Abstract
The molecular mechanism in pancreatic β cells underlying hyperlipidemia and insulin insufficiency remains unclear. Here, we find that the fatty acid-induced decrease in insulin levels occurs due to a decrease in insulin translation. Since regulation at the translational level is generally mediated through RNA-binding proteins, using RNA antisense purification coupled with mass spectrometry, we identify a novel insulin mRNA-binding protein, namely, DDX1, that is sensitive to palmitate treatment. Notably, the knockdown or overexpression of DDX1 affects insulin translation, and the knockdown of DDX1 eliminates the palmitate-induced repression of insulin translation. Molecular mechanism studies show that palmitate treatment causes DDX1 phosphorylation at S295 and dissociates DDX1 from insulin mRNA, thereby leading to the suppression of insulin translation. In addition, DDX1 may interact with the translation initiation factors eIF3A and eIF4B to regulate translation. In high-fat diet mice, the inhibition of insulin translation happens at an early prediabetic stage before the elevation of glucose levels. We speculate that the DDX1-mediated repression of insulin translation worsens the situation of insulin resistance and contributes to the elevation of blood glucose levels in obese animals.
INTRODUCTION
Insulin, which is produced by pancreatic β cells, plays a pivotal role in the maintenance of blood glucose homeostasis. Insulin is synthesized as a precursor termed preproinsulin that is converted to proinsulin by cleavage of the signal peptide in the endoplasmic reticulum. Then, proinsulin is processed into mature insulin by protein convertases in secretory granules (1). Insulin synthesis and secretion are tightly regulated by circulating nutrients such as glucose, fatty acids and amino acids (1). The regulation of insulin by glucose has been extensively studied, and both transcriptional and posttranscriptional mechanisms have been revealed (2,3). At the transcriptional level, glucose can increase the expression of insulin transcription factors (PDX1 and MAFA) and induce the binding of these insulin transcription factors to the insulin promotor to upregulate insulin expression (4–6). At the posttranscriptional regulation level, several RNA-binding proteins, namely, HuD, PDI and PTBP1, have been identified that regulate insulin expression via binding to the untranslated region (UTR) of insulin mRNA (7–9). HuD binds to a 22-nucleotide segment of the Ins2 5′-UTR and is required for glucose-induced insulin translation (7). This segment mostly overlaps with a conserved element termed the preproinsulin glucose element (ppIGE) in the 5′-UTR of rat Ins2 mRNA, which is required for glucose-induced insulin translation (10). PDI (protein-disulfide isomerase) regulates glucose-stimulated insulin biosynthesis through its interaction with the 5′-UTR of insulin mRNA and with PABP, which is a poly (A)-binding protein (8). PTBP1 (polypyrimidine tract-binding protein 1) associates with the pyrimidine-rich sequence in the 3′-UTR of insulin mRNA and contributes to its high stability (9,11,12). Glucose stimulation induces the nucleocytoplasmic translocation of PTBP1, which then binds to insulin mRNA and upregulates its translation (13).
It has been well recognized that prolonged exposure to free fatty acids (FFAs) impairs insulin secretion in vitro (14,15) as well as in vivo in both animals (16) and humans (17). However, the mechanism underlying the regulation of insulin expression by FFAs remains unclear. In this study, we identified a novel insulin mRNA-binding protein, namely, DDX1, that is responsible for the palmitate (PA)-induced inhibition of insulin translation. DDX1 belongs to the DEAD box helicase superfamily, which consists of 37 members in humans (18). DEAD box helicases have central and, in many cases, essential physiological roles in cellular RNA metabolism, including transcription, translation, ribosome biogenesis, RNA splicing, RNA export and RNA decay. For example, DDX2A (eIF4A1), DDX2B (eIF4A2), DDX3, DDX4 and DDX6 are required for translation (19–23); and DDX48 (eIF4A3) is an essential part of the exon junction complex (EJC), which affects RNA metabolism (24). DDX1 has been proposed to play multiple functions in RNA metabolism (25), tRNA splicing (26), microRNA maturation (27), the repair of transcriptionally active regions of the genome (28) and dsRNA sensing (29).
Here, we reported a novel function of DDX1 in the regulation of translation and demonstrated that DDX1 can directly bind to insulin mRNA. Upon FFA stimulation, DDX1 is phosphorylated and dissociates from insulin mRNA, thereby decreasing insulin translation.
MATERIALS AND METHODS
Cell culture and treatment
INS-1 cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere with 5% CO2 at 37°C. Palmitate was dissolved at 100 mM in 0.1 M sodium hydroxide to make a stock solution. The palmitate stock solution was diluted in culture medium to which fatty acid-free BSA had been added, in a 1:19 molar ratio of palmitate to fatty acid-free BSA, to prepare BSA-conjugated palmitate. Cells were incubated with the BSA-conjugated palmitate at 0.5 mM for 24 hr.
Plasmids
The shDDX1 plasmid and a control shRNA plasmid (scrambled) with an RFP (Red Fluorescence Protein) marker were synthesized by GenePharma (Shanghai, China).
The HA-DDX1 (WT/S295D/S295A) plasmids were constructed as follows: HA-DDX1 (WT) was amplified from the cDNA of INS-1 cells using a forward primer bearing a HA tag and a reverse primer. To generate the HA-DDX1 (S295D or S295A) mutants, HA-WT-DDX1 was used as a template for site-directed mutagenesis by overlapping PCR. Briefly, a pair of reverse complementary primers that result in the mutation of serine (S) to aspartic acid (D) or alanine (A) at position 295 were designed, and two truncated DDX1 mutants were amplified from the HA-DDX1 (WT) fragment using two pairs of primers (pair 1: HA-DDX1-up and either DDX1-S295D-down or DDX1-S295A-down; pair 2: either DDX1-S295D-up or DDX1-S295A-up and HA-DDX1-down). Then, by using two truncated DDX1 mutant fragments as templates, the complete DDX1 (S295D or S295A) mutant sequence was amplified by the HA-DDX1-up primer and the HA-DDX1-down primer. Finally, the HA-DDX1 (WT/S295D/S295A) PCR fragments were cloned into the pQCXIP vector.
The EGFP-P2A-DDX1 (WT/S295D) plasmids were constructed as follows: The EGFP-P2A-DDX1 (WT) and EGFP-P2A-DDX1 (S295D) fragments were generated by overlapping PCR as described above with two pairs of primers (pair 1: EGFP-P2A-DDX1-up and DDX1-down; pair 2: DDX1-up and EGFP-P2A-DDX1-down). Then, these two fragments were cloned into the pCDH-CMV-MCS-EF1-Puro (CD510B-1) vector. A list of the primers used for these analyses is shown in Supplemental Table S1.
Construction of the stable DDX1 knockdown and overexpression cell lines and the DDX1 (WT, S295D phospho-mutant and S295A nonphospho-mutant)-rescued INS-1 cell lines
The DDX1 knockdown INS-1 cell line was constructed with shDDX1 plasmids. Briefly, the shRNA plasmid targeting DDX1 and a control shRNA plasmid (scrambled) were used to transfect INS-1 cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and positive cells were selected with G418 for 2 weeks. RFP-positive cells were sorted with flow cytometry to generate stable cell lines.
The DDX1-overexpressing INS-1 cell line was constructed with the retroviral pQCXIP plasmid. Briefly, retrovirus was packaged by transfecting PlatE cells with the HA-DDX1 plasmids and two packaging plasmids (vsvg and phit). At 48 h posttransfection, the virus-containing supernatant was collected, filtered and used to infect INS-1 cells. The infected cells were selected and maintained with puromycin to generate stable cell lines.
The DDX1 (WT, S295D phospho-mutant and S295A nonphospho-mutant)-rescued INS-1 cell lines were constructed with the lentiviral pCDH-CMV-MCS-EF1-Puro plasmid and the retroviral pQCXIP plasmid. Briefly, lentivirus was packaged by transfecting 293T cells with the EGFP-P2A-DDX1 (WT/S295D) plasmids and two packaging plasmids (vsvg and Δ3.1). At 48 h posttransfection, the virus-containing supernatant was collected, filtered and used to infect shDDX1 cells. The infected cells were selected with puromycin for 1 week. EGFP-positive cells were sorted with flow cytometry to generate stable cell lines.
For the retroviral pQCXIP plasmid-transfected cell lines, retrovirus was packaged by transfecting PlatE cells with the HA-DDX1 (WT or S295D or S295A) plasmids and two packaging plasmids (vsvg and phit). At 48 h posttransfection, the virus-containing supernatant was collected, filtered and used to infect shDDX1 cells. The infected cells were selected and maintained with puromycin to generate stable cell lines.
Real-time quantitative PCR
Total RNA was extracted directly from cells or from RIP and ChIRP samples using TRIzol (Invitrogen, Carlsbad, CA, USA) as per the manufacturer's instructions. After reverse transcription (RT) using random hexamer and oligo-dT primer mixes and SuperRT reverse transcriptase (CWBio, Beijing, China) as per the manufacturer's instructions. Quantitative PCR was carried out with a 1:20 dilution of cDNA and 2× SYBR Green PCR Mix in combination with 10 mM specific primers (listed in Supplementary Table S2). The Gapdh primer was used for normalization, and ΔCt was calculated to derive the relative expression levels.
Western blotting
Whole-cell lysates prepared using RIPA buffer containing a proteinase inhibitor (Sigma-Aldrich, St. Louis, MO, USA) were separated by SDS-PAGE and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were incubated with primary antibodies against DDX1 (Proteintech, Wuhan, China), proinsulin (CCI-3, HyTest, Turku, Finland), β-actin (Sigma-Aldrich, St. Louis, MO, USA), PTBP1 (Proteintech, Wuhan, China), eIF4B (Proteintech, Wuhan, China), eIF3A (Proteintech, Wuhan, China) and HA (Thermo Fisher Scientific, Waltham, MA, USA), followed by the appropriate HRP-conjugated secondary antibodies (Sungene Biotech, Tianjin, China), and protein expression was detected with enhanced luminescence reagents (GE Healthcare, Piscataway, NJ, USA).
RNA antisense purification with mass spectrometry (RAP-MS)
The isolation of RNA-binding proteins by insulin antisense probes was carried out as described previously (30). Briefly, cells were crosslinked with 3% formaldehyde and lysed in a buffer containing 50 mM Tris–Cl (pH 7.0), 10 mM EDTA, 1% SDS, 1% proteinase inhibitor and 0.1% RNase inhibitor for 30 min on ice. The cell lysate was sonicated for 10 min on ice and was then centrifuged at 10 000 × g for 15 min at 4°C. The supernatants were incubated in a 2× volume of hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris–Cl (pH 7.0), 1 mM EDTA, 15% formamide, 1% proteinase inhibitor and 0.1% RNase inhibitor) containing either 100 pmol/ml biotinylated insulin antisense probes or LacZ antisense probes overnight at 37°C. Antisense oligo probes were designed using the online probe designer at singlemoleculefish.com (sequences are listed in Supplementary Table S3). The sample mix was incubated with C-1 streptavidin magnetic beads (100 μl/100 pmol probe) for 30 min at room temperature and were then washed 5 times with a wash buffer (2× NaCl and sodium citrate, 0.5% SDS, 1% proteinase inhibitor and 0.1% RNase inhibitor). For RT-qPCR, the precipitates were incubated with 20 U of RNase-free DNase I (15 min at 37°C) to remove DNA and were digested further with 0.5 mg/ml Proteinase K at 55°C for 15 min to remove proteins. The RNA was further analyzed by RT-qPCR. For MS and western blot analyses, the precipitates were incubated with SDS buffer containing 0.5% β-ME at 95°C for 30 min. Proteins were separated by SDS-PAGE and subjected to silver staining. The protein bands specifically pulled down by the insulin antisense probes were analyzed by LC–MS/MS.
IP of RNP complexes
IP of endogenous RNP complexes from whole-cell extracts was carried out as described previously (7). Briefly, cells were lysed in a buffer containing 20 mM Tris–HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.5% NP-40 and 1% proteinase inhibitor for 30 min on ice and centrifuged at 10 000 × g for 15 min at 4°C. The supernatants were incubated with protein A/G magnetic beads coated with an anti-DDX1 antibody or control IgG (Beyotime, Shanghai, China) overnight at 4°C. Then, the beads were washed three times with NT2 buffer (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40). For RT-qPCR, the precipitates were incubated with 20 U of RNase-free DNase I (15 min at 37°C) to remove DNA and were incubated further with 0.1% SDS/0.5 mg/ml Proteinase K at 55°C for 15 min to remove proteins. The RNA isolated by IP was further analyzed by RT-qPCR. For MS and western blot analyses, the DDX1 complexes were treated with or without RNase A for 30 min at 37°C and were then washed 3 times with NT2 buffer. The precipitates were incubated in SDS buffer containing 0.5% β-ME at 95°C for 30 min. The DDX1-specific bands pulled down by IP were identified by SDS-PAGE followed by silver staining and were analyzed by MS.
Immunofluorescence detection of DDX1 distribution
To detect the subcellular distribution of DDX1, INS-1 cells treated with or without PA were fixed, permeabilized, blocked, and incubated with anti-DDX1 antibodies (Abcam, Cambridge, UK, 1:100 dilution) and then with secondary antibodies (Invitrogen, Carlsbad, CA, USA, 1:200 dilution) and DAPI (Invitrogen, Carlsbad, CA, USA) for nuclear staining. Fluorescence images were acquired using a confocal laser scanning microscope (FV1200, Olympus, Tokyo, Japan).
Pancreata from mice fed a regular diet and mice fed a high-fat diet were fixed with 4% PFA at 4°C for 24 h. The fixed tissues were soaked sequentially in solutions of 70%, 85%, 95% and 100% alcohol and then in dimethylbenzene; the tissues were then embedded in paraffin. Five micrometer sections were prepared with a microtome. The sections were pretreated using a heat-mediated antigen retrieval buffer with sodium citrate (pH 6, epitope retrieval solution) for 20 min and were then blocked with 5% goat serum. The sections were incubated with primary antibodies at 4°C overnight. The primary antibodies used in this study were guinea pig anti-insulin (prepared by our lab, 1:200 dilution) and rabbit anti-DDX1 (Abcam, Cambridge, UK, 1:100 dilution). Incubation with primary antibodies was followed by incubation with secondary antibodies (Invitrogen, Carlsbad, CA, USA, 1:200 dilution) and DAPI (Invitrogen, Carlsbad, CA, USA) for nuclear staining. Fluorescence images were acquired using a confocal laser scanning microscope (FV1200, Olympus, Tokyo, Japan).
Nascent translation assay
INS-1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 50 μM β-mercaptoethanol, 1 mM sodium pyruvate, and penicillin/streptomycin. After starvation in isoleucine-free medium for 1 h, cells were labeled with pulse medium containing [13C, 15N]-isoleucine (Sigma-Aldrich, St. Louis, MO, USA) and 16.7 mM glucose for 20 min at 37°C. Cells were harvested, washed, sonicated, and subjected to acetonitrile precipitation for the analysis of insulin species by MS. Samples were analyzed on an LC-ESI-Q Exactive MS in MS scan mode to detect ions in a range of 950–1400 m/z; the mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in 90% acetonitrile (B). The peak of proinsulin 1 with a charge of 7+ was extracted using Xcalibur 2.2 software (Thermo Fisher Scientific, Waltham, MA, USA), and the peak areas of proinsulin 1 were calculated for quantitative analysis, while the protein content in the cells was used for normalization.
ELISA detection of insulin
For detection of insulin content, cells transfected with scrambled shRNA and shDDX1 were lysed with RIPA buffer supplemented with proteinase inhibitor and were then centrifuged at 10 000 × g for 15 min at 4°C. The supernatant was assayed with an insulin ELISA kit (Shibayagi, Shibukawa, Japan).
Fractionation of polyribosomes
INS-1 cells were preincubated with 100 μg/ml cycloheximide for 30 min and lysed with a polysome extraction buffer containing 10 mM HEPES, 300 mM KCl, 5 mM MgCl2, 0.5% NP-40 and 100 μg/ml cycloheximide. Cytoplasmic lysates were fractionated by ultracentrifugation (36 000 rpm for 200 min) through 10–50% linear sucrose gradients and were divided into 12 fractions; RNA was extracted from each fraction and used for RT-qPCR analysis.
Stability of mRNA
To measure the effect of DDX1 silencing on regulating the stability of insulin mRNA, the scramble- and shDDX1-transfected cells were treated with the transcription inhibitor actinomycin D at 10 μg/ml for 0, 2, 4, 6 and 8 h. RNA was extracted and used for RT-qPCR analysis.
Stability of protein
To measure the effect of PA treatment or DDX1 silencing on regulating the stability of the proinsulin protein, the scramble- and shDDX1-transfected cells were treated with 100 μg/ml cycloheximide for 0, 1, 2, 4, 6 and 8 h. Total protein was extracted from whole-cell lysates with RIPA buffer containing proteinase inhibitor and was used for western blot analysis.
Gene Ontology (GO) enrichment analysis
GO and pathway analyses were performed using the WebGestalt online database (31). A total of 16 011 genes that are expressed in INS-1 cells were obtained from the RNA-seq data and used as the reference set (32), and the Benjamini–Hochberg multiple comparisons test was used to adjust for multiple comparisons. Functional category terms with a false discovery rate (FDR) of <0.05 were considered significant hits.
Animals
Mice were housed in groups of three to five at 22–24°C with a 12 h light/12 h dark cycle. Animals had access to water ad libitum. All experiments were approved by the Animal Care Committee at the Institute of Biophysics (License No. SYXK2016-19).
For the high-fat diet (HFD) experiments, weight-matched male C57BL/6N mice were used. At 4 weeks of age, the animals were fed either a regular chow diet (D12450B, Research Diets, USA) or a HFD (D12492, Research Diets, USA) in which 21% of the calories came from carbohydrates; 19%, protein and 60%, fat. Random blood glucose was assessed by test strips (Roche Diagnostics, Mannheim, Germany).
Islet isolation
Pancreatic islets were isolated by collagenase digestion as described previously (33). Briefly, the pancreas was inflated by instilling 5 ml of Hanks’ buffered saline solution (HBSS) containing 0.5 mg/ml collagenase P (Roche Applied Science, Mannheim, Germany) through the pancreatic duct. The pancreas was harvested and incubated in a water bath at 37°C for 25 min. The digested pancreas was rinsed with HBSS, and islets were separated on a Ficoll density gradient. After three washes with HBSS, the islets were manually isolated under a dissection microscope.
Statistics
Data are expressed as the means ± SEMs, and the statistical significance of differences was assessed using a two-tailed t-test when the data met the assumptions of the t-test. P values <0.05 were considered significant.
RESULTS
Insulin translation is inhibited in vitro and in vivo after FFA treatment
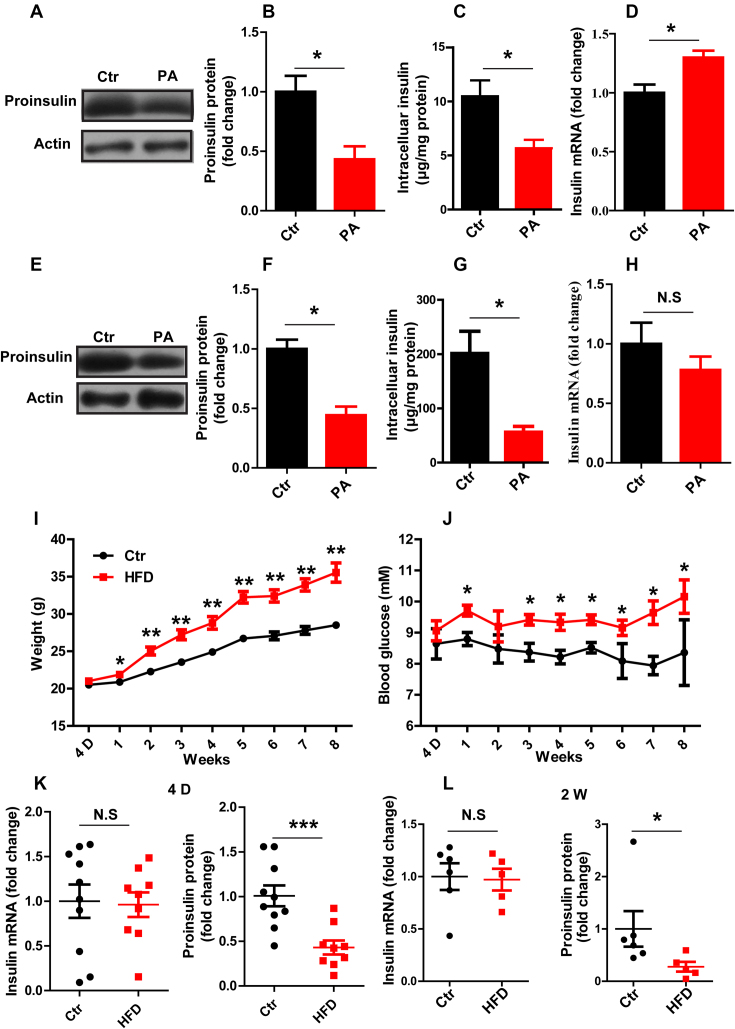
Whereas previous studies mainly focused on the effect of glucolipotoxicity on insulin regulation in β cells (34–38), we were interested in a scenario of high FFA and normal glucose levels, resembling the in vivo conditions in high-fat diet (HFD) animals or in obese humans with euglycemia before the onset of diabetes. To this end, INS-1 cells were treated with 0.5 mM saturated FFA, PA, for 24 h. PA treatment induced a significant reduction in both the proinsulin (Figure 1A and B) and intracellular insulin levels (Figure 1C) without changing the proinsulin protein stability (Supplementary Figure S1), consistent with previous results showing that PA inhibited insulin secretion and reduced the intracellular insulin content (14,15,39). In addition, we found that the level of insulin mRNA was not decreased but instead was mildly increased by PA treatment in INS-1 cells (Figure 1D), suggesting that the decrease in insulin biosynthesis is likely caused by a mechanism at the translational level. We further tested another unsaturated FFA, oleate (OA). Interestingly, whereas OA alone did not inhibit proinsulin synthesis, combination treatment with OA and PA, which resembles the more mixed FFA composition of the diet, caused much greater inhibition of insulin biosynthesis than did PA alone (Supplementary Figure S2). Next, we proceeded to verify this phenomenon in isolated mouse islets. When islets were treated with PA for 24 h, again we observed a significant reduction in insulin at the protein level but not at the mRNA level (Figure 1E–H). Together, these data confirmed that prolonged exposure to PA can inhibit insulin biosynthesis in both INS-1 β cells as well as mouse islets and suggested a possible mechanism at the translational level.
Figure 1.
Insulin translation is inhibited in vitro and in vivo after FFA treatment. (A, E) Western blotting was used to determine proinsulin expression. (B, F) The fold change in proinsulin expression was determined by normalization to actin expression. (C, G) ELISA was used to detect the intracellular insulin content. (D, H) RT-qPCR was used to determine the insulin mRNA level in Control (Ctr) and PA-treated (PA) INS-1 cells (A–D) and in mouse islets (E–H) in vitro (n ≥ 3). The weight (I) and random blood glucose levels (J) in control (Ctr) and high-fat diet (HFD) mice were monitored for 8 weeks (n = 15/15). RT-qPCR was used to determine the insulin mRNA level, and western blotting was used to determine proinsulin protein expression at different time points, namely, 4 days (K) and 2 weeks (L). The results are shown as the means ± SEMs. Significance was determined by a two-tailed unpaired t-test; *P < 0.05; **P < 0.01; N.S, no significance.
We went on to verify whether the PA-induced repression of insulin biosynthesis could happen in vivo in obese mice fed a HFD starting at 4 weeks of age. After mice were fed the HFD for one week, their weight and blood glucose level increased significantly compared to the weight and blood glucose level of those fed a chow diet (Figure 1I and J). We then assessed insulin expression at both the mRNA and protein levels after HFD feeding for 4 days and again at 2 weeks. Interestingly, at 4 days, when there was no significant increase in blood glucose, the level of insulin mRNA was comparable in the HFD and chow diet groups, while the proinsulin protein level was markedly decreased (Figure 1K). This decrease in insulin at the protein level, but not the mRNA level, continued to 2 weeks of HFD feeding (Figure 1L). Together, these results suggest that FFA treatment mediates inhibition of insulin expression at protein levels both in vitro and in vivo.
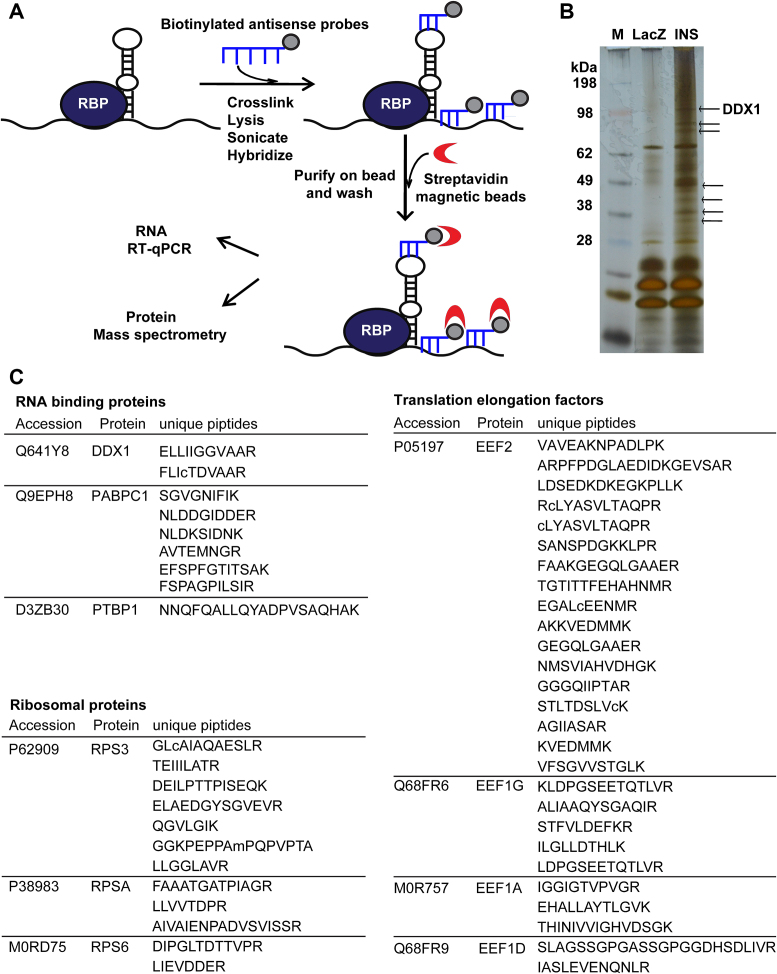
RNA pull-down assay identifies a novel insulin mRNA-binding protein DDX1, that dissociates from insulin mRNA after PA treatment
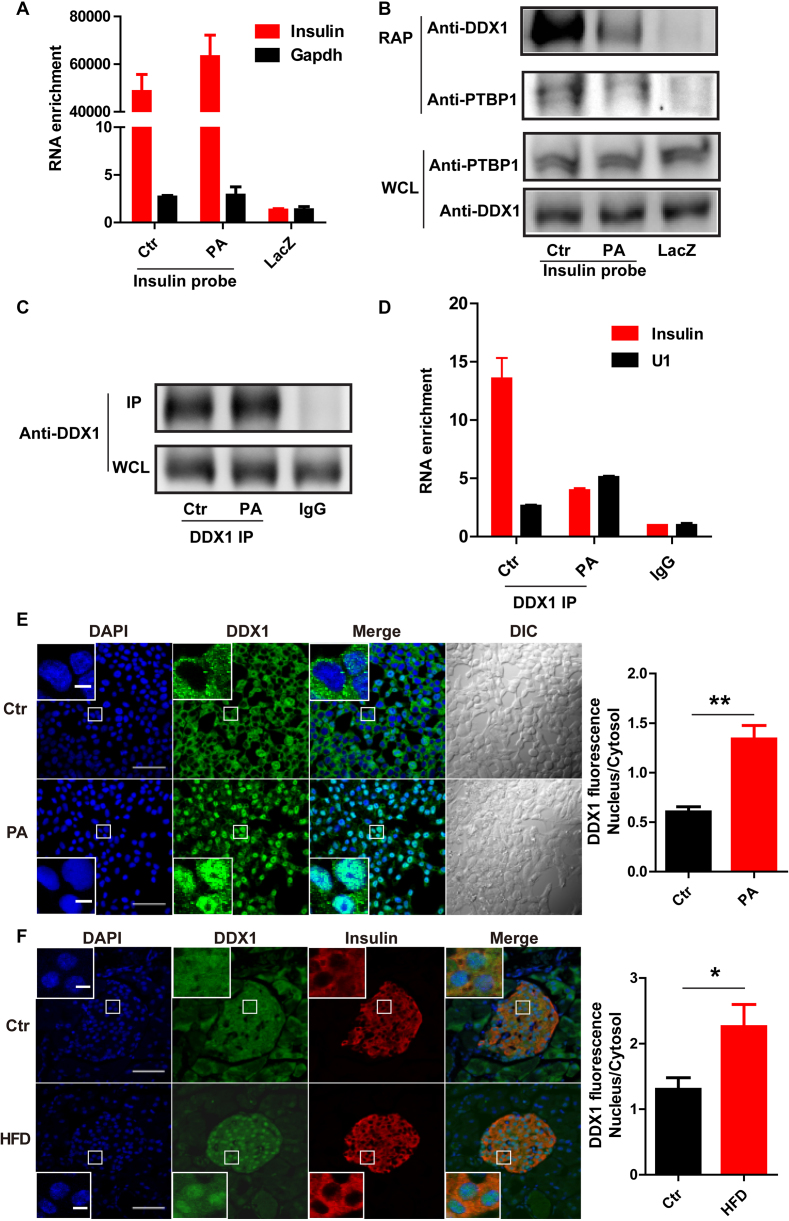
Since regulation at the protein but not mRNA level is generally mediated through RNA-binding proteins, we sought to identify the insulin mRNA-binding proteins that regulate insulin translation in response to PA treatment by RNA antisense purification coupled with mass spectrometry (RAP-MS) in INS-1 cells (30). After crosslinking by formaldehyde, the endogenous insulin mRNA was enriched by biotinylated insulin antisense probes. The mRNA-associated proteins were separated by polyacrylamide gel electrophoresis (PAGE), and the enriched bands were analyzed using liquid-chromatography tandem mass spectrometry (LC–MS/MS) (Figure 2A). As shown in Figure 3A, insulin mRNA was enriched over 40 000 times by insulin antisense probes compared to the enrichment by LacZ antisense probes, and several insulin mRNA-enriched protein bands were visible after silver staining (Figure 2B). Among these bands, seven (indicated by the arrows in Figure 2B) were selected for further MS identification. Most of the proteins identified by LC–MS/MS were ribosomal proteins and translation elongation factors, for example, RPS3 and EEF2; two known insulin mRNA-binding proteins, namely, PTBP1 and PABPC1, were also found (Figure 2C) (8,9), thus verifying the specificity of our pull-down assay. Interestingly, we identified a novel insulin mRNA-binding protein that belongs to the DEAD box helicase superfamily, namely, DDX1. Subsequent western blot analysis confirmed that the DDX1 and PTBP1 proteins were pulled down by the insulin antisense probes but not by the control LacZ probes. PA treatment significantly reduced DDX1 protein expression when even higher levels of insulin mRNA were pulled down (Figure 3A and B). To exclude the possibility that some of the interacting proteins are the result of protein-RNA reassociations occurring in the lysate, we performed the RAP experiment including noncross-linked cells as a better control. The result (Supplementary Figure S3) showed that only insulin antisense probes could pull down DDX1 in cross-linked cells. These results indicate that DDX1 is a novel insulin mRNA-binding protein and is sensitive to PA treatment. Conversely, to further confirm the interaction between insulin mRNA and DDX1, we performed UV crosslinking RNA immunoprecipitation (RIP) with an anti-DDX1 antibody. Insulin mRNA was significantly enriched ∼12-fold by anti-DDX1 RIP compared to the enrichment attained by IgG RIP, while much less insulin mRNA was bound to DDX1 upon PA treatment even with the same amount of DDX1 protein pulled down by the DDX1 antibody (Figure 3C and D). As a control, the nonspecific pull-down of the housekeeping snRNA U1 was not reduced by PA treatment. Together, these findings strongly indicate that DDX1 directly binds to insulin mRNA and that PA treatment can induce the dissociation of DDX1 from insulin mRNA.
Figure 2.
RAP-MS identifies the insulin mRNA-binding proteins. (A) A schematic represented the multiple steps of RAP-MS. (B) Insulin mRNA-protein complexes were identified by incubating biotinylated insulin mRNA antisense probes with protein extracts from INS-1 cells. The arrows indicate seven bands that were pulled down by the biotinylated insulin mRNA antisense probes; these bands were then identified by MS. M denotes marker proteins to the left. (C) A list of insulin mRNA-associated proteins identified by RAP-MS analysis in INS-1 cells.
Figure 3.
PA treatment induces the dissociation of DDX1 from insulin mRNA. (A) RT-qPCR was used to determine the enrichment of insulin and Gapdh mRNA levels by binding to insulin antisense probes with and without PA treatment and by binding to LacZ antisense probes as a control. (B) Western blotting was used to determine the levels of DDX1 and PTBP1 pulled down by insulin antisense probes with and without PA treatment and by LacZ antisense probes as a control. (C) Immunoprecipitation (IP) of DDX1 by an anti-DDX1 antibody with and without PA treatment in INS-1 cells. (D) RT-qPCR was used to determine the levels of insulin and Gapdh mRNA co-IPed with a DDX1 antibody in the presence and absence of PA treatment. IgG and U1 snRNA were used as controls. The results are shown as the means ± SEMs (n = 3). (E) Fluorescence staining reflects the nuclear translocation of DDX1 in INS-1 cells with PA treatment. INS-1 cells under normal conditions or PA stimulation were subjected to immunofluorescence with a specific antibody against DDX1 (green). Nuclei were stained with DAPI (blue). Original picture scale bar, 50 μm; enlarged picture scale bar, 5 μm. The average fluorescence intensity of the nucleus was divided by that of the cytosol; the ratio was markedly increased by PA treatment (right panel). The results are shown as the means ± SEMs (n = 3). Significance was determined by a two-tailed unpaired t-test; **P < 0.01. (F) Fluorescence staining reflects the nuclear translocation of DDX1 in pancreatic islets of HFD mice. C57BL/6N mice were fed a regular chow diet (Ctr) or a high-fat diet (HFD) for 1 week (n/n = 4/4). The pancreas was isolated and embedded in paraffin. The paraffin sections were dewaxed and subjected to antigen retrieval followed by immunostaining with specific antibodies against DDX1 (green) and insulin (red). Nuclei was stained with DAPI (blue). Original picture scale bar, 50 μm; enlarged picture scale bar, 5 μm. The results are shown as the means ± SEMs (n = 4 animals per treatment). Significance was determined by a two-tailed unpaired t-test; *P < 0.05.
The subcellular localization of DDX1 was further studied by immunofluorescence. Under normal conditions, DDX1 mainly localized to the cytoplasm. Interestingly, after PA stimulation, a significant amount of DDX1 translocated into the nucleus (Figure 3E). Next, we proceeded to verify this phenomenon in islets of HFD mice in vivo. After the mice were fed a HFD for 1 week, we again observed significant translocation of DDX1 into the nucleus (Figure 3F).
Knockdown or overexpression of DDX1 affects insulin translation
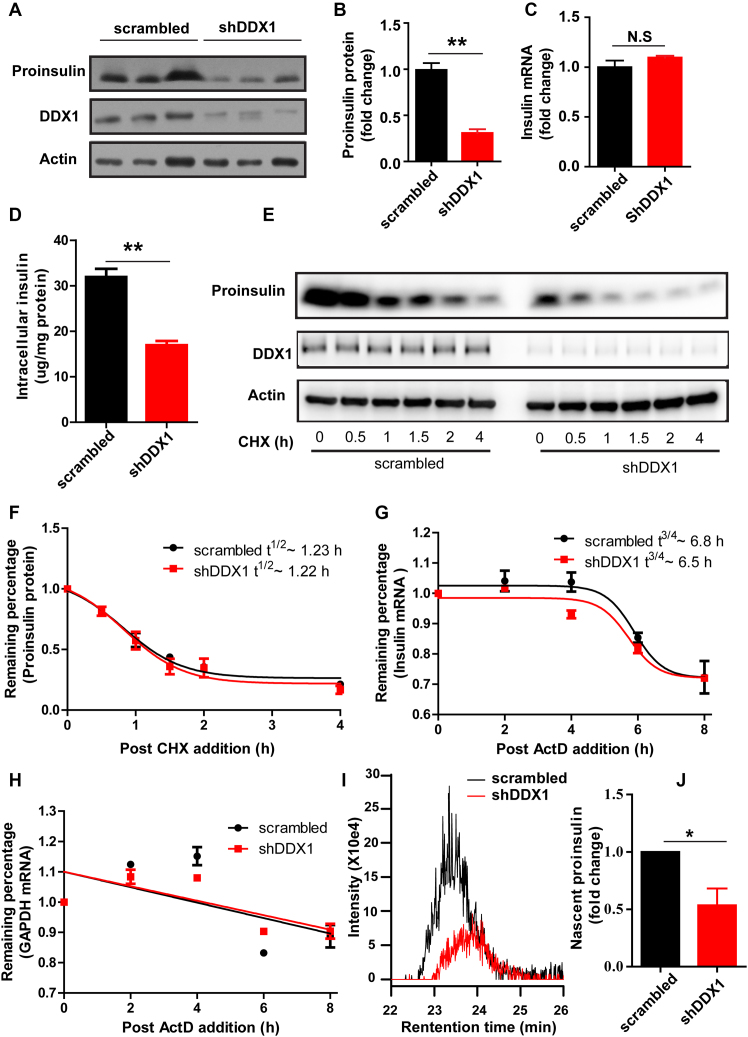
To determine whether DDX1 is required for the translation of the insulin protein, we constructed a knockdown INS-1 cell line (shDDX1) by stably expressing short hairpin RNA (shRNA) targeting DDX1. We verified that the DDX1 level was significantly lower in the shDDX1 cells than in the cells expressing scrambled shRNA (scrambled) (Figure 4A). The knockdown of DDX1 caused a significant reduction in the proinsulin protein level (Figure 4A and B) but not in the insulin mRNA level (Figure 4C). The reduction of intracellular insulin content in the shDDX1 cells was further confirmed by ELISA assay (Figure 4D). To assess whether the reduced levels of proinsulin are due to accelerated mRNA or protein degradation, we monitored the degradation of proinsulin mRNA and protein. The results showed that the knockdown of DDX1 did not change the degradation rate of the proinsulin protein or insulin mRNA (Figure 4E–H), indicating that the knockdown of DDX1 decreases proinsulin level by decreasing the proinsulin translation. To gain more evidence of whether DDX1 is required for the translation of insulin, we monitored the nascent proinsulin synthesis in INS-1 cells. The scrambled and shDDX1 cells were starved for 1 h before being labeled with [13C, 15N]-isoleucine for 20 min in 16.7 mM glucose. Then, the level of [13C, 15N]-labeled proinsulin was measured by LC–MS. We verified that the [13C, 15N]-labeled proinsulin level was reduced by 50% in the shDDX1 cells compared with that in the scrambled cells (Figure 4I and J). On the other hand, the overexpression of DDX1 in INS-1 cells increased the proinsulin level significantly (Supplementary Figure S4A and B), while the insulin mRNA level remained unchanged (Supplementary Figure S4C). Taken together, these results suggest that DDX1 is required for the regulation of insulin translation.
Figure 4.
Knockdown of DDX1 affects insulin translation. (A–D) Western blotting was used to determine proinsulin expression in INS-1 cells stably expressing shRNA against DDX1 (shDDX1) and in INS-1 cells stably expressing scrambled shRNA (scrambled) as a control (A). The fold change in proinsulin expression was calculated by normalization to actin expression (B). RT-qPCR was used to determine the insulin mRNA levels (C). ELISA was used to detect the intracellular insulin content (D). (E, F) To assess whether the knockdown of DDX1 affected the stability of proinsulin, scrambled and shDDX1 cells were treated with cycloheximide to block nascent protein synthesis, and the stability of the proinsulin protein was assessed by western blotting (E). The fold change in proinsulin expression was calculated by normalization to actin expression (F). (G, H) To assess whether the knockdown of DDX1 affected the stability of insulin mRNA, scrambled and shDDX1 cells were treated with the transcription inhibitor actinomycin D, and RT-qPCR was used to determine the remaining insulin (G) and Gapdh (H) mRNA levels. (I, J) LC/MS was used to detect nascent proinsulin biosynthesis in scrambled and shDDX1 stable cell lines (I). The nascent proinsulin level was normalized to the total protein level (J). The results are shown as the means ± SEMs (n = 3). Significance was determined by a two-tailed unpaired t-test; *P < 0.05; **P < 0.01; N.S, no significance.
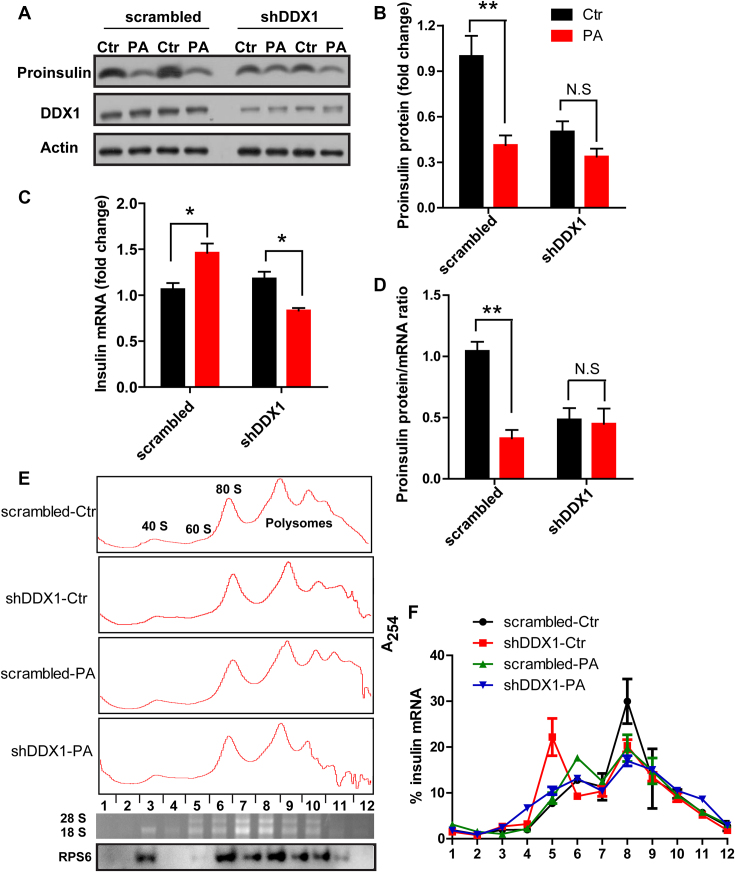
Knockdown of DDX1 eliminates the PA-induced repression of insulin translation
To determine whether DDX1 plays a role in the PA-induced repression of insulin translation, we tested whether PA could decrease the insulin level in the absence of DDX1. Indeed, PA induced only a slight decrease in the proinsulin level in shDDX1 cells, in contrast to the marked decrease in the proinsulin protein level that it induced in the scrambled cells (Figure 5A and B). As the insulin mRNA level was increased in the scrambled cells but was slightly reduced in the shDDX1 cells by PA treatment (Figure 5C), we further analyzed the ratio of proinsulin protein/mRNA. We found that PA treatment induced highly significant inhibition of the proinsulin protein/mRNA ratio in control cells, but not changed in the shDDX1 cells (Figure 5D).
Figure 5.
Knockdown of DDX1 eliminates the PA-induced repression of insulin translation. (A–D) Western blotting was used to determine proinsulin expression (A). Proinsulin expression was normalized to actin expression (B). RT-qPCR was used to determine the insulin mRNA level (C). The ratio of proinsulin protein/mRNA (D) in stable scrambled and shDDX1 cell lines treated with and without PA (n = 4). (E, F) Polysome analysis of insulin mRNA. (Up) lysates prepared from scrambled and shDDX1 cells treated with or without PA were fractionated through sucrose gradients to generate polysome profiles, (Bottom) the 28S and 18S rRNA distribution in the polysome profile was confirmed by the agarose gel, and the 40S ribosomal protein S6 distribution in the polysome profile was confirmed by western blotting (E). The relative distribution of insulin mRNA on polysome gradients was analyzed by RT-qPCR, and the insulin mRNA level is presented as the percentage of total mRNA (F) (n = 3). The results are shown as the means ± SEMs. Significance was determined by a two-tailed unpaired t-test; *P < 0.05; **P < 0.01; N.S, no significance.
More direct evidence that DDX1 mediated the PA-induced repression of insulin translation was obtained by polysome analysis. Cytoplasmic extracts from scrambled and shDDX1 INS-1 cells treated with or without PA were fractionated through sucrose gradients, with the lightest components sedimenting at the top in fractions 1–2, the small (40S) ribosomal subunit in fraction 3, large (60S) ribosomal subunit in fractions 4–5 and monosomes (80S) in fractions 6–7. Larger polysomes, ranging from the low- to high-molecular-weight polysomes (LMWPs and HMWPs), progressively sediment in fractions 8–12 (Figure 5E). In all the cells, the polysome-associated insulin mRNA peak was in fraction 8. When DDX1 was silenced or when cells were stimulated with PA, the polysome-associated insulin mRNA peak was decreased, and the ribosomal subunit and monosome peak was increased. In shDDX1 cells treated with PA, the polysome-associated insulin mRNA peak was slightly decreased compared with that in shDDX1 cells under normal conditions (Figure 5F). Together, these data indicate that silencing DDX1 or stimulating cells with PA can repress insulin translation and that silencing DDX1 eliminates the PA-induced repression of insulin translation.
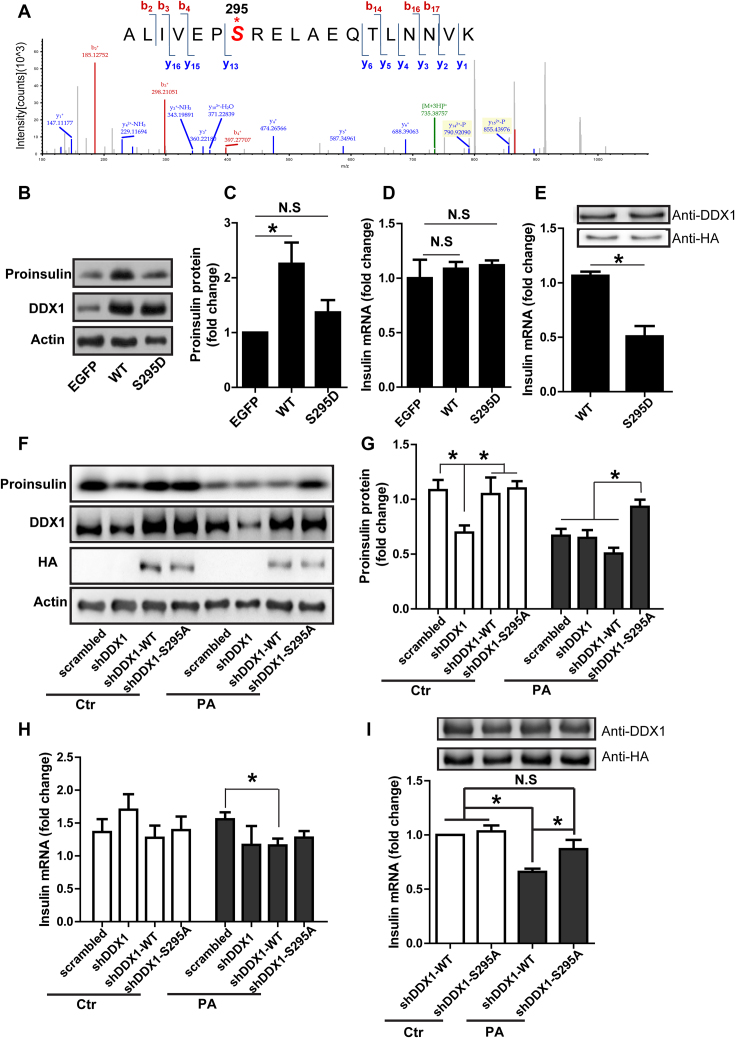
PA induces the phosphorylation of DDX1 at amino acid S295 that is required for the repression of insulin translation
It has been reported that the posttranslational modification of RNA-binding proteins would affect their binding capability (27). To verify whether posttranslational modification of DDX1 occurred upon PA stimulation, we enriched DDX1 and identified its modification by mass spectrometry. Under normal condition and under stimulation with PA, the phosphorylation of DDX1 at S481 was detected (data not shown). This site was reported by other phosphoproteomic studies (40,41). However, we identified a novel phosphorylation site of DDX1 at S295; phosphorylation at this site was present only after PA stimulation (Figure 6A). This result verifies that PA stimulation does cause the phosphorylation of DDX1.
Figure 6.
The PA-induced phosphorylation of DDX1 at S295 is responsible for insulin translation repression. (A) The mass spectra showing DDX1 phosphorylation at S295 after PA stimulation. (B–D) Western blotting was used to determine proinsulin expression in shDDX1 cells overexpressing WT or S295D-mutant DDX1 (B). Proinsulin expression was normalized to actin expression (C). RT-qPCR was used to determine the insulin mRNA level in shDDX1 cells overexpressing WT or S295D-mutant DDX1 (D) (n = 7). (E) RIP-PCR was used to determine the amount of insulin mRNA pulled down by HA-DDX1 (WT) or HA-DDX1 (S295D) (n = 4). (F-H) Western blotting was used to determine proinsulin expression in shDDX1 cells overexpressing WT or S295A-mutant DDX1 with or without PA treatment (F). Proinsulin expression was normalized to actin expression (G). RT-qPCR was used to determine the insulin mRNA level in shDDX1 cells overexpressing WT or S295A-mutant DDX1 with or without PA treatment (H) (n = 3). (I) RIP-PCR was used to determine the amount of insulin mRNA pulled down by HA-DDX1 (WT) or HA-DDX1 (S295A) with or without PA treatment (n = 3). The results are shown as the means ± SEMs. Significance was determined by a two-tailed unpaired t-test; *P < 0.05; N.S., no significance.
To illustrate the effect of DDX1 phosphorylation on insulin translation, we examined whether the overexpression of wild-type (WT) and mutant proteins mimicking phosphorylated DDX1 would restore the proinsulin content in shDDX1 cells. The results showed that the overexpression of WT DDX1 could increase the proinsulin content in shDDX1 cells (Figure 6B and C) without affecting the level of insulin mRNA (Figure 6D). In contrast, the overexpression of S295D phospho-mutants of DDX1 failed to restore the proinsulin content at the same level of insulin mRNA (Figure 6B–D). Furthermore, the UV crosslinking RIP-PCR results confirmed that while the same amount of the WT and S295D DDX1 protein was pulled down, much less insulin mRNA was bound to the S295D phospho-mutant (Figure 6E). The result indicated that the overexpression of S295D phospho-mutants of DDX1 could repress insulin translation. On the other hand, we examined whether the overexpression of wild-type (WT) and mutant proteins mimicking nonphosphorylated DDX1 would eliminate the PA-induced decrease in proinsulin translation in shDDX1 cells. The results showed that the overexpression of both WT DDX1 and the S295A nonphospho-mutant could restore the proinsulin content in shDDX1 cells (Figure 6F and G). Under treatment with PA, the overexpression of WT DDX1 in cells could not restore the proinsulin content; however, the overexpression of the S295A nonphospho-mutant DDX1 in cells could restore the proinsulin content (Figure 6F and G). In all cases, the level of insulin mRNA was not changed significantly except for a slight decrease in shDDX1 cells expressing WT DDX1 as compared with the scrambled cells (Figure 6H). Furthermore, the UV crosslinking RIP-PCR results confirmed that while the same amount of the WT and S295A DDX1 proteins was pulled down, the same amount of insulin mRNA was bound to WT DDX1 and S295A nonphospho-mutant DDX1. When cells were treated with PA, the same amount of the WT and S295A DDX1 proteins were pulled down, but more insulin mRNA was bound to the S295A nonphospho-mutant (Figure 6I). This result indicated that the overexpression of the DDX1 S295A nonphospho-mutant could eliminate the PA-induced repression of insulin translation and the dissociation of the DDX1-insulin mRNA complex. Taken together, these results suggest that the S295 site of DDX1 is critical for the binding of DDX1 to insulin mRNA and hence for the translation of insulin mRNA.
DDX1 may associate with the translation initiation complex to regulate translation
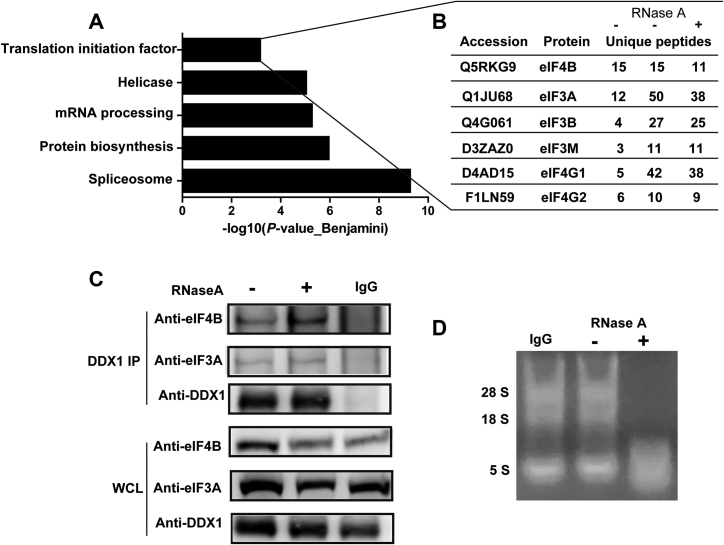
To further explore the detailed mechanism by which DDX1 regulates translation, we sought to identify DDX1-interacting proteins by co-immunoprecipitation (IP). We enriched the endogenous DDX1-interacting proteins and separated them by SDS-PAGE for subsequent identification by LC–MS/MS. RNase A was used to exclude some proteins that interacted with DDX1 depending on the presence of RNA.
In order to identify DDX1-interacting proteins with high confidence, we selected overlapping proteins from two separate DDX1 IP experiments without RNase A treatment (RNase A-) and one with RNase A treatment (RNase A+) (Supplemetary Figure S5). For the RNase A- case, 359 and 539 DDX1-interacting proteins were identified by mass spectrometry, respectively. In addition, 475 DDX1-interacting proteins were identified for the RNase A+ case. There are 106 overlapping proteins between these three groups (Supplementary Figure S5C, Supplementary data 1). Gene Ontology (GO) analysis of the 106 overlapping proteins showed a significant enrichment of proteins in the processes of spliceosome, protein biosynthesis, mRNA processing, helicase, and translation initiation factor (Figure 7A). These results are consistent with the previously reported role of DDX1 as a tRNA splicing factor (26). Several subunits of the eukaryotic translation initiation complex were detected at high abundance, such as the translation initiation factors eIF4B and the eIF3 family, along with scaffold proteins such as eIF4G1 and eIF4G2 (Figure 7B). The interaction between DDX1 and eIF4B as well as that between DDX1 and eIF3A was further validated through co-IP experiments followed by western blotting; these associations did not depend on the presence of RNA because RNase A treatment failed to disrupt the interaction between DDX1 and either eIF4B or eIF3A (Figure 7C and D). Taken together, these data suggest that DDX1 may interact with the translation initiation complex to regulate translation.
Figure 7.
DDX1 interacts with the translation initiation complex. (A) GO analysis of DDX1-interacting proteins. (B) Summary of the subunits of the translation initiation complex identified from the DDX1 co-IP experiment followed by MS analysis. (C) Western blotting was used to verify two subunits of the translation initiation complex pulled down by a DDX1 antibody with or without RNase A treatment (n = 3). (D) The RNase A activity was confirmed on an agarose gel.
DISCUSSION
Obesity is a major factor leading to diabetes. Previously, studies about the relationship between obesity and diabetes have been focused on insulin resistance and the adaptive responses of β cells to elevated levels of blood glucose. These adaptive responses include a compensatory stage of increased insulin expression (42) and β cell mass (43,44) in response to high glucose, followed by a decompensation stage of β cell dedifferentiation (45) or apoptosis (46,47) caused by glucolipotoxicity. However, the influence of fatty acids on the β cell response before the onset of glucose elevation has been less studied. Here, we identified a novel RNA-binding protein, namely, DDX1, that regulates insulin translation upon binding to insulin mRNA. The PA-mediated dissociation of the DDX1–insulin mRNA complex downregulates insulin translation. We provide evidence that the downregulation of insulin translation occurs at a very early stage, before the elevation of glucose, in HFD-fed mice. We speculate that the DDX1-mediated repression of insulin translation acts counter to the higher demand for insulin synthesis and secretion imposed by insulin resistance, which then leads to the elevation of blood glucose in obese animals.
DDX1 is a member of the DEAD box family of RNA helicases, which has been implicated in a variety of biological processes, including 3′-end processing of mRNA, DNA repair, microRNA processing, tRNA maturation and mRNA transport. It is interesting to identify a novel function for DDX1 in promoting the translation of insulin mRNA. We provide evidence that DDX1 interacts with eIF4B (Figure 7B and C), which is a well-established factor for translation initiation (48). eIF4B is an RNA-binding protein that stimulates eIF4A (DDX2) helicase activity, thus enabling eIF4A to unwind more stable duplexes. We also identified eIF4G1/2 as binding partners of DDX1 (Figure 7D). eIF4G is a scaffolding protein that bridges eIF4E and eIF4A to form the eIF4F complex. Then, eIF4F binds to the 5′-UTR of mRNA and recruits the 43S preinitiation complex. Indeed, our mass spectrometry data suggest that DDX1 could pull down components of the 43S complex, i.e. eIF3A, eIF3B and eIF3M (Figure 7B). We confirmed by western blotting that DDX1 and insulin mRNA interact with eIF3A, thus suggesting that DDX1 is required for the recruitment of the 43S preinitiation complex. DDX1 has been shown to be upregulated in various tumors (49–52), suggesting a pathological role for DDX1 in cancer cell proliferation. However, the physiological role of DDX1 has remained elusive. Our data illustrate that DDX1 is responsible for the repression of translation of insulin mRNA in pancreatic β cells in response to fatty acid overload and thus suggest a nutrient-sensing role for DDX1 in regulating hormone secretion. Our results thus extend the physiological role of DEAD box helicase proteins to metabolism regulation.
The modification of RNA-binding proteins is important for their RNA-binding capability and function (53,54). We identified the phosphorylation sites in DDX1 protein in response to PA treatment in INS-1 cells. We found that the phosphorylation of S295 is critical for the binding of DDX1 to insulin mRNA and hence the regulation of insulin translation. Our in silico analysis predicted that ATM kinase could likely induce phosphorylation at the S295 residue of DDX1. Indeed, it has been reported that ATM kinase can interact with DDX1 and phosphorylate DDX1 at S373 and S667 after DNA damage (27). Therefore, it needs to be verified whether PA could activate ATM kinase, which could then phosphorylate DDX1 at S295.
It has been reported that a HFD impairs insulin secretion in vivo as early as 1 week after inception (43,55,56). We confirmed this early downregulation of insulin in HFD mice (Figure 1K and L) and revealed its underlying molecular mechanism, which is partly attributed to the DDX1-mediated inhibition of the translation of insulin. The physiological significance of this downregulation remains to be demonstrated, but it may reflect a feedback response of insulin signaling to balance lipid storage and degradation (57–59). On the other hand, fatty acid overload also causes insulin resistance, which means that more insulin is required for glucose disposal. This paradox leads to an increase in blood glucose at a prediabetic stage in HFD mice. The increase in blood glucose then feeds back to β cells and upregulates insulin expression at the transcriptional level. We speculate that the DDX1-mediated inhibition of insulin translation was one of the reasons for the insulin insufficiency at an early prediabetic stage in HFD animals. It is thus intriguing to test whether the overexpression of DDX1 or the expression of DDX1 phospho-mutants in obese mice will delay the onset of hyperglycemia or not. If so, DDX1 will emerge as a promising target for developing an antidiabetic therapy for use in obese subjects.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of the Institute of Biophysics Core Facilities, in particular, Zhensheng Xie and Jifeng Wang for MS operation, Zhenwei Yang for qPCR, Junying Jia for technical support with flow cytometry, Yan Teng for technical support with confocal imaging.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Basic Research Project of China [2015CB910303]; National Key Research and Development Program [2016YFC0903301]; National Natural Science Foundation of China [31770891, 31730054]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDA12030101]. Funding for open access charge: National Key Basic Research Project of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dodson G., Steiner D.. The role of assembly in insulin's biosynthesis. Curr. Opin. Struct. Biol. 1998; 8:189–194. [DOI] [PubMed] [Google Scholar]
- 2. Jahr H., Schroder D., Ziegler B., Ziegler M., Zuhlke H.. Transcriptional and translational control of glucose-stimulated (pro)insulin biosynthesis. Eur. J. Biochem. 1980; 110:499–505. [DOI] [PubMed] [Google Scholar]
- 3. Leibiger B., Wahlander K., Berggren P.O., Leibiger I.B.. Glucose-stimulated insulin biosynthesis depends on insulin-stimulated insulin gene transcription. J. Biol. Chem. 2000; 275:30153–30156. [DOI] [PubMed] [Google Scholar]
- 4. Melloul D., Ben-Neriah Y., Cerasi E.. Glucose modulates the binding of an islet-specific factor to a conserved sequence within the rat I and the human insulin promoters. Proc. Natl. cad. Sci. U.S.A. 1993; 90:3865–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kataoka K., Han S.I., Shioda S., Hirai M., Nishizawa M., Handa H.. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 2002; 277:49903–49910. [DOI] [PubMed] [Google Scholar]
- 6. Melloul D., Marshak S., Cerasi E.. Regulation of insulin gene transcription. Diabetologia. 2002; 45:309–326. [DOI] [PubMed] [Google Scholar]
- 7. Lee E.K., Kim W., Tominaga K., Martindale J.L., Yang X., Subaran S.S., Carlson O.D., Mercken E.M., Kulkarni R.N., Akamatsu W. et al. RNA-binding protein HuD controls insulin translation. Mol. Cell. 2012; 45:826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kulkarni S.D., Muralidharan B., Panda A.C., Bakthavachalu B., Vindu A., Seshadri V.. Glucose-stimulated translation regulation of insulin by the 5′ UTR-binding proteins. J. Biol. Chem. 2011; 286:14146–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tillmar L., Welsh N.. Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tract-binding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Mol. Med. 2002; 8:263–272. [PMC free article] [PubMed] [Google Scholar]
- 10. Wicksteed B., Uchizono Y., Alarcon C., McCuaig J.F., Shalev A., Rhodes C.J.. A cis-element in the 5′ untranslated region of the preproinsulin mRNA (ppIGE) is required for glucose regulation of proinsulin translation. Cell Metab. 2007; 5:221–227. [DOI] [PubMed] [Google Scholar]
- 11. Wicksteed B., Herbert T.P., Alarcon C., Lingohr M.K., Moss L.G., Rhodes C.J.. Cooperativity between the preproinsulin mRNA untranslated regions is necessary for glucose-stimulated translation. J. Biol. Chem. 2001; 276:22553–22558. [DOI] [PubMed] [Google Scholar]
- 12. Tillmar L., Carlsson C., Welsh N.. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. J. Biol. Chem. 2002; 277:1099–1106. [DOI] [PubMed] [Google Scholar]
- 13. Knoch K.P., Bergert H., Borgonovo B., Saeger H.D., Altkruger A., Verkade P., Solimena M.. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 2004; 6:207–214. [DOI] [PubMed] [Google Scholar]
- 14. Olofsson C.S., Collins S., Bengtsson M., Eliasson L., Salehi A., Shimomura K., Tarasov A., Holm C., Ashcroft F., Rorsman P.. Long-term exposure to glucose and lipids inhibits glucose-induced insulin secretion downstream of granule fusion with plasma membrane. Diabetes. 2007; 56:1888–1897. [DOI] [PubMed] [Google Scholar]
- 15. Hoppa M.B., Collins S., Ramracheya R., Hodson L., Amisten S., Zhang Q., Johnson P., Ashcroft F.M., Rorsman P.. Chronic palmitate exposure inhibits insulin secretion by dissociation of Ca(2+) channels from secretory granules. Cell Metab. 2009; 10:455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mason T.M., Goh T., Tchipashvili V., Sandhu H., Gupta N., Lewis G.F., Giacca A.. Prolonged elevation of plasma free fatty acids desensitizes the insulin secretory response to glucose in vivo in rats. Diabetes. 1999; 48:524–530. [DOI] [PubMed] [Google Scholar]
- 17. Carpentier A., Mittelman S.D., Bergman R.N., Giacca A., Lewis G.F.. Prolonged elevation of plasma free fatty acids impairs pancreatic beta-cell function in obese nondiabetic humans but not in individuals with type 2 diabetes. Diabetes. 2000; 49:399–408. [DOI] [PubMed] [Google Scholar]
- 18. Linder P., Jankowsky E.. From unwinding to clamping - the DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011; 12:505–516. [DOI] [PubMed] [Google Scholar]
- 19. Iwasaki S., Floor S.N., Ingolia N.T.. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016; 534:558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van der Meulen J. et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014; 513:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soto-Rifo R., Rubilar P.S., Limousin T., de Breyne S., Decimo D., Ohlmann T.. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012; 31:3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferguson S.B., Blundon M.A., Klovstad M.S., Schupbach T.. Modulation of gurken translation by insulin and TOR signaling in Drosophila. J. Cell Sci. 2012; 125:1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Arribas-Layton M., Chen Y., Lykke-Andersen J., Sen G.L.. DDX6 Orchestrates mammalian progenitor function through the mRNA degradation and translation pathways. Mol. Cell. 2015; 60:118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giorgi C., Yeo G.W., Stone M.E., Katz D.B., Burge C., Turrigiano G., Moore M.J.. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007; 130:179–191. [DOI] [PubMed] [Google Scholar]
- 25. Bleoo S., Sun X., Hendzel M.J., Rowe J.M., Packer M., Godbout R.. Association of human DEAD box protein DDX1 with a cleavage stimulation factor involved in 3′-end processing of pre-MRNA. Mol. Biol. Cell. 2001; 12:3046–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Popow J., Jurkin J., Schleiffer A., Martinez J.. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014; 511:104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Han C., Liu Y., Wan G., Choi H.J., Zhao L., Ivan C., He X., Sood A.K., Zhang X., Lu X.. The RNA-binding protein DDX1 promotes primary microRNA maturation and inhibits ovarian tumor progression. Cell Rep. 2014; 8:1447–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L., Monckton E.A., Godbout R.. A role for DEAD box 1 at DNA double-strand breaks. Mol. Cell. Biol. 2008; 28:6413–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Z., Kim T., Bao M., Facchinetti V., Jung S.Y., Ghaffari A.A., Qin J., Cheng G., Liu Y.J.. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011; 34:866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu C., Quinn J., Chang H.Y.. Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 2012; 61:e3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J., Duncan D., Shi Z., Zhang B.. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41:W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong W., Li Z., Zhou M., Xu T., Wang Y.. DDX1 regulates alternative splicing and insulin secretion in pancreatic beta cells. Biochem. Biophys. Res. Commun. 2018; 500:751–757. [DOI] [PubMed] [Google Scholar]
- 33. Hou J., Li Z., Zhong W., Hao Q., Lei L., Wang L., Zhao D., Xu P., Zhou Y., Wang Y. et al. Temporal transcriptomic and proteomic landscapes of deteriorating pancreatic islets in type 2 diabetic rats. Diabetes. 2017; 66:2188–2200. [DOI] [PubMed] [Google Scholar]
- 34. Hagman D.K., Latour M.G., Chakrabarti S.K., Fontes G., Amyot J., Tremblay C., Semache M., Lausier J.A., Roskens V., Mirmira R.G. et al. Cyclical and alternating infusions of glucose and intralipid in rats inhibit insulin gene expression and Pdx-1 binding in islets. Diabetes. 2008; 57:424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kelpe C.L., Moore P.C., Parazzoli S.D., Wicksteed B., Rhodes C.J., Poitout V.. Palmitate inhibition of insulin gene expression is mediated at the transcriptional level via ceramide synthesis. J. Biol. Chem. 2003; 278:30015–30021. [DOI] [PubMed] [Google Scholar]
- 36. Hagman D.K., Hays L.B., Parazzoli S.D., Poitout V.. Palmitate inhibits insulin gene expression by altering PDX-1 nuclear localization and reducing MafA expression in isolated rat islets of Langerhans. J. Biol. Chem. 2005; 280:32413–32418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ritz-Laser B., Meda P., Constant I., Klages N., Charollais A., Morales A., Magnan C., Ktorza A., Philippe J.. Glucose-induced preproinsulin gene expression is inhibited by the free fatty acid palmitate. Endocrinology. 1999; 140:4005–4014. [DOI] [PubMed] [Google Scholar]
- 38. Jacqueminet S., Briaud I., Rouault C., Reach G., Poitout V.. Inhibition of insulin gene expression by long-term exposure of pancreatic beta cells to palmitate is dependent on the presence of a stimulatory glucose concentration. Metabolism. 2000; 49:532–536. [DOI] [PubMed] [Google Scholar]
- 39. Maris M., Robert S., Waelkens E., Derua R., Hernangomez M.H., D’Hertog W., Cnop M., Mathieu C., Overbergh L.. Role of the saturated nonesterified fatty acid palmitate in beta cell dysfunction. J. Proteome Res. 2013; 12:347–362. [DOI] [PubMed] [Google Scholar]
- 40. Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A. et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010; 3:ra3. [DOI] [PubMed] [Google Scholar]
- 41. Rigbolt K.T., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., Blagoev B.. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 2011; 4:rs3. [DOI] [PubMed] [Google Scholar]
- 42. Gonzalez A., Merino B., Marroqui L., Neco P., Alonso-Magdalena P., Caballero-Garrido E., Vieira E., Soriano S., Gomis R., Nadal A. et al. Insulin hypersecretion in islets from diet-induced hyperinsulinemic obese female mice is associated with several functional adaptations in individual beta-cells. Endocrinology. 2013; 154:3515–3524. [DOI] [PubMed] [Google Scholar]
- 43. Mosser R.E., Maulis M.F., Moulle V.S., Dunn J.C., Carboneau B.A., Arasi K., Pappan K., Poitout V., Gannon M.. High-fat diet-induced beta-cell proliferation occurs prior to insulin resistance in C57Bl/6J male mice. Am. J. Physiol. Endocrinol. Metab. 2015; 308:E573–E582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tschen S.I., Dhawan S., Gurlo T., Bhushan A.. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009; 58:1312–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laybutt D.R., Glandt M., Xu G., Ahn Y.B., Trivedi N., Bonner-Weir S., Weir G.C.. Critical reduction in beta-cell mass results in two distinct outcomes over time. Adaptation with impaired glucose tolerance or decompensated diabetes. J. Biol. Chem. 2003; 278:2997–3005. [DOI] [PubMed] [Google Scholar]
- 46. Hennige A.M., Ranta F., Heinzelmann I., Dufer M., Michael D., Braumuller H., Lutz S.Z., Lammers R., Drews G., Bosch F. et al. Overexpression of kinase-negative protein kinase Cdelta in pancreatic beta-cells protects mice from diet-induced glucose intolerance and beta-cell dysfunction. Diabetes. 2010; 59:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sauter N.S., Schulthess F.T., Galasso R., Castellani L.W., Maedler K.. The antiinflammatory cytokine interleukin-1 receptor antagonist protects from high-fat diet-induced hyperglycemia. Endocrinology. 2008; 149:2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pelletier J., Graff J., Ruggero D., Sonenberg N.. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015; 75:250–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Godbout R., Li L., Liu R.Z., Roy K.. Role of DEAD box 1 in retinoblastoma and neuroblastoma. Future Oncol. 2007; 3:575–587. [DOI] [PubMed] [Google Scholar]
- 50. Tanaka K., Okamoto S., Ishikawa Y., Tamura H., Hara T.. DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene. 2009; 28:2142–2151. [DOI] [PubMed] [Google Scholar]
- 51. Germain D.R., Graham K., Glubrecht D.D., Hugh J.C., Mackey J.R., Godbout R.. DEAD box 1: a novel and independent prognostic marker for early recurrence in breast cancer. Breast Cancer Res. Treat. 2011; 127:53–63. [DOI] [PubMed] [Google Scholar]
- 52. Hodgson J.G., Yeh R.F., Ray A., Wang N.J., Smirnov I., Yu M., Hariono S., Silber J., Feiler H.S., Gray J.W. et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009; 11:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rudel S., Wang Y., Lenobel R., Korner R., Hsiao H.H., Urlaub H., Patel D., Meister G.. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011; 39:2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thapar R. Structural basis for regulation of RNA-binding proteins by phosphorylation. ACS Chem. Biol. 2015; 10:652–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Winzell M.S., Ahren B.. The high-fat diet-fed mouse: a model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes. 2004; 53(Suppl. 3):S215–S219. [DOI] [PubMed] [Google Scholar]
- 56. Winzell M.S., Magnusson C., Ahren B.. Temporal and dietary fat content-dependent islet adaptation to high-fat feeding-induced glucose intolerance in mice. Metabolism. 2007; 56:122–128. [DOI] [PubMed] [Google Scholar]
- 57. Titchenell P.M., Lazar M.A., Birnbaum M.J.. Unraveling the Regulation of Hepatic Metabolism by Insulin. Trends Endocrinol. Metab. 2017; 28:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haas J.T., Miao J., Chanda D., Wang Y., Zhao E., Haas M.E., Hirschey M., Vaitheesvaran B., Farese R.V. Jr, Kurland I.J. et al. Hepatic insulin signaling is required for obesity-dependent expression of SREBP-1c mRNA but not for feeding-dependent expression. Cell metabolism. 2012; 15:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Biddinger S.B., Hernandez-Ono A., Rask-Madsen C., Haas J.T., Aleman J.O., Suzuki R., Scapa E.F., Agarwal C., Carey M.C., Stephanopoulos G. et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008; 7:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.