Abstract
Constitutive heterochromatin undergoes a dynamic clustering and spatial reorganization during myogenic differentiation. However the detailed mechanisms and its role in cell differentiation remain largely elusive. Here, we report the identification of a muscle-specific long non-coding RNA, ChRO1, involved in constitutive heterochromatin reorganization. ChRO1 is induced during terminal differentiation of myoblasts, and is specifically localized to the chromocenters in myotubes. ChRO1 is required for efficient cell differentiation, with global impacts on gene expression. It influences DNA methylation and chromatin compaction at peri/centromeric regions. Inhibition of ChRO1 leads to defects in the spatial fusion of chromocenters, and mislocalization of H4K20 trimethylation, Suv420H2, HP1, MeCP2 and cohesin. In particular, ChRO1 specifically associates with ATRX/DAXX/H3.3 complex at chromocenters to promote H3.3 incorporation and transcriptional induction of satellite repeats, which is essential for chromocenter clustering. Thus, our results unveil a mechanism involving a lncRNA that plays a role in large-scale heterochromatin reorganization and cell differentiation.
INTRODUCTION
Constitutive heterochromatin, formed mainly at the gene-poor regions of pericentromeres and telomeres, undergoes massive reorganization during early embryogenesis, organogenesis and terminal differentiation of muscle and brain (1–3). Although clustering and reorganization of constitutive heterochromatin are supposed to function as a driver of nuclear organization, the detailed mechanisms and the relevance to gene regulation remain elusive (4,5).
Constitutive heterochromatin is characterized by high density of repetitive DNA elements and strong enrichment of trimethylation of histone H3 lysine 9 (H3K9me3). H3K9me3, mediated by Suv39H1/2 histone methyltransferases (HMTs), has an initial role for constitutive heterochromatin formation, by serving as a binding platform for different isoforms (α, β, γ) of HP1 (6). HP1 in turn recruits DNA methyltransferases (DNMTs) or Suv420 HMTs. CpG methylation by DNMTs and subsequent recruitment of methyl-CpG binding proteins contribute to constitutive heterochromatin stability (7,8). Suv420H2, the HMT for H4K20me3, is essential for telomere homeostasis (9,10) and required for the recruitment of the cohesin complex to the pericentromeric regions for proper chromosome segregation (11).
In addition to dedicated protein components, local ncRNAs transcribed from the repetitive DNA elements of the pericentromere or rDNA regions participate in the chromatin compaction of their own origin. For example, pericentromeric major satellite (MajS) repeat-derived RNA transcripts contribute to the condensation of constitutive heterochromatin by mediating pericentromeric localization of HP1 (12). DAXX/ATRX is a histone chaperone that is responsible for the deposition of histone variant H3.3 at pericentromeric and telomeric regions (13,14). While H3.3 deposition in euchromatin by HIRA complex contributes to active transcription (15–17), H3.3 by DAXX/ATRX is involved in heterochromatinization through RNA polymerase II (pol II)-dependent transcription of satellite repeats (13,18), which in turn promotes HP1 association and H3K9me3 enrichment, further enhancing a positive feedback loop for chromatin compaction. Although the mechanism of heterochromatin formation at repressive domains is relatively well investigated, the mechanism for the large-scale reorganization associated with cell differentiation is still unknown.
To understand the dynamic reorganization of constitutive heterochromatin domains, we have analyzed myogenesis as a model system. Myogenesis is characterized by progressive ‘clustering’ of chromocenters that form a large heterochromatin compartment (19,20). Here, we describe the discovery of lncRNA ChRO1 that mediates constitutive heterochromatin reorganization during myogenesis. In myotubes (MT), ChRO1 brought DAXX/H3.3 to ATRX to form a stable ATRX/DAXX/H3.3 complex at chromocenters for satellite RNA elevation, which led to spatial fusion of chromocenters into large repressive compartments and cell differentiation. Our results unveil a novel lncRNA, necessary for large-scale nuclear organization and cell differentiation, through interaction with H3.3-specific histone chaperone complex.
MATERIALS AND METHODS
Cell culture and RNAi assays
The murine myoblast cell line C2C12 was obtained from the American Type Culture Collection (ATCC), and maintained at low confluency (<50%) in Dulbecco’s-modified Eagle’s medium (DMEM) containing 10% (v/v) fetal bovine serum (growth medium, GM) at 37°C with 5% CO2. For differentiation of myoblasts into MT, GM was replaced by DMEM containing 2% (v/v) horse serum (differentiation medium, DM) when myoblasts reached 80% confluency. HEK293T cells (from ATCC) were maintained in DMEM supplemented with 10% (v/v) fetal bovine serum. For the analysis of protein stability of DAXX, 20 μM of MG132 was treated for 24 h. Transfection assays for RNAi were carried out using Lipofectamine RNAimax (Invitrogen), according to the manufacturer’s instruction. C2C12 cells were transfected with siRNAs in myoblast states, and differentiated into MT for the indicated times. siRNAs against ChRO1 were designed using the The BLOCK-iT™ RNAi Designer, and synthesized by GenePharma. The sequences of siRNAs are listed in Supplementary Table S1.
Antisense oligonucleotides (ASOs) against satellite repeat RNA
Antisense oligonucleotides (ASOs) targeting major or minor satellite repeat RNAs were synthesized by the Cosmo Genetech (Korea), as described in the previous report (21). Sequences of ASOs are demonstrated in Supplementary Table S1. One day after plating, C2C12 were transfected with ASOs (0.1 μM) using Lipofectamine 2000 (Invitrogen) and then shifted to DM medium (0-day). Cells were further incubated for 3 days before analysis.
RNA-FISH
For RNA-FISH, C2C12 were grown in 4-well culture slides (Falcon), and in the case of MT, differentiated for 3–7 days. Cells were washed once in cold phosphate-buffered saline (PBS), and fixed with 4% (w/v) formaldehyde and 10% acetic acid in PBS at RT for 10 min. After washing with PBS twice, cells were permeabilized with 70% ethanol by incubation at 4°C for overnight. Cells were rehydrated in 2× SSC (300 mM NaCl, 30 mM sodium citrate, pH 7.0) containing 50% formamide at RT, for 5 min. After rehydration, cells were hybridized with 0.25 μM of CAL Fluor Red 610-labeled oligonucleotide probe set (Stellaris Biosearch technologies), specific for ChRO1, in 2× SSC, 10% dextran sulfate, 2 mM vanadyl-ribonucleoside complexes, 0.02% RNase-free bovine serum albumin, 40 μg of yeast tRNA and 50% formamide, for overnight at 37°C. After hybridization, slides were washed twice for 30 min at 37°C with 2× SSC and 50% formamide. Nuclei were stained with DAPI (20 μg/ml, Invitrogen).
Rapid Amplification of cDNA Ends (RACE)
3′ RACE and 5′ RACE for identification of the exact location of ChRO1a ends, were performed using 3′ or 5′ RACE System for Rapid Amplification of cDNA Ends kit (Invitrogen). Primers for RACE are listed in the Supplementary Table S1.
RNA extraction and RT-qPCR
RNA was purified by NucleoSpin® RNA (Macherey-Nagel), according to the manufacturer's instruction. Purified RNA (2 μg) was used for cDNA synthesis with cDNA synthesis kit (Thermo Fisher Scientific). RNA from mice tissues was extracted from various tissues of four, 8-week-old male C57BL6/J mice purchased from Daehan BioLink (DBL). Quantitative polymerase chain reaction (qPCR) was performed using the CFX96 system (Biorad) and KAPA SYBR FAST Master Mix (KAPA Biosystem). Relative quantification of PCR products was done by the 2ΔΔCT (Livak) method. RNA level was normalized by GAPDH unless otherwise mentioned in the figure legends. The primers used for qRT-PCR are listed in the Supplementary Table S1.
Plasmid construction and stable cell lines
For synthesis of ChRO1a-cloned plasmid, PCR was performed with cDNA synthesized from RNAs purified from MT using primers specific to ChRO1a as shown in Supplementary Table S2. The PCR product was incorporated into TA vector (RBC). After confirmation of sequences, BamHI/EcoRI digested TA-ChRO1a was cloned into pBabe-puro to establish the ChRO1a over-expressing stable C2C12 cell line. Preparation of ChRO1 shRNA plasmids was performed as described previously (15) using oligomers containing ChRO1-specific shRNA sequences as listed in Supplementary Table S1. To generate ChRO1 shRNA stable cell line, lentiviral particles containing ChRO1 shRNA was used to infect C2C12, followed by selection with puromycin (2 μg/ml, Biomax). H3.1 Flag-HA, H3.3 Flag-HA or ChRO1a over-expressing stable C2C12 cell lines were also established in the same way that ChRO1 shRNA cell line was produced.
Myogenic conversion of NIH3T3
NIH3T3, mouse fibroblast was converted to muscle lineage by expression of MyoD as previously described (22). In detail, NIH3T3 fibroblasts were treated with retrovirus containing pBabe puro MyoD. Thus infected NIH3T3 cells were selected with puromycin (2 μg/ml, Biomax) for 1 week. To induce muscle differentiation, media was changed from GM to DM [DMEM containing 2% (v/v) horse serum, 1× Insulin-Transferin-Selenium (ITS) supplement (Sigma-Aldrich) and 2 μg/ml puromycin (Biomax)] for 3 days.
Deletion of the ChRO1 promoter using CRISPR/Cas9 system
For ChRO1 knockout, a part of ChRO1 promoter was deleted using CRISPR/Cas9 system. The pSpCas9(BB)-2A-GFP(PX458) carrying wild-type Cas9 was from Addgene (#48138; http://www.addgene.org) (23). Two different sgRNA oligos were designated using the CRISPR Design Tool (http://crispr.mit.edu/). Cloning of sgRNAs into pSpCas9(BB)-2A-GFP was performed as described previously (23). After sgRNA cloning into Cas9 containing plasmid, C2C12 or embryonic stem cell line (E14) were transfected with 2 μg of each sgRNA plasmid using lipofectamine 2000 (Invitrogen), according to manufacturer’s instruction. GFP-positive cells were collected for sorting into a 96-well cell culture plate with a BD cell sorter Aria™ (BD Biosciences) followed by 24-h incubation. After 10 days, genomic DNA was extracted and amplified using a specific primer set to confirm genomic knockout of the specific region. Primers used for sgRNAs and confirmation of genomic knockout are shown in Supplementary Table S1.
Muscle regeneration after cardiotoxin injury
Female C57BL/6J mice were purchased from Orient Bio. To examine skeletal muscle regeneration, mice, blindly randomized into groups, were anesthetized with a 1 - 2% Isoflurane, followed by 50 μl of 10 μM cardiotoxin (CTX) (Sigma-Aldrich) injection into the tibialis anterior (TA) muscles. The TA muscles were harvested at 0, 2, 4, 7 and 14 days after injury. Total RNA prep, cDNA synthesis and qPCR were performed as mentioned above. Primers are listed in the Supplementary Table S1.
Self-renewal assay
Murine embryonic stem cells (E14) were trypsinized to single cell suspension and plated on 0.1% gelatin-coated 6-well culture plates. After 7 days, the culture medium was aspirated and cells washed with PBS. Cells were fixed with 4% paraformaldehyde at room temperature for 2 min, followed by rinsing with PBS twice. Cells were stained with Fast Red Violet LB Salt (Sigma-Aldrich) and Naphthol A S-BI-phosphate (Santa Cruz Biotechnology) at room temperature for 15 min.
Nuclear fractionation
Nuclear fractionation was performed as described previously (24) with little modification for cytosol isolation. In detail, cytosol was isolated by buffer A (100 mM HEPES-KOH, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05% NP40, pH 7.9) with a protease inhibitor cocktail and centrifuged at 3000 rpm, 4°C, for 10 min. Supernatant was kept as ‘cytoplasmic fraction’. The steps for nuclear fractionation into nucleoplasm, and chromatin fraction were previously described (24).
Western blot analysis
Cells were washed twice with cold PBS and lysed by lysis buffer (20 mM Tris–HCl pH 7.5, 1 mM ethylenediaminetetraacetic acid, 150 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors and phosphatase inhibitors. Lysates were sonicated (Bioruptor) and centrifuged (13 000 rpm, 4°C) for 10 min. Protein concentration was determined and equal amounts of protein were separated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane before conventional western blotting experiments using primary antibodies listed in Supplementary Table S2.
Immunofluorescence and fusion index
Cells were fixed with 4% (w/v) paraformaldehyde, and permeabilized with 0.5% Triton X-100 in PBS. Cells were blocked with 2% (w/v) bovine serum albumin in PBS, and incubated with the primary antibodies shown in Supplementary Table S2. Alexa 594-conjugated anti rabbit goat IgG and Alexa 488-conjugated anti mouse goat IgG (Invitrogen) were used as secondary antibodies. DNA in the nuclei was detected by DAPI staining (20 μg/ml, Molecular Probes). Immunofluorescence was examined using an LSM 700 Confocal microscopy (Carl Zeiss), and images were processed by ZEN software for calculation of the correlation coefficient R between DAPI and red signals. The number of chromocenters was manually counted by ImageJ with images that were taken by Z-stack. For analysis of percentage of myosin heavy chain (MyHC)-positive cells, the number of nuclei co-stained with MyHC was counted in at least 10 random microscopic fields. The percentage of MyHC-positive cells was calculated by dividing the number of MyHC positive nuclei by the total nuclei. Fusion index was calculated by the ratio of nuclei that appear as more than two in one cell to the total number of nuclei.
Chromatin immunoprecipitation (ChIP)
C2C12 cells were cross-linked with 1% formaldehyde at RT for 10 min. Crosslinking was quenched by incubation in 0.125 M glycine for 5 min at RT. After three washing steps on ice with PBS, cells were lysed and sonicated for 45 min at 0°C to produce DNA fragments ranging between 200 and 1000 bp (Bioruptor). After centrifugation, 2% of lysate was de-crosslinked for 4 h at 65°C followed by RNaseA treatment for 1 h at 50°C. Agarose gel electrophoresis to confirm the size range of DNA fragments and quantification was performed with the de-crosslinked DNA. Antibodies and beads were added to the 25 μg of chromatin samples and incubated overnight at 4°C with rotation. After washing serially with low salt, high salt, LiCl buffer and TE buffer, immunoprecipitated chromatin was de-crosslinked for 4 h at 65°C followed by RNaseA treatment, and then DNA was purified using PCR purification kit (QIAGEN). We performed ChIP-qPCR using primers targeting peri/centromere in MT (25). The relative amount of immunoprecipitated DNA was calculated as the percentage of input DNA. Primers used for ChIP are listed in the Supplementary Table S1.
Formaldehyde-Associated Isolation of Regulatory Elements (FAIRE) assay
Formaldehyde-Associated Isolation of Regulatory Elements (FAIRE) assay was performed as described previously (26), with modification. In detail, cells were cross-linked with 1% formaldehyde for 5 min at RT, and quenched by incubation with 0.125 M glycine for 5 min at RT with shaking. After three washes with PBS, cells were lysed and sonicated for 30 min at 0°C (Bioruptor). After centrifugation, 10% input from supernatant was taken and de-crosslinked with RNaseA and proteinase K. DNA from the remaining cell lysate was purified by phenol–chloroform–isoamylalcohol extraction before de-crosslinking. DNA was then de-crosslinked with RNase and proteinase K treatment at 65°C overnight. Final purification was done after overnight de-crosslink, and FAIRE enrichment was analyzed by qPCR using primers targeting major satellite and minor satellite repeats. See the Supplementary Table S1 for primers.
Detection of CpG methylation at pericentromere
CpG methylation of pericentromere in C2C12 was analyzed by digestion of genomic DNA with HpyCH4IV, followed by qPCR with primers targeting pericentromeric region as shown in Supplementary Table S1 (19). Genomic DNA from C2C12 was extracted by phenol, and purified by ethanol precipitation. Equal volume of genomic DNAs (5 μg) were digested with 10 units of HpyCH4IV for overnight, leaving 0.1 μg of genomic DNA without digestion, for input. Enzyme-digested genomic DNA was purified, and analyzed with two primer sets: targeting digestion-free region (without ACGT) and ACGT containing region. Relative HpyCH4IV was determined by comparison between % input of PCR products of digestion-free region and the ACGT containing region.
RNA pull-down, chromatin isolation by RNA purification (ChIRP) and RNA immunoprecipitation (RNA-IP)
RNA pull-down and chromatin isolation by RNA purification (ChIRP) using biotinylated antisense (AS) oligonucleotides against ChRO1 were performed as previously described (27,28) with modifications. 5′ biotinylated AS nucleotides (20-mers) targeting ChRO1 or LacZ for negative control were synthesized by Integrated DNA Technologies, of which sequences shown in the Supplementary Table S1. RNA purification after pull-down was performed by using NucleoSpin® RNA (Macherey-Nagel) according to the clean-up protocol of manufacturer’s instruction. The relative amount of enriched RNA was demonstrated as the percentage of input RNA. For the calculation of relative enrichment, normalization by the percentage input of AS-LacZ was carried out. For ChIRP, DNA purification was performed after pull-down using PCR purification kit (QIAGEN). Quantitative PCR using primers targeting pericentromere and centromere was carried out. ChRO1 enriched DNA was represented as the relative enrichment, with normalization by the % input of AS-LacZ. RNA-IP was performed using indicated antibodies as previously described with modifications (29). After immunoprecipitation, equal volumes of purified RNAs were used for cDNA synthesis by cDNA synthesis kit (Thermo), and qPCR was performed. The relative amount of immunoprecipitated RNA was represented as the percentage of input RNA, and then finally relative enrichment was obtained from division by Flag-, HA-, or IgG-immunoprecipitated ChRO1. Supplementary Table S1 lists the primers used for RNA-pull down, ChIRP, and RNA-IP.
Conserved domain search
To identify conserved regions, we aligned transcribed sequences of human ChRO1 against mouse ChRO1. We took the sliding-window approaches. For the first approach, we made a 200 nt-long window on each human RNA sequence and shifted the window by 40 nt to align against the whole length of the transcribed mouse RNA sequence. We used MATLAB function localalign, which returns local optimal and suboptimal alignments between two sequences. We found highly concordant search results and further analyzed the alignments by applying the following steps: (i) retain alignments only if the alignment score is >20 or the ratio of identical matches is >80%, (ii) remove duplicate alignments among isoforms of RNAs based on merged isoforms of RNA list, (iii) remove alignments if the aligned regions in human and mouse RNAs are not in same order of exons on their transcribed sequences and (iv) retain the best alignment if there are multiple alignments for the same region.
Motif search in conserved regions
To determine which regulatory motifs are over-represented in conserved regions with respect to background non-conserved regions, we identified all possible ungapped 8-mers in conserved domains and computed their frequency. In the list of motifs, we found the presence of repeats that are consisted of a single nucleotide or dimer repeated for the entire 8-mer. This phenomenon is common in genomic sequences and generally is associated with non-functional components, and thus, these were filtered out. To assess the statistical significance of the computed frequency for the over-represented motifs, we generated random sequences according to the nucleotide composition of the original sets of sequences. The frequencies for the random 8-mers were computed, and the distribution of the frequencies was approximated by the extreme value distribution. We used the MATLAB function gevfit to compute the maximum likelihood estimation of the extreme value distribution. We then overlaid a scaled version of its probability density function, computed using MATLAB function gevpdf, with the histogram of the frequency of the random 8-mer sequences. We repeated this process 100 times for bootstrapping and calculated the P-value. We concluded that the over-representation of the 8-mer motifs in conserved domain is statistically significant if the P-value estimate is <1.0E-4.
Consensus motifs and de novo motif discovery
To identify consensus motifs, 47 enriched 8-mers were phylogenetically clustered into 10 groups. We used the MATLAB function seqlinkage to construct phylogenetic tree from pairwise distances. We then used the MATLAB function seqlogo to identify consensus motifs and their weight matrix for the clustered 8-mer(s) in each group. We found known transcription factors per 10 identified consensus that have aligned part of sequence with the consensus by using the MEME suite.
Hi-C topological associated domains and loops
We used the JuiceBox (http://www.aidenlab.org/juicebox/) (30) to visualize Hi-C topological associated domains and loops. The Hi-C GM12878 cell data downloaded from GEO (GSE63525).
Statistical analysis
All experiments were performed at least three times independently under similar conditions, unless otherwise specified in the figure. Results were demonstrated as mean values with error bars representing standard errors of means. Statistical analysis was carried out using GraphPad Prism v7.0 (GraphPad Software) and Microsoft Excel 2007 (Microsoft) to assess the differences between experimental groups. Statistical significance between two groups was analyzed by unpaired, two-tailed Student’s t-test. One-way analysis of variance (ANOVA) was performed for comparison of multiple groups. F-values and Degree of Freedom were stated in the figure legend where ANOVA was used for determination of statistical significance. P < 0.05 were considered to be statistically significant. Determined P-values were indicated in the figures.
Ethical compliance
All animal experiments were approved by the Institutional Animal Care and Research Advisory Committee at Sungkyunkwan University School of Medicine Laboratory Animal Research Center.
Detailed methods about microarray analysis and RNA-seq analysis are provided in the supplementary file.
RESULTS
ChRO1 is a lncRNA expressed in terminally differentiated muscle cells
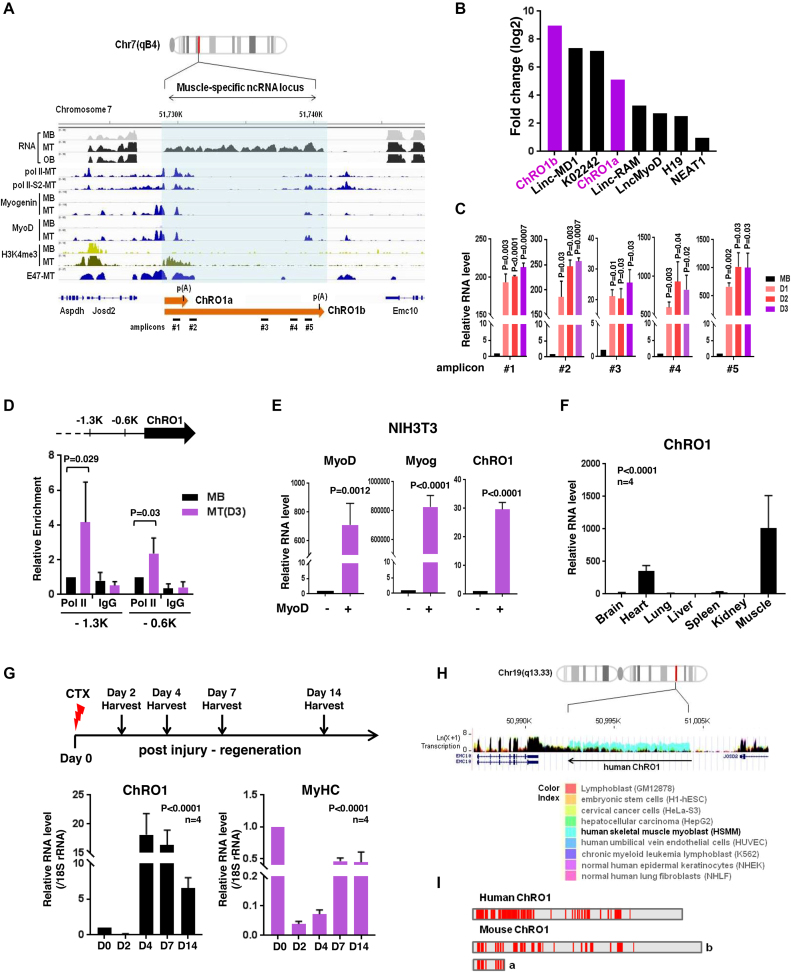
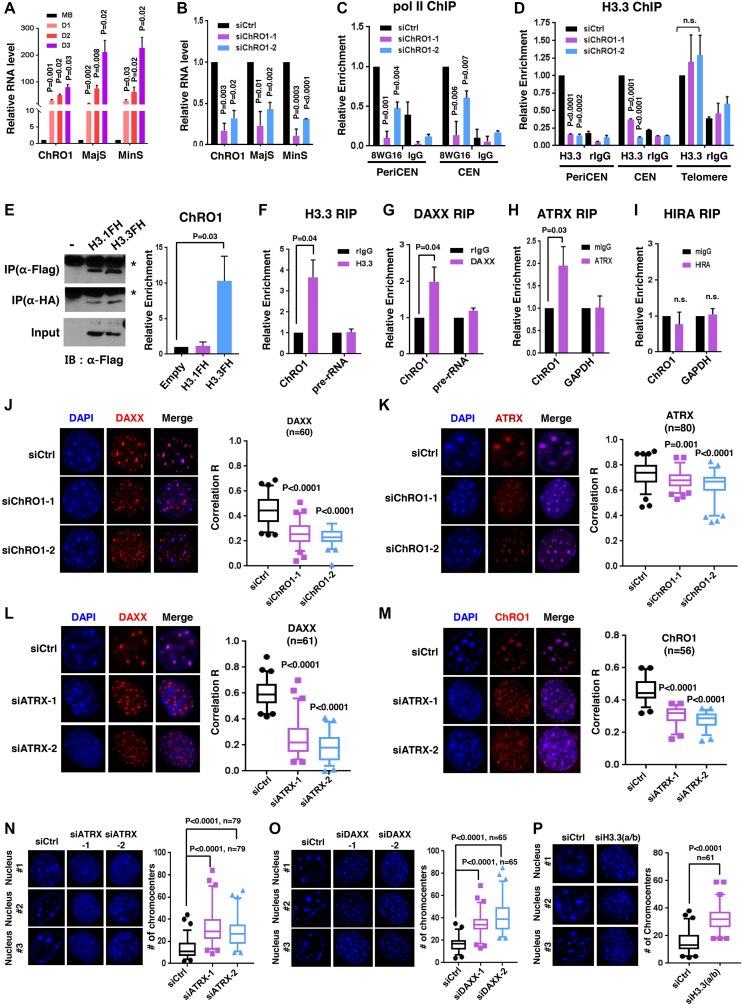
To search for novel lncRNAs that function as myogenic regulators, we performed RNA-seq with poly(A)-selected RNAs from proliferating C2C12 myoblasts and cells that had been differentiated into either MT or osteoblasts (Supplementary Figure S1A–E). Among differentially expressed genes, 194 ncRNAs, significantly upregulated during myogenesis, were further explored. We ranked the top 40 most highly induced lncRNAs, and individually analyzed them by filtering with features of H3K4-H3K36 methylation and a loss-of-function analysis using siRNAs (Supplementary Figure S1F and G). These efforts led us to discover a previously unannotated lncRNA, hereinafter termed ChRO1 (Chromatin reorganization 1). ChRO1 was located at chr 7, and highly expressed in MT, but not in myoblasts or osteoblasts. The promoter region of ChRO1 gene was enriched by pol II and myogenic regulatory factors, MyoD and Myogenin, in a cell-type specific manner (Figure 1A). ChRO1 was produced as two isoforms, ChRO1a and ChRO1b. We cloned ChRO1a using the rapid amplification of complementary DNA ends (RACE), which yielded an unspliced 1337 nt transcript with a genomically encoded poly (A) site. ChRO1b was estimated as an ∼11 000 nt transcript. Both ChRO1a and b were induced as robustly as other well-known myogenic lncRNAs during muscle differentiation (Figure 1B) (31–35). As expected, quantitative RT-qPCR using primer pairs that span the ChRO1 locus (Figure 1C) and the ChIP assay using pol II antibody (Figure 1D) showed that ChRO1 was transcribed by pol II and dramatically induced during myoblast differentiation.
Figure 1.
ChRO1 is a muscle-specific lncRNA induced during myogenesis (A) Schematic representation of the ChRO1 genomic locus. Occupancies of myogenic transcription factors are shown. MB, myoblasts; MT, myotubes; OB, osteoblasts; Pol II, RNA polymerase II; Pol II-S2, Serine 2 phosphorylated RNA polymerase II. (B) Fold changes of lncRNA levels during myogenesis were calculated from FPKM. (C) ChRO1 RNA was measured by RT-qPCR under growth (myoblasts, MB) or differentiation condition (Day 1, 2, 3) using amplicon primers as shown in (A). (D) Occupancy of pol II (8WG16) on the 0.6K and 1.3K upstream regions from the ChRO1 transcription start site in MB and MT, differentiated for 3 days (D3). (E) RT-qPCR for MyoD, Myogenin and ChRO1 in myogenesis-induced NIH3T3. (F) RT-qPCR for ChRO1 in various mouse tissues. RNA level was normalized by 18S rRNA. n = 4 biological replicates, one-way ANOVA, P < 0.0001, Degree of Freedom = 6, F-value = 15.83. (G) RT-qPCR of ChRO1 and MyHC during muscle regeneration after injection of CTX into TA muscle of mice. RNA level was normalized by 18S rRNA, n = 4. One-way ANOVA, P < 0.0001, Degree of Freedom = 4, F-value = 64.12 for ChRO1 and 107.7 for MyHC. (H) Expression of ChRO1 in human cells. GRCh27/hg19 was used as a human genome reference. (I) Conserved regions identified by sliding-window local alignment are shown at bottom. Identical sequence alignments (conserved domains) between human and mouse ChRO1 are represented in red.
To ascertain whether ChRO1 is a bona fide muscle-specific lncRNA, we showed that when NIH3T3 fibroblast cells were acutely converted into the muscle lineage by MyoD, ChRO1 was strongly induced (Figure 1E). Furthermore, ChRO1 was highly enriched in cardiac and skeletal muscle of adult mouse (Figure 1F), and its level was promptly induced after injury followed by the recovery profile of MyHC during muscle regeneration (Figure 1G). In added support, analysis of public data sets of RNA-seq from various cells indicated that ChRO1 expression was highly specific to muscle (Supplementary Figure S2A).
Intriguingly, comparative analysis identified a syntenic human ortholog located on chr 19, whose expression profile also showed preferential enrichment in muscle cells (Figure 1H). Notably, mouse and human ChRO1 exhibited a significant level of sequence conservation (local alignment score > 20 or ratio of direct alignment per nucleotide >80%) (Figure 1I). Further searching for repeat elements revealed a ‘CTCCTC’ motif (Monte Carlo P-value 6.37E-06), and related phylogenic variants that are often recognized by various zinc finger proteins (Supplementary Figure S2B). ChRO1 might be one of the tapRNAs that were recently characterized (36), as it is situated at the borders of topologically associating domains (TADs) (Supplementary Figure S2C). Collectively, our data show that ChRO1 is a muscle-specific lncRNA with highly conserved regions between human and mouse, whose expression is upregulated upon myoblast differentiation.
ChRO1 is localized at chromocenters, and associated with myotube-specific heterochromatin compartmentalization
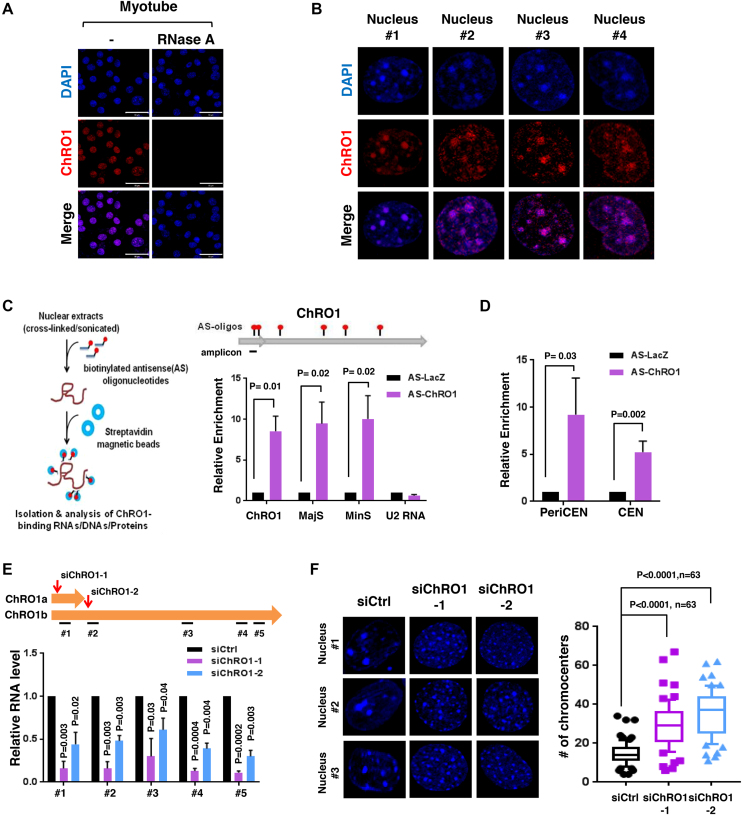
Biochemical fractionation of MT revealed that ChRO1 localized in the nucleus (Supplementary Figure S3A) and stably associated with chromatin (Supplementary Figure S3B). To further characterize, we performed RNA-FISH with fluorescence-labeled oligonucleotide probes recognizing ChRO1. This effort confirmed that ChRO1 was detected only in the nuclei of MT, while it disappeared following RNase treatment (Figure 2A). Furthermore, surprisingly, ChRO1 typically appeared as 5–20 large bright foci in the nucleus that co-localized with DAPI-dense foci (Figure 2B). As DAPI foci represent pericentromeric heterochromatin (PCH), the major constituents of constitutive heterochromatin compartments in mouse (37), our data indicated that ChRO1 preferentially localized at PCH domains in MT nuclei. To confirm the focal localization of ChRO1 at PCH, we performed an RNA pull-down assay using ChRO1-specific and LacZ-specific AS oligonucleotides as a control. RT-qPCR analysis showed that ChRO1 was specifically pulled down by AS-ChRO1 with co-precipitation of pericentromeric and centromeric repeat-derived RNAs, MajS and MinS (Figure 2C). This interaction was specific, as U2 snRNA was not coprecipitated. It also coprecipitated peri/centromeric repeat DNAs, demonstrating that ChRO1 was associated with the peri/centromeric chromocenters (Figure 2D).
Figure 2.
ChRO1 is enriched at chromocenters and associated with heterochromatin reorganization (A) RNA-FISH for ChRO1 in MT (Day 7). RNase A was either treated or not. Scale bars, 50 μm. (B) Representative MT nuclei of RNA-FISH analysis. (C) Experimental scheme of RNA pull-down for analysis of ChRO1-interacting DNA/RNA by using biotinylated ChRO1-AS oligonucleotides (AS-ChRO1) (left). RT-qPCR of ChRO1, MajS, MinS and U2 RNAs after RNA pull-down (right). AS-LacZ (AS oligonucleotides targeting LacZ) was used for a negative control. (D) DNA was analyzed by qPCR with primers for pericentromere (PeriCEN) and centromere (CEN) after ChIRP. (E) RT-qPCR of ChRO1 with amplicons to confirm ChRO1 knockdown using two specific siRNAs. (F) DAPI-staining of ChRO1-depleted MT (left). Number of chromocenters per MT nuclei is shown (right).
During myogenesis, chromocenters consist of PCH domains fused together to form chromocenter clusters, which are easily recognized by an increase in size and reduction of the number of DAPI foci (19,20). Because ChRO1 was localized at the chromocenters, we were particularly interested in exploring whether ChRO1 might be involved in the reorganization of PCH domains during myogenesis. To test this, we used two independent ChRO1-specific siRNAs: siChRO1-1, designed to target both isoforms, and siChRO1-2 specific for ChRO1b. C2C12 cells were treated with siRNAs and allowed to differentiate for 3 days. ChRO1 was efficiently downregulated by both siRNAs (Figure 2E). Strikingly, our data showed that DAPI foci appeared scattered and diffusely distributed upon ChRO1 knockdown with an increase of the number of DAPI foci, implying that chromocenter clustering was impaired with the ablation of ChRO1 (Figure 2F). Altogether, our data suggest that ChRO1 resides in MT chromocenters and might have a role in chromocenter clustering.
ChRO1 affects myogenesis
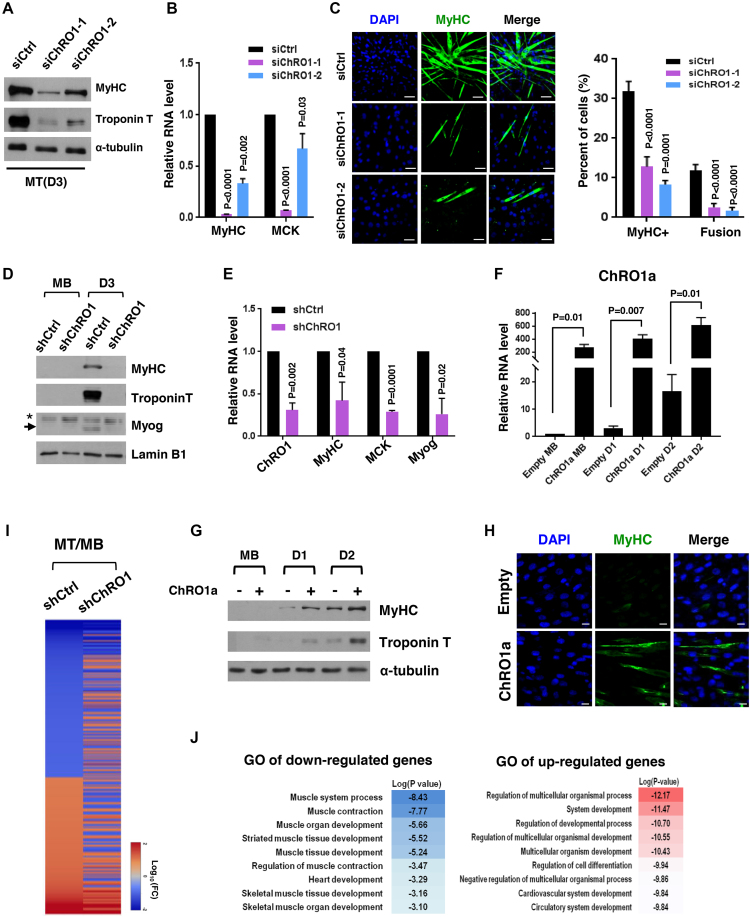
We next explored the role of ChRO1 in myogenic differentiation by modulation of its expression through RNAi or ChRO1a overexpression. C2C12 myoblasts were treated with ChRO1 siRNAs and shifted to differentiation medium. Differentiation status was monitored by the expression of muscle markers. When ChRO1 was depleted, both protein and mRNA levels of myogenic markers (MyHC, MCK and Troponin T) were significantly decreased in MT differentiated for 3 days, compared to siRNA control (Figure 3A and B). In addition, immunostaining data confirmed that MyHC+ population and the multi-nucleated cell fusion index were obviously reduced by ChRO1 knockdown (Figure 3C). We also established C2C12 clones stably expressing ChRO1-specific shRNA. Although these cells had no growth defect for myoblast proliferation (Supplementary Figure S4A), they showed significant impairment of chromocenter clustering (Supplementary Figure S4B) and marked reduction in MyHC, MCK, Myogenin and Troponin T levels, when shifted to differentiation medium (Figure 3D and E). In addition, the effect of ectopic expression of ChRO1 was determined by expression of ChRO1a that carries a part of the conserved sequences. RT-qPCR analysis demonstrated that ChRO1a was indeed efficiently overexpressed in C2C12 cells throughout differentiation (Figure 3F). Overexpression of ChRO1a led to high levels of muscle markers (Figure 3G) and increased population of MyHC+ cells (Figure 3H). Furthermore, it induced chromocenter clustering, even in growth state (Supplementary Figure S4C), and led to the accelerated expression of MyHC and Troponin T, when cells were grown to high cell density (Supplementary Figure S4D).
Figure 3.
Muscle differentiation is impaired by ChRO1 depletion. (A) Western blot for MyHC and Troponin T in 3-day differentiated MT after siRNA treatment. α-tubulin was used as a loading control. (B) RT-qPCR for MyHC and muscle creatin kinase (MCK) in siRNA-treated C2C12 that was differentiated for 3 days. (C) Representative immunofluorescence for MyHC and DAPI staining. Scale bars, 50 μm (left). MyHC-positive cells and fusion index in MT (right). (D) Western blots for MyHC, Troponin T and Myogenin in either shCtrl or shChRO1 MB and MT (Day 3). Lamin B1 was used as a loading control. *: non-specific band, arrow indicates Myogenin. (E) RT-qPCR for ChRO1, MyHC, MCK and Myogenin in shCtrl or shChRO1 MT (Day 3). (F) RT-qPCR of ChRO1a in empty or ChRO1a over-expressing C2C12 cells in growing (MB) or differentiation (Day 1, Day 2) medium. n = 4 independent experiments. (G) Western blots for MyHC and Troponin T with ChRO1a over-expressing MB and MT (Day 1, Day 2). α-tubulin was used as a loading control. (H) MyHC immunostaining of 1 day-differentiated C2C12 cells over-expressing ChRO1a. Scale bars, 50μm. (I) Differentially expressed genes of shCtrl or shChRO1 C2C12 (Fold Change > 2, P < 0.05). Data are shown as log 10 of fold change. (J) Gene ontology analysis of ChRO1-depleted cells, shown in log P-value.
Many lncRNAs function as cis-acting regulators of their neighboring genes (38). We therefore tested whether this might be true of ChRO1 as a mode of action. Analysis of genes located within 100 kb from the ChRO1 site showed that their expression was hardly affected by ChRO1 depletion (Supplementary Figure S5A), suggesting the possibility that ChRO1 might act in trans. We also established ChRO1 knockout by deleting the promoter proximal MyoD binding sites using the CRISPR/Cas9 system in a background of C2C12 or ES cells (Supplementary Figure S5B). These results consistently showed that myogenic differentiation was greatly affected by ChRO1 knockout (Supplementary Figure S5C and D), whereas trans-differentiation into osteoblasts (Supplementary Figure S5E) or the pluripotency and differentiation potential of ES cells (Supplementary Figure S5F–I) were completely normal. Our data suggest that ChRO1 is required for the terminal differentiation of myoblasts in a lineage-specific manner.
In an attempt to gain more insight on a global scale, we performed microarray analysis to compare control and shChRO1 cells at both myoblast and MT states. Many genes differentially regulated during myogenesis (MT/MB) were greatly affected by ChRO1 knockdown (Figure 3I). A total of 635 genes were identified as either up- or downregulated by ChRO1 knockdown. GO analysis further revealed that many affected genes included a large portion of genes involved in muscle development and its regulation (Figure 3J). In addition, the motif analysis to predict transcription factors associated with the affected genes (MSigDB v6.0), revealed important myogenic regulators such as MyoD, Mef2a and E12/E47 (Supplementary Figure S5J). Taken together, our results demonstrate that ChRO1 functions as a positive regulator of muscle differentiation, with a broad impact on myogenic gene expression.
ChRO1 is involved in heterochromatin compaction through relocalization of chromatin factors to chromocenters
As ChRO1 resides predominantly at PCH chromocenters in MT, we next asked whether ChRO1 could affect chromatin compaction. To do this, nucleosome-free MajS and MinS repeats were analyzed by FAIRE assay (26,39). These regions changed into more condensed chromatin state with differentiation (Supplementary Figure S6A). However, ChRO1 knockdown resulted in decondensation, while its over-expression caused further compaction of chromatin in myoblasts and D1 MT (Supplementary Figure S6B–D). Consistently, CpG methylation of pericentromere was affected by ChRO1 (Supplementary Figure S6E–G), demonstrating that ChRO1 was involved in DNA methylation and heterochromatin compaction at peri/centromeric domains during muscle differentiation.
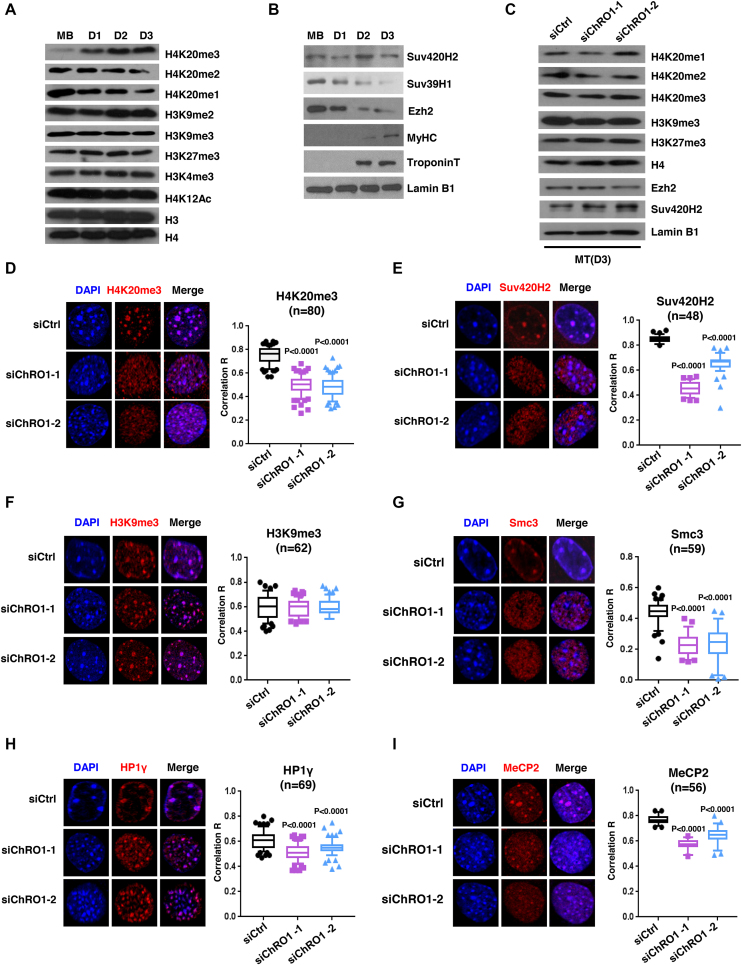
Based on the data above and its requirement for differentiation-associated chromocenter clustering, we hypothesized that ChRO1 might display a pro-myogenic function by facilitating constitutive heterochromatin reorganization. Accordingly, we first sought any chromatin signatures that parallel the myogenic progress, particularly focusing on the histone marks related to heterochromatin function. The global levels of histone methylation of H3K4, H3K9 or H3K27 were not significantly changed during myoblast differentiation, however H4K20me3 was prominently induced, while H4K20me1 and H4K20me2 were decreased (Figure 4A). Furthermore, in contrast with Suv39H1 or Ezh2 that declined with myogenesis, as reported previously (40,41), Suv420H2 was maintained throughout myogenic differentiation (Figure 4B), implying that differentiation-associated chromocenter clustering might be accompanied by gain of H4K20me3 mediated by Suv420H2. Next, we examined the effect of ChRO1 on the levels of histone methylation and HMTs. Surprisingly, neither histone methylation nor HMTs were significantly affected by ChRO1 knockdown (Figure 4C), which suggested that the pro-myogenic activity of ChRO1 might not be necessarily related to the control of the total amount of H4K20me3 or Suv420H2.
Figure 4.
ChRO1 is responsible for heterochromatin compaction via facilitating localization of chromatin factors to chromocenters. (A) Western blots for various histone modifications in MB and MT (Day 1–3). (B) Western blots for Suv420H2, Suv39H1, Ezh2, MyHC and Troponin T in MB and MT (Day 1–3). (C) Western blots for histone modifications and HMTs in ChRO1-depleted MT (Day 3). H4 and Lamin B1 are the loading controls. (D–I) Representative MT nuclei of immunostaining (left) and correlation R (right) are shown for H4K20me3, Suv420H2, Smc3, H3K9me3, HP1γ and MeCP2, respectively, in ChRO1-depleted or control MT.
Since the global level of histone modifications was not changed by ChRO1, we speculated that ChRO1 would be necessary for proper localization of histone marks or chromatin factors in the nuclear compartment. In order to examine this, we initially tested the relative distribution of repressive histone marks and heterochromatin factors upon differentiation, by immunofluorescence. The correlation coefficient R was applied to quantitatively determine the spatial colocalization of each target with DAPI foci. Consistent with previous reports (42), the focal enrichment of H3K9me3 and HP1γ to chromocenter clusters was modestly but significantly increased in MTs (Supplementary Figure S7A and B). Whereas, H4K20me3, Suv420H2 and Smc3 (cohesin subunit) were robustly enriched at chromocenters in MTs (Supplementary Figure S7C–E), which was also revealed by ChIP assay (Supplementary Figure S7F and G). Our data suggest that global relocalization of heterochromatin factors occurred with clustering of chromocenters in MTs.
These results prompted us to test whether ChRO1 might be required for the enrichment of H4K20me3 and other chromatin factors at chromocenters in MTs. Strikingly, C2C12 cells with ChRO1 knockdown exhibited complete mislocalization of H4K20me3 from chromocenters to nucleoplasm (Figure 4D), which was again confirmed by ChIP assay (Supplementary Figure S7H and I). Consistently, chromocenter localization of Suv420H2 was significantly affected by ChRO1 depletion (Figure 4E), although localization of H3K9me3 was largely unchanged (Figure 4F). In addition, ChRO1 knockdown resulted in nuclear diffusion of Smc3 from chromocenter foci without affecting protein levels (Figure 4G and Supplementary Figure S7J). Likewise, relocalization of HP1γ and MeCP2 to chromocenters was modestly but significantly impaired by ChRO1 knockdown (Figure 4H and I). Therefore, our data suggest that ChRO1 plays an important role in chromocenter loading of heterochromatin-associated factors, especially for the steps downstream of Suv39H/H3K9me3.
ChRO1 regulates satellite RNAs through ATRX/DAXX-dependent deposition of H3.3
In order to further investigate the mechanism we assessed the MajS and MinS repeat RNAs that were reported to be induced during myogenesis (43). Both RNA transcripts were indeed strongly induced in MTs (Figure 5A). However, they were significantly reduced by ChRO1 knockdown, while were increased by ChRO1a over-expression (Figure 5B and Supplementary Figure S8A), implying a potential link between ChRO1 and local ncRNAs.
Figure 5.
ChRO1 mediates satellite RNA accumulation through ATRX/DAXX/H3.3 localization at peri/centromeric chromocenters. (A) RT-qPCR for MajS and MinS RNA transcripts in MB and MT (Day 1–Day 3). (B) RT-qPCR for MajS and MinS in siRNA-treated C2C12. (C) ChIP-qPCR demonstrating pol II occupancy at peri/centromeres in ChRO1-depleted MTs (Day 3). (D) ChIP-qPCR for H3.3 occupancy at indicated genomic regions in ChRO1-depleted MTs (Day 3). (E) Western blots for IP of Flag/HA tagged H3s using antibodies against Flag or HA (left panel). Detection of ChRO1 by RT-qPCR following H3 IP (right panel). (F–I) RT-qPCR for ChRO1 after IP of endogenous H3.3, DAXX, ATRX or HIRA in MTs (Day 3). IP/Input of ChRO1 was normalized by the value of IgG. Pre-rRNA or GAPDH was used as a negative control. (J and K) Representative immunofluorescence and correlation R for DAXX and ATRX in ChRO1-depleted MTs (Day 3). (L) Representative immunofluorescence and correlation R for DAXX in ATRX-depleted MTs (Day 3). (M) RNA-FISH for ChRO1 in ATRX-depleted MTs (Day 3). Correlation R for ChRO1 in ATRX-depleted MTs is shown. (N–P) DAPI staining for ATRX-, DAXX- or H3.3-depleted MTs (Day 3). The number of chromocenters per nucleus is demonstrated.
Pol II and ATRX/DAXX-dependent deposition of H3.3 are known to be necessary for the initial synthesis of satellite RNAs (13,44). Therefore we examined by ChIP whether the occupancy of pol II and H3.3 at peri/centromeric regions was affected by ChRO1. Pol II and H3.3 normally increase at these regions with differentiation (Supplementary Figure S8B and C), but were significantly reduced by ChRO1 knockdown (Figure 5C and D), demonstrating that the satellite RNA transcription by pol II and H3.3 incorporation were dependent on ChRO1. Next, we examined whether ChRO1 could play a role in ATRX/DAXX/H3.3-mediated H3.3 deposition at chromocenters. To this end, we performed RIP using C2C12 cells stably expressing the epitope-tagged H3.1 or H3.3 (Flag/HA). Despite both H3.1 and H3.3 being immunoprecipitated with similar efficiency, in striking contrast, ChRO1 was co-precipitated with H3.3 (Figure 5E). This variant-specificity was further confirmed by RIP with endogenous proteins, showing that ChRO1 specifically interacted with ATRX/DAXX/H3.3, but not with HIRA/H3.3 (Figure 5F–I). We analyzed the interaction domain and found that the N-terminal domain of DAXX was involved in ChRO1 binding (Supplementary Figure S8D).
Salsman et al. reported that DAXX switches its position from the PML nuclear bodies to the PCH compartment during myogenesis (45). Consistently, we also observed the significant induction of chromocenter localization with DAXX during myogenesis (Supplementary Figure S8E), whereas ATRX was associated with chromocenters both in MB and MT without changing its position (Supplementary Figure S8F). These suggest that repositioning of DAXX to chromocenters is likely to be regulated by ChRO1. Indeed, recruitment of DAXX to chromocenters was significantly affected by ChRO1 knockdown (Figure 5J). The DAXX protein level, but not mRNA level, was also partially affected (Supplementary Figure S8G–I). However, interestingly, ATRX comparatively remained co-localized with DAPI foci with reduction of correlation R, upon ChRO1 knockdown, indicating that ATRX remained associated with PCH even when chromocenter clustering was impeded (Figure 5K). These data suggest that ATRX may be involved in ChRO1/DAXX/H3.3 complex loading as a binding platform. Indeed, as expected, siRNA knockdown of ATRX resulted in redistribution of DAXX from chromocenters and mislocalization of ChRO1 as a dispersed pattern (Figure 5L and M), implying that ATRX may function as a major determinant for placing ChRO1/DAXX/H3.3 at these particular genomic loci.
Next, we investigated whether focal enrichment of ChRO1/ATRX/DAXX and H3.3 incorporation directly affect the chromocenter reorganization and muscle differentiation. Indeed, siRNA knockdown of ATRX or DAXX resulted in defective chromocenter clustering and poor myogenic differentiation (Figure 5N and O; Supplementary Figure S9A–D). Furthermore, direct targeting of H3.3 by the mixture of siRNAs that deplete both H3f3a and H3f3b led to reduced chromocenter clustering and defective cell differentiation (Figure 5P; Supplementary Figure S9E and F). In addition, knockdown of ATRX, DAXX and H3.3 all resulted in reduced satellite RNA levels (Supplementary Figure S9G–I). Combined, these data support the idea that ChRO1 cooperates with ATRX to localize DAXX/H3.3 to chromocenters, which is important for H3.3 incorporation, satellite RNA transcription and higher-order heterochromatin reorganization.
Satellite repeat RNAs are involved in large-scale reorganization of constitutive heterochromatin during myogenesis
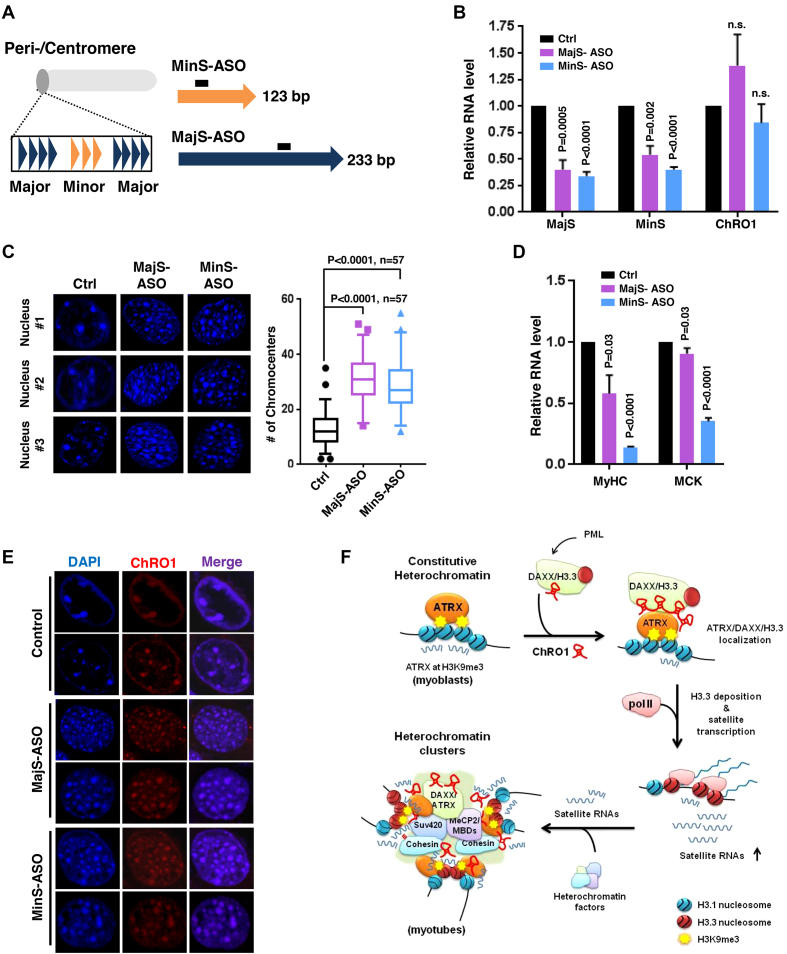
As satellite repeat RNAs were elevated via ChRO1-mediated enrichment of ATRX/DAXX/H3.3 at chromocenters, we tried to investigate whether these local ncRNAs perform a function for chromocenter clustering during myogenesis. To address this, we attempted to deplete MajS and MinS RNAs using antisense RNA/DNA oligonucleotides (ASO), which promote degradation of target RNAs via endogenous RNase H (Figure 6A). C2C12 cells were treated with MajS or MinS-ASOs when cells were induced for differentiation and incubated for 3 days before analysis. Both MajS and MinS were downregulated by ASO treatment with an efficiency of 40–50%, while the level of ChRO1 remained unaffected (Figure 6B). Interestingly, knockdown of MajS and MinS repeat RNAs led to marked increase of the number of chromocenters compared to mock control (Figure 6C). Concomitant with this, mRNA levels of MyHC and MCK were significantly reduced in ASO-treated MT (Figure 6D), indicating that muscle differentiation as well as chromocenter clustering was significantly affected upon satellite RNA knockdown. Next, we investigated whether MajS and MinS RNAs were required for ChRO1 localization at chromocenters. The RNA-FISH data for ChRO1 in MajS- or MinS-ASO-treated MTs show that ChRO1 remained stably associated with chromocenters, albeit fallen apart (Figure 6E), indicating that satellite RNAs were dispensable for ChRO1 localization. These results suggest that ChRO1/ATRX/DAXX/H3.3 recruitment proceeds satellite transcription, and these local RNAs are critical players in heterochromatin reorganization and muscle differentiation.
Figure 6.
Satellite repeat RNAs are involved in chromocenter reorganization during muscle differentiation. (A) Schematic of the structure of the peri/centromeric region that consists of MajS and MinS repeats. The location of antisense oligonucleotides (ASOs) for targeting satellite RNAs is demonstrated. (B) RT-qPCR for MajS, MinS and ChRO1 RNAs in ASO-treated MTs (Day 3). n.s.: not significant. (C) DAPI-staining of nuclei from MTs treated with MajS- or MinS-specific ASOs (left). The number of chromocenters is shown at right. (D) RT-qPCR for MyHC and MCK in MajS- or MinS-depleted MTs (Day 3). (E) RNA-FISH for ChRO1 in MajS- or MinS-ASO treated MTs (Day 3). (F) Schematic of ChRO1-mediated large-scale reorganization of constitutive heterochromatin during myogenesis.
DISCUSSION
In this study, we report the identification and comprehensive characterization of previously unknown lncRNA ChRO1, which is highly expressed in terminally differentiated muscle cells and specifically localized in the differentiation-associated heterochromatic nuclear compartments. Upon differentiation, ChRO1 recruits and stabilizes the ATRX/DAXX/H3.3 complex at chromocenters, which is responsible for H3.3 incorporation for pol II-mediated transcription of satellite RNAs and clustering of chromocenters (Figure 6F). This PCH/CH-based chromatin reorganization is necessary for appropriate execution of gene expression for progression into the terminal myogenic differentiation path. Our current model also points to satellite RNAs as essential components for chromocenter clustering and myogenesis. In comparison with other scaffold RNAs (46,47), ChRO1 is unique, as it does so, in part, through transcriptional induction of satellite repeats with the binding capacity toward H3.3-specific histone chaperone.
ATRX/DAXX is proposed to play a role in genomic stability through regulation of repetitive regions such as telomeres and pericentromeres (48–50), where the ADD (ATRX-DNMT3-DNMT3L) domain of ATRX is implicated in H3K9me3/H3K4me0 binding to recruit this complex to appropriately marked genomic loci (51,52). ATRX also interacts with HP1 via domain adjacent to ADD, these domains all provide ATRX the ability to instruct where H3.3 should be incorporated along the genome. Whereas, DAXX has SIM (SUMO-interacting motif) domains responsible for DAXX/H3.3 association with PML nuclear bodies via SUMO-mediated interaction (53). Although ATRX/DAXX-dependent H3.3 deposition pathway is suggested to operate from PML bodies to H3K9me3 marked genomic regions, the details of molecular interplay and dynamics are still not known (45,54). Nonetheless the fact that ATRX and DAXX/H3.3 emerge on chromocenter clusters during myogenesis implies that heterochromatin reorganization is a high priority of muscle cells for the cell differentiation. In this regard, ChRO1 serves as a regulatory RNA for the relocalization of DAXX/H3.3 and stable formation of ATRX/DAXX/H3.3 complex at chromocenters in myogenic cells. Our interaction domain analysis further revealed that the ChRO1–DAXX interaction involves N-terminal ∼180 residues of DAXX that include the SIM and the ATRX-interacting DHB (DAXX helical bundle) domains, which suggest that sumoylation could be an additional component of dynamic relocalization.
Satellite repeat RNAs function exclusively in cis for the establishment of heterochromatin structure (55,56). In fission yeast, these RNAs are amplified by Dicer and RNA-dependent RNA polymerase, guide Clr4/Suv39H and Swi6/HP1 to pericentromeres via RITS (RNA-induced transcriptional silencing) complex (57). Although the contribution of yeast-like RNAi system is less clear in higher eukaryotes, satellite RNAs are still critical players in PCH/CH formation, as they promote stable association of Suv39H1/2 and HP1 at these regions through RNA:DNA and RNA:protein interaction (12,58,59). In higher eukaryotes, variant histone H3.3 is essential for pol II-dependent satellite RNA transcription, and crucial for co-transcriptional silencing of these regions (13,54,60,61). From the observation that chromocenter clustering was accompanied by robust induction of satellite RNAs, these repeat RNAs are likely excellent players for large-scale chromatin compaction, by hosting a broader range of chromatin factors, in addition to SUV39H/HP1. A recent report on the engagement of satellite RNAs in the clustering of pericentromeres into chromocenters during development further supports our hypothesis (62). In this regard, it is noteworthy that ChRO1 contributes to the amplification of satellite RNAs at the transcriptional level, in part by complementing an RNAi-mediated heterochromatin silencing pathway (18). However, to understand the detailed mechanisms in chromocenter clustering, a full list of satellite RNA- and ChRO1-interacting factors must be provided.
Constitutive heterochromatin reorganization occurs during cellular differentiation process, and is expected to dramatically affect the nuclear environment of many developmentally associated genomic loci. However, it is still unanswered whether heterochromatin reorganization is a cause, or a consequence, of cell differentiation or gene regulation. Regarding this, our data demonstrate that PCH/CH reorganization caused by ChRO1/ATRX/DAXX/H3.3 is critical for muscle gene activation and myoblast differentiation, although the detailed mechanism of how heterochromatin reorganization affects muscle differentiation or gene regulation must be provided. Collectively, our study sheds light on the regulatory mechanism of the differentiation-associated large-scale heterochromatin formation through the identification of ChRO1.
DATA AVAILABILITY
The microarray and RNA-seq datasets have been deposited in Gene Expression Omnibus under accession number GSE104304 and GSE120044, respectively. We took advantage of the ChIP-seq or transcriptome analysis data from the ENCODE data coordination center (63–65).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Frank Furnari at University of California-San Diego for DAXX constructs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Individual Basic Researcher Program [2018R1D1A1B07048056 to E.-J.C., 2017R1D1A1B03035883 to J.P.]; Advanced Research Center Program [NRF-2010-0029359 to E.-J.C.]; National Creative Research Laboratory Program [2012R1A3A2048767 to H.-D.Y.]; NRF-2012-Fostering Core Leaders of the Future Basic Science Program through the National Research Foundation of Korea [2012H1A8003093 to J.P.]. Funding for open access charge: Advanced Research Center Program through NRF Korea [NRF-2010-0029359 to E.-J.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Politz J.C.R., Scalzo D., Groudine M.. Something silent this way forms: the functional organization of the repressive nuclear compartment. Annu. Rev. Cell Dev. Biol. 2013; 29:241–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L. et al. . LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013; 152:584–598. [DOI] [PubMed] [Google Scholar]
- 3. Fussner E., Djuric U., Strauss M., Hotta A., Perez-Iratxeta C., Lanner F., Dilworth F.J., Ellis J., Bazett-Jones D.P.. Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 2011; 30:1778–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guetg C., Lienemann P., Sirri V., Grummt I., Hernandez-Verdun D., Hottiger M.O., Fussenegger M., Santoro R.. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010; 29:2135–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Padeken J., Mendiburo M.J., Chlamydas S., Schwarz H.J., Kremmer E., Heun P.. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol. Cell. 2013; 50:236–249. [DOI] [PubMed] [Google Scholar]
- 6. Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T.. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001; 410:116–120. [DOI] [PubMed] [Google Scholar]
- 7. Lehnertz B., Ueda Y., Derijck A.A.H.A., Braunschweig U., Perez-Burgos L., Kubicek S., Chen T., Li E., Jenuwein T., Peters A.H.F.M.. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003; 13:1192–1200. [DOI] [PubMed] [Google Scholar]
- 8. Rothbart S.B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A.I., Barsyte-Lovejoy D., Martinez J.Y., Bedford M.T., Fuchs S.M. et al. . Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 2012; 19:1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T.. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004; 18:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blasco M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007; 8:299–309. [DOI] [PubMed] [Google Scholar]
- 11. Hahn M., Dambacher S., Dulev S., Kuznetsova A.Y., Eck S., Wörz S., Sadic D., Schulte M., Mallm J.P., Maiser A. et al. . Suv4-20h2 mediates chromatin compaction and is important for cohesion recruitment to heterochromatin. Genes Dev. 2013; 27:859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maison C., Bailly D., Roche D., Montes de Oca R., Probst A. V, Vassias I., Dingli F., Lombard B., Loew D., Quivy J.-P. et al. . SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat. Genet. 2011; 43:220–227. [DOI] [PubMed] [Google Scholar]
- 13. Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A.. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010; 24:1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lewis P.W., Elsaesser S.J., Noh K.-M., Stadler S.C., Allis C.D.. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:14075–14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang J.-H., Song Y., Seol J.-H., Park J.Y., Yang Y.-J., Han J.-W., Youn H.-D., Cho E.-J.. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harada A., Okada S., Konno D., Odawara J., Yoshimi T., Yoshimura S., Kumamaru H., Saiwai H., Tsubota T., Kurumizaka H. et al. . Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 2012; 31:2994–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song Y., Seol J.H., Yang J.H., Kim H.J., Han J.W., Youn H.D., Cho E.J.. Dissecting the roles of the histone chaperones reveals the evolutionary conserved mechanism of transcription-coupled deposition of H3.3. Nucleic Acids Res. 2013; 41:5199–5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santenard A., Ziegler-Birling C., Koch M., Tora L., Bannister A.J., Torres-Padilla M.-E.. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 2010; 12:853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brero A., Easwaran H.P., Nowak D., Grunewald I., Cremer T., Leonhardt H., Cardoso M.C.. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 2005; 169:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi M.H., Palanichamy Kala M., Ow J.R., Rao V.K., Suriyamurthy S., Taneja R.. GLP inhibits heterochromatin clustering and myogenic differentiation by repressing MeCP2. J. Mol. Cell Biol. 2018; 10:161–174. [DOI] [PubMed] [Google Scholar]
- 21. Ideue T., Cho Y., Nishimura K., Tani T.. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells. 2014; 19:528–538. [DOI] [PubMed] [Google Scholar]
- 22. Russo S., Tomatis D., Collo G., Tarone G., Tatò F.. Myogenic conversion of NIH3T3 cells by exogenous MyoD family members: dissociation of terminal differentiation from myotube formation. J. Cell Sci. 1998; 111:691–700. [DOI] [PubMed] [Google Scholar]
- 23. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Girard C., Mouaikel J., Neel H., Bertrand E., Bordonné R.. Nuclear localization properties of a conserved protuberance in the Sm core complex. Exp. Cell Res. 2004; 299:199–208. [DOI] [PubMed] [Google Scholar]
- 25. Iwata-Otsubo A., Dawicki-McKenna J.M., Akera T., Falk S.J., Chmátal L., Yang K., Sullivan B.A., Schultz R.M., Lampson M.A., Black B.E.. Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 2017; 27:2365–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simon J.M., Giresi P.G., Davis I.J., Lieb J.D.. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012; 7:256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L., McClanahan P., Hendrickson D.G., Sauvageau M., Kelley D.R. et al. . Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014; 21:198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chu C., Quinn J., Chang H.Y.. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 2012; 61:e3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knuckles P., Vogt M.A., Lugert S., Milo M., Chong M.M.W., Hautbergue G.M., Wilson S.A., Littman D.R., Taylor V.. Drosha regulates neurogenesis by controlling Neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012; 15:962–969. [DOI] [PubMed] [Google Scholar]
- 30. Durand N.C., Robinson J.T., Shamim M.S., Machol I., Mesirov J.P., Lander E.S., Aiden E.L.. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016; 3:99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sunwoo H., Dinger M.E., Wilusz J.E., Amaral P.P., Mattick J.S., Spector D.L.. Men ϵ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009; 19:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M., Tramontano A., Bozzoni I.. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011; 147:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kallen A.N., Zhou X.B., Xu J., Qiao C., Ma J., Yan L., Lu L., Liu C., Yi J.S., Zhang H. et al. . The imprinted H19 LncRNA Antagonizes let-7 MicroRNAs. Mol. Cell. 2013; 52:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gong C., Li Z., Ramanujan K., Clay I., Zhang Y., Lemire-Brachat S., Glass D.J.. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell. 2015; 34:181–191. [DOI] [PubMed] [Google Scholar]
- 35. Yu X., Zhang Y., Li T., Ma Z., Jia H., Chen Q., Zhao Y., Zhai L., Zhong R., Li C. et al. . Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017; 8:14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amaral P.P., Leonardi T., Han N., Viré E., Gascoigne D.K., Arias-Carrasco R., Büscher M., Pandolfini L., Zhang A., Pluchino S. et al. . Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol. 2018; 19:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guenatri M., Bailly D., Maison C., Almouzni G.. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 2004; 166:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guil S., Esteller M.. Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 2012; 19:1068–1075. [DOI] [PubMed] [Google Scholar]
- 39. Douet J., Corujo D., Malinverni R., Renauld J., Sansoni V., Posavec Marjanović M., Cantariño N., Valero V., Mongelard F., Bouvet P. et al. . MacroH2A histone variants maintain nuclear organization and heterochromatin architecture. J. Cell Sci. 2017; 130:1570–1582. [DOI] [PubMed] [Google Scholar]
- 40. Mal A.K. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006; 25:3323–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V.. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004; 18:2627–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sdek P., Oyama K., Angelis E., Chan S.S., Schenke-Layland K., MacLellan W.R.. Epigenetic regulation of myogenic gene expression by heterochromatin protein 1 alpha. PLoS One. 2013; 8:e58319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Terranova R., Sauer S., Merkenschlager M., Fisher A.G.. The reorganisation of constitutive heterochromatin in differentiating muscle requires HDAC activity. Exp. Cell Res. 2005; 310:344–356. [DOI] [PubMed] [Google Scholar]
- 44. Quénet D., Dalal Y.. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife. 2014; 3:e03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salsman J., Rapkin L.M., Margam N.N., Duncan R., Bazett-Jones D.P., Dellaire G.. Myogenic differentiation triggers PML nuclear body loss and DAXX relocalization to chromocentres. Cell Death Dis. 2017; 8:e2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Long Y., Wang X., Youmans D.T., Cech T.R.. How do lncRNAs regulate transcription. Sci. Adv. 2017; 3:eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rinn J.L., Chang H.Y.. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012; 81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wong L.H., McGhie J.D., Sim M., Anderson M.A., Ahn S., Hannan R.D., George A.J., Morgan K.A., Mann J.R., Choo K.H.A.. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010; 20:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Voon H.P.J., Wong L.H.. New players in heterochromatin silencing: Histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic Acids Res. 2015; 44:1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morozov V.M., Gavrilova E.V., Ogryzko V.V., Ishov A.M.. Dualistic function of Daxx at centromeric and pericentromeric heterochromatin in normal and stress conditions. Nucleus. 2012; 3:276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dhayalan A., Tamas R., Bock I., Tattermusch A., Dimitrova E., Kudithipudi S., Ragozin S., Jeltsch A.. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum. Mol. Genet. 2011; 20:2195–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eustermann S., Yang J.C., Law M.J., Amos R., Chapman L.M., Jelinska C., Garrick D., Clynes D., Gibbons R.J., Rhodes D. et al. . Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat. Struct. Mol. Biol. 2011; 18:777–782. [DOI] [PubMed] [Google Scholar]
- 53. Lin D.Y., Huang Y.S., Jeng J.C., Kuo H.Y., Chang C.C., Chao T.T., Ho C.C., Chen Y.C., Lin T.P., Fang H.I. et al. . Role of SUMO-Interacting Motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell. 2006; 24:341–354. [DOI] [PubMed] [Google Scholar]
- 54. Corpet A., Olbrich T., Gwerder M., Fink D., Stucki M.. Dynamics of histone H3.3 deposition in proliferating and senescent cells reveals a DAXX-dependent targeting to PML-NBs important for pericentromeric heterochromatin organization. Cell Cycle. 2014; 13:249–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maison C., Bailly D., Peters A.H.F.M., Quivy J.-P., Roche D., Taddei A., Lachner M., Jenuwein T., Almouzni G.. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 2002; 30:329–334. [DOI] [PubMed] [Google Scholar]
- 56. Probst A.V., Okamoto I., Casanova M., Marjou El, F. Le Baccon, Almouzni G.. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell. 2010; 19:625–638. [DOI] [PubMed] [Google Scholar]
- 57. Grewal S.I.S., Elgin S.C.R.. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007; 447:399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Velazquez Camacho O., Galan C., Swist-Rosowska K., Ching R., Gamalinda M., Karabiber F., De La Rosa-Velazquez I., Engist B., Koschorz B., Shukeir N. et al. . Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. Elife. 2017; 6:e25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Johnson W.L., Yewdell W.T., Bell J.C., McNulty S.M., Duda Z., O’Neill R.J., Sullivan B.A., Straight A.F.. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. Elife. 2017; 6:e25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rapkin L.M., Ahmed K., Dulev S., Li R., Kimura H., Ishov A.M., Bazett-Jones D.P.. The histone chaperone DAXX maintains the structural organization of heterochromatin domains. Epigenet. Chromatin. 2015; 8:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morozov V.M., Gavrilova E.V., Ogryzko V.V., Ishov A.M.. Dualistic function of Daxx at centromeric and pericentromeric heterochromatin in normal and stress conditions. Nucleus. 2012; 3:276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Casanova M., Pasternak M., ElMarjou F., LeBaccon P., Probst A.V., Almouzni G.. Heterochromatin reorganization during early mouse development requires a Single-Stranded noncoding transcript. Cell Rep. 2013; 4:1156–1167. [DOI] [PubMed] [Google Scholar]
- 63. Johnson D.S., Mortazavi A., Myers R.M., Wold B.. Genome-Wide mapping of in vivo protein-DNA interactions. Science. 2007; 316:1497–1502. [DOI] [PubMed] [Google Scholar]
- 64. Jiang L., Schlesinger F., Davis C.A., Zhang Y., Li R., Salit M., Gingeras T.R., Oliver B.. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011; 21:1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B.. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008; 5:621–628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray and RNA-seq datasets have been deposited in Gene Expression Omnibus under accession number GSE104304 and GSE120044, respectively. We took advantage of the ChIP-seq or transcriptome analysis data from the ENCODE data coordination center (63–65).