Abstract
Polymorphisms in untranslated regions (UTRs) of disease-associated mRNAs can alter protein production. We recently identified a genetic variant in the 3′UTR of the TNFSF13B gene, encoding the cytokine BAFF (B-cell-activating factor), that generates an alternative polyadenylation site yielding a shorter, more actively translated variant, BAFF-var mRNA. Accordingly, individuals bearing the TNFSF13B variant had higher circulating BAFF and elevated risk of developing autoimmune diseases. Here, we investigated the molecular mechanisms controlling the enhanced translation of BAFF-var mRNA. We identified nuclear factor 90 (NF90, also known as ILF3) as an RNA-binding protein that bound preferentially the wild-type (BAFF-WT mRNA) but not BAFF-var mRNA in human monocytic leukemia THP-1 cells. NF90 selectively suppressed BAFF translation by recruiting miR-15a to the 3′UTR of BAFF-WT mRNA. Our results uncover a paradigm whereby an autoimmunity-causing BAFF polymorphism prevents NF90-mediated recruitment of microRNAs to suppress BAFF translation, raising the levels of disease-associated BAFF.
INTRODUCTION
Cellular responses to internal and external signals are potently controlled by post-transcriptional gene regulatory processes, including pre-mRNA splicing and mRNA maturation, transport, stability, and translation (1). Accordingly, all cellular events, including proliferation, apoptosis, senescence, differentiation, and the cellular response to immune stimuli, are strongly regulated by post-transcriptional factors. Among the most prominent post-transcriptional regulators are RNA-binding proteins (RBPs) and microRNAs; their influence is often elicited through specific sites in 3′-untranslated regions (UTRs) through which RBPs and microRNAs bind mRNAs and modulate mRNA turnover and/or translation (2–4). Immune responses, both physiologic and disease-associated, are strongly regulated by RBPs and microRNAs (5,6).
The large and heterogeneous family of autoimmune diseases including type-I diabetes, rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and multiple sclerosis (MS), is characterized by abnormal immune responses to normal body antigens mediated by specific immune cells and their products, leading to inflammation and tissue injury. Over the last decade, genome-wide association studies (GWAS) have uncovered several strong associations between genetic variants and risk of autoimmune disease (7). In a recent GWAS analysis of the Sardinian population, we uncovered the association of MS and SLE with a genetic variant in the 3′UTR of the TNFSF13B gene, which encodes the cytokine B-cell-activating-factor (BAFF), a member of the tumor necrosis factor superfamily (8). The causal variant was found to be an insertion-deletion (GCTGT > A, [GCTG/-] rs200748895 and [T/A] rs374039502 in the 1000 Genomes Project-variants data set) where ‘A’, the minor, risk-associated variant allele that is found with a frequency of 0.265 in Sardinians (‘BAFF-var’), created an upstream alternative polyadenylation site (APA). The resulting transcript, BAFF-var mRNA, was more actively translated than the long (wild-type) BAFF-WT mRNA, and led to higher production of soluble (s)BAFF. The short variant was more actively expressed partly because it lacked a site of repression by microRNA miR-15a; higher sBAFF in turn increased circulating B cells and immunoglobulins, raising the risk of MS and SLE (8). BAFF is mainly produced by monocytes, macrophages, neutrophils and dendritic cells (9); it binds to B-cell receptors and promotes B cell proliferation and differentiation (10–12). BAFF overexpression resulted in abnormal B cell-mediated autoimmune responses resulting in an increased risk of MS and SLE (8). Interestingly, the observed differences in sBAFF production and downstream phenotypes were only partially due to changes in the levels of BAFF-var mRNA compared with BAFF-WT mRNA (8), suggesting that both mRNA levels and translation rate were regulated.
Here, we investigated the molecular factors that regulate BAFF translation selectively in a polymorphism-dependent manner. Using different approaches to identify BAFF mRNA-associated molecules and their impact on BAFF production, we found that the RBP NF90 (ILF3) potently repressed BAFF translation from BAFF-WT mRNA but not from BAFF-var mRNA in THP-1 cells, a human monocytic leukemia line. Further results revealed that NF90 suppressed BAFF production by promoting the interaction of microRNA miR-15a with BAFF-WT mRNA. Our findings suggest that NF90 represses BAFF translation by recruiting miR-15a to BAFF-WT mRNA, and that the polymorphic variant BAFF-var, having lost the NF90 binding site, is now constitutively translated, producing higher levels of BAFF known to cause abnormal immune responses and promote disease.
MATERIALS AND METHODS
Cell culture and transfections
THP-1 human acute monocytic leukemia cells were cultured in RPMI-1640 medium and human cervical carcinoma HeLa cells were cultured in Dulbecco's modified essential medium, each supplemented with 10% fetal bovine serum and antibiotics. EBV-transformed B-lymphoblastoid cell lines from genotyped healthy human donors from Sardinia were chosen so as to include both BAFF-WT and BAFF-var alleles. Lymphoblastoid cell lines (LCL) were cultured in RPMI-1640 medium supplemented with 14% heat-inactivated fetal calf serum, antibiotics, 1% sodium pyruvate and 1% l-glutamine.
Control small interfering RNA (Ctrl siRNA) was from Qiagen, and the siRNAs directed to NF90, AGO1, AGO2, AGO3 and AGO4 were from Santa Cruz Biotechnology; miRNA mimics and inhibitors were purchased from Thermo Fisher Scientific and transfected at a concentration of 50 nM. Plasmids and small RNAs were transfected using Lipofectamine-2000 (Invitrogen) or electroporation using Amaxa (Lonza).
To generate THP-1 cells expressing constitutively lower levels of NF90, ready-to-use lentiviral particles expressing NF90 shRNA (containing three target-specific constructs that encode 19- to 25-nt short hairpin RNAs (shRNAs) designed to knock down NF90) or scramble control shRNA were purchased from Santa Cruz Biotechnology. Particles were added to THP-1 cells in the presence of polybrene (8 μg/ml). Puromycin (0.5 μg/ml) was added to the culture medium 1 week later to select for cells stably expressing integrated vectors. Pooled NF90-silenced populations were used for experiments.
EGFP-BAFF 3′UTR reporter constructs and Amaxa transfection
The pEGFP-C1 vector, expressing the EGFP coding region, was engineered to express the full-length (WT) 3′UTR of BAFF mRNA (pEGFP-C1-BAFF-WT 3′UTR) or the shorter variant (var) 3′UTR of BAFF mRNA (pEGFP-C1-BAFF-var 3′UTR). Plasmid pEGFP-C1-BAFF-WT 3′UTR was further modified by introducing a deletion of the NF90 site (pEGFP-C1-BAFF 3′UTR(ΔNF90), eliminating both miR-15a sites [pEGFP-C1-BAFF-WT 3′UTR (miR-15a mut)], and creating a mutant in which both miR-15a sites were moved [pEGFP-C1-BAFF-WT 3′UTR (new miR-15a)]. A naturally occurring stop codon located 50 nt after the beginning of the 3′UTR was maintained in all the EGFP expression vectors. Constructs were verified by sequencing. Reporter assays were carried out in THP-1 cells on 12-well plates. Cells expressing normal (Ctrl shRNA) or reduced levels (NF90 shRNA) of NF90 were transfected with 500 ng of either reporter construct, or empty vector using Amaxa (Lonza). Twenty-four hours after transfection, EGFP expression was measured by western blot analysis and the relative protein levels quantified by densitometry and normalized to the levels of a housekeeping protein (e.g. HSP90, ACTB).
RNA isolation and RT-qPCR analysis
RNA was isolated from cells using the TriPure isolation reagent (Roche) and the PureLink RNA mini Kit (Life Technologies) following the manufacturers’ protocols. Total RNA was reverse-transcribed into cDNA using Maxima reverse transcriptase (Thermo Fisher) and random hexamers and analyzed by quantitative (q)PCR analysis using SYBR Green mix (Kapa Biosystems) and gene-specific primers listed in Supplementary Table S2. The relative mRNA expression levels were calculated by the 2-ΔCt method. miRNAs were reverse-transcribed using MicroRNA first-strand synthesis (Clontech), and quantified by qPCR analysis using U6 as internal control and the primers listed in Supplementary Table S2.
Biotin pulldown and mass spectrometry (MS) analyses
To synthesize biotinylated RNAs corresponding to the BAFF 3′UTR and control RNAs, PCR fragments were prepared using forward primers that contained the T7 RNA polymerase promoter sequence [(T7), CCAAGCTTCTAATACGACTCACTATAGGGAGA] and reverse primers listed in Supplementary Table S2. After purifying the DNA template, biotinylated transcripts were synthesized using MaxiScript T7 kit (Ambion); whole-cell lysates (200 μg) from THP-1 cells were incubated with 1 μg of biotinylated RNA for 30 min at room temperature (13). Complexes were isolated with Streptavidin-coupled Dynabeads (Invitrogen) and proteins in the pulldown material were analyzed using western blot analysis and MS.
For MS analysis, peptide mixtures from each sample were loaded onto a peptide trap cartridge and eluted onto a reversed-phase PicoFrit column (New Objective, Woburn, MA, USA). Eluted peptides were ionized and sprayed into the mass spectrometer, using a Nanospray Flex Ion Source ES071 (Thermo Scientific). The LC/MS/MS analysis of samples were carried out using a Thermo Scientific Q-Exactive hybrid Quadrupole-Orbitrap Mass Spectrometer and a Thermo Dionex UltiMate 3000 RSLCnano System. Proteins were identified using the Thermo Proteome Discoverer 1.4.1 platform. Database search against public human protein database from NCBI was performed through the Proteome Discoverer 1.4.1 platform.
Immunoprecipitation of ribonucleoprotein (RNP) complexes (RIP analysis)
For RIP analysis, lysates of THP-1 cells were prepared in PEB buffer (10 mM Hepes, 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0.5% IGEPAL, 2 mM DTT, 50 U/ml RNase out and protease inhibitors); after clarification, lysates were incubated (2 h, 4°C) with a suspension of protein-A Sepharose beads precoated with 5 μg of anti-NF90 or mouse IgG (BD Transduction Laboratories). Following incubation, the beads were washed with NT2 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% IGEPAL) and subsequently treated with RNase-free DNase I (15 min, 30°C), and washed with NT2 buffer. RNA enrichments were analyzed by RT-qPCR using the primers listed in Supplementary Table S2.
Co-immunoprecipitation (IP) analysis
For co-IP analysis, THP-1 lysates were prepared in PEB buffer followed by 2 freeze/thaw cycles. After clarification and preclearing, lysates were incubated for 16 h at 4°C with protein-A Sepharose beads precoated with 5 μg of anti-NF90, anti-AGO2 or IgG (BD Transduction Labs). After incubation, the beads were washed in NT2 buffer for 5 min, with rotation. Proteins were detected by western blot analysis.
Western blot analysis and ELISA
Whole-cell lysates were prepared in RIPA buffer, and proteins were separated using electrophoresis in SDS-containing polyacrylamide gels (SDS-PAGE). Following transfer, nitrile membranes were incubated for 16 h with primary antibodies recognizing NF90 (BD Transduction Laboratories), BAFF, AGO3, AGO4 (Millipore), AGO1, AGO2 (Abcam), HSP90 α/β, GAPDH, HSP70, or EGFP (Santa Cruz Biotechnology). Secondary antibodies conjugated with horseradish peroxidase (HRP) (Kindle Biosciences) were detected by enhanced chemiluminescence (Kevik). The levels of soluble BAFF were evaluated using an ELISA Kit (Adipogene).
Polysome gradients
For polysome analysis, THP-1 or HeLa cells were incubated with 0.1 mg/ml cycloheximide for 10 min, and cytoplasmic extracts were fractionated through a linear sucrose gradient [10–50% (w/v)] as previously reported (14). Fractions were collected using a fraction collector and monitored by optical density measurement (A254) (Brandel). The RNA in each fraction was isolated and the relative enrichment was quantified using RT-qPCR analysis and the primers listed in Supplementary Table S2. The polysome data were displayed using the conventional representation of mRNAs on polysome gradients. The analysis was performed multiple times, preparing fresh gradients each time, and collecting RNA from each fraction for RT-qPCR quantification. The % mRNA in each fraction was then plotted relative to the total levels of that mRNA in the entire gradient. Given the variability in relative distribution among experimental repeats, mainly due to the fact that the gradients are not always identical, representative experiments are displayed in the main text; independent experiments are shown in Supplementary Figure S4B and C.
Biotinylated miRNA pulldown
THP-1 cells were transfected with biotinylated miR-15a or Ctrl (60 nM each) using Lipofectamine 2000. Forty-eight h later, cells were washed with PBS twice and lysed in 1 ml lysis buffer [20 mM Tris (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.3% NP-40, 100 U of RNase OUT (Invitrogen), protease inhibitor (Roche)], and incubated on ice for 10 min. After clearing the cytoplasmic lysate by centrifugation at 12 000 × g (10 min), steptavidin-coated Dynabeads (100 μl) were added and further incubated (4 h, 4°C with rotation). Beads were washed four times with 1 ml ice-cold lysis buffer, RNA was isolated using TriPure isolation solution, and the enrichment of BAFF mRNA was measured by RT-qPCR analysis and normalized to the levels of ACTB mRNA or 18S rRNA.
Statistical analysis
Data are presented as the means ± standard error of the means (SEM). Significance was tested using two-tailed Student's t test. P< 0.05 was considered significant.
RESULTS
BAFF-var mRNA is more actively translated
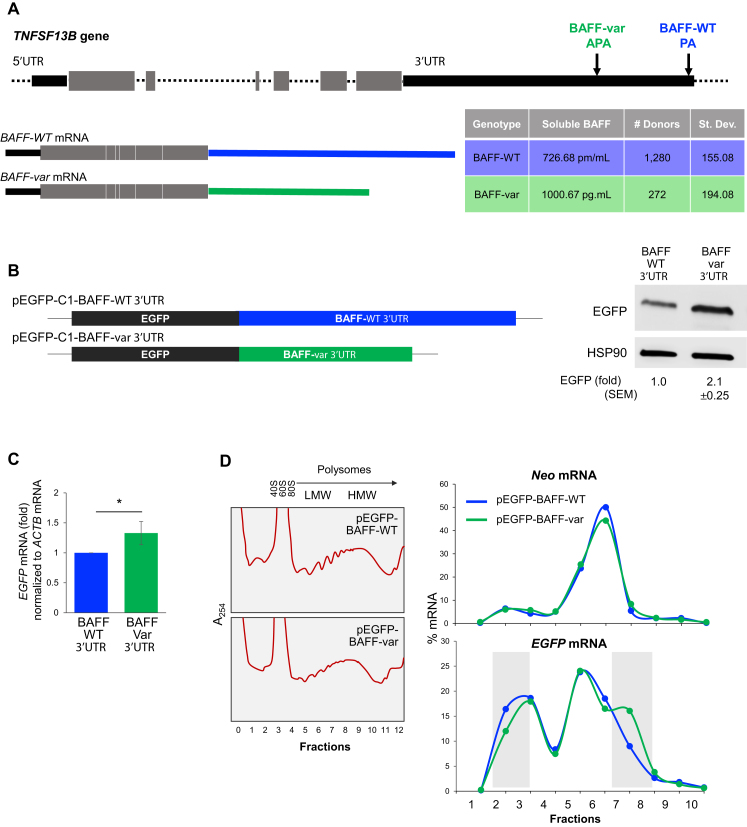
By GWAS analysis, we recently reported an association between the TNFSF13B gene, encoding the cytokine BAFF, with autoimmune diseases MS and SLE (8). Depicted in Figure 1A is the identified variant, an insertion-deletion in the 3′UTR, GCTGT>A, where the minor variant ‘A’, the risk allele, created an upstream alternative polyadenylation (APA) signal that gave rise to a shorter transcript, BAFF-var mRNA. Using serum from Sardinian participants, we found that individuals with both BAFF-WT alleles (n = 1280) expressed far lower sBAFF levels (726.7 pg/ml) than individuals with both BAFF-var alleles (n = 272, 1000.7 pg/ml) with an associated P-value of 8.89e−82 (Figure 1A). Interestingly, these differences were only partially due to changes in the levels of BAFF-var mRNA compared with BAFF-WT mRNA (8), suggesting regulation at both levels, increased mRNA levels and increased translation.
Figure 1.
Differential regulation of BAFF from BAFF-var and BAFF-WT mRNAs. (A) Top, schematic of the TNFSF13B gene, polyadenylation site (PA), and alternative polyadenylation site (APA) generated due to a polymorphic insertion-deletion. Bottom, mRNAs generated from TNFSF13B according to polyadenylation site usage [BAFF-WT mRNA (blue) and BAFF-var mRNA (green)], as well as relative circulating soluble BAFF proteins levels measured from the Sardinia cohort by ELISA in plasma from individuals homozygous for BAFF-WT (1280) or for BAFF-var (272) (8). (B) Left, reporter constructs derived from pEGFP-C1, carrying the BAFF-WT (top) and BAFF-var 3′UTR (bottom). Right, 24 h after transfecting HeLa cells with the constructs, western blot analysis was used to assess EGFP levels; HSP90 was assessed as a loading control. Signals were quantified using ImageJ. (C) EGFP mRNA levels in each transfection group, as measured by RT-qPCR analysis. (D) Left, 24 h after transfecting HeLa cells with the constructs in panel (B), cytoplasmic lysates were fractionated through sucrose gradients, and RNA was extracted from fractions 1 and 2 (free RNA/RNPs), fractions 3 and 4 [ribosomal subunits (40S, 60S) and monosomes (80S)], fractions 5–7 [low-molecular-weight (LMW) polysomes], and fractions 8–10 [high-molecular weight (HMW) polysomes]; fractions 11 and 12 typically had no detectable RNA and were discarded. Right, relative distribution of Neo mRNA, expressed from the pEGFP expression vectors and included as a control mRNA, and the EGFP-BAFF-WT and EGFP-BAFF-var mRNAs, expressed from the respective vectors, as measured by RT-qPCR analysis and represented as the percentage of total mRNA in the gradient. Shaded rectangles highlight regions of rightward shift in the distribution for EGFP-BAFF-var mRNA on sucrose gradient compared to EGFP-BAFF-WT mRNA. Results shown are representative of three independent experiments. In (B, C), data are the means and standard errors of the means (SEM) of three or more independent experiments (*P < 0.05; significance was determined by Student's t-test).
To investigate the mechanism through which higher levels of BAFF were produced from BAFF-var mRNA, we prepared reporter constructs that expressed chimeric mRNAs spanning the EGFP (enhanced green fluorescence protein) coding region (black) and the 3′UTR from either the BAFF-WT (blue) or BAFF-var (green): plasmids pEGFP-C1-BAFF-WT 3′UTR and pEGFP-C1-BAFF-var 3′UTR (Figure 1B). The impact of each 3′UTR was tested by transfecting HeLa cells with each reporter construct; 24 h later, western blot analysis of EGFP levels revealed that greater than twofold higher EGFP was expressed from the variant reporter (pEGFP-C1-BAFF-var 3′UTR) (Figure 1B, right). These differences did not completely arise from EGFP mRNA levels, assessed by reverse transcription (RT) followed by quantitative (q)PCR analysis (Figure 1C), as the variant transcript (EGFP-BAFF-var 3′UTR mRNA) was only 1.3-fold more abundant than the WT transcript (EGFP-BAFF-WT 3′UTR mRNA). These results agreed with our earlier findings (8) suggesting that BAFF production increased through both higher mRNA abundance and enhanced translation. A numerical measure of translation status known as the ‘translation index’ is calculated as the ratio of differences in protein levels relative to differences in the corresponding mRNA levels. For the reporters studied in Figures 1B and C, the abundance ratio of mRNAs produced from each vector (EGFP-BAFF-var 3′UTR mRNA relative to EGFP-BAFF-WT 3′UTR mRNA) is 1.3, while the ratio of EGFP proteins produced from each vector is 2.1. Thus, the relative translation index for these two constructs, 1.6, is consistent with the notion that translation is enhanced.
To test directly the possibility of regulated translation, we assessed the rate of translation of each chimeric mRNA by measuring the sizes of polysomes associated with each mRNA. Twenty-four hours following transfection, cytoplasmic preparations from transfected HeLa cells were fractionated through sucrose gradients, yielding fractions that lacked ribosome components (fractions 1 and 2), fractions with ribosome subunits 40S and 60S, as well as single monosomes (80S) (fractions 3 and 4), and fractions with low-molecular-weight (LMW) and high-molecular-weight (HMW) polysomes (fractions 5–7 and 8–10, respectively). As shown in Figure 1D, there were no major differences in polysome profiles between the transfection groups. After isolating RNA from each fraction, the levels of EGFP-BAFF-var and EGFP-BAFF-WT mRNAs were measured by RT-qPCR analysis and represented as a percent of that mRNA in each fraction relative to the total amount of mRNA in that sample. As shown in Figure 1D (right) and Supplementary Figure S4B, there was a modest but consistent shift between the distributions of the two chimeric mRNAs: compared to the distribution of EGFP-BAFF-WT mRNA, EGFP-BAFF-var mRNA shifted rightward, revealing that it associated with larger polysomes (a shift that was consistent with more active translation), and exhibited a second peak at fraction 7, indicating that a subset of larger polysomes formed on the EGFP-BAFF-var mRNA. By contrast, no such shifts were observed when measuring the levels of control neomycin-resistance (Neo) mRNA, expressed constitutively from the pEGFP-C1 reporter backbone, confirming that the two transfection groups shared similar rates of global protein translation, and further supporting the notion that translation of EGFP from the EGFP-BAFF-var chimeric mRNA was selectively enhanced. The relative distribution of the transcript encoding the housekeeping protein β-Actin (ACTB mRNA) was also comparable between the transfection groups (not shown). Together, these results indicate that compared to the 3′UTR of BAFF-WT mRNA, the shorter 3′UTR of BAFF-var mRNA conferred more robust translation of BAFF.
NF90 preferentially binds BAFF-WT 3′UTR
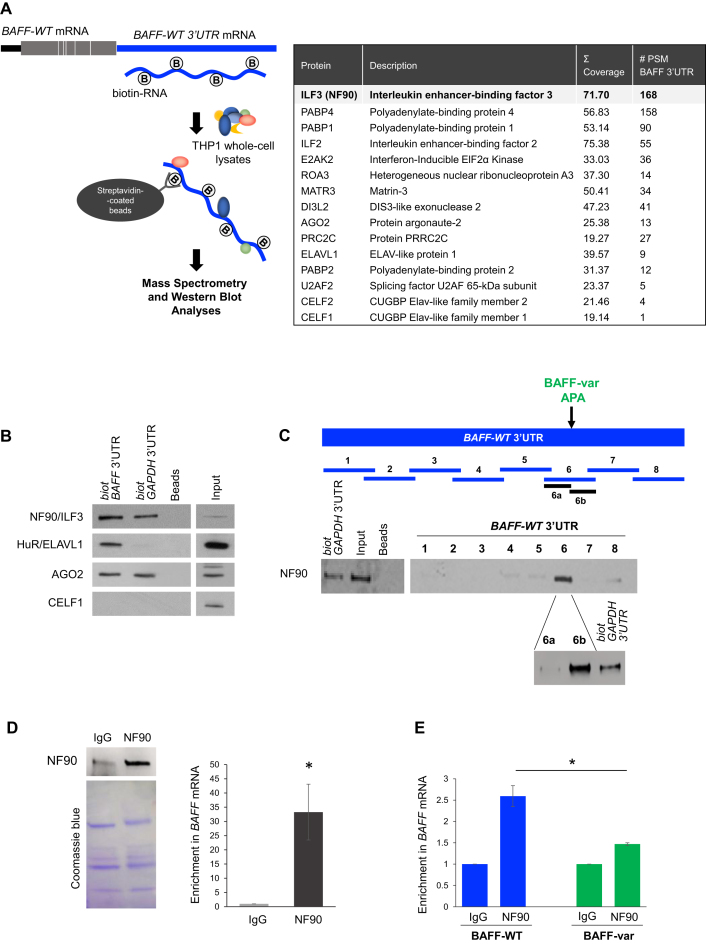
To search for factors that might modulate BAFF translation, we employed an affinity pulldown strategy to identify RBPs interacting with BAFF 3′UTR. Biotinylated BAFF 3′UTR was synthesized in vitro (Materials and Methods) and incubated with whole-cell extracts prepared from THP-1 cells, an immortalized monocytic leukemia line chosen because monocytes and macrophages are the main producers of BAFF. Ribonucleoprotein (RNP) complexes were pulled down using streptavidin beads and the interacting proteins were identified by mass spectrometry (MS) analysis (Figure 2A, left); control beads were analyzed in parallel. Among the many proteins detected in the pulldown materials, the most enriched RBP was the interleukin enhancer-binding factor 3 (ILF3, also known as nuclear factor 90, NF90) (Figure 2A, table). Following validation by western blot analysis of several RBPs identified in the MS screen, including HuR/ELAVL1, AGO2, and CELF1, NF90 was found to display most extensive binding to BAFF 3′UTR (Figure 2B). GAPDH 3′UTR, previously found to bind NF90 (not shown), was included as a positive control.
Figure 2.
NF90 binds distal to the APA in BAFF 3′UTR. (A) Left, schematic representation of BAFF-WT mRNA and the biotinylated full-length 3′UTR prepared for pulldown analysis using streptavidin beads to identify THP-1 proteins with affinity for BAFF 3′UTR. After pulldown, the material was washed and the associated proteins were size-fractionated by SDS-PAGE and analyzed by mass spectrometry (MS) analysis (right, Materials and Methods, Supplementary Table S1). Those RBPs enriched in biotin-BAFF 3′UTR relative to beads are listed, along with the number of peptides recovered for each RBP in the MS analysis (right). (B) Western blot analysis was performed to verify the specific association of biotin-BAFF 3′UTR with several RBPs (NF90/ILF3, HuR/ELAVL1, AGO2, CELF1) identified by MS analysis. (C) To identify the region of NF90 binding on BAFF mRNA, small biotin-RNA fragments spanning the 3′UTR were generated (fragments 1 through 8), incubated with THP-1 lysate, and subjected to pulldown followed by western blot analysis to identify the RNAs that associated with NF90. Fragment 6 was subdivided into fragments 6a and 6b, and each was assayed similarly for interaction with NF90. (D) The association of NF90 with BAFF mRNA was tested by ribonucleoprotein immunoprecipitation (RIP) analysis using anti-NF90 antibody. Left, after IP using anti-NF90 or IgG antibodies, the presence of NF90 in the IP material was assayed by western blot analysis (top), and equal loading was monitored by staining with Coomassie blue (bottom). Following RNA extraction, the enrichment of BAFF mRNA in NF90 IP samples was assessed by RT-qPCR analysis using BAFF-specific primers, normalized to the levels of UBC mRNA in each sample, and plotted relative to background BAFF mRNA levels in the IgG IP samples. (E) RIP analysis was subsequently performed in whole-cell lysates of LCL cells prepared from donors who were homozygotic for either BAFF-WT or BAFF-var. NF90 IP and RT-qPCR detection of BAFF mRNA were performed as described in panel 2D. Data in (B, C) are representative of at least 3 experiments; data in (D,E) are the means and SEM from at least 3 independent experiments (*P < 0.05; significance was determined by Student's t-test).
Finer mapping of the region(s) of interaction of NF90 with BAFF 3′UTR was conducted by preparing biotin-RNA segments spanning the BAFF 3′UTR. After incubation with THP-1 whole-cell lysates, pulldown analysis using streptavidin beads followed by NF90 western blot analysis (including the GAPDH 3′UTR RNA as a positive control) revealed that NF90 associated most avidly with segment 6, which spanned the APA; GAPDH 3′UTR was again included as a positive control. Following the division of RNA 6 into fragments 6a and 6b, fragment 6b was found to show stronger association. Interestingly, this fragment is located immediately downstream of the APA, supporting the notion that NF90 selectively binds the BAFF-WT mRNA isoform (Figure 2C).
To gain independent evidence that endogenous NF90 interacts with endogenous BAFF mRNA, we carried out RNP immunoprecipitation (RIP) analysis in THP-1 cells employing an anti-NF90 antibody and IgG in control IP reactions; NF90 levels in each IP sample were monitored by western blot analysis (Figure 2D). RNA was then extracted and subjected to RT-qPCR analysis using BAFF mRNA-specific primers. As shown in Figure 2D, BAFF mRNA associated with endogenous NF90 in THP-1 cells, as demonstrated by the enrichment in BAFF mRNA in NF90 RIP samples compared with IgG IP (Figure 2D); enrichments were normalized to the levels of ubiquitin C (UBC) mRNA in each IP reaction.
Additional evidence that NF90 bound the long BAFF mRNA was obtained using lymphoblastoid cells generated from donors from the SardiNIA cohort who were confirmed to have either BAFF-WT or BAFF-var genotypes. RIP analysis in these cells revealed that endogenous NF90 associated with BAFF mRNA significantly more in BAFF-WT cells (which expressed only BAFF-WT mRNA) compared with BAFF-var cells, which expressed high levels (∼70% for individuals homozygous for BAFF-var) of the shorter, BAFF-var mRNA (Figure 2E). Only BAFF-WT cells reached the 2-fold enrichment that is generally used as the cut-off to identify bona fide interactions. BAFF-var cells showed a 1.5-fold enrichment likely the result of (i) weak binding of the proximal half of the BAFF mRNA (fragments 1–5 in Figure 2C) and (ii) the presence in BAFF-var cells of a mixture of BAFF-WT mRNA (∼30%) and BAFF-var mRNA (∼70%).
Taken together, these data indicate that NF90 preferentially binds the 3′UTR of the BAFF-WT mRNA over the BAFF-var mRNA.
NF90 suppresses BAFF translation
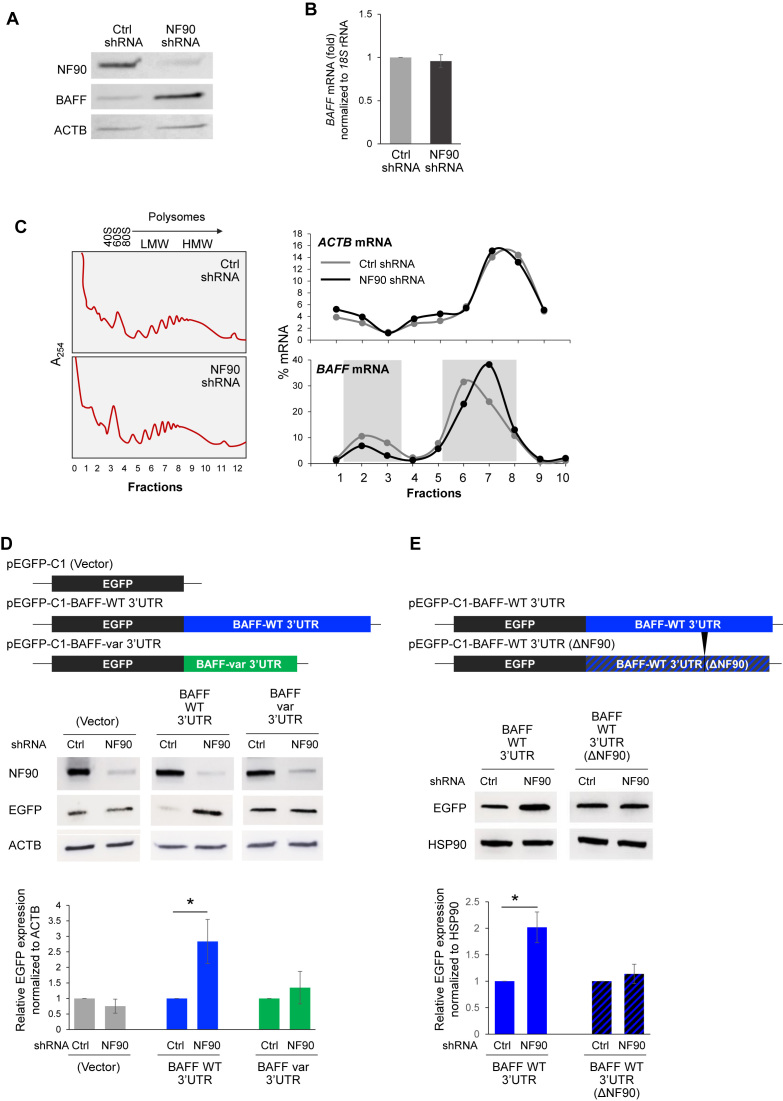
To investigate if NF90 regulates BAFF production, we generated a THP-1 line (pooled population) stably expressing NF90 shRNA, along with a scrambled shRNA control line. Interestingly, the reduction in NF90 levels in THP-1 cells led to higher BAFF protein production (Figure 3A) and higher soluble BAFF (not shown); no significant changes in BAFF mRNA levels were observed after lowering NF90 abundance in THP-1 cells (Figure 3B). The hypothesis that NF90 might reduce BAFF mRNA translation without affecting its stability was in agreement with earlier reports that NF90 represses the translation of several cytokines without altering the half-lives of the respective mRNAs (13,15).
Figure 3.
NF90 represses BAFF translation. (A, B) Western blot analysis (A) and BAFF mRNA (B) produced by THP-1 cell populations with stably reduced NF90 levels (NF90 shRNA) and control cells (Ctrl shRNA). (C) The relative distribution of BAFF mRNA (and ACTB mRNA, encoding the housekeeping protein β-Actin) in polysomes prepared from the THP-1 populations described in (A) was assessed by RT-qPCR analysis as described in Figure 1D. Shaded rectangles highlight the rightward shift in the distribution of BAFF mRNA in sucrose gradients in the NF90 shRNA group compared to the Ctrl shRNA group. (D) Schematic of the EGFP reporter constructs bearing the BAFF-WT 3′UTR, the BAFF-var 3′UTR, or no 3′UTR (Vector) used for reporter analysis. Twenty-four hours after transfecting the THP-1 cells described in panel (A) with each EGFP vector, EGFP expression levels were assessed by western blot analysis (top). EGFP signals from three experiments were analyzed using imageJ and plotted relative to Ctrl shRNA in each transfection group (bottom). (E) Schematic of EGFP reporter constructs bearing the BAFF-WT 3′UTR, or the BAFF-WT 3′UTR(ΔNF90) 3′UTR. Twenty-four hours after transfecting THP-1 cells as described in (A) with each vector, EGFP expression levels were assessed by western blot analysis. EGFP signals from three experiments were quantified using imageJ and plotted relative to Ctrl shRNA in each transfection group. Data in (B, D, E) are the means and SEM from at least 3 independent experiments (*P < 0.05; significance was determined by Student's t-test).
To test this hypothesis, we monitored the distribution of BAFF mRNA in polysome gradients prepared from control and NF90-silenced THP-1 cells. As shown in a representative experiment (Figure 3C, left), global polysome profiles were similar between the two cell lines. However, the distribution of BAFF mRNA along the gradient, as determined by RT-qPCR analysis, shifted rightward when NF90 was silenced, yielding relatively lower BAFF mRNA levels in unbound fractions (e.g. fractions 2 and 3) and relatively higher in polysomal fractions (e.g. fractions 6 and 7). This pattern was indicative of BAFF mRNA forming larger polysomes and hence being more actively translated (Figure 3C, right). ACTB mRNA, encoding the housekeeping protein β-Actin, was found distributed with comparable patterns regardless of NF90 status (Figure 3C, right and Supplementary Figure S4C). To validate this result using a different approach, THP-1 cells were transiently transfected with siRNA directed to NF90 or control siRNA; analysis of polysomes from these cells was also consistent with NF90 suppressing BAFF mRNA translation (Supplementary Figure S1A–D).
To further investigate the regulation of BAFF production by NF90, and to address whether NF90 affected differentially the translation of BAFF-WT and BAFF-var mRNAs, we studied the influence of NF90 on the production of EGFP from reporter constructs bearing BAFF-WT or BAFF-var 3′UTRs (Figure 1B) in THP-1 cells with NF90 stably silenced (described in Figure 3A). Twenty-four hours after transfection of THP-1 cells with the EGFP reporters shown in Figure 3D, the levels of EGFP produced were assessed by western blot analysis. In NF90-silenced cells, the levels of EGFP synthesized from a chimeric mRNA bearing the BAFF-WT 3′UTR were >2-fold higher than those synthesized from cells expressing normal NF90 levels or from cells expressing BAFF-var 3′UTR (or no BAFF sequences, ‘Vector’) (Figure 3D, right). These findings indicate that NF90 represses BAFF translation by acting upon the full-length BAFF 3′UTR allele (WT) but does not repress BAFF production from the shorter BAFF 3′UTR allele (var).
To test directly whether NF90 regulates BAFF production, we generated a vector that expressed a BAFF 3′UTR lacking the NF90 binding site [pEGFP-BAFF 3′UTR (ΔNF90) (Figure 3E)]. BAFF 3′UTR WT and ΔNF90 reporters were transfected into THP-1 cells in which NF90 was stably silenced, and EGFP expression was analyzed 24 h later. As shown in Figure 3E, EGFP production from EGFP-BAFF 3′UTR(ΔNF90) mRNA failed to respond to change in NF90 abundance, supporting the notion that NF90 binding to BAFF 3′UTR was required to regulate BAFF expression.
Collectively, our results indicate that NF90 represses BAFF production and secretion by lowering BAFF mRNA translation. They further reveal that NF90 actively represses the translation of full-length BAFF mRNA (BAFF-WT mRNA), but not the short variant (BAFF-var mRNA) lacking the site of NF90 binding.
NF90 represses BAFF translation by promoting the association of miR-15a with BAFF mRNA
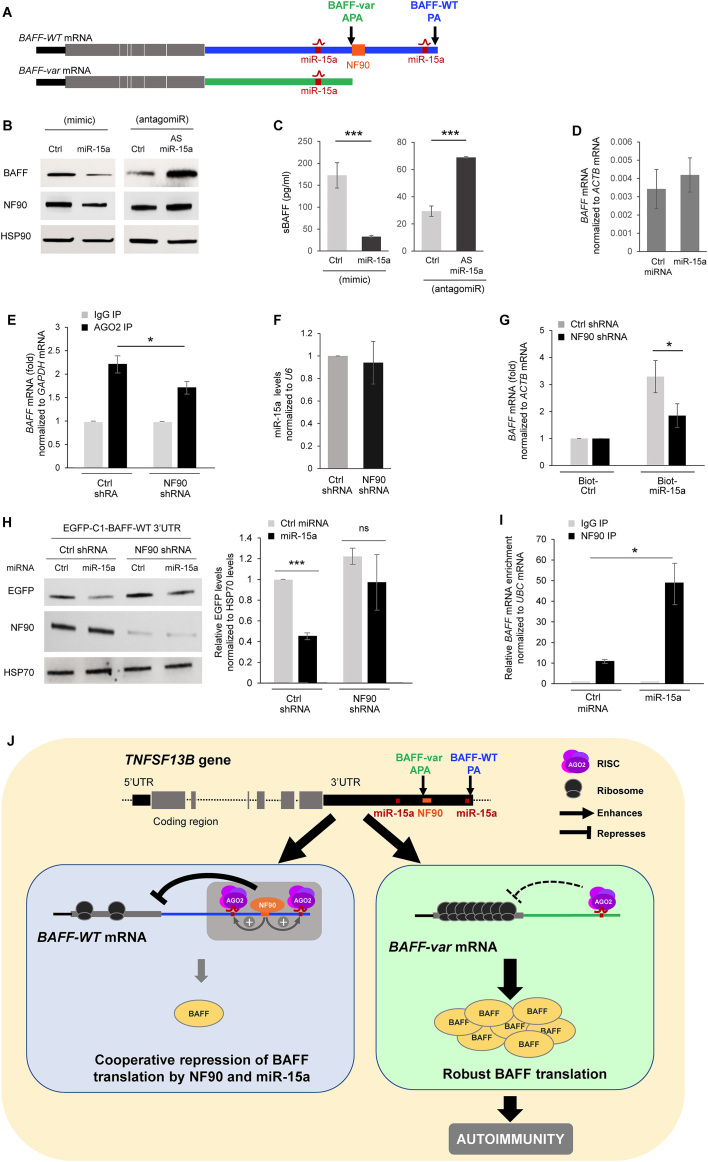
We hypothesized that the translational repression elicited by NF90 might be linked to the previously identified inhibition of BAFF production by miR-15a (8). Given that there is one miR-15a site on BAFF-var mRNA and two on BAFF-WT mRNA (Figure 4A), BAFF-var mRNA was more abundant constitutively and produced higher levels of BAFF protein, as previously discovered by cotransfection of HeLa cells with miR-15a and pmirGLO-BAFF 3′UTR constructs (8). Indeed, only BAFF-WT mRNA associated with NF90 also had a miR-15a site downstream of the APA (Figure 4A). Thus, by 48 h after transfecting THP-1 cells (which bear two BAFF-WT alleles and hence only express BAFF-WT mRNA) with miR-15a mimic to enhance miR-15a levels (Supplementary Figure S1E), BAFF production and secretion were reduced, while transfection of a miR-15a antagomiR [antisense (AS) miR-15a] to suppress miR-15a function increased BAFF production and secretion (Figure 4B and C). miR-15a overexpression did not affect BAFF mRNA levels significantly (Figure 4D). Interestingly, overexpressing miR-15a reduced NF90 levels while antagonizing miR-15a elevated NF90 levels (Figure 4B).
Figure 4.
NF90 cooperates with miR-15a to suppress BAFF translation. (A) Schematic of BAFF-WT and BAFF-var mRNAs, with the location of miR-15a and NF90 indicated. (B, C) The levels of BAFF, NF90 and sBAFF produced by THP-1 cells were assessed by western blot analysis (B) or ELISA (C) 48 h after overexpressing miR-15a by transfecting a miR-15a mimic (left) or neutralizing miR-15a by transfecting a miR-15a antagomir (right). (D) BAFF mRNA levels in THP-1 cells transfected with miR-15a mimic as in panel (B) were measured 48 h later by RT-qPCR analysis. (E) AGO2 RIP analysis was performed in cells expressing normal (Ctrl shRNA) or reduced (NF90 shRNA) NF90 levels to identify AGO2-bound BAFF mRNA. (F) The steady-state levels of miR-15a in the cells described in panel (E) were assessed by RT-qPCR analysis. (G) The cells described in (E) were transfected with biotinylated miR-15a or a biotinylated control RNA; 48 h later, cells were lysed and the levels of BAFF mRNA enrichment in the biot-miR-15a pulldown were assessed by RT-qPCR analysis. (H) THP-1 cells described in (E) were co-transfected with a chimeric reporter construct expressing EGFP-BAFF-WT 3′UTR mRNA and either Ctrl miRNA or miRNA-15a mimic; 24 h later, EGFP expression levels were assessed by western blot analysis (left). EGFP signals were quantified using ImageJ and plotted (right). (I) Forty-eight hours after transfecting THP-1 cells with Ctrl miRNA or miR-15a mimic, the interaction of NF90 with BAFF mRNA was assessed by RIP followed by RT-qPCR analysis. (J) Model summarizing the results of this study. The wild-type TNFSF13B gene uses the constitutive polyadenylation site (blue, PA) to give rise to BAFF-WT mRNA (left), bearing a long 3′UTR with two miR-15a sites and an NF90 binding site; the cooperative actions of these factors contribute to maintaining low BAFF translation levels and overall BAFF production. The variant TNFSF13B gene uses with high frequency an upstream polyadenylation site (green, APA) that gives rise to a shorter transcript, BAFF-var mRNA (right), bearing a shorter 3′UTR that lacks the distal miR-15a site and the NF90 binding site; consequently, BAFF-var mRNA is translated at aberrantly higher levels and results in the production of higher BAFF in individuals bearing this variant sequence (8). In (C, D, G, H, I), the data represent the means and SEM from three independent experiments (*P < 0.05, ***P< 0.001; significance was determined by Student's t-test).
To test if miR-15a and NF90 actions were functionally linked, we first investigated, in THP-1 cells, if the microRNA-binding AGO protein family associated with BAFF mRNA differently depending on NF90 levels. Since MS analysis (Table in Figure 2A) indicated that AGO2, but not other AGO proteins, bound BAFF mRNA, we concentrated our analysis on AGO2. The interaction of other AGO proteins with BAFF mRNA, and the possible regulation of BAFF expression by other AGO proteins was analyzed (Supplementary Figures S2 and S3, Supplementary text). We observed interaction between AGO2 and NF90, as determined by co-IP analysis in THP-1 cells (Supplementary Figure S2C), and further established that the interaction was RNA-dependent (not shown). As shown in Figure 4E, in THP-1 cells constitutively expressing lower NF90 levels, the interaction of AGO2 with BAFF mRNA, as assessed by RIP analysis using an anti-AGO2 antibody (Materials and Methods), was lower than in control cells, indicating that NF90 increased the interaction of microRNA-AGO2 complexes with BAFF mRNA. Control shRNA and NF90 shRNA populations of THP-1 cells had similar levels of miR-15a (Figure 4F).
Next, we assayed whether the binding of miR-15a to BAFF mRNA was altered in cells with reduced NF90 levels. Forty-eight hours after transfection of biotinylated miR-15a, lysis, and pulldown using streptavidin-coated beads, significantly more BAFF mRNA was recovered in THP-1 cells with normal NF90 levels (Ctrl shRNA) than in THP-1 cells with silenced NF90 (Figure 4G). Furthermore, cells in which NF90 was silenced displayed an impaired ability to lower BAFF production by miR-15a, as assessed by measuring EGFP production from the reporter construct pEGFP-C1-BAFF-WT 3′UTR (Figure 4H). As expected, baseline production of EGFP from this construct increased in the NF90 shRNA group, since reducing NF90 levels de-repressed EGFP production driven by the BAFF-WT 3′UTR. A direct role of miR-15a in regulating BAFF mRNA was further confirmed by using a pEGFP-BAFF 3′UTR construct in which the miR-15a binding sites were mutagenized; this vector was refractory to the repressive effects of miR-15a (Supplementary Figure S4A). Interestingly, NF90 binding to BAFF mRNA increased when miR-15a was overexpressed in THP-1 cells (Figure 4I), supporting the notion that miR-15a facilitated BAFF mRNA-NF90 interaction (Figure 4I).
Taken together, these findings indicate that the presence of NF90 contributed to the interaction of BAFF-WT mRNA with miR-15a to elicit BAFF repression (Figure 4J).
DISCUSSION
We have identified NF90 as an RBP that associates with the most common BAFF mRNA (BAFF-WT mRNA) and reduces BAFF translation. The shorter variant BAFF-var mRNA lacks the NF90 site and is thus more actively translated, leading to enhanced BAFF production in THP-1 cells (Figure 4J). The differential impact of NF90 on BAFF production recapitulates the effects reported for miR-15a, which selectively repressed BAFF-WT mRNA but not BAFF-var mRNA (8). Importantly, the interaction of miR-15a with BAFF-WT mRNA was enhanced by NF90, suggesting a cooperative mode of action whereby NF90 binding to BAFF mRNA facilitates miR-15a recruitment and translational repression of BAFF mRNA (Figure 4J). Interestingly, this cooperation may be further solidified by the miR-15a-promoted interaction of NF90 with BAFF mRNA (Figure 4I). The reduction of NF90 levels by miR-15a (Figure 4B) may represent a compensatory mechanism to keep the suppression of BAFF production by miR-15a in check; additional investigation is needed to fully understand this regulatory loop.
The BAFF-WT 3′UTR is longer than the BAFF-var 3′UTR. Long 3′UTRs generally enable additional regulation of the mRNA, influencing both their stability and translational efficiency (16). The presence of additional regulatory motifs drive the assembly of RNP complexes that regulate mRNA abundance, as well as the time and location of protein production (17). Here, the cooperative binding of miR-15a and NF90 to BAFF-WT mRNA that suppressed BAFF translation was lost in the shorter BAFF-var mRNA. Additional studies are needed to identify other possible cis elements driving BAFF expression through the short and long 3′UTRs.
There are several reports of coordinated actions of RBPs and microRNAs on a given mRNA, as reviewed (18,19). This coordination can occur sequentially (if the microRNA and the RBP bind one after the other) or simultaneously (if they bind at distant or proximal locations at the same time). Proximal interactions may be competitive, if the binding of one displaces or prevents the binding of the other, or cooperative, if one enhances the binding of the other, as observed here for NF90 and miR-15a. Functionally, both factors may have a negative impact on an mRNA (e.g. both reduce stability or translation), or both may have a positive effect; in each case, their joint impacts might then be additive or synergistic. If the factors have opposite effects, then the net impact on the target mRNA can be more complex and will depend on their relative availabilities at a given place and time.
However, only a handful of cooperative interactions between RBPs and microRNAs affecting mRNA fate have been reported. For example, binding of RBP PUM1 to target p27 mRNA opened the local RNA secondary structure, allowing miRNAs miR-221 and miR-222 to bind and suppress the target mRNAs (20). Similarly, binding of RBP HuR to a long noncoding RNA (lincRNA-p21) promoted let-7 association with lincRNA-p21 and triggered lincRNA-p21 degradation (21). In this regard, the discovery that NF90 promoted the suppressive actions of miR-15a represents a novel mechanism of action for NF90, with potentially important implications for immune gene expression programs.
Generated by alternative splicing of the ILF3 gene, NF90 is a ubiquitous protein that is localized primarily in the nucleus but is also present in the cytoplasm. Although NF90 is capable of binding DNA and regulating transcription, as reviewed by Castella et al. (22), its ability to bind single- and double-stranded RNA has established NF90 as a major regulator of mammalian and viral RNA metabolism. NF90 was found to stabilize several mRNAs, including those that encode p21/CDKN1A, interleukin 2 (IL2), vascular endothelial growth factor (VEGF), and mitogen-activated protein kinase phosphatase 1 (MKP-1/DUSP1), typically by binding their 3′UTRs (22). NF90 also repressed the translation of several mRNAs, including those that encode cytokines and chemokines essential for inflammation and senescence-associated secretory phenotype (SASP) observed in many primary cells that have ceased division in response to telomere attrition or have encountered other sublethal damage. In proliferating cells, high levels of NF90 helped suppress the translation of SASP factors MCP-1/CCL2, GROa/CXCL1, IL6 and IL8, while in senescent cells, which expressed very low levels of NF90, translation of the same SASP factors was strongly elevated (13,15). In the context of senescence, SASP and proinflammatory cytokines, it is interesting to note that NF90 suppressed the processing of several primary microRNAs, including pri-let-7, thereby reducing the levels of the potent growth-inhibitory microRNA let-7 (23,24) and potentially delaying the onset of a senescent phenotype.
Given that BAFF is a vital cytokine for B cells, the regulatory paradigm described here strengthens the involvement of NF90 in controlling immunity and the production of cytokines (13,15,22). In this regard, NF90 was proposed to have a direct role in regulating Psoriasis vulgaris (25), an autoimmune disease influenced by abnormal B cell function (26). In sum, NF90 influences the immune response by controlling the production of immune factors including BAFF.
The finding that NF90 functions jointly with miR-15a to suppress BAFF translation critically advances our understanding of the molecular mechanisms underlying our earlier results that the BAFF-var genotype elevated increased soluble BAFF, increasing the risk of autoimmunity (8). The translation of BAFF-WT mRNA (bearing two miR-15a sites, one on each side of the APA) was strongly suppressed by miR-15a, while BAFF-var mRNA translation (bearing only one miR-15a site located 5′ of the APA) was less affected. BAFF-var mRNA lacks one miR-15a binding site and therefore it is subject to less repression and produces more BAFF protein constitutively. Since only BAFF-WT mRNA can jointly harbor both NF90 and two miR-15a, only BAFF-WT mRNA would be expected to be subject to robust cooperative repression. By contrast, BAFF-var mRNA would be refractory to repression and would be more actively translated, giving rise to high levels of secreted BAFF as measured in SardiNIA donors with the BAFF-var genotype, who show high propensity to autoimmune disorders such as MS and SLE, linked to elevated BAFF concentrations (8). Our results also point to new therapeutic strategies which might be devised to strengthen NF90 binding to BAFF mRNA to repress BAFF translation, raise miR-15a levels, or both, as we seek to intervene therapeutically to reduce the levels of factors that trigger or sustain autoimmunity states.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIA-IRP, NIH; [N01-AG-1-2109 and HHSN271201100005C]; Italian Foundation for Multiple Sclerosis [FISM 2011/R/13 and 2015/R/09]; Fondazione di Sardegna (ex Fondazione Banco di Sardegna, Prot. U1301.2015/AI.1157.BE Prat. 2015-1651). The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Orphanides G., Reinberg D.. A unified theory of gene expression. Cell. 2002; 108:439–451. [DOI] [PubMed] [Google Scholar]
- 2. Glisovic T., Bachorik J.L., Yong J., Dreyfuss G.. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008; 582:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krol J., Loedige I., Filipowicz W.. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010; 11:597–610. [DOI] [PubMed] [Google Scholar]
- 5. Ivanov P., Anderson P.. Post-transcriptional regulatory networks in immunity. Immunol. Rev. 2013; 253:253–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bisogno L.S., Keene J.D.. RNA regulons in cancer and inflammation. Curr. Opin. Genet. Dev. 2017; 48:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kochi Y. Genetics of autoimmune diseases: perspectives from genome-wide association studies. Int. Immunol. 2016; 28:155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steri M., Orrù V., Idda M.L., Pitzalis M., Pala M., Zara I., Sidore C., Faà V., Floris M., Deiana M. et al. . Overexpression of the Cytokine BAFF and autoimmunity risk. N. Engl. J. Med. 2017; 376:1615–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schneider P., Tschopp J.. BAFF and the regulation of B cell survival. Immunol. Lett. 2003; 88:57–62. [DOI] [PubMed] [Google Scholar]
- 10. Gross J.A., Johnston J., Mudri S., Enselman R., Dillon S.R., Madden K., Xu W., Parrish-Novak J., Foster D., Lofton-Day C. et al. . TACI and BCMA are receptors for a TNF homologue implicated in B-cell autoimmune disease. Nature. 2000; 404:995–999. [DOI] [PubMed] [Google Scholar]
- 11. Mackay F., Schneider P.. Cracking the BAFF code. Nat. Rev. Immunol. 2009; 9:491–502. [DOI] [PubMed] [Google Scholar]
- 12. Rauch M., Tussiwand R., Bosco N., Rolink A.G.. Crucial role for BAFF-BAFF-R signaling in the survival and maintenance of mature B cells. PLoS One. 2009; 4:e5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuwano Y., Pullmann R. Jr, Marasa B.S., Abdelmohsen K., Lee E.K., Yang X., Martindale J.L., Zhan M., Gorospe M.. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010; 38:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panda A.C., Martindale J.L., Gorospe M.. Polysome fractionation to analyze mRNA distribution profiles. Biol. Protoc. 2017; 7:e2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tominaga-Yamanaka K., Abdelmohsen K., Martindale J.L., Yang X., Taub D.D., Gorospe M.. NF90 coordinately represses the senescence-associated secretory phenotype. Aging. 2012; 4:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tanguay R.L., Gallie D.R.. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996; 16:146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doran G. The short and the long of UTRs. J. RNAi Gene Silencing. 2008; 4:264–265. [PMC free article] [PubMed] [Google Scholar]
- 18. Ciafrè S.A., Galardi S.. microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol. 2013; 10:935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ho J.J., Marsden P.A.. Competition and collaboration between RNA-binding proteins and microRNAs. Wiley Interdiscip. Rev. RNA. 2014; 5:69–86. [DOI] [PubMed] [Google Scholar]
- 20. Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A., Elkon R., Agami R.. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010; 12:1014–1020. [DOI] [PubMed] [Google Scholar]
- 21. Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M.. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012; 47:648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Castella S., Bernard R., Corno M., Fradin A., Larcher J.C.. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip. Rev. RNA. 2015; 6:243–256. [DOI] [PubMed] [Google Scholar]
- 23. Sakamoto S., Aoki K., Higuchi T., Todaka H., Morisawa K., Tamaki N., Hatano E., Fukushima A., Taniguchi T., Agata Y.. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell. Biol. 2009; 29:3754–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masuda K. NF90 in posttranscriptional gene regulation and MicroRNA biogenesis. Int. J. Mol. Sci. 2013; 14:17111–17121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stuart P.E. Genome-wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. Am. J. Hum. Genet. 2015; 97:816–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hayashi K. Gemcitabine enhances rituximab‐mediated complement‐dependent cytotoxicity to B cell lymphoma by CD20 upregulation. Cancer Sci. 2016; 107:682–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.