Abstract
Human ARTD2 (or PARP2) is an ADP-ribosyltransferase, which is catalytically activated by binding to damaged DNA. ARTD2 subsequently ADP-ribosylates itself and other proteins, initiating a cascade of events leading to DNA repair. In contrast to ARTD1, the founding member of the enzyme family, ARTD2 does not have specialized zinc-fingers for detecting DNA damage. The domain organization of ARTD2 includes disordered N-terminus, WGR and catalytic domains. However, the N-terminus of ARTD2 is not strictly required for the DNA dependent activity. While it is known that ARTD2 requires the WGR domain for efficient DNA binding and subsequent catalytic activation, the mechanism of DNA damage detection and subsequent catalytic activation are not completely understood. Here, we report crystal structures of ARTD2 WGR domain bound to double-strand break mimicking DNA oligonucleotides. Notably, the crystal structures revealed DNA binding mode of ARTD2 involving DNA end to end interaction. Structures demonstrate how ARTD2 recognizes nicked DNA, how it interacts with the 5′-phosphate group, and how it can mediate joining of DNA ends in vitro. Extensive mutagenesis of the ARTD2-DNA interface combined with activity, binding, and stoichiometry measurements demonstrate that the WGR domain is the key for DNA break detection.
INTRODUCTION
Human diphtheria toxin-like ADP-ribosyltransferases (ARTDs) also known as poly(ADP-ribose) polymerases (PARPs) participate in many biological processes such as DNA repair, transcriptional control, and chromatin remodeling. There are 17 protein modifying ARTDs in human genome involved in regulating multiple aspects of cellular activities (1). ARTD1, ARTD2 and ARTD3 detect cellular DNA damage, resulting in the hydrolysis of NAD+, poly-ADP-ribosylation of proteins and subsequent recruitment of DNA damage repair factors (1–3). The role of ARTDs in DNA repair has made them promising therapeutic targets as ARTD inhibitors can suppress DNA repair and sensitize cancer cells to DNA damaging agents (4).
ARTD2 was discovered based on the residual ADP-ribosylation activity observed in mouse embryonic fibroblast deficient in ARTD1 (3). It functions in DNA damage detection and subsequent recruitment of DNA repair factors to the site of DNA damage in an ADP-ribosylation dependent manner (1,5,6). ARTD2 ADP-ribosylation activity upon the detection of damaged DNA is important in the remodeling of chromatin structure as it ADP-ribosylates histones (6–8). In addition, hyper-sensitivity of ARTD2−/− mice to gamma irradiation has been reported (9). Presently, it is widely accepted that ARTD2 has a major role in the repair of single strand break repair (SSBR) and a very recent study has shown a fast association of the enzyme to nicked DNA compared with blunt end DNA (10). ARTD2 has been shown to interact with AP site containing DNA also highlighting a role of the enzyme in SSBR (11).
ARTD1 consists of three zinc fingers responsible for DNA binding, BRCT1 domain, WGR domain and a C-terminal catalytic domain. Despite being activated by DNA, human ARTD2 and ARTD3 lack the zinc fingers and a BRCT domain, but they contain a WGR domain and a catalytic domain. In contrast to ARTD1, which is activated by various DNA oligonucleotides, ARTD2 and ARTD3 are activated by specific DNA structures and require especially 5′-phosphorylated DNA ends for robust activation (12,13). We showed earlier that WGR domain of ARTD2 is essential for DNA binding and activation of the enzyme, and that DNA binds between WGR and catalytic domains (13). The actual mechanism how WGR domain coordinates DNA binding is still unclear. A recent study reported that WGR domain is also critical for DNA binding in ARTD3 (14).
Here, we have used a combination of X-ray crystallography, structure-based mutagenesis, and biophysical/biochemical methods in order to understand how ARTD2 detects DNA damage at molecular level. Studies revealed that ARTD2 binds DNA breaks through the WGR domain and interacts with DNA on both sides of the DNA break. Supported by biochemical data we propose key amino acids required for DNA binding at the damage site and residues that can potentially mediate activation signals to the catalytic domain.
MATERIALS AND METHODS
Cloning, protein expression and purification
The cloning of the DNA constructs coding for ARTD2FL isoform 1 (residues 1–583) and the individual domain constructs (ARTD2WGR residues 90–218, ARTD2WGR+CAT; residues 90–583) has been previously described (13). Mutagenesis of the ARTD2FL enzyme (residue 1–583) was done using the QuickChange Protocol and sequencing of the genes were done using the automated sequencer in Biocenter core facility, University of Oulu, Finland. Expression and purification of proteins were done as described (13). In brief, proteins were expressed in Escherichia coli and purifications were done in three-step chromatography system (IMAC, heparin affinity and size exclusion chromatography).
Fluorescence activity assay
Activity assay of ARTD2 proteins were done as reported earlier (13,15) using dumbbell DNA carrying a nick with 5′-phosphate, double-stranded palindromic DNA breaks with 5′-phosphate, double-stranded palindromic DNA breaks without 5′-phosphate, double-stranded DNA breaks carrying five nucleotide overhang with 5′-phosphate (Supplementary Table S1 and Supplementary Figure S1). Measurements were done in quadruplicates and the incubation time was 1 h for ARTD2FL and point mutants, 2 h for ARTD1FL, ARTD1WGRCAT and ARTD2WGR+CAT. Enzyme concentrations in the reaction wells were 50 nM for ARTD1WGRCAT, ARTD2FL and the different point and deletion mutants, and 25 nM for ARTD1FL. The reaction buffer contained 150 mM NaCl to mimic physiological salt concentration.
Fluorescence polarization (FP)
FP was performed as previously described using double-stranded DNA model with 5′-phosphate (13). The DNA was tagged with 5′-fluorescein at one of the 5′-end and 5′-phosphate at the other end. Measurements were done in triplicates and the buffer was 50 mM HEPES, pH 7.5, 150 mM NaCl, and 0.5 mM TCEP. It was confirmed with the same assay that fluorescein alone does not bind to ARTD2FL. For details on the DNA structure used see Supplementary Table S1 and Supplementary Figure S1.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as described previously (13). Briefly, ARTD2FL, ARTD2FL mutants, ARTD2WGR+CAT and ARTD1WGR+CAT were separately mixed with dumbbell DNA carrying a nick with 5′-phosphate (DNA-2) or double-stranded palindromic DNA breaks with 5′-phosphate (DNA-5, DNA-5, DNA-7) at the ratio of 2:1 (Protein:DNA) for 30 min in 50 mM HEPES, pH 8.0, 300 mM NaCl, 5% glycerol and 0.5 mM TCEP. The complexes were analyzed using 0.5% agarose. Staining and visualization was done using GelRed (Biotium) and gel imaging system (Bio-Rad), respectively.
Analysis of ARTD2 DNA interaction with surface plasmon resonance spectroscopy (SPR)
The kinetics and affinity of the binding of ARTD2FL and mutants to DNA breaks were studied using SPR with a Biacore T-200 (GE-Healthcare) at 25°C with 20 mM HEPES, pH 7.5, 300 mM NaCl, 3 mM EDTA and 0.05% surfactant P-20 (HBS-EP). 300 mM NaCl and 0.05% surfactant P-20 were used to alleviate the unspecific interactions encountered during the test experiment when 150 mM NaCl and 0.005% surfactant P-20 was used. 20-base pair double-stranded DNA breaks with or without 5′-phosphate tagged with 5′-biotin at another end were immobilized on different flow cells of a CM4 chip (GE-Healthcare) resulting in 750 RU signal.
For assessing the binding of ARTD2FL and variants to DNA, multi cycle kinetics analyses were carried out. Enzymes were injected at eight different concentrations (0, 0.781, 1.5625, 3.125, 6.26, 12.5, 50, 100 and 400 nM) to the chip at the flow rate of 30 μl/min, injection time of 3 min, dissociation time of 10 min and stabilization time of 5 min. In the experiment with ARTD2WGR+CAT, 4000, 2000, 500, 125, 62.5, 31.25, 15, 625 and 0 nM protein concentrations were used. For each sample, there were always replicates for at least three out of the eight concentrations. In between injection of the different protein concentrations, regeneration of the chip was done using three times 1 min injection of 0.1% SDS.
Kinetics analyses were done using the Biacore T200 Evaluation Software. Previous studies have shown that upon DNA binding, ARTD2 as well as ARTD1 and ARTD3 undergo conformational changes leading to the catalytic activation. We tested fitting using the 1:1 binding model (with chi2 values of 11.9 and 7.16 for ARTD2FL and ARTDWGRCAT respectively), but two-state binding model provided a much better fit with reasonable chi2 values (2.76 and 0.67 for ARTD2FL and ARTDWGRCAT respectively), so it was used for final analysis. The calculation of the KD by the software was based on the equation KD = kd1/ka1 × (kd2/(kd2+ka2)).
Isothermal titration calorimetry (ITC)
Binding of ARTD2FL to nicked and blunt end DNAs were analyzed by ITC using MicroCal iTC200 (Marvin). Titration was done at 25°C in 50 mM HEPES, pH 7.5, 400 mM NaCl, 0.5 mM TCEP and 1 mM EDTA. 150 μM of hairpin nicked DNA (DNA-1) and hairpin blunt DNA (DNA-3) were titrated to 10 μM ARTD2FL. Data were analyzed with origin 7 (OriginLAB).
Size exclusion chromatography - static light scattering (SEC-SLS)
DNA–ARTD2 complexes in solution were studied using static light scattering in 25 mM HEPES, pH 7.5, 400 mM NaCl and 0.5 mM TCEP. Protein (35 μM) was mixed with DNA (35 μM) and separated with superdex S200 increase (GE Healthcare) and analyzed using an SLS instrument (Wyatt Technology). Mass determination was done using ASTRA (Wyatt Technology).
Crystallization
With the 5′-phosphorylated double-strand DNA (DNA-5) ARTD2WGR complex, initial crystallization hit was obtained in 20% (v/v) polypropylene glycol 400 and 10% 1-propanol, 300 μM protein and 300 μM DNA at 4°C. The crystals diffracted to ∼10 Å. After series of optimization using same protein and DNA concentration, different buffer pH, 2-methyl 2,4 pentanediol or 2,5-hexanediol or 1,3-butanediol or 1,4-dioxane, 2-propanol or tert-butanediol or 1,3-propanediol, diffracting crystals of 2.8–3.5 Å were obtained. The best diffracting crystals were obtained in 12% (v/v) polypropylene glycol 400 and 1% 1-propanol and 0.1 M sodium acetate pH 4.7 and at 4°C.
The crystals of non-phosphorylated double strand DNA (DNA-6) in complex with ARTD2WGR was obtain at 4°C in 25% (w/v) PEGMME 5000 and 0.1 M sodium acetate at pH 4.9. The protein concentration was 300 μM while the DNA concentration was 150 μM.
Poorly diffracting crystals of the 300μM 5′-phosphorylated double stranded overhang DNA (DNA-9) with 300 μM ARTD2WGR domain was initially obtained in 20% (v/v) sokalan CP7, 0.1 M ammonium sulfate, and 0.1 M HEPES–NaOH at pH 7.0. After series of trials, the diffraction quality was not improved and we rescreened the same complex using PACT Screen (Molecular Dimension) and a crystal diffracting to 3 Å was obtain at 4°C in 0.2 M Na-formate, 20% PEG 3350 and 0.1 M sodium acetate pH 4.7. Prior to flash cooling in liquid nitrogen, the crystal was transferred in all cases to the precipitant solution supplemented with 20% glycerol and 0.25 M NaCl.
Data collection, structure determination and refinement
The crystals of 5′-phosphorylated double-strand DNA–ARTD2WGR complex, non-phosphorylated double strand DNA-WGR complex, 5′-phosphorylated double-stranded overhang DNA–ARTD2WGR complex was collected in i03, i04 and i02 beam lines at Diamond light source (UK), respectively. Diffraction data were processed and scaled using XDS (16). The structure of 5′-phosphorylated double-strand DNA–ARTD2WGR complex was solved using molecular replacement with Phaser MR (17) using a polyalanine model of ARTD2WGR and DNA model generated with Coot (18). First homology model of ARDT2WGR was generated with SWISS-MODEL using ARTD1WGR domain as a template (19) and finally polyalanine model was generated using Chainsaw (20). The crystals of nonphosphorylated double-strand DNA ARTD2WGR complex and 5′-phosphorylated double-stranded overhang DNA–ARTD2WGR complex were solved using ARTD2WGR as a model in molecular replacement. Structure refinement and model building were done using Coot (18) and Refmac5 (21). Data collection and refinement statistics are given in Table 1. The figures of the structures were made using Pymol (22).
Table 1.
Data collection and refinement statistics
| ARTD2WGR + dsDNA | ARTD2WGR + 5′-P dsDNA | ARTD2WGR + 5′-P overhang dsDNA | |
|---|---|---|---|
| Data collection | |||
| Space group | P6322 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 119.27, 119.27, 73.58 | 63.20, 84.41, 96.02 | 68.17, 87.17, 185.34 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 30–2.2 (2.20) | 30–2.8 (2.80) | 20–2.98 (2.98) |
| R merge | 8.10 (155.7) | 10.0 (179.7) | 6.9 (136.6) |
| R meas | 8.0 (155) | 17.6 (169.4) | 9.1 (210) |
| I/δ (I) | 20.4 (1.6) | 11.36 (1.3) | 12.6 (0.9) |
| CC 1/2 | 100 (60) | 100 (40) | 100 (60) |
| Completeness (%) | 99.9 (99.7) | 99.7 (100) | 98.8 (98.9) |
| Redundancy | 9.7 (10.0) | 6.1 (6.7) | 6.4 (6.8) |
| Refinement | |||
| No. of reflections | 157093 | 62820 | 148569 |
| R work/Rfree | 21.40/24.98 | 20.81/27.39 | 22.82/27.53 |
| No.of atoms | 1430 | 2679 | 5012 |
| Protein | 977 | 1855 | 3780 |
| DNA/ion | 414 | 824 | 1232 |
| Water | 39 | – | – |
| B-factors | |||
| Protein | 35.59 | 62.7 | 126.7 |
| DNA/ion | 37.90 | 59.5 | 110.3 |
| Water | 49.11 | - | - |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.010 | 0.010 | 0.014 |
| Bond angles (°) | 1.480 | 1.600 | 1.778 |
| Ramachandran statistics | |||
| Favored region (%) | 98.3 | 93.3 | 92.9 |
| Outliers (%) | 0 | 0 | 0 |
Data were collected from one crystal per condition. Values in parentheses are for highest-resolution shell. dsDNA = DNA-2, 5′-P dsDNA = DNA-5, 5′-P overhang dsDNA = DNA-9).
RESULTS
Crystal structures of ARTD2 WGR domain–DNA complexes
Previously we showed that the N-terminus of ARTD2 is a disordered, high affinity DNA interacting module, and not required for the DNA-dependent activity of the enzyme (13). Therefore, the WGR and the catalytic domains are sufficient for DNA dependent activity. In the earlier studies, DNA binding to the catalytic domain was not detected and it is most likely that the WGR domain represents a minimal domain required for DNA damage detection and an essential domain for the catalytic activation. Here, we have determined crystal structures of ARTD2 WGR domain (residues 90–218) bound to DNA molecules mimicking damaged and activating DNA to 2.3–3.0 Å (Table 1, Figure 1). All the residues of the crystallized constructs are visible in the crystal structures with the exception of the last 13 C-terminal residues. Also, the DNAs used in the crystallization are fully defined (Supplementary Figures S2–S4).
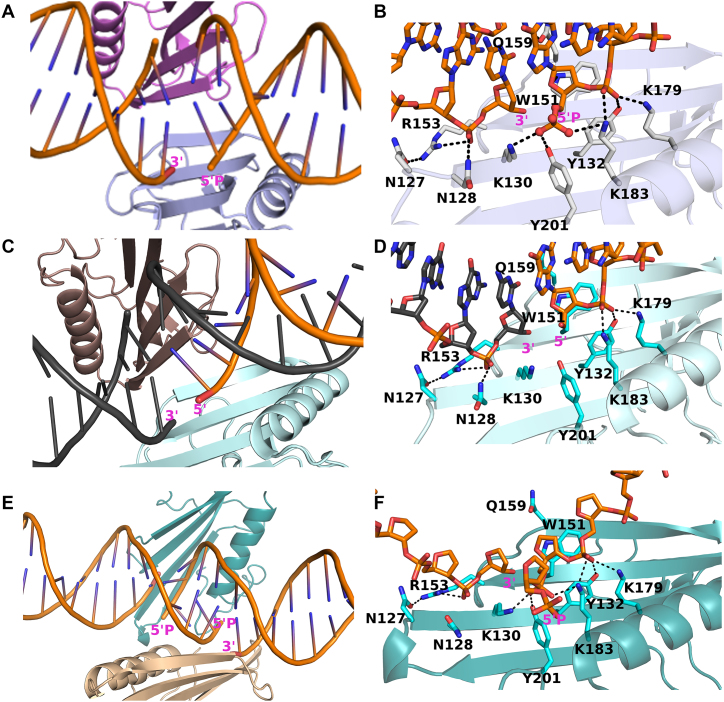
Figure 1.
Structures of ARTD2WGR bound to different DNA breaks. (A) Cartoon representation of ARTD2WGR bound to DNA breaks with 5′-phosphate. Two protein monomers (light blue and pink) and four DNA molecules, representing two double-stranded DNAs were present in the asymmetric unit. (B) Zoomed-in view of the ARTD2WGR bound to DNA breaks with 5′-phosphate. (C) Cartoon representation of ARTD2WGR bound to DNA breaks without 5′-phosphate. In the asymmetric unit, there were a monomeric protein (cyan) and 1 molecule of DNA. Through symmetry, the binding mode is similar to that with the 5′-phosphate DNA. (D) Zoomed-in view of the ARTD2WGR bound to DNA breaks without 5′-phosphate. (E) Cartoon representation of ARTD2WGR bound to DNA breaks with five nucleotides overhangs and 5′-phosphate. Despite the larger asymmetric unit the binding mode of ARTD2WGR to the nicked DNA is similar to the other structures. (F) Zoomed-in view of the ARTD2WGR bound to DNA break with five nucleotides overhang and 5′-phosphate.
The crystal structure of the WGR domain in complex with blunt end DNA with 5′-phosphate (DNA-5; 10 bp) was refined to 2.8 Å resolution. We observed that the DNAs were organized in an end-to-end fashion in the crystal and that WGR was bound to DNA at their contact point, interacting with DNA on both the 3′ and 5′ ends of the break. Due to this arrangement, the binding of a single WGR domain to the break mimics a nicked DNA binding model. In this case, there is a nick on both strands of the DNA due to joining of two blunt ends together. Asymmetric unit of the complex contained four DNA chains (2 dsDNAs) and two protein molecules (Figure 1A, Supplementary Figure S2A). Structural comparison of the two protein molecules and the DNA molecules in the asymmetric unit revealed no significant differences.
Crystal structure of the WGR domain bound to blunt end DNA without 5′-phosphate (DNA-6; 20 bp) was refined to 2.2 Å resolution. In this case the asymmetric unit contained only one protein molecule and one DNA chain. However, the DNA binding mode is very similar to the complex with the 5′-phosphate DNA due to crystal symmetry (Figure 1C, Supplementary Figure S3).
As the described models mimic nicked DNA due to joining of the DNA ends, we tried to break the DNA end-to-end packing by using an overhang 5′-phosphorylated DNA, which also activates the protein. We managed to grow diffracting crystals with one of our DNA oligonucleotides (DNA-9; 10 bp with a five base overhang) with the WGR domain and refined the structure to 2.98 Å resolution. The overhang indeed changed the packing of the molecules but resulted in a DNA break similar to the blunt end models (Figure 1E, Supplementary Figure S4). These three crystal structures allowed us to study the interactions of ARTD2 and DNA in detail.
Recognition of the 5′-phosphorylated DNA break
Compared to non-phosphorylated DNA, the activity of ARTD2 is 25–50-fold higher in the presence of phosphorylated DNA (12,13). Interestingly, the crystal structures with these two DNAs revealed no large differences in the secondary structure elements of the WGR domain and the observable differences between the two structures lie on the specific amino acid residues at the protein–DNA interface. 5′-phosphate is apparently recognized by a tyrosine residue and two lysine residues. The tyrosine interaction (Y201) is in line with the reported ARTD1 based homology model (12) and the positively charged K130 and K183 lie on both sides of the critical tyrosine (Figure 1B). In addition, K183 interacts with the DNA backbone in both DNA-complexes (Figure 1B, D), while Y201 and K130 do not interact directly with the DNA in case of non-phosphorylate DNA (DNA-6) (Figure 1D). WGR domain makes multiple similar interactions with the DNA on both sides of the break in both complexes (Figure 1B, D). K179, Y132, and K183 interact with the phosphate backbone of the DNA and W151 stacks with the ribose sugar at the 5′-end of the DNA. R153 forms hydrogen bonds with the 3′ end phosphate of n – 1 nucleotide and with N127. In addition, Q159 forms a hydrogen bond with the guanidine base and ribose of the preceding nucleotide of the complementary strand (Figure 1B, D).
ARTD2 interaction with 5′-overhang DNA
While it was previously reported that ARTD2 is preferentially activated by 5′-phosphorylated DNA (12,13), it was also reported that overhang DNA is able to activate ARTD2. However, the mechanism employed by ARTD2 in the interaction with 5′-overhang DNA is not known. First, we sought to break the end-to-end organization observed in blunt end DNA complexes by crystallizing WGR domain also in complex with a DNA with an overhang (DNA-9). Surprisingly, this DNA also forms a nick in the crystal structure, where the break is lined by two WGR domains. The overhang is turned away from the binding site and Y201 does not make an interaction with the terminal phosphate group but with the phosphate backbone of the second nucleotide from the 5′ end (Figure 1F). DNA with an overhang is not an especially good activator of ARTD2 and the difference in the enzyme activity in the presence of phosphorylated DNA or non-phosphorylated DNA was not large (Figure 2A) (13). Therefore, direct interaction with the 5′-phosphate group could indeed be missing also in solution. The structure of the overhang DNA-WGR domain complex is strikingly similar with the blunt end complexes demonstrating that the WGR domains individually bind to the DNA breaks on both strands. Notably, the overhangs are not complementary, but despite this, ARTD2 mediates joining of the DNA ends in the crystal leading to distorted non-complementary pairing of the bases (Figure 1E, F, Supplementary Figure S5).
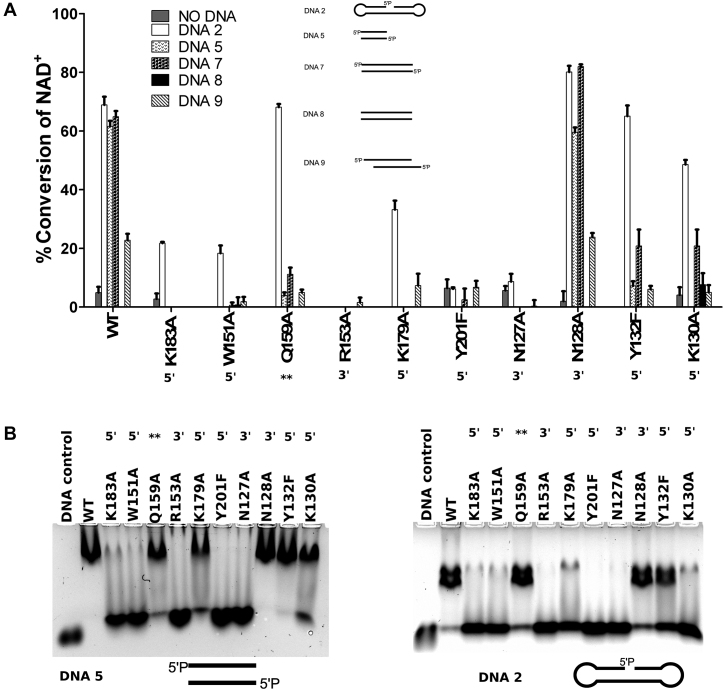
Figure 2.
DNA interaction and activation of ARTD2FL and the different point mutants. (A) The specific DNA dependent activity of full length ARTD2FL and point mutants were measured using fluorescence activity assay. Protein (50 nM) was incubated with 5 μM NAD+ for 1 hour and leftover NAD+ was quantified. DNA was used in 2:1 protein:DNA ratio. Data are presented as means and standard deviation of the quadruplicates. Each DNA was assayed with all the mutants and data is shown for the ones having any NAD+ consumption. (B) Interaction of ARTD2FL and the different point mutants were studied using EMSA. Residues on the 3′ side of the break are labeled with 3′, residues on the 5′ side of the break are labeled with 5′ and residue interacting with a nucleobase is labelled with **.
The role of ARTD2 in the detection of double-strand DNA breaks
In the three structures, there was no distinct WGR-WGR interface and the position of the WGR domain on the DNA is determined by the breaks in the DNA strands. While the organization mimicked nicked DNA binding it also raised a question on whether ARTD2 can bring two DNA ends together in solution. We performed an ITC experiment, which showed 1:1 stoichiometry when hairpin nicked DNA (DNA-1) and dumbbell blunt end DNA (DNA-3) were used (Supplementary Figure S6). The 1:1 stoichiometry observed in the titration with dumbbell DNA is in line with the crystal structures as we expected complex formation by two proteins and two DNA molecules also in solution. In order to support this observation, SEC-SLS experiment was done with the wild type ARTD2WGR+CAT and ARTD2FL together with their complexes with nicked DNA and blunt end DNA. SLS showed that apo ARTD2WGR+CAT and ARTD2FL are monomeric in solution and form stable 1:1 complexes with nicked DNA (Supplementary Table S2). Notably, they can also form some additional higher oligomers with nicked hairpin DNA (Supplementary Figure S8A). In addition, SLS showed that ARTD2WGR+CAT and double-strand DNA with phosphate only at one end (DNA-13) forms a stable 2:2 complex (Supplementary Table S2, Supplementary Figure S7c) and similar behavior was observed for the ARTD2FL (Supplementary Table S2, Supplementary Figure S8A). We also analyzed DNA-7 (20 bp with phosphate at the both 5′ end) with SEC-SLS and several heterogeneous higher oligomeric states were observed (molecular masses were not determined, Supplementary Figure S7D). Taken together, this suggested continuous end-to-end DNA binding by ARTD2 when the DNA is sufficiently long (Supplementary Table S2, Supplementary Figures S7, S9).
WGR domain has a central role in DNA binding and DNA-dependent activation
In order to understand the role of the WGR domain in DNA detection and to study the activation mechanism, we mutated residues of ARTD2FL which formed direct contacts with DNA based on the crystal structures. The wild type enzyme was preferentially activated by 5′-phosphorylated DNA while non-phosphorylated DNA resulted in little or no activation (Figure 2A) as was also previously shown (12,13). All mutations decreased enzymatic activity with the exception of N128A (Figure 2A), which retained wild type –like activity. N128 interacts with a phosphate at the 3′-side of the DNA break. Y201F mutation, which abolishes a hydrogen bond with the 5′-phosphate, had essentially no DNA induced activation and is in agreement with a previous study (12). K130 also coordinates the interaction with 5′-phosphate but K130A was still active, although it had lower activation in case of the DSB models. Residues interacting with the DNA on the 5′-phosphate side of the break (Figure 1B, D, F), Y132A, K183A and K179A had a drastic impact on the enzymatic activity in case of DSB model with or without an overhang, but were still activated by the nick DNA model (Figure 2a). Mutation of residues interacting with 3′-side of the nick (Figure 1B, D, F), R153A and N127A, had negative effects on the activity. R153A and N127A were essentially inactive indicating that their interaction at the 3′-end of the DNA is as important as the interaction at the 5′-end. Also, the interaction at the 3′-end could be vital in mediating the contacts between the DNA, the WGR and possibly the catalytic domain. Mutation of the highly conserved tryptophan of the WGR motif among the DNA dependent ARTDs, W151A, did not entirely inhibit activation by the nick DNA. Q159A mutation showed full activation in case of nick DNA, whereas the enzyme was almost inactive in case of the blunt end models. This finding is intriguing as Q159 does not interact with the nicked DNA strand, but with the complementary strand (Figure 1B, E), which in case of the activity assay does not have a nick.
In order to understand whether the observed changes in activation were due to disruption of the enzymatic activation or due to disruption of DNA binding, we performed electrophoretic mobility shift assays (EMSA). DNA supershift was observed with the wild type enzyme indicating specific binding, whereas the inactivating point mutations K183A, W151A, R153A, Y201F and N127A resulted in a loss of a stable ARTD2 DNA complex (Figure 2B). The pattern of the EMSA is similar in case of a blunt end and nick DNA with some changes in the band intensities. Pattern observed for the nick DNA matches well with the activity data (Figure 2A) with some exceptions. Although K130A and K179A have lower DNA binding in EMSA at 300 mM salt, but were still active in case of a nick DNA at 150 mM salt. Also Q159A and K179A mutants bind to blunt end DNA in EMSA, but showed little if any DNA dependent activation with same blunt end DNA. In order to test the complex formation of the mutants we carried out also SEC-SLS experiments. K179A mutant does not form a DNA-complex at a measurable level according to SLS, while the Q159A showed impaired end-to-end joining as both 2:2 and 1:1 complexes form with blunt end DNA (Supplementary Table S2). This indicates a role of Q159 in end-joining by forming an interaction over the DSB with a base on the complementary strand (Figure 1).
Specificity of ARTD2–DNA interaction
The affinities of ARTD2FL, ARTD2WGR+CAT and ARTD2FL point mutants were first measured using fluorescence polarization assay (FP) with a fluorescein-labelled 5′-phosphorylated blunt end DNA (Supplementary Figure S1 and Supplementary Table S1). Previously, we showed that N-terminus of ARTD2 interacts tightly with DNA (13), and as a result point mutations in the WGR domain do not affect the overall affinity of the protein–DNA-interaction (Table 2). Due to the unspecific binding via the N-terminus, the KD of ARTD2FL and the point mutants were within same range (∼50 nM; 100 mM NaCl). On the contrary, the KD of ARTD2WGR+CAT was significantly higher (5 μM), highlighting the high affinity and non-specificity of the N-terminus to DNA as earlier reported (13). When we repeated the measurements at higher salt concentration for the WT and selected mutated enzymes, the affinities measured were significantly lower at the μM range, but the affinities were all again at the same level (Table 2). This indicates that the DNA–ARTDN interaction is primarily ionic in agreement with the overall positive net charge based on the protein sequence. When salt concentration was increased even more to 300 mM the binding was very weak with estimated affinities at 100 μM range (data not shown).
Table 2.
Binding of ARTD2 to DNA breaks measured from using SPR and FP
| SPR DNA-12 (5′-P) 300 mM NaCl | FP DNA-10 100 mM NaCl | FP DNA-10 150 mM NaCl | ||||||
|---|---|---|---|---|---|---|---|---|
| Proteins | K D (nM) | k a1 (M−1 s−1) | k d1 (s−1) | k a2 (s−1) | k d2 (s−1) | chi2 | K D ± SE (nM) | K D ± SE (nM) |
| WT | 83 | 4.2 × 104 | 0.008 | 0.002 | 0.002 | 2.76 | 41 ± 4.6 | 870 ± 200 |
| R153A | NB | 76 ± 14 | 1900 ± 560 | |||||
| W151A | NB | 66 ± 14 | 1100 ± 340 | |||||
| N127A | NB | 54 ± 6.1 | ND | |||||
| Q159A | 8100 | 1.4 × 103 | 0.033 | 0.002 | 0.001 | 0.07 | 65 ± 15 | 770 ± 260 |
| K183A | NB | 31 ± 7.1 | 2200 ± 630 | |||||
| Y132F | 250 | 7.9 × 104 | 0.02 | 0.007 | 0.001 | 0.06 | 40 ± 5.1 | ND |
| N128A | 88 | 1.3 × 105 | 0.04 | 0.002 | 0.001 | 0.51 | 77 ± 16 | ND |
| Y201F | NB | 70 ± 16 | 890 ± 260 | |||||
| K179A | NB | 62 ± 17 | ND | |||||
| WGRCAT | 940 | 7.2 × 103 | 0.01 | 0.001 | 0.001 | 5000 ± 2400 | >8000 | |
| WT DNA-11 (no 5′-P) | 66 | 5.1 × 104 | 0.015 | 0.005 | 0.001 | 0.06 | ||
NB, no binding detected or signal too low for analysis. ND, not determined.
In order to study specific DNA binding mediated by the WGR domain, we used surface plasmon resonance (SPR) to measure association and dissociation rates of ARTD2FL and ARTD2WGR+CAT using 20-base pair DNA oligonucleotides with or without 5′ -phosphate. DNA was attached to the SPR chip using biotin (Supplementary Table S1 and Supplementary Figure S1) and the measurement was carried out at 300 mM NaCl concentration. The measured KD values are different from the FP measurements, which may be due to a different experimental setup (Table 2). ARTD2FL associates faster with the DNA compared to ARTD2WGR+CAT, and also dissociates slower resulting in a 10-fold lower affinity with the removal of the N-terminus in agreement with the FP data (Table 2, Supplementary Figure S10). The majority of the point mutants completely lost specific interaction with DNA (Table S2, Supplementary Figure S11). Three mutants had measurable binding kinetics. N128A had the same affinity as the wild type enzyme but had both faster association and faster dissociation rates. Y132F results in the disruption of one hydrogen bond with the phosphate backbone and demonstrated slower association and faster dissociation rates and subsequently three times lower affinity. Q159A, which interacts with the complementary strand, had the same affinity as the wild type but the binding kinetics differed as it had much slower association rate and faster dissociation rate. Importantly, Q159A was active only in the case of a nick, and not in case of a blunt end DNA which was used in the SPR. Q159 interacts with a base of the complementary DNA strand (Figure 1B, D) and this nucleotide is missing in the DNA coupled to the SPR chip. Therefore, it is surprising that this residue and the ones on the 3′-side of the nick affect DNA binding. Taken together, Q159 is not strictly required in ARTD2 nicked DNA binding but our finding suggest that the residue might be playing a role in the enzyme localization on blunt end DNA as depicted by the direct interactions seen in the crystal structure.
Distinct DNA binding mode of ARTD2
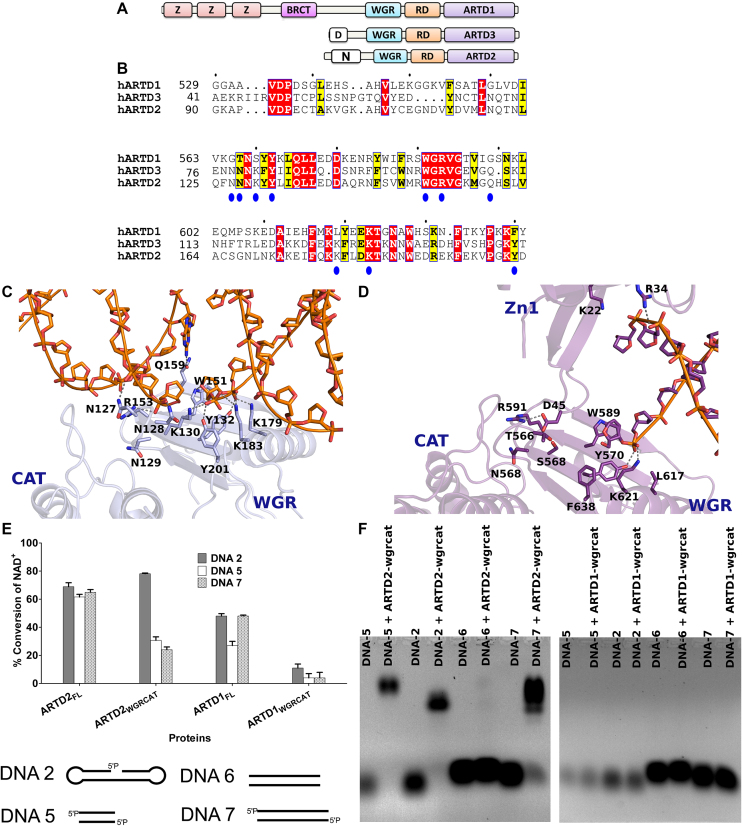
The partial structure of ARTD1 revealed how the zinc finger domains and the WGR domain coordinate blunt end DNA binding (23). In contrast to ARTD1 structure, there are no zinc fingers in ARTD2 (Figure 3A) and we have shown here that ARTD2 utilizes the WGR domain when binding to DNA breaks (Figure 1). WGR domains of ARTD1, ARTD2 and ARTD3 are conserved although there are key amino acid substitutions relevant to the present study (Figure 3B). N127, N128, K130, Q159, K179 and Y201 (ARTD2 numbering) are conserved in ARTD2 and ARTD3 but not in ARTD1. In line with a previous study based on homology model (12), mutation of Y201F to mimic ARTD1 results in a loss of DNA dependent activation and loss of specific DNA binding (Figure 2). N127, which is a glycine in ARTD1, is a crucial residue for DNA dependent activation of ARTD2, while Q159, which is also a glycine in ARTD1, is partially important in the activation of ARTD2 by a blunt end DNA. Based on the structure of ARTD1 in complex with DNA it was shown that R591 of the WGR and D45 of the zinc finger are needed for the domain-domain interaction (Figure 3D). In ARTD2 the corresponding arginine, R153, is mediating directly the interaction between the DNA phosphate backbone and N127 of the WGR domain (Figure 3C).
Figure 3.
Schematic, sequence alignment and structural/functional comparison of ARTD1 and ARTD2. (A) Domain organization of DNA dependent ARTDs. (B) Protein sequence alignment of ARTD1WGR, ARTD2WGR and ARTD3WGR. Identical residues are highlighted in red and similar residues in yellow. (C) Structural representation of ARTD2WGR bound to DNA break with 5′-phosphate (blue). The catalytic domain (CAT) (PDB ID 4PJV) was positioned based on ARTD1 structure seen in panel D. (D) Structural representation of ARTD1 bound to DNA break (purple) (PDB ID 4DQY). WGR, C-terminal catalytic domain (CAT) and Zinc finger (Zn1) are labelled. The superimposed model of ARTD1 and ARTD2 is shown in Supplementary Figure S11. (E) The specific DNA dependent activity of ARTD2FL, ARTD2WGR+CAT, ARTD1FL, ARTD1WGR+CAT were measured using fluorescence activity assay as described in the materials and methods. ARTD2FL at 50 nM was incubated for 1 h, ARTD2WGR+CAT and ARTD1WGR+CAT at 50 nM were incubated for 2 h, and ARTD1FL at 25 nM was incubated for 2 h. The NAD+ concentration in all the experiments was 5 μM. Data are presented as means and standard deviation of the quadruplicates. (F) The interaction of ARTD2WGR+CAT and ARTD1WGR+CAT with DNA were studied using EMSA.
DNA-dependent catalytic activation of ARTD1 depends on the cooperation of amino acid residues located in the zinc finger, WGR, and regulatory domains (Figure 3D). Similar to ARTD1, in our ARTD2-DNA complex structures we observed that R153 mediates an important interaction between the WGR domain and DNA. It interacts with N127, which was also observed for the first time to be a key player in the DNA-dependent activity of the enzyme. Subsequently, we made a homology model to find the position of ARTD2 catalytic domain based on ARTD1 structure and it seems that the interaction by R153 with DNA could be mimicking the interaction of a similar residue in ARTD1 (R591) with Zn1 (D45). The position of ARTD2 R153 suggests that the interaction could be extended to the regulatory domain of the catalytic fragment (RD) of the catalytic fragment in a similar way as in ARTD1 (Figure 3C, D, Supplementary Figure S12). In line with the previous study (12), N129 can also participate in mediating the interaction with the catalytic domain (Figure 3C). Also, based on our previous SAXS study (13), it is possible that the RD domain of ARTD2 can also participate in DNA binding. The position of the ARTD2 catalytic domain based on ARTD2 WGR-DNA complex and ARTD1 structure (PDB code 4DQY) showed a very close proximity of the RD domain to DNA. Specifically, the loop within amino acid 312 and 316 could be involved in the DNA binding of the RD domain (Figure 3C). Taken together, this suggests a potential difference in ARTD2 and ARTD1 DNA binding. While ARTD1 uses different domains in DNA binding, ARTD2 utilizes mainly the WGR domain and possibly the RD domain.
It has been shown that the N-terminal domains of ARTD1 are crucial for its DNA binding and activation (24), but in order to directly compare the ARTD1WGR+CAT and ARTD2 WGR+CAT we tested the activation of ARTD1FL, ARTD1WGR+CAT, ARTD2FL and ARTD2WGR+CAT using nicked DNA and blunt end DNA of different lengths. In our previous study, we have shown ARTD2WGR+CAT is capable of DNA dependent activation and in line with that, ARTD2WGR+CAT was shown here to catalyze DNA dependent NAD+ hydrolysis in a similar way as the full length enzyme (Figure 3E). In contrast, ARTD1WGR+CAT showed only little DNA dependent NAD+ hydrolysis compared to the full-length enzyme (Figure 3E). In addition, ARTD1WGR+CAT was not capable of forming a stable complex with DNA with or without 5′-phosphate and did not result in a band shift on the gel in contrast to ARTD2, highlighting a distinct and central role of ARTD2 WGR domain in DNA damage recognition (Figure 3F).
DISCUSSION
ARTD2 is a DNA repair enzyme and its catalytic activity is highly elevated in response to cellular genotoxic stress. Here we described the structural mechanism of DNA damage detection of ARTD2. A previous study proposed that ARTD1, ARTD2 and ARTD3 might share a similar activation mechanism with distinct DNA damage detection mechanisms due to the differences in their domain organization (12). Compared with ARTD1, the in vitro activity of ARTD2 is rapidly elevated only in the presence of 5′-phosphorylated DNA but not DNA without the 5′-phosphate (12,13). We elucidated the structure of ARTD2 WGR domain bound to three different DNAs: DNA with 5′-phosphate, DNA without 5′-phosphate, and DNA with a five nucleotide overhang with a 5′-phosphate. Interestingly, our high resolution structures revealed that ARTD2 can bring two DNA ends together in vitro creating a nick shielded by the WGR domain. The structures also highlight the differences to the reported DNA damage detection of ARTD1 (23). We have shown that DNA in all crystal structures interacts in an end-to-end manner with two WGR monomers in opposite direction on both sides of the formed nick. While this most likely explains the high preference of ARTD2 toward nicked DNA (10) it is also possible that ARTD2 can utilize this feature in mediating DSBR. In the event of DSBR, this newly discovered feature of ARTD2 is in agreement with the recent report on the role of enzyme in DSBR (25). Previous study that compared activation of ARTD2 with different DNAs without 5′-phosphate, showed that 5′-overhang DNA was a more efficient activator of ARTD2 than blunt end DNA (26). Our structure showed that in the overhang DNA, ARTD2 residues Y201, K183 were not making interactions with the 5′-phosphate rather with the phosphate backbone (Figure 1F, Supplementary Figure S5) explaining why phosphorylation of an overhang does not really enhance the activation of the enzyme (13). It has been reported that addition of a gap to the DNA damage model decreased the activation of ARTD2 in comparison to a nick (12). The crystal structures have now demonstrated that WGR domain makes interactions also on the 3′-side of the nick and lack of those interactions in the gap could provide an explanation for the ARTD2 preference for the nicked DNA.
The crystal structures showed that ARTD2 can detect DNA breaks and cooperatively coordinate end-to-end interaction of two DNA ends in vitro. Structures suggest possible stoichiometries of ARTD2 upon detecting SSB and DSB. In the case of SSB ARTD2 appears to detect the damage site and form a 1:1 protein:DNA complex while in the case of DSBs it forms a 2:2 protein:DNA complex (Supplementary Figure S9). In agreement with this, our isothermal calorimetry studies show that ARTD2 binds to nicked or blunt end DNA with a 1:1 stoichiometry. The 1:1 stoichiometry observed when blunt DNA was used could be related to the 2:2 protein–DNA complex as observed in the crystal structures. Similarly, using SEC-SLS, we observed that ARTD2 can mediate continuous end-to-end joining of longer DNAs resulting in the formation of higher order oligomers (Supplementary Figures S7, S9). This is in agreement with our previous study where we showed that ARTD2 can form different oligomeric states with DNA (13). In order to control the formation of higher oligomers, we used a double-strand DNA oligonucleotide (DNA-13) with phosphate at one of the 5′ends to determine the molecular mass of the complex in solution. As expected the SEC-SLS showed molecular masses fitting for two molecules of ARTD2 and two dsDNAs. The mass determined with nick DNA complex was in agreement with one ARTD2 and one hairpin nicked DNA. The analysis indicates that while ARTD2 is preferentially activated by nicked DNA, it can potentially also participate and in some way mediate DSB detection and repair.
In vivo, the N-terminus of ARTD2 is required for nuclear localization but not required for the localization of the enzyme to the DNA damage site (27). Previously, we reported that ARTD2 N-terminus non-specifically interacts with DNA (13). FP measurements revealed that the deletion of N-terminus drastically lowers DNA-binding affinity, while any construct with an intact N-terminus has an equally high affinity to all DNA models. However, SPR analysis demonstrated that the full-length enzyme associates faster with DNA and dissociates slower than the N-terminal truncation mutant (Table 2). Notably, the binding kinetics of ARTD2 with phosphorylated and nonphosphorylated DNA demonstrate the key role of the 5′-phosphorylation in the enzyme DNA binding and activation (Table 2 and Supplementary Figure S10). Similarly, we observed stable binding to the DNAs only in the presence of 5′-phosphate with EMSA (Figure 3F).
In ARTD1 different domains cooperatively coordinate DNA binding (23). ARTD2, on the other hand, relies on the WGR domain in the specific DNA damage detection (Figures 1 and 3C) although there is some cooperativity with the catalytic fragment (13). In agreement, mutations of the key residues of the WGR domain prevented stable association of the enzyme with DNA (Figure 2 and Supplementary Figure S11). Some of these residues (Y201 and W151) were previously predicted based on ARTD1 structure (12). Y201 directly interacts with the 5′-phosphate and a mutation of this residue to phenylalanine yielded an inactive enzyme with no ability to specifically bind to the 5′-phosphorylated DNA. While R153 is well-conserved in ARTD1 (R591), ARTD2 and ARTD3 (R103), it was not known how this residue in ARTD2 participates in DNA binding. In ARTD1 it mediates domain-domain contact between the WGR and zinc finger 1 domains (23). Other previously unidentified residues important for DNA binding were N127, K130, K179 and Q159.
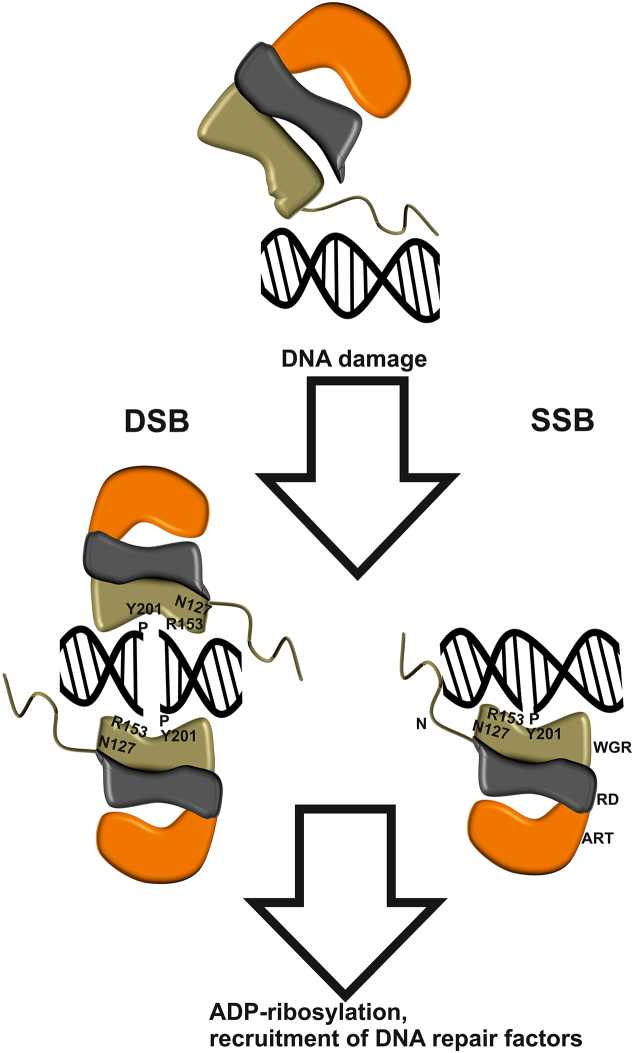
R153 interacts with DNA and N127, and based on the structural comparison of ARTD2 and ARTD1 (Figure 3 and Supplementary Figure S12), these residues could be essential for the domain-domain contacts between WGR and catalytic domain and may act together as a switch to turn on the catalytic activity. R153A and N127A mutants do not form stable DNA complexes and these mutations disrupted the activity of the enzyme in the presence of nicked DNA (mimicking SSB) and blunt end DNA (mimicking DSB). Therefore, the crystal structures and modelling together with biophysical and biochemical data allowed us to draw a simplified diagram on DNA damage detection and activation of ARTD2 (Figure 4). Without DNA damage, the interaction between ARTD2 and DNA is mainly facilitated by the disordered N-terminus, which potentially allows ARTD2 to scan the DNA for damage. Upon binding to the DNA damage, ARTD2 becomes less flexible based on our previous SAXS data and forms a tight complex with DNA (13). In the case of SSB, ARTD2 recognizes the site of the damage and R153 and N127 of the WGR domain and also N129 (12) could, based on ARTD1 homology, participate in transmitting the activation signal to the catalytic domain through domain-domain contact while also interacting with the 3′-end of the nicked DNA. In the case of DSB, the observed binding mode is the same, but two ARTD2 enzymes will be required at the DNA break. This complex can be formed with or without the 5′-phosphate group, but only in the former case it leads to an efficient complex formation and poly-ADP-ribosylation.
Figure 4.
Schematic representation of the mechanism of ARTD2 DNA damage detection. In the absence of DNA damage, ARTD2 is capable of unspecific DNA interaction through the disordered N-terminus (N). WGR domain detects a damage site and locks the enzyme to DNA. If the DNA contains a 5′-phosphate ADP-ribosylation is induced leading to a cascade of events and initiation of the repair. In case of DSB ARTD2 is capable of joining the DNA-ends. Key residues for 5′-phosphate detection (Y201) and 3′-detection and possibly domain contacts (R153, N127) are labelled.
Finally, ARTD2 depletion has been reported to sensitize cells to alkylating agents and ionizing irradiation (5), implicating a possible role of the enzyme in SSBs repair. ARTD2 is also known to interact with various DNA repair factors that are involved in SSB repair (5). We proposed earlier that ARTD2 would dimerize at the DNA end (13) and act as a monomer in case of a nick. Based on the new high resolution structural data this model could still be valid although ARTD2 also appears to facilitate DNA end joining. ARTD2 has also been implicated in DSB repair through homologous recombination and non-homologous end joining (25,28). The biochemical and structural data presented here provide possible mechanism for ARTD2 to function in both SSB repair and in DSB repair and potentially participate in the end joining process of the latter. Recently it was discovered that ARTD2 has ADP-ribosylation independent roles in DSB repair pathway selections (25) and it is an intriguing possibility that the end-joining we observed in vitro would contribute to the repair pathway selection in cells. In case of ARTD2 binding to phosphorylated DNA, a cascade of DNA repair is initiated through the enzymatic labelling of the site with poly-ADP-ribose. It could be that ARTD2 activity is only switched on when the damage site is processed by endonucleases producing a 5′-phosphorylated DNA end (29). On the other hand, in the case of overhang DNA interaction of Y201 with the phosphate backbone instead of phosphate at the DNA 5′-end appears to promote enzymatic activation. The structural and functional studies of DNA damage recognition presented here provide a molecular basis for studying the ARTD2 in the context of DNA repair.
DATA AVAILABILITY
Atomic coordinates and structure factors of the WGR–DNA complexes have been deposited to Protein Data Bank under accession numbers 6F1K, 6F5B and 6F5F.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Local Contacts at DLS for providing assistance in using the beamlines. The use of the facilities and expertise of the Biocenter Oulu Protein Crystallography core facility, a member of Biocenter Finland and Instruct-FI, is gratefully acknowledged. We want to thank also Dr Hongmin Tu of the Biocenter Oulu Proteomics and Protein analysis core facility for support with biophysical measurements.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academy of Finland [287063, 294085 to L.L., 266922 to T.H.]; Jane and Aatos Erkko Foundation; Orion Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Schreiber V., Dantzer F., Amé J.-C., de Murcia G.. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006; 7:517–528. [DOI] [PubMed] [Google Scholar]
- 2. Javle M., Curtin N.J.. The role of PARP in DNA repair and its therapeutic exploitation. Br. J. Cancer. 2011; 105:1114–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amé J.-C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Höger T., Murcia J.M., de Murcia G.. PARP-2, A novel mammalian DNA Damage-dependent Poly(ADP-ribose) polymerase. J. Biol. Chem. 1999; 274:17860–17868. [DOI] [PubMed] [Google Scholar]
- 4. Lord C.J., Ashworth A.. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017; 355:1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schreiber V., Amé J.-C., Dollé P., Schultz I., Rinaldi B., Fraulob V., Ménissier-de Murcia J., de Murcia G.. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002; 277:23028–23036. [DOI] [PubMed] [Google Scholar]
- 6. Hanzlikova H., Gittens W., Krejcikova K., Zeng Z., Caldecott K.W.. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017; 45:2546–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dantzer F., Mark M., Quenet D., Scherthan H., Huber A., Liebe B., Monaco L., Chicheportiche A., Sassone-Corsi P., De M. et al. . Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:14854–14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer-Ficca M.L., Scherthan H., Bürkle A., Meyer R.G.. Poly(ADP-ribosyl)ation during chromatin remodeling steps in rat spermiogenesis. Chromosoma. 2005; 114:67–74. [DOI] [PubMed] [Google Scholar]
- 9. Farrés J., Martín-Caballero J., Martínez C., Lozano J.J., Llacuna L., Ampurdanés C., Ruiz-Herguido C., Dantzer F., Schreiber V., Villunger A. et al. . Parp-2 is required to maintain hematopoiesis following sublethal γ-irradiation in mice. Blood. 2013; 122:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sukhanova M.V., Abrakhi S., Joshi V., Pastre D., Kutuzov M.M., Anarbaev R.O., Curmi P.A., Hamon L., Lavrik O.I.. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Res. 2016; 44:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kutuzov M.M., Khodyreva S.N., Ilina E.S., Sukhanova M.V., Amé J.-C., Lavrik O.I.. Interaction of PARP-2 with AP site containing DNA. Biochimie. 2015; 112:10–19. [DOI] [PubMed] [Google Scholar]
- 12. Langelier M.-F., Riccio A.A., Pascal J.M.. PARP-2 and PARP-3 are selectively activated by 5′ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1. Nucleic Acids Res. 2014; 42:7762–7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Obaji E., Haikarainen T., Lehtiö L.. Characterization of the DNA dependent activation of human ARTD2/PARP2. Sci. Rep. 2016; 6:34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grundy G.J., Polo L.M., Zeng Z., Rulten S.L., Hoch N.C., Paomephan P., Xu Y., Sweet S.M., Thorne A.W., Oliver A.W. et al. . PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2BGlu2. Nat. Commun. 2016; 7:12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Narwal M., Fallarero A., Vuorela P., Lehtiö L.. Homogeneous screening assay for human tankyrase. J. Biomol. Screen. 2012; 17:593–604. [DOI] [PubMed] [Google Scholar]
- 16. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arnold K., Bordoli L., Kopp J., Schwede T.. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006; 22:195–201. [DOI] [PubMed] [Google Scholar]
- 20. Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 2008; 41:641–643. [Google Scholar]
- 21. Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The PyMOL Molecular Graphic System Schrödinger, LLC.
- 23. Langelier M.-F., Planck J.L., Roy S., Pascal J.M.. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012; 336:728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ali A.A.E., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P.O., Hassler M., Ladurner A.G., Pearl L.H., Oliver A.W.. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012; 19:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fouquin A., Guirouilh-Barbat J., Lopez B., Hall J., Amor-Guéret M., Pennaneach V.. PARP2 controls double-strand break repair pathway choice by limiting 53BP1 accumulation at DNA damage sites and promoting end-resection. Nucleic Acids Res. 2017; 45:12325–12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kutuzov M.M., Khodyreva S.N., Amé J.-C., Ilina E.S., Sukhanova M.V., Schreiber V., Lavrik O.I.. Interaction of PARP-2 with DNA structures mimicking DNA repair intermediates and consequences on activity of base excision repair proteins. Biochimie. 2013; 95:1208–1215. [DOI] [PubMed] [Google Scholar]
- 27. Riccio A.A., Cingolani G., Pascal J.M.. PARP-2 domain requirements for DNA damage-dependent activation and localization to sites of DNA damage. Nucleic Acids Res. 2016; 44:1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ghosh R., Roy S., Kamyab J., Danzter F., Franco S.. Common and unique genetic interactions of the poly(ADP-ribose) polymerases PARP1 and PARP2 with DNA double-strand break repair pathways. DNA Repair (Amst.). 2016; 45:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nishino T., Morikawa K.. Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene. 2002; 21:1206135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors of the WGR–DNA complexes have been deposited to Protein Data Bank under accession numbers 6F1K, 6F5B and 6F5F.