and known and novel eye genes. Interestingly, we observed that Ey could target multiple components of the same pathway, which might enhance its control of these pathways during eye development. In addition to protein-coding genes, we discovered Ey also targeted non-coding RNAs, which represents a new regulatory mechanism employed by Ey. These findings suggest that Ey could use multiple molecular mechanisms to regulate target gene expression and pathway function, which might enable Ey to exhibit a greater flexibility in controlling different processes during eye development.
INTRODUCTION
Transcription factors play crucial roles in cell fate specification during development. Many such factors are often highly conserved in function and DNA-binding sequence specificity across species. Since different combinations of transcription factors can be employed to determine specific cell fates, identifying key transcription factors for each cell type and investigating how they regulate downstream genes and pathways have been a major focus of study in the molecular genetics of development.
One of the pivotal transcription factors controlling retina cell fate specification in Drosophila is encoded by the eyeless (ey) gene. Ey is a paired domain (PD)- and homeodomain (HD)-containing protein that is both essential and sufficient for initiating Drosophila eye development (1). Eye development can be completely abolished or severely impaired in flies with loss-of-function mutations in ey, although this phenotype is not completely penetrant (2). Detailed examination reveals the absence of differentiated photoreceptors in ey mutant eye discs, indicating that it plays an essential role in early eye development (3). In addition, overexpression of ey is capable of inducing the formation of ectopic eyes on wings, antennae and legs in flies, providing strong evidence that ey is an important regulator sufficient to initiate the entire genetic cascade underlying Drosophila eye development (1). The homolog of ey in vertebrates, Pax6, is highly conserved in several model organisms and humans (4–8). Similar to the phenotype observed in ey mutant flies, Pax6 knock-out mice present a severe eye phenotype (9). In humans, Pax6 mutations are associated with Aniridia, a genetic eye disease (10). Overexpression of vertebrate Pax6 can induce ectopic eye formation in flies (11), indicating that the regulatory network controlling eye development is likely, in part, to be conserved among different species. Therefore, understanding the molecular mechanism of ey function in Drosophila will provide useful insights into Pax6 function in vertebrates.
The expression of ey is tightly associated with the development of the Drosophila visual system. Drosophila adult compound eyes originate from an eye primordium that is marked at embryonic stage 12 by the expression of ey (12,13). During later embryonic stages, the eye primordium continues to grow and forms the eye imaginal disc, which further increases in size during early larval stages when ey is detected throughout the entire eye disc (3). In the early third instar larval stage, retinal cell differentiation begins with the initiation of the morphogenetic furrow (MF) along the posterior margin of the eye disc. The MF progresses anteriorly across the eye imaginal disc leaving differentiating photoreceptors in its wake. Cells anterior to the furrow remain undifferentiated and divide randomly. However, these cells have already committed to a retinal cell fate as anterior eye disc fragments can continue to develop into eye tissue when transplanted into a larval host (14). Cells posterior to the MF undergo a successive differentiation process, which leads to the formation of patterned eye units called ommatidia (15). At this stage, ey is expressed predominantly in cells anterior to the MF, greatly down-regulated in differentiating photoreceptors posterior to the furrow, and increased again along the posterior margin of the eye disc (16). Through further morphological changes during the pupal phase, differentiated eye imaginal discs eventually give rise to the adult compound eye composed of 750–800 mature ommatidia.
A lot of effort has been devoted to characterize how Ey controls Drosophila eye development. However, current available information is still hard to provide a general picture of Ey function. Only a few direct downstream targets of Ey have been reported so far, including eyes absent (eya), sine oculis (so), optix, shifted (shf), atonal (ato) and rhodopsin 1 (rh1) (3,17–19). The majority of these targets are members of a retinal determination (RD) gene network that controls the early stages of eye development in Drosophila. Currently, the RD network contains five core factors, including twin of eyeless (toy), ey, so, eya and dachshund (dac) (20–23). Similar to ey, these RD genes are expressed in eye discs early during development and continue to be expressed in the undifferentiated retina during MF progression. They are also highly conserved among different species, suggesting that the RD network is an evolutionarily conserved pathway required for eye development. toy is at the top of this genetic network and is required for the activation of ey transcription, which in turn induces the expression of so, eya and the further downstream gene dac (24). The expression of so and eya has been shown to be mediated via Ey binding to a consensus binding motif (18,25). However, the RD network is not purely hierarchical, and extensive positive feedback loops and protein–protein interactions have been identified. For instance, So and Eya can form a protein complex that synergistically regulates eye development. Overexpression of either so or eya alone can weakly induce the expression of each other, while co-expression of both genes, though acting downstream of ey, has a synergistic effect on triggering ectopic eye formation and leads to strong ey activation in ectopic eye tissue (26). Similar results are also observed between eya and dac (27). As a result, positive feedback regulation among ey, so, eya and dac maintains high expression levels of these RD genes in undifferentiated cells and locks in the retinal cell fate.
In addition to regulate the RD network, ey genetically interacts with several signaling pathways important for retinal cell differentiation, such as the Decapentaplegic (Dpp), Hedgehog (Hh), Notch (N) and Wingless (Wg) pathways. Dpp and Hh signaling cooperatively direct the initiation and progression of the MF (28,29), while Wg and N signaling patterns the dorsal–ventral axis of the eye disc (30,31). Earlier studies have shown that proper eye development requires cooperation between the RD network and these signaling pathways (32–34). Nevertheless, the molecular mechanism by which ey interacts with these pathways during eye development has not been systematically studied.
Due to technical difficulties, genome-wide studies of endogenous Ey are uneasy to be performed on Drosophila developing eye discs, so an overexpressing system has been used (18,35,36). In order to have a better understanding of the molecular mechanisms by which endogenous Ey interacts with target genes and pathways in the whole genome during eye development, we performed chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-Seq) on developing eye discs to identify regions bound by Ey across the Drosophila genome. We observed a much higher frequency of Ey binding at the promoter compared to the non-promoter regions and showed that Ey promoter binding was correlated with the active transcription of target genes. We also characterized Ey binding at non-promoter regions and identified a set of putative enhancers bound by Ey in the Drosophila genome. Using an integrative analysis, 311 putative direct downstream targets of Ey were identified, which form an extensive genetic network, including many known eye genes and well-established patterning pathways, such as Dpp, Hh, Wg and N pathway. In particular, we noticed that Ey might have a stronger control of certain signaling pathways via binding to several components of the same pathway. Besides known genes and pathways, we also identified novel eye genes and biological functions that have not been associated with Ey. In addition to protein-coding genes, we observed that Ey could also target non-coding RNAs, which reveals another novel regulatory mechanism employed by Ey during eye development.
MATERIALS AND METHODS
ChIP-Seq and data analysis
Four hundred Canton S third instar larval eye-antenna discs were dissected and crosslinked with 1% formaldehyde for 15 min and quenched by 125 mM Glycine for 5 min at room temperature. After washing with phosphate buffered saline, discs were lysed in lysis buffer (50 mM Hepes potassium (K-Hepes, pH 7.8), 140 mM NaCl, 1 mM ethylene glycol - bis(2-aminoethylether) - N,N,N',N'-tetraacetic acid (EGTA), 1 mM ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 0.1% Na-deoxycholate, protease inhibitors) and sonicated with a Branson sonifier to shear chromatin to ∼200–400 bp. The chromatin lysate was precleared with protein A sepharose (GE healthcare, #17-5280-01) and incubated with antibody at 4°C overnight. The rabbit anti-Ey antibody used for the ChIP-seq experiment was kindly provided by Dr U. Walldorf. The specificity of this antibody for Ey has been confirmed in the lab and also by prior studies (16,37,38). Protein A sepharose was added to the lysate for 3 h to capture chromatin complexes. After three washes with lysis buffer, once with high salt wash buffer (50 mM K-Hepes (pH 7.8), 500 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, protease inhibitors) and once with TE (10 mM Tris (pH 8.0), 1 mM EDTA), chromatin complexes were eluted with 1% sodium dodecyl sulphate, 10 mM Tris (pH 8.0) and 1 mM EDTA at 65°C for 10 min, followed by reverse-crosslinking at 65°C overnight. ChIP for PolII and histone modifications were performed as previously described (39,40) with following antibodies: anti-RNA PolII (Upstate, clone 8W16G), H3K4me1 (Abcam, ab8895) and H3K27Ac (Abcam, ab4729). ChIP samples were purified using Qiagen Polymerase chain reaction (PCR) purification kit and sequenced based on the manufacturer’s instructions (Illumina, San Diego, CA).
Raw sequencing reads were mapped to the Drosophila reference genome dm3 with Burrows-Wheeler Aligner (BWA-MEM) in Unipro UGENE software using default parameters (41,42). The aligned reads were then used for peak-calling by Model-based Analysis of ChIP-seq (MACS; http://liulab.dfci.harvard.edu/MACS/) with default settings (43). Eyeless peaks were annotated using FlyBase version 5.57 with in-house Perl script and visualized using IGV (https://www.broadinstitute.org/igv/). Peak/gene list overlap analysis was performed using BEDTOOLS (https://github.com/arq5x/bedtools2) (44). Motif scans were performed using FIMO from the MEME Suite (http://meme.nbcr.net/meme/cgi-bin/fimo.cgi) (45,46). Gene Ontology (GO) analysis was performed using the DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/) (47).
In vivo enhancer reporter assay
Genomic regions flanking potential enhancers were PCR amplified (0.5–2 kb) and subcloned into the pH-Stinger-attB reporter vector (48). Transgenic flies were generated using PhiC31-mediated integration (49). Third instar larval eye discs were dissected and examined for reporter gene expression.
Isoform-specific RT-PCR
RNA was extracted from third instar larval eye discs using Trizol (Invitrogen, 15596-026) and purified with the RNeasy mini kit (Qiagen, 74104). Reverse transcription was performed according to manufacturer’s instructions (Invitrogen, 18080-051). Sequences of isoform-specific primers used for PCR are: estimated glomerularfiltration rate (EGFR)-A-FW: 5′-cacaagctcatcggtcagcaa-3′; EGFR-A-RV: 5′-ctctgaggttccggtaatgatgtt-3′; EGFR-B-FW: 5′-gagtcaccattcccaccagtc-3′; EGFR-B-RV: 5′-ccgatc-tctgaggttccggt-3′; CG9328-A-FW: 5′-accctcgtcgaatatctccagt-3′; CG9328-B-FW: 5′-acagaacaggc-agaaattcct-3′; CG9328-RV:5′-atcatcgttccggccgcgtt-3′; IP3K1-A-FW:5′-gacaacatcggactcaagca-3′; IP3K1-A-RV:5′-caaagtaggcgggcactatc-3′; IP3K1-B-FW: 5′-caacggagtgaggatcgttt-3′; IP3K1-B-RV:5′-atctgagcggtctgtggttc-3′; control-FW: 5′-gccggcagttcgaacgtata-3′; control-RV: 5′-aacgagtc-atcacctccgc-3′.
In situ hybridization
Complementary DNAs were obtained from the Drosophila Genomics Resource Center (DGRC) and used as templates for generating RNA probes. RNA probes were translated using SP6, T3 and T7 RNA polymerases and labeled with digoxigenin (DIG) using the DIG RNA labeling mix (Roche, 11277073910). Freshly dissected eye-antennal discs were subjected to in situ hybridization as previously described (18).
RESULTS
Genome-wide profiling of Eyeless-binding sites in developing larval eye discs
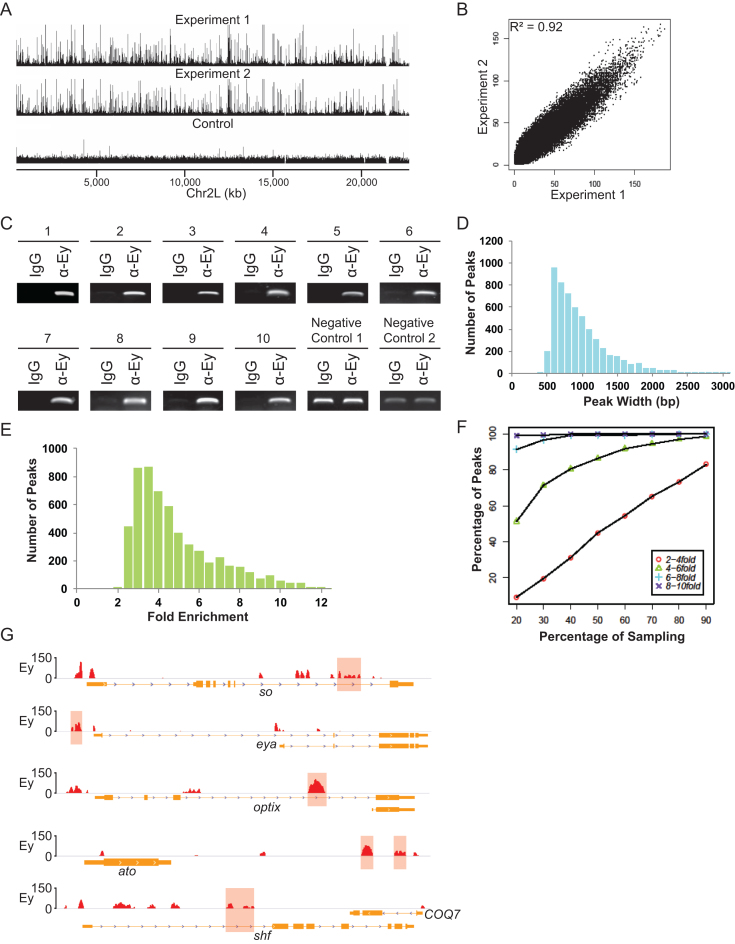
We performed genome-wide profiling of endogenous Ey-binding sites in the third instar larval eye discs using chromatin immunoprecipitation (ChIP) with an antibody against Ey followed by high-throughput next generation sequencing. The ChIP-Seq procedure was optimized so that high-quality sequencing data were obtained from 400 freshly dissected eye-antennal imaginal discs. Two independent ChIP-Seq experiments were performed, generating a total of ∼22.5 million uniquely mapped short reads. Analyses showed that Ey ChIP-Seq data were highly reproducible, as indicated by a correlation coefficient of 0.92 between the results from the two independent experiments (Figure 1A and B). In addition, our initial peak calling revealed 5185 and 6065 individual peaks from our two Ey ChIP-seq datasets, with 4977 common peaks, further indicating that the two ChIP-seqs are highly reproducible. To validate the accuracy of the ChIP-Seq data, ChIP-PCR was performed on a set of 20 randomly selected regions using ChIP samples and all examined regions show significant enrichment of Ey binding (Figure 1C and data not shown). Therefore, a genome-wide Ey-binding profile of third instar larval eye imaginal discs was successfully generated.
Figure 1.
Genome-wide profiling of Ey-binding regions using ChIP-Seq. (A) A snapshot of Ey ChIP-Seq profiles on chromosome 2L from two independent experiments. (B) Plot showing that the two independent Ey ChIP-Seq experiments are highly reproducible. (C) ChIP-PCR validation of Ey ChIP-Seq results. Enrichment of Ey binding at 10 randomly selected regions identified by ChIP-Seq is shown. Two representative non-Ey binding regions are shown as negative controls. (D) The peak width distribution of Ey-binding regions detected by ChIP-Seq. (E) The range of fold enrichment of Ey peaks across the whole genome. (F) Computer simulation of sequencing depth of Ey ChIP-Seq. (G) Ey ChIP-Seq peaks show Ey binds to previously known Ey-binding regions. The reported Ey-binding regions are marked with red shaded region. The Ey ChIP peaks are normalized to the negative ChIP control.
Ey-binding peaks were observed with an average width of ∼930 bp (Figure 1D). We found that the fold enrichment (relative to the negative control) of all Ey-binding sites ranged from 1.8 to 12.1 (P = 10−6), with a median value of ∼4-fold (Figure 1E). To assess the sensitivity of the experimental approach, computer simulations were conducted to examine the efficiency of recovering Ey-enriched peaks at different sequencing depths. We found that our sequencing depth achieved high sensitivity in detecting Ey-bound regions with >4-fold enrichment, as the number of enriched peaks reached saturation at this level (Figure 1F). The fold enrichment of Ey binding at previously reported target regions was also examined (Figure 1G). Among all sites, the lowest Ey enrichment was observed at the gene shf, with an enrichment of 3.49-fold. This relatively low enrichment of Ey binding at shf may be due to the restricted expression pattern of shf at the disc margin anterior to the MF (18). Therefore, combining computer simulations and manual examination, we used 3.45-fold enrichment as the cut-off, leading to the identification of a total of 3562 putative Ey-bound regions throughout the Drosophila genome of third instar larval eye imaginal discs (Supplementary Table S1).
Eyeless is highly enriched at the promoter regions of actively transcribed genes involved in development and transcription regulation
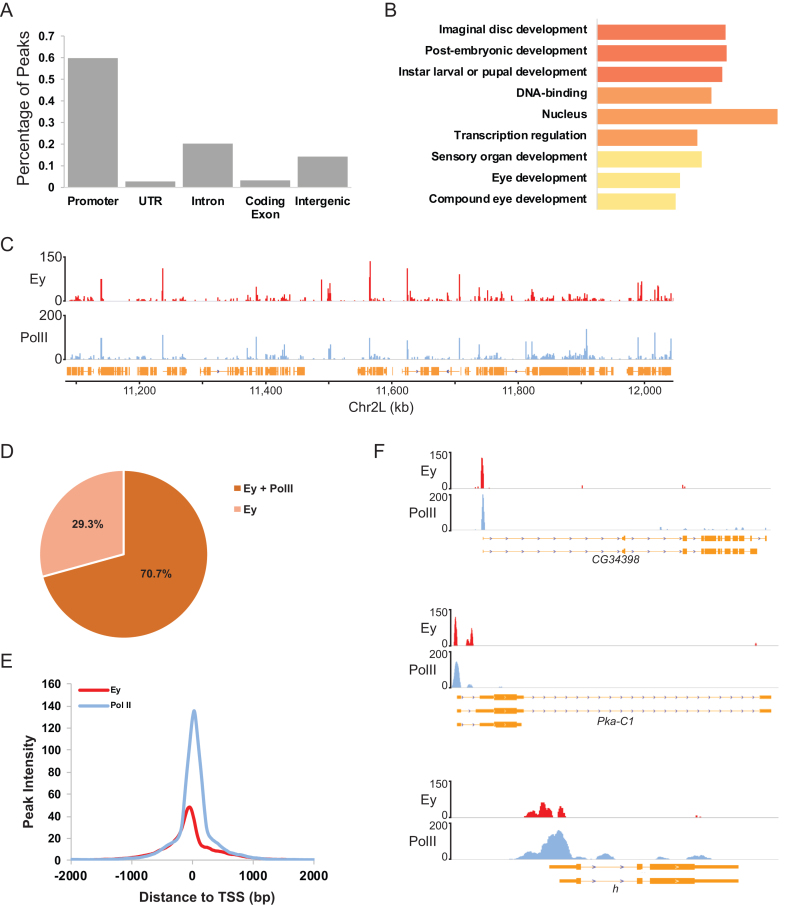
Ey-bound genomic regions were categorized based on their positions relative to annotated transcripts into five classes: the promoter (150 bp ± transcription start site [TSS]), untranslated regions (UTRs), introns, coding exons and intergenic regions. More than half of all Ey peaks (59.6%) were observed at the promoter region. Binding at intergenic and intronic regions accounted for ∼14.1 and 20.3% of all Ey peaks in the genome, respectively (Figure 2A). In contrast, a paucity of Ey binding was observed within coding exons, consistent with the notion that gene regulatory elements are usually enriched in non-coding regions.
Figure 2.
Ey binding at promoters is associated with active transcription of developmental or transcription-related genes. (A) Classification of Ey binding in the genome based on genomic features. (B) GO analysis of genes with Ey binding at the promoters. The top three groups enriched at three function categories are shown. (C) Snapshot of ChIP-Seq profiles at chromosome 2R showing that Ey and PolII ChIP-Seq profiles are highly correlated. (D) Pie chart showing the percentage of all Ey peaks that overlap with PolII bound regions. Ey binding is enriched at promoters with PolII occupancy. (E) Plot showing the distance between the ChIP-seq peaks of Ey and PolII at the nearest annotated TSS. (F) Examples showing Ey and PolII ChIP-Seq profiles around TSSs.
In total, 2124 Ey-binding peaks were mapped to 2894 promoter regions in the Drosophila genome. We found that the binding of Ey at promoters was very selective, occurring at only ∼15% of promoters in the Drosophila genome. GO analysis showed that genes with Ey binding at their promoters were highly enriched for imaginal disc development, DNA binding and transcription regulation (Figure 2B). In order to further characterize Ey binding at promoters, we used ChIP-Seq to generate a genome-wide profile of RNA polymerase II (PolII) binding in third instar larval eye-antennal discs. Interestingly, we observed that Ey and PolII peaks were highly correlated at promoters and a majority of Ey-associated promoters were also bound by PolII (Figure 2C and D). Ey-binding profiles at these promoters were further compared with that of PolII. In contrast to the PolII profiles, with summits 30 bp downstream of the TSS, respectively, the highest enrichment of Ey binding was observed ∼50 bp upstream of the TSS (Figure 2E). We found that the Ey-binding profile, although with lower peak intensity, is very similar to that of PolII near promoters. Therefore, we examined the Ey and PolII profiles at the individual gene level and found that Ey peaks were indeed highly coincident with PolII peaks at promoters (Figure 2F). Given the high correlation of Ey and PolII binding both locally and genomically, it raises the possibility that Ey may act in concert with PolII at promoter regions.
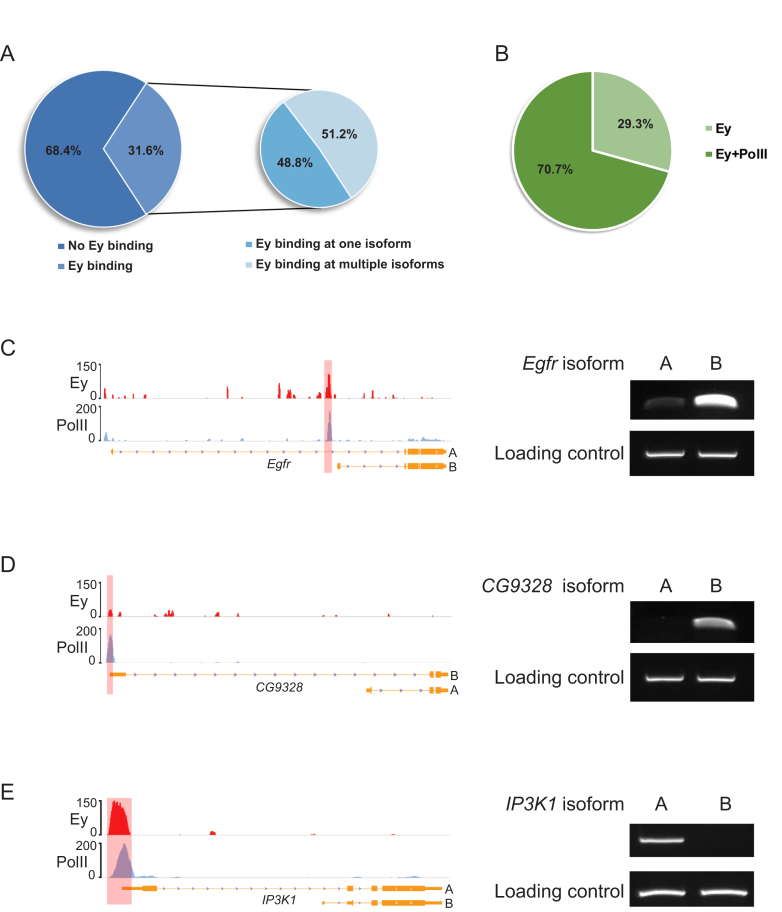
The selective binding of Ey can be observed not only at different genes, but also at promoters of different isoforms of the same gene. Based on the current FlyBase gene annotation, a total of 6941 genes have multiple isoforms, 3659 of which have multiple TSSs. Among these genes, 1157 (∼31.6%) showed Ey binding at their promoter regions. About 48.8% of these genes showed Ey binding at the promoter of only one isoform, while the remaining 51.2% showed Ey binding at two or more isoform promoters (Figure 3A). As expected, the vast majority of the promoters bound by Ey were positive for PolII. Among 1914 promoters of genes with multiple TSSs bound by Ey, ∼70.7% of them were occupied by PolII (Figure 3B). This observation suggests that active transcription occurs at these specific isoforms. This was tested by isoform-specific RT-PCR on several randomly selected genes with differential Ey promoter binding. Indeed, a strong correlation between the transcription of isoform, as inferred from PolII ChIP-Seq signals, and the enrichment of Ey at the corresponding promoter was observed (Figure 3C–E). Taken together, these results suggest that Ey promoter binding is very specific and frequently observed at genes involved in development and/or transcription regulation.
Figure 3.
Ey binding at promoters is associated with active transcription of genes at the isoform level. (A) Ey can selectively bind the promoters of specific isoforms of a gene. The left pie chart shows the fraction of genes with multiple isoforms that are bound by Ey at the promoters. The right chart shows the fraction of genes with Ey binding to the promoters of multiple isoforms versus only a single isoform. (B) Pie chart showing percentage of isoform specific Ey peaks that coincide with PolII peaks. (C–E) Examples showing the isoforms bound by Ey are transcribed in third larval instar eye discs and corresponding ChIP-Seq profiles of Ey and PolII at the promoters.
Genome-wide screening of enhancers bound by Ey in the developing eye
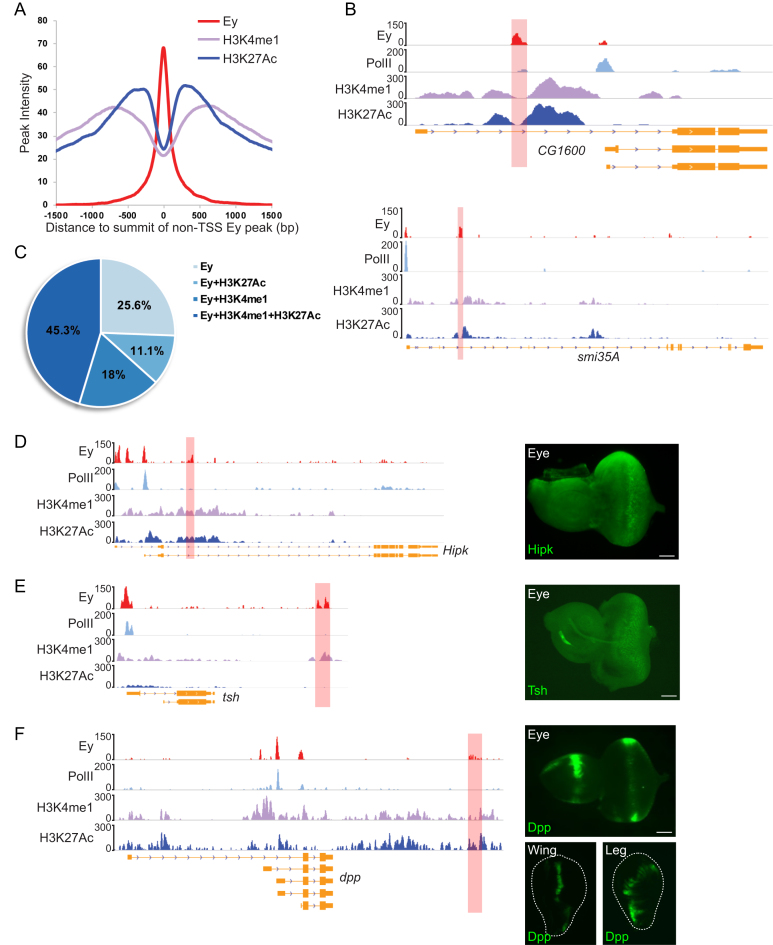
In addition to promoters, 1321 Ey peaks occurred in UTRs, introns and intergenic regions (termed non-TSS regions). We checked Ey enhancers identified in previous studies and found they were well-represented in our ChIP-Seq peaks (Figure 1G). In order to search for more potential Ey-associated regulatory regions in the genome, we performed ChIP-Seq of two enhancer-associated histone modification marks: H3K4me1 and H3K27Ac (40,50,51). We found that Ey binding frequently occurred between two peaks of a histone modification profile (Figure 4A and B). In particular, 56.4% of Ey-binding peaks were flanked by H3K27Ac peaks, while 63.3% had similar patterns of H3K4me1 modification (Figure 4C). In total, 598 Ey-bound non-TSS regions associated closely with both H3K4me1 and H3K27Ac modifications.
Figure 4.
Identification of Ey-associated enhancers using ChIP-Seq. (A) The ChIP-Seq peak profiles of H3K4me1 and H3K27Ac around Ey-binding peaks at non-TSS regions. The peak intensities of H3K4me1 and H3K27Ac are plotted against their distance to the center of Ey peaks. (B) Examples showing predicted enhancers based on Ey binding and histone modification marks. The red shaded areas mark the identified Ey-associated enhancers. (C) The majority of Ey binding at non-TSS regions is associated with H3K4me1 and/or H3K27Ac modification. The percentage of Ey peaks with particular histone marks are indicated. (D–F) Validation of predicted Ey-associated enhancers using in vivo reporter assays. Examples showing GFP reporter expression driven by enhancers of hipk, tsh and dpp identified by Ey ChIP-Seq. The ChIP-Seq profiles of Ey and other chromatin marks are shown. Enhancers tested in the study are marked with red shaded area; scale bar: 50 μm.
To maximize the possibility of locating potential Ey-associated enhancers, a search for Ey-binding motif was also performed, resulting in the identification of an additional 78 peaks that contained Ey-binding motif and were marked with either H3K4me1 or H3K27Ac modification, but not both. These regions were combined with 598 Ey-bound regions with both histone modifications, and hence a list of 676 putative Ey-associated enhancers was generated (Supplementary Table S2). From this list, we found that previously reported Ey-bound enhancers were well-represented in Ey ChIP-Seq profiles. For instance, our ChIP-Seq identified an Ey peak close to the promoter of eya, which overlaps with an eye enhancer published by an earlier study (52). We also observe Ey peaks covering the deletions associated with so5 and so7, two Ey-bound enhancers identified in the so locus via yeast one-hybrid analysis (25). These findings suggested that our ChIP-Seq protocol was reliable and sensitive for identifying putative Ey-associated enhancers.
Among Ey-bound non-TSS regions with both H3K4me1 and H3K27Ac modifications, ∼20% contain an Ey-binding motif, implying that a significant portion of Ey binding might be mediated through its interacting proteins. In addition, we did not observe a significant difference in genomic locations of Ey peaks in relation to the binding motif (Supplementary Figure). Prior studies have shown that members of RD network could function in protein complexes to regulate downstream gene expression (26,27). We therefore examined if Ey-bound peaks contained binding motifs of other RD factors. Binding motifs have been identified for Toy and So, so we started our analyses with these RD factors. Indeed, we found that Toy- and/or So-binding motifs could be identified in ∼32% (154/480) of putative Ey-enhancer regions without an Ey-binding motif and ∼40% (78/196) of regions with an Ey-binding motif, suggesting that interactions between Ey and Toy/So may occur at these enhancers (Supplementary Table S3).
In order to test the function of putative enhancers, we used an in vivo reporter system in which genomic regions spanning potential Ey-associated enhancers were used to drive the expression of a GFP reporter with a basal promoter. Among 19 regions selected for the reporter assay, 15 can activate GFP expression and 11 can drive reporter gene expression in the eye disc (Supplementary Table S4). For example, Hipk encodes a serine-threonine kinase and plays a role in regulating N and Wg signaling pathways (53,54). We identified a novel Ey-bound enhancer located within the second intron of Hipk and found that this enhancer was sufficient to activate GFP expression mimicking endogenous Hipk expression in third instar larval eye discs (Figure 4D) (55). We also found several enhancers that directed reporter gene expression partially mimicking the endogenous expression pattern of target genes. For instance, the transcription of tsh can be detected only anterior to the MF or both anterior and posterior to the MF, depending on the expression system used (56). We identified an Ey-associated enhancer downstream of tsh, which directed reporter gene expression mainly at the central regions anterior to the MF and in several rows of differentiated photoreceptors close to the MF (Figure 4E). In addition, several identified enhancers could drive reporter gene expression in eye discs as well as in other imaginal discs or the brain. For instance, a ∼2.2 kb Ey-bound enhancer was identified downstream of the 3′ end of dpp. Although much shorter than the reported dpp disc enhancer (57), the Ey-bound enhancer was sufficient to drive reporter gene expression not only in eye discs but in wing and leg discs as well (Figure 4F).
Ey regulates a broad network of eye genes and retinal development pathways
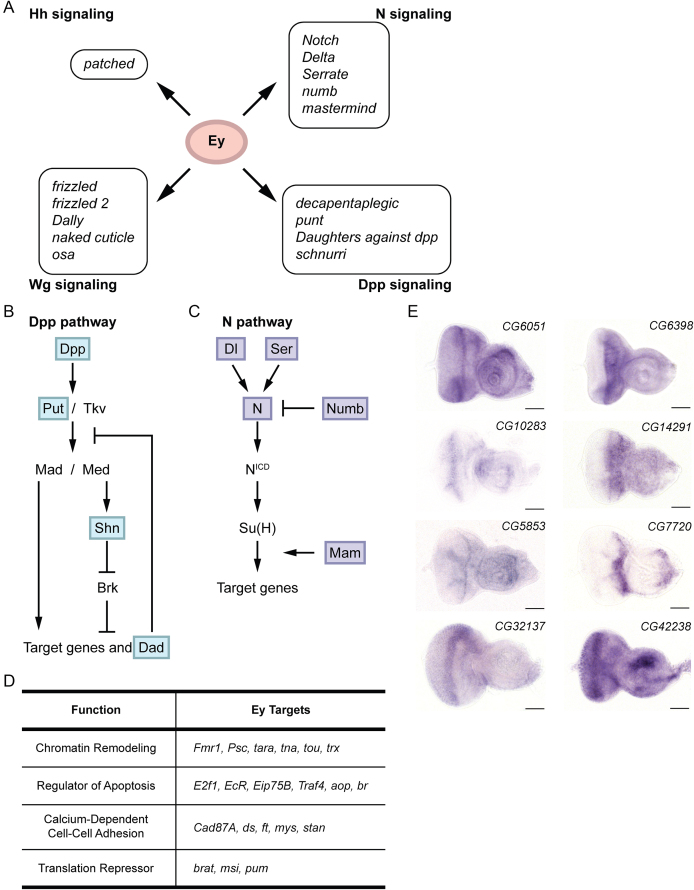
Genes with Ey-binding sites can be divided into three categories, depending on whether they show Ey binding at the promoter, non-promoter regions or both. In order to identify genes that are under tighter regulation by Ey, we focused on genes with Ey binding at both the promoter and the non-TSS regions. By incorporating gene expression profiles in the developing eye based on RNA-Seq data (58), a total of 311 genes were identified as strong candidates under tight Ey regulation (Supplementary Table S5). These Ey targets are highly enriched of developmental proteins, transcription factors, and include many repressors, receptors, kinases and signaling proteins as well (Supplementary Table S6). Especially, we found that many members of several signaling pathways known to be required for Drosophila eye development, including Dpp, Hh, N and Wg pathways, are Ey targets (Figure 5A). In Dpp signaling, Dpp is a diffusible protein and functions as a ligand. Upon binding to Dpp, the receptors Tkv or Put trigger the phosphorylation of Mad and Med. Phosphorylated Mad and Med then enter the nucleus and regulate the expression of downstream genes, such as Dad (59). Interestingly, Ey targeted several key members of this essential pathway, including dpp, put, dad and shn, which represented the ligand, receptor, downstream targets and regulators (Figure 5B). Like Dpp, the N pathway is another signaling cascade with multiple members targeted by Ey. These include N (receptor), Ser and Dl (ligands), mam (co-activator) and numb (inhibitor) (Figure 5C). These findings suggest that targeting multiple components with different functions of the same signaling cascade may be a mechanism that allows Ey to control the function of an essential pathway at different levels during eye development. Besides the above pathways, Ey targets were also found in mitogen-activated protein kinase (MAPK), PI3 kinase and Cadherin signaling pathways. In addition, some Ey targets were enriched in biological or regulatory processes that had not been tightly associated with Ey previously, such as programmed cell death, stem cell development, chromatin remodeling and translation regulation (Figure 5D and Supplementary Table S7). These findings could provide useful insights not only into the cellular events that were under the control of Ey during eye development but also new molecular mechanisms by which Ey regulated target genes as well.
Figure 5.
Identification of Ey targets in developing eye discs. (A) Ey targets identified in the N, Dpp, Wg and Hh signaling pathways are listed. (B and C) Ey targets in the Dpp and N pathways. Identified Ey targets are marked with shaded boxes. (D) Examples of new pathways and cellular events enriched in GO analysis of Ey targets. (E) Expression of novel Ey targets in developing eye discs as detected by in situ hybridization; scale bar: 50 μm.
Although many Ey targets are known eye genes, some genes are novel and have not been studied in Drosophila eye development. Therefore, we used in situ hybridization to validate whether these genes are expressed in developing eye discs. The transcripts of all tested genes were detected in eye discs, suggesting these genes may play novel roles during eye development and are under direct regulation by Ey (Figure 5E). Together with known eye genes and pathways, Ey targets identified by ChIP-Seq form a broad genetic network that controls important cellular events required for Drosophila eye development.
Ey may regulate non-coding RNAs in the Drosophila genome
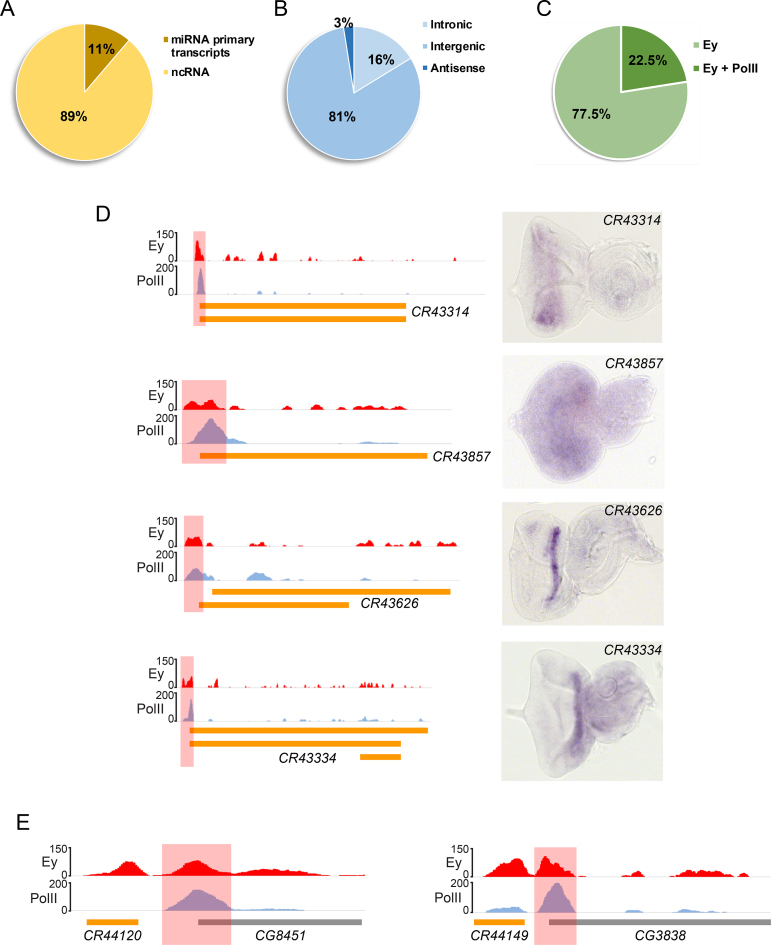
In addition to protein-coding genes, we discovered that Ey also bound non-coding RNA (ncRNA) loci. Due to the complexity of Drosophila genome, we only focused on Ey peaks that could be assigned only to ncRNAs. After filtering, we identified 87 Ey peaks that are associated with 80 ncRNAs (Supplementary Table S8). Eleven percent of these non-protein-coding genes contain microRNAs (miRNAs) within the gene body and therefore are annotated as miRNA primary transcripts or have that potential (Figure 6A). Although the majority of identified ncRNAs are located within intergenic regions of the Drosophila genome, some are found within introns of coding genes. Only two ncRNAs completely overlap with its corresponding coding gene but in an opposite transcriptional direction, and therefore are classified as antisense ncRNAs (Figure 6B).
Figure 6.
Ey binds non-coding RNAs in the genome. (A) A portion of Ey-bound ncRNAs are miRNA primary transcripts. (B) Classification of Ey-bound ncRNAs based on the location in the genome. (C) Some ncRNA-associated Ey peaks are enriched with PolII binding. (D) In situ hybridization showed the expression of Ey-bound ncRNAs in the developing eye disc. Corresponding ChIP-Seq profiles are also shown, with red shaded areas marking Ey peaks; scale bar: 50 μm. (E) Examples showing a bimodal Ey-binding profile at some ncRNAs close to the promoter of adjacent coding genes. The coding gene-associated Ey peaks are marked as red shaded areas.
Like protein-coding genes, we observed PolII signals associated with Ey peaks at the promoters of 22.5% ncRNAs (Figure 6C), indicating that these ncRNAs are actively transcribed in developing eye discs. We therefore performed in situ hybridization assay and found all examined ncRNAs were expressed in eye discs. Some of them exhibited very specific expression patterns (Figure 6D). Interestingly, a previous study has showed that loss of function of CR43314, an ncRNA identified in our study, could affect the development of interommatidial bristles (IOB) in the eye (60). We found that the promoter of CR43314 had strong Ey and PolII (Figure 6D), indicating that Ey might be able to regulate IOB development via controlling the transcription of this ncRNA.
Among ncRNAs with only Ey-binding peaks but no PolII signals, ∼40% are very close to the promoter of a protein-coding gene. We found that such protein-coding genes often had their own Ey peak at the promoter and therefore presented a distinctive bimodal distribution of Ey peaks around TSSs (Figure 6E). Although the detailed mechanisms are still unknown, Ey binding at ncRNAs around the promoters of coding genes could be another new way of regulating protein-coding genes. Since non-coding RNAs exhibit a variety of regulatory functions and participate in many important cellular events (61), these results suggest that regulation of ncRNAs by Ey is an important mechanism employed by Ey, when controlling target gene expression during Drosophila eye development.
DISCUSSION
ChIP-Seq identified endogenous Ey-binding sites in the Drosophila genome
In this study, we have generated, to our knowledge, the first genome-wide binding profile of endogenous Ey in developing Drosophila eye discs. Compared to previous studies, our ChIP-Seq results showed that more than half of Ey binding occurred at promoter regions in the Drosophila genome. Upon closer examination, we found that Ey binding at promoter regions was very selective, which was reproducibly distinct even at different isoforms of the same gene. To further characterize Ey binding at promoters, we integrated ChIP-Seq profiles of PolII, an active promoter mark, and found that majority of genes with Ey binding at their promoters were transcribed. GO analysis demonstrated that these genes were enriched for imaginal disc development and transcription regulation, indicating that Ey preferentially targeted a select group of genes via binding to promoter regions.
We also investigated which factor(s) might determine Ey selective binding at promoters. Drosophila promoters can be classified into different categories depending on the composition of core promoter components (62). After motif analysis, however, we did not discover a particular promoter motif to be enriched for Ey-bound promoters (data not shown). We also tested whether Ey binding at promoters was driven by its own or other novel binding motifs, but were unable to enrich a motif specifically for Ey binding at promoters. Interestingly, we found that Ey promoter peaks exhibited a high resemblance to PolII profile, which had also been observed for some general transcription factors associated with PolII in the promoter initiation complex (63). This suggests that Ey and PolII interact at the promoter and regulate gene transcription together. Although the molecular mechanisms remain further investigation, Ey binding at promoters, especially its interaction with PolII, might be an underappreciated regulatory mechanism for Ey as a transcription factor.
The second most common type of Ey binding occurred at intergenic and intronic regions, which accounted for ∼34% of Ey peaks in the genome. We found that previously reported Ey-bound enhancers were well-represented in Ey ChIP-Seq profiles. However, we found that Ey binding alone was insufficient to provide accurate information for predicting enhancer regions. Therefore, we integrated Ey-binding profile, motif location and histone modification marks for Ey-associated enhancer prediction. We noticed that the majority of Ey peaks were frequently located between two histone modification peaks. Previous studies showed that genomic regions flanked by two histone modification peaks corresponded to nucleosome-free or open chromatin regions and were known to often contain functional regulatory elements (64–66). Therefore, we further focused on Ey-bound regions with this specific histone modification pattern, which led to the identification of 676 putative enhancers regulated by Ey in the Drosophila genome.
We used an in vivo reporter assay to examine the function of predicted enhancer regions. About 79% of tested enhancers are functional and ∼58% are active in the eye. Interestingly, we found several enhancers that are ‘ubiquitous’ and can direct reporter gene expression in the eye and other tissues where Ey is not expressed. In the Drosophila genome, some regulatory elements are bound by multiple transcription factors with different expression patterns (67). Our results suggest that Ey may bind to non-eye specific enhancers that are also responsible for the expression of corresponding targets in non-retinal tissues under the control of other transcription factors.
We found that ∼30% of putative Ey-enhancer regions contained an Ey-binding motif, which led us to suspect that other proteins might be involved in Ey binding at enhancers. We first started our search among RD genes, since several RD genes have been shown to form protein complexes (26,27) and play cooperative roles in regulating target gene expression (19,37). Indeed, we found that Toy and/or So binding motifs could be identified in putative Ey-enhancer regions with or without an Ey-binding motif, which indicates that the cooperation among Ey, Toy and So is dynamic and occurs at many sites in the genome. Via partnering with different proteins, Ey might be able to achieve a greater flexibility in targeting downstream genes. Although we only tested Toy and So in this study, it is very likely that other factors might also interact with Ey at different enhancers. First, the binding motifs of other RD factors are unknown, so motif analyses could not be performed on these genes. Second, there were still Ey enhancer binding that could not be explained by motif analyses. Therefore, ChIP-Seq studies of other RD factors or DNA-binding proteins in developing eye discs will help to address this question in the future.
Ey may use multiple mechanisms to regulate genes and signaling pathways during eye development
We identified Ey targets in developing eye discs based on Ey binding via ChIP-Seq. We found that mechanisms by which Ey regulates target gene expression might differ depending on whether Ey only binds promoters, putative enhancers or both. Specific genes or isoforms with Ey promoter binding are most often expressed in the eye and Ey-bound enhancers are able to direct target gene expression in the eye, indicating that these genes might play a role in eye development. We mainly focused on genes with Ey occupancy at both the promoter and potential enhancers in this study, considering they may be under stronger Ey control. Indeed, many genes known to be important for fly eye development are well-represented in this group of targets. For instance, so and eya, two core factors in the RD network, have Ey binding at both promoters and enhancers. Interestingly, we also observed Ey binding at both the promoter and a potential enhancer of toy. toy is thought to act genetically upstream of ey, since toy can ectopically induce ey, but not vice versa (23). We find that toy transcription can be induced in non-eye tissue with ey ectopic expression, according to published microarray data (18). However, the induction is to a lesser degree than that of so and eya, suggesting that ey alone might not be able to completely activate toy expression. Our findings imply that Ey might cooperate with other proteins to regulate gene expression. Via further investigation, we find that so and toy might be two potential factors that act together with ey to maintain toy expression in the eye (unpublished data). Like so and eya, dac is another RD factor regulated by ey, with Ey binding at both the promoter and a reported enhancer, 5EE (68). Interestingly, we did not observe enrichment of histone modification marks at 5EE, but at a second reported enhancer, 3EE, suggesting 3EE might be the major enhancer of dac expression in third instar larval eye discs. A prior study also showed that the expression of both 3EE and 5EE reporters are absent in so1 mutant eye discs. We therefore examined So binding at these two enhancers in our So ChIP-Seq data (69). We find So binds to both 5EE and 3EE, suggesting that dac expression might be mainly regulated by So in the third instar larval eye. In addition, 5EE is activated very early during eye development (67). Therefore, it is likely that Ey binding at the 5EE enhancer of dac occurs very early during eye development, which might facilitate the recruitment of So to dac at a later developmental stage.
We noticed that expression patterns of some Ey targets do not completely overlap with Ey in the eye. Although Ey is mainly expressed anterior to MF, it is maintained at a very low level in the posterior region and is up-regulated at the posterior margin of eye discs (16). However, the function of Ey posterior to the MF has not been fully characterized. Since there is no systematic information regarding posterior genes in the fly eye, we searched for Ey targets whose expression was significantly changed in ato1 eye discs (18), considering that ato is expressed in and posterior to the MF and genes downstream of ato are likely to expressed in the same region. By combining expression profiles of posterior eye discs (70), 33 Ey targets are predicted to be expressed preferentially at the MF or in the posterior region of eye discs (Supplementary Table S9). Further investigation of Ey regulation on potential posterior genes will provide a better picture of Ey function during eye development.
Ey has been regarded as an activator according to studies of known Ey downstream targets. We identified 188 genes whose expression is significantly induced in non-eye imaginal discs upon Ey ectopic expression (18). About 31% (59/188) of these genes show Ey binding (Supplementary Table S10). Besides regulating gene expression via binding to target genes at different regulatory elements, our ChIP-Seq results suggest that Ey may be regulating gene expression via an indirect way. For instance, we found that Ey can also bind non-coding RNAs, including miRNA primary transcripts, suggesting that Ey might act as a potential repressor for some genes via indirectly regulating the corresponding non-coding RNAs.
Furthermore, many Ey targets are members of signaling pathways essential for eye development. We observe that Ey may have a stronger influence on some pathways by targeting multiple components of the same pathway. Intriguingly, components with opposite roles in the same pathway are often both targeted. One potential explanation is that Ey may selectively activate or inhibit a particular signaling pathway by controlling the expression of its positive or negative regulators in different types of cells. Because the expression patterns of different components may not be the same, ChIP-Seq performed on a smaller number of cells with a more uniform developmental status and transcriptome profile may provide more specific information regarding Ey regulation of these essential pathways in eye discs. In addition to known patterning pathways, we found some new pathways or cellular events that might also be regulated by Ey. Further investigation of how these new pathways are integrated with other patterning pathways would provide insightful information of Ey function during eye development.
A proposed model of Ey function
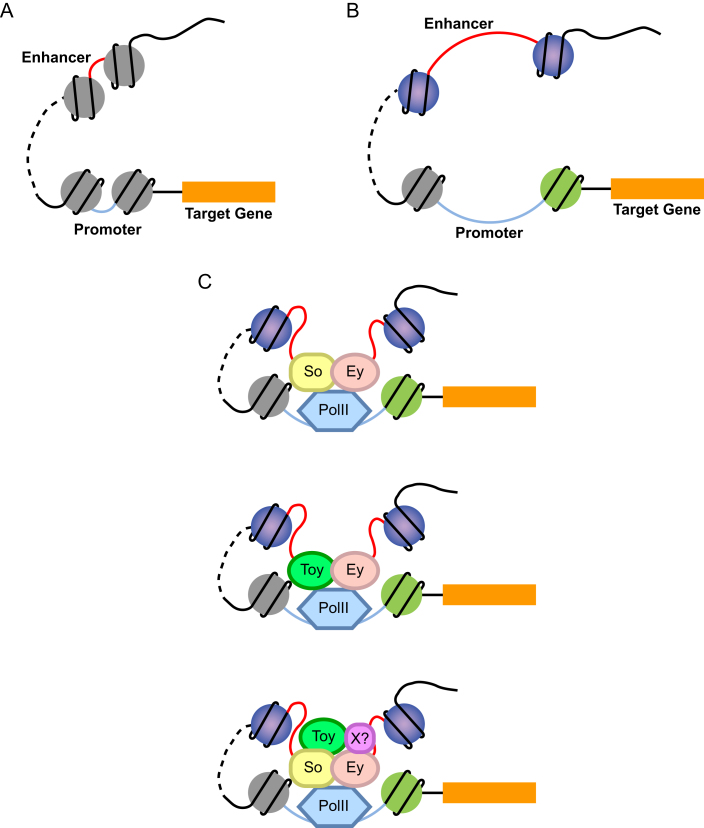
Based on our findings, we propose a model where Ey functions in a dynamic protein complex to modulate target gene expression during Drosophila eye development (Figure 7). This Ey-associated regulatory protein complex may consist of additional RD factors, PolII holoenzyme, chromatin-remodeling proteins or transcription regulatory proteins. Depending on different genes and cellular contexts, the protein complex can change its composition accordingly and regulate gene expression by binding to promoter and/or enhancer regions. Ey binding at promoters or enhancers can be achieved either by direct binding or via interaction with other transcription factors or regulatory proteins. For genes that are more tightly regulated by Ey, Ey may be involved in both promoter and enhancer recognition.
Figure 7.
A model of Ey function in transcription regulation. (A) In the absence of transcription, both promoters and enhancers are in a ‘closed’ status without any prominent active chromatin signatures. (B) Histones of nucleosomes near promoter and enhancer regions are specifically modified, which exposes regulatory regions and allows transcription factor and/or polymerase binding. (C) Ey binding at the promoter or enhancer regions can be mediated by its DNA-binding domain(s) or via interaction with other transcription factors or regulatory proteins in a dynamic protein complex. This Ey-associated protein complex may interact with PolII transcription machinery at the promoter and thus regulate target gene expression. So and Toy were shown as Ey potential binding partners that may also include other proteins yet to be characterized.
Ey contains two DNA-binding domains, a PD and a HD, and one transactivation domain at the C terminus. Both DNA-binding domains contain helix-loop-helix DNA-binding motifs and are connected by a linker region. It has been shown that an intact Ey protein is necessary for its function during development (71). Although misexpression of ey without the PD is insufficient to rescue the eye phenotype of ey mutant flies (72), studies on vertebrate Pax gene family members revealed different types of cooperation between the HD and two sub-domains of the PD, suggesting the involvement of both domains in recognizing different sets of DNA sequences (73). In addition to interacting with its own HD, the Pax6 PD is able to interact with itself and even HDs of other proteins. Moreover, coexpressing both Pax6 and HD-containing genes can increase the transactivation capability of Pax6 (74). These findings further support our model that Ey might act in a regulatory protein complex in vivo and also indicates which type of proteins might be able to interact with Ey. In our study, we show that So might be a potential binding partner of Ey. So contains one HD that mediates DNA binding and a SIX domain for protein–protein interaction (20,26). However, So does not have a transactivation domain, and therefore requires a coactivator for its proper function. Previously, So was shown to form a protein complex with Eya and activate target genes via the DNA-binding domain of So and the transactivation domain of Eya. Considering the potential interaction between the PD and HD, it is possible that So may interact with Ey via PH-HD cooperation and rely on the transactivation domain of Ey for regulating target gene expression. This may also explain the coincidence of So- and Ey-binding motifs in many Ey peaks. Furthermore, since So binds Eya via the SIX domain (26) and its interaction with Ey may be very likely mediated by HD domain, Eya may also be included in the same complex as Ey and So and add another level of regulation. Therefore, Ey could bind target sequences via its own DNA-binding domain and/or its binding partners, depending on which domains are involved in the protein–protein interaction. For binding partners without transactivation capability, the regulatory protein complex may rely on the transactivation domain of Ey for controlling gene expression, while for those partners with activation domains, the transactivation domain of Ey may or may not be involved in gene regulation.
Ey acts at the top of RD network that controls Drosophila eye development. Prior studies tended more toward a model where Ey initiated the RD network by activating a few downstream targets that further regulated more downstream genes. However, based on the Ey-binding profile presented in this study, Ey seems to target a large number of genes outside of the RD network that are also important for eye development. Eye development is a complicated yet highly organized process. Considering the function and expression patterns of different Ey targets, it would be difficult for Ey alone to precisely regulate hundreds of genes during this process. Thus, the interaction with other binding partners could dramatically expand the capability of Ey in targeting downstream genes and in regulating the developmental process.
DATA AVAILABILITY
All ChIP-seq data generated in this study are uploaded to Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) as accession number GSE112868.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Yiyun Chen for performing the ChIP-Seq, ChIP-PCR, RT-PCR, in situ hybridization experiments, and assisting with the ChIP-Seq data analyses. Dr Uwe Walldorf for kindly providing us with the anti-Ey antibody.
Author contributions: K.Y. and F.W. conducted analyses on ChIP-Seq data. Y.L. contributed to imaginal discs dissection for ChIP, and together with K.W., conducted the in vivo reporter assay. The manuscript was prepared by K.Y. and F.W., and reviewed by R.C. and G.M.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 EY011232 to G.M., R01 EY016853 to R.C.]; Keck Center of Gulf Coast Consortia [5 R90 DA023418-03]. Funding for open access charge: National Institutes of Health [R01 EY011232].
Conflict of interest statement. None declared.
REFERENCES
- 1. Halder G., Callaerts P., Gehring W.J.. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995; 267:1788–1792. [DOI] [PubMed] [Google Scholar]
- 2. Lindsley D.L., Zimm G.G.. The Genome of Drosophila Melanogaster. 1992; San Diego: Academic Press. [Google Scholar]
- 3. Halder G., Callaerts P., Flister S., Walldorf U., Kloter U., Gehring W.J.. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998; 125:2181–2191. [DOI] [PubMed] [Google Scholar]
- 4. Ton C.C., Hirvonen H., Miwa H., Weil M.M., Monaghan P., Jordan T., van Heyningen V., Hastie N.D., Meijers-Heijboer H., Drechsler M. et al. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991; 67:1059–1074. [DOI] [PubMed] [Google Scholar]
- 5. Martin P., Carriere C., Dozier C., Quatannens B., Mirabel M.A., Vandenbunder B., Stehelin D., Saule S.. Characterization of a paired box- and homeobox-containing quail gene (Pax-QNR) expressed in the neuroretina. Oncogene. 1992; 7:1721–1728. [PubMed] [Google Scholar]
- 6. Puschel A.W., Gruss P., Westerfield M.. Sequence and expression pattern of pax-6 are highly conserved between zebrafish and mice. Development. 1992; 114:643–651. [DOI] [PubMed] [Google Scholar]
- 7. Quiring R., Walldorf U., Kloter U., Gehring W.J.. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994; 265:785–789. [DOI] [PubMed] [Google Scholar]
- 8. Hirsch N., Harris W.A.. Xenopus Pax-6 and retinal development. J. Neurobiol. 1997; 32:45–61. [PubMed] [Google Scholar]
- 9. Hogan B.L., Horsburgh G., Cohen J., Hetherington C.M., Fisher G., Lyon M.F.. Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J. Embryol. Exp. Morphol. 1986; 97:95–110. [PubMed] [Google Scholar]
- 10. Jordan T., Hanson I., Zaletayev D., Hodgson S., Prosser J., Seawright A., Hastie N., van Heyningen V.. The human PAX6 gene is mutated in two patients with aniridia. Nat. Genet. 1992; 1:328–332. [DOI] [PubMed] [Google Scholar]
- 11. Gehring W.J. The genetic control of eye development and its implications for the evolution of the various eye-types. Int. J. Dev. Biol. 2002; 46:65–73. [PubMed] [Google Scholar]
- 12. Chang T., Mazotta J., Dumstrei K., Dumitrescu A., Hartenstein V.. Dpp and Hh signaling in the Drosophila embryonic eye field. Development. 2001; 128:4691–4704. [DOI] [PubMed] [Google Scholar]
- 13. Green P., Hartenstein A.Y., Hartenstein V.. The embryonic development of the Drosophila visual system. Cell Tissue Res. 1993; 273:583–598. [DOI] [PubMed] [Google Scholar]
- 14. Lebovitz R.M., Ready D.F.. Ommatidial development in Drosophila eye disc fragments. Dev. Biol. 1986; 117:663–671. [DOI] [PubMed] [Google Scholar]
- 15. Kumar J.P. Building an ommatidium one cell at a time. Dev. Dyn. 2012; 241:136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Atkins M., Jiang Y., Sansores-Garcia L., Jusiak B., Halder G., Mardon G.. Dynamic rewiring of the Drosophila retinal determination network switches its function from selector to differentiation. PLoS Genet. 2013; 9:e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sheng G., Thouvenot E., Schmucker D., Wilson D.S., Desplan C.. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev. 1997; 11:1122–1131. [DOI] [PubMed] [Google Scholar]
- 18. Ostrin E.J., Li Y., Hoffman K., Liu J., Wang K., Zhang L., Mardon G., Chen R.. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006; 16:466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Q., Zhang T., Jemc J.C., Chen Y., Chen R., Rebay I., Pignoni F.. Onset of atonal expression in Drosophila retinal progenitors involves redundant and synergistic contributions of Ey/Pax6 and So binding sites within two distant enhancers. Dev. Biol. 2014; 386:152–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheyette B.N., Green P.J., Martin K., Garren H., Hartenstein V., Zipursky S.L.. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994; 12:977–996. [DOI] [PubMed] [Google Scholar]
- 21. Bonini N.M., Bui Q.T., Gray-Board G.L., Warrick J.M.. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997; 124:4819–4826. [DOI] [PubMed] [Google Scholar]
- 22. Shen W., Mardon G.. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997; 124:45–52. [DOI] [PubMed] [Google Scholar]
- 23. Czerny T., Halder G., Kloter U., Souabni A., Gehring W.J., Busslinger M.. twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol. Cell. 1999; 3:297–307. [DOI] [PubMed] [Google Scholar]
- 24. Kumar J.P. Signalling pathways in Drosophila and vertebrate retinal development. Nat. Rev. Genet. 2001; 2:846–857. [DOI] [PubMed] [Google Scholar]
- 25. Niimi T., Seimiya M., Kloter U., Flister S., Gehring W.J.. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999; 126:2253–2260. [DOI] [PubMed] [Google Scholar]
- 26. Pignoni F., Hu B., Zavitz K.H., Xiao J., Garrity P.A., Zipursky S.L.. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997; 91:881–891. [DOI] [PubMed] [Google Scholar]
- 27. Chen R., Amoui M., Zhang Z., Mardon G.. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997; 91:893–903. [DOI] [PubMed] [Google Scholar]
- 28. Chanut F., Heberlein U.. Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development. 1997; 124:559–567. [DOI] [PubMed] [Google Scholar]
- 29. Dominguez M., Hafen E.. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 1997; 11:3254–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dominguez M., de Celis J.F.. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998; 396:276–278. [DOI] [PubMed] [Google Scholar]
- 31. Heberlein U., Borod E.R., Chanut F.A.. Dorsoventral patterning in the Drosophila retina by wingless. Development. 1998; 125:567–577. [DOI] [PubMed] [Google Scholar]
- 32. Chen R., Halder G., Zhang Z., Mardon G.. Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development. 1999; 126:935–943. [DOI] [PubMed] [Google Scholar]
- 33. Kango-Singh M., Singh A., Henry Sun Y.. Eyeless collaborates with Hedgehog and Decapentaplegic signaling in Drosophila eye induction. Dev. Biol. 2003; 256:49–60. [DOI] [PubMed] [Google Scholar]
- 34. Firth L.C., Baker N.E.. Retinal determination genes as targets and possible effectors of extracellular signals. Dev. Biol. 2009; 327:366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Michaut L., Flister S., Neeb M., White K.P., Certa U., Gehring W.J.. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:4024–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nfonsam L.E., Cano C., Mudge J., Schilkey F.D., Curtiss J.. Analysis of the transcriptomes downstream of Eyeless and the Hedgehog, Decapentaplegic and Notch signaling pathways in Drosophila melanogaster. PLoS One. 2012; 7:e44583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Punzo C., Seimiya M., Flister S., Gehring W.J., Plaza S.. Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development. 2002; 129:625–634. [DOI] [PubMed] [Google Scholar]
- 38. Oros S.M., Tare M., Kango-Singh M., Singh A.. Dorsal eye selector pannier (pnr) suppresses the eye fate to define dorsal margin of the Drosophila eye. Dev. Biol. 2010; 346:258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J. 3rd, Gingeras T.R. et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005; 120:169–181. [DOI] [PubMed] [Google Scholar]
- 40. Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007; 39:311–318. [DOI] [PubMed] [Google Scholar]
- 41. Okonechnikov K., Golosova O., Fursov M., team U.. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012; 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- 42. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 48. Barolo S., Carver L.A., Posakony J.W.. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. BioTechniques. 2000; 29:726. [DOI] [PubMed] [Google Scholar]
- 49. Venken K.J., He Y., Hoskins R.A., Bellen H.J.. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006; 314:1747–1751. [DOI] [PubMed] [Google Scholar]
- 50. Heintzman N.D., Hon G.C., Hawkins R.D., Kheradpour P., Stark A., Harp L.F., Ye Z., Lee L.K., Stuart R.K., Ching C.W. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009; 459:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zimmerman J.E., Bui Q.T., Liu H., Bonini N.M.. Molecular genetic analysis of Drosophila eyes absent mutants reveals an eye enhancer element. Genetics. 2000; 154:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee W., Swarup S., Chen J., Ishitani T., Verheyen E.M.. Homeodomain-interacting protein kinases (Hipks) promote Wnt/Wg signaling through stabilization of beta-catenin/Arm and stimulation of target gene expression. Development. 2009; 136:241–251. [DOI] [PubMed] [Google Scholar]
- 54. Lee W., Andrews B.C., Faust M., Walldorf U., Verheyen E.M.. Hipk is an essential protein that promotes Notch signal transduction in the Drosophila eye by inhibition of the global co-repressor Groucho. Dev. Biol. 2009; 325:263–272. [DOI] [PubMed] [Google Scholar]
- 55. Blaquiere J.A., Lee W., Verheyen E.M.. Hipk promotes photoreceptor differentiation through the repression of Twin of eyeless and Eyeless expression. Dev. Biol. 2014; 390:14–25. [DOI] [PubMed] [Google Scholar]
- 56. Pan D., Rubin G.M.. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:15508–15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Blackman R.K., Sanicola M., Raftery L.A., Gillevet T., Gelbart W.M.. An extensive 3′ cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development. 1991; 111:657–666. [DOI] [PubMed] [Google Scholar]
- 58. Naval-Sanchez M., Potier D., Haagen L., Sanchez M., Munck S., Van de Sande B., Casares F., Christiaens V., Aerts S.. Comparative motif discovery combined with comparative transcriptomics yields accurate targetome and enhancer predictions. Genome Res. 2013; 23:74–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Affolter M., Basler K.. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat. Rev. Genet. 2007; 8:663–674. [DOI] [PubMed] [Google Scholar]
- 60. Hilgers V., Bushati N., Cohen S.M.. Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol. 2010; 8:e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cech T.R., Steitz J.A.. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014; 157:77–94. [DOI] [PubMed] [Google Scholar]
- 62. Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006; 34:5943–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rhee H.S., Pugh B.F.. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012; 483:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. He H.H., Meyer C.A., Shin H., Bailey S.T., Wei G., Wang Q., Zhang Y., Xu K., Ni M., Lupien M. et al. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010; 42:343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bonn S., Zinzen R.P., Girardot C., Gustafson E.H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczynski B., Riddell A., Furlong E.E.. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 2012; 44:148–156. [DOI] [PubMed] [Google Scholar]
- 66. Vierstra J., Wang H., John S., Sandstrom R., Stamatoyannopoulos J.A.. Coupling transcription factor occupancy to nucleosome architecture with DNase-FLASH. Nat. Methods. 2014; 11:66–72. [DOI] [PubMed] [Google Scholar]
- 67. mod E.C., Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L. et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010; 330:1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pappu K.S., Ostrin E.J., Middlebrooks B.W., Sili B.T., Chen R., Atkins M.R., Gibbs R., Mardon G.. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005; 132:2895–2905. [DOI] [PubMed] [Google Scholar]
- 69. Jusiak B., Wang F., Karandikar U.C., Kwak S.J., Wang H., Chen R., Mardon G.. Genome-wide DNA binding pattern of the homeodomain transcription factor Sine oculis (So) in the developing eye of. Genom. Data. 2014; 2:153–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Baig J., Chanut F., Kornberg T.B., Klebes A.. The chromatin-remodeling protein Osa interacts with CyclinE in Drosophila eye imaginal discs. Genetics. 2010; 184:731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Clements J., Hens K., Merugu S., Dichtl B., de Couet H.G., Callaerts P.. Mutational analysis of the eyeless gene and phenotypic rescue reveal that an intact Eyeless protein is necessary for normal eye and brain development in Drosophila. Dev. Biol. 2009; 334:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Punzo C., Kurata S., Gehring W.J.. The eyeless homeodomain is dispensable for eye development in Drosophila. Genes Dev. 2001; 15:1716–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jun S., Desplan C.. Cooperative interactions between paired domain and homeodomain. Development. 1996; 122:2639–2650. [DOI] [PubMed] [Google Scholar]
- 74. Mikkola I., Bruun J.A., Holm T., Johansen T.. Superactivation of Pax6-mediated transactivation from paired domain-binding sites by dna-independent recruitment of different homeodomain proteins. J. Biol. Chem. 2001; 276:4109–4118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ChIP-seq data generated in this study are uploaded to Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) as accession number GSE112868.