Abstract
N 6-methyldeoxyadenosine (6mA) is a well-characterized DNA modification in prokaryotes but reports on its presence and function in mammals have been controversial. To address this issue, we established the capacity of 6mA-Crosslinking-Exonuclease-sequencing (6mACE-seq) to detect genome-wide 6mA at single-nucleotide-resolution, demonstrating this by accurately mapping 6mA in synthesized DNA and bacterial genomes. Using 6mACE-seq, we generated a human-genome-wide 6mA map that accurately reproduced known 6mA enrichment at active retrotransposons and revealed mitochondrial chromosome-wide 6mA clusters asymmetrically enriched on the heavy-strand. We identified a novel putative 6mA-binding protein in single-stranded DNA-binding protein 1 (SSBP1), a mitochondrial DNA (mtDNA) replication factor known to coat the heavy-strand, linking 6mA with the regulation of mtDNA replication. Finally, we characterized AlkB homologue 1 (ALKBH1) as a mitochondrial protein with 6mA demethylase activity and showed that its loss decreases mitochondrial oxidative phosphorylation. Our results show that 6mA clusters play a previously unappreciated role in regulating human mitochondrial function, despite 6mA being an uncommon DNA modification in the human genome.
INTRODUCTION
DNA methylation is a common epigenetic modification where underlying genomic sequences are methylated without altering their coding capacity. The most well-characterized type of human DNA methylation is 5-methyldeoxycytosine (5mC) that regulates genomic imprinting, X-chromosome inactivation and transposon silencing, which are all essential for normal growth and development (1,2). Compared to 5mC, less is known about the importance of 6mA in the field of mammalian epigenetics. That is because 6mA was previously believed to exist only in prokaryotes, unicellular eukaryotes and plants where its functions vary from regulating DNA replication and repair to gene expression and host genome defence (3,4). However, recent reports have demonstrated the presence and functionality of 6mA in various animal species, including mammals (5–12). In mouse prefrontoral cortex during stress, 6mA presents on long interspersed nuclear element 1 (LINE1) transposons and is associated with LINE1 repression (9). In mouse embryonic stem cells, 6mA is enriched in young and active LINE1 transposons and acts to silence these transposons (10). 6mA enrichment on active LINE1s was also recently shown to be conserved in human cells (11). However, another recent report found that 6mA was not enriched in transposons but rather in exon-coding regions, and was also associated with transcriptional activation (12). This was despite the fact that both human studies utilized single-molecule real-time (SMRT) sequencing technology for mapping 6mA.
The ability to accurately and precisely sequence 6mA genome-wide can facilitate the functional characterization of human 6mA and help resolve the conflicting results thus far. While there currently exists methods for 6mA sequencing, they all exhibit various disadvantages, such as poor resolution (6mA-DNA-immunoprecipitation-sequencing or 6mA-DIP-seq (5–10)) or sequence motif limitations (DpnI-assisted-6mA-sequencing or DA-6mA-seq (13)). When sequencing 6mA in bacterial genomes where 6mA density is high, SMRT-sequencing is preferred because it can achieve single-nucleotide-resolution (14). However, 6mA identification via SMRT-sequencing is fraught with a high false discovery rate in mammalian genomes where the absolute abundance of 6mA has been shown to be low (11,15,16). This problem is further exacerbated by the high 5mC density within the human genome, which complicates the analysis needed to accurately identify 6mA sites (11,17). In order to overcome these complications and accurately identify 6mA using SMRT-sequencing, a sufficiently high coverage is necessary. While this is easily achieved when sequencing megabase scale bacterial genomes, it becomes extremely costly and time-consuming when sequencing and analyzing at the magnitude of gigabase scale human genomes. As such, it is also impractical to sequence multiple samples or replicates using this method.
In our work, we optimized an alternative 6mA sequencing strategy to map 6mA in human genomes at single nucleotide resolution. In doing so, we discovered and characterized novel mitochondrial chromosomal-wide 6mA clusters as well as a putative mitochondrial 6mA reader and eraser. This led to new insights on how human 6mA can regulate mitochondrial DNA (mtDNA) replication and mitochondrial function. Finally, we also addressed recent debates over the location, abundance and function of human 6mA (11,12).
MATERIALS AND METHODS
Bacterial strains
Dam+ UTI89 Escherichia coli (E. coli), Dam+ Salmonella Typhimurium 14028s and Dam- S. Typhimurium 14028s were all cultivated overnight at 37°C in L-Broth.
Tissue culture
ATCC HEK293T CRL-3216 cells were cultivated in a sterile 5% CO2 incubator at 37°C, and in Dulbecco's Modified Eagle Media supplemented with 10% Fetal Bovine Serum and 1% penicillin/streptomycin, and used within passage number 3–15. HEK293T cells designated for any genomic extractions were always subjected to MycoAlert Plus Mycoplasma kit (Lonza LT07) and occasionally to EZ-polymerase chain reaction (PCR) Mycoplasma Detection kit (Biological Industries 20-700-20) to verify that they were mycoplasma-free. For DNA extraction, cells were trypsinized and centrifuged in cold phosphate buffered saline twice at 200 g for 5 min at 4°C. Cells were frozen in liquid nitrogen and stored at −80°C before DNA extraction.
Purification of intact mitochondria for mtDNA extraction
mtDNA was purified by adapting a previously developed protocol (18). About 20 million cells were first lysed by douncing with a 2 ml tissue mortar/pestle set (Fisher Scientific K885300-0002). Intact mitochondria were then purified according to manufacturer’s instructions using the Mitochondria Isolation Kit for Culture Cells (Thermofisher 89874). The intact mitochondria were further treated with 400U DNase I (Sigma D5025) in 200 μl modified DNAse I buffer (5 mM HEPES buffer pH 7.4, 210 mM mannitol, 70 mM sucrose, 10 mM MgCl2, 5 mM CaCl2, 0.2% bovine serum albumin and 1× Complete ethylenediaminetetraacetic acid (EDTA)-free protease inhibitor cocktail) for 30 min at 37°C to digest away any DNA outside of mitochondria. Mitochondria were then centrifuged at 12 000 g for 5 min at 4°C. The mitochondrial pellet was washed with a 1:1 mixture of reagent A plus reagent C from the kit, re-centrifuged at 12 000 g for 5 min at 4°C, decanted then frozen in liquid nitrogen and stored at −80°C before DNA extraction.
DNA extraction
Bacteria, HEK293T cells or intact HEK293T mitochondria were lysed overnight in SNET lysis buffer (400 mM NaCl, 1% sodium dodecylsulphate (SDS), 20 mM Tris pH 8, 5 mM EDTA, 200 μg/ml Proteinase K) at 56°C with shaking at 1000 rpm. Genomic DNA was purified from cell lysate using Phenol-chloroform-Isoamyalcohol (25:24:1) and precipitated with 50% isopropanol. The DNA pellet was washed once with 100% ethanol and once with 70% ethanol before being reconstituted in IDTE buffer (10 mM Tris pH 8, 0.1 mM EDTA).
Oligonucleotide annealing
Complementary + and − strand oligonucleotides (Supplementary Table S1) were first mixed at a final concentration of 45 μM each in 1× annealing buffer (50 mM NaCl, 10 mM Tris pH 7.4). The mixture was incubated in boiling water and left to cool gradually to below 30°C.
6mACE-seq library preparation
Genomic DNA in 130 μl IDTE was first sonicated to ∼150 bp (Covaris M220) and treated with RNAseA/T1 (Ambion AM2286) for 1 h at 37°C then purified using 1.8× volume Agencourt AMpure XP beads (Beckman A63880). Five micrograms of sonicated and RNA-free genomic DNA was then incubated with 4 μg anti-6mA antibody in 250 μl 1× IP buffer (150 mM NaCl, 10 mM Tris pH 7.4, 0.1% IGEPAL) overnight at 4°C. Unless otherwise stated, the anti-6mA antibody used was the rabbit polyclonal anti-6mA antibody (Synaptic Systems 202003). This antibody-genomic DNA mixture was split into 50 μl aliquots on ice and crosslinked with 0.15 J/cm2 254 nm ultraviolet (UV) radiation six times before being mixed with 40 μl decanted BSA-blocked Protein-A-dynabeads (Life Technologies 10002) for 1.5 h at 4°C. Beads bound with crosslinked genomic DNA were then washed with 500 μl of the following cold buffers in this order: Wash buffer 1 (1M NaCl, 50 mM HEPES-KOH pH 7.4, 1% Triton X-100, 0.1% Sodium Deoxycholate, 2 mM EDTA), Wash buffer 2 (0.5M NaCl, 50 mM HEPES-KOH pH 7.4, 1% IGEPAL, 0.1% Sodium Deoxycholate, 2 mM EDTA), Wash buffer 3 (1% Sodium Deoxycholate, 25 mM LiCl, 10 mM Tris pH 8, 1% Triton X-100, 2 mM EDTA), TE buffer (10 mM Tris pH 8, 1 mM EDTA) and finally 10 mM Tris pH 8. Genomic DNA was then subjected to progressive on-bead reactions as follows: end-repair with 6U T4 DNA polymerase (NEB M0203) at 12°C for 30 min, end-phosphorylation with 30U T4 PNK (NEB M0201) and T4 DNA ligase buffer (NEB B0202) at 37°C for 30 min, reverse adapter ligation with 1500U T4 DNA ligase (NEB M0202) and 202.5 pmol reverse adapter (Supplementary Table S1) at 25°C for 2 h, fill-in DNA synthesis with 20U Phi29 DNA polymerase (NEB M0269), a second round of end-phosphorylation using the same conditions, Lamda exonuclease digestion with 20U Lambda exonuclease (NEB M0262) at 37°C for 30 min and finally RecJf exonuclease digestion with 90U RecJf (NEB M0264) at 37°C for 30 min. The same bead-wash steps were carried out between each reaction. DNA was then eluted off beads with elution buffer (0.5% SDS, 0.2 M NaCl, 25 mM Tris pH 8, 2 mM EDTA) and treated with 8U proteinase K (Thermo Scientific EO0491) overnight at 65°C with shaking at 1000 rpm. The eluted single-stranded DNA (ssDNA) was ethanol-precipitated before undergoing the following reactions as follows: Annealing of PG162 primer (Supplementary Table S1) to the reverse adapter and primer extension with 20U Phi29 polymerase at 30°C for 30 min, forward adapter ligation with 1000U T4 DNA ligase and 30.15 pmol forward adapter (Supplementary Table S1) at 25°C for 2 h, double-stranded DNA (dsDNA) purification with 1.7× volume Ampure XP beads and amplification with Phusion High-fidelity PCR mastermix (NEB M0530) and Truseq PCR primers. Bacterial and human libraries were respectively amplified for 13 and 18 cycles. Finally, primer-dimer and adapter-dimers were removed with 0.8× volume AMpure XP beads before undergoing sequencing on the Illumina Miseq or Nextseq single-end platforms.
6mACE-seq data analysis
Single-end fastq sequences were trimmed of any 3′ adapter sequences using Cutadapt (19) before being mapped using Bowtie2 (20) allowing for at most 1 mismatch to either the human genome version hg38, S. typhimurium 14028S genome, UTI89 E. coli genome or PG482 (Supplementary Table S1). Afterward, the coordinates for the first nucleotide and the total number of associated mapped reads were consolidated and used for 6mA site-calling: a potential site was under consideration only if it possessed at least 1 read in all replicates. For each potential site, the probability of reads beginning at that site was calculated using a negative binomial distribution based on the maximum-likelihood estimates of the parameters determined from the counts of read-starts in a 1000 nt window centered around the site. Probabilities at each candidate site across replicates were then combined via Fisher’s method for combining P-values. Multiple-testing correction using the Benjamini–Hochberg procedure was implemented and sites where the false discovery rate <0.01 were deemed significant. Significant 6mA sites were aligned against coding gene regions (UCSC hg38 GENCODE v24) or repetitive sequences (UCSC hg38 RepeatMasker) to respectively calculate the observed proportion of 6mAs that mapped to various regions of non-transposable protein-coding genes or to various transposon subfamilies. For generating consensus 6mA motifs using Weblogo3 (21), the 21 nt sequences of significant 6mA sites +/− 10nt was used as input. For generating consensus 6mA motifs using the MEME-ChIP (22) comprehensive motif analysis, significant 6mA sites were first filtered for only those that possess an A at the actual 6mA site before using the 21 nt sequences of these 6mA sites +/− 10 nt as input.
RESULTS
6mACE-seq identifies 6mA at stranded single-nucleotide resolution
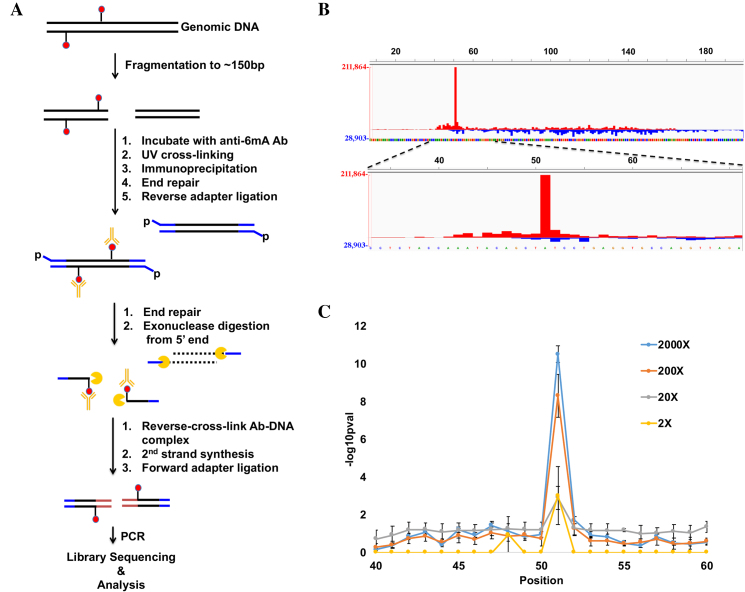
For sequencing 6mA, we prioritized sequencing resolution and thus, leveraged on past strategies that have reported relatively high resolution sequencing of chromatin and DNA methylation (23,24). Briefly, anti-6mA antibodies are first crosslinked onto 6mA-containing dsDNA fragments, which are thus protected from subsequent 5′-to-3′ exonuclease treatment. Exonuclease degradation of all genomic DNA other than regions proximal to the protected 6mA sites is what confers the high resolution of 6mA locations (Figure 1A).
Figure 1.
6mACE-seq maps 6mA site in synthetic 6mA-dsDNA. (A) Flowchart documenting the procedure for 6mACE-seq. (B) Counts of 6mACE-seq read start-sites mapped to a 200 bp synthetic dsDNA (PG482/PG483, Supplementary Table S1) with a single 6mA at position 51 of the plus strand. Red and blue represent 6mACE-seq reads that map respectively to the + or − strand. (C) Significance (-log10P-val) at varying coverage was calculated at each position of the dsDNA from (B), and repeated over 100 simulated trials. Average significance +/− SD was then plotted for the plus strand from positions 40 to 60.
We first tested this method by using a synthetic 200 bp dsDNA containing a single 6mA site on one strand. While we expected sequencing reads to begin proximally upstream of the known 6mA site, our sequencing results revealed that the start of most 6mACE-seq reads coincided exactly with the single 6mA site (Figure 1B). This demonstrates that exonucleases are able to digest away DNA up to and including the nucleotide just 5′ of 6mA nucleotides, which results in a single-nucleotide-resolution map of 6mA sites. This also means that the strand location of 6mA sites is preserved. This is in contrast to previous reports of this strategy only achieving non-stranded ∼33 bp 6mA mapping resolution (24). As such, we term our refined method for both the preparation and single-nucleotide-resolution analysis of such 6mA-sequencing libraries as 6mA-Crosslinking-Exonuclease-sequencing (6mACE-seq). We then set out to determine the minimal coverage required for accurate detection of 6mA with 6mACE-seq. Using a bootstrap method, we were able to identify a significant peak specifically at the single 6mA site at 2000× coverage (Figure 1C). Furthermore, we could still specifically identify the location of 6mA within the synthetic dsDNA even at 2× coverage (Figure 1C).
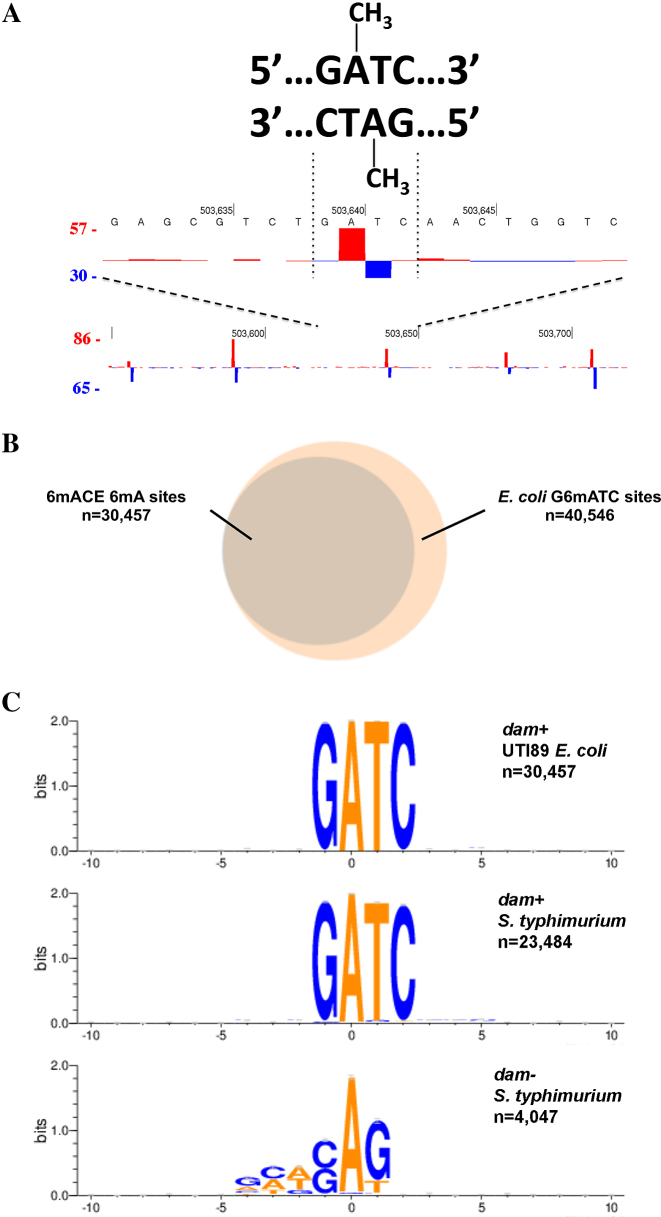
6mACE-seq accurately maps bacterial 6mA without a motif bias
We next set out to test how 6mACE-seq performs on genomic DNA by sequencing 6mA sites in UTI89 E. coli and S. typhimurium bacteria, where exact positions of 6mA sites have already been well-established (14,25). In E. coli, we observed that the start of 6mACE reads generally formed a bipolar pattern coinciding with the adenine nucleotides on both strands of a palindromic GATC sequence (Figure 2A), as expected of G6mATC motifs that are fully methylated on both strands. On a genome-wide scale, 6mACE-seq could identify ∼74% of the 40 546 known G6mATC sites significantly (False Discovery Rate, FDR<0.01) using only ∼1.8 million reads (Figure 2B). In S. typhimurium, we first focused on the region upstream of the opvAB operon, which contains four palindromic GATC sequences that had previously been characterized as methylated or unmethylated (25). As expected, we only observed bipolar 6mACE-read-start patterns on the two methylated GATC palindromes (green arrows) but not on the two unmethylated GATC palindromes (black arrows) (Supplementary Figure S1a). We next tested if different antibodies affected the accuracy of 6mACE by performing 6mACE-seq to map S. typhimurium 6mA using anti-6mA antibodies from different sources. While our default 6mACE antibody (Synaptic Systems 202003) could identify ∼61% of the known G6mATC sites significantly (FDR<0.01), the two other antibodies tested either yielded high false-positives (Synaptic Systems 202111) or high false-negatives (Abcam 151230) (Supplementary Figure S1b-d). Therefore, we continued using the default antibody for all remaining experiments.
Figure 2.
6mACE-seq maps 6mA sites in multiple bacterial genomes. (A) Counts of 6mACE-seq read start-sites at five representative fully-methylated G6mATC sites within the UTI89 E. coli genome. The format follows that of (Figure 1B). (B) Venn diagram showing overlap in 6mA sites identified by 6mACE-seq and all G6mATC sites within the UTI89 E. coli genome. (C) Weblogo representations of consensus 6mA motifs constructed from significant (FDR<0.01) 6mA sites identified using 6mACE-seq performed on genomes of dam+ UTI89 E. coli (top panel), dam+ S. typhimurium (middle panel) and dam- S. typhimurium (bottom panel).
We subsequently used all significantly identified 6mA sites to construct a consensus 6mA motif and were able to reconstruct the canonical ‘G6mATC’ motif that is known to be targeted by the Dam methylase in E. coli and S. typhimurium (Figure 2C) (3). In order to confirm the specificity of 6mACE-seq for 6mA sites, we also sequenced dam−S. typhimurium. While we did not observe the same ‘G6mATC’ motif in dam− S. typhimurium due to an absence of the Dam methylase, we instead found a ‘CAG6mAG’ motif that is characteristic of a type III methylase in Salmonella serovars (Figure 2C) (26). Similarly, besides ‘G6mATC’, 6mACE-seq of UTI89 E. coli also identified the ‘CC6mANNNNNNNCTTC’ motif that is characteristic of a type I methylase in E. coli (Supplementary Figure S2) (27). Together, these results revealed two additional advantages of 6mACE-seq in that it is able to detect uncommon 6mA sites and it also has no sequence bias.
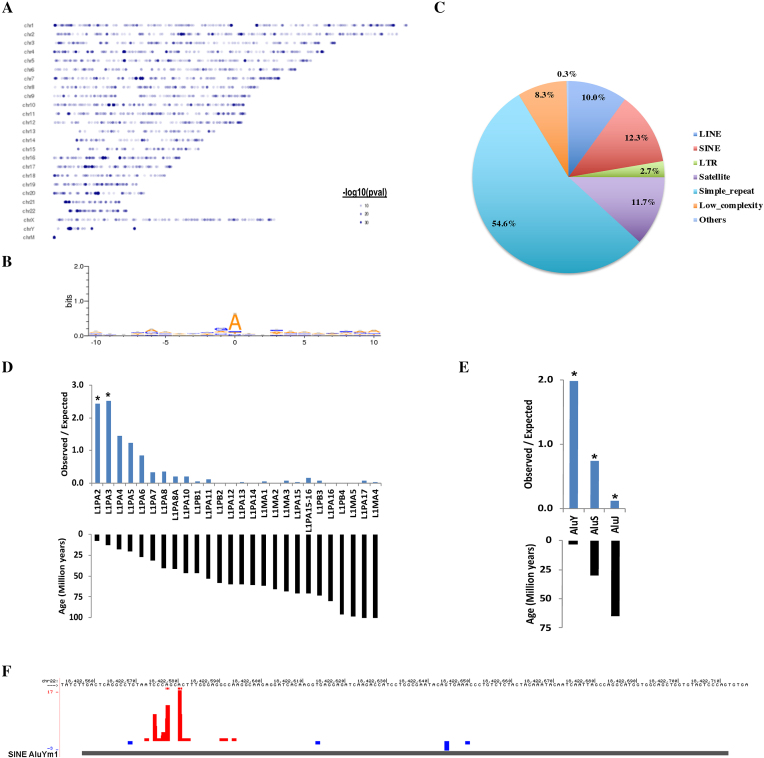
6mACE-seq maps human 6mA site enrichment in young and active LINE and SINE subfamilies.
We proceeded to map 6mA sites in human HEK293T cells and identified ∼14 000 significant 6mA sites throughout the human genome (Figure 3A, FDR <0.01) possessing a moderate ‘6mA’ motif (Figure 3B), with a comprehensive motif analysis identifying ‘AATGG’ repeats as the prevalent motif (Supplementary Figure S3a). Of the ∼16% of 6mA sites aligned to non-transposable protein-coding genes (Supplementary Figure S3b), there was a preference against intronic regions and to a greater extent, both the CDS and 3′ untranslated region (UTR) (Supplementary Figure S3c). Instead, 6mA appeared preferentially localized towards the promoter, which we designated as the 1 kbp region directly upstream of the transcription start site (Supplementary Figure S3c).
Figure 3.
6mACE-seq identifies 6mA enrichment in young and active human LINEs and SINEs. (A) Plot of genome-wide location of 6mA sites by chromosome. Each dot represents a single 6mA site and the colour intensity corresponds to the statistical significance (-log10P-val) of the 6mA site determined from 6mACE-seq reads. (B) Consensus 6mA motif constructed using all significant (FDR<0.01) 6mA sites across the human genome (C) Pie-chart displaying proportion of 6mA sites mapped to various repetitive genomic elements. (D and E) Enrichment of 6mA sites in various LINE (D) or SINE (E) subfamilies, quantified as the ratio of Observed proportion/Expected proportion of 6mA sites in each transposon subfamily. Age of each LINE1 subfamily and SINE subfamily are plotted accordingly. * represents P-value<10-19 calculated using a 2-tailed test for population proportion. (F) Counts of 6mACE-seq read start-sites in a representative AluYm1 SINE element. The format follows that of (Figure 1B) with additional horizontal red bars denoting statistically significant 6mA sites (FDR<0.01) mapped to the positive strand.
In comparison to the minority of 6mA sites associated with protein-coding genes, up to ∼60% of 6mA sites were associated with various repetitive sequences. Out of these, around half were in simple repeat regions with the next largest representations being in retrotransposons (Figure 3C). Previous studies had found 6mA to be enriched in younger and more active mouse (10) and human (11) LINE1 transposons. Therefore, we classified our identified human LINE1-associated-6mAs according to the age of the various transposon subfamilies (28). Concordant with past studies, 6mACE-seq identified 6mA sites to be similarly enriched towards the younger and more active L1PA subfamilies than the L1PB or L1MA subfamilies (Figure 3D), which reinforces the accuracy of 6mACE-seq in mapping genome-wide 6mA in the human genome. Although mouse 6mA did not exhibit any enrichment for particular short interspersed nuclear element (SINE) subfamilies, we also classified human SINE-associated-6mAs by SINE subfamily age (29). This revealed a similar and clear trend for human 6mA to be enriched on the younger and more active SINE AluY subfamilies than the older AluS and AluJ subfamilies (Figure 3E and F). Taken together, these findings demonstrate 6mACE-seq’s ability to both validate pre-established as well as identify novel human 6mA patterns.
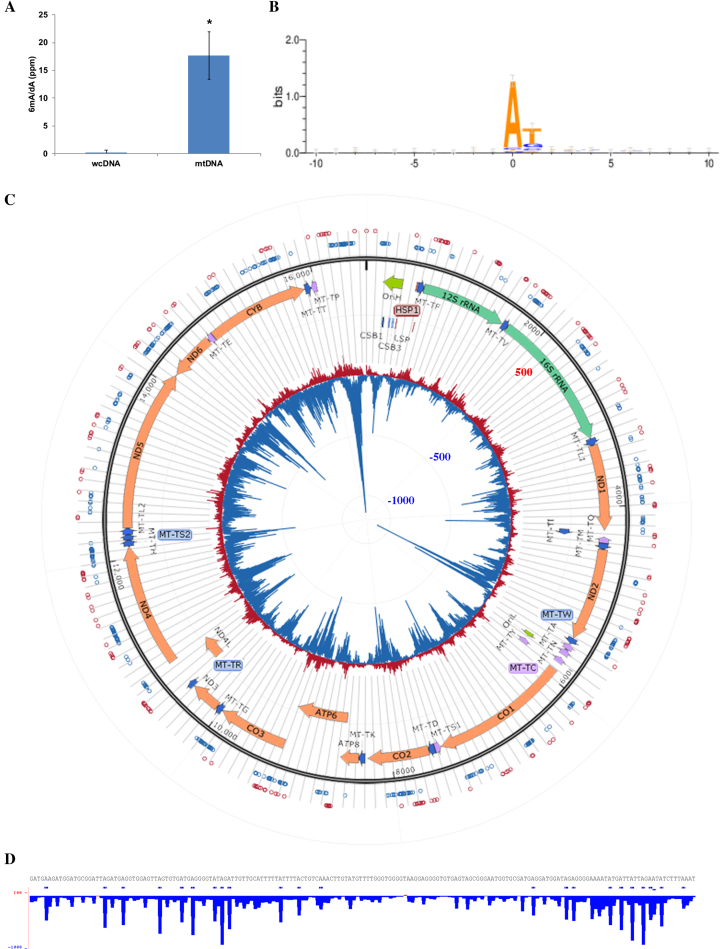
Contrary to other reports (9–11) and our findings, a recent study used SMRT sequencing to map human blood lymphocyte 6mA and found 6mA to be enriched in exon-coding regions rather than in transposable regions (12). One possible reason for this discrepancy is the difference in cell-types sequenced. Another possibility relates to the level of 6mA in the human genome. Using ultra-high-performance liquid-chromatography coupled with tandem mass spectrometry (UHPLC-MS/MS) of nucleosides, we quantified only <1 ppm 6mA/dA within whole-cell DNA (wcDNA) (Figure 4A), consistent with previous quantifications of mammalian wcDNA 6mA levels (15,16). Given that low 6mA levels coupled with high 5mC levels have been shown to confound SMRT sequencing-based 6mA detection (11,17), this is a possible cause for SMRT sequencing being unable to detect 6mA enrichment in transposons in certain scenarios (12).
Figure 4.
6mA is enriched in mtDNA and exhibit strand-asymmetric clustering patterns. (A) UHLPC-MS/MS quantification of 6mA/dA in whole-cell DNA versus purified mtDNA. Data represents average of at least three biological replicates +/− SD. Samples were digested with degradase plus. * represents T-test P < 0.001. (B) Weblogo representation of consensus 6mA motif constructed using all significant (FDR<0.01) 6mA sites across the human mitochondrial genome. (C) Map of mitochondrial 6mA. Inner layer: counts of 6mACE-seq read start-sites following the same format as (Figure 1B). Middle layer: open reading frames of mitochondrial chromosome. Outer layer: significant (FDR<0.01) 6mA sites (hollow circles) determined using raw 6mACE-seq reads. Red and blue represent 6mA sites mapped to the mtDNA + or − strands respectively. (D) Counts of 6mACE-seq read start-sites within chrM:5339–5520. The format follows that of (Figure 1B) with horizontal blue bars denoting statistically significant 6mA sites (FDR<0.01) mapped to the negative heavy strand. Printed genomic sequence corresponds to the negative heavy strand.
6mACE-seq uncovers asymmetrical 6mA clusters in the human mitochondrial genome
Besides LINEs and SINEs, we also searched for similar enrichments of 6mA in other regions of the human genome and were intrigued to find that 6mA site density is >8000 times higher in the mitochondrial genome than in the nuclear genome (Supplementary Figure S4a). We verified this result using 6mA-DIP coupled with qPCR (6mA-DIP-qPCR), which showed a similar 6mA enrichment for the mitochondrial genome (Supplementary Figure S4b). Similarly, using UHPLC-MS/MS, we found that 6mA is indeed more enriched within purified mtDNA than in total wcDNA (Figure 4A and Supplementary Figure S4c).
Across the mitochondrial genome, these 6mA sites generally exhibited a ‘6mAT’ dinucleotide motif that consists of a strong ‘6mA’ followed by a moderate ‘T’ (Figure 4B). Upon a closer observation, we noticed 6mA sites to be arranged throughout the entire circular chromosome with no bias towards any particular messenger RNA, transfer RNA, ribosomal RNA, origin of replication or promoter sequence (Figure 4C). Instead, identified 6mA sites were arranged in chromosome-wide clusters, each spanning hundreds of bases. While opposite strand clusters would often overlap partially, we noted that mitochondrial 6mA sites were generally asymmetrically methylated. Interestingly, there was also a clear preference for 6mACE-seq reads to map to the negative heavy strand than to the positive light strand (Figure 4C and D). Given the enrichment of 6mA in the mitochondria as well as its unique asymmetrical patterns, we focused subsequent efforts on characterizing the function of mitochondrial 6mA.
6mA destabilizes dsDNA and recruits SSBP1
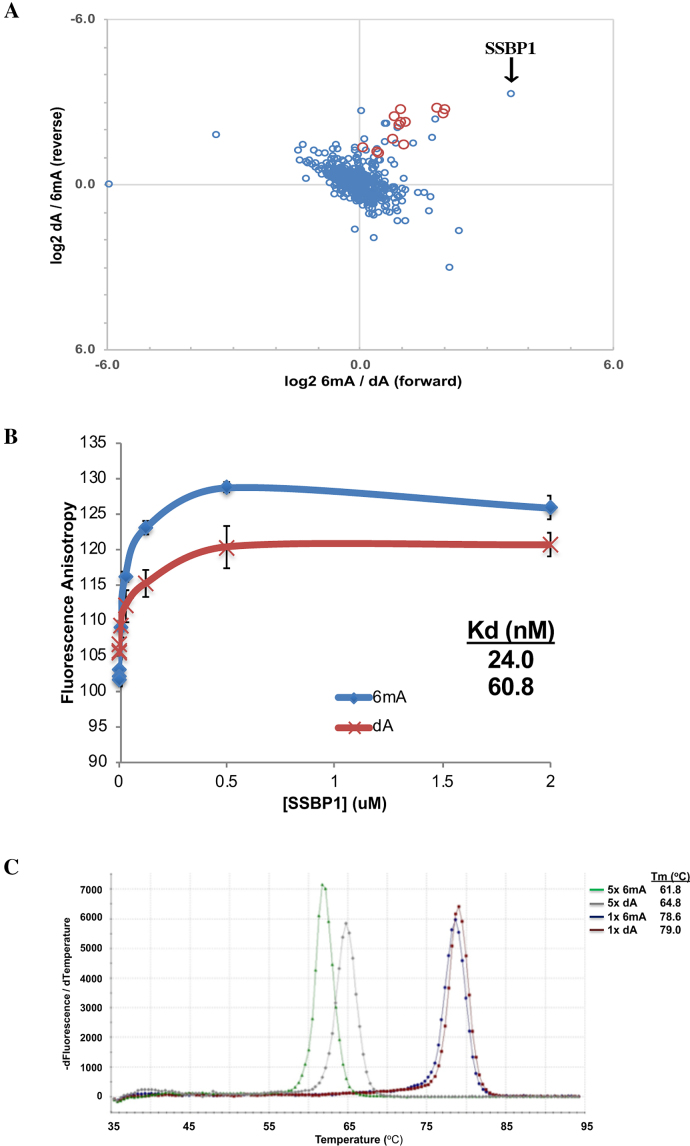
Since DNA modifications often exert their regulatory effect by promoting the binding of specific DNA-binding proteins, we set out to identify mitochondrial 6mA-binding proteins. HEK293T cells were first subjected to stable isotope labelling by amino acids in cell culture (SILAC), then mitochondrial protein lysates were extracted for DNA-pulldowns using asymmetrically-methylated (6mA-bait) or non-methylated (dA-bait) dsDNA baits corresponding to an identified mitochondrial 6mA sequence (Supplementary Table S1). We incubated immobilized 6mA-bait and dA-bait respectively with ‘Heavy’ and ‘Light’ SILAC-labelled lysates for the ‘forward’ pulldown as well as a label-swap for the ‘reverse’ pulldown. The associated proteins were then identified using quantitative mass-spectrometry based proteomics. Of the 753 pulled down in our experiment, 19 proteins preferentially bound to the 6mA-bait than to the dA-bait by at least 2-fold (Figure 5A and Supplementary Table S2). This list contains HNRNP and YTH-domain proteins, which are known to preferentially bind RNA with N6-methyladenosine (m6A) modification (Figure 5A, red circles) (30). This indicates that we were at least successful at pulling down proteins that are recruited by the m6A modification, which itself shows strong structural similarity with 6mA.
Figure 5.
Dense 6mA destabilizes dsDNA structure to potentially promote binding of mitochondrial SSBP1. (A) SILAC-based pulldown from HEK293T mitochondrial extract using dsDNA bait (Supplementary Table S1). Data are plotted as ratio of 6mA/dA binding (forward pulldown) on X-axis and ratio of dA/6mA binding (label-swapped reverse pulldown) on Y-axis. Red circles represent known N6-methyladenosine RNA-binding proteins (YTH family and HNRNP family proteins). Blue circles represent all other SILAC-detected candidates. (B) Fluorescence anisotropy curve of fluorescent-tagged dsDNA probes (Supplementary Table S1) with (blue) or without (red) five 6mAs, incubated with varying concentrations of recombinant SSBP1. Data represents average of three technical replicates +/− SD. (C) Melting curve of dsDNA containing 5, 1 or 0 6mA sites (Supplementary Table S1), plotted as the negative first derivative of fluorescence against temperature. Data represents average of three technical replicates.
Our top candidate for 6mA-binding proteins is single-stranded DNA-binding protein 1 (SSBP1), which we confirmed orthogonally via fluorescence anisotropy (Figure 5b). Given that SSBP1 binds preferentially to ssDNA (31) and also binds along the heavy-strand of the mitochondrial chromosome (32), coinciding with where 6mA clusters are enriched (Figure 4C and D), we postulated that mitochondrial 6mA might recruit SSBP1 by first promoting dsDNA melting. This is quite plausible given that 6mA had previously been shown to modify the structure of dsDNA (33). The pulldown of HNRNP and YTH-domain proteins, which bind single-stranded oligonucleotides also alludes to the dsDNA 6mA baits transitioning into a mixture of dsDNA and ssDNA, further suggesting that 6mA might promote dsDNA melting. Using a dsDNA melting temperature assay, we found that presence of a single 6mA resulted only in a 0.4°C decrease in the melting temperature (Figure 5C). However, when using a dsDNA containing five 6mAs, which betters mimics the clustering of 6mA in the mitochondrial genome, we observed a 3.0°C decrease in the melting temperature (Figure 5C). This 6mA-density dependent decrease in dsDNA melting temperature shows that 6mA clusters can potentially promote melting of the dsDNA structure of the mitochondrial chromosome, which might be a mechanism for 6mA-mediated recruitment of SSBP1 to the mitochondrial heavy strand.
Mitochondrial ALKBH1 is a 6mA demethylase that regulates mitochondrial function
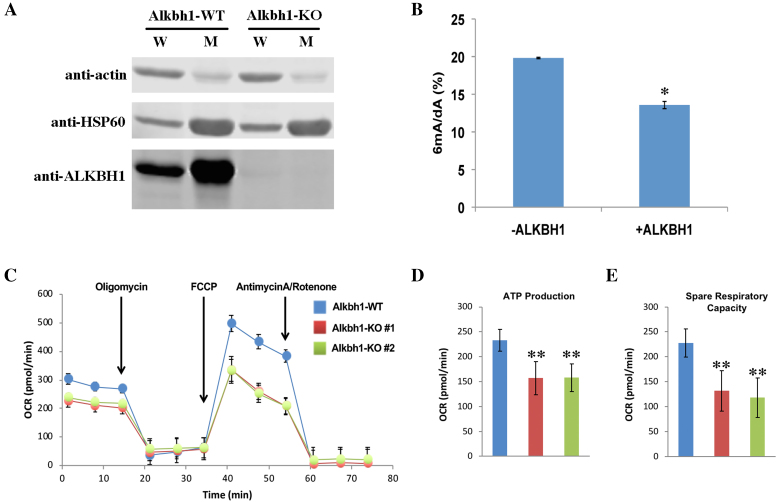
We next searched for human cellular factors that can metabolize mitochondrial 6mA. Previous studies have identified AlkB homologue 1 (ALKBH1), a Fe2+/2-oxoglutarate-dependent dioxygenase, as a 6mA demethylase in mammals (10, 12). However, other studies disagree on ALKBH1’s cellular localization and have also reported alternative demethylation substrates such as N1-methyladenosine and 3-methylcytosine (34–37). In order to resolve these conflicts and determine whether ALKBH1 is a mitochondrial 6mA demethylase, we first assessed whether ALKBH1 localizes to the mitochondria. Comparison of anti-ALKBH1 western blotting of whole-cell versus mitochondrial lysates showed a visible ∼43.8 kDa protein band in the whole-cell lysate that was intensified in the mitochondrial lysate (Figure 6A). We also used CRISPR-Cas9 to knock out the Alkbh1 gene and verified the identity of this mitochondrial-enriched protein band as ALKBH1, confirming that AKLBH1 is a mitochondrial-localized protein (Figure 6A). Next, we generated recombinant ALKBH1 protein to assess its ability to demethylate 6mA in vitro (Supplementary Figure S5a). We used a synthetic 6mA-containing oligonucleotide corresponding to an identified mitochondrial 6mA sequence (Supplementary Table S1) and incubated it with recombinant ALKBH1. This led to ∼30% demethylation of the oligonucleotide as quantified by UHPLC-MS/MS (Figure 6B). Previous reports identified H231A and D233A as gene mutations that disrupt the catalytic domain of ALKBH1 by abrogating its ability to bind iron (10,12,35). We therefore generated the H231A/D233A catalysis-dead ALKBH1 mutant protein (ALKBH1-CD) and confirmed that it no longer demethylates 6mA, proving that wild-type ALKBH1 catalyzes the demethylation of 6mA (Supplementary Figure S5b,c). Expectedly, ALKBH1-knockout (ALKBH1-KO) cells also exhibited slightly higher mitochondrial 6mA levels than cells expressing wild-type ALKBH1 (ALKBH1-WT) (Supplementary Figure S5d).
Figure 6.
ALKBH1 is a mitochondrial protein with 6mA demethylase activity and its loss disrupts mitochondrial function. (A) Western blotting of ALKBH1 in ALKBH1-WT and ALKBH1-KO whole-cell (W) or mitochondrial (M) lysate. Actin and HSP60 are used as cytoplasmic/non-mitochondrial and mitochondrial loading markers. (B) UHPLC-MS/MS quantification of resultant 6mA/dA within 6mA-containing ssDNA (Supplementary Table S1) after incubation without or with recombinant ALKBH1. Data represents average of three biological replicates +/− SD. * T-test P < 0.0001. Samples were digested with 20× diluted NPAP (see ‘Materials and Methods’ section). (C) Plot of oxygen consumption rate, OCR (pmol/min) against time during the Seahorse Mito-Stress test assay of ALKBH1-WT versus two separate ALKBH1-KO clones. Time points for administering of all three drugs used in this assay are as denoted. Data represents average of at least three biological replicates +/− SD. (D and E) ATP production (D) and SRC (E) calculated from OCR measurements in (C). Data represents average of at least three biological replicates +/− SD. ** T-test P < 0.00001. Sample legend follows that of (C).
Given the localization of 6mA and ALKBH1 in the mitochondria, we tested whether ALKBH1 loss affects the mitochondria’s primary role of oxidative phosphorylation (OXPHOS) by subjecting ALKBH1-KO cell lines to a Mito-Stress test assay. Briefly, oxygen consumption rate (OCR) of cells are measured as they are subjected to various drug treatments that inhibit various aspects of the oxidative respiratory pathway. By comparing OCR differences between ALKBH1-WT and ALKBH1-KO cells, we found that loss of ALKBH1 resulted in ∼30 and ∼45% respective reduction in basal adenosine triphosphate (ATP) production and spare respiratory capacity (SRC) (Figure 6C, D and E). This suggests a role for the 6mA demethylase ALKBH1 in maintaining wild-type levels of mitochondrial OXPHOS.
DISCUSSION
While SMRT sequencing can map 6mA at single-nucleotide-resolution, it takes at least 75 runs on the Pacbio Sequel machine to achieve the minimal ∼150× coverage necessary to generate a single replicate of a full human 6mA methylome map (11). Without such coverage, SMRT sequencing-based detection of 6mA in the human genome is complicated by low 6mA density and high 5mC density, resulting in high false discovery rates (11,17). Here, we present 6mACE-seq as an alternative method that can sequence genome-wide 6mA at stranded single-nucleotide-resolution. We demonstrate this first by utilizing 6mACE-seq to accurately identify the 6mA site in a synthesized dsDNA molecule containing a single 6mA. Second, we used 6mACE-seq to identify 6mA sites within multiple well-characterized E. coli methylomes, demonstrating that 6mACE-seq is not biased for 6mA in any particular motif context. Therefore, 6mACE-seq exhibits an advantage over another single-nucleotide-resolution 6mA-sequencing method, DA-6mA-Seq that relies on the DpnI restriction enzyme, limiting its utility only to 6mA sites within G6mATC motifs (13). Third, 6mACE-seq generated a human genome-wide map that validated the 6mA enrichment within young LINE1 subfamilies, which was previously reported in mouse and human cells (9–11). This reinforces the accuracy of 6mACE-seq to detect genome-wide 6mA in a gigabase-scale eukaryotic genome. Coincidentally, our use of UV-crosslinking to improve upon the 6mA sequencing resolution of 6mA-DIP-seq parallels the use of UV-crosslinking in miCLIP to improve upon the m6A sequencing resolution of m6A-RIP-seq (38,39). Improved m6A resolution has facilitated studies about the function of m6A in RNA regulation (40). Therefore, we anticipate that precise 6mA mapping via 6mACE-seq will likewise facilitate studies of 6mA in DNA regulation.
A recent study reported a substantial ∼570 ppm 6mA/dA in HEK293T wcDNA as quantified via LC-MS/MS (12). However, they used a combination of NucleaseP1, Phosphodiesterase and Alkaline Phosphatase (NPAP) to process the genomic DNA. Some of these enzymes are expressed in fungi or bacteria, and can contribute contaminating levels of 6mA (16,41,42). In fact, we observed a similar contamination issue and switched to only using 20× diluted NPAP or other commercial enzymes (Supplementary Figure S6 and ‘Materials and Methods’). Given the low 6mA levels in human genomes reported by others (11) and us (Figure 4A), it is vital to utilize methods that enrich for human 6mA sites so as to help improve 6mA detection accuracy via sequencing. This can be accomplished through enzymatic enrichment (DA-6mA-seq (13)) or direct 6mA immunoprecipitation (6mA-DIP-seq (5–10) and 6mACE-seq). The choice of antibody used in 6mACE-seq is also vital for establishing high specificity of 6mA mapping (Supplementary Figure S1b-d). This is especially relevant in the human genome with low 6mA abundance, where the choice of antibody used seems to affect the mapping of 6mA sites more so than in the bacterial genome (Supplementary Figure S7a,b).
Using a variety of orthogonal approaches including 6mACE-seq, 6mA-DIP-qPCR and UHPLC-MS/MS, we demonstrated that 6mA is more enriched in mtDNA than in nuclear DNA (nDNA). Notably, 6mA sites are more than 8000× denser in mtDNA than in nDNA (Supplementary Figure S4a). This mitochondrial enrichment was observed both for 6mA sites mapped with 6mACE-seq using a second independent anti-6mA antibody (Supplementary Figure S7c), as well as for 6mA sites mapped commonly by two independent antibodies (Supplementary Figure S7d). Together, these support the possibility that 6mA may play specific physiological roles in human mitochondria. According to the strand displacement model of asynchronous mtDNA replication (32), the original light strand first serves as a template for synthesizing a new heavy strand. Synthesis of the new light strand is delayed and SSBP1 binds along the original single-stranded heavy strand to prevent random initiation of light strand synthesis. Using 6mACE-seq, we found 6mA clusters to be asymmetrically enriched on the heavy strand, suggesting a potential role for 6mA to mediate asynchronous mtDNA replication. This is supported by the subsequent identification of SSBP1 as a putative 6mA-binding protein. Furthermore, we showed that a high density of 6mA also promotes dsDNA melting. Therefore, one working model is that 6mA clusters within the heavy strand can potentially promote melting of the double-stranded mitochondrial genome. This might in turn promote SSBP1 recruitment to the heavy ssDNA strand during earlier phases of asynchronous mtDNA replication (31).
Finally, we characterized ALKBH1 as a mitochondrial protein that can demethylate 6mA. ALKBH1 was previously identified as a nuclear 6mA demethylase in both mouse and human cells (10,12). Given that we instead observed a clear localization of ALKBH1 to the mitochondria, it seems ALKBH1 might exhibit different cellular localizations depending on the cell type or growth condition being tested. We also found that ALKBH1 loss reduces mitochondrial function, specifically in basal ATP production and SRC, which is a measure of the cell’s ability to cope with increased energetic demands. Since ALKBH1 possesses only a modest capacity to demethylate 6mA, it is plausible that ALKBH1 regulates mitochondrial OXPHOS by demethylating only a few specific 6mA sites or clusters within mtDNA, which in turn affects mtDNA replication or expression of mtDNA-encoded OXPHOS factors. Despite being an uncommon human DNA modification, our study provides the first link between human 6mA and the regulation of mitochondrial function.
DATA AVAILABILITY
Data were deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE119094. The custom Python scripts used in this study are available on request from the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Goh, Burkholder and Quake laboratories for feedback and discussions. We also thank J. Ho and E. Guccione for help on mitochondrial purification, and P. Ahl and J. Connolly for help with mitochondrial stress assays. HEK293T CRL-3216, UTI89 E. coli strain, salmonella strains and SSBP1 tetramer were gifts respectively from S. Xue, S. Chen, J. Casadesus and M. Falkenberg.
Author contributions: W.S.S.G. conceived the study. C.W.Q.K., Y.T.G., J.D.W.T, S.P.N. and W.S.S.G. performed the experiments. S.B.N. and W.S.S.G. performed bioinformatics analysis. S.B.N., J.G., Y.-G.G., S.R.Q. and W.F.B. assisted in experimental design. W.S.S.G wrote the manuscript with input from all authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biomedical Research Council (A*STAR) Young Investigator Grant [1610151037 to W.S.S.G]; Genome Institute of Singapore [to W.S.S.G]. Funding for open access charge: Genome Institute of Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1. Smith Z.D., Meissner A.. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013; 14:204–220. [DOI] [PubMed] [Google Scholar]
- 2. Jaenisch R., Bird A.. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003; 33:245–254. [DOI] [PubMed] [Google Scholar]
- 3. Wion D., Casadesús J.. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat. Rev. Micro. 2006; 4:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo G.-Z., Blanco M.A., Greer E.L., He C., Shi Y.. DNA N(6)-methyladenine: a new epigenetic mark in eukaryotes. Nat. Rev. Mol. Cell Biol. 2015; 16:705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Greer E.L., Blanco M.A., Gu L., Sendinc E., Liu J., Aristizábal-Corrales D., Hsu C.-H., Aravind L., He C., Shi Y.. DNA Methylation on N(6)-Adenine in C. elegans. Cell. 2015; 161:868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J. et al. N(6)-methyladenine DNA modification in Drosophila. Cell. 2015; 161:893–906. [DOI] [PubMed] [Google Scholar]
- 7. Koziol M.J., Bradshaw C.R., Allen G.E., Costa A.S.H., Frezza C., Gurdon J.B.. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 2015; 23:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J., Zhu Y., Luo G.-Z., Wang X., Yue Y., Wang X., Zong X., Chen K., Yin H., Fu Y. et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 2016; 7:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yao B., Cheng Y., Wang Z., Li Y., Chen L., Huang L., Zhang W., Chen D., Wu H., Tang B. et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun. 2017; 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M. et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016; 532:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu S., Beaulaurier J., Deikus G., Wu T.P., Strahl M., Hao Z., Luo G., Gregory J.A., Chess A., He C. et al. Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 2018; 28:1067–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiao C.-L., Zhu S., He M., Chen D., Zhang Q., Chen Y., Yu G., Liu J., Xie S.-Q., Luo F. et al. N6-Methyladenine DNA modification in the human genome. Mol. Cell. 2018; 71:306–318. [DOI] [PubMed] [Google Scholar]
- 13. Luo G.-Z., Wang F., Weng X., Chen K., Hao Z., Yu M., Deng X., Liu J., He C.. Characterization of eukaryotic DNA N6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun. 2016; 7:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang G., Munera D., Friedman D.I., Mandlik A., Chao M.C., Banerjee O., Feng Z., Losic B., Mahajan M.C., Jabado O.J. et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 2012; 30:1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ratel D., Ravanat J.-L., Charles M.-P., Platet N., Breuillaud L., Lunardi J., Berger F., Wion D.. Undetectable levels of N6-methyl adenine in mouse DNA: Cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 2006; 580:3179–3184. [DOI] [PubMed] [Google Scholar]
- 16. Schiffers S., Ebert C., Rahimoff R.. Quantitative LC–MS provides no evidence for m6dA or m4dC in the genome of mouse embryonic stem cells and tissues. Angew. Chem. 2017; 56:11268–11271. [DOI] [PubMed] [Google Scholar]
- 17. Schadt E.E., Banerjee O., Fang G., Feng Z., Wong W.H., Zhang X., Kislyuk A., Clark T.A., Luong K., Keren-Paz A. et al. Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 2013; 23:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higuchi Y., Linn S.. Purification of all forms of HeLa cell mitochondrial DNA and assessment of damage to it caused by hydrogen peroxide treatment of mitochondria or cells. J. Biol. Chem. 1995; 270:7950–7956. [DOI] [PubMed] [Google Scholar]
- 19. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011; 17:3–12. [Google Scholar]
- 20. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rhee H.S., Pugh B.F.. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011; 147:1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu Y., Luo G.-Z., Chen K., Deng X., Yu M., Han D., Hao Z., Liu J., Lu X., Dore L.C. et al. N(6)-methyldeoxyadenosine marks active transcription start sites in chlamydomonas. Cell. 2015; 161:879–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cota I., Bunk B., Spröer C., Overmann J., König C., Casadesús J.. OxyR-dependent formation of DNA methylation patterns in OpvAB OFFand OpvAB ONcell lineages of Salmonella enterica. Nucleic Acids Res. 2016; 44:3595–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pirone-Davies C., Hoffmann M., Roberts R.J., Muruvanda T., Timme R.E., Strain E., Luo Y., Payne J., Luong K., Song Y. et al. Genome-Wide methylation patterns in salmonella enterica Subsp. enterica serovars. PLoS One. 2015; 10:e0123639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kasarjian J.K.A., Kodama Y., Iida M., Matsuda K., Ryu J.. Four new type I restriction enzymes identified in Escherichia coli clinical isolates. Nucleic Acids Res. 2005; 33:e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khan H., Smit A., Boissinot S.. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006; 16:78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E.. Active Alu retrotransposons in the human genome. Genome Res. 2008; 18:1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miralles Fusté J., Shi Y., Wanrooij S., Zhu X., Jemt E., Persson Ö., Sabouri N., Gustafsson C.M., Falkenberg M.. In Vivo occupancy of mitochondrial Single-Stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014; 10:e1004832-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gustafsson C.M., Falkenberg M., Larsson N.-G.. Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 2016; 85:133–160. [DOI] [PubMed] [Google Scholar]
- 33. Diekmann S. DNA methylation can enhance or induce DNA curvature. EMBO J. 1987; 6:4213–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ougland R., Lando D., Jonson I., Dahl J.A., Moen M.N., Nordstrand L.M., Rognes T., Lee J.T., Klungland A., Kouzarides T. et al. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells. 2012; 30:2672–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G. et al. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016; 167:1–30. [DOI] [PubMed] [Google Scholar]
- 36. Westbye M.P., Feyzi E., Aas P.A., Vågbø C.B., Talstad V.A., Kavli B., Hagen L., Sundheim O., Akbari M., Liabakk N.-B. et al. Human AlkB Homolog 1 is a mitochondrial protein that demethylates 3-Methylcytosine in DNA and RNA. J. Biol. Chem. 2008; 283:25046–25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kawarada L., Suzuki T., Ohira T., Hirata S., Miyauchi K., Suzuki T.. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017; 45:7401–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R.. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016; 537:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo G.-Z., He C.. DNA N6-methyladenine in metazoans: functional epigenetic mark or bystander. Nat. Struct. Mol. Biol. 2017; 24:503–506. [DOI] [PubMed] [Google Scholar]
- 42. Mondo S.J., Dannebaum R.O., Kuo R.C., Louie K.B., Bewick A.J., LaButti K., Haridas S., Kuo A., Salamov A., Ahrendt S.R. et al. Widespread adenine N6-methylation of active genes in fungi. Nat. Genet. 2017; 49:964–968. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE119094. The custom Python scripts used in this study are available on request from the corresponding author.