Abstract
The active 3D conformation of the spliceosome's catalytic U2/U6 RNA core is stabilised by a network of secondary and tertiary RNA interactions, but also depends on spliceosomal proteins for its formation. To determine the contribution towards splicing of specific RNA secondary and tertiary interactions in the U2/U6 RNA core, we introduced mutations in critical U6 nucleotides and tested their effect on splicing using a yeast in vitro U6 depletion/complementation system. Elimination of selected RNA tertiary interactions involving the U6 catalytic triad, or deletions of the bases of U6-U80 or U6-A59, had moderate to no effect on splicing, showing that the affected secondary and tertiary interactions are not required for splicing catalysis. However, removal of the base of U6-G60 of the catalytic triad completely blocked splicing, without affecting assembly of the activated spliceosome or its subsequent conversion into a B*-like complex. Our data suggest that the catalytic configuration of the RNA core that allows catalytic metal M1 binding can be maintained by Protein–RNA contacts. However, RNA stacking interactions in the U2/U6 RNA core are required for productive coordination of metal M2. The functional conformation of the U2/U6 RNA core is thus highly buffered, with overlapping contributions from RNA–RNA and Protein–RNA interactions.
INTRODUCTION
Nuclear pre-mRNA splicing is catalysed by the spliceosome and proceeds via two sequential transesterification reactions. In step 1 the 2′-OH of the pre-mRNA branch site (BS) adenosine attacks the phosphodiester bond at the 5′ splice site (ss) to produce the cleaved 5′ exon and the branched intron lariat-3′ exon. In step 2, the 3′-OH group of the 5′ exon attacks the phosphodiester bond at the 3′ ss leading to exon ligation and excision of the intron lariat. The spliceosome is a highly dynamic molecular machine that undergoes multiple structural and compositional rearrangements, including extensive changes in its RNA–RNA network, that are triggered by at least eight conserved DEAH/D-box ATPases or RNA helicases (1,2).
Spliceosomes assemble de novo for each round of splicing by the ordered interaction of five snRNPs and numerous splicing factors with the pre-mRNA (1). Initially, U1 and U2 snRNP interact with the 5′ ss and the BS, respectively, yielding the A complex. Then the preformed U4/U6.U5 tri-snRNP, in which the U4 and U6 snRNAs are base paired, binds, leading to the pre-B complex. A short helix between the 3′ end of U6 and 5′ end of U2 snRNA (U2/U6 helix II) is formed at this stage. After stable tri-snRNP integration, the B complex is generated. In the latter the 5′ ss has been transferred from U1 to the U6 snRNA ACAGA box by the action of the Prp28 RNA helicase (3). Spliceosome activation is then initiated by the Brr2 helicase, which unwinds the U4/U6 duplex, displacing U4 from the spliceosome. During activation, U4/U6 proteins are released, while many other proteins are newly recruited or stably integrated, including the Prp19 complex (NTC), yielding the Bact complex. In the latter, the U6 snRNA has undergone major rearrangements, leading to formation of the U6 internal stem-loop (ISL), and new base pairing interactions with U2 that generate U2/U6 helices Ia and Ib (1,2). Collectively, these structures form the catalytic U2/U6 RNA core of the spliceosome. Subsequent catalytic activation by the Prp2 RNA helicase (yielding the B* complex) is a prerequisite for the branching reaction (i.e. step 1), which also requires the splicing factors Cwc25 and Yju2, and generates the C complex. Rearrangements by the Prp16 RNA helicase lead to the removal of the branched step 1 product from the catalytic centre, forming the C* complex, which is activated for the second catalytic step. Following recruitment of the step 2 factors Slu7, Prp18, and Prp22, exon ligation occurs, yielding the P complex. The ligated exons are then released by the action of Prp22 and the resulting intron lariat spliceosome (ILS) is then dismantled (1).
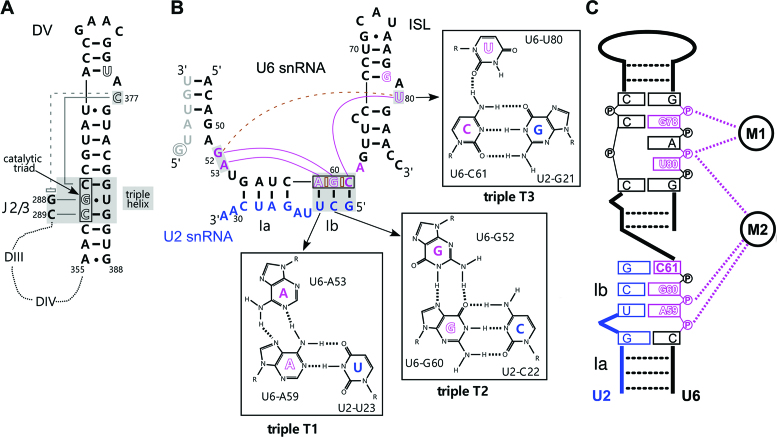
Nuclear pre-mRNA splicing and the autocatalytic excision of group II introns follow the same chemical pathway. Biochemical and structural studies of group II introns revealed that domain V (DV, Figure 1A) is the most functionally important region of group II introns and, together with nucleotides of the junction between domains 2 and 3 (J2/3), forms the reactive centre of the intron (4). Three conserved nucleotides in the lower DV stem (denoted the catalytic triad) together with nucleotides in the upper part of the stem, including a conserved bulged nucleotide, bind via their 5′ phosphate groups two divalent cations that play a key role in splicing catalysis. Base-triple interactions between the CGC catalytic triad and the bulged DV nucleotide or nucleotides of the J2/3 linker lead to the formation of an RNA triple helix, which is additionally stabilized by base stacking interactions. By supporting the 3D conformation of DV, this triple helix (also called catalytic triplex) aids in positioning the two catalytic metals within the 4 Å distance that is required for catalysis (4–6).
Figure 1.
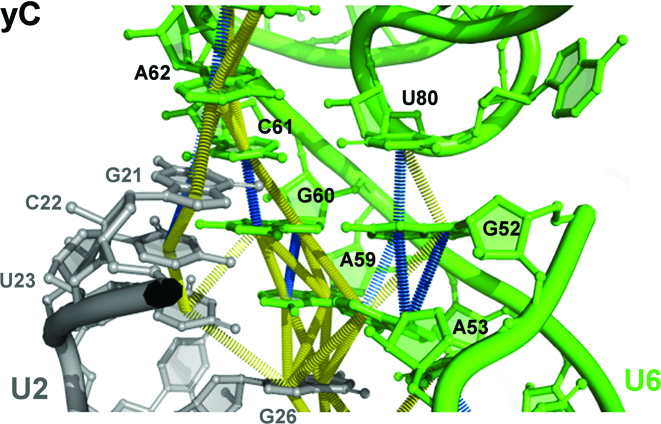
Structure of the core RNA–RNA network in group IIC introns and catalytically-active yeast spliceosomes. (A) Schematic diagram of the RNA core of a group IIC intron (6) highlighting domain V (DV) and the triple interations of the bulged C and the J2/3 linker. Catalytic triad nucleotides are boxed and nucleotides forming the triple helix are indicated by gray shading. Nucleotides coordinating the catalytic metals are shown in white, with metal M1 coordination by C377 and U375, and M2 by C358 and G359. The dashed line with an open rectangle indicates a stacking interaction. The dotted lines indicate the highly folded intron domains III and IV. (B) RNA–RNA network in the catalytically-active yeast spliceosome. The U6 and U2 snRNAs are colored black and blue, respectively, and intron nucleotides near the 5′ ss (G+1 is circled) are grey. Hoogsteen interactions are indicated by purple lines and stacking interactions in the catalytic triad AGC (boxed) are indicated by open brown bricks. Nucleotides forming the triple helix are indicated by gray shading. Dashed line indicates the base stacking interaction between U80 and G52. Metal binding nucleotides are white with purple borders. (C) Schematic diagram of metal M1 and M2 coordination by the phosphate backbone of nucleotides of the U6 ISL and catalytic triad.
Functional and structural studies have revealed that a homologous core structure is formed by the U2/U6 RNA core in the spliceosome (Figure 1B), with U6 playing a key catalytic role. Initially, biochemical studies in yeast demonstrated that pre-mRNA splicing is catalysed by the U6 snRNA (7), and that the chemistry involves metal coordination by U6 nucleotides G78/U80 and A59/G60 (Figure 1C) (7,8). Further, yeast genetic studies (9) and biochemical studies in the human spliceosome (10) showed that a catalytic triplex is formed in the U2/U6 RNA core. Recent cryo-EM studies confirmed that a conserved, U2/U6 RNA core structure with a catalytic triplex is present in yeast and human Bact (11–14), C (15–17) and C* spliceosomal complexes (18–21), as well as the yeast P complex (22–24) and the ILS (25,26). The spliceosomal triple helix is composed of the three base triples (Figure 1B; denoted T1, T2, and T3) formed between U6 nucleotides A53, G52 and the bulged U80, and the Hoogsteen edges of the U6 AGC catalytic triad that is paired with U2 nucleotides in the U2/U6 helix Ib (Figure 1B). Base stacking interactions are also observed between nucleotides of the U6 catalytic AGC triad, and between U6-G52 and U6-U80, which likely aid in stabilizing the 3D structure of the catalytic triplex.
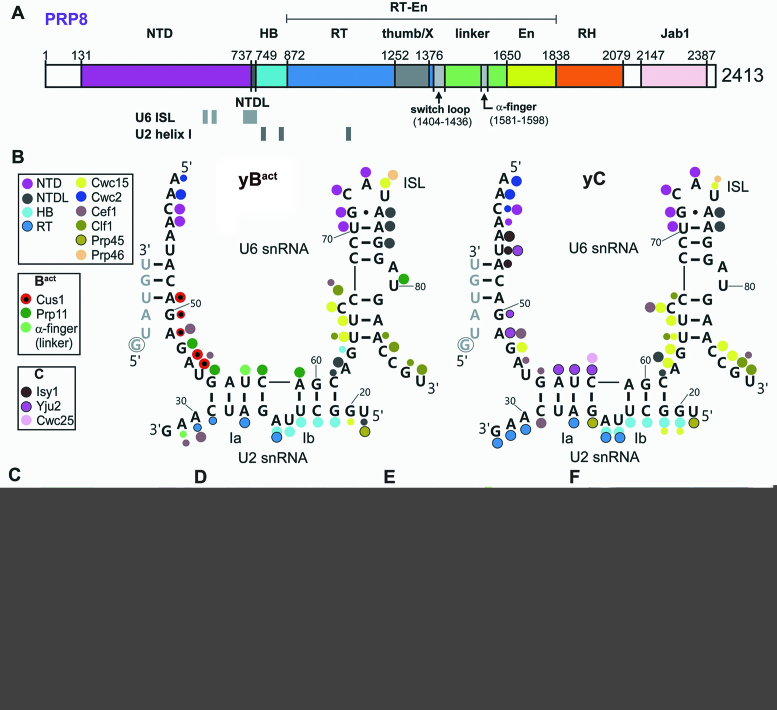
The 3D architecture of the catalytic RNA core of group II introns and spliceosomes is very similar (27), supporting the idea that the two systems are homologous and evolutionarily related (28–30). However, a fundamental difference is that the group II intron catalytic core is embedded in a complex network of RNA–RNA interactions that are essential for splicing catalysis (5,6,31). In contrast, in the spliceosome the U2/U6 RNA core is embedded in a large protein cavity, formed mainly by the Prp8 protein and proteins of the NTC (26,32,33). Prp8 is a large, highly conserved scaffolding protein that contains several functionally important domains (Figure 2A). Crosslinking studies previously showed that Prp8 interacts with all of the chemically reactive groups of the pre-mRNA (i.e., the 5′ss, BS, 3′ss) (34), and these interactions were subsequently confirmed by cryo-EM of various spliceosomal complexes. In Bact and subsequently formed spliceosomal complexes (12,13,15,16,19,20), the U6 ISL is bound by the Prp8 NTD and NTDL. The nearby PRP8 HB domain accommodates the U2 strand of U2/U6 helix Ib and the α-finger of the PRP8 Linker (also called the 1585-loop in yeast) further stabilizes the catalytic centre by interacting with U2 nucleotides between U2/U6 helix I and the U2/BS helix. Consistent with studies showing that the yeast NTC plays a role in stabilizing the association of U6 with the spliceosome (35), U6 nucleotides comprising the ISL, U2/U6 helix I or ACAGAGA sequence are also contacted by NTC or so-called NTC-related proteins in Bact and/or catalytically-active spliceosomes. Thus, in the spliceosome, proteins appear to play a major role in stabilising the 3D structure of the catalytic U2/U6 RNA network.
Figure 2.
RNP architecture of the catalytic core of the spliceosome. (A) Schematic of the domain organization of PRP8 in S. cerevisiae. Regions binding the U6 ISL and U2/U6 helix I are indicated below. NTD, N-terminal domain; NTDL, NTD linker; HB, helical bundle; RT, reverse transcriptase-like; En, endonuclease-like; RH, RNase H-like; Jab1, Jab1/MPN-like. (B) Protein–RNA contacts in the U2/U6 RNA core of the S. cerevisiae Bact (PDB ID: 5GM6 (13)) or and C complex (PDB ID: 5GMK (16)) (see also Supplementary Table S3). Large dots indicate highly favourable geometry for hydrogen bonding (3 stars in Supplementary Table S3) and small dots indicate less favourable geometry for hydrogen bonding (1 star in Supplementary Table S3). (C) Space filling model of proteins or PRP8 domains forming the U2/U6 RNA core binding pocket (with the RNA excluded) in the S. cerevisiae C complex (PDB ID: 5GMK (16)). For orientation, the position of catalytic metal M1 is shown as a brown dot (also in panels D–F). Prp8 domains are coloured as in (A). Other proteins are coloured as in (B) and (C). (D) Space filling model with surface potential of the proteins and protein domains shown in panel (C). The electrostatic potential was calculated using the pyMOL vacuum electrostatics program. Charges are shown as a heat map, with blue for positive and red for negative potential (±5 kT). (E) Space filling model of the U2/U6 RNA network, including the U2/BS helix, in the yeast C complex. Intron nucleotides, light yellow; U2 snRNA (aside from those comprising U2/U6 helix 1b which is black), light green; U6 snRNA, dark green except for the ISL (dark yellow) or nucleotides in helix Ib (black). (F) Fit of the U2/U6 RNA network into the protein space filling model from panel C, where the top layer of protein has been cut away. The cutaway plane is in grey and the regions of the U2/U6 RNA network (compare to panel (E)) which are below the plane are darkened.
Consistent with the latter idea, some secondary RNA interactions in the U2/U6 RNA core appear to be dispensable. For example, yeast genetic studies revealed that changes in U2 bases in U2/U6 helix Ib, which contains the U6 catalytic triad, are less deleterious for splicing than changes in U6 bases (36). Moreover, more recent genetic studies showed that helix Ib formation is not essential for splicing in a wild-type (wt) AGC context (37). As the AGC catalytic triad is involved in the binding of catalytic metal M2 (7), the apparent dispensability of base pairing in helix Ib raises the question of how stable metal binding is achieved. In addition, we recently showed that the N7 position of U6-G60 is accessible to methylation by dimethylsulfate in the spliceosomal B* complex (38). This suggested that the central triple T2 was destabilised before step 1 of splicing, and thus that catalytic triplex formation is unexpectedly dynamic during pre-mRNA splicing.
Here, we have investigated the contribution of Hoogsteen base pairing and the stacking of the base triples and catalytic triad nucleotides to splicing, by assaying the function of synthetic, mutant U6 RNA molecules in an in vitro U6 depletion/complementation system (7,39). This allowed us to dissect interactions essential for catalytic triplex formation in atomic detail not attainable via in vivo genetic investigations. We find that most hydrogen bonds involved in the Hoogsteen interactions were not essential. Remarkably, the base moieties of U6-U80, U6-G78 and U6-A59 were also not essential for splicing and therefore, by extension, for metal coordination. However, the stacking of U6-G60 with neighbouring bases in the catalytic triad was required. The results demonstrate that while RNA tertiary interactions support the functional conformation of the U2/U6 RNA core, some of these interactions are not essential and can be compensated for by protein–RNA interactions in their absence. The functional conformation of the U2/U6 RNA core is thus highly buffered, with overlapping contributions from RNA–RNA and protein–RNA interactions.
MATERIALS AND METHODS
Materials
Oligodeoxynucleotides (Supplementary Table S1) were obtained from Eurofins Genomics (Ebersberg, Germany). The 7cG60 deaza-purine containing oligoribonucleotide (Supplementary Table S2) was from IBA (Göttingen, Germany). All other oligoribonucleotides containing atomic mutations or abasic sites were from Axolabs (Kulmbach, Germany). All RNA oligonucleotides were HPLC purified by the manufactures (Axolabs and IBA). Axolabs additionally used ESI-MS to determine the quality of the HPLC purified oligonucleotides, and the experimental mass of the final product was routinely within 1–2 Da of the theoretical, expected mass value. The flanking unmodified oligoribonucleotides were either from Axolabs, IBA or Dharmacon (GE Healthcare).
Yeast whole cell extract preparation
Whole cell yeast extracts were prepared as described previously (12). For standard in vitro splicing, extracts were prepared from the Saccharomyces cerevisiae strain BJ2168 (MATa, leu2, trp1, ura3, prb1-1122, pep4-3, prc1-407, gal2). For spliceosome assembly analysis, extracts were prepared from the S. cerivisiae strain 3.2.AID/CRL2101 (MATalpha, prp2-1, ade2, his3, lys2-801, ura3) (40), where the prp2-1 mutation leads to stalling of spliceosome assembly at the at Bact stage after initial heat inactivation of the extract at 35°C.
RNA preparation
MS2 actin pre-mRNA was purified as described (41). Yeast U6 snRNAs containing N7-deaza modifications and abasic mutations were prepared by splinted ligation (42) with T4 DNA ligase (NEB) and a U6(19–108) DNA splint that bridges three U6 snRNA fragments, with the desired mutations located in the central fragment (Supplementary Table S2). To monitor ligation and recovery, the central U6 fragments were trace-labelled with γ-[32P] ATP (Perkin Elmer) and polynucleotide kinase (NEB). Full-length U6 snRNAs were routinely purified on 6% polyacrylamide/8M urea denaturing gels.
U6 depletion and complementation
Endogenous U6 snRNA depletion, reconstitution with synthetic U6 snRNA molecules, and splicing complementation assays were as described (39), except that 12 nM of synthetic U6 snRNA was used. A standard splicing reaction (5 μl) contained 60 mM K-phosphate buffer, pH 7.25, 0.3% (w/v) PEG8000, 2.5 mM MgCl2, 2 mM spermidine, 2 mM ATP, 40% of dialyzed wt extract, and 1–2 nM 32P-labelled actin pre-mRNA and was incubated at 23°C for the indicated times. Radiolabelled pre-mRNA and splicing products were isolated, analysed by denaturing PAGE, and visualized with a Phosphorimager (GE Healthcare). Three independent experiments were quantified using ImageQuantTL (GE Healthcare Life Sciences). After correcting for background and the different number of labels per RNA species, the percent of mRNA or intron lariat-3′ exon as a fraction of the total RNA was determined for each lane. Numbers were normalized to the amount of the respective RNA species present at the 60 min time point in the wt U6 reaction and plotted against time.
Affinity purification of yeast spliceosomes
To analyse the effect of the U6-G60ab mutation on spliceosome assembly, dialyzed prp2-1 extract was first incubated at 35°C for 30 min to heat-inactivate Prp2. U6 snRNA was then depleted and reconstitution was subsequently performed by adding the synthetic U6-G60ab RNA to 6 nM concentration. 32P-labelled actin pre-mRNA, pre-incubated with a 30-fold molar excess of MS2-MBP fusion protein at 4°C for 30 min, was added directly to the U6-G60ab reconstitution mixture. Standard in vitro splicing was initiated by adding ATP to 2 mM and incubating at 23°C for 45 min. The reaction was loaded onto a amylose-agarose column (New England Biolabs) pre-equilibrated with GK75 buffer (20 mM HEPES, pH 7.25, 75 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, pH 8.0). The column was washed with GK75 buffer, and spliceosomes were eluted with GK75 containing 12 mM maltose and directly loaded onto a linear 10–30% (v/v) glycerol gradient in GK75 buffer. Centrifugation was at 60 000 rpm for 1 h and 47 min (488 576 × g) in a Sorvall TH660 rotor. Gradients were harvested manually and the distribution of 32P-labelled RNA was determined by Cherenkov counting. Peak fractions containing Bact complexes were pooled and half of the Bact complexes were used to reconstitute the B* complex by addition of a 20-fold excess of recombinant Prp2 and Spp2 proteins, and 2 mM ATP and subsequent incubation at 23°C for 1 h (41). Both the Bact and B* complexes were then subjected to another round of gradient centrifugation and fractionation as described above.
Mass spectrometry and western blotting
Proteins in the gradient fractions were separated by SDS-PAGE and identified by mass spectrometry as described previously (43). For western blotting, proteins of affinity-purified complexes were separated by SDS-PAGE, transferred to a nitrocellulose membrane (Protran, Whatman), which was then probed with antibodies against Prp19 (44) and Cwc24 (45). Bound antibody was detected using an ECL detection kit (GE Healthcare).
Computational analysis of structure files
Cryo-EM structures were analysed and visualised with pyMOL (Schrödinger, LLC). Protein–RNA contacts were determined from available PDB structures using the contact program from the CCP4 suite (46), and then extracting the unique contacts of core proteins with U2 and U6 snRNA in the yeast Bact (PDB ID: 5GM6 (13)), C (PDB ID: 5GMK (16)), and C* (PDB ID: 5WSG (20)) complexes (Supplementary Table S3). The amino acid contacts for Supplementary Figure S5B were extracted from Supplementary Table S3. The inner surface of the protein cavity accommodating the U2/U6 RNA core was determined by CASTp (47). Proteins or regions of proteins that contact the RNA core were submitted as one ensemble to the CASTp server (http://sts.bioe.uic.edu/castp/). The largest protein pocket found accommodated the U2/U6 RNA. The unique residues lining the catalytic RNA core protein cavity were extracted from the pocket file and used to display the interior of the protein cavity (Figure 2C) and calculation of its electrostatic surface (Figure 2D, F). Stacking interactions in the U2/U6 RNA core were determined with the Arpeggio server (48) at http://biosig.unimelb.edu.au/arpeggioweb, using coordinates extracted from the yeast C complex (PDB ID: 5GMK).
RESULTS
The catalytic U2/U6 RNA core is tightly embedded in protein in the spliceosome
Cryo-EM studies revealed that the catalytic U2/U6 RNA core is embedded in a protein cavity formed by multiple spliceosomal proteins. We analysed in detail amino acids that contact U6 and U2 nucleotides within the U2/U6 catalytic core of the yeast Bact, C and C* complexes using the protein contact program from the CCP4 suite (see Supplementary Table S3 for interaction details). As shown schematically in Figure 2B, for the yeast Bact and C complexes, the vast majority of nucleotides in the U2/U6 core are contacted by protein. While most protein–RNA interactions in the RNP core are conserved between Bact, C and C* complexes (Supplementary Table S3, Figure 2B), dynamic contacts are observed in some cases. For example, the U2-associated Cus1 and Prp11 proteins contact the U2/U6 core in the Bact complex, but are released/destabilized from the spliceosome during the transformation of Bact into a C complex. In the latter, several Cus1 and Prp11 U6 contacts are replaced by Cef1 and the step 1 factor Yju2. Amino acids from the N-terminal domain (NTD), NTD linker (NTDL) and helical bundle (HB) of Prp8, as well as from Cwc15, Yju2 (Cwc16) and Cef1 are major contributors to the inner surface of the protein cavity accommodating the U2/U6 RNA core in the yeast C complex (Figure 2C). Analysis of the surface potential of this cavity shows that it is highly basic and thus well-suited to accommodate RNA (Figure 2D). Docking of the U2/U6 RNA into a space filling model reveals how tightly the RNA is embedded in this protein pocket and underscores the likely contribution by proteins in stabilizing the 3D conformation of the U2/U6 RNA core (Figure 2E, F).
Disruption of Hoogsteen hydrogen bonds in triples T2 and T1 has little effect on splicing
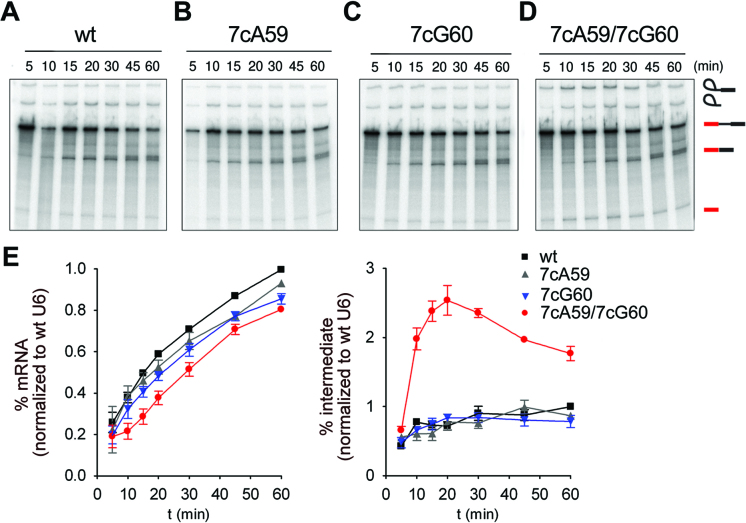
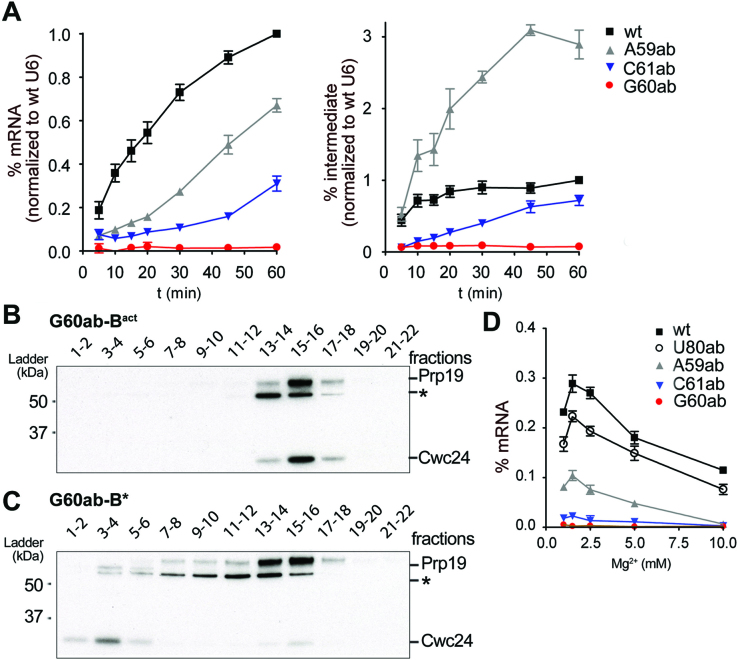
The tight fit of the U6 ISL, U2/U6 helix Ia/b and the catalytic triplex and their highly positively-charged protein environment raised the question whether some RNA–RNA interactions may be dispensable for the formation of a catalytically active RNA conformation that binds the catalytic metal ions in an orientation compatible for splicing. We thus assayed the contribution of RNA tertiary interactions within the U2/U6 RNA core to pre-mRNA splicing in vitro. For this purpose, we constructed mutant U6 snRNA molecules that contained atomic mutations predicted to disrupt specific hydrogen bonds in the catalytic triplex network, or site-specific abasic lesions that additionally would disrupt base stacking interactions. We then assayed for splicing with each U6 mutant over the course of 60 min after adding them to S. cerevisiae extract depleted of endogenous U6 by RNAse H digestion. Initially we focused on the hydrogen bonds involving the N7 positions of G60 and A59 in triples T2 and T1 (Figure 1B; boxed in Supplementary Figure S1). Disruption of the N7 hydrogen bond between U6-G60 and U6-G52 in T2, by replacing nitrogen at the N7 position of G60 with carbon (U6-7cG60), reduced splicing efficiency after 60 min to 85% relative to wt U6 (Figure 3C and E). The equivalent disruption of the N7 hydrogen bond at A59 in triple T1 (U6-7cA59) had even less effect on splicing (93% efficiency; Figure 3B, E). The splicing efficiency (80%) after disruption of both N7 hydrogen bonds (U6-7cA59/7cG60) was comparable to the effect observed with the single U6-7cG60 mutant. However, a significant increase in splicing intermediates (3.5-fold at the 20 min time point, Figure 3E) was observed, suggesting that there is a step 2 defect. As step 1 was still efficiently catalysed, this double mutation does not appear to substantially affect catalytic metal coordination by U6, but may instead affect RNA–RNA or protein–RNA interactions important for highly efficient step 2 catalysis (see Discussion).
Figure 3.
The Hoogsteen interactions of U6 nucleotides A59 and G60 are not essential for splicing. (A–E) Kinetics of splicing of actin pre-mRNA in U6-depleted yeast extract supplemented with different synthetic U6 snRNAs. Splicing performed with (A) wild-type U6 (wt), (B) N7-deaza-A59 U6 (7cA59), (C) N7-deaza-G60 U6 (7cG60) or (D) N7-deaza-A59/N7-deaza-G60 U6 (7cA59/7cG60). The positions of the pre-mRNA, and of the splicing intermediates and products are indicated on the right. (E) Quantification of spliced mRNA production (left panel) and intron lariat-3′ exon (denoted as intermediate) production (right panel). The average amount of mRNA or intermediate present (plus the standard error) at each time point was calculated from three independent experiments. The production of mRNA at all time points was normalized to the amount of mRNA produced at the 60 min time point with wt U6. Similarly, lariat-3′ exon production (intermediate) was normalized to the amount of splicing intermediate produced at the 60 min time point with wt U6.
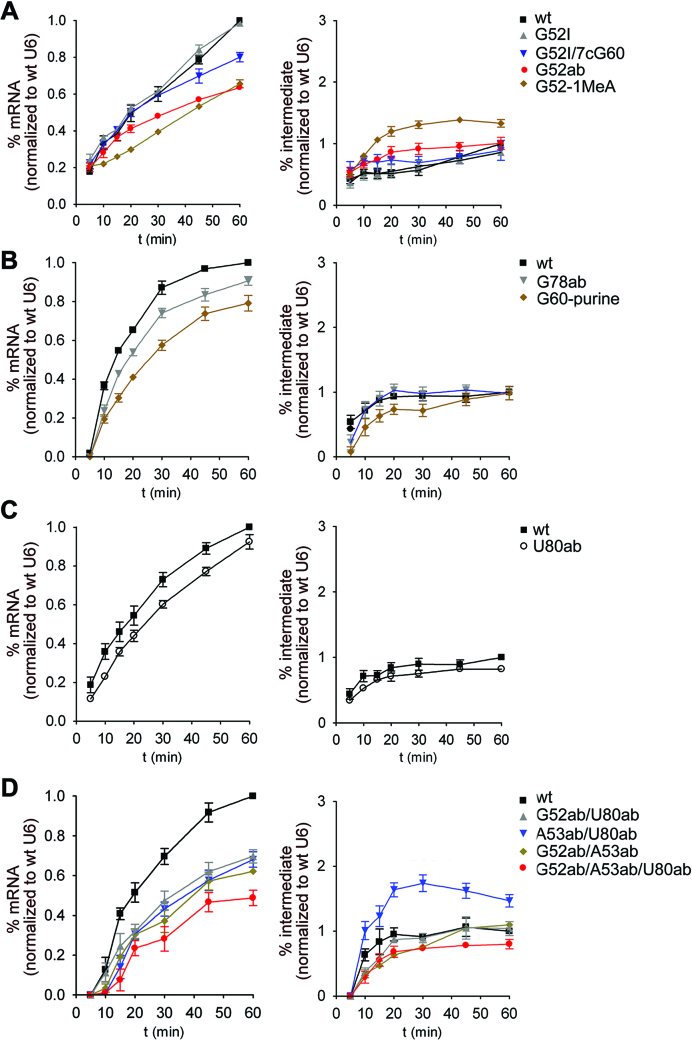
We next analysed the hydrogen bond network of triple T2 in more detail. Replacing G52 with inosine (U6-G52I), which abolishes formation of the other hydrogen bond on the Hoogsteen edge of U6-G60 in T2, had no effect on splicing (98% efficiency; Figure 4A, Supplementary Figure S2). Disruption of both hydrogen bonds (G52I/7cG60) had a mild effect on splicing (80% efficiency; Figure 4A). Given the mild effects that disruption of single or double hydrogen bonds have, we replaced the guanine with a purine at U6-G60 (G60-purine), which disrupts base pairing with U2-C22 in T2 (Figure 1B; Supplementary Figure S1). With this mutant, splicing efficiency was 79% after 60 min (Figure 4B), without any second step effect, demonstrating that base pair formation between U6-G60 and U2-C22 is not essential for splicing. Finally, we tested a U6-G52-1MeA mutant that alters the hydrogen bonding network of T2, by inserting a bulky methyl group at the N7 position, thereby increasing the C1′-C1′ distance between U6-G52 (now A52) and U6-G60. The splicing with this mutant was reduced to 65% (Figure 4A) and it showed an accumulation of the lariat-3′ exon intermediate (2-fold increase at the 30 min time point), indicating that the bulky methyl group hinders step 2 of splicing. Magnesium coordination by this U6 mutant is probably not substantially affected, as step 1 proceeds normally. Instead, protein interactions involving U6-G52 and surrounding nucleotides (Figure 2B), that potentially ensure efficient step 2 catalysis, may be altered as discussed in detail below.
Figure 4.
Limited effects on splicing of atomic or abasic mutations of U6 nucleotides in the triple helix structure. (A–D) Quantification of the kinetics of mRNA and intermediate production after performing splicing with indicated synthetic U6 snRNA mutants. The corresponding splicing assays are shown in Supplementary Figure S2 and quantification was as in Figure 3. The mutants in panels A-D were all analysed in one experimental series to ensure that the quantification relative to the wt U6 was reliable. The average amount of mRNA or intron lariat-3′ exon (denoted as intermediate) present (plus the standard error) at each time point was calculated from three independent experiments, with the exception of panel D which was derived from two independent experiments.
Disruption of stacking interactions and hydrogen bonds via abasic mutations
We next tested U6 snRNAs containing abasic sites to determine whether base stacking interactions in the triples are essential for splicing. As shown in Figure 5, U6-U80 and U6-G52, as well as the catalytic triad nucleotides, are involved in base stacking interactions within the catalytic triplex (49). In addition to lacking all hydrogen bonding properties, abasic mutants are predicted to display a more flexible phosphate backbone because of the role of the stacking interactions in ensuring the proper configuration of the phosphate backbone. We first focussed on U6-G52 and U6-U80, as they stack onto one another and are involved in tertiary interactions with the U6-G60 (triple T3) and U6-C61 (triple T2), respectively, but not in base pairing interactions. Removal of the guanosine base of U6-G52 (U6-G52ab) resulted in a substantially lower splicing efficiency (63%, Figure 4A), whereas deletion of the uridine base at U6-U80 (U6-U80ab) led to a milder reduction in splicing (80% efficiency; Figure 4C). The less efficient splicing observed with U6-G52ab suggests that disruption of the U6 G52/U80 stack is not the sole effect of this mutation. Indeed, in contrast to U6-U80, U6-G52 is cradled by both Cwc15 and Cef1 in the yeast C complexes (Figure 2B, Supplementary Table S3), and thus removal of its base may also affect Protein–RNA interactions important for efficient catalysis. An essential function of U6-U80 together with U6-G78 is to coordinate metal M1 via their 5′ phosphate groups (7) (Figure 1C). Splicing with the U6-G78ab mutant was nearly as efficient as with wt U6 (90% splicing; Figure 4B), indicating that deletion of the base of U6-G78 does not affect M1 coordination.
Figure 5.
Base stacking interactions within the spliceosomal U2/U6 RNA core. Close-up of the base stacking interactions of the AGC triad nucleotides and U6-U80 in the U2/U6 RNA core of the yeast C complex (PDB ID: 5GMK). Stacks are shown as towering disks between the involved centres of the nucleobases. Base stacking geometries are classified according to (60,61), with fully stacked bases in blue, and staggered stacked bases in yellow. The stacking interactions, in particular the staggered stacks, correspond to real densities in the C complex EM density map (EMD-9525; (16)).
We next generated U6 mutants with combinations of two or three bases removed from the three triple nucleotides, U6-G52, U6-A53 and U6-U80 (Figure 1B). Mutants with two abasic residues (A53ab/U80ab, G52ab/U89ab, G52ab/A53ab) supported splicing with an average efficiency of only ∼60% when compared to the wt U6 (Figure 4D). In addition, an accumulation of splicing intermediates was observed with U6-A53ab/U80ab, indicating that there is a second step defect. Generally, the effects were not additive, as the double mutants exhibited splicing efficiencies corresponding approximately to that of the worst performing single mutant within the error margin (Supplementary Table S4). Surprisingly, splicing with a triple abasic U6 mutant (U6-A53ab/G52ab/U80ab) was still observed, although the efficiency of splicing (47%, Figure 4D, Supplementary Table S4) was lower than that observed with the mutants containing for example the single U6-G52ab or the double U6-G52/U80ab mutation. Taken together, these results suggest that the characteristic fold of the backbone containing the U6-U80 bulge that is required to coordinate the catalytic metal ions is not dependent on several of the secondary and tertiary interactions of the bases involved, as the RNA backbone appears to fold correctly for the most part in the absence of bases at positions U6-U80 or U6-G78. Further, elimination of the bases of the U6 nucleotides A53, G52 and U80 did not completely block splicing, suggesting that they are not absolutely essential for productive coordination of the catalytic metal ions. Thus, specific hydrogen bonds and base stacking interactions in the catalytic triplex are not absolutely required for stabilizing the active conformation of the U2/U6 RNA core, indicating that additional stabilization is supplied by spliceosomal proteins.
Base stacking involving the catalytic triad nucleotide U6-G60 is required for splicing
We next investigated the contribution of hydrogen bonding and base stacking involving the bases of the catalytic triad nucleotides. U6-A59 engages in base stacking interactions with U6-G60 and U2-G26 of U2/U6 helix Ia and U6-C61 stacks with U6-G60 and U6-A62, whereas U6-G60 has stacking interactions with all triple T1 bases (U6-A53/A59/U2-U23), as well as with U6-C61 (Figure 5). The catalytic triad nucleotide G60 is not involved in Protein–RNA interactions, whereas C61 is contacted by the NTDL of Prp8 in Bact and C, and A59 is contacted by Prp11 only in the Bact complex (Figure 2B; Supplementary Table S3). Elimination of the base of U6-A59 (U6-A59ab) led to a moderate reduction in splicing (67% efficiency) (Figure 6A, Supplementary Figure S2E). As this mutation also led to an accumulation of the intron lariat-3′ exon intermediate (3-fold at the 45 min time point), we conclude that step 2 is compromised. A59 is not contacted by protein in catalytically-active C/C* spliceosomes (Figure 2B, Supplementary Table S3), and neither its base pairing interaction with U2-U23 (37), nor its Hoogsteen base pairing with U6-A53 (Figure 3B) are essential for splicing. Thus, the reduction in step 2 could be due to the disruption of base stacking interactions involving A59 (see Discussion).
Figure 6.
Stacking of the base of U6-G60 is required for splicing catalysis, but not for Bact and Prp2-mediated B* complex formation. (A) Quantification of the splicing kinetics of mRNA and intermediate production for different abasic mutants of the catalytic triad. Splicing assays are shown in Supplementary Figure S2 and quantification was as in Figure 3. (B and C) The U6-G60 abasic (G60ab) mutant allows for assembly of Bact complexes and their conversion into a B*-like complex. Bact complexes containing the U6-G60ab mutant were assembled in prp2-1 yeast extract and purified Bact complexes were analysed on a gradient directly (B) or (C) after incubation with Prp2 and Spp2 proteins to form B* (41). Gradient fractions (indicated above) of Bact (B) and B* (C) complex preparations were probed simultaneously with anti-Prp19 and anti-Cwc24 antibodies. (D) Magnesium dependence of splicing with select U6 mutants. Production of mRNA after 20 min was quantified as in Figure 3 at the different magnesium concentrations indicted. In panels A and D, the average amount of mRNA or intron lariat-3′ exon (denoted as intermediate) present (plus the standard error) at each time point was calculated from three independent experiments.
In contrast to A59, when the base of U6-G60 was eliminated (U6-G60ab), both steps of pre-mRNA splicing were completely blocked (Figure 6A, Supplementary Figure S2E). To determine whether the observed inhibition is due to a defect in spliceosome assembly as opposed to splicing catalysis, we analysed spliceosome formation on actin pre-mRNA in the presence of wt U6 or the U6-G60ab mutant. To this end, we affinity purified wt or U6-G60ab Bact complexes, employing a heat-treated extract from yeast cells harbouring the prp2-1 mutation, which allows for Bact assembly, but blocks catalytic activation of the Bact complex (41). Gradient centrifugation revealed that the purified wt and U6-G60ab Bact complexes have identical sedimentation properties (Supplementary Figure S3A, B). Mass spectrometry further confirmed that their protein compositions are essentially identical (Supplementary Table S5). Most importantly, the purified U6-G60ab Bact complexes could be transformed into B*-like complexes upon incubation with Prp2, Spp2 and ATP as evidenced by (i) a shift in migration on glycerol gradients to a slightly lower S value than Bact spliceosomes (Figure 6B,C, compare peak of Prp19 protein; Supplementary Figure S3), which is typical for B* complexes (41); and (ii) the release of the majority of the Cwc24 protein (compare Figure 6B and C; Supplementary Figure S3), which is another hallmark of successful Prp2-mediated remodelling of the spliceosome (41,50). However, unlike bona fide B* complexes, these B*-like complexes are not catalytically-active. Our results with the U6-G60-purine mutant (Figure 4B) showed that base pairing with U6-G60 is not essential. As G60 is not contacted by protein (Figure 2B), one or more stacking interactions involving the base of U6-G60 appear to be essential for splicing catalysis, but not for spliceosome assembly or formation of a B*-like complex.
In addition to A59, U6-G60 stacks with C61 of the catalytic triad. Splicing was not completely abolished in the presence of a U6-C61ab mutant, but the reduction in both catalytic steps was substantial (31% mRNA production efficiency after 60 min; Figure 6A). Given that the removal of hydrogen bonds between U6-C61 and U6-U80 in triple T3 had little effect on splicing (U6-U80ab mutant; Figure 4C), and that base-pairing of U6-C61 to U2-G20 is not essential for splicing (36,51), the poor splicing activity of the U6-C61ab mutant suggests that, similar to the situation with U6-G60, stacking interactions involving the base of U6-C61 are essential for efficient splicing. However, the requirement cannot be as strict as for U6-G60, as residual splicing still occurs. U6-C61 is stacked between U6-G60 and U6-A62, and is not involved in any other base stacking interactions (Figure 5). To address which of these stacks is important, we prepared a U6-A62ab mutant that disrupts its stacking with U6-C61. Splicing of the U6-A62ab mutant was indistinguishable from wt U6 (100% splicing efficiency; Supplementary Figures S2D and S4), demonstrating that the stack of U6-C61 and U6-A62 is dispensable for splicing. As the elimination of the base of U6-G60 abolished splicing, but the A59 abasic mutation led to only a moderate reduction in splicing, either additional stacking interactions involving the base of G60 (e.g. with U6-A53) must be important, or the simultaneous elimination of multiple base stacking interactions involving U6-G60 is required to abolish splicing.
Elimination of the bases of U6 nucleotides coordinating catalytic metals does not appear to alter their affinity for Mg+2
Some of the abasic lesions described above were at nucleotides that directly bind catalytic metals via their 5′ phosphates (U80ab, A59ab, and G60ab) and the loss of splicing activity could be due to a lower affinity for Mg2+. To investigate whether the lesions influenced the strength of metal interactions, we assayed splicing with these U6 mutants, as well as U6-C61ab) over a range of magnesium concentrations. Splicing was assayed after a 20 min incubation at 1, 1.5, 2.5, 5 and 10 mM magnesium (Figure 6D). For wt U6, an optimum was detected at 1.5 mM. All of the single-site abasic mutants that still supported splicing (U80ab, A59ab and C61ab) showed the same optimum and a reduction in splicing at higher magnesium concentrations. The splicing efficiencies observed with all of the U6 mutants tested were not improved by increasing the magnesium concentration, suggesting that these single-site abasic mutations did not affect the affinity of U6 for magnesium.
DISCUSSION
The Hoogsteen interactions involving the catalytic triad are not essential for splicing
Our results show that the Hoogsteen interactions involving nucleotides of the catalytic triad are not essential for splicing. The hydrogen bonds in triples T3, T2, and T1 could be eliminated without dramatically affecting the efficiency of splicing (Figure 3). Further, the bases of U6-G78 and U6-U80 could be eliminated without significantly disrupting splicing (Figure 4). This was unexpected as U6-U80 is part of triple T3 (Figure 1B), which is thought to play an essential function in positioning U6-U80 close to the U6-C61/U2-G21 base pair and acts as a platform for the U6-U80/U6-G52 stacking interaction (9). As U6-G52 is located directly adjacent to the U6 AGAGA/5′ss helix, this configuration is thought to tether the emerging catalytic centre to the 5′ ss during the B to Bact transition. Moreover, by analogy to group II introns, triple T3 is thought to play an essential architectural role in correctly positioning metal M1 at U6-U80. Our results indicate that in vitro, the RNA tertiary interactions involving the base of U6-U80 are not essential for pre-mRNA splicing and thus for the positioning of metal M1. Similarly, the mutants designed to eliminate the hydrogen bonds to U6-G60 in triple T2, including the U6-G60-purine mutant (Figure 4), all supported splicing, although in some cases (i.e. with U6-G52ab) a moderate drop in efficiency (63% of the wt U6 level) was observed. It is thus unlikely that hydrogen bonding to the Hoogsteen edge of U6-G60 contributes significantly to splicing. For triple T1, disruption of one of the Hoogsteen hydrogen bonds between A53 and A59 (7cA59) had essentially no effect, whereas a step 2 defect was observed with the U6-A59ab mutant (Figure 6A). The latter shows that coordination of metal M2 by U6-A59 does not strictly require the base of U6-A59 and, by extension, a base pair with U2 snRNA at this position, at least not for the first catalytic step.
In addition to not changing splicing rates significantly, these U6 mutations (and some others discussed below) also do not lead to any new rate-limiting steps prior to splice site cleavage. For example, U4/U6.U5 tri-snRNP assembly also does not become rate-limiting despite major modifications to critical U6 nucleotides that base pair to U4 snRNA. Formation of the U2/U6 catalytic RNA core occurs during the initial activation of the spliceosome, and based on genetic experiments in yeast, its proper formation appears to be proofread by the DExD/H-box ATPase Prp2 during the final catalytic activation step, just prior to step 1 of splicing (52). The aforementioned U6 mutations tested here also do not appear to trigger discard of the activated spliceosomes that are assembled due to suboptimal formation of the catalytic core, at least not in vitro. However, the effects of these U6 mutations may become rate limiting and thus more severe in vivo, where a large number of diverse pre-mRNAs must be efficiently spliced and where the splicing machinery is functionally linked to other cellular processes, such as transcription.
The importance of base stacking within the catalytic triad
The U6-G60ab mutation led to a complete block in both steps of splicing, but not in spliceosome assembly (Figure 6). That is, this mutated U6 snRNA was efficiently assembled into a Bact complex and allowed Prp2-induced catalytic activation to generate a B*-like complex, with a protein composition and sedimentation behaviour indistinguishable from a B* complex containing wt U6. In agreement with previous genetic observations (36,37), base pairing of U6-G60 with U2-C22 was not required for efficient splicing (U6-G60-purine mutant, Figure 4B), and our data additionally showed that the triple interactions at the Hoogsteen edge of U6-G60 are also not required. The purine base therefore has an essential role, although the base is not directly involved in coordinating catalytic metal M2. As G60 is not contacted by proteins in Bact, C and C* complexes (Figure 2B, Supplementary Table S3), we conclude that the G60 base engages in critical base stacking interactions within the catalytic triad that cannot be compensated for by protein–RNA interactions. This stacking is likely required for rigid stabilisation of the phosphate backbone to facilitate correct and stable coordination of metal M2. In addition, the overall integrity of the catalytic triplex is probably dependent on G60 base stacking. Consistent with the importance of base stacking within the catalytic triad, the elimination of the bases of U6-A59 or U6-C61 which flank G60, substantially reduced splicing efficiency without, however, completely blocking the reaction (Figure 6). This suggests that the simultaneous disruption of multiple base stacking interactions involving U6-G60 leads to the observed block in splicing.
A number of studies were performed previously on group II introns using nucleotide analogue modification interference (53–56). One of these studies (55) demonstrated a requirement for the phosphates of the catalytic triad, using phosphorothioate substitutions in a group II intron derived from the S. cerevisiae cytochrome oxidase subunit I mitochondrial gene. Site-specific modifications of D5 in a trans-splicing system derived from the same intron, showed that an inosine substitution at the central G of the AGC catalytic triad is non-functional in splicing (57). This result differs substantially from our findings here, where we show that a purine substitution of U6-G60 has no discernible effect on pre-mRNA splicing (Figure 4B).
The two catalytic metal binding sites differ in their dependence on RNA–RNA tertiary interactions
The metal M1 binding site is located in a characteristic S-shaped backbone between U6-G78 and U6-U80 (6). Our data indicate that this structure can form in the absence of the bases of U6-G78 or U6-U80, or of the stack between U6-G52 and U6-U80. This implies that the S-shaped backbone is still generated during folding of the U6 ISL during Bact formation, even in the absence of different subsets of RNA–RNA interactions involving these nucleotides. Thus, protein–RNA contacts apparently can substitute for RNA tertiary interactions to form a productive 3D conformation of the RNA backbone required for M1 binding. The U6 ISL and tertiary interactions involving U6-U80 are first formed after U6 is released from the U4/U6 duplex during Bact formation. Their formation appears to coincide with the establishment of protein–RNA contacts during insertion of the U6 ISL into the RNA binding pocket formed by Prp8. Except for one phosphate backbone contact to U6-U80 by Prp11 in the yeast Bact complex (Figure 2B and Supplementary Table S3), there are no protein contacts to U6 nucleotides G77 to A82 in the yeast Bact, C and C* complexes. Likely, the S-shape of the RNA backbone is induced by protein contacts with the RNA elements neighbouring the bulge in the U6 ISL, which involve Prp8 and also primarily Cwc15 and Clf1 (Figure 2B, Supplementary Table S3). Productive formation of the metal M2 binding site, in contrast, is strictly dependent on base stacking interactions involving the U6-G60 base, which includes stacks with the bases of A59 and C61 of the catalytic triad, and U6-A53 (Figure 5). Our data show that the M2 binding site is not dependent on Hoogsteen interactions or U2 base pairing involving the AGC of the catalytic triad. The importance of G60 stacking with the other catalytic triad bases may provide an explanation for previous results from yeast genetic experiments showing asymmetric requirements for helix Ib, with U2 base pairing being dispensable (36,37). As we have analysed the effects on the splicing of only the actin pre-mRNA substrate, and the spliceosome assembles on each intron, with the intron itself potentially playing a structural role, it is possible that the RNA–RNA contacts that help coordinate metal M1 are more important for the splicing of other pre-mRNAs.
The U2/U6 RNA core is embedded in a protein cavity forming the spliceosome's RNP core
Except for stacking of U6-G60 in the catalytic triad, our data show that the requirements for RNA structural determinants, i.e. W/C base pairing and Hoogsteen interactions, are surprisingly relaxed within the U2/U6 RNA core. This is especially true for the evolutionarily conserved Hoogsteen interactions involving the catalytic triplex (see also above) and likely reflect the intimate contacts that proteins make with these catalytically important RNA regions within the spliceosome. The catalytic triplex of the spliceosome has distinct structural and catalytic faces (Supplementary Figure S5B) (27), a general property of RNA catalysts, initially proposed for group II introns (57). The structural part of the catalytic triplex, which is not involved in metal binding, is tightly embedded in the protein cavity formed by Prp8 and other spliceosomal proteins. In particular, the RNA backbone around U6-G52 is cradled by lysines 165 and 166 of Cef1, and histidine 5 of Cwc15 contacts the N2 and N3 positions of U6-G52 on its sugar edge (Supplementary Figure S5 and Supplementary Table S3). The catalytically important phosphate-ribose backbone of the catalytic triplex, which coordinates M1 and M2, is not contacted by any protein side chains. The space facing the catalytic side of the catalytic triplex is populated by the RNA substrate and other RNA structural elements. Thus, there is only limited space available in the protein cavity to accommodate the U2/U6 RNA core of the spliceosome, in particular the catalytic triplex. This may in turn ensure the productive folding of the U2/U6 RNA core even after disruption of one or more RNA–RNA interactions during Bact formation.
The results of extensive yeast genetic analyses of catalytic triplex mutants in vivo (9) led to the conclusion that only isosteric mutants of the base triples support growth in vivo. This is entirely consistent with the more recently obtained structural evidence for a tight fitting protein cavity that could only accommodate mutant triples with isosteric geometries when compared to the wt triples. Additionally, only isosteric mutants would preserve the important stack involving the catalytic triad. Our studies suggest that non-isosteric mutants of the base triples also function in splicing, at least in vitro. However, as we only tested their effects on actin pre-mRNA, we cannot rule out that other pre-mRNAs may have a more stringent requirement for specific hydrogen bonds and base stacking interactions in the RNA catalytic core. Numerous pre-mRNAs must be spliced efficiently in yeast cells in order to support their growth, and thus cell growth may be a more stringent read-out. However, the mutations studied here cannot be assayed in vivo.
Potential mechanisms whereby some U6 mutations lead to a step 2 defect
Some U6 mutations led to an accumulation of splicing intermediates, indicating that step 2 is in some way compromised. As catalytic step 1 occurs, these step 2 defects do not appear to be due to non-productive coordination of the catalytic metals M1 and M2. Instead, it is likely that they alter/disrupt RNA–RNA or RNA–protein interactions/rearrangements important for efficient step 2 catalysis. Prior to step 2, the branched intron structure (BIS) generated during step 1 of splicing must be displaced from the catalytic centre of the spliceosome to allow juxtapositioning of the step 2 reactants, the 3′OH of the 5′ exon and the 3′ss. During the C to C* complex transition, Cwc25 and Yju2 are destabilized, and Isy1 is repositioned (15,16,19,20). This allows the BIS to move ca 20 Å away from the catalytic centre, creating space for the 3′ exon to dock near the 5′ exon and for the recruitment of the step 2 factors Slu7 and Prp18. During C* formation, the Prp8 RH domain (Prp8RH) rotates by 70 degrees and its β-hairpin is inserted between the U2/BS helix and the U6 ACAGA/5′ss helix, thereby stabilizing the conformation of the BIS in C* (15,16,19,20). The Prp17 WD40 domain also moves about 70 Å, contacting PRP8RH and the extended U2/BS helix. The step 2 factors Slu7 and Prp18 also interact with PRP8RH, stabilizing its rotated conformation. Thus, together with the Prp8 RH domain, Prp17, Prp18 and Slu7 likely aid in stabilizing the step 2 conformation of the spliceosome. The movement of the BIS is accompanied by a large-scale movement of the 3′ domain of the U2 snRNP; this is likely achieved by rotation around the U2 nucleotides located between the BS interacting region and nts that base pair with U6 to form U2/U6 helix Ia (18,20).
The double-deaza mutant (U6-7cA59/7cG60) led to the accumulation of splicing intermediates (Figure 3). Model studies have shown that deaza substitutions in a helix negatively affect helix stability by reducing stacking, hydration and/or cation interaction (58,59). The double-deaza mutant would therefore be predicted to lead to weakened base stacking, suggesting that the A59/G60 stack in the U2/U6 RNA core (Figure 5) might have a stabilising effect on the structural transitions between steps 1 and 2 of splicing. Consistent with this, the U6-A59ab mutant, which should completely disrupt A59 base stacking interactions also exhibited a step 2 defect. Although no accumulation of splicing intermediates was observed with the U6-G60ab mutant, the first step was abolished, precluding the accumulation of splicing intermediates. Potentially, the stack neutralizes or buffers the structural strain on the RNA backbone that develops when the U2/BS helix and the 3′ domain of the U2 snRNP undergo a concerted movement around U2-A30 (U2-A29 in human spliceosomes), which is adjacent to U2/U6 helix Ia of the central U2/U6 RNA core stack.
The spatial limitations within the protein cavity that binds the U2/U6 RNA core may explain the effect on splicing observed with the U6-G52-1MeA mutant. The U6-G52ab mutant demonstrated that the base at this position is not essential for splicing. However, insertion of the 1-methyl-A at U6-G52 leads to a step 2 defect, as indicated by the increase in splicing intermediates and corresponding loss of mRNA product. With U6-G52-1MeA, the gap between U6-G52 and U6-G60 is widened due to the presence of the bulky methyl group, which would therefore lead to repositioning of U6-G52 and likely alter the interaction of its backbone with Cef1, which is observed in the C complex (Supplementary Figure S5). The cryo-EM structures show that the movement of the BIS has no effect on the relative position of the U6 ACAGA box/5′ ss helix, but results in a repositioning of the phosphate ribose backbone of U6-G50, A51, by ∼3 Å, and a splaying out of the bases G50 and A51 (Supplementary Figure S5A). The latter may be hindered when the bulky methyl group is present on G52 due to the tighter fit of U6-G52-1MeA in the Prp8/NTC protein cavity. The C to C* complex transition might also be impeded if the U6-G52 interaction with Cef1 is altered, as Cef1 interacts with Isy1 and Yju2 in the C complex, and later with Prp17 in the C* complex (15,16,19,20). Alternatively or in addition, the bulky methyl group may distort other contacts in the RNP core and hinder the transformation of the C complex into C*.
Proteins likely play an evolutionarily conserved role in stabilizing the U2/U6 RNA core
Comparison of the 3D structures of human and yeast Bact C and C* complexes reveals that the catalytic U2/U6 RNA network in human spliceosomes adopts a 3D structure that closely resembles that found in S. cerevisiae spliceosomes (17). Furthermore, the catalytic U2/U6 RNA network is embedded in a very similar protein environment in both organisms. Indeed, in several cases the same conserved protein regions contact equivalent RNA sites/regions of the U2/U6 RNA network. For example, conserved regions of the Prp8 NTD and NTDL interact with the upper stem and loop of the U6 ISL, with Prp8′s NTDL extending across the major groove of the upper stem of the U6 ISL, and amino acids of the Prp8 HB domain contact the backbone of the U2 strand of U2/U6 helix Ib in a similar manner in both yeast and human spliceosomes (11–21). Thus, Prp8 appears to act as a chaperone that actively helps to fold the U2 and U6 snRNAs into a catalytically active conformation both in higher and lower eukaryotes. Finally, human homologs of the yeast NTC and NTC-related proteins make similar contacts with the U2/U6 RNA core as their yeast counterparts. For example, the human counterparts of Cwc15 (hCwc15) and Prp46 (PLRG1) contact the equivalent nucleotide in the loop of the U6 ISL. Thus, the structure of the catalytic RNP core is for the most part conserved between yeast and human splicesomes. It is thus highly likely that the requirement for some RNA–RNA secondary and tertiary interactions within the U2/U6 core, and the dispensability of others that we observe in the yeast spliceosome also applies to higher eukaryotes.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Jana Schmitzová for the generous gift of recombinant proteins, Zbigniew Warkocki and Patricia Fabrizio for advice concerning the spliceosome reconstitution system, Henning Urlaub for mass spectrometry, and Cohue Peña for the initial characterization of the U6-7cG60 mutant. We thank Eric Westhof for suggesting to distort the triplex with the U6-1MeA mutant and Berthold Kastner for helpful discussions of the manuscript and figures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society [to R.L.]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wahl M.C., Will C.L., Lührmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 2. Staley J.P., Guthrie C.. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998; 92:315–326. [DOI] [PubMed] [Google Scholar]
- 3. Staley J.P., Guthrie C.. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol. Cell. 1999; 3:55–64. [DOI] [PubMed] [Google Scholar]
- 4. Pyle A.M. The tertiary structure of group II introns: implications for biological function and evolution. Crit. Rev. Biochem. Mol. Biol. 2010; 45:215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robart A.R., Chan R.T., Peters J.K., Rajashankar K.R., Toor N.. Crystal structure of a eukaryotic group II intron lariat. Nature. 2014; 514:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toor N., Keating K.S., Taylor S.D., Pyle A.M.. Crystal structure of a self-spliced group II intron. Science. 2008; 320:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fica S.M., Tuttle N., Novak T., Li N.S., Lu J., Koodathingal P., Dai Q., Staley J.P., Piccirilli J.A.. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013; 503:229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yean S.L., Wuenschell G., Termini J., Lin R.J.. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000; 408:881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fica S.M., Mefford M.A., Piccirilli J.A., Staley J.P.. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat. Struct. Mol. Biol. 2014; 21:464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anokhina M., Bessonov S., Miao Z., Westhof E., Hartmuth K., Lührmann R.. RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J. 2013; 32:2804–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haselbach D., Komarov I., Agafonov D.E., Hartmuth K., Graf B., Dybkov O., Urlaub H., Kastner B., Lührmann R., Stark H.. Structure and conformational dynamics of the human spliceosomal B(act) complex. Cell. 2018; 172:454–464. [DOI] [PubMed] [Google Scholar]
- 12. Rauhut R., Fabrizio P., Dybkov O., Hartmuth K., Pena V., Chari A., Kumar V., Lee C.T., Urlaub H., Kastner B. et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016; 353:1399–1405. [DOI] [PubMed] [Google Scholar]
- 13. Yan C., Wan R., Bai R., Huang G., Shi Y.. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science. 2016; 353:904–911. [DOI] [PubMed] [Google Scholar]
- 14. Zhang X., Yan C., Zhan X., Li L., Lei J., Shi Y.. Structure of the human activated spliceosome in three conformational states. Cell Res. 2018; 28:307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galej W.P., Wilkinson M.E., Fica S.M., Oubridge C., Newman A.J., Nagai K.. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016; 537:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wan R., Yan C., Bai R., Huang G., Shi Y.. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science. 2016; 353:895–904. [DOI] [PubMed] [Google Scholar]
- 17. Zhan X., Yan C., Zhang X., Lei J., Shi Y.. Structure of a human catalytic step I spliceosome. Science. 2018; 359:537–545. [DOI] [PubMed] [Google Scholar]
- 18. Bertram K., Agafonov D.E., Liu W.T., Dybkov O., Will C.L., Hartmuth K., Urlaub H., Kastner B., Stark H., Lührmann R.. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017; 542:318–323. [DOI] [PubMed] [Google Scholar]
- 19. Fica S.M., Oubridge C., Galej W.P., Wilkinson M.E., Bai X.C., Newman A.J., Nagai K.. Structure of a spliceosome remodelled for exon ligation. Nature. 2017; 542:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yan C., Wan R., Bai R., Huang G., Shi Y.. Structure of a yeast step II catalytically activated spliceosome. Science. 2017; 355:149–155. [DOI] [PubMed] [Google Scholar]
- 21. Zhang X., Yan C., Hang J., Finci L.I., Lei J., Shi Y.. An atomic structure of the human spliceosome. Cell. 2017; 169:918–929. [DOI] [PubMed] [Google Scholar]
- 22. Wilkinson M.E., Fica S.M., Galej W.P., Norman C.M., Newman A.J., Nagai K.. Postcatalytic spliceosome structure reveals mechanism of 3′-splice site selection. Science. 2017; 358:1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu S., Li X., Zhang L., Jiang J., Hill R.C., Cui Y., Hansen K.C., Zhou Z.H., Zhao R.. Structure of the yeast spliceosomal postcatalytic P complex. Science. 2017; 358:1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bai R., Yan C., Wan R., Lei J., Shi Y.. Structure of the post-catalytic spliceosome from Saccharomyces cerevisiae. Cell. 2017; 171:1589–1598. [DOI] [PubMed] [Google Scholar]
- 25. Wan R., Yan C., Bai R., Lei J., Shi Y.. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell. 2017; 171:120–132. [DOI] [PubMed] [Google Scholar]
- 26. Yan C., Hang J., Wan R., Huang M., Wong C.C.L., Shi Y.. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015; 349:1182–1191. [DOI] [PubMed] [Google Scholar]
- 27. Hang J., Wan R., Yan C., Shi Y.. Structural basis of pre-mRNA splicing. Science. 2015; 349:1191–1198. [DOI] [PubMed] [Google Scholar]
- 28. Cech T.R. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986; 44:207–210. [DOI] [PubMed] [Google Scholar]
- 29. Sharp P.A. On the origin of RNA splicing and introns. Cell. 1985; 42:397–400. [DOI] [PubMed] [Google Scholar]
- 30. Sharp P.A. Five easy pieces. Science. 1991; 254:663. [DOI] [PubMed] [Google Scholar]
- 31. Marcia M., Pyle A.M.. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012; 151:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017; 18:655–670. [DOI] [PubMed] [Google Scholar]
- 33. Galej W.P., Toor N., Newman A.J., Nagai K.. Molecular mechanism and evolution of nuclear pre-mRNA and group II intron splicing: insights from cryo-electron microscopy structures. Chem. Rev. 2018; 118:4156–4176. [DOI] [PubMed] [Google Scholar]
- 34. Grainger R.J., Beggs J.D.. Prp8 protein: at the heart of the spliceosome. RNA. 2005; 11:533–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan S.P., Kao D.I., Tsai W.Y., Cheng S.C.. The Prp19p-associated complex in spliceosome activation. Science. 2003; 302:279–282. [DOI] [PubMed] [Google Scholar]
- 36. Madhani H.D., Guthrie C.. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992; 71:803–817. [DOI] [PubMed] [Google Scholar]
- 37. Hilliker A.K., Staley J.P.. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004; 10:921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bao P., Hobartner C., Hartmuth K., Lührmann R.. Yeast Prp2 liberates the 5′ splice site and the branch site adenosine for catalysis of pre-mRNA splicing. RNA. 2017; 23:1770–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fabrizio P., Abelson J.. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990; 250:404–409. [DOI] [PubMed] [Google Scholar]
- 40. Yean S.L., Lin R.J.. U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction. Mol. Cell. Biol. 1991; 11:5571–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Warkocki Z., Odenwalder P., Schmitzova J., Platzmann F., Stark H., Urlaub H., Ficner R., Fabrizio P., Lührmann R.. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 2009; 16:1237–1243. [DOI] [PubMed] [Google Scholar]
- 42. Moore M.J., Sharp P.A.. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993; 365:364–368. [DOI] [PubMed] [Google Scholar]
- 43. Fabrizio P., Dannenberg J., Dube P., Kastner B., Stark H., Urlaub H., Lührmann R.. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009; 36:593–608. [DOI] [PubMed] [Google Scholar]
- 44. Fourmann J.B., Schmitzova J., Christian H., Urlaub H., Ficner R., Boon K.L., Fabrizio P., Lührmann R.. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013; 27:413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schneider C., Agafonov D.E., Schmitzova J., Hartmuth K., Fabrizio P., Lührmann R.. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3′ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLos Genet. 2015; 11:e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J.. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006; 34:W116–W118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jubb H.C., Higueruelo A.P., Ochoa-Montano B., Pitt W.R., Ascher D.B., Blundell T.L.. Arpeggio: a web server for calculating and visualising interatomic interactions in protein structures. J. Mol. Biol. 2017; 429:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fica S.M., Nagai K.. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol. 2017; 24:791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ohrt T., Prior M., Dannenberg J., Odenwalder P., Dybkov O., Rasche N., Schmitzova J., Gregor I., Fabrizio P., Enderlein J. et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012; 18:1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hilliker A.K., Mefford M.A., Staley J.P.. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007; 21:821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wlodaver A.M., Staley J.P.. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA. 2014; 20:282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boudvillain M., de Lencastre A., Pyle A.M.. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature. 2000; 406:315–318. [DOI] [PubMed] [Google Scholar]
- 54. Boudvillain M., Pyle A.M.. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J. 1998; 17:7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chanfreau G., Jacquier A.. Catalytic site components common to both splicing steps of a group II intron. Science. 1994; 266:1383–1387. [DOI] [PubMed] [Google Scholar]
- 56. Chanfreau G., Jacquier A.. An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J. 1996; 15:3466–3476. [PMC free article] [PubMed] [Google Scholar]
- 57. Konforti B.B., Abramovitz D.L., Duarte C.M., Karpeisky A., Beigelman L., Pyle A.M.. Ribozyme catalysis from the major groove of group II intron domain 5. Mol. Cell. 1998; 1:433–441. [DOI] [PubMed] [Google Scholar]
- 58. Ganguly M., Wang F., Kaushik M., Stone M.P., Marky L.A., Gold B.. A study of 7-deaza-2′-deoxyguanosine 2′-deoxycytidine base pairing in DNA. Nucleic Acids Res. 2007; 35:6181–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Forconi M., Benz-Moy T., Gleitsman K.R., Ruben E., Metz C., Herschlag D.. Exploring purine N7 interactions via atomic mutagenesis: the group I ribozyme as a case study. RNA. 2012; 18:1222–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chakrabarti P., Bhattacharyya R.. Geometry of nonbonded interactions involving planar groups in proteins. Prog. Biophys. Mol. Biol. 2007; 95:83–137. [DOI] [PubMed] [Google Scholar]
- 61. Singh J., Thornton J.M.. The interaction between phenylalanine rings in proteins. FEBS Lett. 1985; 191:1–6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.